Abstract
Previously, we reported that the fluorapatite (FA)–modified polycaprolactone (PCL) nanofiber could be an odontogenic/osteogenic inductive tissue-engineering scaffold by inducing stem cell differentiation and mineralization. The present study aimed to explore which of the signal pathways affected this differentiation and mineralization process. The Human Signal Transduction PathwayFinder RT2 Profiler PCR Array was used to analyze the involvement of potential signal transduction pathways during human dental pulp stem cell (DPSCs) osteogenic differentiation induced by FA-modified PCL nanofiber scaffolds. Based on the results, perturbation studies of the signaling pathways hedgehog, insulin, and Wnt were performed. Moreover, the autophagy process was studied, as indicated by the expression of the microtubule-associated protein 1 light chain 3A/B-II (LC3-II) and the cell osteogenic phenotypic changes. In a comparison of the cells grown on PCL + FA scaffolds and those on PCL-only scaffolds, the transcript expression of BMP2, BMP4, FOXA2, PTCH1, WNT1, and WNT2 (PCR array–labeled signal proteins of the hedgehog pathway); CEBPB, FASN, and HK2 (PCR array–labeled signal proteins of the insulin pathway); and CCND1, JUN, MYC, TCF7, and WISP1 (PCR array–labeled signal proteins of the Wnt pathway) doubled at day 14 when obvious cell osteogenic differentiation occurred. Phenotypically, in all the perturbation groups at day 14, ALP activity, OPN, and autophagy marker LC3-II expression were coincidently decreased. Consistently, no positive alizarin red staining or von Kossa staining was observed in the specimens from these perturbation groups at day 28. The results showed that when obvious cell differentiation occurred at day 14 on PCL + FA control groups, the inhibition of the hedgehog, insulin, and Wnt pathways significantly decreased DPSC osteogenic differentiation and mineralization. The osteogenic differentiation of DPSCs grown on FA-modified PCL scaffolds appeared to be positively modulated by the hedgehog, insulin, and Wnt signal pathways, which were coordinated with and/or mediated by the cell autophagy process.
Keywords: signal transduction, tissue engineering, biomaterial(s), osteogenesis, tissue scaffolds, autophagy
Introduction
In the discipline of tissue engineering, stem cells, biomimetic scaffolds, and cellular inductive differentiation molecules are 3 recognized crucial factors needed for therapeutic success (Langer and Vacanti 1993; Ripamonti and Reddi 1997; Polykandriotis et al. 2010). The scaffolds should mimic the extracellular matrix to support the proliferation and differentiation of the stem cells. However, most scaffolds currently support only cellular proliferation, lacking the capability of specific cell lineage induction. To induce cellular differentiation, many proteins and peptides have been used as cellular inductive molecules; nevertheless, the stable release of these proteins and peptides is difficult to achieve. Occasionally, these applied proteins and peptides have resulted in immunologic reactions (Nie and Wang 2007; Anderson et al. 2008).
Fluorapatite (FA) is an inorganic material that has shown significant differentiation-inducing ability without any immunologic reaction. The ordered FA crystals forming a 2-dimensional cell culture surface can induce dental pulp stem cells (DPSCs) and adipose-derived stem cell differentiation into osteogenic cells (Liu et al. 2010; Liu et al. 2012; Wang et al. 2012). In our previous study, we found that the FA-modified polycaprolactone (PCL) nanofiber 3-dimensional scaffold could support in vitro the growth, differentiation, and mineralization of DPSCs to a greater degree than ordered FA 2-dimensional surfaces (Guo et al. 2014).
The process of stem cell odontogenic/osteogenic differentiation is complex. Many mechanical and biological signals are either synergistic or antagonistic in this process. Previous studies showed that the specific ligands initially need to be identified by transmembrane receptors and then certain signal pathways are activated through a multistep cascade. These pathway signal factors—for example, bone morphogenetic proteins (BMPs), transforming growth factor β, hedgehog, Wnt, fibroblast growth factors, platelet-derived growth factors, and insulin-like growth factors (IGFs)—in turn activate transcription factors that are translocated into the nucleus of cells. The phosphorylated transcription factors then interact with specific DNA regions, working as a promoter, enhancer, or inhibitor. Finally, the target protein will be expressed, sealing the cell’s fate of being either an osteoblast or odontoblast. During these complex processes, many factors are shared by the pathways, resulting in the pathway signals being coordinated or counteracted (Katoh 2008; Varjosalo et al. 2008; Forbes et al. 2010; Li et al. 2011; Cenciarelli et al. 2014; Bajinting and Ng 2017).
Autophagy is an evolutionarily conserved homeostatic process in which special cytoplasm substrates are delivered to lysosomes for degradation. Through autophagy, the cell restructures to differentiate or replace the damaged organelles. It has been reported that autophagy interacts with many signal pathways and plays important roles during stem cell differentiation and mineralization (Li et al. 2016; Ghosh and Kapur 2017; Humbert et al. 2017; Johnson and Tee 2017).
In this study, we aimed to discover which pathways were involved in the process of DPSC differentiation induced by the FA-modified PCL nanofiber scaffolds.
Materials and Methods
Coating of FA on PCL Nanofiber Surfaces
The PCL nanofiber scaffolds were modified by FA crystals at 37 °C under ambient conditions (Guo et al. 2014). Briefly, the PCL nanofiber scaffolds were immersed in a solution with 0.10M HEDTA-Ca, 0.06M KH2PO4, and 0.02M KF and incubated at 37 °C under ambient pressure condition for 1 d. After FA crystal formation, the scaffolds were carefully rinsed with phosphate-buffered saline (PBS; GIBCO, Invitrogen), dried in air, observed by scanning electron microscope (SEM), and stored in desiccators.
Cell Culture and Seeding
DPSCs were a gift from Dr. S. Shi (University of Southern California, Los Angeles; Gronthos et al. 2002). The cells were cultured in Dulbecco’s modified Eagle’s medium (GIBCO) under standard culture conditions. The medium was supplemented with 10% fetal bovine serum (GIBCO), 100 U/mL of penicillin, and 100 mg/mL of streptomycin (GIBCO). The medium was changed every 2 d.
The prepared scaffolds were rinsed in PBS for 5 times and preincubated in the medium at 37 °C for 2 h before cell seeding. DPSCs at passage 3 were seeded on each scaffold at a density of 1 × 105 cells/scaffold for all experiments. The scaffold is circular with a base area of 2 cm2. After cell attachment, the medium was replenished. The medium was changed every 2 d, and the cells were cultured for up to 28 d.
SEM Observation
After 28 d, the cell scaffolds were observed by SEM. Briefly, the samples were washed twice with PBS, fixed with 4% paraformaldehyde for 45 min, serially dehydrated to 100% ethanol, dried in desiccators, and observed with a Philips XL30 FEG SEM (FEI Company).
RNA Isolation and Reverse Transcription
At days 7 and 14, total cellular RNA was extracted from DPSCs grown on the PCL + FA and PCL-only scaffolds by Trizol (Invitrogen) according to the manufacturer’s protocols. For converting the total RNA into cDNA for RNA array samples, the RT2 First Stand cDNA Synthesis and the SuperScript II Reverse Transcriptase (Invitrogen) kits were used following manufacturer’s protocols.
Human Signal Transduction PathwayFinder RT2 Profiler PCR Array
The Human Signal Transduction PathwayFinder RT2 Profiler PCR Array (Qiagen) was used to analyze the involvement of potential signal transduction pathways. According to the manufacturer’s protocols, RT2 SYBR Green PCR Master Mix (Qiagen) was used in the real-time polymerase chain reaction (RT-PCR) of the ViiA 7 system with 3 replicates from each group (Life Tech). Duplicate 96-well array plates were used in the PCR array analyses. Data were analyzed by the Qiagen online data analysis tool. A similar methodology was adopted in our previous publications (Liu et al. 2010; Liu et al. 2012; Li et al. 2016).
Pathway Perturbation
Based on the results of the PCR array, a perturbation study of 3 signaling pathways was performed: cyclopamine (inhibitor of hedgehog), PPP (inhibitor of insulin), and FH535 (inhibitor of Wnt). Briefly, 0.1μM cyclopamine, 0.02μM PPP, and 0.2μM FH535 were added respectively to the medium on the following day after cell seeding. The medium and inhibitors were changed every 2 d. The cells were cultured for up to 28 d.
Real-time Polymerase Chain Reaction
RT-PCR quantitation was carried out by the ViiA 7 system (Life Tech). The osteogenesis relative gene markers runx2 and spp1 were used for the quantitative detection with 3 replicates from each group. The TaqMan Universal PCR Master Mix Kit (Applied Biosystems) was used following the manufacturer’s protocol. The target gene expressions were normalized with the housekeeping gene β-actin. Relative gene expression values were calculated by ∆∆CT-based fold-change calculations.
Alkaline Phosphatase Activity
On 7 and 14 d, alkaline phosphatase (ALP) activities of DPSCs in the different groups were measured with SensoLyte pNPP Alkaline Phosphatase Assay Kit (AnaSpecCA) following the manufacturer’s protocol. The ALP activity was calculated with the OD405 values and normalized with its corresponding total protein content, according to the standard curve.
Western Blot
After culturing for 14 d, the cell scaffolds were lysed in NP-40 protein lysis buffer. The extracted proteins were separated with 8% to 15% SDS-PAGE gels and transferred electrophoretically to a nitrocellulose membrane. The membrane was incubated with anti-human β-catenin, OPN, and LC3 antibody (1:1,000) at 4 °C overnight and further incubated with secondary antibody IgG (1:10,000) for 2 h before development. β-actin was used as a loading control. In our previous studies, the applied antibodies were thoroughly checked to ensure no nonspecific binding and false-positive results (Guo et al. 2014; Li et al. 2016). The PCL + FA group without inhibitors was used as the control to compare the effects of inhibitors on the expression of relevant markers. Relative band densities were calculated by determining the ratio of target proteins to β-actin with the ImageJ program (National Institutes of Health).
Alizarin Red Staining and Osteogenesis Quantitative Assay
After culturing for 21 and 28 d, the cells and scaffolds were fixed and stained with alizarin red (Osteogenesis Assay Kit; Millipore) according to the manufacturer’s protocol. Then quantification was carried out by measuring the OD405 values of extracted alizarin red from each stained specimen.
Von Kossa Staining
After culturing for 21 and 28 d, the cells and scaffolds were fixed, hydrated, stained with silver nitrate in the dark at 37 °C for 1 h, and then exposed to bright light for color development. ImageJ was used to compare the staining intensity of specimens from different groups.
Statistical Analysis
Statistical analysis was carried out with 1-way analysis of variance and Tukey’s post hoc test for all quantification studies except for the PCR array and the RT-PCR analyses, and significance was set at P < 0.05. These quantification studies were carried out by performing 3 separate experiments with 5 replicate samples in each group.
Results
The expression of hedgehog, insulin, and Wnt signal molecules dramatically changed in the PCL + FA–induced DPSC differentiation process.
The Human Signal Transduction PathwayFinder RT2 Profiler PCR Array was used to compare the gene expression of the PCL and PCL + FA groups at 7 and 14 d. It showed that several functional gene groupings changed dramatically (Table). For the hedgehog pathway, at day 14, BMP2, BMP4, FOXA2, PTCH1, WNT1, and WNT2 expression more than doubled in the PCL + FA groups versus the PCL-only scaffolds. Between the 14- and 7-d data in the PCL + FA group, BMP2 expression increased >7 times, while EN1 and FOXA2 expression decreased by more than half. For Wnt pathway genes, between the PCL + FA and PCL groups at day 14, CCND1, JUN, MYC, TCF7, and WISP1 expression more than doubled. In the PCL + FA group, at day 14 versus day 7, JUN and MYC expression more than doubled, while VEGFA decreased by more than half. For the insulin pathway, at day 14 between the PCL + FA and PCL groups, CEBPB, FASN, and HK2 expression was >4 times greater, while LEP expression decreased more than half. The trend was same for the expression of these molecules at days 14 and 7 in the PCL + FA group (Fig. 1). According to these data, the hedgehog, insulin, and Wnt pathways were chosen for further perturbation experimentation.
Table.
Fold Changes of Representative Signal Molecules Expressed by the Dental Pulp Stem Cells Grown on PCL and PCL + FA Scaffolds for 7 and 14 d.
| Symbol | Unigene | RefSeq | Description | Fold Change |
|---|---|---|---|---|
| PCL + FA vs. PCL scaffolds a | ||||
| BAX | Hs.624291 | NM_004324 | BCL2-associated X protein | 2.07 |
| BCL2 | Hs.150749 | NM_000633 | B-cell CLL/lymphoma 2 | 3.30 |
| BCL2L1 | Hs.516966 | NM_138578 | BCL2-like 1 | 4.57 |
| BIRC3 | Hs.127799 | NM_001165 | Baculoviral IAP repeat containing 3 | 4.96 |
| BMP2 | Hs.73853 | NM_001200 | Bone morphogenetic protein 2 | 6.45 |
| BMP4 | Hs.68879 | NM_130851 | Bone morphogenetic protein 4 | 3.37 |
| CCL20 | Hs.75498 | NM_004591 | Chemokine (C-C motif) ligand 20 | 2.04 |
| CCND1 | Hs.523852 | NM_053056 | Cyclin D1 | 2.07 |
| CD5 | Hs.58685 | NM_014207 | CD5 molecule | 2.70 |
| CDKN1B | Hs.238990 | NM_004064 | Cyclin-dependent kinase inhibitor 1B (p27, Kip1) | 2.28 |
| CEBPB | Hs.719041 | NM_005194 | CCAAT/enhancer binding protein (C/EBP), beta | 4.80 |
| CXCL9 | Hs.77367 | NM_002416 | Chemokine (C-X-C motif) ligand 9 | 2.70 |
| EGR1 | Hs.708393 | NM_001964 | Early growth response 1 | 3.28 |
| FASLG | Hs.2007 | NM_000639 | Fas ligand (TNF superfamily, member 6) | 2.70 |
| FASN | Hs.83190 | NM_004104 | Fatty acid synthase | 4.75 |
| FN1 | Hs.203717 | NM_002026 | Fibronectin 1 | 2.22 |
| FOS | Hs.25647 | NM_005252 | FBJ murine osteosarcoma viral oncogene homolog | 11.61 |
| FOXA2 | Hs.155651 | NM_021784 | Forkhead box A2 | 2.70 |
| HK2 | Hs.591588 | NM_000189 | Hexokinase 2 | 4.53 |
| HOXA1 | Hs.67397 | NM_005522 | Homeobox A1 | 2.70 |
| ICAM1 | Hs.643447 | NM_000201 | Intercellular adhesion molecule 1 | 2.55 |
| IL2 | Hs.89679 | NM_000586 | Interleukin 2 | 2.70 |
| IL4 | Hs.73917 | NM_000589 | Interleukin 4 | 2.70 |
| IL8 | Hs.624 | NM_000584 | Interleukin 8 | 3.76 |
| IRF1 | Hs.436061 | NM_002198 | Interferon regulatory factor 1 | 4.21 |
| JUN | Hs.696684 | NM_002228 | Jun proto-oncogene | 3.47 |
| KLK2 | Hs.515560 | NM_005551 | Kallikrein-related peptidase 2 | 2.70 |
| LTA | Hs.36 | NM_000595 | Lymphotoxin alpha (TNF superfamily, member 1) | 2.53 |
| MDM2 | Hs.733536 | NM_002392 | Mdm2 p53 binding protein homolog (mouse) | 2.98 |
| MMP7 | Hs.2256 | NM_002423 | Matrix metallopeptidase 7 (matrilysin, uterine) | 2.70 |
| MYC | Hs.202453 | NM_002467 | V-myc myelocytomatosis viral oncogene homolog (avian) | 4.08 |
| NRIP1 | Hs.155017 | NM_003489 | Nuclear receptor interacting protein 1 | 2.41 |
| PRKCA | Hs.708867 | NM_002737 | Protein kinase C, alpha | 2.09 |
| PRKCE | Hs.580351 | NM_005400 | Protein kinase C, epsilon | 2.97 |
| PTCH1 | Hs.494538 | NM_000264 | Patched 1 | 3.50 |
| SELE | Hs.82848 | NM_000450 | Selectin E | 2.70 |
| SELPLG | Hs.591014 | NM_003006 | Selectin P ligand | 2.15 |
| TANK | Hs.132257 | NM_004180 | TRAF family member-associated NFKB activator | 4.78 |
| TCF7 | Hs.573153 | NM_003202 | Transcription factor 7 (T-cell specific, HMG-box) | 2.59 |
| TERT | Hs.492203 | NM_198253 | Telomerase reverse transcriptase | 2.70 |
| TNF | Hs.241570 | NM_000594 | Tumor necrosis factor | 2.70 |
| TP53 | Hs.740601 | NM_000546 | Tumor protein p53 | 2.04 |
| WISP1 | Hs.492974 | NM_003882 | WNT1 inducible signaling pathway protein 1 | 3.36 |
| WNT1 | Hs.248164 | NM_005430 | Wingless-type MMTV integration site family, member 1 | 2.70 |
| WNT2 | Hs.567356 | NM_003391 | Wingless-type MMTV integration site family member 2 | 2.20 |
| LEP | Hs.194236 | NM_000230 | Leptin | −3.49 |
| PMEPA1 | Hs.517155 | NM_020182 | Prostate transmembrane protein, androgen induced 1 | −2.04 |
| RBP1 | Hs.529571 | NM_002899 | Retinol binding protein 1, cellular | −2.59 |
| PCL + FA scaffold: 14 d vs. 7 d b | ||||
| BIRC3 | Hs.127799 | NM_001165 | Baculoviral IAP repeat containing 3 | 2.34 |
| BMP2 | Hs.73853 | NM_001200 | Bone morphogenetic protein 2 | 7.86 |
| BRCA1 | Hs.194143 | NM_007294 | Breast cancer 1, early onset | 2.04 |
| CEBPB | Hs.719041 | NM_005194 | CCAAT/enhancer binding protein (C/EBP), beta | 4.85 |
| CXCL9 | Hs.77367 | NM_002416 | Chemokine (C-X-C motif) ligand 9 | 2.38 |
| FASN | Hs.83190 | NM_004104 | Fatty acid synthase | 7.20 |
| GADD45A | Hs.80409 | NM_001924 | Growth arrest and DNA-damage-inducible, alpha | 4.08 |
| HK2 | Hs.591588 | NM_000189 | Hexokinase 2 | 4.52 |
| ICAM1 | Hs.643447 | NM_000201 | Intercellular adhesion molecule 1 | 2.59 |
| IRF1 | Hs.436061 | NM_002198 | Interferon regulatory factor 1 | 3.08 |
| JUN | Hs.696684 | NM_002228 | Jun proto-oncogene | 3.79 |
| MMP7 | Hs.2256 | NM_002423 | Matrix metallopeptidase 7 (matrilysin, uterine) | 2.06 |
| MYC | Hs.202453 | NM_002467 | V-myc myelocytomatosis viral oncogene homolog (avian) | 2.42 |
| TP53 | Hs.740601 | NM_000546 | Tumor protein p53 | 2.51 |
| BCL2 | Hs.150749 | NM_000633 | B-cell CLL/lymphoma 2 | −27.55 |
| BCL2L1 | Hs.516966 | NM_138578 | BCL2-like 1 | −2.04 |
| CCL20 | Hs.75498 | NM_004591 | Chemokine (C-C motif) ligand 20 | −3.05 |
| CD5 | Hs.58685 | NM_014207 | CD5 molecule | −22.08 |
| CSF2 | Hs.1349 | NM_000758 | Colony stimulating factor 2 (granulocyte-macrophage) | −2.64 |
| CYP19A1 | Hs.260074 | NM_000103 | Cytochrome P450, family 19, subfamily A, polypeptide 1 | −2.20 |
| EN1 | Hs.271977 | NM_001426 | Engrailed homeobox 1 | −2.02 |
| FASLG | Hs.2007 | NM_000639 | Fas ligand (TNF superfamily, member 6) | −2.96 |
| FOS | Hs.25647 | NM_005252 | FBJ murine osteosarcoma viral oncogene homolog | −2.83 |
| FOXA2 | Hs.155651 | NM_021784 | Forkhead box A2 | −23.70 |
| GREB1 | Hs.467733 | NM_014668 | Growth regulation by estrogen in breast cancer 1 | −31.75 |
| HOXA1 | Hs.67397 | NM_005522 | Homeobox A1 | −2.84 |
| HSP90AA2 | Hs.523560 | NM_001040141 | Heat shock protein 90kDa alpha (cytosolic), class A member 2 | −1.86 |
| IL1A | Hs.1722 | NM_000575 | Interleukin 1, alpha | −42.19 |
| IL2 | Hs.89679 | NM_000586 | Interleukin 2 | −6.83 |
| IL4 | Hs.73917 | NM_000589 | Interleukin 4 | −3.08 |
| KLK2 | Hs.515560 | NM_005551 | Kallikrein-related peptidase 2 | −22.83 |
| LEP | Hs.194236 | NM_000230 | Leptin | −19.34 |
| LTA | Hs.36 | NM_000595 | Lymphotoxin alpha (TNF superfamily, member 1) | −4.65 |
| MMP10 | Hs.2258 | NM_002425 | Matrix metallopeptidase 10 (stromelysin 2) | −2.03 |
| NAIP | Hs.654500 | NM_004536 | NLR family, apoptosis inhibitory protein | −3.67 |
| NOS2 | Hs.709191 | NM_000625 | Nitric oxide synthase 2, inducible | −5.27 |
| PECAM1 | Hs.376675 | NM_000442 | Platelet/endothelial cell adhesion molecule | −2.95 |
| PTGS2 | Hs.196384 | NM_000963 | Prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) | −5.43 |
| RBP1 | Hs.529571 | NM_002899 | Retinol binding protein 1, cellular | −2.15 |
| TERT | Hs.492203 | NM_198253 | Telomerase reverse transcriptase | −1.69 |
| TNF | Hs.241570 | NM_000594 | Tumor necrosis factor | −94.34 |
| VEGFA | Hs.73793 | NM_003376 | Vascular endothelial growth factor A | −2.42 |
FA, fluorapatite; PCL, polycaprolactone.
Fold changes of the signal molecules expressed by the cells grown on PCL + FA scaffold relative to PCL scaffold at 14 d.
Fold changes of the signal molecules expressed by the cells grown on PCL + FA scaffold for 14 d relative to 7 d.
Figure 1.
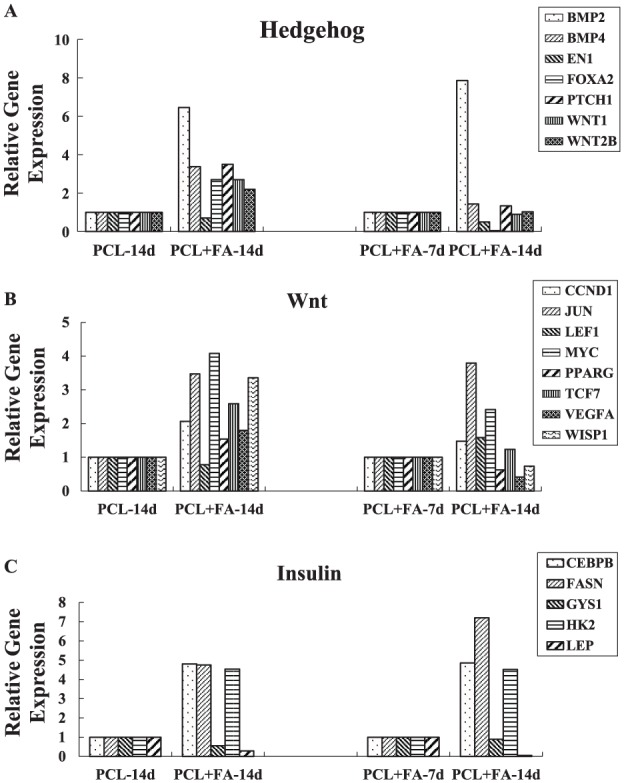
Relative gene expression of the dental pulp stem cells grown on PCL and PCL + FA scaffolds for 7 and 14 d: (A) hedgehog, (B) Wnt, and (C) insulin pathway signal molecules. The relative gene expression was calculated by comparing the cells grown on PCL + FA scaffold and PCL scaffold at day 14 and the cells grown on PCL + FA scaffold between days 14 and 7. FA, fluorapatite; PCL, polycaprolactone.
The perturbation for hedgehog, insulin, and Wnt signal pathways blocked autophagy, and the PCL + FA induced the DPSC differentiation process.
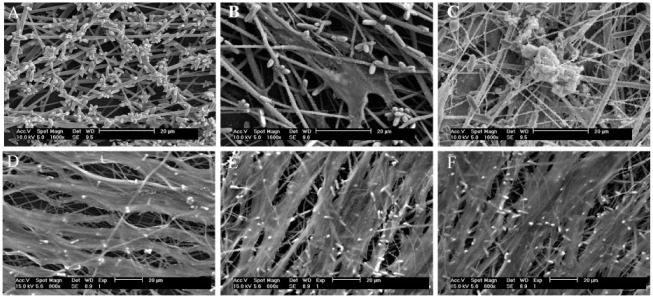
In these experiments, the administered specific inhibitors strongly disrupted the DPSC differentiation process. SEM observation revealed no nodules in the perturbation groups, whereas obvious nodules were found on the FA-modified PCL scaffolds without perturbation (Fig. 2).
Figure 2.
Scanning electron microscope observation of cell-mediated mineral nodules. (A) Observation of FA-modified PCL nanofiber scaffold. Observation of dental pulp stem cells grown on PCL + FA scaffolds for (B) 7 d and (C) 28 d. At 28 d, densely deposited mineral nodules were observed. Observation of dental pulp stem cells grown on PCL + FA scaffolds for 28 d with specific pathway inhibitors: (D) cyclopamine (the inhibitor of hedgehog), (E) FH535 (the inhibitor of Wnt), and (F) PPP (the inhibitor of insulin). No mineral nodules were seen on the surfaces. FA, fluorapatite; PCL, polycaprolactone.
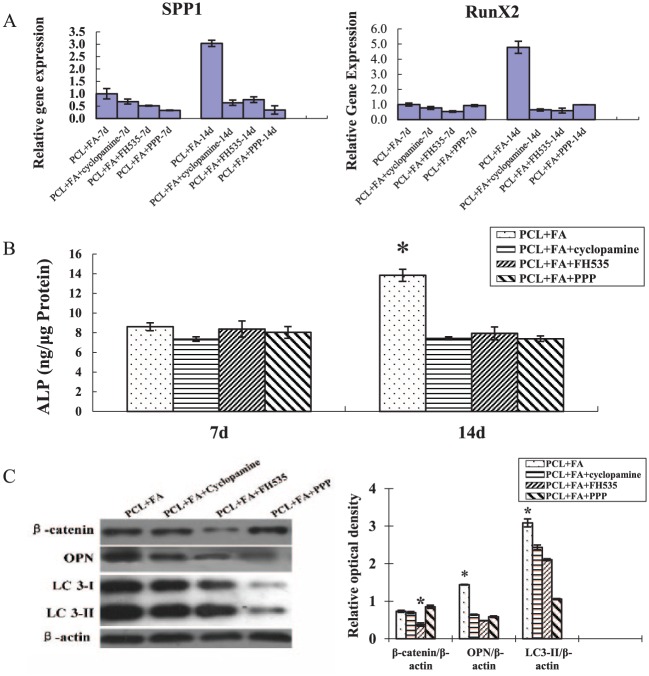
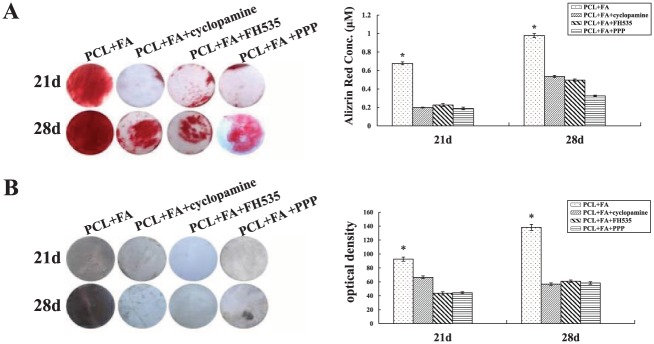
On day 14, the RT-PCR data showed that the expression of runx2 and spp1 decreased significantly in the perturbation groups. Concurrently, ALP activity and OPN and LC3-II expression decreased significantly (Fig. 3). On days 21 and 28, alizarin red and von Kossa staining intensity was also significantly reduced (Fig. 4).
Figure 3.
The effects of specific pathway inhibitors on the osteogenic gene expression and alkaline phosphatase (ALP) and autophagy activity of the dental pulp stem cells (DPSCs) grown on PCL + FA scaffolds. (A) Real-time polymerase chain reaction quantification of the osteogenic gene expression of DPSCs grown on PCL + FA scaffolds for 7 and 14 d. (B) Quantification of ALP activity of the DPSCs grown on PCL + FA scaffolds for 7 and 14 d. *P < 0.05. (C) Western blot of β-catenin, OPN, LC3-I/II, and β-actin expression of DPSCs grown on PCL + FA scaffolds for 14 d. The group of PCL + FA without inhibitors was used as the control to compare the effects of inhibitors on the expression of relevant markers. The optical band density of the expression of each protein was normalized with its corresponding β-actin. *P < 0.05. Values are presented as mean ± SD. FA, fluorapatite; PCL, polycaprolactone.
Figure 4.
The effects of specific pathway inhibitors on the mineralization of the dental pulp stem cells grown on PCL + FA scaffolds for 21 and 28 d. (A) Alizarin red staining and quantitative analysis. (B) Von Kossa staining and the staining intensity quantitative analysis. *P < 0.05. Values are presented as mean ± SD. FA, fluorapatite; PCL, polycaprolactone.
Discussion
An integral part of the tissue engineering of mineralized tissues is the modification of scaffolds to mimic the extracellular matrix of the defective tissues (Caliari et al. 2015; Lee et al. 2015; Weisgerber et al. 2015). Our previous studies showed that 1) ordered FA crystal coatings and FA-modified PCL nanofiber scaffolds can induce the DPSCs and adipose-derived stem cells to differentiate into odontogenic/osteogenic cells that can produce extracellular matrix for further mineralization and 2) mechanistically, autophagy appears to be essentially involved in this differentiation process (Wang et al. 2012; Guo et al. 2014; Li et al. 2016). In this study, our data indicated that signal pathways (i.e., hedgehog, insulin, and Wnt) were involved in DPSC differentiation and mineralization.
The hedgehog pathway is important in directing growth and tissue patterning, especially in skeletal formation and osteoblast development. Overexpression of SHhN in periosteal-derived mesenchymal progenitor cells was shown to improve bone defect reconstruction by enhancing stem cell survival, differentiation, and revascularization (Huang et al. 2014). Other research revealed a relationship between hedgehog and autophagy by showing that Gli1 knockdown could induce apotosis and autophagy through regulation of mTOR phosphorylation (Sun et al. 2014). Similarly, in the present study, after the hedgehog pathway was blocked by cyclopamine, autophagy and cell differatiation and mineralization greatly decreased (Figs. 2D, 3, 4).
Among WNT signals, Smad4 and β-catenin, through canonical Wnt signaling, were shown to regulate osteoblast proliferation and differentiation to enhance bone formation (Salazar et al. 2013). Through canonical (β-catenin) and noncanonical pathways, Wnt11 is actively involved in osteoblast differentiation and fracture healing (Friedman et al. 2009). Accordingly, in our study, the WNT pathway representative transcripts—CCND1, JUN, MYC, TCF7, and WISP1—were upregulated in the DPSC osteogenic differentiation induced by the PCL + FA scaffolds. Not surprising, this upregulation concurrently occurred with upregulation of the DPSC osteogenic differentiation markers, osteogenic phenotypic changes, and eventual mineral nodule formation (Figs. 1C, 3, 4). Autophagic pathways are involved in cell survival and death. Of interest to us, a feedback loop between Wnt signaling and autophagy was identified in recent studies. This loop may involve mTOR, phosphoinositide 3-kinase (PI3-K), protein kinase B (Akt), AMP activated protein kinase (AMPK), silent mating type information regulation 2 homolog 1 (Saccharomyces cerevisiae; SIRT1), and Wnt1-inducible signaling pathway protein 1 (WISP1) cascade (Sato et al. 2010; Maiese et al. 2012; Kimura et al. 2013; Murahovschi et al. 2015; Maiese 2016). In our studies, we also found that WNT and autophagy were both involved in the DPSC osteogenic differentiation and mineralization induced by FA-modified PCL scaffolds. The inhibition of the WNT pathway prevented autophagy as well as DPSC differentiation and mineralization (Figs. 2E, 3, 4).
Secreted by β cells of the pancreas, insulin is a potent anabolic hormone in stem cell differentiation and bone formation (Wu et al. 2017). Interestingly, at the early stage of osteogenic differentiation of MC-3T3 cells (days 3 to 6), IGF-I and IGFBP-2 activated the AMPK pathway and enhanced the phosphorylation of ULK-1 S555 (a known AMPK phosphorylation site in ULK-1 that is necessary to assemble the essential components of autophagosome) and the expression of beclin-1 and LC3-II, which indicated the stimulation of autophagy. This process is important for the osteoblast to acquire the energy source for respiration. However, at the late stage (after day 9), IGF-I and IGFBP-2 significantly decreased, with a significant reduction of AMPK T172 expression, ULK-1 S555 phosphorylation, and beclin-1 and LC3-I/II expression, which suggested the attenuation of autophagosome formation (Xi et al. 2016). One of our previous studies also showed that the expression of autophagic marker LC3-II protein reached its peak value at day 7 and then decreased and diminished slowly at day 21 (Li et al. 2016). In this study, we found that insulin signals were involved in the DPSCs differentiation induced by PCL + FA scaffolds, which were coordinated with and/or mediated by the cell autophagy process (Figs. 2F, 3, 4).
Cellular differentiation involves many cell signal pathways, which are well established and strongly supported by many in vivo and in vitro studies. Our data showed that during DPSC differentiation induced by the PCL + FA scaffold, hedgehog, insulin, and Wnt signaling protein expressions changed dramatically. Furthermore, perturbation of the hedgehog, insulin, and Wnt signaling pathways inhibited osteogenic phenotype expression and subsequent mineralization, as well as autophagic LC3-II expression. Therefore, we draw the conclusion that the hedgehog, insulin, and Wnt signaling pathways are involved in the DPSC differentiation induced by the PCL + FA scaffold and may activate the osteogenesis process through autophagic modulation. In future applications, precisely controlled hard tissue regeneration could be accomplished through targeted modulation of individual and/or synchronized osteogenesis-related signal pathways, combined with aimed regulation of autophagic process. Although the WNT, hedgehog, and insulin signaling cascades and the autophagic process are associated with each other, the exact signal cross-talking mechanism needs further studies.
Author Contributions
T. Guo, contributed to conception, design, data acquisition, analysis, and interpretation, drafted and critically revised the manuscript; G. Cao, contributed to design, data acquisition, and analysis, drafted and critically revised the manuscript; Y. Li, contributed to design, data acquisition, and interpretation, drafted the manuscript; Z. Zhang, contributed to design and data analysis, critically revised the manuscript; J.E. Nör, contributed to data interpretation, critically revised the manuscript; B.H. Clarkson, contributed to conception and data interpretation, critically revised the manuscript; J. Liu, contributed to conception, design, data analysis, and interpretation, drafted and critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Footnotes
This work was supported by the Department of Cariology, Restorative Sciences, and Endodontics at the University of Michigan School of Dentistry. Support was also provided by the National Natural Science Foundation of China (81500872), Natural Science Foundation of Jiangsu Province (BK20161389), Young Medical Talent Foundation of Jiangsu Province (QNRC2016906), and Six Talent Peaks Project in Jiangsu Province (2016-WSW-093), National Postdoctoral Foundation of China (2016M593040).
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
References
- Anderson JM, Rodriguez A, Chang DT. 2008. Foreign body reaction to biomaterials. Semin Immunol. 20(2):86–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajinting A, Ng HL. 2017. Recombinant expression in E. coli of human FGFR2 with its transmembrane and extracellular domains. PeerJ. 5:e3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caliari SR, Weisgerber DW, Grier WK, Mahmassani Z, Boppart MD, Harley BA. 2015. Collagen scaffolds incorporating coincident gradations of instructive structural and biochemical cues for osteotendinous junction engineering. Adv Healthc Mater. 4(6):831–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenciarelli C, Marei HE, Zonfrillo M, Pierimarchi P, Paldino E, Casalbore P, Felsani A, Vescovi AL, Maira G, Mangiola A. 2014. PDGF receptor alpha inhibition induces apoptosis in glioblastoma cancer stem cells refractory to anti-notch and anti-EGFR treatment. Mol Cancer. 13:247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes K, Souquet B, Garside R, Aplin JD, Westwood M. 2010. Transforming growth factor-{beta} (TGF{beta}) receptors I/II differentially regulate TGF{beta}1 and IGF-binding protein-3 mitogenic effects in the human placenta. Endocrinology. 151(4):1723–1731. [DOI] [PubMed] [Google Scholar]
- Friedman MS, Oyserman SM, Hankenson KD. 2009. Wnt11 promotes osteoblast maturation and mineralization through R-spondin 2. J Biol Chem. 284(21):14117–14125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh J, Kapur R. 2017. Role of mTORC1-S6K1 signaling pathway in regulation of hematopoietic stem cell and acute myeloid leukemia. Exp Hematol. 50:13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronthos S, Brahim J, Li W, Fisher LW, Cherman N, Boyde A, DenBesten P, Robey PG, Shi S. 2002. Stem cell properties of human dental pulp stem cells. J Dent Res. 81(8):531–535. [DOI] [PubMed] [Google Scholar]
- Guo T, Li Y, Cao G, Zhang Z, Chang S, Czajka-Jakubowska A, Nor JE, Clarkson BH, Liu J. 2014. Fluorapatite-modified scaffold on dental pulp stem cell mineralization. J Dent Res. 93(12):1290–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Tang M, Yehling E, Zhang X. 2014. Overexpressing sonic hedgehog peptide restores periosteal bone formation in a murine bone allograft transplantation model. Mol Ther. 22(2):430–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humbert M, Federzoni EA, Tschan MP. 2017. Distinct TP73-DAPK2-ATG5 pathway involvement in ato-mediated cell death versus atra-mediated autophagy responses in apl. J Leukoc Biol. 102(6):1357–1370. [DOI] [PubMed] [Google Scholar]
- Johnson CE, Tee AR. 2017. Exploiting cancer vulnerabilities: mTOR, autophagy, and homeostatic imbalance. Essays Biochem. 61(6):699–710. [DOI] [PubMed] [Google Scholar]
- Katoh M. 2008. Wnt signaling in stem cell biology and regenerative medicine. Curr Drug Targets. 9(7):565–570. [DOI] [PubMed] [Google Scholar]
- Kimura R, Okouchi M, Kato T, Imaeda K, Okayama N, Asai K, Joh T. 2013. Epidermal growth factor receptor transactivation is necessary for glucagon-like peptide-1 to protect PC12 cells from apoptosis. Neuroendocrinology. 97(4):300–308. [DOI] [PubMed] [Google Scholar]
- Langer R, Vacanti JP. 1993. Tissue engineering. Science. 260(5110):920–926. [DOI] [PubMed] [Google Scholar]
- Lee JC, Pereira CT, Ren X, Huang W, Bischoff D, Weisgerber DW, Yamaguchi DT, Harley BA, Miller TA. 2015. Optimizing collagen scaffolds for bone engineering: effects of cross-linking and mineral content on structural contraction and osteogenesis. J Craniofac Surg. 26(6):1992–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Klim JR, Derda R, Courtney AH, Kiessling LL. 2011. Spatial control of cell fate using synthetic surfaces to potentiate tgf-beta signaling. Proc Natl Acad Sci U S A. 108(29):11745–11750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Guo T, Zhang Z, Yao Y, Chang S, Nör JE, Clarkson BH, Ni L, Liu J. 2016. Autophagy modulates cell mineralization on fluorapatite-modified scaffolds. J Dent Res. 95(6):650–656. [DOI] [PubMed] [Google Scholar]
- Liu J, Jin T, Chang S, Czajka-Jakubowska A, Zhang Z, Nör JE, Clarkson BH. 2010. The effect of novel fluorapatite surfaces on osteoblast-like cell adhesion, growth, and mineralization. Tissue Eng Part A. 16(9):2977–2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Wang X, Jin Q, Jin T, Chang S, Zhang Z, Czajka-Jakubowska A, Giannobile WV, Nör JE, Clarkson BH. 2012. The stimulation of adipose-derived stem cell differentiation and mineralization by ordered rod-like fluorapatite coatings. Biomaterials. 33(20):5036–5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiese K. 2016. Novel nervous and multi-system regenerative therapeutic strategies for diabetes mellitus with mtor. Neural Regen Res. 11(3):372–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiese K, Chong ZZ, Wang S, Shang YC. 2012. Oxidant stress and signal transduction in the nervous system with the PI 3-K, Akt, and mTOR cascade. Int J Mol Sci. 13(11):13830–13866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murahovschi V, Pivovarova O, Ilkavets I, Dmitrieva RM, Docke S, Keyhani-Nejad F, Gogebakan O, Osterhoff M, Kemper M, Hornemann S, et al. 2015. WISP1 is a novel adipokine linked to inflammation in obesity. Diabetes. 64(3):856–866. [DOI] [PubMed] [Google Scholar]
- Nie H, Wang CH. 2007. Fabrication and characterization of PLGA/HAp composite scaffolds for delivery of BMP-2 plasmid DNA. J Control Release. 120(1-2):111-121. [DOI] [PubMed] [Google Scholar]
- Polykandriotis E, Popescu LM, Horch RE. 2010. Regenerative medicine: then and now—an update of recent history into future possibilities. J Cell Mol Med. 14(10):2350–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripamonti U, Reddi AH. 1997. Tissue engineering, morphogenesis, and regeneration of the periodontal tissues by bone morphogenetic proteins. Crit Rev Oral Biol Med. 8(2):154–163. [DOI] [PubMed] [Google Scholar]
- Salazar VS, Zarkadis N, Huang L, Watkins M, Kading J, Bonar S, Norris J, Mbalaviele G, Civitelli R. 2013. Postnatal ablation of osteoblast Smad4 enhances proliferative responses to canonical Wnt signaling through interactions with beta-catenin. J Cell Sci. 126(Pt 24): 5598–5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato A, Sunayama J, Matsuda K, Tachibana K, Sakurada K, Tomiyama A, Kayama T, Kitanaka C. 2010. Regulation of neural stem/progenitor cell maintenance by PI3K and mTOR. Neurosci Lett. 470(2):115–120. [DOI] [PubMed] [Google Scholar]
- Sun Y, Guo W, Ren T, Liang W, Zhou W, Lu Q, Jiao G, Yan T. 2014. Gli1 inhibition suppressed cell growth and cell cycle progression and induced apoptosis as well as autophagy depending on ERK1/2 activity in human chondrosarcoma cells. Cell Death Dis. 5:e979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varjosalo M, Bjorklund M, Cheng F, Syvanen H, Kivioja T, Kilpinen S, Sun Z, Kallioniemi O, Stunnenberg HG, He WW, et al. 2008. Application of active and kinase-deficient kinome collection for identification of kinases regulating hedgehog signaling. Cell. 133(3):537–548. [DOI] [PubMed] [Google Scholar]
- Wang X, Jin T, Chang S, Zhang Z, Czajka-Jakubowska A, Nör JE, Clarkson BH, Ni L, Liu J. 2012. In vitro differentiation and mineralization of dental pulp stem cells on enamel-like fluorapatite surfaces. Tissue Eng Part C Methods. 18(11):821–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisgerber DW, Caliari SR, Harley BA. 2015. Mineralized collagen scaffolds induce hMSC osteogenesis and matrix remodeling. Biomater Sci. 3(3):533–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Wang C, Miao X, Wu Y, Yuan J, Ding M, Li J, Shi Z. 2017. Age-related insulin-like growth factor binding protein-4 overexpression inhibits osteogenic differentiation of rat mesenchymal stem cells. Cell Physiol Biochem. 42(2):640–650. [DOI] [PubMed] [Google Scholar]
- Xi G, Rosen CJ, Clemmons DR. 2016. IGF-I and IGFBP-2 stimulate AMPK activation and autophagy, which are required for osteoblast differentiation. Endocrinology. 157(1):268–281. [DOI] [PMC free article] [PubMed] [Google Scholar]