Abstract
High-risk human papillomavirus (HPV) is a major risk factor for oral and pharyngeal cancers (OPCs), yet the detailed mechanisms by which HPV promotes OPCs are not understood. Forkhead box M1B (FoxM1B) is an oncogene essential for cell cycle progression and tumorigenesis, and it is aberrantly overexpressed in many tumors. We previously showed that FoxM1B was the putative target of an epithelial-specific transcription factor, Grainyhead-like 2 (GRHL2). In the current study, we demonstrate that HPV type 16 (HPV-16) E6 induces FoxM1B in human oral keratinocytes (HOKs) and tonsillar epithelial cells (TECs) in part through GRHL2. FoxM1B was barely detectable in cultured normal human oral keratinocytes (NHOKs) and progressively increased in immortalized HOKs harboring HPV-16 genome (HOK-16B) and tumorigenic HOK-16B/BaP-T cells. Retroviral expression of HPV-16 E6 and/or E7 in NHOKs, TECs, and hypopharyngeal carcinoma cells (FaDu) revealed induction of FoxM1B and GRHL2 by the E6 protein but not E7. Both GRHL2 and FoxM1B were strongly induced in the epidermis of HPV-16 E6 transgenic mice and HPV+ oral squamous cell carcinomas. Ectopic expression of FoxM1B led to acquisition of transformed phenotype in HOK-16B cells. Loss of FoxM1B by lentiviral short hairpin RNA vector or chemical inhibitor led to elimination of tumorigenic characteristics of HOK-16B/BaP-T cells. Luciferase reporter assay revealed that GRHL2 directly bound and regulated the FoxM1B gene promoter activity. Using epithelial-specific Grhl2 conditional knockout mice, we exposed wild-type (WT) and Grhl2 KO mice to 4-nitroquinolin 1-oxide (4-NQO), which led to induction of FoxM1B in the tongue tissues and rampant oral tumor development in the WT mice. However, 4-NQO exposure failed to induce tongue tumors or induction of FoxM1B expression in Grhl2 KO mice. Collectively, these results indicate that HPV-16 induces FoxM1B in part through GRHL2 transcriptional activity and that elevated FoxM1B level is required for oropharyngeal cancer development.
Keywords: carcinogenesis, oncogenes, oropharyngeal neoplasms, tongue, epithelial cells, gene knockout technique
Introduction
FoxM1B is a member of the Forkhead box (Fox) transcription factor family known for regulating cell cycle–related gene expression to promote tumor progression (Barger et al. 2015; Kelleher and O’Sullivan 2016). Procarcinogenic role of FoxM1B has been well documented in the literature; Gemenetzidis et al. (2009) showed overexpression of FoxM1B in the early stage of oral squamous cell carcinomas (OSCCs) and in normal human oral keratinocytes (NHOKs) under the influence of nicotine and that aberrant upregulation of FoxM1B elicited tumorigenic effects by induction of genetic instability. Similarly, FoxM1B was found overexpressed in 76.9% of cervical squamous cell carcinoma (SCC) specimens, as well as in precancerous lesions, demonstrating its pathogenic role in the early stage of cervical cancers (Chan et al. 2008).
Grainyhead-like 2 (GRHL2) is one of the known mammalian homologues of Drosophila Grainyhead (GRH) involved in epithelial regeneration and function (Stramer and Martin 2005). Our earlier study identified GRHL2 as a novel pro-oncogene, which modulates oral cancer formation (Chen et al. 2016). GRHL2 is also linked with recurrence of tumors in hepatocellular carcinomas, lung cancer, and breast cancer formation (Tanaka et al. 2008; Dompe et al. 2011; Xiang et al. 2012). GRHL2 overexpression in NIH3T3 cells led to tumorigenic conversion of cells (Werner et al. 2013). In addition, we have demonstrated that GRHL2 plays a unique role in the control of cellular proliferation through transcriptional regulation of its target genes, including hTERT, PCNA, and FoxM1B (Chen et al. 2010, 2012).
A recent study showed that FoxM1B was induced by human papillomavirus (HPV) infection through MZF1/NKX2-1 transcription factor in various cancer cells and required for HPV-associated tumorigenesis (Chen et al. 2014). Interestingly, NKX2-1 is a direct transcriptional target of GRHL2 and forms a positive transcriptional regulatory loop in lung epithelial cells for the coordination of epithelial morphogenesis (Varma et al. 2012). In the current study, we investigated the role of GRHL2 in FoxM1B induction by HPV type 16 (HPV-16). Our data indicate that HPV-16 E6 elevated the expression of both GRHL2 and FoxM1B in human keratinocytes and that GRHL2 directly regulated FoxM1B expression through promoter binding. This regulatory relationship was validated in Grhl2 conditional knockout (cKO) and HPV transgenic mouse models. We also found that FoxM1B elevation by HPV alone was not sufficient for tumorigenic phenotype, which required further elevation of FoxM1B by transgene overexpression. Treatment of mice with a chemical carcinogen, 4-nitroquinolin 1-oxide (4-NQO), led to tongue tumor development in wild-type (WT) mice but not in Grhl2 cKO mice, which lacked the expression of FoxM1B. These data demonstrate that HPV-16 initiates oral carcinogenesis, in part, through elevation of GRHL2 and FoxM1B, which are required for tumorigenic phenotype of cells.
Experimental Procedures
Cells and Cell Culture
Primary NHOKs and tonsillar epithelial cells (TECs) were prepared from keratinized oral epithelial tissues and normal tonsil tissues after tonsillectomies, respectively, according to the methods described elsewhere (Kang et al. 2000). Human OSCC cell lines (SCC4, SCC9, HEp2, BaP-T, and SCC15) and HOK-16B cells were cultured according to the methods previously described (Kang et al. 2007, 2009). Exogenous GRHL2, HPV-16 E6, HPV-16 E7, and HPV-16 E6/E7 were transduced into rapidly proliferating cells by retroviral vectors. The infected cells were selected with 200 µg/mL G418 (Sigma-Aldrich) for 2 wk. Ectopic expression of FoxM1B in HOK-16B was induced through transducing retroviral vectors expressing FoxM1B (pBabe-FoxM1B). The infected cells were selected with 1 µg/mL puromycin (Sigma) for 1 wk. GRHL2 or FoxM1B was knocked down with the lentiviral vectors (LV-GRHL2i or pLKO-FoxM1Bi) expressing short hairpin RNA (shRNA) targeting GRHL2 and FoxM1B sequences, respectively.
Mouse Models
HPV-16 E6, HPV-16 E7 mice were bred in the animal house facility under standard conditions (24°C, 50% ± 5% humidity) with a 12-h light/dark cycle. The animals were given standard feed and water ad libitum.
Epithelial tissue–specific Grhl2 cKO mice (denoted K14/Grhl2 cKO) were generated by crossing Grhl2 floxed (fl/fl) mice with the K14-CreERT mice (Jackson Laboratory) (Walentin et al. 2015). Grhl2 cKO was achieved through intraperitoneal (IP) administration of tamoxifen (Tmx) (75 mg/kg) into 8-wk-old K14/Grhl2 cKO mice for 7 consecutive days to induce homozygous deletion of Grhl2 in epithelial tissues. To induce oral tumors, Grhl2 WT (without Tmx) and Grhl2 KO mice were exposed to 4-NQO (Sigma-Aldrich) diluted in drinking water to the final concentration of 30 µg/mL for 16 wk, followed by 6 wk of normal drinking water, as described previously (Czerninski et al. 2009).
Real-time Quantitative Reverse Transcription Polymerase Chain Reaction
Biopsies from mouse dorsal skin were punched and immediately snap frozen in liquid nitrogen and stored at –80°C until further processing. Total RNA was isolated using the TRIzol Reagent (Thermo Fisher Scientific) and used for reverse transcription (RT) to generate complementary DNA (cDNA). Quantitative polymerase chain reaction (qPCR) was performed in triplicates for each sample with LC480 SYBR Green I Master using universal cycling conditions in Light Cycler 480 (Roche).
Western Blotting
Whole-cell extracts (WCEs) from the cultured cells and mouse skin tissues were isolated using protein lysis buffer. WCEs were then fractionated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to an Immobilon protein membrane (Millipore). The membranes were incubated successively with the primary and the secondary antibodies and exposed to the chemiluminescence reagent (Amersham Pharmacia Biotech) for signal detection.
Gene Promoter Luciferase Assay
Promoter region of FoxM1B (–1000 to +54) was cloned into pGL3B-Luc reporter plasmid (Promega), expressing firefly luciferase. The promoter-luciferase constructs were transfected into 293 cells or SCC4 with or without GRHL2 loss using Lipofectin 2000 Reagent (Invitrogen), along with pRL-SV40 containing Renilla luciferase. The gene promoter activity, reflected by the firefly luciferase activity, was determined as the mean of at least triplicates per experiment after normalizing against Renilla luciferase activity.
Immunohistochemistry and Immunofluorescence Staining Analyses
Mouse skin and human OSCC tissues were fixed in 4% (wt/vol) paraformaldehyde for 24 h. Samples were embedded in paraffin, sectioned, and stained as described previously (Chen et al. 2012). For immunofluorescence staining (IFS), cells were cultured in a Nunc Lab-Tek II Chamber Slide System (ThermoFisher Scientific) to reach 70% to 80% confluence, fixed in 2% paraformaldehyde for 20 min. Cells were sequentially incubated with the primary and the secondary antibodies. Slides were mounted in Prolong Gold w/DAPI (Invitrogen). Images were captured on an Olympus epifluorescence inverted microscope.
Antibodies
The following primary antibodies were used in this study: glyceraldehyde 3-phosphate dehydrogenase (GAPDH), FoxM1B, Cyclin A, Cyclin B1, HELLS, p53, p21WAF1, and p16INK4A from Santa Cruz Biotech; p-RbSer807/811, p-AKTSer473, and p-p38Thr180/182 from Cell Signaling Technology; and GRHL2 antibody from Abnova. Horseradish peroxidase (HRP)–conjugated secondary antibodies were obtained from Santa Cruz Biotech.
Statistical Analysis
Statistical analysis was performed using the Student’s t test (2-tailed) for the qPCR gene expression, luciferase activity assay, and spheroid formation experiments. P values <0.05 were considered significant. All numerical data were expressed as mean ± SD.
Results
GRHL2 and FoxM1B Expression Are Regulated by HPV-16 E6 in NHOK
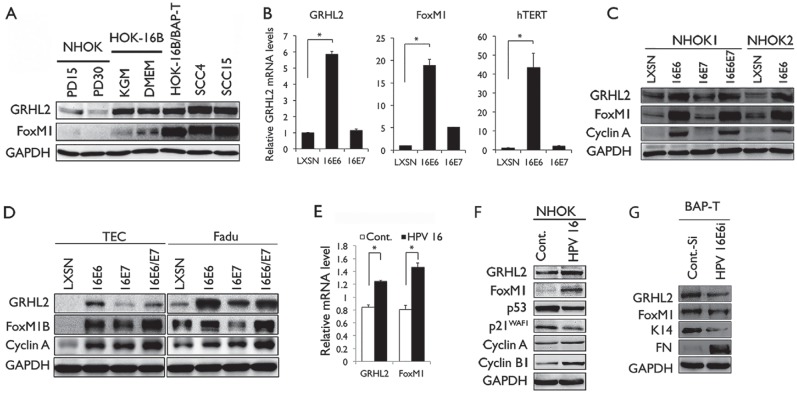
To determine the roles of GRHL2 and FoxM1B in HPV-associated oral carcinogenesis, we surveyed the changes in GRHL2 and FoxM1B levels in the multistep oral carcinogenesis model (Park et al. 1995). This model consists of NHOK, immortalized nontumorigenic HOK harboring HPV-16 DNA (HOK-16B), and tumorigenic HOK cell line (HOK-16B/BaP-T) (Park et al. 1995; Kang and Park 2001). GRHL2 and FoxM1B expression levels were weakly detectable in NHOK but elevated in HOK-16B (Fig. 1A). Further elevation of FoxM1B level was noted in HOK-16B/BaP-T. There was strong induction of GRHL2 and its targets (e.g., hTERT, FoxM1B, and Cyclin A) in NHOK infected with the retroviral vector expressing HPV-16 E6 but not HPV-16 E7 (Fig. 1B, C). Since HPV+ oropharyngeal cancers (OPCs) are frequently found in tonsils (Ringström et al. 2002; Hocking et al. 2011), we sought to determine the expression of GRHL2 and FoxM1B in TECs. Etopic overexpression of HPV-16 E6 in primary TECs strongly induced GRHL2, FoxM1B, and its target protein expression, while HPV-16 E7 did not (Fig. 1D). Similarly, HPV-16 E6 expression in the hypopharyngeal cell carcinoma cell line (FaDu) led to induction of GRHL2 and FoxM1B. Infection of primary NHOK with HPV-16 virus particles, prepared according to a published method (Schowalter et al. 2011), also led to notable induction of GRHL2, FoxM1B, and the Cyclins A and B1 (Fig. 1E, F). Knockdown of HPV-16 E6 in HOK-16B/BaP-T cells with siRNA led to diminution of GRHL2, FoxM1B, and their target protein levels (Fig. 1G). These data suggest that HPV-16 E6 contributes to elevation of GRHL2 and FoxM1B, while HPV-16 E7 does not.
Figure 1.
Human papillomavirus type 16 (HPV-16) E6 induces GRHL2 and FoxM1B expression. (A) Western blotting was performed for GRHL2 and FoxM1B using whole-cell extracts (WCEs) from normal human oral keratinocytes (NHOKs) with different population doubling (PD15 and PD30), HOK-16B cultured in keratinocyte growth medium (KGM) or Dulbecco’s modified Eagle’s medium, and oral squamous cell carcinoma (OSCC) cell lines (HOK-16B/BaP-T, SCC4, and SCC15). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as loading control. (B) GRHL2, FoxM1B, and hTERT messenger RNA (mRNA) levels were determined by quantitative reverse transcription polymerase chain reaction (qRT-PCR) in NHOKs infected by HPV-16 E6 or HPV-16 E7. Bars indicate standard deviation and * indicates statistical significance (P < 0.05), compared with the mean values of the control group (LXSN). (C) Western blotting was performed for GRHL2, FoxM1B, and Cyclin A in 2 strains of NHOKs expressing HPV-16 E6 or HPV-16 E7. (D) GRHL2, FoxM1B, and Cyclin A protein levels in tonsillar epithelial cells (TECs) and FaDu cells infected by HPV-16 E6, HPV-16 E7, or 16E6/E7. (E, F) NHOK cells were infected with HPV-16 virus, and then qRT-PCR was performed to detect gene expression of GRHL2 and FoxM1B and Western blotting was executed for determining the protein levels of GRHL2, FoxM1B, and its target proteins, Cyclin A and Cyclin B1. P53 and P21WAF1 were used as markers of HPV-16 virus infection. (G) HOK-16B/BaP-T cells were infected with HPV-16 E6 small interfering RNA to knockdown HPV-16 E6, and then Western blot was performed to determine protein levels of GRHL2, FoxM1B, and GRHL2 target proteins K14 and FN. GAPDH was used as loading control. Each experiment was repeated 3 times.
HPV-16 E6 Upregulates GRHL2 and FoxM1B Expression in Transgenic Mice and HPV+ OSCC Cells
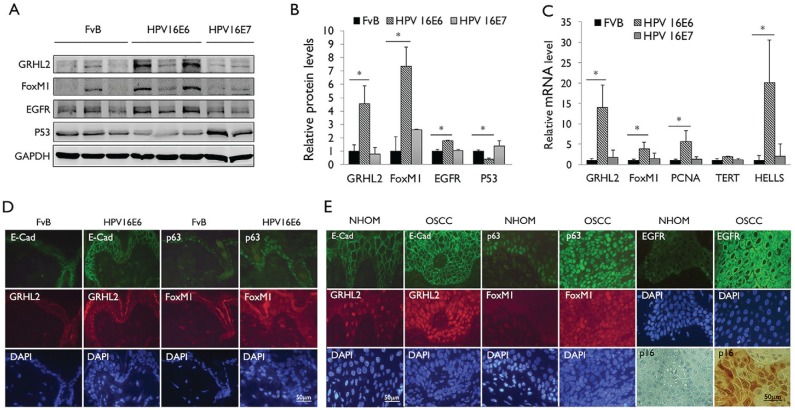
We confirmed the induction of GRHL2 and FoxM1B expression by HPV-16 E6 using K14–HPV-16 E6 or K14–HPV-16 E7 transgenic mice, which allow for expression of HPV-16 E6 or HPV-16 E7 in an epithelial tissue–specific manner (Herber et al. 1996; Song et al. 1999). GRHL2 and FoxM1B protein levels were notably induced in the epidermal cells of HPV-16 E6 mice but not HPV-16 E7 mice (Fig. 2A, B). p53 expression level was reduced in HPV-16 E6 transgenic mice, confirming the active expression of E6. In accordance to the protein levels, the messenger RNA (mRNA) expressions of GRHL2, FoxM1B, and the downstream genes (e.g., PCNA and HELLS) were significantly induced in the HPV-16 E6 transgenic mice, while no congruent changes were noted in the HPV-16 E7 mice (Fig. 2C). With in situ IFS for GRHL2 and FoxM1B, we confirmed induced levels of these proteins, as well as their targets, E-cadherin and p63, in the epidermis of HPV-16 E6 mice compared with the FvB control mice (Fig. 2D). FoxM1B and GRHL2 protein levels were also determined in situ in archived samples of normal human oral mucosa (NHOM) and HPV+ OSCCs by IFS. FoxM1B was almost undetectable in NHOM, while the OSCC samples showed distinct GRHL2 and FoxM1B staining, as well as enhanced E-cadherin and p63 staining (Fig. 2E). p16 immunoreactivity was shown as a marker for HPV infection (Liu et al. 2015). Collectively, the above data indicate that HPV-16 E6 induces aberrant expression of GRHL2 and FoxM1B in epithelial cells in vivo, which may alter the cellular proliferation and differentiation, as well as promote carcinogenesis.
Figure 2.
Human papillomavirus type 16 (HPV-16) E6 induces GRHL2 and FoxM1B expression in epithelial tissue in vivo. (A) Western blotting was performed with epidermal tissues isolated from wild-type mice (FvB) and HPV-16 E6 or HPV-16 E7 transgenic mice for GRHL2, FoxM1B, EGFR, and p53. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as loading control. (B) Western blotting signals were quantitated by densitometric analysis and plotted with the mean values for wild-type mice (FvB). Bar indicates mean (SD), and * indicates P < 0.05. (C) Quantitative reverse transcription polymerase chain reaction (qRT-PCR) was performed with total RNAs isolated from the epidermis of wild-type mice (FvB) and HPV-16 E6 or HPV-16 E7 mice. Experiments were performed in triplicates. Bars indicate standard deviation. * indicates statistically significant change (P < 0.05) compared with wild-type (WT) mice. (D) Skin tissues from HPV-16 E6 transgenic mice were stained for GRHL2, E-cadherin, FoxM1B, and p63 by immunofluorescence staining (IFS). Elevated GRHL2 and FoxM1B were noted in the epidermis of HPV-16 E6 transgenic mice. (E) Immunofluorescence staining was performed with normal human oral mucosa (NHOM) and HPV+ oral squamous cell carcinoma (OSCC) tissues for GRHL2, E-cadherin, p63, and FoxM1B antibody. Numbers of patients with HPV+ OSCC or healthy donors for NHOM were 10. Representative staining results are shown. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI).
GRHL2 Regulates FoxM1B Expression through Activating Its Cognate Gene Promoter
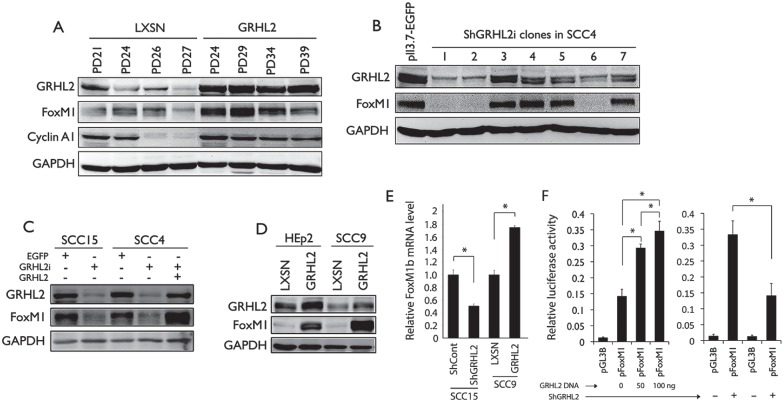
In the next experiments, we determined whether GRHL2 directly regulated FoxM1B expression. GRHL2 overexpression via retroviral vector led to increased expression of FoxM1B and Cyclin A, allowing for extended replicative capacity in normal human keratinocytes (NHK) (Fig. 3A). Positive correlation was noted between GRHL2 and FoxM1B levels among the SCC4 clones in which endogenous GRHL2 was knocked down to various levels (Fig. 3B). Likewise, depletion of endogenous GRHL2 in SCC4 and SCC15 cells led to FoxM1B suppression (Fig. 3C, E), while reexpression of GRHL2 in SCC4 with GRHL2 knockdown restored the FoxM1B level (Fig. 3C). GRHL2 overexpression in HEp2 and SCC9 cells led to strong induction of FoxM1B (Fig. 3D, E), confirming the regulatory role of GRHL2 on FoxM1B expression. We previously showed direct binding of GRHL2 on the proximal promoter region of FoxM1B (Chen et al. 2012). Using FoxM1B promoter luciferase construct, we showed that GRHL2 increased the FoxM1B promoter activity in a dose-dependent manner (Fig. 3F); conversely, when GRHL2 was knocked down in SCC4, there was concomitant reduction of the FoxM1B promoter activity.
Figure 3.
GRHL2 is an upstream regulator of FoxM1B. (A) Normal human oral keratinocytes (NHOKs) stably transduced with retrovirus plasmid expressing GRHL2 and the empty vector (LXSN) at various population doubling (PD) were assessed for change in protein expression of GRHL2, FoxM1B, and Cyclin A1. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was using as the loading control. (B) GRHL2 expression was knocked down using lentiviral vector (LV-ShGRHL2) in SCC4 followed by single-cell colony selection. As control, SCC4 was infected with lentiviral vector expressing EGFP (LV-EGFP). GRHL2 and FoxM1B levels were examined in different GRHL2 knockdown clones. (C) GRHL2 was knocked down in SCC15 and SCC4 and led to loss of FoxM1B. When GRHL2 was reexpressed in SCC4 through retroviral infection with LXSN-GRHL2 vector, FoxM1B expression was restored. (D) GRHL2 was overexpressed in HEp2 and SCC9 cells, both of which express low level of endogenous GRHL2. Ectopic expression of GRHL2 in these cells led to enhanced FoxM1B expression. (E) GRHL2 was knocked down in SCC15 or overexpressed in SCC9, and quantitative reverse transcription polymerase chain reaction (qRT-PCR) was performed for FoxM1B messenger RNA (mRNA) levels. (F) In the left panel, the FoxM1B promoter assay was performed in 293 cells with various concentrations of luciferase reporter plasmids, in which the luciferase reporter gene is driven by FoxM1B promoter (pFoxM1). In the right panel, FoxM1B promoter assay was performed in SCC4 cells with or without GRHL2 knockdown, using the luciferase reporter contsruct. * indicates statistical significance (P < 0.05). Empty plasmid, pGL3B, served as the negative control. Experiments were performed in triplicates.
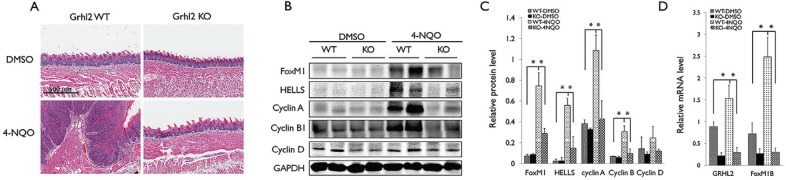
To evaluate the role of GRHL2 in FoxM1B expression in vivo, we generated Grhl2 cKO mice by crossing Grhl2 floxed (fl/fl) with K14-CreERT mice, which led to Grhl2 KO in epithelial tissues. Grhl2 KO in oral epithelium did not elicit gross histological alterations in tongue (Fig. 4A). We then induced tongue tumor development by chronic exposure to 4-NQO using the established protocol (Squarize et al. 2013). 4-NQO exposure led to rampant tumor formation in the wild-type mice, while tumor development was absent in Grhl2 KO mice (Fig. 4A). Western blotting and qRT-PCR revealed strong induction of FoxM1B and its downstream targets (e.g., HELLS, Cyclin A, and Cyclin B1) in the tumor-bearing tongue of the wild-type mice compared to the unexposed control mice (Fig. 4B–D). Also, Grhl2 KO prohibited induction of FoxM1B and its target genes when exposed to 4-NQO. These data indicate that GRHL2 is required for FoxM1B induction and chemical-induced oral carcinogenesis.
Figure 4.
Loss of GRHL2 suppresses FoxM1B expression in 4-nitroquinolin 1-oxide (4-NQO)–induced tongue tumors. (A) Morphological change of tongues from Grhl2 wild-type (WT) and knockout (KO) mice treated without or with 4-NQO (30 µg/mL) for 16 wk to induce oral cancer formation. (B) Western blotting was performed with tongue tissues from Grhl2 WT or KO mice for FoxM1B, Cyclin A, Cyclin B1, and Cyclin D with or without 4-NQO exposure for 16 wk. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as the loading control. (C) Western blotting signals were quantitated by densitometric analysis and plotted with the mean values for Grhl2 WT or KO mice with or without 4-NQO exposures. (D) Quantitative reverse transcription polymerase chain reaction (qRT-PCR) was performed with total RNA isolated tongue tissues. Data were derived from 3 independent experiments, and qRT-PCR assays were performed in triplicates. Error bars indicate standard deviation of the mean; *P < 0.05.
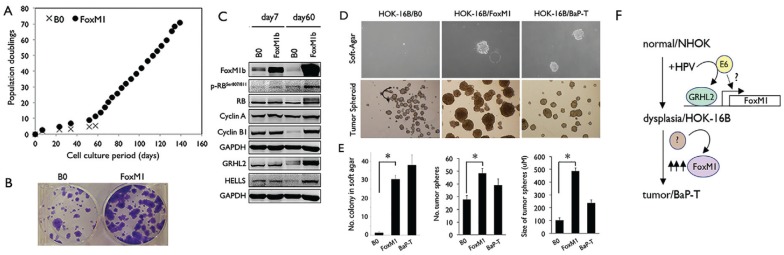
FoxM1B Induces Transformed Phenotype of HPV-Harboring HOKs
To assess the role of FoxM1B in tumorigenic conversion of HPV-containing HOKs, we overexpressed FoxM1B in HOK-16B cells through retroviral vector to generate HOK-16B/FoxM1B or control cells, HOK-16B/B0. Resistance to physiological levels of Ca++ is a transformed phenotype of keratinocytes (Park et al. 1995). After culturing in medium with a physiological level of Ca++ (1.5 mM) for 60 d, HOK-16B/B0 cells underwent terminal differentiation and senescence, while HOK-16B/FoxM1B continued to proliferate and maintained the undifferentiated morphology (Fig. 5A). HOK-16B/FoxM1B cells demonstrated higher colony formation ability, and the cells expressed a higher level of FoxM1B target genes, for example, Cyclin B1, Cyclin A, and HELLS (Fig. 5B, C). HOK-16B/FoxM1B cells exhibited higher capabilities of anchorage-independent growth in soft agar and tumor spheroid formation (Fig. 5D, E), which are transformed phenotypes (Clevers 2011). FoxM1B is also required for maintaining the tumorigenic ability of SCC cells harboring HPV DNA, for example, HOK-16B/BaP-T and Siha (Park et al. 1995; Adler et al. 1997). Targeting FoxM1B in these cells by FoxM1B shRNA or exposure to Siomycin, a FoxM1B inhibitor, led to suppression of FoxM1B target proteins and induction of growth inhibitors, for example, p53, p21, and p27 (Appendix Fig.). Furthermore, loss of FoxM1B diminished the transformed phenotype, including cell proliferation, migration, and tumor spheroid–forming abilities. Collectively, the above data suggest that FoxM1B plays crucial role in HPV-associated oral carcinogenesis, with its induction by HPV-16 E6 partly mediated through GRHL2 activity, yet further elevation of FoxM1B may lead to tumorigenic conversion of HPV-harboring cells.
Figure 5.
FoxM1B promotes malignant transformation of human oral keratinocytes (HOKs) harboring human papillomavirus type 16 (HPV-16). (A) HOK-16B cells were infected with pBabe empty vector (B0) or pBabe-FoxM1B and then selected with puromycin (1 µg/mL). The puromycin-resistant cells were exposed to medium with physiologic Ca++ level (1.5 mM) and maintained in serial subcultures. Their replication kinetics was plotted against time in culture. (B) Colony formation was performed using HOK-16B cells infected with pBabe empty vector (B0) or pBabe-FoxM1B (200 cells per well). (C) Western blotting was performed for FoxM1B, Cyclin A, Cyclin B1, HELLS, GRHL2, and p-RBser807/811 in HOK-16B cells infected with empty vector (B0) or pBabe-FoxM1B exposed to medium with physiologic Ca++ level for 7 d or 60 d. (D) Anchorage-independent growth (AIG) assay was performed with HOK-16B/B0, HOK-16B/FoxM1B, and HOK-16B/BaP-T in soft agar plates. Representative colony sizes were shown after 14 d postseeding in the upper panel. In the bottom panel, tumor spheroid assay was performed with HOK-16B/B0, HOK-16B/FoxM1B, and HOK-16B/BaP-T cells in suspension culture for 7 d. Growth of colonies in soft agar and tumor spheroids in ultra-low attachment plate was visualized under light microscopy. (E) Quantification of the number of colonies formed in the soft agar and the number and size of tumor spheroids formed by HOK-16B/B0, HOK-16B/FoxM1B, and HOK-16B/BaP-T cells in an ultra-low attachment plate. (F) Multistep oral carcinogenesis involves stepwise conversion of normal human oral keratinocytes (NHOKs), immortalized and nontumorigenic cells (HOK-16B), and tumorigenic cells (HOK-16B/BaP-T). HPV infection leads to FoxM1 induction, in part, through GRHL2. During tumorigenic conversion, additional unknown factors may lead to further elevation of FoxM1B, which is required to maintain the transformed phenotype.
Discussion
The current study is focused on the mechanisms that regulate propagation of neoplastic cells harboring the HPV genome rather than the oncogenic mechanism immediately succeeding the cellular entry of HPV. Our data showed that FoxM1B and GRHL2 expression was induced by HPV-16 infection via the E6 oncoprotein. The role of the E6 protein on the regulation of FoxM1B was demonstrated in cultured HOKs by retroviral gene transduction or infection with HPV-16 virus particles. Also, HPV-16 E6 transgenic mice showed elevation of FoxM1B and GRHL2 levels in vivo, while HPV-16 E7 transgenic mice did not. Since GRHL2 was strongly induced by HPV-16 E6, it suggested a role of GRHL2 in the regulation of FoxM1B expression in cells harboring HPV-16 in the progression of the disease. Further study is required to understand the role of the GRHL2/FoxM1B functional axis in HPV-mediated oncogenesis in the oropharynx (e.g., tonsils) in the contextual relationship with surrounding lymphoid tissues. Furthermore, our data indicated that GRHL2 bound to the FoxM1B proximal promoter region to regulate the gene promoter activity. Therefore, GRHL2 appears to regulate FoxM1B gene expression directly by promoter binding and indirectly through other transcription regulators, such as NKX2-1 (Chen et al. 2014).
It appears that GRHL2 is required for FoxM1B activation during OPC development. This was evident in the mouse tongue cancer model exposed to 4-NQO, in which GRHL2 conditional KO abolished FoxM1B expression in tongue tissues with chronic exposure to 4-NQO, resulting in the absence of tumor formation. The 4-NQO–induced tongue cancer model is well established with the standard protocol. Although it is a synthetic carcinogen and not a naturally occurring agent for the oral cavity, the oral lesions induced by 4-NQO and those in OSCCs in humans produce very similar molecular characteristics, for example, elevation of p-Erk1/2, Cox-2, RARβ2, and p16Ink4A and MCM7 for HPV+ OPCs (Osei-Sarfo et al. 2013; Yang et al. 2013). Likewise, our previous studies showed strong expression of GRHL2 in human OSCCs and that knockdown of GRHL2 abolished the epithelial tumor phenotype (Kang et al. 2009; Chen et al. 2016). The current study provided more evidence to support the role of GRHL2 in oral carcinogenesis through its regulation of FoxM1B.
Although HPV infection is a risk factor for OPCs, it alone is not sufficient for tumorigenic conversion of normal oral epithelium (Kang and Park 2001). This has been demonstrated in our multistep oral carcinogenesis model, in which stable transduction of the HPV genome led to cellular immortalization of NHOKs, yet the cells remained nontumorigenic (Park et al. 1991). Tumorigenic conversion of HPV-harboring HOKs required chronic genotoxic challenge with chemical carcinogens for at least 6 mo (Park et al. 1995). However, the mechanism by which chronic genotoxicity led to tumorigenic conversion in HPV-harboring HOKs remained unknown. Our data showed that FoxM1B overexpression in HOK-16B cells induced anchorage-independent growth (AIG) in soft agar, indicating the carcinogenic effects of FoxM1B. It remains a limitation of this experiment that the level of FoxM1 induction in the transduced cells is beyond the physiological level. However, the data also supported the dosage effect of FoxM1B because FoxM1B level was already induced in HOK-16B cells by E6, yet the cells did not acquire the transformed phenotype until exogenous FoxM1B was overexpressed using the retroviral vector. This FoxM1B dosage effect was also reflected in the multistep oral carcinogenesis model, in which the FoxM1B level was further increased in the tumorigenic cells compared to that of HOK-16B, suggesting that tumorigenic conversion was associated with further elevation of FoxM1B (Fig. 5F). Our current data indicate the transcriptional role of GRHL2 on FoxM1B promoter activation, which may be the underlying mechanism of FoxM1B induction by E6. However, additional factors (other than GRHL2) are likely involved in further activation of FoxM1B during tumorigenic conversion as the effect of GRHL2 on FoxM1B promoter was modest at 2- to 3-fold (Fig. 3F). When exogenous FoxM1B was transduced in HOK-16B cells, the cells gained resistance to the physiologic Ca++ level (1.5 mM), which is a transformed phenotype (Park et al. 1995), only after 60 d in culture. Also during this time, there was a sharp increase in the intracellular level of FoxM1B in the transduced cells (Fig. 5C). Thus, it is likely that there was a selection process during the serial cultures for the cells that expressed a higher level of FoxM1B, resulting in a tumor phenotype in those cells at the late passage cultures. Induction of FoxM1B by HPV-16 E6 is clearly a procarcinogenic effect, but additional carcinogenic events leading to further elevation of FoxM1B level are required for tumorigenicity.
The known FoxM1B target genes (e.g., Cyclin A, Cyclin B1, and HELLS) are primarily involved in control of DNA replication and cell division (Leung et al. 2001; Barsotti and Prives 2009; Kelleher and O’Sullivan 2016; Chen et al. 2017), in line with its role in tumor cell proliferation. FoxM1B is repressed by p53, in part through p21/WAF1, allowing the cells to halt cell proliferation (Barsotti and Prives 2009). This may be another mechanism by which HPV-16 E6 led to FoxM1B induction since the E6 protein suppresses p53 expression. Our data also demonstrated an inverse correlation between FoxM1B and p53 levels in HOK-16B/BaP-T cells exposed to Siomycin, a FoxM1B inhibitor (Appendix Fig.); within 1 d of Siomycin treatment, FoxM1B level precipitously decreased, while p53 level was strongly induced after 2 d. This raises a possibility that FoxM1B may reciprocally regulate the p53 level, since the loss of FoxM1B preceded p53 induction in cells exposed to Siomycin. A recent study also showed that loss of FoxM1B by siRNA or thiostrepton induced p53 expression and its downstream apoptotic program in nasopharyngeal carcinomas (NPCs) (Jiang et al. 2014). Thus, the oncogenic effect of FoxM1B in OSCC may be linked to its regulation of p53 function, and further study will be needed to elucidate this effect.
Author Contributions
W. Chen, contributed to conception, design, data acquisition, analysis, and interpretation, drafted and critically revised the manuscript; T. Shimane, contributed to conception and data acquisition, critically revised the manuscript; K. Satoshi, A. Alshaikh, S.Y. Kim, S.H. Chung, K. Walentin, K.M. Schmidt-Ott, contributed to contributed to conception, critically revised the manuscript; R.H. Kim, contributed to conception and data interpretation, critically revised the manuscript; K.H. Shin, N.H. Park, contributed to data interpretation, critically revised the manuscript; M.K. Kang, contributed to conception, design, data analysis, and interpretation, drafted and critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Supplementary Material
Footnotes
A supplemental appendix to this article is available online.
This study was supported in part by the grants (R56DE024593, R03DE024259) from the National Institute of Dental and Craniofacial Research/National Institutes of Health and the UC Cancer Research Coordinating Committee. M.K.K. is also supported by the Jack A. Weichman Endowed Fund. K.M.S.-O. is supported by the German Research Foundation (Deutsche Forschungsgemeinschaft) and the Urological Research Foundation (Stiftung Urologische Forschung).
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
ORCID iD: K.M. Schmidt-Ott  https://orcid.org/0000-0002-7700-7142
https://orcid.org/0000-0002-7700-7142
References
- Adler K, Erickson T, Bobrow M. 1997. High sensitivity detection of HPV-16 in SiHa and CaSki cells utilizing FISH enhanced by TSA. Histochem Cell Biol. 108(4–5):321–324. [DOI] [PubMed] [Google Scholar]
- Barger CJ, Zhang W, Hillman J, Stablewski AB, Higgins MJ, Vanderhyden BC, Odunsi K, Karpf AR. 2015. Genetic determinants of FOXM1 overexpression in epithelial ovarian cancer and functional contribution to cell cycle progression. Oncotarget. 6(29):27613–27627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barsotti AM, Prives C. 2009. Pro-proliferative FoxM1 is a target of p53-mediated repression. Oncogene. 28(48):4295–4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan DW, Yu SY, Chiu PM, Yao KM, Liu VW, Cheung AN, Ngan HY. 2008. Over-expression of FOXM1 transcription factor is associated with cervical cancer progression and pathogenesis. J Pathol. 215(3):245–252. [DOI] [PubMed] [Google Scholar]
- Chen PM, Cheng YW, Wang YC, Wu TC, Chen CY, Lee H. 2014. Up-regulation of FOXM1 by E6 oncoprotein through the MZF1/NKX2-1 axis is required for human papillomavirus–associated tumorigenesis. Neoplasia. 16(11):961–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Dong Q, Shin KH, Kim RH, Oh JE, Park NH, Kang MK. 2010. Grainyhead-like 2 enhances the human telomerase reverse transcriptase gene expression by inhibiting DNA methylation at the 5′-CpG island in normal human keratinocytes. J Biol Chem. 285(52):40852–40863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Xiao Liu Z, Oh JE, Shin KH, Kim RH, Jiang M, Park NH, Kang MK. 2012. Grainyhead-like 2 (GRHL2) inhibits keratinocyte differentiation through epigenetic mechanism. Cell Death Dis. 3:e450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Yi JK, Shimane T, Mehrazarin S, Lin YL, Shin KH, Kim RH, Park NH, Kang MK. 2016. Grainyhead-like 2 regulates epithelial plasticity and stemness in oral cancer cells. Carcinogenesis. 37(5):500–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Liu Y, Ni H, Ding C, Zhang X, Zhang Z. 2017. FoxM1 overexpression promotes cell proliferation and migration and inhibits apoptosis in hypopharyngeal squamous cell carcinoma resulting in poor clinical prognosis. Int J Oncol. 51(4):1045–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H. 2011. The cancer stem cell: premises, promises and challenges. Nat Med. 17(3):313–319. [DOI] [PubMed] [Google Scholar]
- Czerninski R, Amornphimoltham P, Patel V, Molinolo AA, Gutkind JS. 2009. Targeting mammalian target of rapamycin by rapamycin prevents tumor progression in an oral-specific chemical carcinogenesis model. Cancer Prev Res (Phila). 2(1):27–36. [DOI] [PubMed] [Google Scholar]
- Dompe N, Rivers CS, Li L, Cordes S, Schwickart M, Punnoose EA, Amler L, Seshagiri S, Tang J, Modrusan Z, et al. 2011. A whole-genome RNAi screen identifies an 8q22 gene cluster that inhibits death receptor-mediated apoptosis. Proc Natl Acad Sci U S A. 108(43):E943–E951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemenetzidis E, Bose A, Riaz AM, Chaplin T, Young BD, Ali M, Sugden D, Thurlow JK, Cheong SC, Teo SH, et al. 2009. FOXM1 upregulation is an early event in human squamous cell carcinoma and it is enhanced by nicotine during malignant transformation. PLoS One. 4(3):e4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herber R, Liem A, Pitot H, Lambert PF. 1996. Squamous epithelial hyperplasia and carcinoma in mice transgenic for the human papillomavirus type 16 E7 oncogene. J Virol. 70(3):1873–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hocking JS, Stein A, Conway EL, Regan D, Grulich A, Law M, Brotherton JM. 2011. Head and neck cancer in Australia between 1982 and 2005 show increasing incidence of potentially HPV-associated oropharyngeal cancers. Br J Cancer. 104(5):886–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Wang P, Chen L, Chen H. 2014. Down-regulation of FoxM1 by thiostrepton or small interfering RNA inhibits proliferation, transformation ability and angiogenesis, and induces apoptosis of nasopharyngeal carcinoma cells. Int J Clin Exp Pathol. 7(9):5450–5460. [PMC free article] [PubMed] [Google Scholar]
- Kang MK, Bibb C, Baluda MA, Rey O, Park NH. 2000. In vitro replication and differentiation of normal human oral keratinocytes. Exp Cell Res. 258(2):288–297. [DOI] [PubMed] [Google Scholar]
- Kang MK, Kim RH, Kim SJ, Yip FK, Shin KH, Dimri GP, Christensen R, Han T, Park NH. 2007. Elevated Bmi-1 expression is associated with dysplastic cell transformation during oral carcinogenesis and is required for cancer cell replication and survival. Br J Cancer. 96(1):126–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang MK, Park NH. 2001. Conversion of normal to malignant phenotype: telomere shortening, telomerase activation, and genomic instability during immortalization of human oral keratinocytes. Crit Rev Oral Biol Med. 12(1):38–54. [DOI] [PubMed] [Google Scholar]
- Kang X, Chen W, Kim RH, Kang MK, Park NH. 2009. Regulation of the hTERT promoter activity by MSH2, the hnRNPs K and D, and GRHL2 in human oral squamous cell carcinoma cells. Oncogene. 28(4):565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher FC, O’Sullivan H. 2016. Foxm1 in sarcoma: role in cell cycle, pluripotency genes and stem cell pathways. Oncotarget. 7(27):42792–42804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung TW, Lin SS, Tsang AC, Tong CS, Ching JC, Leung WY, Gimlich R, Wong GG, Yao KM. 2001. Over-expression of FoxM1 stimulates cyclin B1 expression. FEBS Lett. 507(1):59–66. [DOI] [PubMed] [Google Scholar]
- Liu SZ, Zandberg DP, Schumaker LM, Papadimitriou JC, Cullen KJ. 2015. Correlation of p16 expression and HPV type with survival in oropharyngeal squamous cell cancer. Oral Oncol. 51(9):862–869. [DOI] [PubMed] [Google Scholar]
- Osei-Sarfo K, Tang XH, Urvalek AM, Scognamiglio T, Gudas LJ. 2013. The molecular features of tongue epithelium treated with the carcinogen 4-nitroquinoline-1-oxide and alcohol as a model for HNSCC. Carcinogenesis. 34(11):2673–2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park NH, Gujuluva CN, Baek JH, Cherrick HM, Shin KH, Min BM. 1995. Combined oral carcinogenicity of HPV-16 and benzo(a)pyrene: an in vitro multistep carcinogenesis model. Oncogene. 10(11):2145–2153. [PubMed] [Google Scholar]
- Park NH, Min BM, Li SL, Huang MZ, Cherick HM, Doniger J. 1991. Immortalization of normal human oral keratinocytes with type 16 human papillomavirus. Carcinogenesis. 12(9):1627–1631. [DOI] [PubMed] [Google Scholar]
- Ringström E, Peters E, Hasegawa M, Posner M, Liu M, Kelsey KT. 2002. Human papillomavirus type 16 and squamous cell carcinoma of the head and neck. Clin Cancer Res. 8(10):3187–3192. [PubMed] [Google Scholar]
- Schowalter RM, Pastrana DV, Buck CB. 2011. Glycosaminoglycans and sialylated glycans sequentially facilitate Merkel cell polyomavirus infectious entry. PLoS Pathog. 7(7):e1002161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Pitot HC, Lambert PF. 1999. The human papillomavirus type 16 E6 gene alone is sufficient to induce carcinomas in transgenic animals. J Virol. 73(7):5887–5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squarize CH, Castilho RM, Abrahao AC, Molinolo A, Lingen MW, Gutkind JS. 2013. PTEN deficiency contributes to the development and progression of head and neck cancer. Neoplasia. 15(5):461–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stramer B, Martin P. 2005. Cell biology: master regulators of sealing and healing. Curr Biol. 15(11):R425–R427. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Kanai F, Tada M, Tateishi R, Sanada M, Nannya Y, Ohta M, Asaoka Y, Seto M, Shiina S, et al. 2008. Gain of GRHL2 is associated with early recurrence of hepatocellular carcinoma. J Hepatol. 49(5):746–757. [DOI] [PubMed] [Google Scholar]
- Varma S, Cao Y, Tagne JB, Lakshminarayanan M, Li J, Friedman TB, Morell RJ, Warburton D, Kotton DN, Ramirez MI. 2012. The transcription factors Grainyhead-like 2 and NK2-homeobox 1 form a regulatory loop that coordinates lung epithelial cell morphogenesis and differentiation. J Biol Chem. 287(44):37282–37295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walentin K, Hinze C, Werth M, Haase N, Varma S, Morell R, Aue A, Potschke E, Warburton D, Qiu A, et al. 2015. A Grhl2-dependent gene network controls trophoblast branching morphogenesis. Development. 142(6):1125–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner S, Frey S, Riethdorf S, Schulze C, Alawi M, Kling L, Vafaizadeh V, Sauter G, Terracciano L, Schumacher U, et al. 2013. Dual roles of the transcription factor grainyhead-like 2 (GRHL2) in breast cancer. J Biol Chem. 288(32):22993–23008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang X, Deng Z, Zhuang X, Ju S, Mu J, Jiang H, Zhang L, Yan J, Miller D, Zhang HG. 2012. Grhl2 determines the epithelial phenotype of breast cancers and promotes tumor progression. PLoS One. 7(12):e50781 Erratum in: PLoS One. 2013;8(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Guan B, Men T, Fujimoto J, Xu X. 2013. Comparable molecular alterations in 4-nitroquinoline 1-oxide-induced oral and esophageal cancer in mice and in human esophageal cancer, associated with poor prognosis of patients. In Vivo. 27(4):473–484. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.