Abstract
The repeatability observed across cancers arising in the same tissue can help understand the evolutionary process of tumour initiation. We recently developed a framework to quantify the local malignant adaptation of genetic clones in tissue-specific environments. In this Commentary, we argue that such a 1-dimensional model can be improved by separating its 2 components to obtain a dual scale: local adaptation, dictating proliferation rates in the local environment, and malignant adaptation, influencing the likelihood that a clone becomes cancerous and invasive. Such a change could strengthen our understanding of the population dynamics underlying cancer initiation and assess different evolutionary scenarios.
Keywords: Cancer, genetics, evolution, somatic mutations, bioinformatics
Comment on: Tokutomi N, Moyret-Lalle C, Puisieux A, Sugano S, Martinez P. Quantifying local malignant adaptation in tissue-specific evolutionary trajectories by harnessing cancer’s repeatability at the genetic level. Evol Appl. 2019;12(5):1062-1075. doi:10.1111/eva.12781. PubMed PMID: 31080515. PubMed Central PMCID: PMC6503823. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6503823/.
We recently developed a framework to measure the local malignant adaptation (LMA) of genetic clones in 9 tissues, based on the repeatability of genomic alterations in these cancers.1 Such a framework can help assess the malignant potential of genetic clones and predict their future evolution. This methodology used a single scale to reflect how likely a combination of alterations was to give rise to a cancer in a specific environment (Figure 1). Here, we propose that this methodology can be enhanced by separating the local and malignant dimensions of the adaptation process driving tumourigenesis.
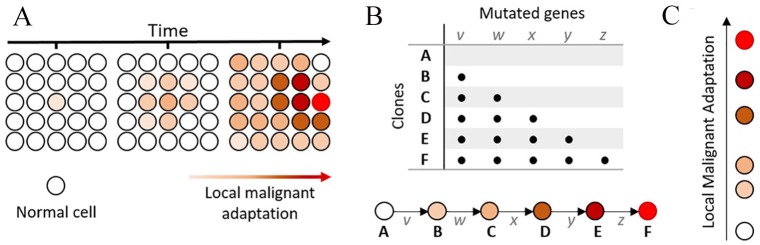
Figure 1.
Local malignant adaptation (LMA): (A) growth of a finite population over time. Cells acquire alterations that similarly affect their adaptation and their probability to become cancerous. (B) Relationship between clones (A to F) and acquisition of diver alterations (v to z) over time. (C) Single-scale LMA.
Cancer is a harmful outcome of the somatic evolutionary process at work in multicellular organisms.2 The human adult organism consists of trillions of cells obtained via successive divisions, each of its tissues being maintained by further divisions of specific stem and stem-like cells.3 The acquisition of (epi)genetic alterations through cell replication generates diversity in these cellular populations, as it does for unicellular organisms. Some of these alterations can then allow cells to escape and/or avoid the organism’s control mechanisms regulating homeostasis, eventually giving rise to highly proliferative and invasive cancer cells.
This evolutionary process of cancer development repeats itself on a tissue-specific basis.1 We harnessed this repeatability to quantify the LMA of a ‘clone’ (a genotype based on the presence of driver alterations) to its tissue-specific environment. To factor in the contribution of each driver alteration and their interplay, our framework was centred on a single assumption: despite different evolutionary trajectories, all clones that gave rise to cancer were similarly adapted to their local tissue. We optimised our LMA calculation based on this assumption, so as to limit the discrepancy between the scores obtained for samples in each of 9 tumour types, using The Cancer Genome Atlas (TCGA) data. Local malignant adaptation score calculations included 3 factors, whose combination was therefore optimised to minimise standard deviation across samples from the same tumour type:
Selective advantage (observed number of alterations compared to expectations);
Self-sufficiency (number of other alterations also present compared to other drivers);
Epistatic interactions (mutations co-occurrence compared to expectations from their respective frequencies).
We used this framework to investigate the adaptation of pre-malignant and metastatic lesions. We reported that pre-malignant skin and colorectal lesions were specifically adapted to their local environment, yet insufficiently to reach a fully invasive phenotype. Our analyses further suggested that genetics helped clones adapt to a tissue of origin, but seemed to have little influence on the adaptation to specific metastatic locations. Finally, we found that combining our framework to network analyses could recapitulate tissue-specific features of tumour initiation.
Our approach focused on clonal mutations, ie, those present in the original transformed cell and inherited by all subsequent generations. Alteration recurrence and co-occurrence patterns in these first cancer cells shed light on the ‘hidden’ dynamics of the evolutionary processes of which they are outcomes. The repeatability observed in these patterns is, however, only informative on the genetic makeup of the clones that did progress to cancer. The genomic alterations in untransformed cells undergoing similar evolutionary pressures in the same individual are still poorly characterised.
There is now evidence that alterations giving rise to cancer are not necessarily the ones providing the highest selective advantage in the normal tissue, at least in the oesophagus.4,5 This highlights that proliferation and colonisation of normal tissue are not necessarily linked to subsequent malignancy. The prevalence of NOTCH1 mutations in the normal epithelium, compared to their scarcity in cancer clones, further suggests a potential protective effect on cancer development despite their proliferative advantage.
To improve our framework, aiming to exploit the repeatability of cancer genotypes, the local and malignant dimensions of adaptation could thus be separated (Figure 2). By uncoupling the local growth advantage that cells acquire from a given alteration from its influence on malignant development, we could better model evolutionary dynamics and simulate different evolutionary trajectories. For instance, the relevance of the following scenarios could be assessed in different tissues:
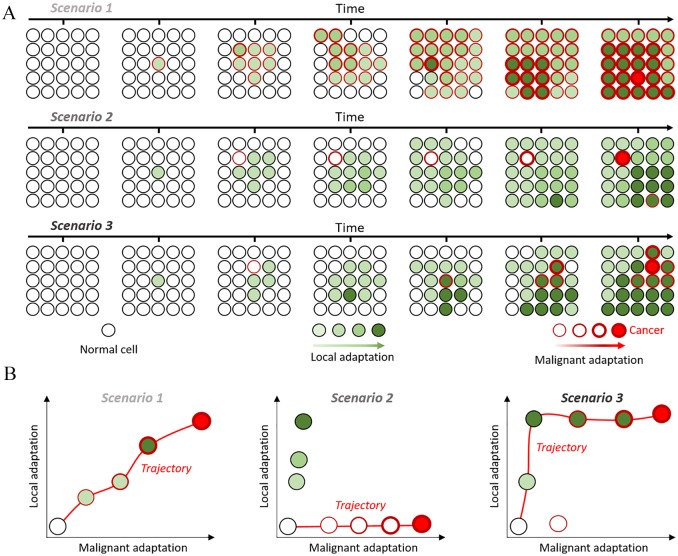
Figure 2.
Uncoupling local and malignant adaptation: (A) growth of a finite population over time under 3 different scenarios. Cells acquire alterations that differently affect their adaptation (green fill) and probability to become cancerous (red outline). (B) Distribution of local and malignant adaptation scores for each clone and the trajectories leading from normal to cancer in red.
Malignant potential linearly follows local adaptation, with the most proliferative cells being most likely to become cancerous (Figure 2, Scenario 1);
Malignant potential and local adaptation are completely uncoupled and cancer develops from rare, low-proliferating clones (Figure 2, Scenario 2);
There is a dynamic interplay between both dimensions, implying that alterations increasing malignant potential occurring in clones with a proliferative advantage are more likely to lead to cancer (Figure 2, Scenario 3). In this scenario and the previous one, clones with malignant potential but low proliferation can be outcompeted by more adapted benign ones.
The NOTCH1 example in the oesophagus for instance appears to fit the second scenario, but it would be interesting to understand the contribution of other alterations and the dynamics of other tissues. Providing reliable answers will, however, require more data on how somatic evolution impacts the genetics of normal cells, on a tissue-specific basis. We already know the most relevant cancer drivers for common tumour types and their frequencies in patient populations. Frequencies of somatic alterations in normal tissue can furthermore allow to assess their selective advantage in homeostatic conditions.6 Discrepancies between pre- and post-cancer frequencies can then help tease out how each alteration impacts the local and malignant aspect of tumour initiation. These data-derived parameters can then serve to accurately simulate the emergence of cancer genotypes within initially normal populations, allowing for heterogeneous growth rates and malignant potential among different clones that each acquire further somatic alterations over time.
We recently provided a framework to make use of the repeated patterns observed in the somatic evolution process underlying cancer formation. In this Commentary, we propose that this framework could be enhanced by splitting the single scale used to measure the adaptation of genetic clones to tissue-specific environments into 2 orthogonal dimensions: local adaptation and malignant adaptation. This endeavour, however, requires additional data on the genetics of somatic evolution in normal tissue, which is a growing field. Computational models built around this dual scale to simulate cancer initiation would more globally encompass the evolutionary dynamics of oncogenesis, harnessing the genetic repeatability observed in normal tissue in addition to the one of cancer-initiating clones. This improvement would not yet compensate previous drawbacks of the framework, such as the absence of epigenetic alterations and interactions with the micro-environment.7,8 Such a framework would however help us better model past evolutionary trajectories, predict probable future ones, and identify opportunities for therapeutic interventions.
Footnotes
Funding:The author(s) received no financial support for the research, authorship, and/or publication of this article.
Declaration of Conflicting Interests:The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: BR and PM designed this commentary and co-wrote the manuscript.
ORCID iD: Pierre Martinez  https://orcid.org/0000-0001-7069-4413
https://orcid.org/0000-0001-7069-4413
References
- 1. Tokutomi N, Moyret-Lalle C, Puisieux A, Sugano S, Martinez P. Quantifying local malignant adaptation in tissue-specific evolutionary trajectories by harnessing cancer’s repeatability at the genetic level. Evol Appl. 2019;12:1062-1075. doi: 10.1111/eva.12781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23-28. [DOI] [PubMed] [Google Scholar]
- 3. Tomasetti C, Vogelstein B. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science. 2015;347:78-81. doi: 10.1126/science.1260825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Martincorena I, Fowler JC, Wabik Aet al. Somatic mutant clones colonize the human esophagus with age. Science. 2018;362:911-917. doi: 10.1126/science.aau3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yokoyama A, Kakiuchi N, Yoshizato Tet al. Age-related remodelling of oesophageal epithelia by mutated cancer drivers. Nature. 2019;565:312-317. doi: 10.1038/s41586-018-0811-x. [DOI] [PubMed] [Google Scholar]
- 6. Balachandran VP, Łuksza M, Zhao JNet al. Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature. 2017;551:512-516. doi: 10.1038/nature24462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rozhok AI, Salstrom JL, DeGregori J. Stochastic modeling reveals an evolutionary mechanism underlying elevated rates of childhood leukemia. Proc Natl Acad Sci U S A. 2016;113:1050-1055. doi: 10.1073/pnas.1509333113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bissell MJ, Radisky D. Putting tumours in context. Nat Rev Cancer. 2001;1:46-54. doi: 10.1038/35094059. [DOI] [PMC free article] [PubMed] [Google Scholar]