Abstract
Context:
Although uterine fibroids (UFs) continue to place a major burden on female reproductive health, the mechanisms behind their origin remain undetermined. Normal myometrial stem cells may be transformed into tumor-initiating stem cells, causing UFs, due to unknown causes of somatic mutations in MED12, found in up to 85% of sporadically formed UFs. It is well established in other tumor types that defective DNA repair increases the risk of such tumorigenic somatic mutations, mechanisms not yet studied in UFs.
Objective:
To examine the putative cause(s) of this stem cell transformation, we analyzed DNA repair within stem cells from human UFs compared to those from adjacent myometrium to determine whether DNA repair in fibroid stem cells is compromised.
Design:
Human fibroid (F) and adjacent myometrial (Myo) stem cells were isolated from fresh tissues, and gene expression relating to DNA repair was analyzed. Fibroid stem cells differentially expressed DNA repair genes related to DNA double- (DSBs) and single-strand breaks. DNA damage was measured using alkaline comet assay. Additionally, DNA DSBs were induced in these stem cells and DNA DSB repair evaluated (1) by determining changes in phosphorylation of DNA DSB-related proteins and (2) by determining differences in γ-H2AX foci formation and relative DNA repair protein RAD50 expression.
Results:
Overall, F stem cells demonstrated increased DNA damage and altered DNA repair gene expression and signaling, suggesting that human F stem cells demonstrate impaired DNA repair.
Conclusions:
Compromised F stem cell DNA repair may contribute to further mutagenesis and, consequently, further growth and propagation of UF tumors.
Keywords: uterine fibroids, stem cells, DNA damage response, DNA repair, genomic instability, MED12
Introduction
Uterine fibroids (UFs), benign myometrial tumors, continue to severely impact female reproductive health and require roughly US$34.4 billion health-care dollars each year in the United States for uterine fibroid–related costs.1 Although benign tumors, UFs can cause severe obstetric- and gynecologic-related complications, including excess uterine bleeding, often causing anemia, infertility, recurrent pregnancy loss, preterm labor, and postpartum hemorrhage.2–4 Currently, an accepted model for the origin of UFs implicates the MED12 gene; it is proposed that somatic UF-causing mutations in this gene, detected in up to 85% of all sporadic human UFs, in “at-risk” myometrial (Myo) stem cells converts them into UF-forming Myo stem cells.5–7 In addition, it has been established that only human UF tissues and corresponding stem cells harbor mutations in the MED12 gene while those of adjacent myometrium do not.8,9
Although the cause of these specific UF-causing MED12 mutations remains unknown, it is well accepted that defects in DNA repair, including pathways relating to DNA double-strand breaks (DSBs) or DNA single-strand breaks (SSBs), in a variety of tissues increase the risk of somatic tumor-forming mutations.10–14 In addition, several studies correlate increased numbers/markers of tissue-specific progenitor cells with an increased risk of genomic instability and even neoplasia; progenitor cells necessitate a high number of mitotic events to maintain the composition of tissue with high turnover or needed remodeling throughout the lifetime of that specific tissue, for example, the myometrium.15–19 This suggests that with increased numbers of progenitor cells, there is an increased risk of random mutations occurring even during normal physiologic processes, such as DNA replication, which contributes thousands of DNA lesions each day. This requires constant clearance of genomic injuries,10,20 and this maintenance of the genome requires sensitive, effective induction of the DNA damage response (DDR), which is achieved by damage sensors, signal transducers, repair effectors, and arrest or death effectors.10 Of note, the most debilitating lesions, DNA DSBs, must be repaired via homologous recombination (HR) or nonhomologous end-joining (NHEJ), requiring a high level of fidelity to maintain genome integrity.10,21 Advances in cancer research attempt to capitalize on the necessity of intact DNA repair for cell survival; chemo- and radiotherapeutic agents create genomic instability in cancer cells to induce cell death, although some robust subpopulations of cancer stem cells evade DNA damage–induced apoptosis.10,21–24
Moreover, reduced expression of several DNA repair genes, suggesting compromised DNA repair, has been indicative of increased cancer prevalence in a variety of tissues, including sex steroid hormone–regulated breast tumors.25–28 Some tissue-specific stem cells demonstrate differential utilization of the various DNA repair mechanisms, with some cancers hijacking DNA repair mechanisms to promote cell survival. Interestingly, however, sex steroid hormone–regulated mammary stem cells (MaSCs) of the breast that are deficient in DNA repair–related Breast cancer 1 (BRCA1) accumulate destabilizing mutations, ultimately increasing their susceptibility to cancer development, and hematopoietic stem cells demonstrating reduced function in DNA repair protein RAD50 (RAD50) and loss of p53 become dysfunctional.24,29,30 Finally, it has been proposed that estrogens, which stimulate cell proliferation in sex hormone–regulated tumors, like UFs, promote additional replication errors.31,32 The combined effects of increased estrogen-influenced DNA replication, greater risk of mutagenesis, and impaired DNA repair in the Myo stem cell population may be implicated in the processes that transform these cells into tumor-forming cells, ultimately leading to UF development and/or propagation.31,32
We have recently shown that several DNA repair genes are downregulated in fibroid tissues and differentiated cells versus those of adjacent myometrium. This suggests that disruption of human DNA repair may affect genomic integrity in the cells from which these UF tumors originate, potentially leading to both initiation and propagation of UF tumor growth and development.33,34 It has yet to be shown, however, whether stem cells within the tumors themselves harbor defects in DNA repair, which would reflect the capacity for these progenitor cells to propagate defects in genome integrity, allowing for further UF tumor growth. Thus, in this work, we aimed to evaluate DNA repair in the human Stro1+/CD44+ fibroid (F) stem cell population versus the Stro1+/CD44+ stem cells of adjacent myometrium to explore whether changes in F stem cells include dysregulation and/or impairment of DNA repair systems.
Materials and Methods
Ethical Approval
Use of human tissue specimens was approved by the institutional review board of Augusta University, and all patients had previously signed a written informed consent form prior to collection of otherwise discarded tissue.
Human uterine myometrial and fibroid tissue sample collection and single-cell suspension collection
Fresh human fibroid and adjacent myometrial samples were obtained via the Augusta University Biorepository from reproductive-age women (31-54 years old) undergoing hysterectomy or myomectomy for either symptomatic UFs or other gynecological disorders, excluding malignancies. These women had not used any hormonal treatment for 3 months prior to the day of surgery (ie, the day of sample collection). Uterine tissue collection includes information on endometrial status (proliferative vs secretory), age, race, body mass index, gravida, and parity according to deidentified records and corresponding pathology reports of collected tissues. Consistently, we collected at least 2 × 2 × 2 cm samples (Supplemental Table S1) from UF-containing uteri, both fibroid tissue and adjacent (>2 cm away from neighboring UF, to eliminate the potential mechanical or hormonal effects of the UF) myometrial tissue samples. In efforts to minimize the effects of topological differences in gene expression that have been observed in larger UF tumors,35 which have also been demonstrated to have quite heterogeneous genomic landscapes,36 we sampled the UF tissue from the center of each tumor.
As described previously,37 fresh myometrial and fibroid fragments were washed, manually, diced into small pieces (∼1 mm3), and digested at 37°C. The digested tissue was filtered to obtain single cell suspensions.
Myometrial and Fibroid Stem Cell Isolation
As progenitor cells have been proposed as an origin and propagators of UFs, we first isolated Stro-1+/CD44+ dual-positive stem cells from fresh human UFs and adjacent normal myometrial tissues from n = 5 patients using antibodies against cell surface markers, Stro-1 and CD44 as described in our previous research.37 Magnetic bead selection was performed according to the manufacturer’s instructions (Life Technologies, Carlsbad, CA, USA), as previously described.37 Briefly, freshly isolated myometrial and fibroid cells were resuspended at a concentration of 1 × 107 cells/mL and incubated first with biotinylated (Molecular Probes, Eugene, OR, USA) anti-CD44 antibody (Supplemental Table S2: MAB7045, R&D Systems, Minneapolis, MN, USA) for isolation of CD44–positive (CD44+) cells from the supernatant; this was then repeated with biotinylated anti-Stro1 antibody (Supplemental Table S2: MAB1038, R&D Systems) to positively select for Stro1+/CD44+ Myo and F stem cells within the first population of CD44+ isolated cells, as Stro1-positive cells are the limiting factor for isolating dual-positive cells. Myo and F stem cells were cultured separately in Dulbecco Modified Eagle Medium: Nutrient Mixture F-12 (DMEM/F12, Sigma, St Louis, Missouri) with 12% fetal bovine serum (FBS, Omega Scientific) and 1% antibiotic–antimycotic (Life Technologies), plated in collagen-coated dishes, and incubated under normal Myo and F stem cell conditions at 37°C, 2% O2, and 5% CO2. For each experiment, cells were cultured in 12% FBS-supplemented DMEM/F12 media.
Cell culture of Stro1+/CD44+ stem cells under variable concentrations of serum-supplemented growth media
In order to assess Myo and F stem cell growth in vitro, dose-dependent cell proliferation assays were performed using different dilutions of FBS-supplemented DMEM/F12 growth media. Stro1+/CD44+ Myo and F stem cells were seeded in triplicate at a density of 937 cells/cm2 and grown in DMEM/F12 media supplemented with 0%, 3%, 6%, or 12% (full concentration) FBS under hypoxic conditions (2% O2, 37°C, 5% CO2). Twenty-four, 48, and 72 hours after seeding, respectively, media were removed from the wells, and 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT reagent, 5 mg/mL in phosphate-buffered saline (PBS), Sigma-Aldrich, St. Louis, Missouri) was added and incubated for an additional 4 hours at 37°C. After this period of time, MTT reagent was blocked with 99.9% dimethyl sulfoxide, (Sigma) and incubated for 15 minutes. The intensity of the resulting purple color per well is proportional to the number of viable cells. Optical density (OD) was measured at 570 nm with a reference filter of 620 nm using a Synergy HT Multi-Mode microplate reader (BioTek Instruments, Inc, Winooski, Vermont). Cell proliferation rates were calculated by subtracting the OD of the control wells from the OD of cell-plated wells, and data were represented as the mean of triplicate wells (per cell type and growth environment for each time point) (standard deviation [SD]).
Genomic DNA Extraction
To evaluate whether MED12 mutations were present in F and Myo stem cells and in respective tissues from which they originated, genomic DNA (gDNA) was isolated from each. DNeasy Blood & Tissue Kit (Qiagen) was used to extract gDNA according to the manufacturer’s protocol. Briefly, a 500 000-cell pellet of F and Myo stem cells from each patient was treated with proteinase K to lyse cells. Respective tissues (∼15 mg) were lysed in lysis buffer and proteinase K to begin DNA extraction.
Polymerase Chain Reaction Amplification and Sanger Sequencing
DNA amplification was performed to produce the 291-bp polymerase chain reaction (PCR) product of interest as described previously.6,38 The DNA fragment was amplified using REDTaq ReadyMix PCR Reaction Mix (Sigma) using MED12 gene-specific primers (Integrated DNA Technologies, Coralville, Iowa); primer sequences for amplification of gDNA for MED12 gene: sense 5′-GCCCTTTCACCTTGTTCCTT-3′ and anti-sense 5′-TGTCCCTATAAGTCTTCCCAACC-3.6,38 Using previously published PCR thermocycler conditions,6 gDNA was subjected to amplification, and postamplification PCR products were purified using traditional methods.6 Mixtures were incubated on ice for 20 minutes, then centrifuged at 13 000 rpm for 15 minutes. Supernatant was aspirated, and each sample’s pellet was washed twice in 80% ethanol (EtOH). Each dried pellet was resuspended in nuclease-free ddH2O to original PCR reaction volume and then diluted, and purified products underwent Sanger sequencing analysis as performed by the Genomics & Proteomics Core Laboratory at Augusta University. Bidirectional sequencing was performed, ending with capillary electrophoresis on a 96-capillary ABI 3730xl DNA Analyzer (ThermoFisher Scientific, Columbia, South Carolina), and PCR products were sequenced using BigDye Terminator v3.1 (ThermoFisher Scientific) and original primers specific to MED12 gene exon 2. Mutations in exon 2 of the MED12 gene were then analyzed manually from the resulting sequence graphs.
Alkaline Comet Assay
Trevigen instructions were followed for CometAssay HT: Reagent Kit for Higher Throughput Single Cell Gel Electrophoresis Assay. Briefly, aliquots of F and Myo stem cells were prepared from n = 5 patients and placed on ice. Cells were washed in PBS, pelleted, and resuspended in cold PBS at ∼1.5 × 105 cell/mL before adding cells to melted LMAgarose (37°C), which was spread in wells on 20-well slides. Samples and controls were run in triplicate. On slides, cells were lysed, DNA unwound and electrophoresed for 30 minutes at 21 V. Slides were then washed in ddH2O and 70% EtOH and dried. SybrGold was added to each well to visualize cells, and cells were imaged using the Loats Automated Comet Assay Scoring System.
DNA Damage PrimePCR Array
Total RNA from human F versus adjacent Myo stem cells (n = 3 patient pairs) was isolated by using TRIzol Reagent (Invitrogen, Carlsbad, California) and reverse transcribed into the first-strand complementary DNA (cDNA) using SuperScript III First-Strand Synthesis System for reverse transcription polymerase chain reaction (Invitrogen, Carlsbad, California). The first strand cDNA synthesis reaction was primed using random hexamers (Invitrogen). cDNA of 11 ng from (n = 3) F and Myo stem cells were loaded per gene into separate DNA Damage Tier 1 R96 plates (BioRad Inc, Hercules, California). Expression levels of 88 total genes involved in DNA damage signaling pathways were detected at the same time with SsoAdvanced Universal SYBR Green Supermix on Bio-Rad CFX96 real-time PCR system. The following thermocycling conditions were used: 1 cycle of 95°C for 10 minutes; 40 cycles of 95°C for 30 seconds, and 60°C for 1 minute. For data analysis, the comparative method (ΔΔCt) was used to calculate relative quantities of nucleic acid sequence through Bio-Rad CFX manager software.
Gene expression analysis by quantitative reverse transcription polymerase chain reaction
To verify the results obtained by the DNA Damage PrimePCR array (Table 2), quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR; Bio-Rad CFX96) was performed in n = 5 patient pairs (to confirm in the original 3 pairs + 3 additional patient pairs) of human F vs Myo stem cells for 5 and 22 genes shown to be consistently up- and downregulated, respectively, among the initial n = 3 patient pairs of F versus Myo stem cells. The messenger RNA (mRNA) levels of these 27 genes were quantified using gene-specific primers (Supplemental Table S3): Upregulated: Downregulated. The mRNA values for each gene were normalized to internal control 18 S ribosomal RNA (rRNA), and the comparative method (ΔΔCt) was used to calculate relative quantities of rRNA and reflected in the log2 fold-change gene expression in F stem cells through Bio-Rad CFX Manager (TM) Software version 3.0. All experiments were performed in triplicates.
Table 2.
Genes Shown by PrimePCR to be Consistently up- or Downregulated in n = 3 Patients.a
| Gene Product (Gene) | Gene Product GO Class (Direct) | Up/Downregulated | Log2 Fold Change ± Standard Deviation (SD) |
|---|---|---|---|
| Cyclin-dependent kinase inhibitor 1A (CDKN1A) | Regulation of cyclin-dependent protein serine/threonine kinase activity, G1/S transition of mitotic cell cycle | Up | 1.12 (1.11) |
| Retinoblastoma-associated protein (RB1) | Cell cycle checkpoint; G1/S transition of mitotic cell cycle | Up | 0.84 (0.21) |
| Ataxia Telangiectasia and Rad3-Related Protein (ATR) | DNA damage checkpoint; protein serine/threonine kinase activity | Up | 0.43 (0.36) |
| E3 ubiquitin-protein ligase Mdm2 (MDM2) | negative regulation of transcription from RNA polymerase II promoter | Up | 0.35 (0.36) |
| X-ray repair cross complementing 4 (XRCC4) | DSB repair via NHEJ | Up | 0.21 (0.16) |
| Cellular tumor antigen p53 (TP53) | Negative regulation of transcription from RNA polymerase II promoter | Down | −2.47 (1.63) |
| Breast cancer 2 (BRCA2) | Double-strand break repair via homologous recombination | Down | −1.71 (0.40) |
| BRCA1-associated RING domain 1 (BARD1) | Regulation of phosphorylation, DNA repair | Down | −1.00 (0.73) |
| TP53-binding protein 1 (TP53BP1) | DNA damage checkpoint | Down | −0.99 (0.88) |
| X-ray repair cross complementing 6 (XRCC6) | DSB repair via NHEJ; telomere maintenance | Down | −0.91 (0.48) |
| Breast cancer 1 (BRCA1) | Double-strand break repair via homologous recombination | Down | −0.89 (0.59) |
| Serine/threonine-protein kinase Chk2 (CHEK2) | DNA damage checkpoint | Down | −0.87 (0.31) |
| Poly [ADP-ribose] polymerase 1 (PARP1) | negative regulation of transcription from RNA polymerase II promoter; nucleotide-excision repair, DNA damage recognition | Down | −0.85 (0.34) |
| Replication protein A 14 kDa subunit (RPA3) | G1/S transition of mitotic cell cycle | Down | −0.82 (0.19) |
| Cell cycle checkpoint protein RAD9, alpha (RAD9A) | DNA replication checkpoint; DNA damage checkpoint | Down | −0.81 (0.10) |
| Serine/threonine-protein kinase Chk1 (CHEK1) | DNA damage checkpoint | Down | −0.79 (0.46) |
| RAD52 homolog (RAD52) | Double-strand break repair via homologous recombination | Down | −0.71 (0.25) |
| Replication factor C subunit 4 (RFC4) | Telomere maintenance via recombination; complexes with PCNA, RAD17 | Down | −0.70 (0.07) |
| DNA-directed RNA polymerase II subunit RPB2 (POLR2B) | mRNA splicing, via spliceosome | Down | −0.60 (0.11) |
| MRE11 Homolog, Double Strand Break Repair Nuclease (MRE11A) | Double-strand break repair via homologous recombination | Down | −0.57 (0.27) |
| Mediator of DNA damage checkpoint protein 1 (MDC1) | Protein binding | Down | −0.49 (0.05) |
| DNA damage binding protein 1 (DDB1) | Nucleotide-excision repair, DNA damage recognition | Down | −0.48 (0.20) |
| Ataxia Telangiectasia-Mutated (ATM) | Double-strand break repair via homologous recombination | Down | −0.45 (0.32) |
| Proliferating cell nuclear antigen (PCNA) | Regulation of transcription involved in G1/S transition of mitotic cell cycle; telomere maintenance via recombination | Down | −0.45 (0.26) |
| Nijmegen Breakage Syndrome 1/Nibrin (NBS1/NBN) | Double-strand break repair via homologous recombination | Down | −0.43 (0.29) |
| DNA repair protein RAD50 (RAD50) | Double-strand break repair via homologous recombination | Down | −0.24 (0.29) |
| Small ubiquitin-related modifier 1 (SUMO1) | Negative regulation of transcription from RNA polymerase II promoter; double-strand break repair via nonhomologous end joining | Down | −0.20 (0.09) |
| Cell cycle checkpoint protein RAD1 (RAD1) | DNA damage checkpoint | NA | NA |
| Cell cycle checkpoint protein RAD17 (RAD17) | DNA replication checkpoint; DNA damage checkpoint | NA | NA |
| E3 ubiquitin-protein ligase RAD18 (RAD18) | Cellular response to DNA damage stimulus | NA | NA |
| DNA repair protein RAD51 (RAD51) | Double-strand break repair via homologous recombination | NA | NA |
| RAD51 associated protein 1 (RAD51AP1) | Double-strand break repair via homologous recombination | NA | NA |
| X-ray repair cross complementing 2 (XRCC2) | Recombinase activity; DSB repair via HR | NA | NA |
Abbreviations: HR, homologous recombination; NHEJ, nonhomologous end-joining; NA, not applicable.
a Used for Gene Expression Validation in n = 5 Human Patients (NA: Not Tested by PrimePCR - Added to qRT-PCR Analysis Due to Function/Association with Other Genes).
Drug Preparation
Bleomycin (Sigma) stock solution was prepared in PBS and then diluted in culturing medium to 15μg/mL in preparation for each experimental treatment.
Bleomycin induction of DNA DSBs in Myo and F stem cells for phosphorylation analysis by Western blot
Myo and F stem cells were cultured to confluency in collagen-coated dishes and then treated with 0 and 15 μg/mL bleomycin for 1 hour.39 Following 1 hour of treatment, bleomycin was removed, and cells were either collected immediately by trypsinization or maintained in normal growth media to recover until the following time points: 1, 6, and 24 hours, at which points media was removed, and cells were trypsinized and collected in the same manner, washed in PBS, and centrifuged at 1000 rpm for 5 minutes. Phosphate-buffered saline supernatant was removed, and pellets were snap-frozen in liquid nitrogen for storage at −80°C until prepared for protein isolation and Western blot analysis.
Protein Expression Analysis by Western Blot
Myo and F stem cell pellets (prepared as described above) were prepared for Western blot analysis as described previously.40 Briefly, cells were lysed and centrifuged. Samples equivalent to 30 μg of protein were separated using 4% to 20% MiniPROTEAN TGX Precast Protein Gels (Bio-Rad, Hercules, California) and transferred to polyvinyl difluoride membranes according to standard procedures. Equal loading and transfer were assessed by reprobing the blots with anti-β-actin antibody (Supplemental Table S2: A5441, Sigma). Membranes were blocked for 1 hour at room temperature in either 5% wt/vol nonfat dry milk or 5% bovine serum albumin (BSA) in 0.1% Tween-supplemented PBS (0.1% PBS-T) per antibody specification. Membranes were then incubated with primary antibodies (Supplemental Table S2): rabbit anti-phospho-ATM (Ser1981), rabbit anti-ATM, rabbit anti-phospho-ATR (Thr1989), rabbit anti-ATR, rabbit anti-phospho-CHK1 (Ser345), mouse anti-CHK1, rabbit anti-phospho-CHK2 (Thr68), mouse anti-CHK2, rabbit anti-phospho-MRE11 (Ser676), rabbit anti-MRE11, rabbit anti-phospho-p95/NBS1 (Ser343), rabbit anti-p95/NBS1, and rabbit anti-RAD50 (all antibodies: Cell Signaling, Danvers, Massachusetts; 1:1000 dilution) overnight at 4°C in either 1% wt/vol nonfat dry milk or 1% BSA in 0.1% PBS-Tween (PBS-T) per antibody specification. Membranes were washed and then incubated with either antirabbit or antimouse **horseradish peroxidase-labeled secondary antibody (1:5000 dilution in 1% wt/vol nonfat dry milk in 0.1% PBS-T; Cell Signaling; Supplemental Table S2). β-actin was used as an internal standard to account for variations in the amount of protein (usually 30 μg) loaded in each lane. Protein bands were detected using an enhanced chemiluminescence system, ChemiDoc XRS+ System (Bio-Rad, Hercules, California, USA). Levels of protein expression were quantified using the Bio-Rad Image Lab (TM) Software 6.0 (Bio-Rad) and normalized to the β-actin levels.
Phospho-H2AX (γ-H2AX) and RAD50 foci induction by bleomycin treatment
Myo and F cells were plated in 6-well plates (50 000 cells/well) and cultured to confluency. As described previously, cells were treated with 0 and 15 μg/mL bleomycin for 1 hour.39 Then, bleomycin was removed, and cells were either fixed immediately with 4% paraformaldehyde (PFA) for 15 minutes at room temperature or maintained in normal growth media to recover until the following time points: 0, 3, 6, and 24 hours, at which points media was removed and cells were fixed in the same manner (4% PFA, 15 minutes, 25°C), washed in PBS, and stored at 4°C until prepared for immunofluorescence staining.
γ-H2AX/RAD50 staining for immunocytochemistry/immunofluorescence
In preparation for staining, cells were permeabilized for 30 minutes with 0.1% Triton X-100 in PBS (0.1% PBS-T). After washing, cells were then blocked for 30 minutes with 1% BSA in 0.1% PBS-T blocking buffer. Staining with the rabbit anti-phospho-histone H2A.X (Ser139; 2577; Cell Signaling) and mouse anti-RAD50 (MA1-23269; ThermoFisher Scientific) antibodies was performed for 1 hour (at a 1:250 and 1:200 dilution, respectively) in blocking buffer. Samples were washed 3 times with PBS and then stained with Goat anti-Rabbit IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 555 (A-11001; ThermoFisher Scientific) and Goat anti-Mouse IgG (H+L) Secondary Antibody, Alexa Fluor 488 (A-11001, ThermoFisher Scientific) for 1 hour in the dark at a 1:1000 dilution in blocking buffer. Samples were again washed 3 times with PBS and stained with 4′,6-diamidino-2-phenylindole (DAPI; Sigma) for nuclear counterstaining. Cells were washed 3 times again, and coverslips were fixed on slides for imaging using Fluoroshield mounting medium (Sigma) and dried overnight in the dark.
Confocal fluorescence imaging of γ-H2AX foci
As previously described,39 for cell quantification and the study of γ-H2AX and RAD50 foci formation in Myo versus F stem cells, confocal images were captured using a Plan apochromat 40×/1.4 oil DIC M27 objective lens (Zeiss 780 Upright Confocal, 1024 x 1024 pixels; Zeiss (Thornwood, NY, USA)) in a blinded manner of 10 representative fields for each cell, treatment, and recovery time point. For each field, 3 images were taken: (1) a DAPI image (405 nm laser, 638 master gain, 28 µm pinhole), (2) a γ-H2AX image (543 nm laser, 765 master gain, 37 μm pinhole), and (3) an RAD50 image (488 nm laser, 684 master gain, 32 μm pinhole). The pictures were saved as 16-bit Zeiss Vision Image ZVI files with no further editing.
Quantification of γ–H2AX foci fluorescence density and RAD50 colocalization
A common method of quantifying γ-H2AX is to count individual foci, either manually or by an automated method.41–43 Thus, with inForm Cell Analysis v.2.3 software (PerkinElmer), individual foci were quantified in cell nuclei, and the standardized threshold for particle detection and quantification was used in a “batch” application for all already acquired images. In addition, quantifying the integrated fluorescence intensity of individual cells has also been used, as it accurately measures γ-H2AX expression when considering the 3-dimensional nature of the nucleus and the inability to visualize discrete foci in a highly damaged cell.44 Thus, using the ImageJ v.1.48 k software (NIH, Bethesda, Maryland) the integrated density (IntDen) was calculated in all images for each nucleus to determine the relative fluorescence density and intensity, corresponding to nuclear γ-H2AX and/or RAD50 expression, after subtracting the background signal of each nucleus. The IntDen is a calculus of the mean stained area times the intensity of stain in each pixel in the area and indicates the total amount of stained material in that area. For both quantification methods, at least 100 cells for each cell and treatment type per patient were analyzed.
Statistical Analyses
For confirmation of PrimePCR data, qRT-PCR data were analyzed using unpaired, one-tailed Student t test (since PrimePCR data provided information on expression directionality) for comparative parametric analysis with a significance level of P value <.05 considered statistically significant. Experiments were performed in triplicate for n = 5 patients, and gene expression results depicted as log2 fold change of F versus Myo stem cells ± standard error of the mean (SEM). Western blot data were analyzed at each untreated or treatment time point by comparing the F:Myo ratio to 1 using a one-sample t test. Experiments were performed in triplicate for each respective F and Myo stem cell pair and results expressed as mean F:Myo ± SEM. Alkaline comet assay data (n = 5 patients) were analyzed for each patient (#1-5) after combining experimental replicate data, using unpaired, two-tailed Student t test and expressed as mean ± SEM. To analyze γ-H2AX and RAD50:γ-H2AX quantification data, nonparametric one-way analysis of variance (ANOVA) Kruskal-Wallis test was utilized for all treatment and recovery time points of Myo and F groups (from n = 5 patients). Nonparametric one-way ANOVA justified post hoc comparisons between group medians, which were conducted using Dunn multiple comparisons test. Data are depicted as median F versus Myo and F:Myo expression, respectively, interquartile range (IQR). Differences were considered significant at P < .05. To analyze RAD50 immunocytochemistry/immunofluorescence (ICC/IF), data parametric one-way ANOVA was utilized followed by Tukey Multiple Comparisons test. Data are depicted as mean F and Myo expression. All of these analyses were completed using GraphPad Prism 7.03.
Results
No Significant Difference in Rates of Proliferation Exists Between Human F Stem Cells and Adjacent Myo Stem Cells
To mitigate any effects that proliferation may have on the DDR and expression of DNA repair-related genes and proteins, the rates of proliferation of both F and Myo stem cells were measured to determine whether FBS supplementation was required to normalize their growth rates prior to completing downstream experimental assays. By MTT assay, the rate of proliferation of F stem cells did not differ significantly versus that of Myo stem cells after 24, 48, and 72 hours when DMEM/F12 media was supplemented with 0%, 3%, or 6%, or 12% FBS (Supplemental Figure S1). Because we have observed previously that our cells grow most optimally in 12% FBS-supplemented DMEM/F12 media and, thus, have completed previous experiments using this condition, this concentration of FBS supplementation was used for all in vitro experiments.
Fibroid Tissues, but not Adjacent Normal Myometrium, From Which Stem Cells are Isolated Contain Point Mutations in MED12
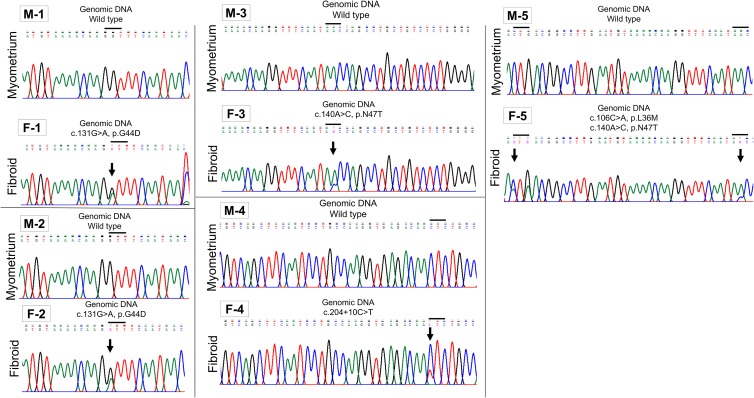
By Sanger sequencing and manual examination of chromatograms, it was determined that 5 of 5 human fibroid tissue samples from which F stem cells were isolated contained mutation(s) in the MED12 gene, while the adjacent normal myometrium did not (Figure 1). One fibroid sample (F-1) had the c.131G>A mutation; another (F-2) also had the c. 131G>A mutation (Figure 1). The third sample (F-3) had a c.140A>C; the fourth (F-4), a c.204+10C>T mutation; and the final sample (F-5) had 2 mutations: c.106C>A and c.140A>C (Figure 1). MED12 mutations were detected in only 1 set of matched F stem cells, those from tissue sample F-5 (c.106C>A, Patient #5) and not in F stem cells from patients #1 to 4 F tissues. Importantly, however, it has been demonstrated previously that cells isolated from MED12-mutated UFs may originally harbor MED12 mutations but can be selected out and lost with passaging in in vitro cultures. This limits the numbers of MED12-mutated tumor cells even after only limited (ie, 3 passages) culturing of cells in vitro.45 Thus, because these F stem cells were isolated from tissues positive for MED12 mutations, the parental F stem cells may have originally harbored mutations in MED12.
Figure 1.
MED12 mutations are detected in fibroid tumor tissue, but not adjacent normal myometrium. Sanger sequencing detected mutations in 5 of the 5 fibroid tumors (F-1, F-2, F-3, F-4, and F-5) from which stem cells were derived. No mutations were detected in any of the 5 matched adjacent normal myometrial tissues (M-1 through M-5). Arrows denote mutated residue(s), and MED12 point mutations are designated as residue (c.), nucleotide substitution (eg, C>T), and resulting (if any) amino acid (p.) substitution.
Human F Stem Cells Demonstrate Increased DNA Damage when Compared to Myo Stem Cells Isolated From Adjacent Normal Myometrium
To determine whether baseline DNA damage differs between F and Myo stem cells, we utilized the alkaline comet assay to compare respective percentage DNA in tail and tail moments, measures of DNA damage. Trevigen Alkaline CometAssay (Trevigen, Gaithersburg, MD, USA) is a single-cell gel electrophoresis assay for evaluating DNA damage in cells.46 Briefly, denatured, cleaved DNA fragments will migrate into the “tail” of a cell “comet” under the influence of an electric field, while undamaged DNA will not migrate as quickly, remaining confined primarily to the nucleoid “head” of the cell “comet.”46,47
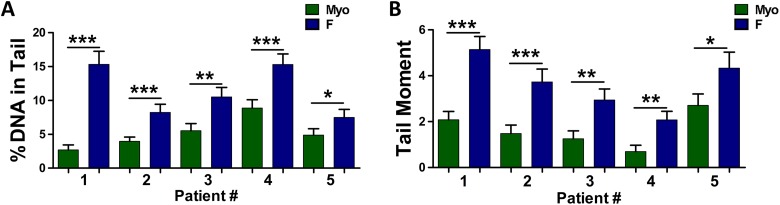
By alkaline comet assay it was seen that DNA damage was significantly increased in F versus Myo stem cells, as reflected by increased mean percentage DNA in tail (standard error of the mean [SEM]) (Figure 2A and Table 1) and increased tail moment (Figure 2B and Table 1) in F stem cells in all 5 patients versus Myo stem cells. Means of combined experimental replicates from F versus Myo stem cells were compared using Student unpaired t test. Because DNA repair activity or potential may be correlated with increased susceptibility of tumorigenic cell transformation, it was imperative to determine whether this baseline difference in DNA damage, specifically DNA DSBs, exists between F and Myo stem cells. Although differential expression of genes and corresponding proteins may highlight differences in regulation of DNA repair, an assay like the alkaline comet assay can inform on DNA repair activity in F and Myo stem cells.
Figure 2.
DNA damage is increased in fibroid (F) stem cells when compared to adjacent myometrial (Myo) stem cells. A, Alkaline comet assay detected significantly higher levels of DNA damage, as reflected by increased % DNA in tail, in F stem cells from n = 5 patients. Lines represent mean % DNA in tail ± standard error of the mean in each patient #1-5, ***P < .0001, **P < .001, *P < .05. B, Alkaline comet assay detected significantly higher levels of DNA damage, as reflected by increased tail moment, in F stem cells from n = 5 patients. Lines represent mean tail moment ± standard error of the mean in each patient, ***P < .0001, **P < .001, *P < .05. Means of combined technical replicates were compared using Student unpaired t test.
Table 1.
By Alkaline Comet Assay, Mean % DNA in Tail and Tail Moment was Increased in each Respective set of F Stem Cells Versus Adjacent Myo Stem Cells in Patients #1-5.
| Mean % DNA in Tail ± SEM | Mean Tail Moment ± SEM | |||||
|---|---|---|---|---|---|---|
| Patient # | F Stem Cells | Myo Stem Cells | P Value | F Stem Cells | Myo Stem Cells | P Value |
| 1 | 15.33% ± 0.74% | 2.68% ± 0.74% | <.0001 | 5.14 ± 0.57 | 2.08 ± 0.36 | <.0001 |
| 2 | 8.22% ± 1.21% | 3.96% ± 0.62% | <.0001 | 3.73 ± 0.56 | 1.48 ± 0.37 | <.0001 |
| 3 | 10.51% ± 1.41% | 5.52% ± 1.06% | <.001 | 2.94 ± 0.47 | 1.25 ± 0.34 | <.001 |
| 4 | 15.30% ± 1.57% | 8.87% ± 1.23% | <.0001 | 2.07 ± 0.38 | 0.69 ± 0.27 | <.001 |
| 5 | 7.49% ± 1.19% | 4.87% ± 0.95% | <.05 | 4.33 ± 0.69 | 2.71 ± 0.50 | <.05 |
Abbreviation: SEM, standard error of the mean.
Fibroid Stem Cells Isolated from Human UFs Demonstrate Overall Decreased Expression of DNA Repair-Related Genes
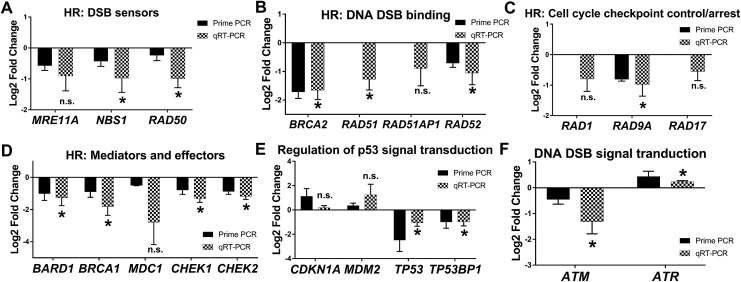
We utilized PrimePCR DNA Damage gene expression arrays to screen 3 pairs of F and adjacent Myo stem cells for differentially expressed DNA repair genes. Analysis of the PrimePCR data suggested that 2 important populations of DNA repair-related genes were found to be up- or downregulated in F stem cells when compared to Myo stem cells among the 88 DNA repair-related genes available in the array (Table 2). Genes consistently up- (n = 5) or down(n = 22) regulated in F stem cells among the 3 patients were selected for further confirmation by qRT-PCR (Table 2). Of those differentially expressed genes, only 1 demonstrated significantly upregulated expression, while more than half of the downregulated genes demonstrated significantly decreased expression in F versus Myo stem cells (Table 2; Figures 3 –5). Six additional genes not tested within the PrimePCR array (Tables 2 and 3) were selected for gene expression analysis by qRT-PCR due to their close involvement in DNA repair with several genes that were significantly decreased in F stem cells.
Figure 3.
Fibroid (F) stem cells differentially express homologous recombination (HR) DNA repair-related genes versus adjacent myometrial (Myo) stem cells. PrimePCR and qRT-PCR validation of genes relating to DNA double-strand break (DSB) repair, specifically HR, (FC, P value). A, DSB sensors: MRE11A (−0.91, P = .06), NBS1 (−0.97, P = .04), and RAD50 (−1.3, P = .01). B, HR DSB binding: BRCA2 (−1.7, P = .004), RAD51 (−1.3; P = .01), RAD51AP1 (−0.9, P = .12), and RAD52 (−1.1; P = .03). C, Cell cycle checkpoint control/arrest: RAD1 (−0.8, P = .06), RAD9A (−0.98; P = .03), and RAD17 (−0.55; P = .0.6). D, HR mediators and effectors: BARD1 (−1.3; P = .03), BRCA1 (−1.8; P = .01), MDC1 (−2.8; P = .06), CHEK1 (−1.2, P = .001), and CHEK2 (−0.9; P = .02). E, p53 signal transduction: CDKN1A (0.18, P = .19), MDM2 (1.2, P = .10), TP53 (−1.1, P = .009), and TP53BP1 (−0.96, P = .03). F, DNA DSB signal transduction: ataxia telangiectasia-mutated (ATM; −1, P = .04) and Ataxia Telangiectasia and Rad3-related protein (ATR;0.23, P = .002). Bars represent log2 fold change expression from PrimePCR (black bars) or quantitative reverse transcription polymerase chain reaction (qRT-PCR) validation (checkered bars) in F (relative to adjacent Myo control stem cells) ± standard error of the mean. Mean log2 fold change was compared to 0 using one-tailed, one-sample t test. *P < .05, n.s. indicates not significant. Genes lacking representation by a black bar were not part of the PrimePCR plates. ATM, Ataxia Telangiectasia-Mutated; BARD1, BRCA1-associated RING domain 1; BRCA1, Breast cancer 1; BRCA2, Breast cancer 2; CHEK1, Serine/threonine-protein kinase Chk1; CHEK2, Serine/threonine-protein kinase Chk2; MDC1, Mediator of DNA damage checkpoint protein 1; MRE11A, MRE11 Homolog, Double Strand Break Repair Nuclease; NBS1/NBN, Nijmegen Breakage Syndrome 1/Nibrin; RAD1, Cell cycle checkpoint protein RAD1; RAD9A, Cell cycle checkpoint protein RAD9, alpha; RAD17, Cell cycle checkpoint protein RAD17; RAD50, DNA repair protein RAD50; RAD51, DNA repair protein RAD51; RAD51AP1, RAD51-associated protein 1; RAD52, RAD52 homolog; TP53, cellular tumor antigen p53; TP53BP1, TP53-binding protein 1; and XRCC2, X-ray repair cross complementing 2.
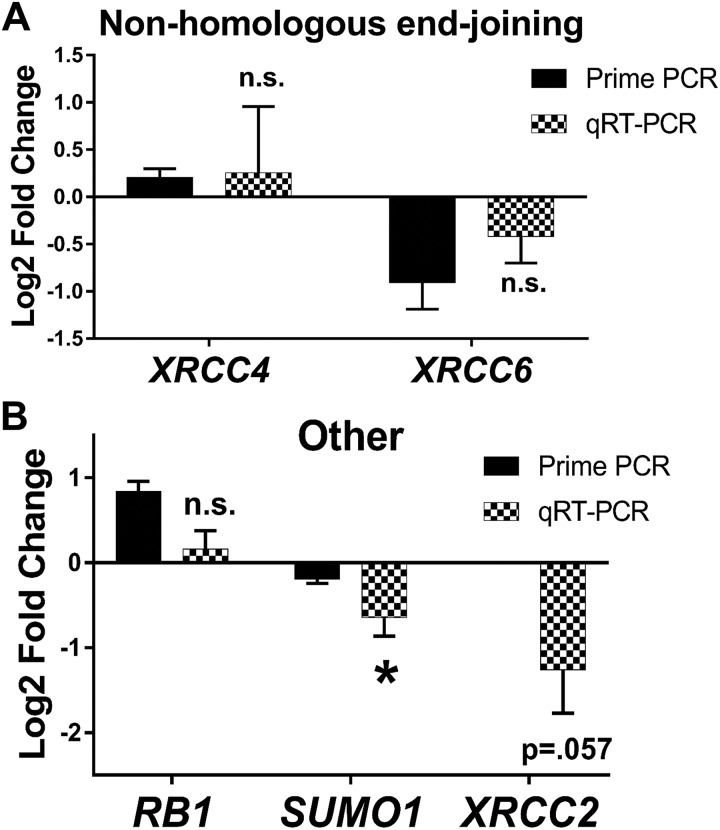
Figure 4.
Fibroid (F) stem cells differentially express DNA double-strand break repair-related genes versus adjacent myometrial (Myo) stem cells. PrimePCR and quantitative reverse transcription polymerase chain reaction (qRT-PCR) validation of genes relating to DNA double-strand break (DSB) repair, specifically nonhomologous end-joining (NHEJ) and other DSB repair–related genes (FC, P value). A, NHEJ: XRCC4 (0.26, P = .40) and XRCC6 (−0.42, P = .10). B, Other DNA DSB repair–related genes: RB1 (0.16, P = .25), SUMO1 (−0.65, P = .02), and XRCC2 (−1.3, P = .03). Bars represent log2 fold change expression from PrimePCR (black bars) or qRT-PCR validation (checkered bars) in F (relative to adjacent Myo control stem cells) ± standard error of the mean. Mean log2 fold change was compared to 0 using one-tailed, 1-sample t test. *P < .05, ns indicates not significant.
Figure 5.
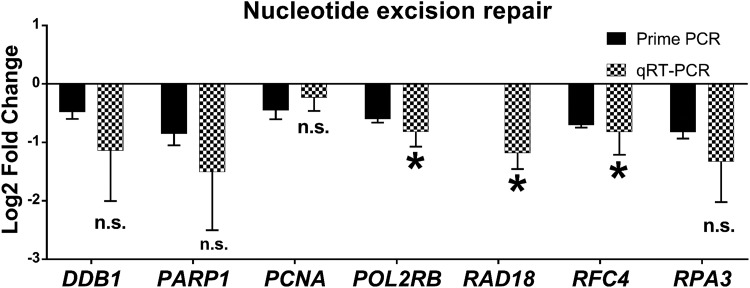
Fibroid (F) stem cells differentially express DNA single-strand break repair–related genes versus adjacent myometrial (Myo) stem cells. PrimePCR and qRT-PCR validation of genes relating to DNA single-strand break (SSB) repair, specifically nucleotide excision repair (FC, P value): DDB1 (−1.1, P = .12), PARP1 (−1.5, P = .10), proliferating cell nuclear antigen PCNA (−0.23, P = .19), POL2RB (−0.82, P = .01), RAD18 (−1.2, P = .004), RFC4 (−0.82, P = .04), RPA3 (−1.3, P = .06). Bars represent log2 fold change expression from PrimePCR (black bars) or qRT-PCR validation (checkered bars) in F (relative to adjacent Myo control stem cells) ± standard error of the mean. Mean log2 fold change was compared to 0 using 1-tailed, 1-sample t test. *P < .05, ns indicate not significant. Genes lacking representation by a black bar were not part of the PrimePCR plates.
Table 3.
Gene Expression (log 2 fold change) in F Stem Cells as Confirmed by qRT-PCR in n = 5 Patients.
| Gene Product (Gene) | Biological Process | Log2 Fold Change ± SEM | P Value | Significanta |
|---|---|---|---|---|
| MRE11 Homolog, Double Strand Break Repair Nuclease (MRE11A) | Homologous recombination (HR): sensors | −0.91 ± 0.48 | .06 | |
| Nijmegen Breakage Syndrome 1/Nibrin (NBS1/NBN) | HR: sensors | −0.97 ± 0.46 | .04 | a |
| DNA repair protein RAD50 (RAD50) | HR: sensors | −0.99 ± 0.29 | .01 | a |
| Breast cancer 2 (BRCA2) | HR: DNA double-strand break (DSB) binding | −1.70 ± 0.33 | .004 | a |
| DNA repair protein RAD51 (RAD51) | HR: DNA DSB binding | −1.30 ± 0.37 | .01 | a |
| RAD51-associated protein 1 (RAD51AP1) | HR: DNA DSB binding | −0.90 ± 0.61 | .12 | |
| RAD52 homolog (RAD52) | HR: DNA DSB binding | −1.10 ± 0.41 | .03 | a |
| BRCA1-associated RING domain 1 (BARD1) | HR: mediators and effectors | −1.30 ± 0.49 | .03 | a |
| Breast cancer 1 (BRCA1) | HR: mediators and effectors | −1.80 ± 0.53 | .01 | a |
| Mediator of DNA damage checkpoint protein 1 (MDC1) | HR: mediators and effectors | −2.80 ± 1.40 | .06 | |
| Serine/threonine-protein kinase Chk1 (CHEK1) | HR: mediators and effectors | −1.20 ± 0.21 | .001 | a |
| Serine/threonine-protein kinase Chk2 (CHEK2) | HR: mediators and effectors | −0.90 ± 0.32 | .02 | a |
| Cell cycle checkpoint protein RAD1 (RAD1) | HR: cell cycle checkpoint control/arrest | −0.80 ± 0.40 | .06 | |
| Cell cycle checkpoint protein RAD9, alpha (RAD9A) | HR: cell cycle checkpoint control/arrest | −0.98 ± 0.38 | .03 | a |
| Cell cycle checkpoint protein RAD17 (RAD17) | HR: cell cycle checkpoint control/arrest | −0.55 ± 0.30 | .06 | |
| Ataxia Telangiectasia-mutated (ATM) | DNA DSB signal transduction | −1.00 ± 0.48 | .04 | a |
| Ataxia Telangiectasia and Rad3-Related Protein (ATR) | DNA DSB signal transduction | 0.23 ± 0.04 | .002 | a |
| Cyclin-dependent kinase inhibitor 1A (CDKN1A) | Regulation of p53 signal transduction | 0.18 ± 0.18 | .19 | |
| E3 ubiquitin-protein ligase Mdm2 (MDM2) | Regulation of p53 signal transduction | 1.20 ± 0.86 | .10 | |
| Cellular tumor antigen p53 (TP53) | Regulation of p53 signal transduction | −1.10 ± 0.27 | .009 | a |
| TP53-binding protein 1 (TP53BP1) | Regulation of p53 signal transduction | −0.96 ± 0.36 | .03 | a |
| X-ray repair cross complementing 4 (XRCC4) | Nonhomologous end-joining (NHEJ) | 0.26 ± 0.70 | .40 | |
| X-ray repair cross complementing 6 (XRCC6) | Nonhomologous end-joining (NHEJ) | −0.42 ± 0.28 | .10 | |
| Retinoblastoma-associated protein (RB1) | Other DNA repair-related processes | 0.16 ± 0.22 | .25 | |
| Small ubiquitin-related modifier 1 (SUMO1) | Other DNA repair-related processes | −0.65 ± 0.22 | .02 | a |
| X-ray repair cross complementing 2 (XRCC2) | Other DNA repair-related processes | −1.30 ± 0.51 | .03 | a |
| DNA damage binding protein 1 (DDB1) | Nucleotide excision repair (NER) | −1.10 ± 0.87 | .12 | |
| Poly [ADP-ribose] polymerase 1 (PARP1) | NER | −1.50 ± 1.00 | .10 | |
| Proliferating cell nuclear antigen (PCNA) | NER | −0.23 ± 0.23 | .19 | |
| DNA-directed RNA polymerase II subunit RPB2 (POLR2B) | NER | −0.82 ± 0.26 | .01 | a |
| E3 ubiquitin-protein ligase RAD18 (RAD18) | NER | −1.20 ± 0.27 | .004 | a |
| Replication factor C subunit 4 (RFC4) | NER | −0.82 ± 0.39 | .04 | a |
| Replication protein A 14 kDa subunit (RPA3) | NER | −1.30 ± 0.70 | .06 |
a p < 0.05.
Genes and their respective fold-change values confirmed by qRT-PCR for the n = 5 patients were categorized by biological processes as shown in Table 3: HR–sensors; HR–DNA DSB binding; HR–mediators and effectors; HR–cell cycle checkpoint control/arrest; DNA DSB signal transduction; and regulation of p53 signal transduction (Figure 3); genes relating to NHEJ and other DNA repair-related genes (Figure 4). Also shown in Table 3 are those genes related to DNA SSB repair, specifically nucleotide excision repair (NER; Figure 5).
Overall, F stem cells showed decreased expression of DNA DSB repair-related genes, specifically those involved in HR DDR and signal transduction: Ataxia Telangiectasia-Mutated (ATM), BRCA1-associated RING domain 1 (BARD1), BRCA1, Breast cancer 2 (BRCA2), Serine/threonine-protein kinase Chk1 (CHEK1), Serine/threonine-protein kinase Chk2 (CHEK2), Mediator of DNA damage checkpoint protein 1 (MDC1), MRE11 Homolog, Double Strand Break Repair Nuclease (MRE11A), Nijmegen Breakage Syndrome 1/Nibrin (NBS1/NBN), Cell cycle checkpoint protein RAD1, Cell cycle checkpoint protein RAD9, alpha, Cell cycle checkpoint protein RAD17, RAD50, DNA repair protein RAD51 (RAD51), RAD51-associated protein 1 (RAD51AP1), RAD52 homolog (RAD52), cellular tumor antigen p53 (TP53), TP53-binding protein 1, and X-ray repair cross complementing 2 (Figure 3).
Fibroid Stem Cells Show Decreased Expression of Total DNA Repair-Related Proteins and Differential Phosphorylation Versus Adjacent Myo Stem Cells, Suggesting an Altered DDR
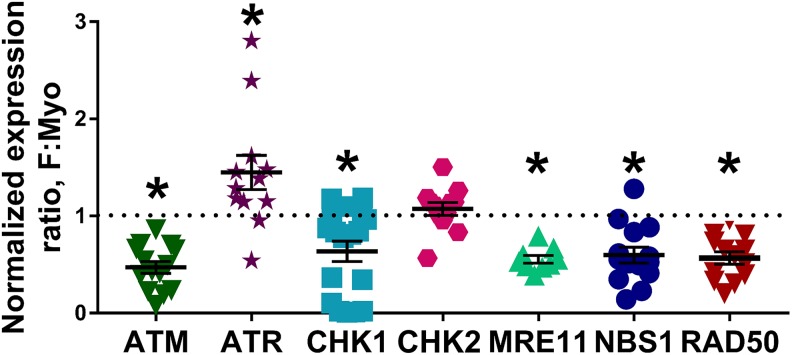
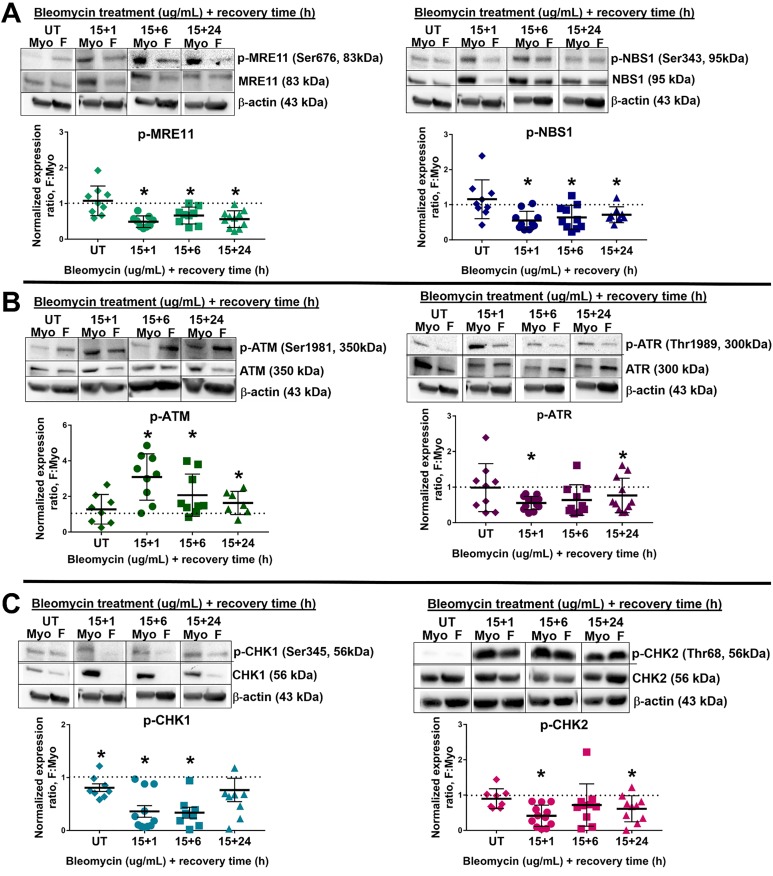
Because a notable number of genes related to DNA DSB repair, specifically HR, were downregulated in F stem cells, several proteins relating to HR were analyzed to determine how F stem cell DDR differed from Myo stem cells: phosphorylated-ATM (p-ATM) and ATM, phosphorylated-Ataxia Telangiectasia and Rad3-related protein (p-ATR, Ser1989) and ATR, phosphorylated-MRE11 Homolog, Double Strand Break Repair Nuclease (p-MRE11, Ser676) and MRE11, phosphorylated-Nijmegen Breakage Syndrome 1 (p-NBS1, Ser343) and NBS1, RAD50 Homolog, Double Strand Break Repair Protein (RAD50), phosphorylated-Checkpoint Kinase 1 (p-CHK1, Ser345) and CHK1, phosphorylated-Checkpoint Kinase 2 (p-CHK2, Thr68), and CHK2. Notably, in untreated cells, several DNA repair proteins were decreased in F stem cells (mean F: Myo stem cell expression ratio [standard error of the mean], P value; a ratio less than 1 indicates a decrease in protein expression in F): total ATM (0.47 ± 0.06; P < .0001), total CHK1 (0.64 ± 0.11; P = .002), total MRE11 (0.55 ± 0.04; P < .0001), total NBS1 (0.60 ± 0.08; P = .0002), and total RAD50 (0.57 ± 0.06; P < .0001), with a significantly increased mean expression ratio of total ATR (1.45 ± 0.18; P = .03), and no difference in total CHK2 expression (1.07 ± 0.07; P = .29), suggesting F stem cells’ ability to initiate and propagate the DDR may be disrupted (Figures 6 and 7). Of note, increased total ATR expression may be attributed to a compensatory mechanism as a result of markedly decreased expression of total CHK1. Even more interesting, however, were the differences in phosphorylation of ATM, ATR, CHK1, CHK2, MRE11, and NBS1 following DNA DSB induction. To experimentally induce DNA DSBs in Myo and F stem cells to study DNA DSB repair protein activation and repair efficiency, we utilized the radiomimetic, bleomycin, as a tool compound for inducing DNA DSBs. This method and others, including the use of other DNA DSB inducers, such as radiation, hydrogen peroxide (H2O2), or etoposide, are used widely to understand the effects of DNA DSB induction in in vitro cell culture experiments.43,48–52 Using these chemical or radiological inducers of DNA DSBs, we can study how much damage occurs when such an exogenous insult is made in vitro, if/how DNA DSBs are repaired (using γ-H2AX as a marker of DNA DSBs), and if/which proteins that respond to DNA DSB formation are activated and further activate downstream effector proteins.49–51 After challenging the 5 pairs of F and Myo human stem cells with bleomycin (15 µg/mL) for 1 hour, F stem cells demonstrated significantly higher mean p-ATM expression (mean F: Myo expression ratio ± standard error of the mean; P value) after 1 hour (3.09 ± 0.43; P = .0013), 6 hours (2.07 ± 0.40; P = .03), and even 24 hours (1.63 ± 0.25; P = .04) of recovery in normal growth media versus control Myo stem cells; this may indicate the DDR is effectively initiated in F stem cells and is exaggerated, indicating greater DNA damage as well (Figure 8). Interestingly, however, phosphorylation, corresponding to activation of the proteins, of all other 5 DNA repair proteins was significantly (P < .05) decreased in F stem cells following DNA DSB induction after 1, 6, and 24 hours of recovery, respectively: p-ATR: (0.55 ± 0.05; P < .0001), (0.64 ± 0.14; P = .03), and (0.77 ± 0.15; P = .14); p-CHK1: (0.36 ± 0.11; P = .0002), (0.34 ± 0.10; P = .0003), and (0.76 ± 0.22; P = .31); p-CHK2: (0.42 ± 0.09; P < .0001), (0.72 ± 0.19; P = .17), and (0.61 ± 0.12; P = .009); p-MRE11: (0.49 ± 0.05; P < .0001), (0.66 ± 0.08; P = .003), and (0.56 ± 0.07; P = .0001); and p-NBS1: (0.55 ± 0.08; P = .0004), (0.64 ± 0.11; P = .009), and (0.72 ± 0.08; P = .009; Figure 8).
Figure 6.
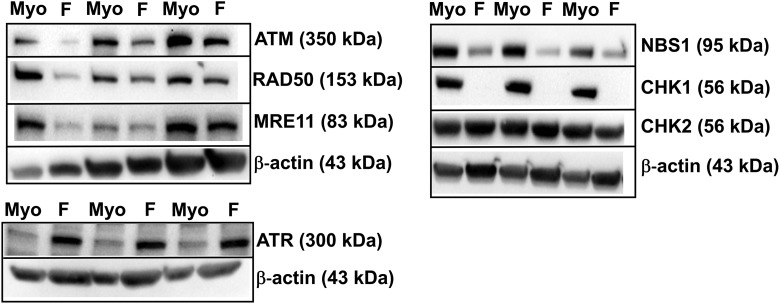
DNA repair-related protein expression, specifically relating to homologous recombination (HR) DNA damage response (DDR) initiation and propagation, is disrupted in fibroid (F) stem cells. By Western blot, in n = 5 patients, baseline expression in untreated F stem cells was decreased in several DNA DSB repair–related proteins (F: Myo expression): ATM, CHK1, MRE11, NBS1, and RAD50. Expression of ATR was increased in F stem cells. Expression of CHK2 was not different in F versus Myo stem cells. Blot images represent 3 different pairs of Myo/F stem cells from 3 of the n = 5 patients. ATM indicates Ataxia Telangiectasia-Mutated; CHK1, checkpoint kinase 1; MRE11A, MRE11 Homolog, Double Strand Break Repair Nuclease; NBS1, Nijmegen Breakage Syndrome 1; RAD50, DNA repair protein RAD50.
Figure 7.
Fibroid (F) stem cells demonstrated significantly decreased expression of proteins related to DNA homologous recombination (HR) repair and the DNA damage response (DDR) relative to adjacent myometrial (Myo) stem cells. By densitometry quantification of the results obtained by Western blot, in n = 5 patients, baseline expression in untreated F stem cells was significantly decreased in several DNA DSB repair–related proteins (mean F: Myo expression, P value): ataxia telangiectasia-mutated (ATM; 0.47, P < .0001), CHK1 (0.64, P = .003), MRE11 (0.55, P < .0001), NBS1 (0.60, P = .0003), and RAD50 (0.57, P < .0001). Expression of ATR was significantly increased in F stem cells (1.45, P = .03). Expression of CHK2 was not significantly different in F versus Myo stem cells (1.07, P = .29). For each DNA DSB repair-related protein, the mean F: Myo expression ratio was compared to 1 using a one-sided, one-sample t test. Lines represent mean F: Myo protein expression ratio (n = 5 patients) ± standard error of the mean. *P < .05, n.s. indicates not significant. ATM indicates Ataxia Telangiectasia-Mutated; MRE11A, MRE11 Homolog, Double Strand Break Repair Nuclease; NBS1, Nijmegen Breakage Syndrome 1; RAD50, DNA repair protein RAD50.
Figure 8.
Altered initiation of DNA damage response (DDR) in fibroid (F) stem cells: decreased DDR signal amplification and transduction following DNA double-strand break (DSB) induction. A, Western blots and corresponding densitometry of sensors of DNA DSBs, p-MRE11 (Ser676), MRE11, p-NBS1 (Ser343), and NBS1: untreated (UT), bleomycin-treated + 1, 6, and 24 hours of recovery in n = 4 patients. B, Western blots and corresponding densitometry of signal-transducing kinases, phosphorylated-Ataxia Telangiectasia Mutated (p-ATM; Ser1981), ATM, p-ATR (Ser1989), ATR: UT, bleomycin-treated + 1, 6, and 24 hours of recovery in n = 4 patients. C, Western blots and corresponding densitometry of effector kinases, p-CHK1 (Ser345), CHK1, p-CHK2 (Thr68), CHK2: UT, bleomycin-treated + 1, 6, and 24 hours of recovery in n = 4 patients. For each time point, the mean expression ratio in F: Myo was compared to 1 using a 2-sided 1-sample t test. Lines represent means ± standard error of the meam. *P < .05. p-CHK1 indicates phosphorylated-Checkpoint Kinase 1; p-MRE11, phosphorylated-MRE11; p-NBS1, phosphorylated-Nijmegen Breakage Syndrome 1; p-ATR, phosphorylated-Ataxia Telangiectasia.
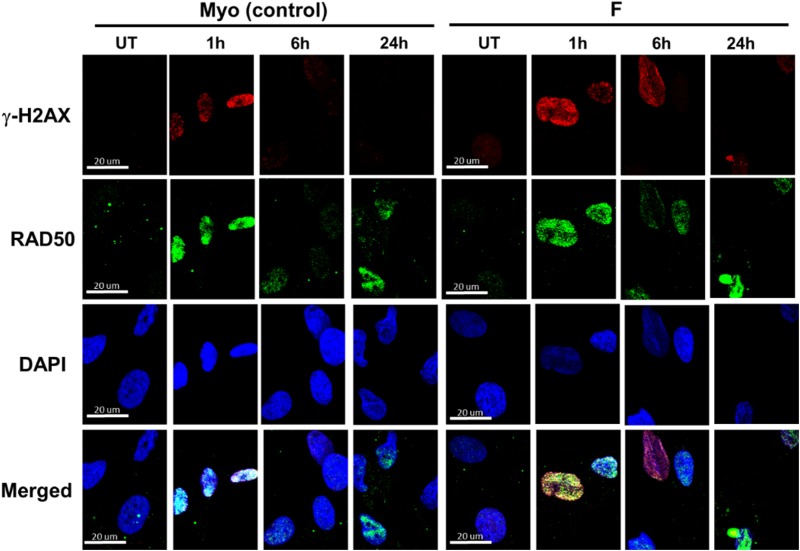
Human F Stem Cells Demonstrate Increased DNA DSBs, Decreased Recruitment of RAD50 DNA Repair Protein, and Decreased DNA DSB Repair Following Induction of DSBs with Bleomycin
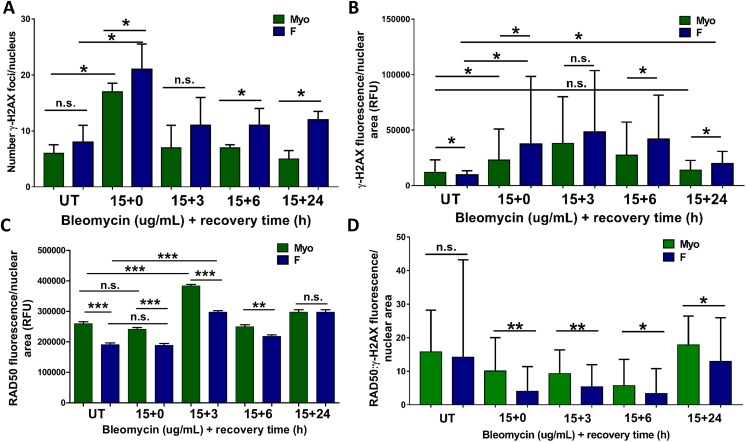
Phosphorylation of histone H2A.X at Serine 139 (a.k.a. γ–H2AX) occurs rapidly in response to DNA DSB formation.43,50,53,54 This phosphorylation is dependent on DNA repair kinases, such as ATM or ATR, or the MRE11-NBS1-RAD50 (MRN) complex, which is also activated early in response to DNA DSBs.51,53,55,56 Thus, utilizing γ-H2AX as a marker of DNA DSBs, we can determine how much DNA DSB damage has occurred in the stem cells and also follow the kinetics of how rapidly/if the DNA damage is repaired and resolved as the γ-H2AX foci disappear. We also examined recruitment of RAD50 to the nucleus following DNA DSB induction as a proxy for MRN complex activation, which is required for downstream activation of DNA DSB repair proteins. Thus, quantification of γ-H2AX foci and RAD50 immunofluorescence (Figures 10 and 11) was completed to determine whether a functional difference exists between F and Myo stem cells’ DNA repair capacity. In untreated F stem cells, although there was a small but significant (P < .0001) lower median fluorescence intensity of γ-H2AX foci per nucleus in F stem cells, 9564 (IQR: 6055-13335) when compared to Myo control cells, 11743 (IQR: 6841-23245) (Figure 11B), the mean (±SEM) number of γ-H2AX foci per nucleus was significantly greater in F (8.8 ± 1.2) versus Myo (6 ± 0.71) stem cells (Figure 11A). Interestingly, both F and Myo stem cells demonstrated increased γ-H2AX foci fluorescence immediately following induction of DSBs with bleomycin (0 hour of recovery in normal growth media), but F stem cells demonstrated higher levels of γ-H2AX foci fluorescence than Myo stem cells, 37529 (IQR: 15542-98329) and 22997 (IQR: 10961-50895), respectively, P < .0001 (Figure 11B). There were also significantly higher numbers of γ-H2AX foci in F (22 ± 2.2) versus Myo (15 ± 2.1) stem cell nuclei, indicating that bleomycin was inducing more DSBs in F stem cells; this shows an increase in γ-H2AX foci formation in both F and Myo cells versus their untreated counterparts as expected (Figure 11A). Interesting, however, is that in F versus Myo stem cell nuclei, neither the median integrated γ-H2AX foci fluorescence (48277 [IQR: 23867-103642] vs. 38048 [IQR: 21951-80044], P > .9999] nor the mean quantities of γ-H2AX foci (12 ± 2.2 vs 8.4 ± 1.1; P = .81), significantly differed from that of Myo stem cell nuclei following 3 hours of recovery (Figure 11B). This suggests that equally large numbers of DNA DSBs were eventually induced in both F and Myo stem cell nuclei. More importantly, even given 24 hours of recovery time in normal growth media, median integrated γ-H2AX foci fluorescence in F stem cell nuclei remained significantly elevated, 19606 (IQR: 12359-30761), when compared to both untreated F stem cells, 9564 (IQR: 6055-13335), P < .0001 and untreated Myo stem cells, 11743 (IQR: 6841-23245), P < .0001 and DNA DSB-induced Myo stem cells 13990 (IQR: 7499-22644), P < .0001 (Figure 11B); of note, no significant difference in median integrated γ-H2AX foci fluorescence existed between Myo stem cells allowed to recover 24 hours post-DNA DSB induction versus untreated Myo stem cells: 13990 (IQR:7499-22644) versus 11743 (IQR: 6841-23245), P > .9999 (Figure 11B). After 24 hours of recovery in normal media, γ-H2AX foci fluorescence in Myo stem cells returned to equal that of untreated Myo stem cells, suggesting effective, complete DNA DSB repair occurs as γ-H2AX foci are eliminated, while after the same 24 hours recovery time period, γ-H2AX foci fluorescence remained significantly elevated in F stem cells, suggesting less efficient or perhaps ineffective DNA DSB repair mechanisms in F stem cells (Figure 11B).
Figure 10.
Increased DNA damage and decreased recruitment of RAD50 and repair of double-strand breaks (DSB) following DSB induction. Immunocytochemistry/immunofluorescence (ICC/IF) demonstrated greater numbers of DNA DSBs (γ-H2AX foci reflect DNA DSBs) and decreased RAD50 nuclear recruitment in F versus Myo stem cells following DNA DSB induction by bleomycin treatment. Scale bars = 20 μm.
Figure 11.
Increased DNA damage, decreased RAD50 nuclear recruitment, and decreased double-strand break (DSB) repair following DSB induction, as measured by densities of γ-H2AX foci and RAD50 fluorescence. A, Numbers of γ-H2AX foci/nucleus prior to and following DSB induction by bleomycin. Mean quantities γ-H2AX foci/nucleus were compared using parametric 1-way analysis of variance (ANOVA) followed by Tukey Multiple Comparisons. Each bar represents mean ± standard error of the mean. *P < .05, n.s. indicate not significant. B, γ-H2AX foci fluorescence density quantified by ImageJ “RawIntDen” measurements. Median integrated fluorescence densities were compared using nonparametric Kruskal Wallis followed by multiple comparisons. Each bar represents median, interquartile range (IQR). *P < .0001, n.s. indicate not significant. C, RAD50 fluorescence density quantified by ImageJ “RawIntDen” measurements. Mean integrated fluorescence densities were compared using parametric one-way ANOVA followed by Tukey Multiple Comparisons. Each bar represents mean ± standard error of the mean (SEM). ***P < .0001, **P < .001, *P < .05, n.s. indicates not significant. D, Ratios of RAD50:γ-H2AX foci fluorescence density per nuclear area quantified by ImageJ “RawIntDen” measurements. Median ratios were compared using nonparametric Kruskal Wallis followed by multiple comparisons. Each bar represents median, interquartile range (IQR). *P < .05, **P < .0001.
Altogether, these data suggest F stem cells have an increased the rate of induction of DNA DSBs when challenged by bleomycin versus healthy control Myo stem cells but a decreased rate of repair of these DNA DSBs, which may ultimately contribute to increased mutation and/or decreased ability to repair probable mutations in DNA or DNA DSBs, leading to further propagation and growth of human UFs.
Discussion
In efforts to advance the development of more specific targets for medicinal and potentially preventative treatments of UFs, research has begun to focus on putative origins of UFs. Beginning with the emergence of MED12 mutations that are thought to contribute to UF-forming cell transformation, and examining more closely the role F stem cells may play in UF development, evidence suggests these cells, although present in quantities less than those found in surrounding myometrium, are essential for steroid hormone-dependent UF growth.8,57–59 It is unclear, however, what mechanisms are involved which cause the dysregulation of said stem cells leading to UF development. Normal tissue-specific stem cells across multiple organ systems are noted repeatedly for their role in regenerating tissue for biostructural and physiological homeostasis.60 The functions of these special cells can be severely disrupted, however, as a result of genome instability.24,29,30,60 DNA damage can alter gene function, leading to aberrant gene expression or inactivation of tumor suppressors, which could override normal proliferation signaling, leading to cancer-like growth.60
Although explicit indications, for example, genetic mutations, of compromised DNA repair may be observed in a variety of cancer types, including mutations in BRCA1 and BRCA2 of hormone-regulated breast and ovarian cancers,61,62 even some baseline DNA damage in tumor or protumorigenic tissue (vs that of normal tissue) can indicate inherent potential defects in DNA repair.63–65 In our system, for example, by Alkaline CometAssay, human F stem cells demonstrated an increase in baseline DNA damage, reflective in increased percentage DNA in tail and tail moments (Figure 2). Although the absolute amount of DNA damage in these F stem cells was considered “normal” when normalized to healthy control cells, the significant increase in DNA damage in F stem cells suggests inherent defects in DNA repair, which when aggravated by endogenous or exogenous stressors may initiate tumorigenesis.
Normal cellular processes, such as transcription and replication, may produce endogenous DNA damage, including free radicals, SSBs generated in DNA replication, and collapsed replication forks. When left unmanaged and unrepaired, these insults can generate costly DNA DSBs and ultimately genome instability.20,60,66,67 In DNA replication, for example, SSBs will be converted into a DSB on one of the sister chromatids.67 If unrepaired, as in the case of a DNA repair system with decreased HR DNA repair capacity, these seemingly small errors in DNA replication and synthesis may lead to collapsed replication forks and suddenly become mutagenic events.20,67,68 Additionally, constitutive loss of DNA repair activity is not required for mutagenesis to occur; rather, if even error-prone DNA repair is overwhelmed intermittently, DNA repair “signatures” associated with various DNA repair deficiencies can emerge.67
Support for this inherent inability to repair DNA DSBs was evidenced by delayed DNA DSB repair in F stem cells when examined using ICC/IF(Figures 10 and 11). Although Myo stem cells demonstrated ability to repair DNA DSBs, reflected in the nonsignificant difference in levels of γ-H2AX, after 24 hours of recovery versus untreated cells, there remained a significant increase in γ-H2AX foci in F stem cells after 24 hours versus their untreated counterparts (Figure 11A and B). Several studies have shown that delayed disappearance of γ-H2AX foci following DNA damage initiation indicates less/slower DNA DSB repair and is further emphasized by increased chromosomal abnormalities and instability.41,42,69 These findings suggest compromised DNA repair in F stem cells.
Disrupted DNA repair in F stem cells was also observed in the altered expression of a large proportion of DNA DSB repair-related genes. Of note, all 3 genes whose products comprise the MRN complex necessary for HR DDR initiation, MRE11A, NBS1, and RAD50, were downregulated in F stem cells (Figure 3A). Those genes relating to DSB binding (BRCA2, RAD51, RAD51AP1, and RAD52) and HR mediators and effectors (BARD1, BRCA1, MDC1, CHEK1, and CHEK2) were also downregulated, and ATM and ATR, DNA DSB signal transduction genes, were altered as well (Figure 3B, D and F). Thus, even if downstream effector proteins can compensate for minute defects in MRN, additional alterations in these signaling pathways could affect the overall efficacy of DNA DSB repair in F stem cells, as we observed in decreased expression and disrupted activation of proteins involved in HR DNA DSB repair (Figures 6, 7 and 8).
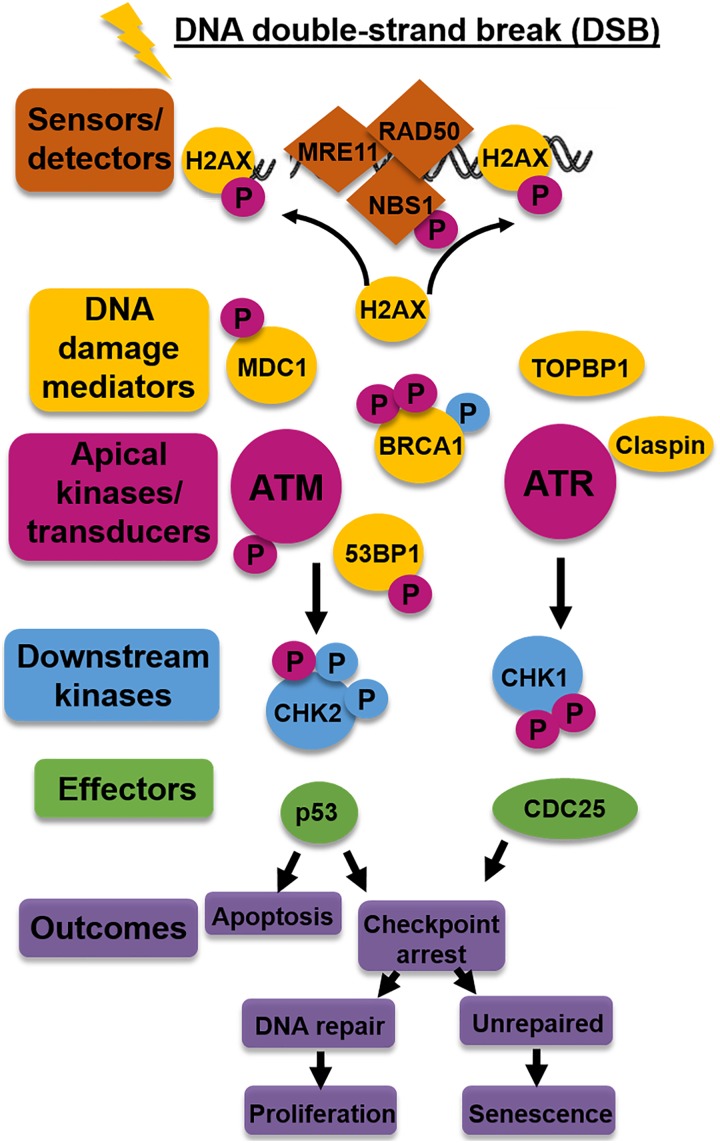
Activation of these proteins regulate the intricate processes of DDR initiation and recruitment of downstream effector proteins that are necessary for DNA DSB repair to occur (Figures 8 and 9). In F stem cells, it appears ATM activation was increased following DNA DSB induction, suggesting the DDR was, in fact, initiated, since ATM will be phosphorylated at Ser1981, by either autophosphorylation or phosphorylation by the MRN complex (Figure 9). Interestingly, however, p-MRE11 and p-NBS1 demonstrated decreased expression (Figure 8A and B), and RAD50 recruitment to the site of DNA DSBs (marked by increased γ-H2AX localization) was reduced relative to the amount of DNA damage following DNA DSB induction (Figures 10 and 11). These changes may point to a source of decreased activation and decreased binding of downstream mediators and effector DNA repair proteins, including ATR, CHK1, and CHK2 (Figure 9), since overall phosphorylation of MRE11 and NBS1 (sensors), ATR (signal transducer), and CHK1 and CHK2 (effector proteins/mediators) was decreased in F stem cells (Figure 8). In addition, autophosphorylation or phosphorylation by the MRN complex is vital for further phosphorylation of CHK1 and CHK2 kinases, which regulate p53 signaling, ultimately affecting pathways involved in cell cycle arrest, apoptosis, and cell senescence.70–72
Figure 9.
Key proteins involved in the DNA damage response (DDR) following DNA double-strand breaks (DSBs). A simplified representation of the key protein involvement in the DDR that is initiated, amplified, and propagated in response to DNA DSBs and the outcomes of each signaling pathway. Color identifiers correspond to proteins categorized by the type (eg, gold indicates DNA damage mediators), while “P” designates a phosphorylated protein. Magenta “P” is indicative of ATM/ATR-mediated phosphorylation; light blue “P,” CHK2/CHK1-mediated phosphorylation.
Although determining the amount of γ-H2AX foci fluorescence in F and Myo stem cells provides insight into the status of DNA DSB repair function, determining the expression of nuclear RAD50 protein relative to the amount of γ-H2AX foci fluorescence may elucidate additional mechanistic information about the DDR in F versus Myo stem cells following DNA DSB induction with bleomycin, since RAD50 rapidly colocalizes with γ-H2AX at the site of DNA DSBs to initiate recruitment of other DNA repair proteins.50,55 Overall, in both untreated and treated cells, there was a significantly decreased expression of nuclear RAD50 in F stem cells versus Myo stem cells; in untreated cells, for example, mean (±SEM) integrated RAD50 fluorescence was significantly decreased in F stem cells (190688 ± 5822) versus Myo stem cells (259662 ± 6348), P < .0001. (Figure 11C). Interestingly, however, there was no significant difference in the baseline median RAD50:γ-H2AX expression ratio between F and Myo stem cells, 14.0 (IQR: 6.4-43.0) and 16.0 (IQR: 7.1-28.0), P > .9999 (Figure 11D), suggesting that both F and Myo stem cells may maintain appropriate relative expression of RAD50 protein in the absence of genomic insult. Of note, however, this ratio changed drastically following DNA DSB induction with bleomycin.
Although both cell types demonstrated a decrease in the RAD50–γ-H2AX expression ratio following DNA DSB induction, F stem cells demonstrated significantly decreased RAD50:γ-H2AX expression after DNA DSB induction after 0 hour recovery time versus normal Myo stem cells: 4.0 (IQR: 1.6-11.0) and 10.0 (IQR: 2.7-20.0); P < .0001 (Figure 11D). This significant decrease in F stem cells is maintained after 3 hours and 6 hours of recovery in normal growth medium (5.3 [IQR: 2.6-12.0] vs 9.3 [IQR: 4.6-16.0]), P < .0001 and (3.3 [IQR: 1.9-11.0] vs 5.7 [IQR: 2.6-14.0]); P = .002, respectively (Figure 11D). Notably, although the median RAD50:γ-H2AX expression ratio was increased in both F and Myo stem cells after 24 hours of recovery, it remained significantly less in F stem cells, 13.0 (IQR: 6.0-26.0) versus adjacent normal Myo stem cells, 18.0 (IQR: 9.8-26), P = .04 (Figure 11D). This suggests that F stem cells may not adequately express and recruit RAD50 to the sites of DNA DSB relative to the amount of DNA damage that is occurring in the nucleus and, thus, may not properly initiate DNA repair, as a result (Figure 12).
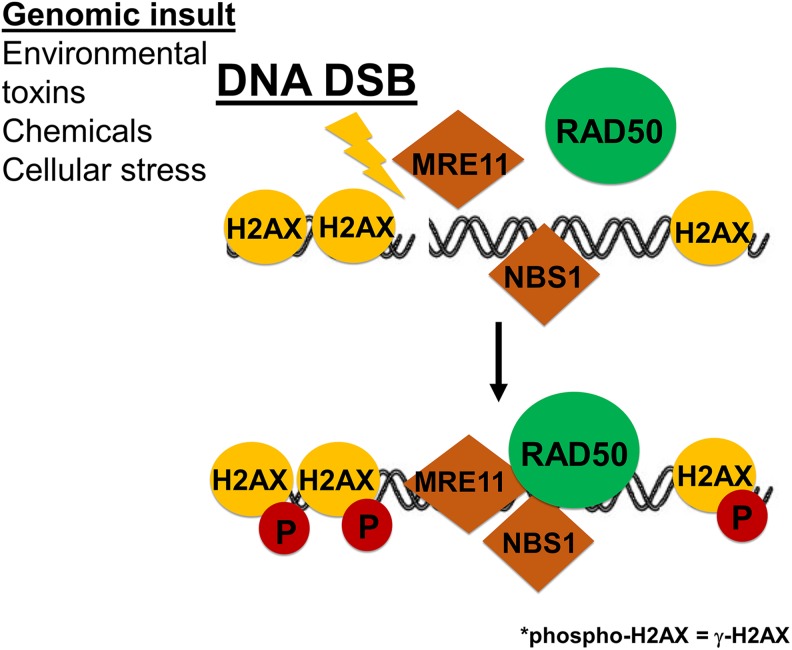
Figure 12.
MRE11-RAD50-NBS1 (MRN) complex activation and H2AX phosphorylation in response to DNA souble-strand break (DSBs). A simplified representation of RAD50 recruitment to the site of a DNA DSB in initiating the DNA damage response (DDR).
These data suggest activation of ATM may be occurring independent of MRN complex function and may instead be undergoing autophosphorylation or activation by the DNA–PKcs–Ku complex, which has been shown to regulate ATM activation in the absence of a fully functional MRN complex. This would account for maintenance of phosphorylation of H2AX at the site of DSBs, which were elevated in F stem cells.73 Altogether, these data suggest that when challenged by genomic stress, F stem cells may be unable to activate the DDR effectively.
Without proper activation of these critical proteins and kinases, therefore, DNA DSBs may experience, at least, delayed repair, leading to untold consequences that may be the ultimate origins for tumorigenic events, including further transformation of seemingly normal stem cells. Decreased nuclear localization of RAD50, a critical component of the MRN complex and the DDR, per amount of induced DNA damage in F stem cells demonstrate further impairment in the DDR. Altogether, these impairments in DNA repair may lead either to the emergence of MED12 in myometrium-derived tumor-forming stem cells or to further amplification of the effects of MED12 if mutagenesis occurs by another mechanism, causing ongoing UF burden in the female human body.8,9,67,74
Conclusion
Because these defects in DNA DSB repair have been discovered in the stem cell population that is thought to be the source of UF tumors, these impairments provide novel insight into potential causes underlying the origins of MED12 mutations and/or the impact these impairments may have in stem cells originating from existing MED12-mutated UF tissues and cells. Although significant, the findings in this study should be further validated in additional human samples to account for variability. Further studies are also needed for understanding how these mutations may not only emerge due to defective DNA repair but also affect the fibroid and Myo tissue microenvironment that is created as a result.
Supplemental Material
Supplemental Material, Suppl_figure_and_Tables for A Preliminary Study: Human Fibroid Stro-1+/CD44+ Stem Cells Isolated From Uterine Fibroids Demonstrate Decreased DNA Repair and Genomic Integrity Compared to Adjacent Myometrial Stro-1+/CD44+ Cells by Lauren E. Prusinski Fernung, Ayman Al-Hendy and Qiwei Yang in Reproductive Sciences
Acknowledgments
The authors would like to thank Ms. Archana Laknaur and Ms. Lelyand Stone for their help in collecting human tissues and for providing some technical assistance. The authors thank the Augusta University (AU) Department of Obstetrics and Gynecology, AU Clinical Trials Office and AU Biorepository and Central Repository for Biorepository Alliance of GA – Oncology (BRAG-Onc) for their assistance in recommending patients, consenting patients, and obtaining fibroid and myometrial tissues, respectively.
Authors’ Note: Lauren Prusinski Fernung conceived and designed the studies, provided study material, completed experiments, collected and assembled data, analyzed and interpreted data, wrote the manuscript.
Ayman Al-Hendy conceived studies, provided financial support, provided study material, approved final manuscript.
Qiwei Yang conceived and designed parts of the study, provided financial support, provided study material, revised the manuscript. All data supporting this study are provided either in full in the results section of this paper or as supplementary information accompanying this article.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was conducted at Augusta University (AU, Augusta, GA) and was supported by the National Institutes of Health: National Institute of Environmental Health Sciences grant R01 ES028615 (AA) and the National Institute of Child Health and Human Development grant F30 HD089585 (LPF); the AU Start-up Package from the Department of Obstetrics and Gynecology; and the AU Intramural Grants Program (QY) .
ORCID iD: Lauren E. Prusinski Fernung, BS  https://orcid.org/0000-0002-6522-3315
https://orcid.org/0000-0002-6522-3315
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Cardozo E Clark AD, Banks NK, Henne MB, Stegmann BJ, Segars JH. The estimated annual cost of uterine leiomyomata in the United States. Am J Obstet Gynecol. 2012;206(3):211.e211–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cook J, Davis B, Cai S, Barrett J, Conti C, Walker C. Interaction between genetic susceptibility and early-life environmental exposure determines tumor-suppressor-gene penetrance. Proc Natl Acad Sci U S A. 2005;102(24):8644–8649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lee H, Norwitx E, Shaw J. Contemporary management of fibroids in pregnancy. Rev Obstet Gynecol. 2010;3(1):20–27. [PMC free article] [PubMed] [Google Scholar]
- 4. Lethaby A, Vollenhoven B. Fibroids (uterine myomatosis, leiomyomas) BMJ Clin Evid. 2011;pii: 0814 [PMC free article] [PubMed] [Google Scholar]
- 5. Markowski D, Bartnitzke S, Löning T, Drieschner N, Helmke B, Bullerdiek J. MED12 mutations in uterine fibroids–their relationship to cytogenetic subgroups. Int J Cancer. 2012;131(7):1528–1536. [DOI] [PubMed] [Google Scholar]
- 6. Halder S, Laknaur A, Miller J, Layman LC, Diamond M, Al-Hendy A. Novel MED12 gene somatic mutations in women from the Southern United States with symptomatic uterine fibroids. Mol Genet Genomics. 2015;290(2):505–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mehine M, Kaasinen E, Mäkinen N, et al. Characterization of uterine leiomyomas by whole-genome sequencing. N Engl J Med. 2013;369(1):43–53. [DOI] [PubMed] [Google Scholar]
- 8. Ono M, Qiang W, Serna V, et al. Role of stem cells in human uterine leiomyoma growth. PLoS One. 2012;7(5): e36935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bulun S. Uterine fibroids. N Engl J Med. 2013;369(14):1344–1355. [DOI] [PubMed] [Google Scholar]
- 10. Louka M, Boutou E, Bakou V, et al. DNA damage response/repair in cancer stem cells—potential vs. controversies In: Chen C, ed. Biochemistry, Genetics and Molecular Biology » “Advances in DNA Repair”. London, UK: InTech Open; 2015;415–444. [Google Scholar]
- 11. Bonner A, Redon C, Dickey J, et al. gH2AX and cancer. Nat Rev Cancer. 2008;8(12):957–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lavin M, Delia D, Chessa L. ATM and the DNA damage response. EMBO Reports. 2006;7(2):154–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sung P, Klein H. Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat Rev Mol Cell Biol. 2006;7(10):739–750. [DOI] [PubMed] [Google Scholar]
- 14. Branzei D, Foiani M. Regulation of DNA repair throughout the cell cycle. Nat Rev Mol Cell Biol. 2008;9(4):297–308. [DOI] [PubMed] [Google Scholar]
- 15. Tomasetti C, Vogelstein B. Cancer Etiology. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science. 2015;347(6217):78–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Albanes D, Winick M. Are cell number and cell proliferation risk factors for cancer? J Natl Cancer Inst. 1988;80(10):772–774. [DOI] [PubMed] [Google Scholar]
- 17. Tomatis L. Cell proliferation and carcinogenesis: a brief history and current view based on an IARC workshop report. International Agency for Research on Cancer. Environ Health Perspect. 1993;101(Suppl 5):149–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Atkinson RL, Yang WT, Rosen DG, et al. Cancer stem cell markers are enriched in normal tissue adjacent to triple negative breast cancer and inversely correlated with DNA repair deficiency. Breast Cancer Res. 2013;15(5): R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Baker AM, Graham TA. Revealing human intestinal stem cell and crypt dynamics. Molecular & Cellular Oncology. 2014;1(3) e970069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Blokzijl F, Ligt JD, Jager M, et al. Tissue-specific mutation accumulation in human adult stem cells during life. Nature. 2016;538 (7624):260–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Maugeri-Sacca M, Bartucci M, Maria RD. DNA damage repair pathways in cancer stem cells. Mol Cancer Ther. 2012;11:1627–1636. [DOI] [PubMed] [Google Scholar]
- 22. Pawlowska E, Blasiak J. DNA repair—a double-edged sword in the genomic stability of cancer cells—the case of chronic myeloid leukemia. J Mol Sci. 2015;16(11):27535–27549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dietlein F, Thelen L, Reinhardt H. Cancer-specific defects in DNA repair pathways as targets for personalized therapeutic approaches. Trends Genet. 2014;30(8):326–339. [DOI] [PubMed] [Google Scholar]
- 24. Blanpain C, Mohrin M, Sotiropoulou P, Passegue E. DNA-damage response in tissue-specific and cancer stem cells. Cell Stem Cell. 2011;8(1):16–29. [DOI] [PubMed] [Google Scholar]
- 25. Kappil M, Liao Y, Terry M, Santella R. DNA repair gene expression levels as indicators of breast cancer in the breast cancer family registry. Anticancer Res. 2016;36(8):4039–4044. [PMC free article] [PubMed] [Google Scholar]
- 26. Batar B, Guven G, Eroz S, Bese N, Guven M. Decreased DNA repair gene XRCC1 expression is associated with radiotherapy-induced acute side effects in breast cancer patients. Gene. 2016;582(1):33–37. [DOI] [PubMed] [Google Scholar]
- 27. Ribeiro E, Ganzinelli M, Andreis D, et al. Triple negative breast cancers have a reduced expression of DNA repair genes. PLoS One. 2013;8(6): e66243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Matta J, Morales L, Ortiz C, et al. Estrogen receptor expression is associated with DNA repair capacity in breast cancer. PLoS One. 2016;11(3): e0152422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu S, Ginestier C, Charafe-Jauffret E, et al. BRCA1 regulates human mammary stem/progenitor cell fate. Proc Natl Acad Sci. 2008;105(5):1680–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kenyon J, Gerson S. The role of DNA damage repair in aging of adult stem cells. Nucleic Acids Res. 2007;35(22):7557–7565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Iso T, Watanabe T, Iwamoto T, Shimamoto A, Furuichi Y. DNA damage caused by bisphenol a and estradiol through estrogenic activity. Biol Pharm Bull. 2006;29(2):206–210. [DOI] [PubMed] [Google Scholar]
- 32. Kitagishi Y, Kobayashi M, Matsuda S. Defective DNA repair systems and the development of breast and prostate cancer (Review). Int J Oncol. 2012;42(1):29–34. [DOI] [PubMed] [Google Scholar]
- 33. Yang Q, Elam L, Laknaur A, et al. Altered DNA repair genes in human uterine fibroids are epigeniticallly regulated via EZH2 histone methyltransferase. ASRM Abstracts. 2015;104(3, Supplement): e1–e387. [Google Scholar]
- 34. Yang Q, Nair S, Laknaur A, Ismail N, Diamond M, Al-Hendy A. The polycomb group protein EZH2 impairs DNA damage repair gene expression in human uterine fibroids. Biol Reprod. 2016;94(3):1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wei J-J, Zhang X-M, Chiriboga L, Yee H, Perle MA, Mittal K. Spatial differences in biologic activity of large uterine leiomyomata. Fertil Steril. 85(1):179–187. [DOI] [PubMed] [Google Scholar]
- 36. Yatsenko SA, Mittal P, Wood-Trageser MA, et al. Highly heterogeneous genomic landscape of uterine leiomyomas by whole exome sequencing and genome-wide arrays. Fertil Sterl. 2017;107(2):457–466.e459. [DOI] [PubMed] [Google Scholar]
- 37. Mas A, Nair S, Laknaur A, Simon C, Diamond M, Al-Hendy A. Stro-1/CD44 as putative human myometrial and fibroid stem cell markers. Fertil Steril. 2015;104(1):225–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Makinen N, Mehine M, Tolvanen J, et al. MED12, the mediator complex subunit 12 gene, is mutated at high frequency in uterine leiomyomas. Science. 2011;334(6053):252–255. [DOI] [PubMed] [Google Scholar]
- 39. Prusinski Fernung LE, Yang Q, Kumari A, Sakamuro D, Mas A, Al-Hendy A. Endocrine disruptor exposure during development increases incidence of uterine fibroids by altering DNA repair in myometrial stem cells. Biol Reprod. 2018; In Press doi:10.1093/biolre/ioy097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Halder SK, Beauchamp RD, Datta PK. Smad7 induces tumorigenicity by blocking TGF-β-induced growth inhibition and apoptosis. Exp Cell Res. 2005;307(1):231–246. [DOI] [PubMed] [Google Scholar]
- 41. Mariotti L, Pirovano G, Savage K, et al. Use of the γ-H2AX assay to investigate DNA repair dynamics following multiple radiation exposures. PLoS One. 2013;8(11):e79541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schmid T, Zlobinskaya O, Multhoff G. Differences in phosphorylated histone H2AX foci formation and removal of cells exposed to low and high linear energy transfer radiation. Curr Genomics. 2012;13(6):418–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dickey J, Redon C, Nakamura A, Baird B, Sedelnikova O, Bonner W. H2AX: functional roles and potential applications. Chromosoma. 2009;118(6):683–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kreienkamp R, Croke M, Neumann MA, et al. Vitamin D receptor signaling improves Hutchinson-Gilford progeria syndrome cellular phenotypes. Oncotarget. 2016;7(21):30018–30031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Markowski DN, Tadayyon M, Bartnitzke S, Belge G, Helmke BM, Bullerdiek J. Cell cultures in uterine leiomyomas: rapid disappearance of cells carrying MED12 mutations. Genes Chromosomes Cancer. 2014;53(4):316–323. [DOI] [PubMed] [Google Scholar]
- 46. Trevigen I. CometAssay(R) HT: reagent kit for higher throughput single cell gel electrophoresis assay. In: Trevigen, ed. (Trevigen, Gaithersburg) 2012;Vol # 4252-040-K2012:1-18.
- 47. Azqueta A, Slyskova J, Langie S, Gaivao ION, Collins A. Comet assay to measure DNA repair: approach and applications. Front Genet. 2014;5(288). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Benitez-Bribiesca L, Sanchez-Suarez P. Oxidative damage, bleomycin, and gamma radiation induce different types of DNA strand breaks in normal lymphocytes and thymocytes. A comet assay study. Ann N Y Acad Sci. 1999;887:133–149. [DOI] [PubMed] [Google Scholar]
- 49. Harper JV, Anderson JA, O’Neill P. Radiation induced DNA DSBs: contribution from stalled replication forks? DNA Repair. 2010;9(8):907–913. [DOI] [PubMed] [Google Scholar]
- 50. Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol: CB. 2000;10(15):886–895. [DOI] [PubMed] [Google Scholar]
- 51. Kim JS, Krasieva TB, Kurumizaka H, Chen DJ, Taylor AM, Yokomori K. Independent and sequential recruitment of NHEJ and HR factors to DNA damage sites in mammalian cells. J Cell Biol. 2005;170(3):341–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Olive P, Banath J. Detection of DNA double-strand breaks through the cell cycle after exposure to X-rays, bleomycin, etoposide and 125IdUrd. Int J Radiat Biol. 1993;64(4):349–358. [DOI] [PubMed] [Google Scholar]
- 53. Burma S, Chen B, Murphy M, Kurimasa A, Chen D. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J Biol Chem. 2001;276(45):42462–42467. [DOI] [PubMed] [Google Scholar]
- 54. Kuo L, Yang L. γ-H2AX—a novel biomarker for DNA double-strand breaks. In Vivo. 2008;22(3):305–309. [PubMed] [Google Scholar]
- 55. Lavin M, Kozlov S, Gatei M, Kijas A. ATM-dependent phosphorylation of all three members of the mrn complex: from sensor to adaptor. Biomolecules. 2015;5(4):2877–2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Uziel T, Lerenthal Y, Moyal L, Andegeko Y, Mittelman L, Shiloh Y. Requirement of the MRN complex for ATM activation by DNA damage. EMBO J. 2003;22(20):5612–5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Moravek M, Bulun S. Endocrinology of uterine fibroids: steroid hormones, stem cells, and genetic contribution. Curr Opin Obstet Gynecol. 2015;27(4):276–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mas A, IC I, Gil-Sanchis C, et al. Identification and characterization of the human leiomyoma side population as putative tumor-initiating cells. Fertil Steril. 2012;98(3):741–751.e746. [DOI] [PubMed] [Google Scholar]
- 59. Chang H, Senaratne T, Zhang L, et al. Uterine leiomyomas exhibit fewer stem/progenitor cell characteristics when compared with corresponding normal myometrium. Reprod Sci. 2010;17(2):158–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Behrens A, Deursen JV, Rudolph K, Schumacher B. Impact of genomic damage and ageing on stem cell function. Nat Cell Biol. 2014;16(3):201–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Institute NC. BRCA1 and BRCA2: cancer risk and genetic testing. Genetics. 2015; https://www.cancer.gov/about-cancer/causes-prevention/genetics/brca-fact-sheet.
- 62. Miki Y, Swensen J, Shattuck-Eidens D, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266(5182):66–71. [DOI] [PubMed] [Google Scholar]
- 63. Smith T, Miller M, Lohman K, Case L, Hu J. DNA damage and breast cancer risk. Carcinogenesis. 2003;24(5):883–889. [DOI] [PubMed] [Google Scholar]
- 64. Buchynska L, Brieieva O, Glushchenko N, Vorobyova L, Bilyk O. DNA repair deficiency in peripheral blood lymphocytes of endometrial cancer patients with a family history of cancer. BMC Cancer. 2014;14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Shahidi M, Mozdarani H, Mueller W. Radiosensitivity and repair kinetics of gamma-irradiated leukocytes from sporadic prostate cancer patients and healthy individuals assessed by alkaline comet assay. Iran Biomed J. 2010;14(3):67–75. [PMC free article] [PubMed] [Google Scholar]
- 66. Gent DV, Hoeijmakers J, Kanaar R. The DNA double-stranded break connection. Nature Rev Genet. 2001;2(3):196–206. [DOI] [PubMed] [Google Scholar]
- 67. Tubbs A, Nussenzweig A. Endogenous DNA damage as a source of genomic instability in cancer. Cell. 2017;168(4):644–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Helleday T, Eshtad S, Nik-Zainal S. Mechanisms underlying mutational signatures in human cancers. Nature Rev Genet. 2014;15(9):585–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Borras-Fresneda M, Barquinero J, Gomolka M, et al. Differences in DNA repair capacity, cell death and transcriptional response after irradiation between a radiosensitive and a radioresistant cell line. Sci Rep. 2016;1(6):27043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zannini L, Delia D, Buscemi G. CHK2 kinase in the DNA damage response and beyond. J Mol Cell Biol. 2014;6(6):442–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ou Y, Chung P, Sun T, Shieh S. . p53 C-terminal phosphorylation by CHK1 and CHK2 participates in the regulation of DNA-damage-induced C-terminal acetylation. Mol Biol Cell. 2005;16(4):1684–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. III DO, Mayo L. Emerging non-canonical functions and regulation by p53: p53 and stemness. Int J Mol Sci. 2016;17(12):1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hartlerode A, Morgan M, Wu Y, Buis J, Ferguson D. Recruitment and activation of the ATM kinase in the absence of DNA damage sensors. Nat Struct Mol Biol. 2015;22(9):736–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Maruyama T, Miyazaki K, H HM, Ono M, Uchida H, Yoshimura Y. Review: human uterine stem/progenitor cells: implications for uterine physiology and pathology. Placenta 2013;34: S68–S72. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material, Suppl_figure_and_Tables for A Preliminary Study: Human Fibroid Stro-1+/CD44+ Stem Cells Isolated From Uterine Fibroids Demonstrate Decreased DNA Repair and Genomic Integrity Compared to Adjacent Myometrial Stro-1+/CD44+ Cells by Lauren E. Prusinski Fernung, Ayman Al-Hendy and Qiwei Yang in Reproductive Sciences