Abstract
Calcium and phosphorus homeostasis is achieved by interplay among hormones, including 1,25(OH)2D3 (1,25D), parathyroid hormone, and fibroblast growth factor 23 (FGF23), and their interactions with other proteins. For example, mutations in dentin matrix protein 1 (DMP-1) result in increased FGF23 and hypophosphatemic rickets. 1,25D is reported to modulate FGF23; thus, we hypothesized that 1,25D may be involved in modulating DMP-1 in an intermediary step. Murine cementoblasts (OCCM-30) and osteocyte-like cells (MLO-Y4 and MLO-A5), known to express DMP-1, were used to analyze effects of 1,25D on DMP-1 expression in vitro. DMP-1 mRNA levels decreased by 50% (p < .05) in the presence of 1,25D in all cell types, while use of a vitamin D receptor (VDR) agonist (EB1089) and antagonist (23S,25S)-DLAM-2P confirmed that VDR pathway activation was required for this response. Further analysis showed that histone deacetylase recruitment was necessary, but neither protein kinase A nor C pathways were required. In conclusion, our results support the hypothesis that 1,25D regulates DMP-1 expression through a VDR-dependent mechanism, possibly contributing to local changes in bone/tooth mineral homeostasis.
Keywords: Vitamin D receptor, phosphate homeostasis, osteogenesis, SIBLING family, fibroblast growth factor 23, phosphate regulating endopeptidase homolog
Introduction
The active form of vitamin D, 1α,25(OH)2D3 (1,25D), plays a central role in regulating mineralization of skeletal and dental tissues, signaling through the vitamin D receptor (VDR). VDR and its binding partner, retinoid X receptor, recognize vitamin D response elements (VDREs) in the promoter region of 1,25D-responsive genes, modulating numerous biological pathways. 1,25D can also produce rapid, nongenomic (i.e., VDRE-independent) responses linked to calcium (Ca2+) fluxes and activation of protein kinase C (PKC). Several mineralized tissue genes are 1,25D targets, including Spp1 (gene for osteopontin), Bglap (osteocalcin), Lrp5 (low-density lipoprotein–related protein 5), Tnfsf11 (receptor activator of NF-κB ligand), and Tnfsf11b (osteoprotegerin) (Haussler et al., 2012; Pike et al., 2012).
1,25D and 2 other hormones—parathyroid hormone (PTH) and fibroblast growth factor 23 (FGF23)—control circulating Ca2+ and phosphate (Pi) levels and, subsequently, mineralization (Haussler et al., 2012; Kolek et al., 2005). FGF23 directly antagonizes 1,25D by decreasing its activating hydroxylation step in the kidney and increasing its catabolism, secondarily restricting absorption/reabsorption of Ca2+ and Pi (Haussler et al., 2012; Shimada et al., 2004). 1,25D upregulates FGF23 expression in osteocytes, providing a feedback loop between these factors (Barthel et al., 2007; Haussler et al., 2012). PTH stimulation of FGF23 expression closes the loop of the bone-kidney-parathyroid axis of Ca2+ and Pi regulation. However, several aspects of the interactions among 1,25D, PTH, and FGF23 remain unclear. For example, increased levels of circulating FGF23 are observed in hereditary hypophosphatemic rickets, despite causative mutations in disparate genes, including phosphate-regulating endopeptidase homolog, X-linked (PHEX), and dentin matrix protein 1 (DMP-1). Inactivating mutations in PHEX cause X-linked hypophosphatemia (MIM 307800). Mutations in DMP-1 cause type 1 autosomal recessive hypophosphatemic rickets (ARHR1; MIM 241520), remarkably similar to X-linked hypophosphatemia. While it remains unclear how increased FGF23 and hypophosphatemia result from loss of an endopeptidase or matrix protein, proposed mechanisms include a common FGFR1 signaling pathway (Martin et al., 2011) and/or controlling levels of circulating aspartate-rich motif peptides (David et al., 2011).
PHEX and DMP-1 represent potential regulatory links between 1,25D and FGF23. PHEX is downregulated by 1,25D (Ecarot and Desbarats, 1999; Hines et al., 2004); however, attempts to define mechanisms have not been fruitful. Parallel roles for PHEX and DMP-1 in controlling FGF23 suggest that 1,25D may also downregulate DMP-1; yet, the limited studies on this topic are conflicting (Farrow et al., 2009; Ito et al., 2013). We hypothesized that 1,25D-mediated regulation of DMP-1 may provide a link between 1,25D and FGF23, and we tested this using DMP-1 expressing murine cementoblasts (OCCM-30) and osteocyte-like cells (MLO-Y4 and MLO-A5).
Materials & Methods
Cell Lines
An established immortalized murine cementoblast cell line (OCCM-30) (D’Errico et al., 2000) and 2 osteocyte-like cell lines—MLO-A5 preosteocytes and MLO-Y4 mature osteocytes (Kato et al., 1997; Kato et al., 2001)—were used for these studies.
Cell Culture
Cell culture reagents were obtained from Life Technologies (Gaithersburg, MD, USA). Fetal bovine serum (FBS) and calf serum were purchased from Hyclone (Logan, UT, USA). Cell seeding concentration was adjusted for approximately 100% confluence at RNA isolation: OCCM-30 cells at 1 × 104, MLO-Y4 cells at 1.3 × 104, and MLO-A5 cells at 5.3 × 104 cells/cm2. After 24 hrs, medium was changed to DMEM with 2% FBS plus antibiotics for OCCM-30 cells or α-MEM with heat-inactivated 1% FBS and 1% fetal calf serum for osteocyte-like cells. Medium was changed at days 3 and 5, and 1,25D (10 nM) or vehicle (ethanol) plus ascorbic acid (50 µg/mL) was added on day 5. Total RNA was harvested 24 hrs after addition of 1,25D, unless otherwise stated. For time course, RNA was harvested at 3, 6, 18, 24, 48, and 72 hrs after treatment.
To determine if effects of 1,25D on DMP-1 expression were transcription dependent, cells were pretreated for 30 mins with transcription inhibitor actinomycin D (2 µg/mL) before addition of 1,25D. To determine whether effects required de novo protein synthesis, cells were pretreated with cycloheximide (1/10/100 µM) before addition of 1,25D, with RNA harvested 24 hrs later.
To determine involvement of signaling pathways, dose-response experiments were performed with inhibitors GF109203X, U73122, H-89, MDL12,330A, and PD98059 at 1/20/100 µM and SB203580 at 1/15/100 µM, with doses based on previous reports and confirmation of cell viability in our experimental system (Lochner and Moolman, 2006; Tada et al., 2011; Bai et al., 2013; Wu-Zhang and Newton, 2013). Cells were then pretreated for 30 mins with GF109203X (1 µM) or U73122 (20 µM), H-89 (20 µM) or MDL12,330A (20 µM), SB203580 (15 µM), PD98059 (1 µM); PKC/PLC, PKA/AC, MAPK, and MEK/ERK inhibitors, respectively; followed by addition of 1,25D. RNA was harvested 24 hrs later.
To determine involvement of VDR, cells were treated with VDR agonist EB-1089 (10 nM) or antagonist (23S,25S)-DLAM-2P (Ishizuka et al., 2008), with/without 1,25D. Cells were pretreated with (23S,25S)-DLAM-2P for 1 hr before addition of 1,25D. RNA was harvested 24 hrs later.
To determine involvement of histone deacetylases (HDACs), cells were treated with suberoylanilide hydroxamic acid (SAHA) (2 µM; inhibitor of class I and II HDACs), MGCD0103 (0.1 µM; inhibitor of class I HDACs), MC1568 (1 µM; inhibitor of class IIa HDACs), and tubastatin (2 µM; inhibitor of class IIb, HDAC 6). Cells were pretreated with HDAC inhibitors for 30 mins before addition of 1,25D.
1,25D, cycloheximide, actinomycin D, and SAHA were purchased from Sigma Aldrich (St. Louis, MO, USA). EB1089, PD98059, H-89, SB203580, U73122, GF109203X, MDL 12,330A, and MC1568 were purchased from Tocris Bioscience (Bristol, UK). MGCD0103 was obtained from SelleckChem (Houston, TX, USA).
Immunocytochemistry was performed with rat anti-VDR and GAPDH antibodies (Abcam, Cambridge, MA, USA) and anti-rat secondary antibody (Molecular Probes, Eugene, OR, USA).
Real-Time Quantitative Polymerase Chain Reaction
Isolation of total RNA, cDNA synthesis, and quantitative polymerase chain reaction were performed for gene expression analysis on the Roche Lightcycler 480 system (Roche Applied Science, Indianapolis, IN, USA) using intron-spanning primers (Appendix Table) and relative expression analysis from Cp values (the Efficiency method, Roche Applied Science software). One-way analysis of variance and post hoc Tukey or Bonferroni test were used to detect intra- and intergroup gene expression differences. Student t test was used for comparisons between 2 groups (SigmaStat, SSI, San Jose, CA, USA).
Western Blot Analysis
Cells were exposed to 1,25D under serum-free conditions for 48 hrs; 10 µg of total protein was used for SDS-PAGE in a 4% to 12% (w/v) polyacrylamide stacking gel (Invitrogen, Grand Island, NY, USA), then transferred to a nitrocellulose membrane. Rabbit anti-DMP-1 antibody (Huang et al., 2008) and IRDye secondary antibodies (LI-COR, Lincoln, NE, USA) were used. An imaging system (ODYSSEY CLx–LI-COR) with Image Studio Software (LI-COR) was used to quantify proteins.
Results
1,25D Downregulates DMP-1 Expression in Cementoblasts and Osteocyte-like Cells
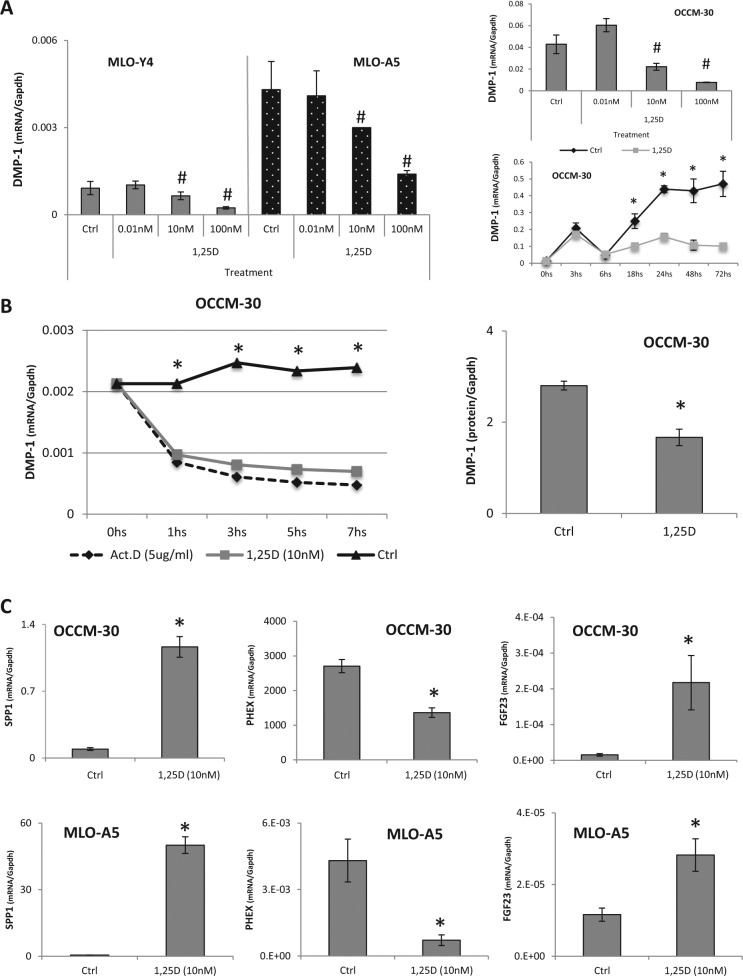
For all cells types used, 1,25D downregulated DMP-1 gene expression in dose-dependent fashion (0.01, 10, 100 nM) by 24 hrs (Fig. 1A). Time course analysis based on OCCM-30 cells treated with 1,25D (10 nM) indicated suppression of DMP-1 mRNA (approximately 3-fold decrease, p < .05) by 18 hrs, which was maintained up to 72 hrs, when the experiment was terminated. Pretreatment of OCCM-30 cells for 30 mins with actinomycin D indicated that 1,25D-mediated decrease of DMP-1 mRNA expression was regulated at the transcriptional level (Fig. 1B) and de novo protein synthesis was not required (determined with cycloheximide; data not shown). 1,25D decreased DMP-1 protein levels by 48 hrs.
Figure 1.
1α,25(OH)2D3 (1,25D) decreases the expression of DMP-1 mRNA and protein. (A) 1,25D downregulates DMP-1 mRNA expression in a dose-dependent manner (0.01, 10, 100 nM) by 24 hrs following treatment of MLO-Y4 and MLO-A5 osteocyte-like cells and OCCM-30 cementoblasts. In addition, 1,25D at 10 nM suppresses DMP-1 mRNA by 18 hrs in OCCM-30 cells, and suppression is maintained up to 72 hrs. (B) 1,25D treatment does not affect DMP-1 mRNA stability in actinomycin D (Act.D)–treated cells compared with the control group, and Western blot data demonstrate significant reduction in DMP-1 protein expression in media obtained from OCCM-30 cells at 48 hrs after 1,25D treatment. (C) Treatment of OCCM-30 and MLO-A5 cells with 10 nM 1,25D significantly regulates vitamin D–responsive genes, including SPP1 (OPN), PHEX, and FGF23 mRNA levels. Experiments were repeated at least 2 times in triplicate with comparable results. #Indicates statistical intergroup differences compared with the control by 1-way analysis of variance, followed by post hoc Bonferroni test (p < .05). *Indicates statistical intergroup difference by the Student t test (p < .05).
1,25D (10 nM) significantly increased OPN/SPP1 (15-fold/ 100-fold), decreased PHEX (2-fold/6-fold), and increased FGF23 (14-fold/2.6-fold) mRNA levels in OCCM-30 and MLO-A5 cells, respectively (Fig. 1C), as previously reported for other cell types (Ecarot and Desbarats, 1999; Hines et al., 2004; Nagata et al., 1994), validating the use of OCCM-30 cells in further experiments.
PKC, PKA, MAPK, and MEK/ERK Pathways are Not Required for 1,25D Effect on DMP-1
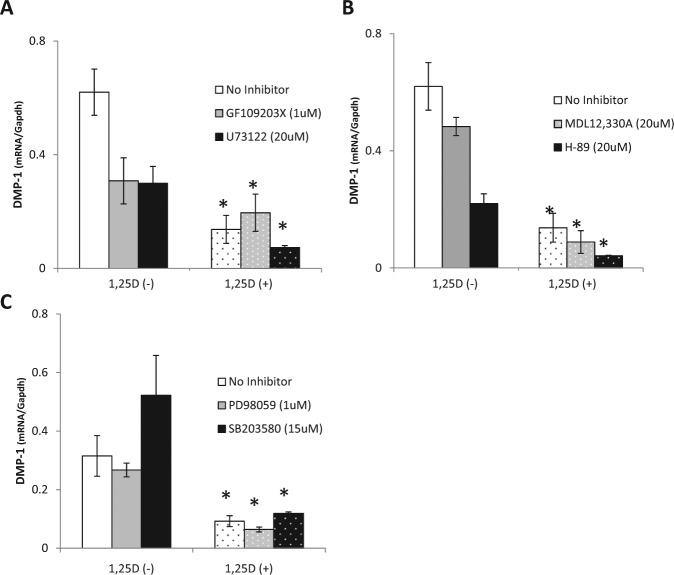
OCCM-30 cells were pretreated with pathway-specific inhibitors before addition of 1,25D (10 nM). Inhibition of PKC, PKA, MAPK, or MEK/ERK signaling failed to block 1,25D-mediated downregulation of DMP-1 (Fig. 2), suggesting that these pathways are not required for observed changes.
Figure 2.
PKC, PKA, MAPK, or MEK/ERK pathways are not required for 1α,25(OH)2D3 (1,25D) effect on DMP-1. OCCM-30 cells were pretreated for 30 mins with (A) 1 µM GF109203X or 20 µM U73122 (PKC/PLC inhibitors), (B) 20 µM H-89 or 20 µM MDL12,330A (PKA/AC inhibitors), (C) 1 µM PD98059 (MEK/ERK inhibitor) or 15 µM SB203580 (MAPK inhibitor). Cells were then treated with 10 nM 1,25D or vehicle (ethanol) and RNA harvested 24 hrs later. 1,25D-mediated downregulation of DMP-1 expression is not blocked by any of the signaling inhibitors evaluated, indicating that these pathways are not required. Experiments were repeated at least 2 times in triplicate with comparable results. *Indicates statistical intergroup difference by the Student t test (p < .05).
Class I HDAC Activity is Required for VDR-mediated Regulation of DMP-1
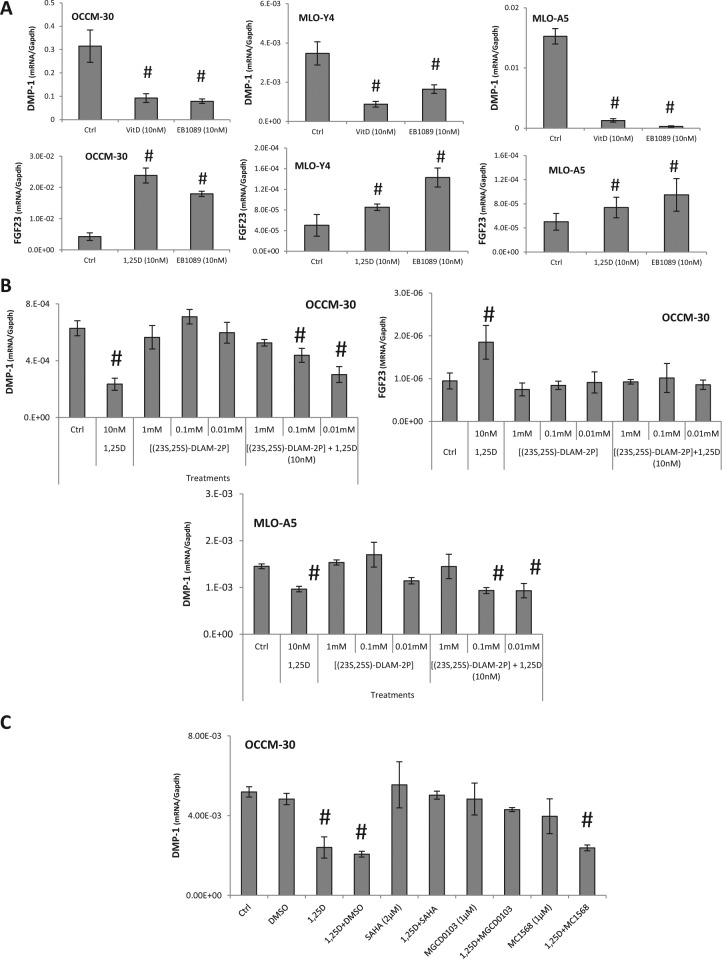
Increased VDR transcripts were noted in 1,25D-treated OCCM-30, MLO-Y4, and MLO-A5 cells (Appendix Fig. 1). Cells exposed to 10 nM EB1089, a VDR agonist, exhibited reduced DMP-1 mRNA and increased FGF23 mRNA at 24 hrs, similar to effects of 1,25D (Fig. 3A). Conversely, addition of (23S,25S)-DLAM-2P, a VDR antagonist, rescued both DMP-1 and FGF23 gene expression in the presence of 1,25D in OCCM-30 and MLO-A5 cells (Fig. 3B).
Figure 3.
Class I histone deacetylase (HDAC) activity is required for vitamin D receptor (VDR)–mediated regulation of DMP-1. (A) Participation of the VDR signaling pathway in 1α,25(OH)2D3 (1,25D)–mediated inhibition of DMP-1 and increase in FGF23 expression was analyzed with a VDR agonist, EB1089, and antagonist, (23S,25S)-N-benzyl-1alpha,25-dihydroxyvitaminD(3)-26,23-lactam [(23S,25S)-DLAM-2P]. Activation of the VDR pathway by EB1089 mimics the effect of 1,25D on DMP-1 and FGF23 expression in OCCM-30, MLO-Y4, and MLO-A5 cells. (B) Inhibition of VDR signaling by (23S,25S)-DLAM-2P blocks the 1,25D-mediated inhibition of DMP-1 expression in MLO-A5 and OCCM-30 cells and FGF23 upregulation in OCCM-30 cells. (C) Application of 2 µM suberoylanilide hydroxamic acid (SAHA), a broad-spectrum HDAC class I and II inhibitor, rescues 1,25D-mediated reduction of DMP-1 mRNA levels in OCCM-30 cells. Treatment with 1 µM MGCD0103 (HDAC class I 1/2/3 inhibitor) mimics the rescue effect of SAHA. However, treatment with 1 µM MC1568 (HDAC class IIa inhibitor) does not inhibit 1,25D-mediated downregulation of DMP1. Experiments were repeated at least 2 times in triplicate with comparable results. #Indicates intergroup statistical differences compared with the control group by 1-way analysis of variance, followed by the Bonferroni test (p < .05).
VDR-dependent transrepression has been shown to require HDAC activity via a heterodimer complex. We determined whether HDAC was required for 1,25D-mediated downregulation of DMP-1 expression. OCCM-30 cells expressed transcripts for the most common forms of HDACs (1-4), and 1,25D did not alter their expression (Appendix Fig. 2).
Application of a broad-spectrum HDAC inhibitor, SAHA, rescued 1,25D- mediated reduction of DMP-1 mRNA levels in OCCM-30 cells (p > .05) (Fig. 3C). Next, OCCM-30 cells were pretreated with class-specific HDAC inhibitors. Class I (HDAC 1/2/3) inhibitor MGCD0103 reproduced SAHA-mediated recovery of DMP-1, while MC1568 (class IIa inhibitor), as well as TH-C106 and Tubastatin (class I, HDAC 1/3 inhibitor and class IIb, HDAC 6 inhibitor, respectively; data not shown), did not block 1,25 D reduction of DMP-1 mRNA. The results suggest that a class I HDAC, likely HDAC 2, is involved in the regulation of DMP-1 expression by 1,25D.
Discussion
Discovery of new factors, hormones, and pathways controlling calcium and phosphate and directing physiologic and pathologic mineralization not only make us appreciate these complex processes but create opportunities for new therapeutic approaches. Our findings here demonstrate that 1,25D decreases DMP-1 levels in murine cementoblasts and osteocyte-like cells via the VDR pathway, possibly contributing to local changes in bone and tooth matrix mineralization and identifying a potential new mechanism for 1,25D regulation of FGF23 and systemic mineral homeostasis.
DMP-1 is a Target Gene Regulated by 1,25D
DMP-1 is an extracellular matrix protein first discovered in dentin, though later studies identified a broader range of expression in bone, with high levels in osteocytes (Huang et al., 2008). Further studies revealed that DMP-1-null mice exhibit severe hypophosphatemia, and biochemical aspects of hypophosphatemic rickets, notably high levels of circulating FGF23 (Liu et al., 2008). Mutations in the human DMP-1 gene were identified as causative for ARHR1, which features elevated FGF23, hypophosphatemia, low (or inappropriately normal) 1,25D, hyperparathyroidism, osteomalacia, and tooth defects.
1,25D-induced transactivation by the VDR is well documented, and positive VDREs in gene promoters are known (Pike et al., 2012). In contrast, mechanisms involved in negative gene regulation by VDR remain poorly understood. DR3-like and E-box-type negative VDRE sequences (nVDREs) have been characterized in the promoter regions of the PTH, PTHrP, and CYP27B1 genes, directly or indirectly binding the VDR as a monomer or the VDR–retinoid X receptor heterodimer, driving transcriptional repression (Murayama et al., 1998; Okazaki et al., 2003). The association of the VDR–retinoid X receptor heterodimer with nVDRE sequences may occur via a bHLH transcription factor referred to as VDIR (Murayama et al., 2004). Existing data support an essential role of HDACs in promoting conformational chromatin changes allowing for transcription to occur (Pike et al., 2012). After demonstrating 1,25D repression of DMP-1 gene and protein expression in cementoblasts and osteocyte-like cells, we confirmed expression of VDR in OCCM-30 cells and demonstrated expression of common HDAC isoforms. Next we demonstrated that the VDR agonist EB-1089 replicated the effects of 1,25D by downregulating DMP-1 transcripts, while the VDR antagonist (23S,25S)-DLAM-2P rescued DMP-1 expression in the presence of 1,25D. Use of broad-spectrum and selective HDAC inhibitors revealed that recruitment of class I HDACs (1/2/3) were required for downregulation of DMP-1. These data provide strong evidence that 1,25D-mediated regulation of DMP-1 is dependent on VDR activation, and further, that gene repression is dependent on HDAC function, as for many genes that are negatively regulated by nuclear hormone receptors (Zhang and Dufau, 2003).
Having established that DMP-1 expression was modulated by a VDR/HDAC-containing complex, we also examined the murine DMP-1 promoter for the presence of nVDRE sequences. Four putative nVDRE sequences were previously identified (Murayama et al., 2004; Tovar Sepulveda and Falzon, 2003), and by gel shift assay, none were found to be involved in the 1,25D-mediated effect on DMP-1 (data not shown). Moreover, promoter studies were inconclusive, similar to studies on 1,25D and the PHEX promoter (Hines et al., 2004). Promoter analysis demonstrated that the only 1,25D-responsive DMP-1 promoter construct contained 4 kb of 5′ flanking DNA, suggesting that the 1,25D-responsive region is located between 2.5 and 4 kb upstream of the transcription start site (Appendix Fig. 3). In the future, we would like to further investigate the role that the promoter region plays in the 1,25D-mediated negative regulation of the DMP-1 gene. Based on these findings, it is possible that 1,25D may indirectly affect DMP-1 expression by regulating the expression of other genes. Future studies using RNAseq approaches will help determine the specific DNA sequences and transcription factor(s) involved in DMP-1 regulation by 1,25D, and ex vivo and in vivo studies may assist us determining direct and indirect mechanisms involved.
Role of DMP-1 in 1,25D-FGF23 Regulation
The results here provide potential new insights on interactions between hormones directing mineral metabolism, 1,25D and FGF23, summarized in a hypothesized model in Fig. 4. While it is established that FGF23 is upregulated by 1,25D, the molecular mechanisms remain to be clarified. A recent report highlighting the ability of combined Pi and 1,25D to increase FGF23 expression in vitro did not evaluate 1,25D alone, leaving its role in the induction unclear (Ito et al., 2013). A direct effect of 1,25D on FGF23 expression has been suggested to be via VDR, with the identification of a putative VDRE sequence in the promoter (Haussler et al., 2012). However, because ongoing protein synthesis seems to be required for the action of 1,25D on FGF23 expression, an intermediary transfactor may participate in the mechanism (Kolek et al., 2005). It has also been suggested that 1,25D may control FGF23 expression by its repressor action on PHEX, and results here suggest that DMP-1 may be involved in parallel fashion. PHEX represses FGF23 expression (Yuan et al., 2008), possibly via FGF receptor signaling (Martin et al., 2011) and/or control of aspartate-rich motif serum levels (David et al., 2011). Hypophosphatemic rickets, caused by increased levels of FGF23, results from mutations in both DMP-1 and PHEX. The mechanisms by which DMP-1 and PHEX affect FGF23 expression remain unclear; however, possibilities include (1) FGF receptor 1 signaling (Martin et al., 2011), (2) potential for DMP-1 to act as a transcription factor (Narayanan et al., 2003; Siyam et al., 2012), (3) alterations in local extracellular matrix that trigger cells to increase FGF23 production, or (4) interactions between PHEX/DMP-1 and yet unknown genes and proteins.
Figure 4.
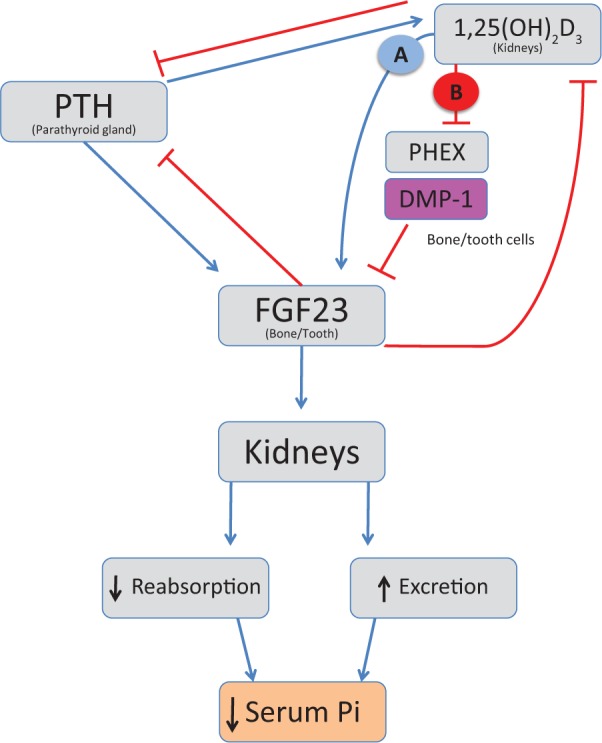
Model for hypothesized 1α,25(OH)2D3 (1,25D) regulation of FGF23 expression, in part mediated through its effects on DMP-1 expression. 1,25D and parathyroid hormone (PTH) have been reported to stimulate FGF23 secretion (blue arrows) controlling inorganic phosphate (Pi) reuptake and excretion by the kidneys. FGF23 suppresses 1,25D and PTH production (red capped lines). Although controversial, 1,25D has been suggested to directly regulate FGF23 by activation of the VDR pathway, leading to gene transactivation through a VDRE sequence in the promoter region of the FGF23 gene (pathway A), and indirectly through its suppression of PHEX (pathway B). We now provide evidence suggesting that the ability of 1,25D to increase FGF23 expression may additionally be linked to its ability to downregulate DMP-1 expression (pathway B), consistent with excess FGF23 observed in ARHR1 resulting from mutations in the DMP1 gene. Effects of 1,25D on DMP1 would also be expected to influence local mineralized tissue properties, such as mineralization, which can affect systemic Ca and Pi homeostasis.
Supplementary Material
Acknowledgments
The authors thank Dr. Lynda Bonewald (University of Missouri–Kansas City) for providing MLO-Y4 and MLO-A5 cells, Dr. Kazuo Nagasawa (Tokyo University of Agriculture and Technology) for providing vitamin D receptor antagonist, Dr. Jian Feng (Baylor College of Dentistry) for providing DMP-1 promoter constructs, and Dr. Chunlin Qin (Baylor College of Dentistry) for providing DMP-1 antibody.
Footnotes
A supplemental appendix to this article is published electronically only at http://jdr.sagepub.com/supplemental.
This research was supported by the Intramural Research Programs of the National Institute of Arthritis and Musculoskeletal and Skin Diseases (MJS) and the National Institute of Dental and Craniofacial Research (MTC).
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
References
- Bai B, He J, Li YS, Wang XM, Ai HJ, Cui FZ. (2013). Activation of the ERK1/2 signaling pathway during the osteogenic differentiation of mesenchymal stem cells cultured on substrates modified with various chemical groups. Biomed Res Int 2013:361906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthel TK, Mathern DR, Whitfield GK, Haussler CA, Hopper HA, 4th, Hsieh JC, et al. (2007). 1,25-Dihydroxyvitamin D3/VDR-mediated induction of FGF23 as well as transcriptional control of other bone anabolic and catabolic genes that orchestrate the regulation of phosphate and calcium mineral metabolism. J Steroid Biochem Mol Biol 103:381-388. [DOI] [PubMed] [Google Scholar]
- David V, Martin A, Hedge AM, Drezner MK, Rowe PS. (2011). ASARM peptides: PHEX-dependent and -independent regulation of serum phosphate. Am J Physiol Renal Physiol 300:F783-F791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Errico JA, Berry JE, Ouyang H, Strayhorn CL, Windle JJ, Somerman MJ. (2000). Employing a transgenic animal model to obtain cementoblasts in vitro. J Periodontol 71:63-72. [DOI] [PubMed] [Google Scholar]
- Ecarot B, Desbarats M. (1999). 1,25-(OH)2D3 down-regulates expression of Phex, a marker of the mature osteoblast. Endocrinology 140:1192-1199. [DOI] [PubMed] [Google Scholar]
- Farrow EG, Davis SI, Ward LM, Summers LJ, Bubbear JS, Keen R, et al. (2009). Molecular analysis of DMP1 mutants causing autosomal recessive hypophosphatemic rickets. Bone 44:287-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haussler MR, Whitfield GK, Kaneko I, Forster R, Saini R, Hsieh JC, et al. (2012). The role of vitamin D in the FGF23, klotho, and phosphate bone-kidney endocrine axis. Rev Endocr Metab Disord 13:57-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines ER, Kolek OI, Jones MD, Serey SH, Sirjani NB, Kiela PR, et al. (2004). 1,25-Dihydroxyvitamin D3 down-regulation of PHEX gene expression is mediated by apparent repression of a 110 kDa transfactor that binds to a polyadenine element in the promoter. J Biol Chem 279:46406-46414. [DOI] [PubMed] [Google Scholar]
- Huang B, Maciejewska I, Sun Y, Peng T, Qin D, Lu Y, et al. (2008). Identification of full-length dentin matrix protein 1 in dentin and bone. Calcif Tissue Int 82:401-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizuka S, Kurihara N, Hiruma Y, Miura D, Namekawa J, Tamura A, et al. (2008). 1alpha,25-Dihydroxyvitamin D(3)-26,23-lactam analogues function as vitamin D receptor antagonists in human and rodent cells. J Steroid Biochem Mol Biol 110:269-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito N, Findlay DM, Anderson PH, Bonewald LF, Atkins GJ. (2013). Extracellular phosphate modulates the effect of 1α,25-dihydroxy vitamin D3 (1,25D) on osteocyte like cells. J Steroid Biochem Mol Biol 136:183-186. [DOI] [PubMed] [Google Scholar]
- Kato Y, Windle JJ, Koop BA, Mundy GR, Bonewald LF. (1997). Establishment of an osteocyte-like cell line, MLO-Y4. J Bone Miner Res 12:2014-2023. [DOI] [PubMed] [Google Scholar]
- Kato Y, Boskey A, Spevak L, Dallas M, Hori M, Bonewald LF. (2001). Establishment of an osteoid preosteocyte-like cell MLO-A5 that spontaneously mineralizes in culture. J Bone Miner Res 16:1622-1633. [DOI] [PubMed] [Google Scholar]
- Kolek OI, Hines ER, Jones MD, LeSueur LK, Lipko MA, Kiela PR, et al. (2005). 1alpha,25-Dihydroxyvitamin D3 upregulates FGF23 gene expression in bone: the final link in a renal-gastrointestinal-skeletal axis that controls phosphate transport. Am J Physiol Gastrointest Liver Physiol 289:G1036-G1042. [DOI] [PubMed] [Google Scholar]
- Liu S, Zhou J, Tang W, Menard R, Feng JQ, Quarles LD. (2008). Pathogenic role of Fgf23 in Dmp1-null mice. Am J Physiol Endocrinol Metab 295:E254-E261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lochner A, Moolman JA. (2006). The many faces of H89: a review. Cardiovasc Drug Rev 24:261-274. [DOI] [PubMed] [Google Scholar]
- Martin A, Liu S, David V, Li H, Karydis A, Feng JQ, et al. (2011). Bone proteins PHEX and DMP1 regulate fibroblastic growth factor Fgf23 expression in osteocytes through a common pathway involving FGF receptor (FGFR) signaling. FASEB J 25:2551-2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama A, Takeyama K, Kitanaka S, Kodera Y, Hosoya T, Kato S. (1998). The promoter of the human 25-hydroxyvitamin D3 1 alpha-hydroxylase gene confers positive and negative responsiveness to PTH, calcitonin, and 1 alpha,25(OH)2D3. Biochem Biophys Res Commun 249:11-16. [DOI] [PubMed] [Google Scholar]
- Murayama A, Kim MS, Yanagisawa J, Takeyama K, Kato S. (2004). Transrepression by a liganded nuclear receptor via a bHLH activator through co-regulator switching. EMBO J 23:1598-1608. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Nagata T, Yokota M, Ohishi K, Nishikawa S, Shinohara H, Wakano Y, et al. (1994). 1 alpha,25-dihydroxyvitamin D3 stimulation of osteopontin expression in rat clonal dental pulp cells. Arch Oral Biol 39:775-782. [DOI] [PubMed] [Google Scholar]
- Narayanan K, Ramachandran A, Hao J, He G, Park KW, Cho M, et al. (2003). Dual functional roles of dentin matrix protein 1: implications in biomineralization and gene transcription by activation of intracellular Ca2+ store. J Biol Chem 278:17500-17508. [DOI] [PubMed] [Google Scholar]
- Okazaki T, Nishimori S, Ogata E, Fujita T. (2003). Vitamin D–dependent recruitment of DNA-PK to the chromatinized negative vitamin D response element in the PTHrP gene is required for gene repression by vitamin D. Biochem Biophys Res Commun 304:632-637. [DOI] [PubMed] [Google Scholar]
- Pike JW, Meyer MB, Bishop KA. (2012). Regulation of target gene expression by the vitamin D receptor: an update on mechanisms. Rev Endocr Metab Disord 13:45-55. [DOI] [PubMed] [Google Scholar]
- Shimada T, Kakitani M, Yamazaki Y, Hasegawa H, Takeuchi Y, Fujita T, et al. (2004). Targeted ablation of Fgf23 demonstrates an essential physiological role of FGF23 in phosphate and vitamin D metabolism. J Clin Invest 113:561-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siyam A, Wang S, Qin C, Mues G, Stevens R, D’Souza RN, et al. (2012). Nuclear localization of DMP1 proteins suggests a role in intracellular signaling. Biochem Biophys Res Commun 424:641-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada H, Nemoto E, Foster BL, Somerman MJ, Shimauchi H. (2011). Phosphate increases bone morphogenetic protein-2 expression through cAMP-dependent protein kinase and ERK1/2 pathways in human dental pulp cells. Bone 48:1409-1416. [DOI] [PubMed] [Google Scholar]
- Tovar Sepulveda VA, Falzon M. (2003). Prostate cancer cell type-specific regulation of the human PTHrP gene via a negative VDRE. Mol Cell Endocrinol 204:51-64. [DOI] [PubMed] [Google Scholar]
- Wu-Zhang AX, Newton AC. (2013). Protein kinase C pharmacology: refining the toolbox. Biochem J 452:195-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan B, Takaiwa M, Clemens TL, Feng JQ, Kumar R, Rowe PS, et al. (2008). Aberrant Phex function in osteoblasts and osteocytes alone underlies murine X-linked hypophosphatemia. J Clin Invest 118:722-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Dufau ML. (2003). Dual mechanisms of regulation of transcription of luteinizing hormone receptor gene by nuclear orphan receptors and histone deacetylase complexes. J Steroid Biochem Mol Biol 85:401-414. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.