Abstract
Amelogenin gene organization varies from 6 exons (1,2,3,5,6,7) in amphibians and sauropsids to 10 in rodents. The additional exons are exons 4, 8, 9, and “4b”, the latter being as yet unidentified in AMELX transcripts. To learn more about the evolutionary origin of these exons, we used an in silico approach to find them in 39 tetrapod genomes. AMEL organization with 6 exons was the ancestral condition. Exon 4 was created in an ancestral therian (marsupials + placentals), then exon 9 in an ancestral placental, and finally exons “4b” and 8 in rodents, after divergence of the squirrel lineage. These exons were either inactivated in some lineages or remained functional: Exon 4 is functional from artiodactyls onward; exon 9 is known, to date, only in rodents, but could be coding in various mammals; and exon “4b” was probably coding in some rodents. We performed PCR of cDNA isolated from mouse and human tooth buds to identify the presence of these transcripts. A sequence analogous to exon “4b”, and to exon 9, could not be amplified from the respective tooth cDNA, indicating that even though sequences similar to these exons are present, they are not transcribed in these species.
Keywords: amelogenin, small exons, evolutionary origin, PCR, enamel, tetrapods
Introduction
Amelogenin, the major protein of forming enamel, mainly plays a role in crystal growth (Robinson et al., 1996; Beniash et al., 2005). Its encoding gene (AMELX = AMEL in non-mammalian species) is composed of 7 exons in mammals, except in rats and mice, in which 2 additional exons (8 and 9) have been found (R Li et al., 1995; W Li et al., 1998). These exons and exon 4 are not present in non-mammalian AMEL. AMELX is subjected to alternative splicing, giving rise to several transcripts and various isoforms. Some of them might possess signaling capabilities. In the mouse, 17 AMELX transcripts have been identified, among which 7 lack exon 7 and end with exons 8 and 9 (R Li et al., 1995; W Li et al., 1998; Bartlett et al., 2006). In 2006, when analyzing the genomic region containing AMELX exon 8, Bartlett and colleagues revealed that this exon was homologous to exon 5. In addition, they found that a small sequence located immediately upstream of exon 8 was identical to the exon 4 sequence, and they referred to this as a putative exon 4b. Exons 4b and 8 were, therefore, generated from the duplication of a gDNA segment containing exons 4 and 5, which was translocated downstream of exon 7. Surprisingly, exon 4 was never found in AMELX transcripts identified in rodent cDNA, and hence hereafter are marked “4b”.
Using in silico investigations, we traced the origin of mammalian-specific AMELX exons 4, “4b”, 8, and 9 through tetrapod evolution. Our analyses provide information on the birth of these exons and led us to wonder whether AMELX exon “4b” is coding in the mouse, and whether exon 9 is present in human AMELX and AMELY transcripts. We addressed these questions using PCR.
Materials & Methods
In silico Searches
In total, 39 sequenced tetrapod genomes [37 mammals, a lizard (Anolis carolinensis), and a frog (Xenopus tropicalis)] were explored for AMEL exons 4, “4b”, 8, and 9. Published sequences were used as a template for localization of the sequences in the genomes by BLAST. The regions potentially housing AMEL exon 4, 1.5 kb between exons 3 and 5, and AMEL exons “4b”, 8, and 9, 20 kb downstream of exon 7, were extracted from each genome (Fig. 1). These regions were screened with UniDPlot (Girondot and Sire, 2010), with human and rodent sequences of the targeted exons as a template. The sequences were validated by means of alignment with human and murine sequences with Se-Al 2.0 (Rambaut, 1996).
Figure 1.
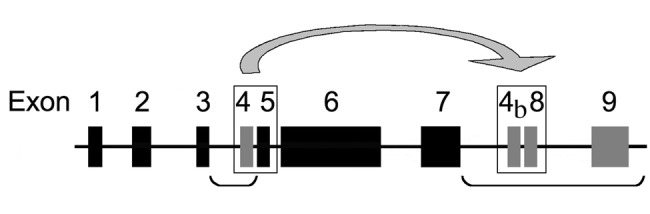
AMELX organization in the mouse genome showing the 10 exons: exons 1 to 9 and the putative exon 4b. Exon 4b and exon 8 were created after the DNA region containing exon 4 and exon 5 was duplicated and translocated downstream of exon 7 (arrow). The regions that were explored in various genomes for exons 4, 4b, 8, and 9 (gray blocks) are indicated with brackets.
References of the studied genomes and AMEL sequences are listed in Appendix 1.
Selective Pressure Analysis by the Hyphy Method
Hyphy software (http://hyphy.org; Pond et al., 2005) was used in the search for selective pressures that acted on exon 4 during evolution (Appendix 2).
PCR
Primers were designed with Primer Premier 5.0 software (PREMIER Biosoft International, Palo Alto, CA, USA) (Appendix 3). PCRs were performed on mouse genomic DNA (gDNA) with primer pair M1, to obtain an accurate sequence of the non-coding genomic region located between exons 7 and 8. This aimed to check whether AMELX exon “4b” sequence was really present in the mouse genome or was an artifact resulting from an incorrect computer-predicted sequence assembly. PCRs were also done on a mouse tooth bud cDNA library with primer pairs M2, M3, and M4, to find transcripts possessing the putative exon “4b”. In humans, utilizing primer pairs H1 and H2, we performed PCR on cDNAs for putative transcripts ending with exon 9 (including a human fetal tooth cDNA library and cDNA samples freshly prepared from fetal human tooth buds, collected under the guidelines of the University of California Committee on Human Research), by using the RNeasy Mini Kit and SuperScript III Reverse Transcriptase Kit. Both in humans and mice, the primers designed for PCRs on cDNA were used to amplify gDNA, to demonstrate the effectiveness of the primer sets.
Results
AMELX Exon 4
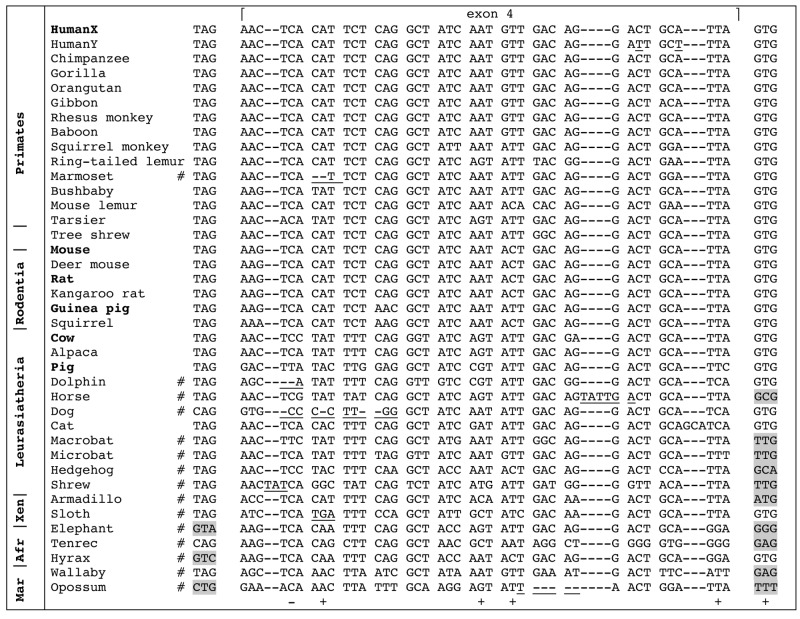
Exon 4 was found in all mammalian genomes, except in the monotreme platypus (Fig. 2). Exon 4 is absent in lizard and frog gDNA. In several mammals [a primate (marmoset) out of 11 species studied, 7 laurasiatherians out of 11, the 2 xenarthrans, the 3 afrotherians, and the 2 marsupials], our analysis indicated that the putative exon 4 was inactivated (see Fig. 2). In contrast, it is possibly coding in primates, the tree shrew, rodents, artiodactyls (cow, alpaca, and pig), the cat, and in human AMELY, in which it possesses correct intronic splice sites and no deleterious mutation (Fig. 2). Selective pressure analysis identified the second residue encoded by exon 4 as being negatively selected (i.e., conserved) and residues 3, 7, 8, and 13, and the intronic splicing site as being positively selected (see Appendix 2).
Figure 2.
Alignment of 37 nucleotide sequences of either functional (i.e., found in cDNA, species indicated in bold), or putatively functional (but no cDNA data), or inactivated (see below) AMELX exon 4 recovered in the genome of representative species of mammalian lineages. Human AMELY exon 4 was included in this alignment; it exhibits only 2 nucleotide substitutions (underlined) and looks functional. The important nucleotides of the donor (left) and acceptor (right) intron splices are indicated on both sides of the alignment. Exon 4 is assumed to be functional in all primates except in the marmoset, in the tree shrew, in rodents, and in a few laurasiatherians. In contrast, exon 4 is inactivated (# = pseudogenic) in numerous lineages; it shows either splice-site-mutated (in gray background) or deleterious mutations (reading frame shift or stop codon, underlined). Selective pressure analysis (Hyphy method, see Appendix 2) identified 1 negatively (-) and 5 positively (+) selected sites. Latin names of species and accession numbers of sequences are indicated in Appendix 1. Afr = afrotherians; Mar = marsupials; Xen = xenarthrans.
AMELX Exon “4b”
In the mouse, sequencing the 2.0-kb genomic region separating exons 7 and 8 provided a sequence identical (not shown) to that available in databases, which means that the gDNA sequence containing exon “4b”, and identical to the sequence around exon 4, is a valid sequence and not an artifact generated during computer-aided assembly of this region.
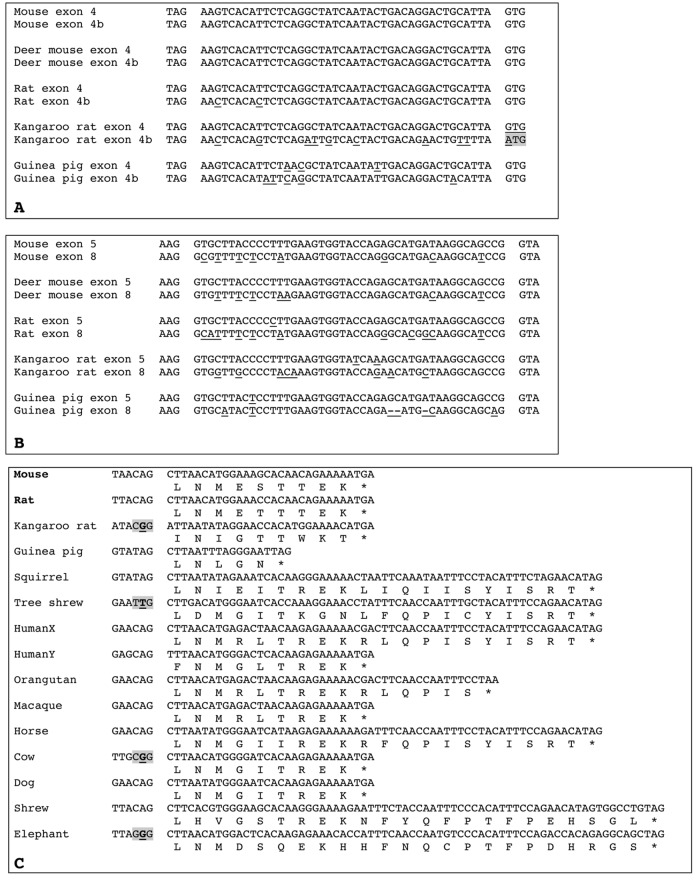
A search for exon “4b” downstream of AMEL exon 7 in all genomic sequences revealed that a homologous sequence is present in murids (mouse, rat, and the 2 related species) and in caviids (guinea pig), while it is absent in sciurids (squirrel) (Fig. 3A). It could be coding in the guinea pig, rat, deer mouse, and mouse, while not coding in the kangaroo rat, in which the splice acceptor site is mutated. The inactivating mutation of exon “4b” in this species could have favored the accumulation of numerous substitutions, as observed in its sequence.
Figure 3.
Alignments of amelogenin exons 4 and 4b, exons 5 and 8, and exon 9. (A) Alignment of AMELX exon 4 with the putative exon “4b” sequence found in 5 rodent genomes. In 4 species, exon “4b” is putatively functional: correct donor (left) and acceptor (right) intron splices and sequence either identical or close to that of exon 4. In the kangaroo rat, exon “4b” is not coding, as indicated by the mutation of the acceptor intron splice (gray background); note that this exon “4b” sequence shows more nucleotide substitutions than in, e.g., mouse and rat sequences. Nucleotide differences between exon 4 and exon “4b” are underlined. Latin names of species are indicated in Appendix 1. (B) Alignment of AMELX exon 5 with exon 8 sequences found in 5 rodent genomes. Mouse and rat sequences are functional. The 3 other exons 8 are putatively functional: correct donor (left) and acceptor (right) intron splices, no deleterious mutations, and sequence close to that of mouse exon 8. Nucleotide differences between exon 5 and exon 8 are underlined. Latin names of species are indicated in Appendix 1. (C) Alignment of functional (i.e., found in mouse and rat AMELX transcripts, in bold), putatively functional (but no cDNA data), or non-coding (i.e., putative intron splice mutated, gray background) AMELX exon 9 (nucleotide and protein sequences) of species representative of various mammalian lineages. Several sequences are putatively functional: correct donor intron splice (shown on the left) and beginning of the sequence similar to that of mouse and rat exon 9. Note the remarkable conservation of the sequence from elephant to mice, and the differences between human AMELX and AMELY sequences. * = stop codon. Latin names of species are indicated in Appendix 1.
However, we failed to PCR amplify exon “4b” from a mouse tooth bud cDNA library.
AMELX Exon 8
Genome exploration downstream of AMEL exon 7 revealed that exon 8 is present, in addition to the mouse and rat, in only 3 other rodent AMELX (Fig. 3B). It was not found in the squirrel and in all non-rodent species, even as pseudogenic. Sequence analysis pointed to several substitutions when compared with the sequences of exon 5, from which it is derived. However, with the intronic splice sites being correct and no deleterious mutation being observed, there is no reason to think that AMELX exon 8 was not coding in the deer mouse, kangaroo rat, and guinea pig (Fig. 3B).
AMELX Exon 9
Screening the region downstream AMEL exon 8 in all genomes not only revealed the presence of exon 9 in all rodent sequences possessing exon 8, but also demonstrated the presence of a sequence with a high percentage of nucleotide identity in all placental AMELX, including human AMELX and AMELY. No sequence similarity was found in marsupial, monotreme, and non-mammalian gDNA.
Alignment of all putative exon 9 against murine sequences showed that most sequences, including human AMELX and AMELY exon 9, possessed a correct intron splice donor site at their 5′ side, along with a stop codon (Fig. 3C). In a few sequences only (e.g., kangaroo rat), the putative intron splice site is mutated, which indicates either that exon 9 is not coding (i.e., independently inactivated in a few species) or that another putative intron splice site is located upstream but is hard to define. The sequence length of exon 9 is variable (e.g., 15 bp in guinea pig, 27 in mouse, 60 in human AMELX, but 27 in AMELY, 69 in shrew), but the nucleotide identity in homologous regions indicates that this exon is probably coding. A putative polyadenylation signal is also at the correct location as compared with rodent sequences (not shown). Given the similarity between AMELX exon 9 sequence in representatives of most placental mammal lineages, i.e., covering a period of 104 million years (Ma) of evolution (Fig. 4), we believe that exon 9 is coding; otherwise, mutations would have accumulated at random.
Figure 4.
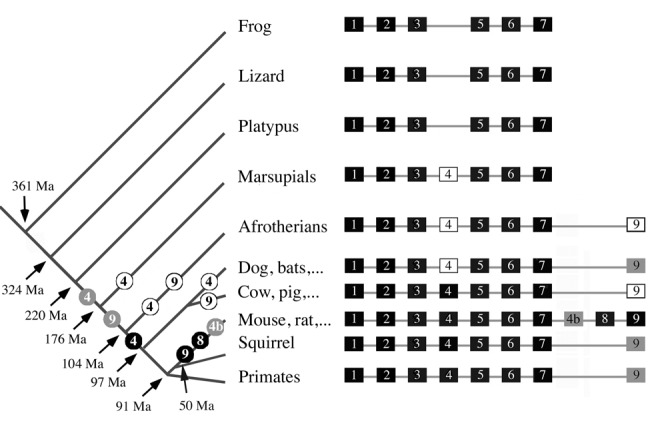
A summary of the story of AMELX small exons during mammalian evolution. On the left: putative location of the recruitment (numbered gray circles) then fixation (numbered black circles) or inactivation (numbered white circles) of AMELX exons 4, “4b”, 8, and 9 during evolution. AMELX exon 4 was recruited first, in an ancestral therian, then exon 9 in an ancestral placental mammal. Exon “4b” and exon 8 were recruited in the lineage leading to murid rodents, around 50 million years (Ma) ago. Once created, these 4 small, mammalian-specific AMELX exons were conserved as either functional (fixed) or not (pseudogenic) in all subsequent mammalian lineages. On the right: For each lineage, AMELX organization is shown with functional exons as black blocks, pseudo-exons as white blocks, and putatively coding exons as gray blocks. Estimated times of divergence: tetrapods from Hedges (2009), amniotes from Shedlock and Edwards (2009), mammals from Madsen (2009), placental mammals from Murphy and Eizirik (2009), and rodents from Adkins et al. (2001) and Huchon et al. (2002).
However, we have not been able to PCR amplify exon 9 from human fetal tooth cDNA.
Discussion
AMELX Exons 4, “4b”, 8, and 9 are Mammalian Innovations
The exploration of 39 tetrapod genomes allowed us to trace the origin of AMEL exons 4, “4b”, 8, and 9 in tetrapod evolution and to demonstrate that they are mammalian-specific AMELX exons. Indeed, these exons are not present in lizard and frog gDNA, confirming previous published AMEL transcript sequences in reptiles (Ishiyama et al., 1998; Delgado et al., 2006; Wang et al., 2006) and amphibians (Toyosawa et al., 1998; Diekwisch et al., 2009). None of these exons was found in the platypus genome, confirming previously sequenced AMEL transcripts (Toyosawa et al., 1998). This means that AMELX was composed of 6 functional exons (1-3, 5-7) in the last common ancestor of therian mammal (placentals + marsupials), i.e., circa 176 Ma ago.
Exon 4 appeared in an ancestral therian, i.e., between 220 and 176 Ma ago, but was not functional, confirming previous cDNA sequencing in the opossum (Hu et al., 1996). It was retained as a functional exon only later in placental evolution (thus, 7 exons for AMELX). Then, exon 9 was recruited in an ancestral placental, but was not retained in several lineages. Eventually, exons “4b” and 8 were created in an ancestral rodent.
Coding or Not Coding Mammalian-specific Exons
In addition to providing information on the timing of the recruitment of these small exons during evolution, our analyses indicated whether these exons are putatively coding in representatives of the mammalian lineages. Indeed, current data concerning AMELX transcript sequences are available in a limited species only.
Exon 4
Exon 4 was identified in the first published human, mouse, rat, cow, and pig AMELX cDNAs (Gibson et al., 1991; Salido et al., 1992; Simmer, 1995). However, this exon is not encoded in the major AMELX isoform (known as A-4) (Veis, 2003). In rodents, the isoform containing the region encoded by exon 4 (called A+4) could have a different function compared with A-4, as suggested by bead implantation in the exposed pulp of rat molars. A-4 induced closure of the root canal, formation of a reparative dentinal bridge, and diffuse mineralization in the mesial part of the pulp chamber. The reaction was weaker after A+4 implantation (Six et al., 2004; Jegat et al., 2007). The positive selection of 5 residues during mammalian evolution means that these residues were fixed (no longer subjected to substitution) during mammalian evolution, suggesting that they have acquired a function important for this region of the protein.
Our results support the following scenario:
(1) Exon 4 appeared in an ancestral therian after a DNA region containing a similarly sized coding exon was duplicated. AMELX exon 5 is the most probable candidate, being close to exon 4 and having the same size. Additional exons are mostly recruited through the duplication of a DNA region within the same gene, as, e.g., for the creation of exons “4b” and 8 in rodent AMELX (see below). A vast majority of such tandem duplications are likely to be involved in mutually exclusive alternative splicing events (Kondrashov and Koonin, 2001; Letunic et al., 2002). Such an alternative splicing is known for exon 4.
(2) Once created from exon 5, the peptide encoded by exon 4 did not improve protein function (as redundant peptide), and it accumulated numerous substitutions until a functional copy was retained by natural selection in, e.g., murids, primates, and artiodactyls; this process is expected after exon duplication, and could explain why sequence homology with exon 5 is no longer recognizable in all species possessing AMELX exon 4.
(3) Additional mutations occurred independently in therian lineages. Mutations affected the splice donor site, resulting in exon 4 inactivation in marsupials, afrotherians, xenarthrans, and some laurasiatherians. Given the short evolutionary period since exon 4 was inactivated, the sequence is still easily recognizable as pseudogenic.
(4) In some placental mammal lineages, exon 4 mutation resulted in a protein sequence somewhat useful for protein function (e.g., useful when included in some particular transcripts), and these changes were fixed after positive selection, which occurred in an ancestor of primates, rodents, and artiodactyls, as revealed by the conserved exon 4 sequence in these species.
Exons “4b” and 8
As mentioned before, exons “4b” and 8 are homologous to exons 4 and 5, respectively (Bartlett et al., 2006). However, exon “4b” has not been identified in the various AMELX transcripts identified thus far in murids.
In the mouse, by sequencing the genomic region separating exons 7 and 8, we answered “no” to the question of exon “4b” being a possible artifact generated during computer-aided assembly of this genomic region. But, how to explain the identical sequences of exons “4b” and 4, and the absence of exon “4b” in AMELX transcripts? If the duplication/translocation event had occurred recently, i.e., in the murine lineage, the resulting exon “4b” could have been non-coding without accumulating mutations during such a short time. However, we found that exons “4b” and 8 were created earlier, after the squirrel lineage diverged, i.e., approximately 50 Ma ago. This is sufficiently long for mutations accumulating at random; otherwise, sequence conservation means that it is subjected to functional constraints, i.e., it is coding. This finding is additionally supported when one considers the few nucleotide substitutions observed in exon “4b” compared with that observed in exon 8…that it is coding; both sequences were duplicated at the same time, but only exon 8 was found in several transcripts. Also, in the kangaroo rat, numerous substitutions were observed, while exon “4b” was inactivated.
However, although these findings indicate that exon “4b” should be coding, at least in rodents, we failed to find AMELX transcripts containing exon “4b” in a cDNA library of murine tooth buds. Either such a transcript is stage-specific during enamel formation or it is to be found in other loci in the mouse tooth. To date, the presence of exon “4b” in rodent AMELX gDNA remains an enigma.
Exon 9
In rodents, AMELX exon 9 encodes 9 residues and a stop codon. It was believed that the translocation of the gDNA region containing exons 4 and 5 downstream of exon 7 has triggered the activation of a downstream sequence, resulting in the expression of exon 9 (Bartlett et al., 2006). Here, we show that the exon 9 sequence was recruited long before rodent differentiation, in a placental mammal ancestor, 176-104 Ma ago. As discussed for exon 4, a gDNA region containing a coding exon was duplicated/translocated downstream of exon 7, mutations were accumulated at random, and fixation or inactivation occurred depending on whether the sequence was subjected to functional constraints.
However, why was exon 9 not found in mammalian AMELX transcripts other than in murids? The high similarity of exon 9 sequence in various mammals indicates that it should be coding, but our attempts to find transcripts including exon 9 in human tooth germs were unsuccessful. It is possible that such transcripts are stage-specific or expressed in other loci, or that human tooth enamel no longer requires the encoded peptide. Our cDNAs were prepared from human fetal tissue younger than 23 wks, when the tooth enamel was still in pre-secretory and early secretory stages. It is possible that exon 9 may not express at a detectable level at these stages. To date, the presence of an exon 9 sequence in non-murine AMELX gDNA also remains an enigma.
Supplementary Material
Footnotes
A supplemental appendix to this article is published electronically only at http://jdr.sagepub.com/supplemental.
This study has been financially supported by grants from the Centre National de la Recherche Scientifique and the Université Pierre et Marie Curie (UMR7138).
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
References
- Adkins RM, Gelke EL, Rowe D, Honeycutt RL. (2001). Molecular phylogeny and divergence time estimates for major rodent groups: evidence from multiple genes. Mol Biol Evol 18:777-791. [DOI] [PubMed] [Google Scholar]
- Bartlett JD, Ball RL, Kawai T, Tye CE, Tsuchiya M, Simmer JP. (2006). Origin, splicing, and expression of rodent amelogenin exon 8. J Dent Res 85:894-899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beniash E, Simmer JP, Margolis HC. (2005). The effect of recombinant mouse amelogenins on the formation and organization of hydroxyapatite crystals in vitro. J Struct Biol 149:182-190. [DOI] [PubMed] [Google Scholar]
- Delgado S, Couble ML, Magloire M, Sire JY. (2006). Cloning, sequencing and expression of the amelogenin gene in two scincid lizards. J Dent Res 85:138-143. [DOI] [PubMed] [Google Scholar]
- Diekwisch TG, Jin T, Wang X, Ito Y, Schmidt M, Druzinsky R, et al. (2009). Amelogenin evolution and tetrapod enamel structure. Front Oral Biol 13:74-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson C, Golub E, Herold R, Risser M, Ding W, Shimokawa H, et al. (1991). Structure and expression of the bovine amelogenin gene. Biochemistry 30:1075-1079. [DOI] [PubMed] [Google Scholar]
- Girondot M, Sire JY. (2010). UniDPlot: a software to detect weak similarities between two DNA sequences. J Bioinform Seq Anal 2:69-74. [Google Scholar]
- Hedges SB. (2009). Vertebrates (Vertebrata). In: The timetree of life. Hedges SD, Kumar S, editors, New York, NY: Oxford University Press, pp. 309-314. [Google Scholar]
- Hu CC, Zhang C, Qian Q, Ryu OH, Moradian-Oldak J, Fincham AG, et al. (1996). Cloning, DNA sequence, and alternative splicing of opossum amelogenin mRNAs. J Dent Res 75:1728-1734. [DOI] [PubMed] [Google Scholar]
- Huchon D, Madsen O, Sibbald MJ, Ament K, Stanhope MJ, Catzeflis F, et al. (2002). Rodent phylogeny and a timescale for the evolution of Glires: evidence from an extensive taxon sampling using three nuclear genes. Mol Biol Evol 19:1053-1065. [DOI] [PubMed] [Google Scholar]
- Ishiyama M, Mikami M, Shimokawa H, Oida S. (1998). Amelogenin protein in tooth germs of the snake Elaphe quadrivirgata, immunohistochemistry, cloning and cDNA sequence. Arch Histol Cytol 61:467-474. [DOI] [PubMed] [Google Scholar]
- Jegat N, Septier D, Veis A, Poliard A, Goldberg M. (2007). Short-term effects of amelogenin gene splice products A+4 and A-4 implanted in the exposed rat molar pulp. Head Face Med 3:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov FA, Koonin EV. (2001). Origin of alternative splicing by tandem exon duplication. Hum Mol Genet 10:2661-2669. [DOI] [PubMed] [Google Scholar]
- Letunic I, Copley RR, Bork P. (2002). Common exon duplication in animals and its role in alternative splicing. Hum Mol Genet 11:1561-1567. [DOI] [PubMed] [Google Scholar]
- Li R, Li W, DenBesten PK. (1995). Alternative splicing of amelogenin mRNA from rat incisor ameloblasts. J Dent Res 74:1880-1885. [DOI] [PubMed] [Google Scholar]
- Li W, Mathews C, Gao C, DenBesten PK. (1998). Identification of two additional exons at the 3’ end of the amelogenin gene. Arch Oral Biol 43:497-504. [DOI] [PubMed] [Google Scholar]
- Madsen O. (2009). Mammals (Mammalia). In: The timetree of life. Hedges SD, Kumar S, editors, New York, NY: Oxford University Press, pp. 459-461. [Google Scholar]
- Murphy WJ, Eizirik E. (2009). Placental mammals (Eutheria). In: The timetree of life. Hedges SD, Kumar S, editors, New York, NY: Oxford University Press, pp. 471-474. [Google Scholar]
- Pond SLK, Frost SD, Muse SV. (2005). HyPhy: hypothesis testing using phylogenies. Bioinformatics 21:676-679. [DOI] [PubMed] [Google Scholar]
- Rambaut A. (1996). Se-Al sequence alignment editor. Found at http://tree.bio.ed.ac.uk/software/seal
- Robinson C, Brookes SJ, Kirkham J, Bonass WA, Shore RC. (1996). Crystal growth in dental enamel: the role of amelogenins and albumin. Adv Dent Res 10:173-179. [DOI] [PubMed] [Google Scholar]
- Salido EC, Yen PH, Koprivnikar K, Yu LC, Shapiro LJ. (1992). The human enamel protein gene amelogenin is expressed from both the X and the Y chromosomes [see comments]. Am J Hum Genet 50:303-316. [PMC free article] [PubMed] [Google Scholar]
- Shedlock AM, Edwards SV. (2009). Amniotes (Amniota). In: The timetree of life. Hedges SD, Kumar S, editors, New York, NY: Oxford University Press, pp. 375-379. [Google Scholar]
- Simmer JP. (1995). Alternative splicing of amelogenins. Connect Tissue Res 32:131-136. [DOI] [PubMed] [Google Scholar]
- Six N, Tompkins K, Septier D, Veis A, Goldberg M. (2004). Recruitment and characterization of the cells involved in reparative dentin formation in the exposed rat molar pulp after implantation of amelogenin gene splice products A+4 and A-4. Oral Biosci Med 1:35-44. [Google Scholar]
- Toyosawa S, O’Huigin C, Figueroa F, Tichy H, Klein J. (1998). Identification and characterization of amelogenin genes in monotremes, reptiles, and amphibians. Proc Natl Acad Sci USA 95:13056-13061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veis A. (2003). Amelogenin gene splice products: potential signaling molecules. Cell Mol Life Sci 60:38-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Fan JL, Ito Y, Luan X, Diekwisch TG. (2006). Identification and characterization of a squamate reptilian amelogenin gene: Iguana iguana. J Exp Zool (Mol Dev Evol) 306(B):393-406. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.