Abstract
ATF6 is an endoplasmic reticulum (ER) membrane-bound transcription factor that regulates various cellular functions. The purpose of this study was to investigate the role of ATF6 in odontoblast differentiation. Rat tooth germs were isolated, changes in gene expression were evaluated over time, and localization of ATF6 was determined by immunohistochemistry. Human dental pulp cells (HDPCs) were cultured with 50 µg/mL ascorbic acid and 5 mmol/L β-glycerophosphate or 100 ng/mL bone morphogenetic protein 2 to induce differentiation. Translocation of ATF6 was observed by immunofluorescence and confocal microscopy. Overexpression of ATF6 was performed with an adenoviral vector. Matrix mineralization was evaluated by alizarin red staining. Immunoreactivity to anti-ATF6 was observed in the odontoblastic layer of the molar tooth germ, and expressions of ATF6, dentin sialophosphoprotein (DSPP) and dentin matrix protein 1 (DMP1) increased gradually during tooth germ development. When HDPCs were cultured in differentiation media, ATF6, DSPP, and DMP1 expression increased with the expression of unfolded protein response (UPR) markers, BiP and CHOP. Immunofluorescence results showed that ATF6 protein moved from cytoplasm to nucleus when cells were exposed to differentiation media. Notably, overexpression of ATF6 increased DSPP and DMP1 expression, alkaline phosphatase (ALP) activity, and matrix mineralization in HDPC cultures. Inhibition of ATF6 decreased ALP activity and mineralization. These results suggest that ER membrane-bound transcriptional factor ATF6 may be involved in odontoblastic differentiation.
Keywords: ER stress, DSPP, matrix mineralization, DMP1, unfolded protein response, tooth development
Introduction
Diverse growth factors, such as bone morphogenetic protein (BMP), fibroblast growth factor (FGF), sonic hedgehog (SHH), and Wnt, are involved in tooth morphogenesis (Jernvall and Thesleff, 2000). Signaling molecules in the SHH, FGF, BMP, and Wnt families appear to regulate the early stages of tooth morphogenesis, and some transcription factors associated with these pathways are necessary for tooth development (Thesleff and Sharpe, 1997).
The dentin-forming odontoblasts and the enamel-forming ameloblasts are unique to the tooth, and they differentiate during the bell stage of tooth development (Thesleff and Sharpe, 1997). Odontoblast differentiation occurs due to reciprocal interactions between the epithelial and mesenchymal tissues (Ruch et al., 1995). Odontoblasts secrete collagenous or non-collagenous extracellular matrix proteins such as dentin sialophosphoprotein (DSPP), which subsequently mineralizes to form the dentin.
Dental pulp tissue contains stem/progenitor cells that have the potential to differentiate into dentin-forming odontoblasts in response to BMPs and other signal molecules. Differentiation of stem/progenitor cells into odontoblasts is required for pulpal wound healing (Nakashima, 2005). BMP2 mediates DSPP gene expression and odontoblastic differentiation via several transcription factors during tooth development (Chen et al., 2008; Oh et al., 2012).
Dentin sialoprotein (DSP) and dentin phosphoprotein (DPP) are synthesized and secreted mainly by odontoblasts. DSP and DPP are secreted as a single protein (DSPP) which is then proteolytically processed to form the individual components found in the dentin matrix. These matrix proteins play a role in dentinogenesis (Butler, 1995, 1998).
DSPP mRNA is abundantly expressed in differentiating odontoblasts and ameloblasts during tooth development, but is weakly detected in osteoblasts (Chen et al., 2009). DSPP knockout mice display tooth defects with dentin hypomineralization that are similar to those of human dentinogenesis imperfecta III (Sreenath et al., 2003). Thus, DSPP plays an important role in dentin mineralization. However, the molecular mechanism of DSPP gene expression is not fully understood.
ATF6 is an endoplasmic reticulum (ER) membrane-bound transcription factor that regulates ER stress-related gene expression. ATF6 is maintained in the ER due to binding of ATF6 to BiP. When unfolded proteins accumulate in the ER lumen, ATF6 is translocated to the Golgi apparatus, where it is converted into its active form. Thus, the active form of ATF6 moves to the nucleus to control the expression of unfolded protein response (UPR)-related genes (Boot-Handford and Briggs, 2010). Metazoan cells express ATF6 protein as a single gene product, whereas mammalian cells express two isoforms of ATF6, ATF6α and ATF6β. In a previous study, ATF6α was reported to function as a critical regulator of ER quality control proteins in mammalian cells (Yamamoto et al., 2007).
BMP2 induces mild ER stress to activate UPR transducers such as ATF4 and PERK (Saito et al., 2011). In addition, BMP2 protein regulates osteocalcin expression and osteoblast differentiation via Runx2-mediated ATF6 gene transcription in osteoblast cells (Jang et al., 2012). This study was undertaken to explore the role of ATF6 in odontoblastic differentiation, and we demonstrated that activation of ATF6 may be partly required for inducing DSPP/DMP1 expression and matrix mineralization.
Materials & Methods
Isolation of Tooth Germs
Forty Sprague-Dawley rats (Damool Science, Daejeon, Korea) were used according to the guidelines of the Chonnam National University Animal Care and Use Committee. The rat pups at post-natal day 3, 6, and 9 were sacrificed, and maxillae including developing tooth germs were removed. The 2nd and 3rd molar tooth germs were surgically isolated by means of a stereomicroscope (Leica, Wetzlar, Germany), and 10 tooth germs per group were pooled for analysis of gene expression.
Immunohistochemistry
The maxillae containing tooth germs were fixed in 4% paraformaldehyde and decalcified in 10% EDTA (pH 7.4) for 6 to 8 wks. The tissues were treated with ethanol dehydration, embedded in paraffin, and cut into 5-µm-thick sections, and immunohistochemical reactions were performed with LSAB+System-HRP (DAKO, Carpinteria, CA, USA) according to the manufacturer’s protocol. Briefly, specimens were de-waxed in xylene, rehydrated in a graded alcohol series, placed in an endogenous peroxide blocker for 10 min, and washed with Tris-buffered saline (TBS). The primary antibodies [anti-ATF6α (1:50, Abcam, Cambridge, UK) and anti-DSP (1:100, Santa Cruz Biotechnology, Santa Cruz, CA, USA)] were incubated at 4°C for 1 hr, followed by the biotin-streptavidin peroxidase complex. The tissues were counterstained with hematoxylin. Negative controls consisted of TBS substituted for ATF6 antibody.
Adenoviruses
The nuclear form of the ATF6 (pcDNA-ATF6) and the dominant-negative (DN) form of ATF6 plasmids were kindly provided by Dr. Ron Prywes (Department of Biological Science, Columbia University, New York). The DN-ATF6 was constructed by PCR amplification of the bZIP domain of ATF6 (Wang et al., 2000). Adenovirus (Ad) encoding the nuclear form of ATF6 (Ad-ATF6) and Ad-DN-ATF6 were constructed by methods described previously (Seo et al., 2008).
Cell Culture
Immortalized human dental pulp cells (HDPCs; Kitagawa et al., 2007) were kindly provided by Professor Takashi Takata of Hiroshima University. Cells were cultured in α-MEM supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin, and 100 µg/mL streptomycin in a humidified atmosphere of 5% CO2 at 37°C. To induce differentiation, we treated cells with 50 µg/mL ascorbic acid and 5 mmol/L β-glycerophosphate, or 100 ng/mL BMP2 (Daewoong Pharmaceutical, Samseong-dong, Korea). For viral infection, the cells were plated in α-MEM containing 10% FBS for 24 hrs, and then the culture medium was changed to a serum-free medium with or without the adenoviral (Ad) vector encoding the nuclear form of ATF6 (Ad-ATF6) or DN-ATF6 (Ad-DN-ATF6).
The same volume of medium containing 10% FBS was added to the cultures 4 hr later. After 20 hrs, the culture medium was replaced with the differentiation-inducing medium. The medium was replaced every other day.
RNA Isolation and RT-PCR Analysis
Total RNA was extracted from the isolated tooth germs of rats or the cultured HDPCs with Trizol reagent (Invitrogen, Carlsbad, CA, USA). cDNA was synthesized with random primers and the AccessQuick™ RT-PCR system (Promega, Madison, WI, USA). PCR primers and reaction conditions are shown in Appendix Tables 1 and 2. The level of gene expression was visualized by ethidium bromide staining. For quantitative analysis, real-time PCR was performed with the StepOnePlus™ Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) and QuantiMix SYBR kit (Philekorea Technology, Daejeon, Korea). Primers and reaction conditions are shown in Appendix Table 2. ATF6 primers were designed from the nucleotide sequence of ATF6α subtype. Quantitative analysis was done by StepOne Software v2.1 (Applied Biosystems).
Western Blot Analysis
Total cell extracts were harvested in a lysis buffer (Cell Signaling Technology, Beverly, MA, USA) and then centrifuged at 12,000 g for 15 min at 4°C. Quantification of total protein was performed with the Lowry protein assay reagent (Bio-Rad Laboratories, Hercules, CA, USA). Proteins were resolved by 10% SDS-PAGE and transferred to a PVDF membrane. After being blocked in 5% milk in TBS with 0.1% Tween 20 (TBST), the membrane was incubated with specific primary antibodies [anti-ATF6α (1:2,000, Abcam), anti-DSP (1:2,000, Santa Cruz Biotechnology), anti-BiP (1:2,000, Cell Signalling), anti-CHOP (1:2,000, Cell Signalling), and anti-DMP1 (1:2,000, Abcam)]. Signals were detected with an enhanced chemiluminescence reagent (Santa Cruz Biotechnology) according to the manufacturer’s instructions.
Immunofluorescence Study
Cells were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 20 min and blocked with 1% bovine serum albumin and 0.5% Triton X-100 in PBS. Cells were incubated with primary antibody (anti-ATF6α, 1:50, Abcam) overnight at 4°C, followed by Alexa 488-conjugated anti-goat IgG treatment (Santa Cruz Biotechnology). Immunoreactivity was detected by LSM confocal microscopy (Carl Zeiss, Oberkochen, Germany). The fluorescence intensity within the nucleus was measured from randomly selected cells by an image analyzer (Carl Zeiss LSM software).
Alkaline Phosphatase (ALP) Staining and Activity
For ALP staining, the cultured cells were fixed with 70% ethanol, rinsed 3 times with deionized water, and then treated with a 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium solution (Sigma-Aldrich, St. Louis, MO, USA) for 15 min. The cells were harvested in a lysis buffer containing 10 mM Tris-Cl (pH 7.4), Triton X-100, and 2 mM phenylmethanesulfonylfluoride, homogenized by sonication, and centrifuged. The lysate was used to measure the ALP activity, as previously reported (Manolagas et al., 1981). The absorbance of the solution was measured with a MultiskanTM GO microplate spectrophotometer (Thermo Scientific, Vantaa, Finland) at a wavelength of 405 nm. The ALP activity was normalized to the total protein content.
Alizarin Red Staining
Cells were fixed as described for ALP staining and treated with a 40 mM alizarin red staining solution (pH 4.2) for 10 min, and then the cell culture plate was photographed. For quantitative analysis, the stains were extracted with 10% cetylpyridinium chloride in 10 mM sodium phosphate solution (pH 7.0) for 15 min, and absorbance was measured at a wavelength of 540 nm with a multiplate reader (Bio-Tek Instruments, Winooski, VT, USA)
Statistical Analyses
All experiments were performed in triplicate. Experimental results are expressed as mean ± standard deviation. Differences between groups were evaluated by analysis of variance (ANOVA), and the Duncan test was used for post hoc analyses with the SPSS 17.0 software program (SPSS, Chicago, IL, USA). p < .05 was considered statistically significant.
Results
Expression of ATF6 during Tooth Development
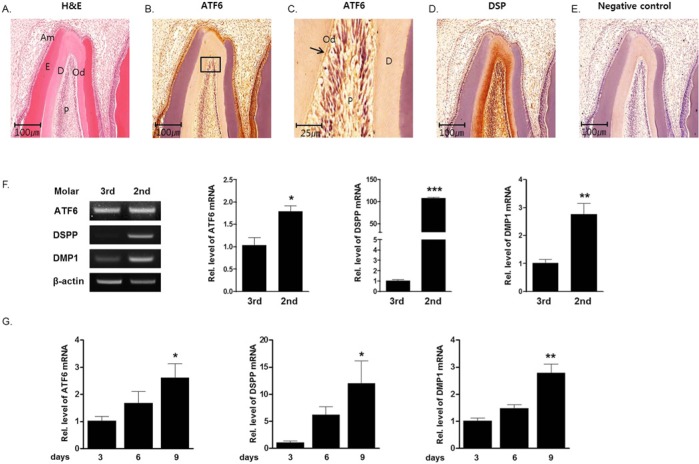
Previously, we identified the developmental stage of the rat tooth germ based on histological analysis (Kim et al., 2009). The 2nd molar tooth germ was at the bell stage, crown formation stage, and root formation stage, at post-natal days 3, 6, and 9, respectively. The 3rd molar tooth germ at post-natal day 9 was at the cap stage. In the present study, expression of ATF6 in the developmental stages of tooth germ was examined. Immunohistochemistry was performed with anti-ATF6 antibody for identification of the localization of ATF6 protein in the tooth germ. Immunoreactivity to anti-ATF6 was observed in odontoblasts and ameloblasts of 2nd molar tooth germs and adjacent osteoblasts. Strong immunoreactivity to anti-DSP was observed in odontoblast, predentin, and dentin layers (Figs. 1A-1E). To determine the involvement of ATF6 in tooth germ development, we evaluated ATF6 mRNA expression using RT-PCR and real-time PCR, according to the development stage of the tooth germ. ATF6 mRNA expression was increased from the cap stage (3rd molar tooth germ at day 9) to the root formation stage (2nd molar tooth germ at day 9) (Figs. 1F, 1G).
Figure 1.
Expression of ATF6, DSP, and DMP1 during tooth development. (A-E) Expression of ATF6 and DSP was histologically evident in the odontoblast layer. (A) Hematoxylin and eosin staining of the 2nd molar tooth germ at post-natal day 9. (B) Immunoreactivity to the anti-ATF6 antibody was observed in the odontoblast layer of the 2nd molar tooth germ. (C) High-power magnification of (B). (D) Immunoreactivity to the anti-DSP antibody was observed in the odontoblast layer of the 2nd molar tooth germ. (E) Negative control. (F, G) Expression profiles of DSPP, DMP1, and ATF6 in rat tooth germs from post-natal days 3 to 9. Total RNA was isolated from the 3rd molar tooth germ at post-natal day 9 (cap stage) and from the 2nd molar tooth germ at post-natal days 3 (bell stage), 6 (crown formation stage), and 9 (root formation stage). (F) RT-PCR analysis and real-time PCR analysis at the cap stage and the root formation stage. During the development stage, DSPP, DMP1, and ATF6 expression was determined by RT-PCR. β-actin was used as an internal control. (G) Real-time PCR analysis at the bell stage, the crown formation stage, and the root formation stage. During the development stage, DSPP, DMP1, and ATF6 expression was determined by real-time PCR. ATF6 mRNA expression at days 6 and 9 increased by 1.7- and 2.5-fold, respectively, when compared with the expression at day 3. Od, odontoblast; Am, ameloblast; P, pulp; D, dentin; E, enamel.
Expression and Activation of ATF6 during Odontoblast Differentiation in vitro
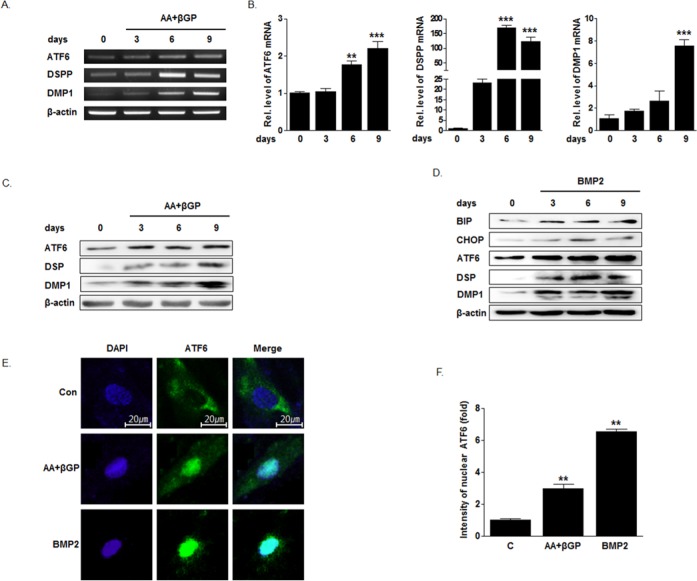
ATF6 mRNA and protein levels were evaluated while HDPCs were cultured in odontogenic differentiation medium. ATF6 mRNA and protein expression were increased in a time-dependent manner, accompanied by an increase in DSPP and DMP1 expression (Figs. 2A-2C). When HDPCs were cultured in BMP2, expressions of ATF6, DSP, and DMP1 were increased. At the same time, the ER stress marker BiP and CHOP expressions were also increased (Fig. 2D).
Figure 2.
Expression of ATF6 mRNA, protein level, and translocation of ATF6 during odontogenic differentiation of human dental pulp cells (HDPCs). HDPCs were cultured with 50 µg/mL ascorbic acid and 5 mM β-glycerophosphate, or 100 ng/mL BMP2. Real-time PCR (RT-PCR), Western blot, and immunofluorescence studies were performed. (A-C) Treatment of ascorbic acid and β-glycerophosphate increased ATF6, DSPP, and DMP1 mRNA and protein expression. (D) BMP2 increased ATF6, DSP, and DMP1, and also BiP and CHOP expression. (E, F) Immunoreactivity of anti-ATF6 was increased in the nucleus when HDPCs were cultured in the ascorbic acid + β-glycerophosphate or BMP2.
To confirm whether ATF6 can be activated as a transcriptional factor during odontoblast differentiation, we examined the nuclear translocation of ATF6 with immunocytochemistry. When cells were exposed to BMP2 and ascorbic acid/β-glycerophosphate, immunoreactivity of anti-ATF6 in the nucleus was increased by more than 6 times (Figs. 2E, 2F). These results suggest that BMP2 also induced ER stress responses to result in cleavage and activation of ATF6 as a transcriptional factor in odontoblastic cells.
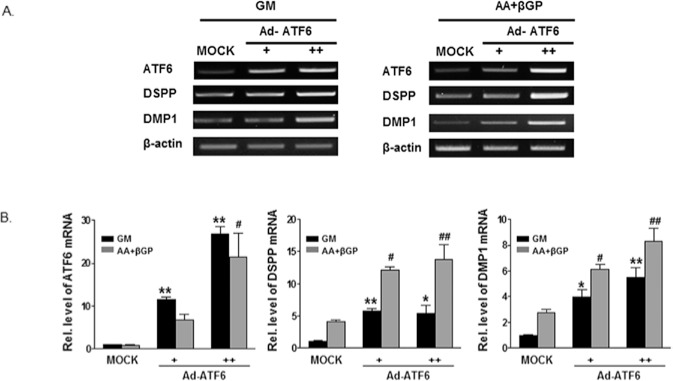
Effect of ATF6 Overexpression on DSPP and DMP1 Expression
Ad-ATF6 was treated into HDPCs, which cultured with growth medium or differentiation medium, and then expressions of DSPP and DMP1 were evaluated. In both culture conditions, overexpression of ATF6 dose-dependently increased the ATF6 mRNA level with the increases in DSPP and DMP1 mRNA expression (Fig. 3).
Figure 3.
Overexpression of ATF6 increased the expression of DSPP and DMP1 in HDPCs. HDPCs were infected with Ad-ATF6 [+ 50 multiplicity of infection (MOI), ++ 100 MOI)] or Ad-GFP (50 MOI) for 24 hrs, and exposed to GM or OM (50 µg/mL ascorbic acid, 5 mM β-glycerophosphate) for 3 days. Cells were harvested for total RNA isolation and RT-PCR or real-time PCR was carried out. Overexpression of ATF6 dose-dependently increased ATF6 mRNA level with the increases in DSPP and DMP1 mRNA expression in RT-PCR (A) and real time PCR (B). GM, growth medium; OM, odontogenic medium.
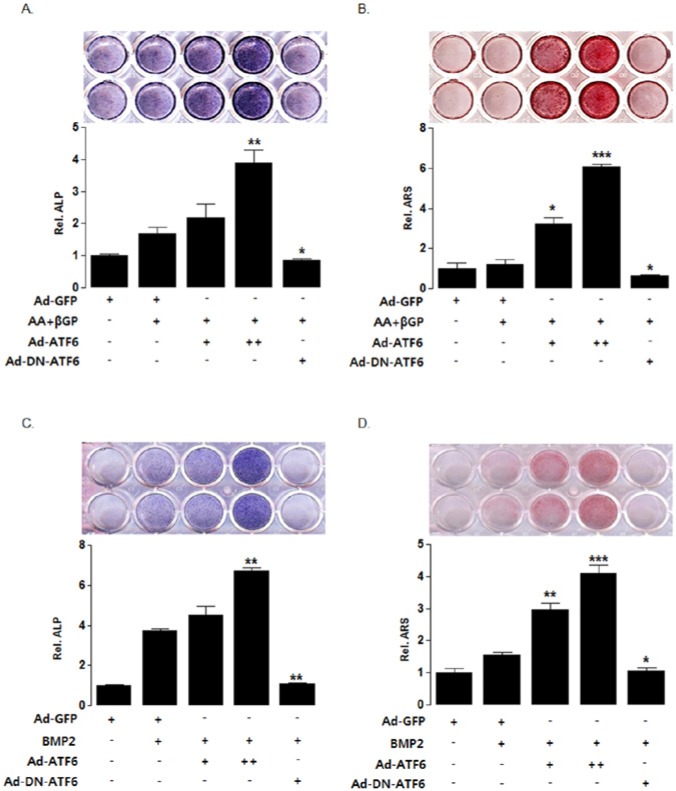
Effect of ATF6 on Mineralization of HDPCs
To examine whether ATF6 functionally regulates the matrix mineralization in HDPCs, we performed ALP staining and alizarin red staining with treatment of Ad-ATF6 or Ad-DN-ATF6. Ad-ATF6 increased ALP staining and mineralized nodule formation; however, inhibition of ATF6 with Ad-DN-ATF6 decreased them, compared with those in the ascorbic acid/β-glycerophosphate control group (Figs. 4A, 4B). BMP2 also induced ALP staining and mineralization nodule formation. ATF6 overexpression enhanced BMP2-induced mineralization, and Ad-DN-ATF6 decreased it (Figs. 4C, 4D)
Figure 4.
ATF6 is involved in the matrix mineralization of HDPCs. (A, C) ALP activity. HDPCs were infected with Ad-ATF6 (+ 50 MOI, ++ 100 MOI), Ad-DN-ATF6 (50 MOI), or Ad-GFP (50 MOI) for 24 hrs and treated with 50 µg/mL ascorbic acid and 5 mM β-glycerophosphate (A) or 100 ng/mL BMP2 (C) for 7 days. ALP stain and enzyme activity was measured as described in Materials & Methods. (B, D) Alizarin red stain. HDPCs were infected with Ad-ATF6 (+ 50 MOI, ++ 100 MOI), Ad-DN-ATF6 (50 MOI), or Ad-GFP (50 MOI) for 24 hrs and treated with 50 µg/mL ascorbic acid and 5 mM β-glycerophosphate (B) or 100 ng/mL BMP2 (D) for 21 days. The cells were stained with an alizarin red solution to evaluate extracellular matrix mineralization. Overexpression of ATF6 enhanced the mineralization effect of odontogenic induction media (ascorbic acid + β-glycerophosphate or BMP2) in HDPCs. Inhibition of ATF6 decreased the mineralization effect induced by ascorbic acid + β-glycerophosphate or BMP2.
Discussion
ATF6 is an ER stress-regulated transmembrane transcription factor that activates the transcription of ER molecular chaperones. Upon ER stress, membrane-bound ATF6 is cleaved and translocates from the ER to the nucleus, where it up-regulates UPR-related genes (Shen et al., 2002). Previously, we showed that ATF6 mediates BMP2-induced osteoblast differentiation and matrix mineralization through regulating osteocalcin expression (Jang et al., 2012). Here, we also observed that ATF6 may be a critical regulator of odontoblast differentiation.
DSPP and DMP1 play important roles in hard tissue development, and they are generally regarded as markers of odontoblast differentiation. Several growth factors and transcription factors, regulating the expression of DSPP and DMP1, are responsible for odontoblast differentiation (Liu et al., 2013). DSPP affects dentin matrix mineralization more significantly than DMP1, acting on both bone and dentin (Suzuki et al., 2012). In the present study, ATF6 protein was expressed in the odontoblastic layer of the rat pulp tissue. In tooth germs, isolated at post-natal days 3, 6, and 9, ATF6 expression increased gradually with DSPP and DMP1 expressions (Fig. 1). In addition, when HDPCs were cultured in ascorbic acid and β-glycerophosphate for inducing odontogenic differentiation, ATF6 expression also increased with DSPP and DMP1 (Figs. 2A-2C). These results suggest that ATF6 might be involved in odontoblast differentiation or dentinogenesis during tooth development.
Based on histology results (Fig. 1A), interestingly, ATF6 was evident in the ameloblasts as well as the osteoblasts in rat maxillae. Additionally, ATF6 expression was detected in human pulpal cells, ameloblast-like cells, and osteoblasts of immortalized cell lineages (Appendix). Differentiating signals still stimulated ATF6 expression in all these cells. These findings suggest that ATF6 or ER-stress signal might affect cell differentiation in mineralizing tissues. The role of ATF6 in the ameloblast differentiation process will be studied in the future.
BMP2 has been well-characterized to induce odontoblast differentiation as well as osteoblast differentiation. Recently, BMP2 has been introduced to stimulate osteoblast differentiation with the induction of mild ER stress. Actually, BMP2 activates ER stress-related transcription factor (or UPR transducers), including ATF4, PERK, and ATF6, to stimulate osteocalcin gene expression (Saito et al., 2011; Jang et al., 2012). In this study, BMP2 also increased the expression of ATF6, DSPP, and DMP1 with increases in the expression of ER stress-related genes, BiP and CHOP (Fig. 2D). Moreover, when HDPCs were cultured in the odontogenic induction medium, nuclear fraction of ATF6 protein increased, indicating that ER-bound ATF6 was cleaved and trafficked into the nucleus (Fig. 2E). These results suggest that BMP2 may also induce mild ER-stress in odontoblasts and activate ATF6 as a transcriptional factor to stimulate odontoblast differentiation.
In the present study, to determine the functional effect of ATF6 on odontoblastic gene expression and matrix mineralization, we did gain- or loss-of-function studies in HDPCs. Overexpression of ATF6 increased DSPP and DMP1 expression, ALP activity, and matrix mineralization (Figs. 3, 4). Inhibition of ATF6 by Ad-DN-ATF6 suppressed the differentiation-related ALP activity and mineralization (Fig. 4). These observations confirm that ATF6 might be involved in the regulation of odontoblast differentiation.
ATF6 protein consists of two subtypes, ATF6α and ATF6β. ATF6α is known to be solely responsible for the transcriptional induction of ER chaperones. Because of this characteristic, in the present study we used the specific PCR primers, antibody, and dominant-negative construct for the ATF6α subtype. Recently, ATF6α knockout mice were generated and were reported to show apparently normal development without analytical results of bone and tooth phenotype in detail (Yamamoto et al., 2007). The findings appear not to be consistent with our results, that inactivation of ATF6α by Ad-DN-ATF6 decreased matrix mineralization in vitro. For the precise role of ATF6 in odontoblast differentiation to be understood, further studies of tooth phenotype in ATF6α knockout mice are needed.
Overall, our study suggests that the ER membrane-bound transcription factor ATF6 might be a novel regulator of odontoblastic differentiation and have potential as a therapeutic target.
Supplementary Material
Footnotes
A supplemental appendix to this article is published electronically only at http://jdr.sagepub.com/supplemental.
This study was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) grant funded by the Ministry of Education, Science and Technology (No. 2012 R1A1A2005002) and by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIP) (No. 2011-0030121).
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
References
- Boot-Handford RP, Briggs MD. (2010). The unfolded protein response and its relevance to connective tissue diseases. Cell Tissue Res 339:197-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler WT. (1995). Dentin matrix proteins and dentinogenesis. Connect Tissue Res 33:59-65. [DOI] [PubMed] [Google Scholar]
- Butler WT. (1998). Dentin matrix proteins. Eur J Oral Sci 106(Suppl 1):204-210. [DOI] [PubMed] [Google Scholar]
- Chen S, Gluhak-Heinrich J, Martinez M, Li T, Wu Y, Chuang HH, et al. (2008). Bone morphogenetic protein 2 mediates dentin sialophosphoprotein expression and odontoblast differentiation via NF-Y signaling. J Biol Chem 283:19359-19370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Gluhak-Heinrich J, Wang YH, Wu YM, Chuang HH, Chen L, et al. (2009). Runx2, osx, and dspp in tooth development. J Dent Res 88:904-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang WG, Kim EJ, Kim DK, Ryoo HM, Lee KB, Kim SH, et al. (2012). BMP2 protein regulates osteocalcin expression via Runx2-mediated Atf6 gene transcription. J Biol Chem 287:905-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jernvall J, Thesleff I. (2000). Reiterative signaling and patterning during mammalian tooth morphogenesis. Mech Dev 92:19-29. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Jeon SK, Kang JH, Kim MS, Ko HM, Jung JY, et al. (2009). Expression of DCC in differentiating ameloblasts from developing tooth germs in rats. Arch Oral Biol 54:563-569. [DOI] [PubMed] [Google Scholar]
- Kitagawa M, Ueda H, Iizuka S, Sakamoto K, Oka H, Kudo Y, et al. (2007). Immortalization and characterization of human dental pulp cells with odontoblastic differentiation. Arch Oral Biol 52:727-731. [DOI] [PubMed] [Google Scholar]
- Liu H, Lin H, Zhang L, Sun Q, Yuan G, Zhang L, et al. (2013). miR-145 and miR-143 regulate odontoblast differentiation through targeting Klf4 and Osx genes in a feedback loop. J Biol Chem 288:9261-9271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolagas SC, Burton DW, Deftos LJ. (1981). 1,25-Dihydroxyvitamin D3 stimulates the alkaline phosphatase activity of osteoblast-like cells. J Biol Chem 256:7115-7117. [PubMed] [Google Scholar]
- Nakashima M. (2005). Bone morphogenetic proteins in dentin regeneration for potential use in endodontic therapy. Cytokine Growth Factor Rev 16:369-376. [DOI] [PubMed] [Google Scholar]
- Oh SH, Hwang YC, Yang H, Kang JH, Hur SW, Jung NR, et al. (2012). SHP is involved in BMP2-induced odontoblast differentiation. J Dent Res 91:1124-1129. [DOI] [PubMed] [Google Scholar]
- Ruch JV, Lesot H, Bègue-Kirn C. (1995). Odontoblast differentiation. Int J Dev Biol 39:51-68. [PubMed] [Google Scholar]
- Saito A, Ochiai K, Kondo S, Tsumagari K, Murakami T, Cavener DR, et al. (2011). Endoplasmic reticulum stress response mediated by the PERK-eIF2(alpha)-ATF4 pathway is involved in osteoblast differentiation induced by BMP2. J Biol Chem 286:4809-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo HY, Kim YD, Lee KM, Min AK, Kim MK, Kim HS, et al. (2008). Endoplasmic reticulum stress-induced activation of activating transcription factor 6 decreases insulin gene expression via up-regulation of orphan nuclear receptor small heterodimer partner. Endocrinology 149:3832-3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Chen X, Hendershot L, Prywes R. (2002). ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev Cell 3:99-111. [DOI] [PubMed] [Google Scholar]
- Sreenath T, Thyagarajan T, Hall B, Longenecker G, D’Souza R, Hong S, et al. (2003). Dentin sialophosphoprotein knockout mouse teeth display widened predentin zone and develop defective dentin mineralization similar to human dentinogenesis imperfecta type III. J Biol Chem 278:24874-24880. [DOI] [PubMed] [Google Scholar]
- Suzuki S, Haruyama N, Nishimura F, Kulkarni AB. (2012). Dentin sialophosphoprotein and dentin matrix protein-1: two highly phosphorylated proteins in mineralized tissues. Arch Oral Biol 57:1165-1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thesleff I, Sharpe P. (1997). Signalling networks regulating dental development. Mech Dev 67:111-123. [DOI] [PubMed] [Google Scholar]
- Wang Y, Shen J, Arenzana N, Tirasophon W, Kaufman RJ, Prywes R. (2000) Activation of ATF6 and an ATF6 DNA binding site by the endoplasmic reticulum stress response. J Biol Chem 275:27013-27020 [DOI] [PubMed] [Google Scholar]
- Yamamoto K, Sato T, Matsui T, Sato M, Okada T, Yoshida H, et al. (2007) Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6 alpha and XBP1. Dev Cell 13:365-376. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.