Abstract
Cleft palate is among the most common birth defects. Currently, only 30% of cases have identified genetic causes, whereas the etiology of the majority remains to be discovered. We identified a new regulator of palate development, protein arginine methyltransferase 1 (PRMT1), and demonstrated that disruption of PRMT1 function in neural crest cells caused complete cleft palate and craniofacial malformations. PRMT1 is the most highly expressed of the protein arginine methyltransferases, enzymes responsible for methylation of arginine motifs on histone and nonhistone proteins. PRMT1 regulates signal transduction and transcriptional activity that affect multiple signal pathways crucial in craniofacial development, such as the BMP, TGFβ, and WNT pathways. We demonstrated that Wnt1-Cre;Prmt1fl/fl mice displayed a decrease in palatal mesenchymal cell proliferation and failure of palatal shelves to reach the midline. Further analysis in signal pathways revealed that loss of Prmt1 in mutant mice decreased BMP signaling activation and reduced the deposition of H4R3me2a mark. Collectively, our study demonstrates that Prmt1 is crucial in palate development. Our study may facilitate the development of a better strategy to interrupt the formation of cleft palate through manipulation of PRMT1 activity.
Keywords: posttranslational modifications, cleft palate, signal transduction, epigenetics, craniofacial biology/genetics, craniofacial anomalies
Introduction
Cleft palate is among the most common congenital defects, affecting nearly 1 in 700 live births (Gorlin et al. 2001; Mossey and Modell 2012). It represents a significant burden to the health care system and demands multidisciplinary treatment starting shortly after birth and extending to adulthood (Schoen et al. 2017). At present, only 30% of cases have identified genetic causes, whereas the etiology of the majority remains to be discovered (Jugessur et al. 2009; Dixon et al. 2011). During palate development, neural crest (NC)–derived cells migrate to the maxillary process and grow downward, flanking the developing tongue to form the palatal shelves. The palatal shelves then elevate, grow toward the midline, and fuse to form the roof of the oral cavity (Chai and Maxson 2006; Bush and Jiang 2012; Lan et al. 2015). Concomitant with palate development, the surrounding craniofacial tissues undergo significant growth and reorganization. During this period of morphogenesis, intrinsic defects in CNC cell migration, palatal shelf growth, elevation, or fusion can perturb palatogenesis and cause cleft palate. Cleft palate can also occur when malformation of surrounding structures impedes the elevation or contact of the palatal shelves at the proper developmental stage (Bush and Jiang 2012).
Palate development is precisely coordinated by a complex network of signaling pathways, including BMP, TGFβ, FGF, SHH, Notch, and WNT (Bush and Jiang 2012; Graf et al. 2016). Signaling activities of these pathways are controlled by posttranslational modification of signal mediators. For example, phosphorylation of Smad1 proteins by BMP-stimulated receptors leads to the transcriptional activation of Smad1 target genes and alterations of cell behavior. We recently identified another type of posttranslational modification, methylation, which controls signaling activity (Xu et al. 2013). Protein methylation on arginine motifs is catalyzed by a group of enzymes called protein arginine methyltransferases (PRMTs) 1 to 9. Among them, PRMT1 is the major methyltransferase, responsible for >85% of the arginine methylation activity in mammalian cells (Tang et al. 2000). Deletion of this enzyme in mice causes lethality shortly after implantation (Pawlak et al. 2000). The roles of PRMT1 have been identified in epigenetic regulation and transcription. PRMT1 methylates histone H4 at arginine 3 (H4R3), generating the H4R3 asymmetric dimethylation mark (H4R3me2a), which is essential for transcriptional activation (Wang et al. 2001). PRMT1 is also involved in multiple signaling pathways that are critical for palate development. PRMT1 is required to initiate BMP-induced signaling via methylation of the inhibitory Smad6 (Xu et al. 2013). PRMT1 also promotes TGFβ signaling through methylation of Smurf2 (Cha et al. 2015). Similarly, PRMT1 induces overactivation of EGF signaling and sustained cell proliferation through methylation of EGFR (Liao et al. 2015). However, PRMT1 negatively regulates the WNT/β-catenin pathway via methylation of Axin, G3BP1/2, and Dvl3 (Bikkavilli et al. 2012; Bikkavilli and Malbon 2011; 2012; Cha et al. 2011). While PRMT1 plays critical roles in the regulation of these signal pathways, whether PRMT1 regulates palate development in vivo is unknown.
In this study, we disrupted PRMT1 function in neural crest cell (NCC)–derived tissues using Wnt1-Cre. NCC-specific deletion of Prmt1 caused cleft palate and craniofacial malformations, including reductions in the size of the head, mandible, and tongue. In the Prmt1-deficient mice, palate mesenchymal cell proliferation was dramatically reduced, and palatal shelves failed to reach the midline, resulting in a complete cleft palate phenotype. We further demonstrated a significant repression of BMP signaling and reduction in histone modification at H4R3me2a, both of which are critical for cell proliferation and cell cycle progression. Our results highlight the physiologic significance of PRMT1 during palate development.
Materials and Methods
Generation of Transgenic Mice
Prmt1fl/fl mice were provided by Dr. Stéphane Richard (McGill University, Canada; Yu et al. 2009). To generate Wnt1-Cre;Prmt1fl/+ and Wnt1-Cre;Prmt1fl/fl mice, Wnt1-Cre mice (Jax 009107) were mated with Prmt1fl/fl mice (Tian et al. 2017). To generate Wnt1-Cre;Prmt1fl/fl;R26R tdTomato mice, Wnt1-Cre;Prmt1fl/+ mice were mated with Prmt1fl/fl;R26RtdTomato/tdTomato mice. Animal usage was approved by the Institutional Animal Care and Use Committee at the University of Southern California.
Detailed materials and methods are included in the Appendix.
Results
NCC-Specific Deletion of Prmt1 Causes Cleft Palate and Multiple Craniofacial Malformations
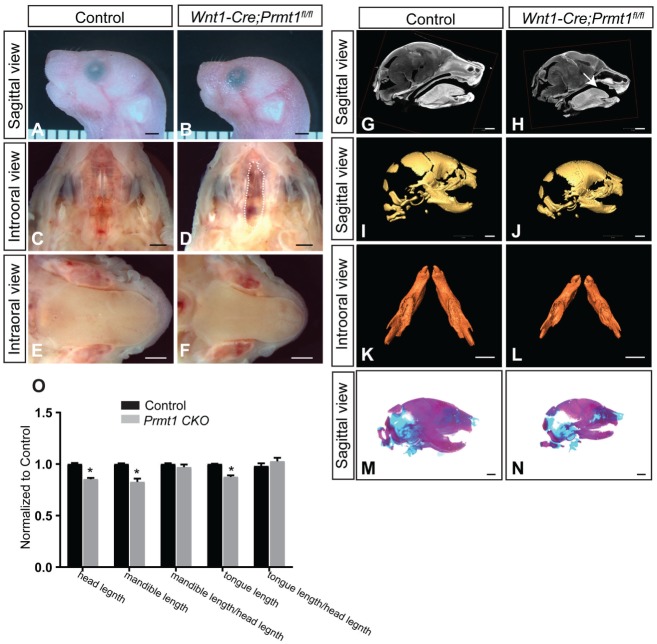
PRMT1 is the major arginine methyltransferase in mammalian cells. Because of its functional importance, loss of Prmt1 leads to lethality early during development, before embryonic day 7.5 (E7.5) in the mouse (Yu et al. 2009). To study the roles of PRMT1 during craniofacial development, we generated an NCC-specific deletion of Prmt1 using a Wnt1-Cre driver. The Wnt1-Cre;Prmt1fl/fl mice died soon after birth and exhibited multiple craniofacial defects, including a complete cleft palate and a smaller head, mandible, and tongue (Fig. 1A–N). With cleft palate, pups cannot create suction to obtain milk (Enomoto et al. 2010; Baek et al. 2011). The mutant newborns with complete cleft palate consistently showed an absence of milk spots (Appendix Fig. 1A, B).
Figure 1.
Wnt1-Cre;Prmt1fl/fl mice exhibit multiple craniofacial malformations at birth. (A, B) Sagittal views of newborn control and Wnt1-Cre;Prmt1fl/fl heads. Intraoral views of the (C, D) palate and (E, F) tongue of control and Wnt1-Cre;Prmt1fl/fl newborn mice. Dashed line in panel D indicates cleft palate. (G, H) Sagittal views of micro–computed tomography scans of control and Wnt1-Cre;Prmt1fl/fl newborn heads. Arrow in panel H indicates cleft palate. Three-dimensional reconstructions from micro–computed tomography scans of control and Wnt1-Cre;Prmt1fl/fl (I, J) heads and (K, L) mandibles. (M, N) Alizarin red/alcian blue staining of control and Wnt1-Cre;Prmt1fl/fl heads. (O) Normalized head length, mandible length, ratio between mandible length and head length, tongue length, and ratio between tongue length and head length. Error bars indicate SD. *P < 0.05. n = 4. Scale bar = 1 mm.
The Prmt1 mutant mice exhibited a complete cleft palate with a 100% penetrance in >60 mice studied, when the palates were analyzed along the anterior-posterior axis at newborn stage (Fig. 1C, D, G, H; Appendix Fig. 1C–H). The heads of Prmt1 mutant mice were smaller at the newborn stage, exhibiting a 14.7% decrease in head length (Fig. 1O). Craniofacial structures surrounding the palate, including the mandible and tongue, also exhibited a reduction in size. We analyzed the mandibles using micro–computed tomography scans, followed by 3-dimensional reconstruction at the newborn stage, and observed a 17% decrease in mandibular length, proportional to the reduction in the head size in Wnt1-Cre;Prmt1fl/fl mice (Fig. 1O). These findings were also confirmed with skeletal staining that showed shorter mandibles (Fig. 1M, N). The Wnt1-Cre;Prmt1fl/fl mice also displayed reduced length of the tongue, which was proportional to the reduction in the head size in Wnt1-Cre;Prmt1fl/fl mice (Fig. 1O). The Wnt1-Cre;Prmt1fl/+ mice were overtly normal (data not shown). These findings suggest that the functions of PRMT1 in NCC-derived tissue are crucial for palate formation and craniofacial development.
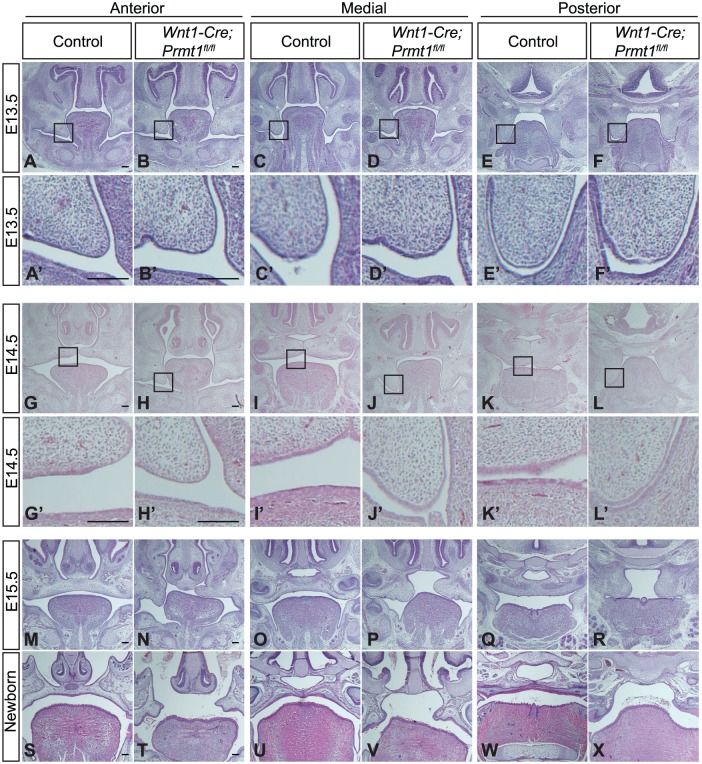
NCC-Specific Deletion of Prmt1 Causes a Developmental Delay and Compromises the Growth of Palatal Shelves
Palate morphogenesis in the Wnt1-Cre;Prmt1fl/fl mutant embryos was analyzed at multiple stages and positions. At E13.5, when palatal shelves of the control embryos grew downward flanking the tongue, the mutants were morphologically similar to the controls (Fig. 2A–F). At E14.5, when palatal shelves of the control mice elevated to reach the midline, the mutant embryos showed a delay in development (Fig. 2G–L) but no adherence to opposing tissues (Fig. 2A′–L′). At E15.5, when palatal shelves of control mice were completing fusion at the midline, palatal shelves of mutant mice had elevated but failed to reach the midline at all positions examined, from anterior, medial, to posterior palate (Fig. 2M–R). At the newborn stage, the mutant mice displayed a complete cleft palate phenotype (Fig. 2S–X). These data suggest that NCC-specific loss of Prmt1 compromised the growth of the palatal shelves.
Figure 2.
Palatal shelves in Wnt1-Cre;Prmt1fl/fl mice display developmental delay and failure to reach the midline. Hematoxylin and eosin staining of control and Wnt1-Cre;Prmt1fl/fl heads at (A–F) embryonic day 13.5 (E13.5), (G–L) E14.5, (M–R) E15.5, and (S–X) newborn stage. Higher-magnification views are shown in panels A′ to L′ to illustrate that the palatal shelves exhibited no adherence to the opposing tissues. Scale bar = 100 μm. n = 3.
Prmt1 Deficiency Impairs Cell Proliferation in Palatal Mesenchyme
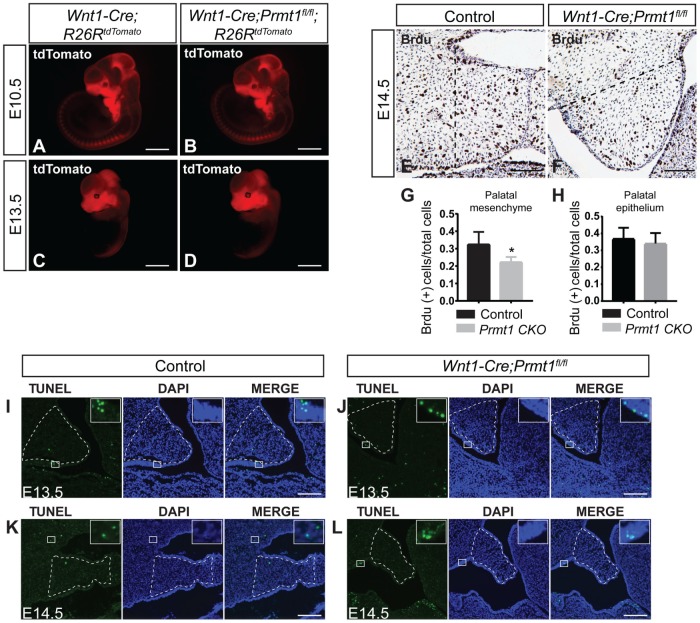
Palatal development is controlled by NCC migration, proliferation, and survival. We investigated whether any of these processes were altered in Wnt1-Cre;Prmt1fl/fl embryos. First, we determined that NCC migration was not impaired. We analyzed the expression profile of neural crest–derived, tdTomato-labeled cells in Wnt1-Cre;R26RtdTomato (control) and Wnt1-Cre;Prmt1fl/fl;R26RtdTomato (mutant) embryos. There was no difference in the expression pattern of tdTomato-labeled cells between control and mutant embryos at E10.5 or E13.5 (Fig. 3A–D), demonstrating that the formation and migration of neural crest–derived mesenchymal progenitors were unaffected. Next, we identified a significant reduction in the proliferation of palatal mesenchymal cells. The majority of palatal mesenchymal cells are derived from NCCs. We found that, in mutant mice, the ratio of BrdU-positive cells was significantly decreased in the NCC-derived palatal mesenchyme but not in the palatal epithelium, indicating a reduction in mesenchymal cell proliferation (Fig. 3E–H). Third, we found that cell apoptosis was not altered. The number of TUNEL-positive cells was very low in the palatal shelves of both groups and the number of TUNEL-positive cells was not different between control and mutant mice at any stage examined (Fig. 3I–L). As positive controls, TUNEL-positive apoptotic cells were observed in Meckel’s cartilage and the mandibular region (Appendix Fig. 3D–G). These results indicate that loss of Prmt1 in NCC-derived tissues impairs palatal mesenchymal cell proliferation without altering cell migration or apoptosis.
Figure 3.
Proliferation of the palatal mesenchymal cells is reduced, whereas neural crest cell migration and cell survival are unaffected in Wnt1-Cre;Prmt1fl/fl mice. tdTomato fluorescent imaging of whole-mount Wnt1-Cre;R26RtdTomato/+ (control) and Wnt1-Cre;Prmt1fl/fl;R26RtdTomato/+ embryos at (A, B) embryonic day 10.5 (E10.5) or (C, D) E13.5. n = 4. BrdU staining (dark brown) of control and Wnt1-Cre;Prmt1fl/fl palatal shelves at E14.5 was (E, F) illustrated and (G, H) quantified. Quantification of BrdU-positive proliferating cells was performed between the dashed line to the tip of palatal shelves, as illustrated in panels E and F. Error bars indicate SD. *P < 0.05. n = 4. For panel G, >300 cells were analyzed per animal. For panel H, >100 cells were analyzed per animal. (I–L) TUNEL staining of E13.5 and E14.5 control and Wnt1-Cre;Prmt1fl/fl palates. High magnification of TUNEL-positive cells in the white square is shown in right upper corner of panels I to L. Dotted lines in panel I to L indicate palatal mesenchyme. Scale bar = 1 mm (A–D), 50 μm (E, F), 100 μm (I–L).
Prmt1 Deficiency Inhibits BMP Signaling Activation and Affects Histone Modification
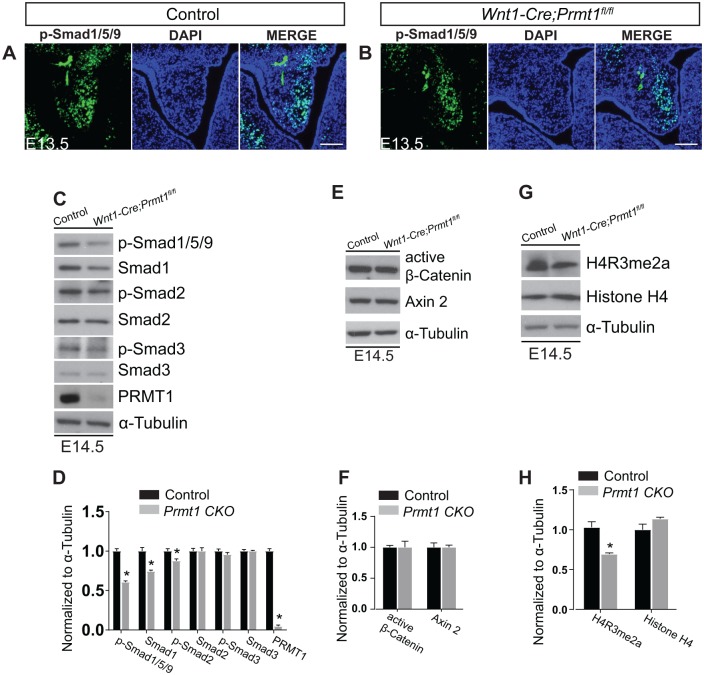
PRMT1-mediated methylation of signaling mediators regulate TGFβ, BMP, and WNT pathways, all of which are critical for palatogenesis and control cell proliferation (Bikkavilli and Malbon 2011; Biggar and Li 2015). We therefore assessed whether BMP, TGFβ, and WNT signaling pathways were altered in Wnt1-Cre;Prmt1fl/fl embryos. We previously reported that PRMT1-mediated methylation controls BMP signaling activation (Xu et al. 2013). Therefore, loss of Prmt1 may impair BMP-induced Smad1/5/9 phosphorylation and reduce the activation of downstream target genes. We examined Smad1/5/9 phosphorylation in palatal shelves from control or Prmt1-deficient mice at E13.5 by immunostaining. In Prmt1-deficient palate, we observed a significant reduction of Smad1/5/9 phosphorylation in the NCC-derived palatal mesenchyme (Fig. 4A, B). These findings were further supported by Western blot analysis at E14.5, in which phospho-Smad1/5/9 decreased in mutant palatal shelves. The total levels of Smad1 also showed a moderate decline in the mutant at E14.5 (Fig. 4C, first and second rows, 4D). PRMT1-mediated methylation is also known to regulate the TGFβ pathway (Cha et al. 2015). We examined Smad2 and Smad3 phosphorylation and noted no change in mutant palatal shelves by immunostaining and Western blot analysis (Appendix Fig. 2A–D; Fig. 4C, third to sixth, rows, 4D). There was a significant decrease in phospho-Smad2, as shown with densitometry (Fig. 4D); however, the magnitude of this change was only 15%. Similarly, activity of the WNT pathway was unchanged in control and mutant palatal shelves (Appendix Fig. 2E, F; Fig. 4E, F).
Figure 4.
BMP activation and H4R3me2a modification are repressed in Wnt1-Cre;Prmt1fl/fl mice. Immunostaining (green) of (A) control and (B) Wnt1-Cre;Prmt1fl/fl palates at embryonic day 13.5 (E13.5) for p-Smad1/5/9. Western blots of control and Wnt1-Cre;Prmt1fl/fl palates at E14.5 for (C) BMP and TGFβ signaling mediators: p-Smad1/5/9, Smad1, p-Smad2, Smad2, p-Smad3, and Smad3. (E) WNT/β-catenin signaling mediators active β-catenin and Axin 2 and (G) epigenetic modification H4R3me2a and histone 4. (D, F, H) Quantification of Western blots from panels C, E, and G, respectively. Data are representative of 3 independent samples for each staining and Western blot. Error bars indicate SD. *P < 0.05. Scale bar = 100 μm.
PRMT1 also regulates histone methylation. PRMT1 methylates H4R3 to generate the H4R3me2a mark that is a prerequisite for histone tail acetylation and an “open chromatin,” which allows transcriptional activation (Strahl et al. 2001; Wang et al. 2001; Huang et al. 2005). We examined H4R3me2a in palatal shelves and identified that Prmt1 deficiency dramatically decreased the H4R3me2a level (Fig. 4G, H). Taken together, our studies indicate that NCC-specific Prmt1 deficiency leads to a significant reduction of BMP-Smad1/5/9 signaling and a decrease in the levels of the H4R3me2a mark.
Prmt1 Deficiency Causes Downregulation of Cyclin D1 Expression
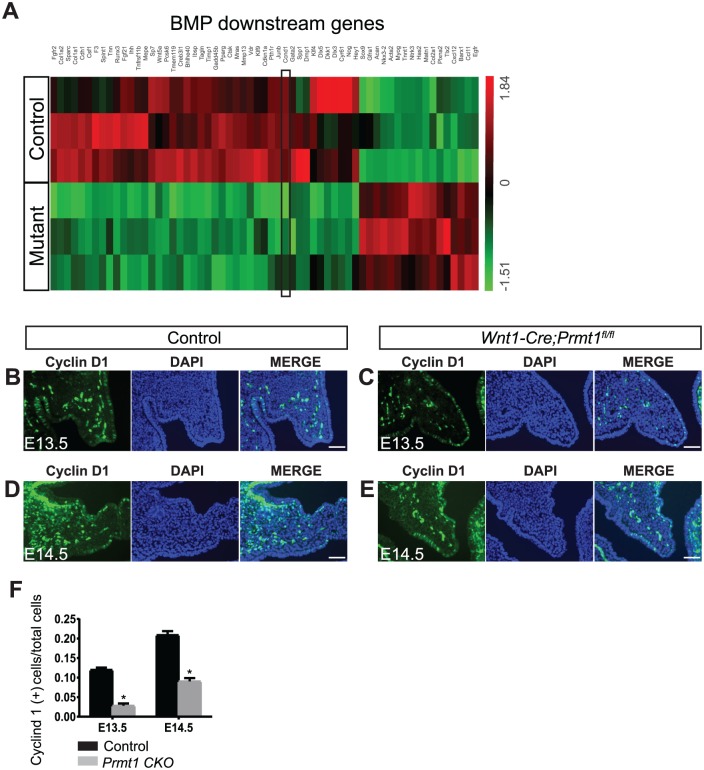
To further define the molecular mechanisms of reduced cell proliferation in the Prmt1-deficient palate, we compared the transcription profile of control and mutant palatal shelves at E14.5 using RNA sequencing and identified 1,782 genes that were differentially expressed. Through Ingenuity Pathway Analysis, we identified the BMP pathway as a significantly inhibited pathway (P = 4.51E-18, Z score = −2.287 for BMP2; P = 2.08E-07, Z score = −1.832 for BMP4) with 61 BMP downstream genes that were differentially expressed in Wnt1-Cre;Prmt1fl/fl palates as compared with controls (Fig. 5A), supporting our findings that BMP signaling was repressed in the mutant palate (Fig. 4A–C). We identified Ccnd1 among the repressed genes downstream of BMP signaling. This gene encodes cyclin D1, which controls palate mesenchyme cell proliferation (Lan and Jiang 2009). To further confirm that Prmt1 deficiency induced downregulation of Ccnd1, we analyzed the expression of cyclin D1 in palatal shelves by immunostaining at E13.5 and E14.5. The ratio of cyclin D1–positive cells was significantly decreased in palatal mesenchyme of mutant mice (Fig. 5B–F), demonstrating that Prmt1 deficiency causes downregulation of cyclin D1 to decrease proliferation.
Figure 5.
Cyclin D1 is downregulated in the palate of Wnt1-Cre;Prmt1fl/fl mice. (A) Heat map of BMP2 and BMP4 downstream genes, which includes 61 genes with significant differences in expression between control and Wnt1-Cre;Prmt1fl/fl (mutant) palates at embryonic day 14.5 (E14.5). n = 3. Black frame indicates Ccnd1. (B–E) Cyclin D1 immunostaining (green) of control and Wnt1-Cre;Prmt1fl/fl palates at E13.5 and E14.5. (F) Quantification of the percentage of cyclin D1–positive cells in palatal mesenchyme. Error bars indicate SD. *P < 0.05. n = 4. Scale bar = 100 μm.
Osteogenic Progenitor Formation in the Mandibles and Muscle Formation in the Tongue Are Unaffected in Wnt1-Cre;Prmt1fl/fl Mice
Cleft palate can arise from intrinsic palatal defects or occur as a consequence of mandible or tongue malformation. To evaluate whether the reduction in mandibular size caused cleft palate in Wnt1-Cre;Prmt1fl/fl mice, we examined whether mandibular malformations restricted palatal growth in Wnt1-Cre;Prmt1fl/fl embryos. From E13.5 until E15.5, when cleft palate was prominent in mutant embryos, mandibular size remained proportional to head length in the mutant mice (Appendix Fig. 3A–I), although the head size and mandible size were 10% to 20% smaller in the mutant mice. The mutant mandible also exhibited a normal symphysis of the Meckel’s cartilage (Appendix Fig. 3J, K). At these stages, cell proliferation and apoptosis in the mandibular primordium were indistinguishable between mutant and control samples (Appendix Fig. 4A–G). We further studied whether Prmt1 deficiency disrupted osteogenic differentiation by analyzing the expression of Sp7, a marker for osteogenic progenitors. At E13.5 and E14.5, there was no significant difference in the expression pattern of Sp7 between control and mutant embryos (Appendix Fig. 4H–K).
Next, to evaluate whether tongue malposition caused cleft palate in Wnt1-Cre;Prmt1fl/fl mice, we examined tongue morphogenesis. NC-derived cells constitute the tongue interstitium and promote mesoderm-derived muscle progenitor differentiation. We evaluated tongue muscle formation using 3 myogenic markers: MyoD for myogenic progenitors, myogenin for differentiated myoblast, and MHC for mature muscle fibers. There was no alteration in the expression of any marker, suggesting that tongue muscle differentiation and patterning were not disrupted (Appendix Fig. 5A–F).
These data demonstrate that the palatal defects are much more pronounced than mandibular and tongue defects in mutant mice, suggesting the palate-intrinsic cause of malformation.
Discussion
In this study, we investigated the function of PRMT1 in craniofacial development, through genetic ablation in postmigratory NC-derived cells using Wnt1-Cre. The loss of Prmt1 resulted in a complete cleft palate. The Wnt1-Cre; Prmt1fl/fl mice died shortly after birth with no milk in the stomach (Appendix Fig. 1A, B), consistent with the notion that cleft palate impairs suckling for milk and is therefore associated with perinatal death (Enomoto et al. 2010; Baek et al. 2011). To our knowledge, this is the first demonstration that arginine methylation by the major arginine methyltransferase PRMT1 is required for palate development.
Protein arginine methylation is a form of posttranslational modifications that is as common as phosphorylation and ubiquitination (Larsen et al. 2016). It is carried out by the PRMT family of enzyme (Bedford and Clarke 2009). Within this family, PRMT1 is the major PRMT that controls the activity of signal pathways through methylation of signaling mediators and epigenetic modifiers. The importance of this methyltransferase is evidenced by the finding that genetic deletion of Prmt1 results in the loss of >85% of arginine methyltransferase activity (Tang et al. 2000; Yu et al. 2009). Although extensive work has been conducted to reveal functions of PRMT1 in cancer, through controlling signal transduction, transcription, RNA processing, and DNA repair, the roles of PRMT1 in craniofacial development were unknown prior to our study.
We previously identified that PRMT1 initiates BMP signaling. PRMT1 methylates the inhibitory Smad, Smad6, at the N-terminal region, which relieves Smad6-mediated inhibition of membrane-bound BMP receptors and initiates Smad1 activation (Xu et al. 2013). Depletion of Prmt1 in mammalian cells or Drosophila leads to the reduction of Smad1 activation (Xu et al. 2013). PRMT1 also modulates other signal pathways in cancer cells. It promotes TGFβ-Smad2/3 signaling through methylation of Smurf2 (Cha et al. 2015) and represses the WNT-βcatenin pathway via methylation of Axin, G3BP1/2, and Dvl3 (Bikkavilli and Malbon 2011, 2012; Cha et al. 2011; Bikkavilli et al. 2012). Because the BMP, TGFβ, and WNT pathways are known to play critical roles during palate development, we analyzed their activity. We observed that in the Prmt1 mutant mice, activation of Smad1/5/9 downstream of BMP was reduced in palatal mesenchymal cells (Fig. 4A–D), which echoes our earlier report (Xu et al. 2013). Smad2, Smad3, and nuclear β-catenin activation were not altered in the palatal shelves of Prmt1 mutant mice (Fig. 4C–F; Appendix Fig. 2A–F). PRMT1 showed more specific signaling effects as compared with the identified roles in cancer cells. This can be explained by several mechanisms. First, the signaling effects of PRMT1 may be controlled by the availability of substrates in palatal mesenchyme. For instance, Smurf2 expression is low in the NC-derived palatal mesenchyme, although it is highly expressed in the palatal epithelium; therefore, NC-specific deletion of PRMT1 may not cause a dramatic change to TGFβ signaling through Smurf2 methylation during palate development (Tateossian et al. 2009). Second, the signaling effects of PRMT1 may be limited by compartmentalization at the subcellular level. These types of regulation have been exemplified by findings on WNT signaling, in which cilia transport the β-catenin cofactor Jbn to distal regions to decrease β-catenin-Jbn complex formation and reduce nuclear import of β-catenin (Lancaster et al. 2011). We reported previously that PRMT1 resides in multiple subcellular compartments and that PRMT1-mediated regulation of BMP signaling is associated with membrane-bound PRMT1 in cultured keratinocytes (Xu et al. 2013). The functional specificity of PRMT1 during palate development may also be controlled by compartmentalization, and the detailed mechanisms await further investigation.
Palate development is a multistep process that involves palatal shelf growth, elevation, adhesion, and fusion (Bush and Jiang 2012). In Prmt1 mutant mice, the mutant palatal shelves failed to elevate at E14.5, indicating a developmental delay. Although palatal shelves of mutant mice elevated at E15.5, they failed to reach the midline. We further showed a reduction of cell proliferation in the palatal mesenchyme, suggesting growth arrest of the palatal shelves. In support of these findings, cyclin D1 expression was dramatically reduced in palatal mesenchymal cells, resulting in cell cycle arrest. The reduction in cyclin D1 expression was first revealed by RNA-sequencing analysis to profile differentially regulated genes in Wnt1-Cre;Prmt1fl/fl palates as compared with controls. Using the Ingenuity Pathway Analysis software for bioinformatic analysis, we further identified the BMP pathway as a significantly inhibited pathway in the mutant palate, lending support to the notion that PRMT1 mediates regulation of BMP signaling. In palatogenesis, the BMP signal promotes cyclin D1 expression through direct and indirect mechanisms (Liu et al. 2003; Lan and Jiang 2009; Bonilla-Claudio et al. 2012; Parada and Chai 2012; Gou et al. 2015). Complementary to these documentations, we identified Ccnd1, the gene encoding cyclin D1, as one of the BMP downstream genes through RNA-sequencing analysis. Therefore, we propose that Prmt1 deficiency in palatal mesenchyme represses BMP signaling, which leads to a reduction in cyclin D1 expression that inhibits proliferation and thereby impairs palatal mesenchyme growth.
The Prmt1 mutant palatal mesenchyme displayed a 31% reduction in cell proliferation at E14.5; however, cell proliferation in the mandibular primordium was not affected at the same developmental stage, although the head size and mandibular length were decreased by 10% to 20% in Prmt1 mutant mice. These data suggest that in different NC-derived tissues, PRMT1-regulated signaling pathways employ distinct developmental kinetics. Further analysis is needed to define distinct PRMT1-regulated signaling mechanisms in various NC-derived tissues.
Taken together, our study revealed critical roles of PRMT1 in craniofacial development, especially palatogenesis. We showed that Prmt1 deficiency in NC-derived cells caused cleft palate and craniofacial malformations in the mandible and tongue. From the clinical perspective, manipulation of PRMT1 activity is potential therapeutic approach to reduce the risk of cleft palate.
Author Contributions
Y. Gou, contributed to conception, design, data acquisition, analysis, and interpretation, drafted and critically revised the manuscript; J. Li, contributed to design, data acquisition, analysis, and interpretation, critically revised the manuscript; O. Jackson-Weaver, J. Wu, T. Zhang, R. Gupta, I. Cho, Y. Chen, M. Li, contributed to data analysis, critically revised the manuscript; T.V. Ho, Y. Chai, contributed to data acquisition and analysis, critically revised the manuscript; S. Richard, contributed to data acquisition, critically revised the manuscript; J. Wang, contributed to conception and design, critically revised the manuscript; J. Xu, contributed to conception, design, data acquisition, analysis, and interpretation, critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Supplemental Material
Supplemental material, DS_10.1177_0022034518785164 for Protein Arginine Methyltransferase PRMT1 Is Essential for Palatogenesis by Y. Gou, J. Li, O. Jackson-Weaver, J. Wu, T. Zhang, R. Gupta, I. Cho, T.V. Ho, Y. Chen, M. Li, S. Richard, J. Wang, Y. Chai and J. Xu in Journal of Dental Research
Acknowledgments
We thank Drs. Hu Zhao, Jifan Feng, Hua Tian, Yuxing Guo, and Ms. Shihong Shi for their support in this project. We also thank Dr. Bridget Samuels for review and editing of the manuscript.
Footnotes
A supplemental appendix to this article is available online.
This work was supported by a research seed grant from the University of Southern California (USC) School of Dentistry (to J.X.), a start-up fund from the Provost at USC (to J.X.), and grants from the National Natural Science Foundation of China (grants 81771114, 81470776; to J.W.).
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
ORCID iDs: Y. Gou  https://orcid.org/0000-0001-8725-3702
https://orcid.org/0000-0001-8725-3702
References
- Baek JA, Lan Y, Liu H, Maltby KM, Mishina Y, Jiang R. 2011. Bmpr1a signaling plays critical roles in palatal shelf growth and palatal bone formation. Dev Biol. 350(2):520–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford MT, Clarke SG. 2009. Protein arginine methylation in mammals: who, what, and why. Mol Cell. 33(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggar KK, Li SS. 2015. Non-histone protein methylation as a regulator of cellular signalling and function. Nat Rev Mol Cell Biol. 16(1):5–17. [DOI] [PubMed] [Google Scholar]
- Bikkavilli RK, Avasarala S, Vanscoyk M, Sechler M, Kelley N, Malbon CC, Winn RA. 2012. Dishevelled3 is a novel arginine methyl transferase substrate. Sci Rep. 2:805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bikkavilli RK, Malbon CC. 2011. Arginine methylation of G3BP1 in response to Wnt3a regulates β-catenin mRNA. J Cell Sci. 124(Pt 13):2310–2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bikkavilli RK, Malbon CC. 2012. Wnt3a-stimulated LRP6 phosphorylation is dependent upon arginine methylation of G3BP2. J Cell Sci. 125(Pt 10):2446–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonilla-Claudio M, Wang J, Bai Y, Klysik E, Selever J, Martin JF. 2012. Bmp signaling regulates a dose-dependent transcriptional program to control facial skeletal development. Development. 139(4):709–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush JO, Jiang R. 2012. Palatogenesis: morphogenetic and molecular mechanisms of secondary palate development. Development. 139(2):231–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha B, Kim W, Kim YK, Hwang BN, Park SY, Yoon JW, Park WS, Cho JW, Bedford MT, Jho EH. 2011. Methylation by protein arginine methyltransferase 1 increases stability of Axin, a negative regulator of Wnt signaling. Oncogene. 30(20):2379–2389. [DOI] [PubMed] [Google Scholar]
- Cha B, Park Y, Hwang BN, Kim SY, Jho EH. 2015. Protein arginine methyltransferase 1 methylates Smurf2. Mol Cells. 38(8):723–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y, Maxson RE. 2006. Recent advances in craniofacial morphogenesis. Dev Dyn. 235(9):2353–2375. [DOI] [PubMed] [Google Scholar]
- Dixon MJ, Marazita ML, Beaty TH, Murray JC. 2011. Cleft lip and palate: understanding genetic and environmental influences. Nat Rev Genet. 12(3):167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto H, Nelson CM, Somerville RP, Mielke K, Dixon LJ, Powell K, Apte SS. 2010. Cooperation of two adamts metalloproteases in closure of the mouse palate identifies a requirement for versican proteolysis in regulating palatal mesenchyme proliferation. Development. 137(23):4029–4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlin RJ, Cohen MM, Jr, Hennekam RC. 2001. Syndromes of the head and neck. Oxford (UK): Oxford University Press. [Google Scholar]
- Gou Y, Zhang T, Xu J. 2015. Transcription factors in craniofacial development: from receptor signaling to transcriptional and epigenetic regulation. Curr Top Dev Biol. 115:377–410. [DOI] [PubMed] [Google Scholar]
- Graf D, Malik Z, Hayano S, Mishina Y. 2016. Common mechanisms in development and disease: BMP signaling in craniofacial development. Cytokine Growth Factor Rev. 27:129–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Litt M, Felsenfeld G. 2005. Methylation of histone H4 by arginine methyltransferase PRMT1 is essential in vivo for many subsequent histone modifications. Genes Dev. 19(16):1885–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jugessur A, Farlie P, Kilpatrick N. 2009. The genetics of isolated orofacial clefts: from genotypes to subphenotypes. Oral Dis. 15(7):437–453. [DOI] [PubMed] [Google Scholar]
- Lan Y, Jiang R. 2009. Sonic hedgehog signaling regulates reciprocal epithelial-mesenchymal interactions controlling palatal outgrowth. Development. 136(8):1387–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan Y, Xu J, Jiang R. 2015. Cellular and molecular mechanisms of palatogenesis. Curr Top Dev Biol. 115:59–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster MA, Schroth J, Gleeson JG. 2011. Subcellular spatial regulation of canonical Wnt signalling at the primary cilium. Nat Cell Biol. 13(6):700–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen SC, Sylvestersen KB, Mund A, Lyon D, Mullari M, Madsen MV, Daniel JA, Jensen LJ, Nielsen ML. 2016. Proteome-wide analysis of arginine monomethylation reveals widespread occurrence in human cells. Sci Signal. 9(443):rs9. [DOI] [PubMed] [Google Scholar]
- Liao HW, Hsu JM, Xia WY, Wang HL, Wang YN, Chang WC, Arold ST, Chou CK, Tsou PH, Yamaguchi H, et al. 2015. PRMT1-mediated methylation of the EGF receptor regulates signaling and cetuximab response. J Clin Invest. 125(12):4529–4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Selever J, Lu MF, Martin JF. 2003. Genetic dissection of Pitx2 in craniofacial development uncovers new functions in branchial arch morphogenesis, late aspects of tooth morphogenesis and cell migration. Development. 130(25):6375–6385. [DOI] [PubMed] [Google Scholar]
- Mossey PA, Modell B. 2012. Epidemiology of oral clefts 2012: an international perspective. Front Oral Biol. 16:1–18. [DOI] [PubMed] [Google Scholar]
- Parada C, Chai Y. 2012. Roles of BMP signaling pathway in lip and palate development. Front Oral Biol. 16:60–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlak MR, Scherer CA, Chen J, Roshon MJ, Ruley HE. 2000. Arginine N-methyltransferase 1 is required for early postimplantation mouse development, but cells deficient in the enzyme are viable. Mol Cell Biol. 20(13):4859–4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoen C, Aschrafi A, Thonissen M, Poelmans G, Von den Hoff JW, Carels CEL. 2017. MicroRNAs in palatogenesis and cleft palate. Front Physiol. 8:165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Briggs SD, Brame CJ, Caldwell JA, Koh SS, Ma H, Cook RG, Shabanowitz J, Hunt DF, Stallcup MR, et al. 2001. Methylation of histone H4 at arginine 3 occurs in vivo and is mediated by the nuclear receptor coactivator PRMT1. Curr Biol. 11(12):996–1000. [DOI] [PubMed] [Google Scholar]
- Tang J, Frankel A, Cook RJ, Kim S, Paik WK, Williams KR, Clarke S, Herschman HR. 2000. PRMT1 is the predominant type I protein arginine methyltransferase in mammalian cells. J Biol Chem. 275(11):7723–7730. [DOI] [PubMed] [Google Scholar]
- Tateossian H, Hardisty-Hughes RE, Morse S, Romero MR, Hilton H, Dean C, Brown SD. 2009. Regulation of TGF-beta signalling by Fbxo11, the gene mutated in the Jeff otitis media mouse mutant. Pathogenetics. 2(1):5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H, Feng J, Li J, Ho TV, Yuan Y, Liu Y, Brindopke F, Figueiredo JC, Magee W, 3rd, Sanchez-Lara PA, et al. 2017. Intraflagellar transport 88 (IFT88) is crucial for craniofacial development in mice and is a candidate gene for human cleft lip and palate. Hum Mol Genet. 26(5):860–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HB, Huang ZQ, Xia L, Feng Q, Erdjument-Bromage H, Strahl BD, Briggs SD, Allis CD, Wong J, Tempst P, et al. 2001. Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science. 293(5531):853–857. [DOI] [PubMed] [Google Scholar]
- Xu J, Wang AH, Oses-Prieto J, Makhijani K, Katsuno Y, Pei M, Yan L, Zheng YG, Burlingame A, Bruckner K, et al. 2013. Arginine methylation initiates BMP-induced smad signaling. Mol Cell. 51(1):5–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Chen T, Hebert J, Li E, Richard S. 2009. A mouse PRMT1 null allele defines an essential role for arginine methylation in genome maintenance and cell proliferation. Mol Cell Biol. 29(11):2982–2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, DS_10.1177_0022034518785164 for Protein Arginine Methyltransferase PRMT1 Is Essential for Palatogenesis by Y. Gou, J. Li, O. Jackson-Weaver, J. Wu, T. Zhang, R. Gupta, I. Cho, T.V. Ho, Y. Chen, M. Li, S. Richard, J. Wang, Y. Chai and J. Xu in Journal of Dental Research