Abstract
Precise and efficient genetic manipulations have enabled researchers to understand gene functions in disease and development, providing a platform to search for molecular cures. Over the past decade, the unprecedented advancement of genome editing techniques has revolutionized the biological research fields. Early genome editing strategies involved many naturally occurring nucleases, including meganucleases, zinc finger nucleases, and transcription activator-like effector-based nucleases. More recently, the clustered regularly interspaced short palindromic repeats (CRISPR) / CRISPR-associated nucleases (CRISPR/Cas) system has greatly enriched genetic manipulation methods in conducting research. Those nucleases generate double-strand breaks in the target gene sequences and then utilize DNA repair mechanisms to permit precise yet versatile genetic manipulations. The oral and craniofacial field harbors a plethora of diseases and developmental defects that require genetic models that can exploit these genome editing techniques. This review provides an overview of the genome editing techniques, particularly the CRISPR/Cas9 technique, for the oral and craniofacial research community. We also discuss the details about the emerging applications of genome editing in oral and craniofacial biology.
Keywords: gene editing, CRISPR, DNA cleavage, DNA repair, genetic therapy, dentistry
Introduction: A Historical View of the Importance of Sequence-Specific DNA Cleavage
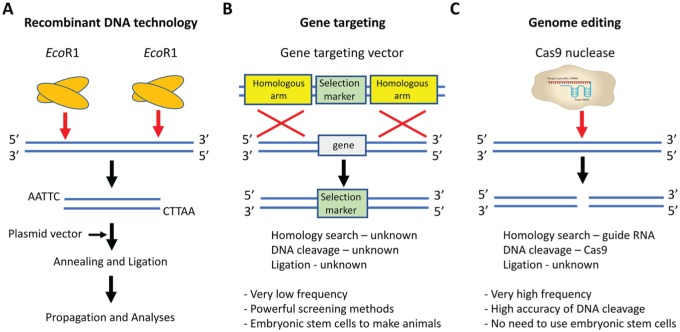
Classically, genetic manipulation methods received rapid attention due to their profound impacts on research opportunities and the highly translational potential to address clinical genetic conditions. The first precise genetic manipulation method was recombinant DNA technology, which encompassed components such as restriction enzymes, plasmid DNA vectors, donor DNA fragments, and DNA ligases. Restriction enzymes such as EcoRI cut the donor and plasmid DNA at defined sites. Ligases such as T4 DNA ligase then rejoin the modified donor and plasmid DNA to facilitate the propagation and analyses of the newly constructed DNA fragments (Fig. 1A). Thanks to the evolutional applications of the restriction enzymes, researchers subverted the nonspecific genetic manipulation method using random shearing. For the first time, specific recognition sites were introduced into the research field to understand the functions of genetic materials.
Figure 1.
A historical view of genetic modification. (A) Recombinant DNA technology. The restriction enzyme EcoRI cuts the donor DNA and plasmid vector. Modified plasmid vector and donor DNA are annealed and ligated to facilitate the propagation and analyses of the newly constructed DNA fragment. (B) Gene targeting. A gene-targeting vector contains a selection marker flanked by 2 homologous arms; the target gene is flanked by the same arms. Through a homologous recombination, the selection marker replaces the target locus. The exact mechanisms of homology search, DNA cleavage, and ligation are poorly understood in gene targeting and are therefore labeled as “unknown.” Gene targeting also presents with low frequency and requires powerful screening methods for embryonic stem cells. (C) Genome editing. Cas9 enzyme and sgRNA containing crRNA and tracrRNA can cleave DNA at the specific sites. In this method, the homology search relies on the guide RNA, and DNA cleavage relies on Cas9 enzyme; the ligation mechanism is still unknown. This genetic manipulation method presents with high frequency and the accuracy of DNA cleavage. It also surpasses the need to use the embryonic stem cells. crRNA, CRISPR RNA; sgRNA, single-guide RNA; tracrRNA, transactivating crRNA.
The second precise genetic manipulation method was the gene-targeting technology. A gene-targeting vector contains a selection marker flanked by 2 homologous arms. The target locus is flanked by the same arms. Through the homologous recombination, the selection marker replaces the target locus, knocking out the gene functions (Fig. 1B). The precise mechanisms of homologous pairing, DNA cleavage, and rejoining in mammalian cells are poorly understood. The subsequent targeting efficiency is quite low (i.e., 1 targeting event out of 1 million cells). Powerful screening strategies for these rare targeting events have therefore become an important key to success. The establishment of embryonic stem cells (ESCs) is a milestone in the screening strategy because researchers can now screen 1 million cultured ESCs other than 1 million fertilized eggs. Those epoch-making technologies generated thousands of genetically modified animals to enrich our understanding of gene functions. Until now, the mouse is one of very few species that were able to generate ESCs. A lack of information about enzymes to specifically and efficiently cleave genomic DNA leads to a long-awaited desire of the field to establish more efficient and precise genetic manipulation tools.
Over the past decade, the discovery of clustered regularly interspaced short palindromic repeats (CRISPR) / CRISPR-associated nucleases (CRISPR/Cas) has represented the third generation of precise genetic manipulation technology, often referred to as genome editing. This CRISPR/Cas technology is to borrow the DNA sequence–dependent DNA endonuclease systems found in the lower organisms to introduce specific double-strand breaks (DSBs) in the host genomes. Endogenous DNA repair pathways succeed DSBs and lead to the desired genetic manipulations. Within the CRISPR/Cas9 system, single-guide RNA (sgRNA) drives the homology search for specific recognition sites, and the Cas9 enzyme leads DNA cleavages (Fig. 1C). Such sequence-specific cleavages greatly improved the precision in genetic manipulation. In 2015, CRISPR/Cas9 was named by Science as the “breakthrough of the year,” given that this system revolutionized the biological research field (Travis 2015). CRISPR/Cas9 offers an easy, inexpensive, and much faster genome-editing strategy. Therefore, as stated by Dr. Jennifer Doudna, who published the first report of CRISPR, CRISPR technology would be “like PCR, a tool in the toolbox.” In line with this worldwide recognition, a race in utilizing and improving the CRISPR technique quickly began. When the keyword “CRISPR” was searched on PubMed, we found that >80% of the articles were published after 2015. This expanding interest has underscored the importance and immediacy of the genome-editing field. The oral and craniofacial field harbors a plethora of diseases and developmental defects that require researchers to explore gene functions on a genome scale and to search for gene therapies. Furthermore, genome editing demonstrates significant translational and clinical potential.
This review provides an overview of major genome-editing nucleases, focusing on the recent advances of CRISPR/Cas9. We also address the applications of genome editing techniques in the oral and craniofacial research. We finally discuss the advantages, limitations, and ethical issues associated with the CRISPR technique. A goal of this work is to stimulate research initiatives in applying genome editing to oral and craniofacial diseases and developmental conditions.
Early Genome Editing Strategies
Prior to the discovery of the CRISPR/Cas system, 3 early genome-editing nucleases had been intensively explored: meganucleases (MNs), zinc finger nucleases (ZFNs), and transcription activator–like effector-based nucleases (TALENs).
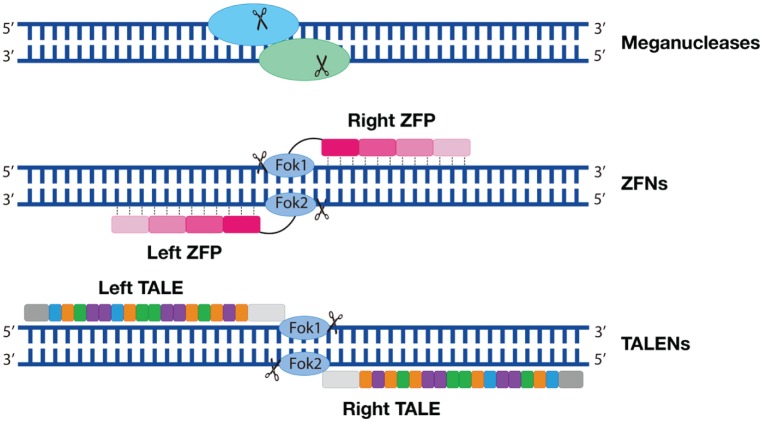
MNs are homing endonucleases with a 14– to 40–base pair (bp) DNA recognition site length; therefore, the recognition sites for MNs are rare in the genome (Fig. 2, top row). MNs create DSBs by mimicking the splicing mechanisms of introns and inteins. However, MNs work most efficiently on a known cleavage site, which might not always exist at the region of interest (Silva et al. 2011). Therefore, engineered MNs have failed to be adopted as a popular genome editing method.
Figure 2.
Engineered early genome editing nucleases. Top row: Meganucleases. The oval shapes symbolize a pair of nucleases; the scissors symbolize DNA cleavage. Middle row: ZFNs are composed of a FokI nuclease domain (oval shapes) and a ZFP domain (a tandem of rectangular shapes). Each ZFP recognizes 3–base pair DNA. Four ZFPs are illustrated on 1 DNA strand. Bottom row: TALENs are composed of a FokI nuclease domain and a tandem of DNA-binding domain (rectangular shapes). Each rectangular shape symbolizes 1 protein repeat that contains 3 amino acids. Each protein repeat binds to 1 nucleotide. TALEN, transcription activator–like effector-based nuclease; ZFN, zinc finger nuclease; ZFP, zinc finger protein.
The drawbacks of MNs leave room for other nucleases to grow. ZFNs are one of them, and they are composed of a DNA-binding motif–zinc finger protein domain and an endonuclease motif–FokI nuclease domain. The zinc finger protein domain contains an array of 3 to 6 zinc finger proteins, each of which recognizes 3-bp DNA; the FokI nuclease domain is responsible for the DNA cleavages. Two ZFNs on the opposite DNA strands dimerize and create DSBs (Gaj et al. 2013; Fig. 2, middle row). Engineering these 3 to 6 zinc finger proteins therefore becomes a key for ZFNs binding to the target DNA sequences. One disadvantage of ZFNs is the lack of known cleavage sites corresponding to the zinc finger proteins. The other disadvantage is that protein engineering is a lengthy and challenging process. Finally, ZFNs exhibit high off-target effects (Gupta and Musunuru 2014).
While optimizing the ZFNs, researchers developed a third genome-editing technique: TALENs. TALENs contain a FokI nuclease domain and a transcription activator-like effector (TALE) domain. The TALE domain is a group of protein repeats, each of which recognizes 1 nucleotide (Fig. 2, bottom row). Each protein repeat has 33– to 35–amino acid repeats, and it is the 12th and 13th amino acids that drive the nucleotide correspondence; therefore, they are referred to as repeat-variable diresidue. Four repeat-variable diresidues—Asn-Asn, Asn-Ile, His-ASP, and Asn-Gly—recognize guanine (G), adenine (A), cytosine (C), and thymine (T), respectively. Because of this feature, TALENs are almost able to target any given DNA sequence, and engineering TALENs is much easier than ZFNs. However, the large size of TALENs makes packaging and delivery into cells difficult, which hinders the broader applications of TALENs (Gupta and Musunuru 2014).
CRISPR System
The early genome editing nucleases rely on protein-DNA interactions to achieve the binding specificities; the CRISPR system simply utilizes RNA-DNA base-pairing mechanisms. CRISPR systems were discovered in bacteria and archaea, as part of the adaptive immunity to defend against foreign DNA invasions (Barrangou and Marraffini 2014). In the bacterial genome, each CRISPR locus consists of a CRISPR gene array and a CRISPR-associated (Cas) gene array (Doudna and Charpentier 2014). The CRISPR array encodes for CRISPR RNA (crRNA) to direct the DNA-binding specificity; the Cas gene arrays are translated into Cas proteins to act as endonucleases. These naturally occurring CRISPR systems have been engineered to facilitate many research endeavors. The most used one is the CRISPR/Cas9 derived from Streptococcus pyogenes (SpCas9). In 2013, the first report of using CRISPR/Cas9 in human cells was considered a “game changer” in the research community (Cong et al. 2013).
Engineered CRISPR/Cas9 System and DNA Repair Pathways
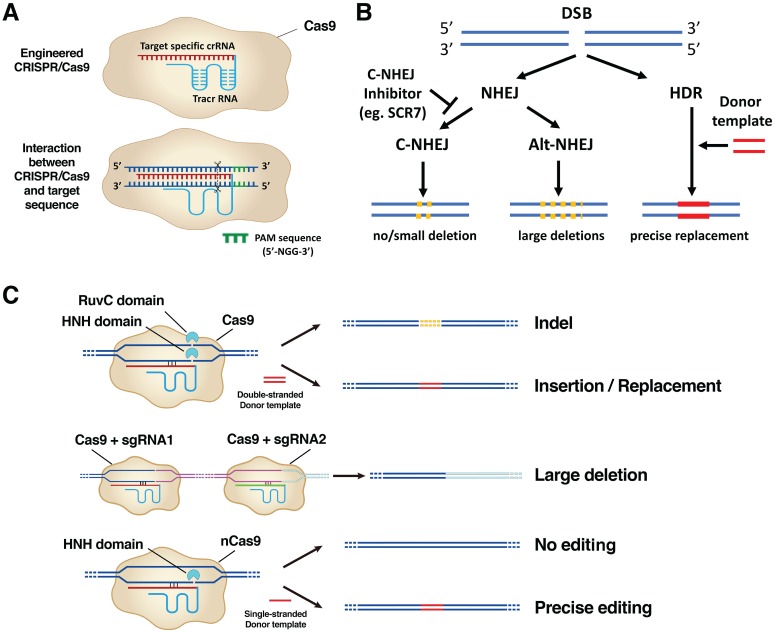
An engineered CRISPR/Cas9 system has 2 components: a single-guide RNA (sgRNA) and a Cas9 enzyme. sgRNA is a fusion of a crRNA and a transactivating crRNA (Jinek et al. 2012). crRNA are ~20-bp RNA sequences that specifically bind to the target DNA region; transactivating crRNAs are fixed RNA sequences that form hairpin loops to stabilize an otherwise unstable single-strand RNA structure (Hsu et al. 2013; Nishimasu et al. 2014; Fig. 3A, top row).
Figure 3.
Mechanisms of the CRISPR/Cas9 system. (A) Top row: Engineered CRISPR/Cas9 is composed of a sgRNA that contains ~20-bp crRNA (red sequence) fused with a tracrRNA (blue sequence) and a Cas9 enzyme component (beige irregular circle). Bottom row: sgRNA binds to the specific target DNA sequences, and Cas9 makes a double-strand break (DSB) at 3 bp upstream of a conserved protospacer-adjacent motif (PAM) region (highlighted in green). Cas9 requires a PAM sequence of 5′-NGG-3′ sequence (N represents any nucleotide base). (B) Repair mechanisms after DSB. Two DNA repair pathways—nonhomologous end-joining (NHEJ) or homology-directed repair (HDR)— kick in after DSB. The NHEJ mechanism is further divided into a canonical (C-NHEJ) or alternative (alt-NHEJ) pathway. C-NHEJ results in no deletion or a small deletion; alt-NHEJ results in the large deletions. Ligase IV inhibitor SCR7 inhibits the C-NHEJ pathway. The HDR pathway is promoted with the presence of a donor template DNA leading to precise replacement of genomic sequences. (C) The application of CRISPR/Cas9 in genome editing. Top row: NHEJ after DSB results in insertion/deletion (indel) mutations; HDR after DSB prompted by a double-stranded donor template DNA results in precise insertion/replacement. Middle row: NHEJ results in a large deletion (pink sequence) when 2 distant DSBs are created by CRISPR/Cas9 systems containing sgRNA1 and sgRNA2. Bottom row: CRISPR/Cas9 with a mutation in the RuvC domain generates nick instead of DSB. DNA repair from a nick with a single-stranded donor template is highly accurate. bp, base pair; crRNA, CRISPR RNA; sgRNA, single-guide RNA; tracrRNA, transactivating crRNA.
In a homology search for the target DNA sequences that form complementary base pairings with the crRNAs, it is critical to find a conserved protospacer-adjacent motif (PAM) sequence at 3′ of the target DNA sequences. The Cas9 enzyme makes DSBs at the exact 3 bp upstream of the PAM sequence (Fig. 3A, bottom row). Different Cas enzymes require specific PAM sequences. The classical 5′-NGG-3′ PAM sequence (N represents any nucleotide followed by 2 guanines) is associated with SpCas9; another Cas enzyme, Cpf1, requires a PAM sequence of 5′-TTTV-3′, where V can be A, C, or G (Gao et al. 2017). Cas9 enzyme adopts a bilobed architecture that contains 2 nuclease domains: RuvC and HNH. Cas9 undergoes conformational changes when it interacts with sgRNA, allowing the target DNA to have access between the 2 lobes of Cas9. Each lobe then engages and nicks 1 DNA strand, therefore generating a composite DSB (Jinek et al. 2012; Jinek et al. 2014; Fig. 3C, top row, left panel).
Two DNA repair pathways then are initiated to correct the DSBs, including a nonhomologous end-joining (NHEJ) pathway or a homology-directed repair (HDR) pathway. The NHEJ mechanism is a “quick-fix” but error-prone DSB repair pathway. The core components of the NHEJ pathway coordinate to sense the DSBs, clean the DNA ends, recruit kinases and ligases (and other factors), and eventually ligate the broken DNA ends (Betermier et al. 2014). NHEJ is further divided into a “canonical” subclass (C-NHEJ) and an “alternative” subclass (alt-NHEJ). C-NHEJ often leads to precise end joining (no deletions) or small deletions; alt-NHEJ results in larger deletions. In contrast to NHEJ, the HDR mechanism leads to more precise DNA repairs promoted by a double-stranded donor template. Depending on the donor templates’ sequences, HDR can either reestablish the original DNA contents or lead to precise replacements such as point mutations or gene insertions. Inhibition of C-NHEJ (e.g., NHEJ-deficient embryos, ligase IV inhibitor SCR7) increases the frequency of alt-NHEJ and HDR events (Hu et al. 2018; Fig. 3B). These DNA repair mechanisms lay the foundation for the biological applications of CRISPR/Cas9.
The NHEJ mechanism results in a small insertion/deletion at a single DSB site or large deletions when 2 distant DSBs are created by CRISPR/Cas9 systems containing 2 sgRNAs (Fig. 3C). The HDR mechanism facilitates CRISPR/Cas9 to create gene insertions or replacements (Fig. 3C), which could be point mutations in coding exons or conditional alleles by inserting loxP sites to the target genes. loxP sites are 34-bp-long recognition sequences that were discovered in P1 bacteriophage. When 2 loxP sites are inserted to the genome to flank the target genes (the process is referred as floxing; these genes are referred to as floxed alleles), specific gene manipulation can occur in combinations with the Cre enzyme that specifically recombines DNA between 2 loP sites. Making floxed alleles/animals is much simplified by the CRISPR/Cas9 technique.
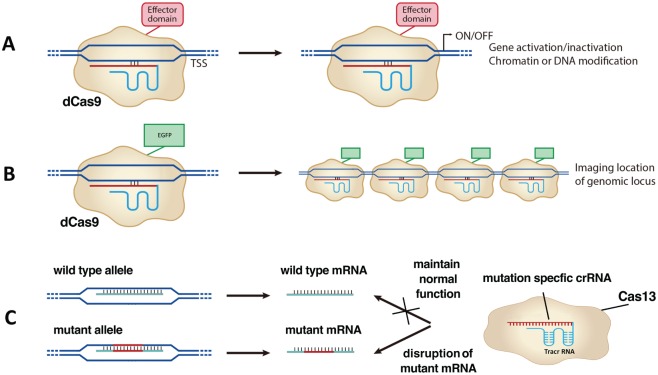
Either naturally occurring or mutant Cas enzymes have been intensively studied, aiming to expand CRISPR’s biological applications. For Cas9, mutations in either the RuvC or HNH domain generate a nickase Cas9 (nCas9), which cuts 1 DNA strand instead of 2 strands (Jinek et al. 2012; Qi et al. 2013). nCas9 edits the genomic sequence with reduced off-target effects, since DNA repairs succeeding nicking have higher fidelities (Ran et al. 2013; Fig. 3C). When mutating RuvC and HNH domains simultaneously, we can utilize a new dead Cas9 (dCas9) to bind to specific DNA sequences without cutting them. Therefore, dCas9 can be fused with effectors such as transcription activator (CRISPRa), repressor (CRISPRi), chromatin, or DNA modifiers to study their transcriptional effects on the gene of interests (Fig. 4A). dCas9 fused with fluorescent tags illuminates specific genomic loci (Fig. 4B). Recently, a newly discovered Cas13 enzyme demonstrated an RNA-editing ability, whereas mRNAs can be knocked down or altered specifically (Abudayyeh et al. 2017; Cox et al. 2017). RNA editing offers additional benefits over DNA editing due to its short-acting and reversible nature, which makes the therapeutic effects safer.
Figure 4.
Application of CRISPR beyond genome editing. (A) dCas9 fused with effector domains reveals gene activation/inactivation or chromatin or DNA modification of these effectors. (B) dCas9 fused with enhanced green fluorescent protein (EGFP) illuminates genomic loci. (C) CRISPR/Cas13 can edit RNA precisely. Mutation-specific crRNA can bind to the mutant mRNA and disrupt its functions.
Applications of CRISPR/Cas9 Preclinically and Clinically
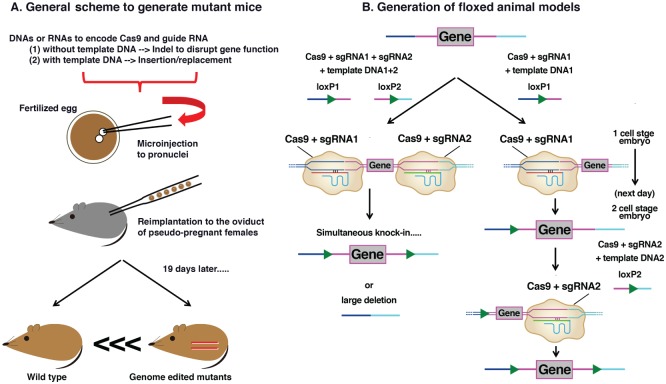
Applications of CRISPR/Cas9 in generating mutant animals have been widely adopted by the research community. The scheme of generating mutant mice involves the microinjection of nucleotides encoding Cas9 and sgRNA into the pronuclei of the fertilized eggs. To generate knockout mice, CRISPR components alone (Cas9 and sgRNA) are adequate to create insertion/deletions to disrupt gene functions through the NHEJ pathway. To generate knock-in mice, additional template DNAs are microinjected simultaneously to permit gene insertion/replacement through the HDR pathway. These genome-edited zygotes are subsequently reimplanted to the oviduct of pseudo-pregnant surrogate mice that will give birth to mutant offspring in 19 gestational days (Fig. 5A). Experimental processes are similar to the ones to generate transgenic mice by random insertion (i.e., no need to screen correctly targeted ESCs). The time length of making mutant animal models is therefore significantly reduced to several weeks, while the traditional gene targeting methods may take 1 to 2 y (Heidenreich and Zhang 2016). Moreover, CRISPR/Cas9 can edit multiple loci simultaneously, which largely increases the editing efficiency. Due to these benefits, CRISPR/Cas9 has led to a considerable number of novel genetically modified animal models, including some unique nonhuman primate models (Niu et al. 2014).
Figure 5.
Applications of CRISPR/Cas9 to generate mutant animals. (A) General scheme to generate mutant mouse models. DNAs or RNAs encoding Cas9 and single-guide RNA (sgRNA) with or without template DNA are microinjected into the pronuclei of the fertilized egg. These modified zygotes are reimplanted to the oviduct of pseudo-pregnant surrogate mice. After 19 gestational days, mutant mice offspring are born. (B) Generation of floxed animal models. The left arm represents the simultaneous method. This method involves coinjecting nucleotides encoding Cas9, 2 sgRNAs, and 2 individual loxP template DNAs to zygote. It simultaneously knocks in 2 loxP alleles to flank the target exon gene. However, large deletions can occur as a result. The right arm represents the sequential method. This method involves coinjecting Cas9, first sgRNA, and first loxP template DNA at 1–cell stage embryo to knock in the first loxP allele. On the next day, the second group of Cas9, second sgRNA, and second loxP template DNA are injected into 2–cell stage embryos, leading to knock-in of the second loxP allele.
CRISPR/Cas9 is useful in generating floxed animal models, therefore resulting in conditional knockout animals. There are 2 methods to make floxed animals (Fig. 5B). The first is a simultaneous method. Coinjecting nucleotides encoding Cas9, 2 sgRNAs, and 2 individual loxP template DNAs to zygote can introduce 2 loxP alleles to flank the target exons. The second is a sequential method. In this model, the first loxP allele is inserted by coinjecting 1 sgRNA and 1 template DNA with Cas9 at the 1–cell stage embryos. At 2–cell stage embryos the following day, the second loxP allele is inserted by using another pair of sgRNA and template DNA with Cas9 (Horii et al. 2017). The sequential method overcomes a frequent deletion of the target exons that may occur in the simultaneous method.
More exciting applications of CRISPR/Cas9 came from preclinical and clinical models. CRISPR/Cas9 demonstrated prominent effectiveness in correcting monogenic diseases such as Duchenne muscular dystrophy through either germline deliveries or direct muscular injections in mice (Long et al. 2014; Xu et al. 2016). In October 2016, the first CRISPR/Cas9 human clinical trial was proposed in China (Cyranoski 2016). Ten patients with lung cancer are planned to receive injections of their own immune cells, in which program death 1 gene (PD1) is edited by CRISPR/Cas9. Primarily a phase 1 trial, this study would be mainly related to the safety side rather than the efficacy side. However, this clinical trial has opened the door for CRISPR/Cas9 to test its clinical applications. At the same time, the first CRISPR clinical trial in the United States was initiated, where both PD1 and T-cell receptor gene (TCR) will be disrupted in cancer patients (ClinicalTrials.gov NCT0279 3856). Motivated by these 2 pioneering trials, many more clinical trials were approved globally in 2017. It is noteworthy that in April 2017, Nanjing University in China started a CRISPR/Cas9 clinical trial in treating patients with aggressive gastric cancer, lymphoma, and nasopharyngeal carcinoma (Normile 2017), which represented the first application of CRISPR/Cas9 in the oral and craniofacial field.
Applications of Genome Editing Techniques in the Oral and Craniofacial Field
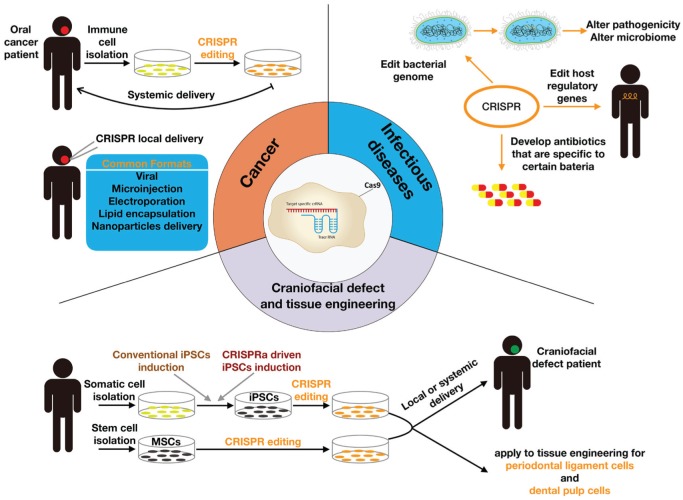
Genome editing techniques have emerged over a period of only a few years; therefore, literature reporting their applications in oral and craniofacial biology is limited. In this section, we focus on the applications of CRISPR/Cas9 in the domains of head and neck cancer (HNC), craniofacial defect/tissue engineering, and infectious diseases. We illustrate in Figure 6 the current applications and potential future applications of CRISPR in oral and craniofacial research.
Figure 6.
Applications of the CRISPR system in oral and craniofacial biology. Cancer: Immune cells are isolated from patients with oral cancer and receive CRISPR genome editing in vitro. Genome-edited immune cells are then systemically delivered back to the patients for cancer therapy. CRISPR components can be locally delivered to the hot spot of the cancer lesion, following various delivery formats. Craniofacial defect and tissue regeneration: Somatic cells are isolated from human subjects. Through either conventional induced pluripotent stem cell (iPSC) induction or CRISPR activation (CRISPRa) iPSC induction, somatic cells become iPSCs. Specific stem cell lineages, such as mesenchymal stem cells (MSCs), can be isolated and receive CRISPR editing. Both stem cells can be delivered either systemically or locally to the patient with craniofacial defects. Genome-edited stem cells also contribute to tissue engineering of periodontal ligament cells or dental pulp cells. Infectious diseases: CRISPR may edit the bacterial genome to alter either pathogenicity or the microbiome; CRISPR may edit host regulatory genes to fight against infections; CRISPR may also contribute to developing pathogen-specific antibiotics.
Head and Neck Cancer
HNC is a common malignant neoplasm, with an estimated incidence of 50,000 cases in the United States in 2017 (American Cancer Society 2017). As a genetically complex disease, cancer arises from a sequence of genetic and epigenetic alterations; therefore, cancer research benefits tremendously from the CRIPSR/Cas9 techniques. As summarized in the Appendix Table, CRISPR/Cas9 has been used to knock out several genes in several HNC cell lines. Those attempts confirm the involvements of fibronectin and LDB1 for cancer cell and invasiveness (Wang et al. 2015; Simonik et al. 2016) and identify novel therapeutic targets such as p75NTR and MUL1-HSPA5 axis (Huang et al. 2017; Kim et al. 2018). However, CRISPR may provide greater breakthroughs in treating HNC. The ongoing clinical trials use CRISPR/Cas9-edited immune cells to fight against cancer cells, which could be applied to treat HNC. Lessons learned from treating monogenic diseases such as Duchenne muscular dystrophy by direct local injection also highlight a possibility of utilizing the same strategy to treat HNC.
Craniofacial Defects and Tissue Engineering
Craniofacial development in vertebrates involves the coordinated patterning of tissues from all 3 germ layers and the neural crest, governed by strict genetic controls. A failure in any of the precise spatiotemporal sequences of events leads to oral and craniofacial anomalies, such as cleft lip and palate (Dixon et al. 2011). Using the CRISPR technique to study the genes involved in craniofacial development enriched the current understandings of how craniofacial defects form. Willems et al. (2015) utilized the CRISPR technique to disrupt wnt-associated gene lrp5 in zebrafish, and found that a lack of this gene led to impaired migrations of cranial neural crestal cells. Through in vitro CRISPR-targeted disruptions, smchd1 and kinesin-1 genes were found to be critical in craniofacial cartilage formation (Shaw et al. 2017; Santos-Ledo et al. 2017). Knockout mice generated by CRISPR/Cas9 revealed the important roles of Golgb1 and Msx1 in tooth and palate development (Lan et al. 2016; Mitsui et al. 2016). CRISPR also facilitates the creations of reporter murine models (i.e., tamoxifen-inducible Pax9-CreER mice, Axin2-mTurquoise2 mice) to investigate craniofacial development (Feng et al. 2016; de Roo et al. 2017).
Various tissue engineering efforts have been given to correct the craniofacial defects. Within the tissue engineering triad, cells (especially stem cells) can be profoundly affected by the advancements in the CRISPR technique. Currently, 2 types of stem cells serve as research tools: one is pluripotent stem cells featuring ESCs and induced pluripotent stem cells (iPSCs); the other is multipotent tissue-specific stem cells that reside in native tissues such as hematopoietic stem cells and mesenchymal stem cells (MSCs). ESCs and iPSCs are ideal targets for CRISPR, as they repopulate easily after the editing (Zhang et al. 2017). CRISPR-edited ESCs contributed greatly to generations of mutant animals. Many craniofacial abnormalities occur early in the embryonic phase, making genetically modifying the embryonic tissues very attractive. However, we understand the controversy associated with this topic. Therefore, substantial preclinical studies are needed prior to clinical applications. CRISPR also participates in generating iPSCs, which can be established by ectopic expression of master transcription factors (i.e., OCT4, SOX2, KFL4, MYC) in somatic cells to activate the pluripotency network. The conventional transfection system associated with iPSC induction presented a low efficiency and a high cytotoxicity. The CRISPR activation system has been proven to boost the endogenous Oct4 or Sox2 expression that is sufficient to trigger the pluripotency reprogramming (Liu et al. 2018). In recent years, MSCs have gained increased attention in clinical therapies for oral and craniofacial diseases. Alveolar bone, periodontal ligament, and dental pulp all have been demonstrated to contain subsets of MSCs. CRISPR/Cas9-edited MSCs could be a valuable tool in correcting oral, periodontal, and craniofacial defects.
Infectious Diseases
Caries and periodontal diseases represent the most prevalent infectious diseases affecting humans. Both diseases are etiologically attributed to bacterial plaque. Naturally occurring CRISPR loci can be found in most human oral microbiota (Rho et al. 2012). Serbanescu et al. (2015) reported that the Streptococcus mutans CRISPR system played a role in preventing the uptake and dissemination of the antibiotic resistance genes. This finding raised a possibility to harness the antibiotic resistance of S. mutans by targeting its CRISPR system. Comparisons of the CRISPR loci within the dental plaque biofilm between healthy and periodontitis subjects revealed that the CRISPR components were more similar to each other within healthy patients, assembling a robust and functional bacterial community to resist the invasion from bacteriophages (Zhou et al. 2015). It is reasonable to predict that the CRISPR system is associated with the equilibrium of the oral microbial community and therefore could be a target to modulate the oral microbiome for disease control. Alternatively, CRISPR may harness the infectious diseases by modifying the host regulatory genes.
In addition to the genetic influence, epigenetic dysregulation has become a putative cause for HNC, craniofacial abnormality, and periodontal diseases (Barros and Offenbacher 2014; Hu et al. 2014; Castilho et al. 2017). As discussed previously, dCas9 fused with epigenetic modifiers can be used to investigate the transcriptional regulations. This methodology could certainly be applied to study the epigenetic regulations in the oral and craniofacial diseases.
Advantages, Limitations, and Ethical Issues of CRISPR/Cas9
CRISPR/Cas9 demonstrates superior simplicity over the early genome-editing nucleases. The CRISPR technique requires only an alteration of its ~20-bp crRNA sequences to adapt to the sequence specificity. Researchers have access to ample commercially available CRISPR-related vectors and bioinformatics tools to design the crRNA sequences and templates (Liu et al. 2015; Park et al. 2015). Another advantage is that CRISPR can simultaneously target multiple genes, highlighting its strength in studying multigenic diseases (Wang et al. 2013).
The major limitation for CRISPR/Cas9 technology is the off-target effects—that is, when nucleases bind to and cut unintended DNA sites, resulting in unwanted genomic changes. Schaefer, Wu, Colgan, et al. (2017) reported that a significantly high number of deleterious mutations were found in 2 CRISPR/Cas9-modified mice, raising concerns of the use of CRISPR/Cas9 in clinical settings. The same group, however, later published a follow-up report to declare no excess mutations in the same samples (Schaefer, Wu, Darbro, et al. 2017). Nonetheless, it is critical to minimize the off-target effects with various strategies (Zhang et al. 2015). Improving the design of sgRNAs remains the most important strategy. Web-based software tools such as the Cas9 Activator Tool and ZiFiT Targeter have been developed to facilitate the designs of the sgRNA sequences and off-target validations (Hsu et al. 2013; Hwang et al. 2013). Titrating Cas9 enzyme concentration and the amount of sgRNA DNA delivery reduces nonspecific targeting as well (Hsu et al. 2013). Kleinstiver et al. (2016) reported that a modified Cas9 enzyme (referred to as high-fidelity Cas9) could result in no detectable off-target effects on a genome-wide scale. Finally, adding a fluorescent reporter to the donor DNA sequence assists in observing whether the donor DNA successfully integrates to the target DNA region. It is equally important to familiarize with assays to identify the off-target effect (Hendel et al. 2015).
CRISPR/Cas9 technology has raised some valid ethical concerns. In the United States, genome editing–related clinical research requires approvals from the Recombinant DNA Advisory Committee of the National Institutes of Health. In June 2016, the advisory committee approved the first proposal to use CRISPR/Cas9 to treat cancer patients at the University of Pennsylvania. One particular concern from the committee was a potential conflict of interest when investigators who had ties to a pharmaceutical company led the clinical trial. However, the immediacy and importance of the trial led to the final approval from the panel (“First-in-Human CRISPR Trial” 2016). The other concern arises from the controversy of germline genome editing. To overcome this barrier, current clinical trials tend to focus on CRISPR-driven cell therapies (Bosley et al. 2015). Scientists will gain more in-depth knowledge of genome editing from the ongoing clinical trials and continue facing new challenges. Therefore, new ethical issues will appear and require more careful scrutiny.
Emerging Evidence and Future Perspectives
More novel and versatile genome-editing systems are under development at an unprecedented speed in the community. In addition to Cas9, many other CRISPR-associated enzymes are being studied. Cpf1 enzyme mimics Cas9 in cutting DNA and has many deactivated forms that can be used in studying the transcriptional effects (Li et al. 2018). Csm6 enzyme shares the similar ribonucleases activity as Cas13 (Niewoehner et al. 2017). Microbial whole genome sequencing and advanced structural biology will greatly facilitate the search for novel CRISPR-associated nucleases. Nevertheless, there are approaches that utilize enzymes to directly convert nucleotide bases on the genome level. Transfer RNA adenosine deaminase editors fused with nCas9 can convert an A-T bp to a G-C pair at the targeted position in the genome without DSB (Gaudelli et al. 2017). It is still necessary to develop fused enzymes to catalyze other conversions, but this finding is certainly a big leap toward the next level of the genome-editing technology that does not require DSBs.
Genome editing techniques, particularly CRISPR/Cas9, have greatly enhanced biological research. There is strong potential to take advantage of this technology to apply it into the oral and craniofacial field. Simple yet precise manipulations of genome editing techniques will enable us to gain deeper insight into the genes involved in oral diseases and craniofacial malformations. The future of applying CRISPR/Cas9 to treat oral and craniofacial disorders is now.
Author Contributions
N. Yu, contributed to conception, design, and data acquisition, drafted the manuscript; J. Yang, contributed to conception and design, drafted the manuscript; Y. Mishina, contributed to conception and design, critically revised the manuscript; W.V. Giannobile, contributed to conception, design, and data interpretation, critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Supplemental Material
Supplemental material, DS_10.1177_0022034518805978 for Genome Editing: A New Horizon for Oral and Craniofacial Research by N. Yu, J. Yang, Y. Mishina and W.V. Giannobile in Journal of Dental Research
Acknowledgments
Special acknowledgements for illustrations and graphic design contributions are given to Victoria Zakrzewski and Kenneth Rieger at the graphic design department of the University of Michigan School of Dentistry.
Footnotes
A supplemental appendix to this article is available online.
The authors received no financial support and declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
ORCID iD: W.V. Giannobile  https://orcid.org/0000-0002-7102-9746
https://orcid.org/0000-0002-7102-9746
References
- Abudayyeh OO, Gootenberg JS, Essletzbichler P, Han S, Joung J, Belanto JJ, Verdine V, Cox DBT, Kellner MJ, Regev A, et al. 2017. RNA targeting with CRISPR-Cas13. Nature. 550(7675):280–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Cancer Society. 2017. Cancer facts and figures. Atlanta (GA): American Cancer Society. [Google Scholar]
- Barrangou R, Marraffini LA. 2014. CRISPR-Cas systems: prokaryotes upgrade to adaptive immunity. Mol Cell. 54(2):234–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros SP, Offenbacher S. 2014. Modifiable risk factors in periodontal disease: epigenetic regulation of gene expression in the inflammatory response. Periodontology 2000. 64(1):95–110. [DOI] [PubMed] [Google Scholar]
- Betermier M, Bertrand P, Lopez BS. 2014. Is non-homologous end-joining really an inherently error-prone process? PLoS Genet. 10(1):e1004086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosley KS, Botchan M, Bredenoord AL, Carroll D, Charo RA, Charpentier E, Cohen R, Corn J, Doudna J, Feng G, et al. 2015. CRISPR germline engineering—the community speaks. Nat Biotechnol. 33(5):478–486. [DOI] [PubMed] [Google Scholar]
- Castilho RM, Squarize CH, Almeida LO. 2017. Epigenetic modifications and head and neck cancer: implications for tumor progression and resistance to therapy. Int J Mol Sci. 18(7):E1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science. 339(6121):819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DBT, Gootenberg JS, Abudayyeh OO, Franklin B, Kellner MJ, Joung J, Zhang F. 2017. RNA editing with CRISPR-Cas13. Science. 358(6366):1019–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyranoski D. 2016. CRISPR gene-editing tested in a person for the first time. Nature. 539(7630):479. [DOI] [PubMed] [Google Scholar]
- de Roo JJD, Breukel C, Chhatta AR, Linssen MM, Vloemans SA, Salvatori D, Mikkers HMM, Verbeek SJ, Staal FJT. 2017. Axin2-mturquoise2: a novel reporter mouse model for the detection of canonical wnt signalling. Genesis. 55(10). [DOI] [PubMed] [Google Scholar]
- Dixon MJ, Marazita ML, Beaty TH, Murray JC. 2011. Cleft lip and palate: understanding genetic and environmental influences. Nat Rev Genet. 12(3):167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna JA, Charpentier E. 2014. Genome editing: the new frontier of genome engineering with CRISPR-Cas9. Science. 346(6213):1258096. [DOI] [PubMed] [Google Scholar]
- Feng J, Jing J, Sanchez-Lara PA, Bootwalla MS, Buckley J, Wu N, Yan Y, Chai Y. 2016. Generation and characterization of tamoxifen-inducible pax9-creer knock-in mice using CRISPR/Cas9. Genesis. 54(9):490–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- First-in-human CRISPR trial. 2016. Nat Biotechnol. 34(8):796. [DOI] [PubMed] [Google Scholar]
- Gaj T, Gersbach CA, Barbas CF., 3rd 2013. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 31(7):397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Cox DB, Yan WX, Manteiga JC, Schneider MW, Yamano T, Nishimasu H, Nureki O, Crosetto N, Zhang F. 2017. Engineered Cpf1 variants with altered PAM specificities. Nat Biotechnol. 35(8):789–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI, Liu DR. 2017. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature. 551(7681):464–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RM, Musunuru K. 2014. Expanding the genetic editing tool kit: ZFNs, TALENs, and CRISPR-Cas9. J Clin invest. 124(10):4154–4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidenreich M, Zhang F. 2016. Applications of CRISPR-Cas systems in neuroscience. Nat Rev Neurosci. 17(1):36–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendel A, Fine EJ, Bao G, Porteus MH. 2015. Quantifying on- and off-target genome editing. Trends Biotechnol. 33(2):132–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horii T, Morita S, Kimura M, Terawaki N, Shibutani M, Hatada I. 2017. Efficient generation of conditional knockout mice via sequential introduction of lox sites. Sci Rep. 7(1):7891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, et al. 2013. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 31(9):827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu N, Strobl-Mazzulla PH, Bronner ME. 2014. Epigenetic regulation in neural crest development. Dev Biol. 396(2):159–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Shi Z, Guo X, Jiang B, Wang G, Luo D, Chen Y, Zhu YS. 2018. Ligase IV inhibitor SCR7 enhances gene editing directed by CRISPR-Cas9 and ssODN in human cancer cells. Cell Biosci. 8:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P, Tong D, Sun J, Li Q, Zhang F. 2017. Generation and characterization of a human oral squamous carcinoma cell line SCC-9 with CRISPR/Cas9-mediated deletion of the p75 neurotrophin receptor. Arch Oral Biol. 82:223–232. [DOI] [PubMed] [Google Scholar]
- Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JR, Joung JK. 2013. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 31(3):227–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 337(6096):816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Jiang F, Taylor DW, Sternberg SH, Kaya E, Ma E, Anders C, Hauer M, Zhou K, Lin S, et al. 2014. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science. 343(6176):1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Kim HJ, Kim HJ, Kim DH, Han JH, Byeon HK, Lee K, Kim CH. 2018. Hspa5 negatively regulates lysosomal activity through ubiquitination of mul1 in head and neck cancer. Autophagy. 14(3):385–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, Joung JK. 2016. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 529(7587):490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan Y, Zhang N, Liu H, Xu J, Jiang R. 2016. Golgb1 regulates protein glycosylation and is crucial for mammalian palate development. Development. 143(13):2344–2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Zeng C, Dong Y. 2018. Design and assessment of engineered CRISPR-Cpf1 and its use for genome editing. Nat Protoc. 13(5):899–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Wei Z, Dominguez A, Li Y, Wang X, Qi LS. 2015. CRISPR-era: a comprehensive design tool for CRISPR-mediated gene editing, repression and activation. Bioinformatics. 31(22):3676–3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Chen M, Liu Y, Qi LS, Ding S. 2018. CRISPR-based chromatin remodeling of the endogenous Oct4 or Sox2 locus enables reprogramming to pluripotency. Cell Stem Cell. 22(2):252–261.e4. [DOI] [PubMed] [Google Scholar]
- Long C, McAnally JR, Shelton JM, Mireault AA, Bassel-Duby R, Olson EN. 2014. Prevention of muscular dystrophy in mice by CRISPR/Cas9-mediated editing of germline DNA. Science. 345(6201):1184–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui SN, Yasue A, Masuda K, Naruto T, Minegishi Y, Oyadomari S, Noji S, Imoto I, Tanaka E. 2016. Novel human mutation and CRISPR/cas genome-edited mice reveal the importance of C-terminal domain of MSX1 in tooth and palate development. Sci Rep. 6:38398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niewoehner O, Garcia-Doval C, Rostol JT, Berk C, Schwede F, Bigler L, Hall J, Marraffini LA, Jinek M. 2017. Type III CRISPR-cas systems produce cyclic oligoadenylate second messengers. Nature. 548(7669):543–548. [DOI] [PubMed] [Google Scholar]
- Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N, Ishitani R, Zhang F, Nureki O. 2014. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 156(5):935–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Y, Shen B, Cui Y, Chen Y, Wang J, Wang L, Kang Y, Zhao X, Si W, Li W, et al. 2014. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell. 156(4):836–843. [DOI] [PubMed] [Google Scholar]
- Normile D. 2017. China sprints ahead in CRISPR therapy race. Science. 358(6359):20–21. [DOI] [PubMed] [Google Scholar]
- Park J, Bae S, Kim JS. 2015. Cas-designer: a web-based tool for choice of CRISPR-Cas9 target sites. Bioinformatics. 31(24):4014–4016. [DOI] [PubMed] [Google Scholar]
- Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. 2013. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 152(5):1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, et al. 2013. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 154(6):1380–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rho M, Wu YW, Tang H, Doak TG, Ye Y. 2012. Diverse CRISPRs evolving in human microbiomes. PLoS Genet. 8(6):e1002441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Ledo A, Garcia-Macia M, Campbell PD, Gronska M, Marlow FL. 2017. Kinesin-1 promotes chondrocyte maintenance during skeletal morphogenesis. PLoS Genet. 13(7):e1006918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer KA, Wu WH, Colgan DF, Tsang SH, Bassuk AG, Mahajan VB. 2017. Unexpected mutations after CRISPR-Cas9 editing in vivo. Nat Methods. 14(6):547–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer KA, Wu W-H, Darbro BW, Colgan DF, Tsang SH, Bassuk AG, Mahajan VB. 2017. Deeper sequencing at unexpected CRISPR/Cas9 off-target sites in vivo: a response to Editas, Intellia, Beacon, Toolgen and others. bioRxiv. p154450. [Google Scholar]
- Serbanescu MA, Cordova M, Krastel K, Flick R, Beloglazova N, Latos A, Yakunin AF, Senadheera DB, Cvitkovitch DG. 2015. Role of the Streptococcus mutans CRISPR-Cas systems in immunity and cell physiology. J Bacteriol. 197(4):749–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw ND, Brand H, Kupchinsky ZA, Bengani H, Plummer L, Jones TI, Erdin S, Williamson KA, Rainger J, Stortchevoi A, et al. 2017. Smchd1 mutations associated with a rare muscular dystrophy can also cause isolated arhinia and Bosma arhinia microphthalmia syndrome. Nat Genet. 49(2):238–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva G, Poirot L, Galetto R, Smith J, Montoya G, Duchateau P, Paques F. 2011. Meganucleases and other tools for targeted genome engineering: perspectives and challenges for gene therapy. Curr Gene Ther. 11(1):11–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonik EA, Cai Y, Kimmelshue KN, Brantley-Sieders DM, Loomans HA, Andl CD, Westlake GM, Youngblood VM, Chen J, Yarbrough WG, et al. 2016. LIM-only protein 4 (LMO4) and LIM domain binding protein 1 (LDB1) promote growth and metastasis of human head and neck cancer (LMO4 and LDB1 in head and neck cancer). PloS One. 11(10):e0164804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travis J. 2015. Making the cut. Science. 350(6267):1456–1457. [DOI] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. 2013. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 153(4):910–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HC, Yang Y, Xu SY, Peng J, Jiang JH, Li CY. 2015. The CRISPR/Cas system inhibited the pro-oncogenic effects of alternatively spliced fibronectin extra domain a via editing the genome in salivary adenoid cystic carcinoma cells. Oral Dis. 21(5):608–618. [DOI] [PubMed] [Google Scholar]
- Willems B, Tao S, Yu T, Huysseune A, Witten PE, Winkler C. 2015. The Wnt co-receptor Lrp5 is required for cranial neural crest cell migration in zebrafish. PloS One. 10(6):e0131768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Park KH, Zhao L, Xu J, El Refaey M, Gao Y, Zhu H, Ma J, Han R. 2016. CRISPR-mediated genome editing restores dystrophin expression and function in mdx mice. Mol Ther. 24(3):564–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XH, Tee LY, Wang XG, Huang QS, Yang SH. 2015. Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol Ther Nucleic Acids. 4:e264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zhang Y, Gao F, Han S, Cheah KS, Tse HF, Lian Q. 2017. CRISPR/Cas9 genome-editing system in human stem cells: current status and future prospects. Mol Ther Nucleic Acids. 9:230–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Zhao H, Zheng J, Gao Y, Zhang Y, Zhao F, Wang J. 2015. CRISPRs provide broad and robust protection to oral microbial flora of gingival health against bacteriophage challenge. Protein Cell. 6(7):541–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, DS_10.1177_0022034518805978 for Genome Editing: A New Horizon for Oral and Craniofacial Research by N. Yu, J. Yang, Y. Mishina and W.V. Giannobile in Journal of Dental Research