Abstract
Zinc finger-homeodomain (ZHD) proteins constitute a plant-specific transcription factor family that play important roles in plant growth, development, and stress responses. In this study, we investigated a total of 10, 17, and 31 ZHD gene members in the peach, Arabidopsis, and apple genome, respectively. The phylogenetic tree divided the identified ZHD genes into 4 subfamilies based on their domain organization, gene structure, and motif distribution with minor variations. The ZHD gene family members were unevenly distributed throughout in apple, peach, and Arabidopsis genomes. Segmental duplication was observed for 14 pairs of genes in apple. Transcript analysis found that ZHD genes mostly expressed in various tissues, particularly in leaves and flowers. Moreover, the transcript of most ZHD genes was significantly affected at different time points in response to various flowering-related exogenous hormones (sugar, gibberellin [GA], and 6-benzylaminopurine [6-BA]), signifying their possible role in the flowering induction in apple. Furthermore, the transcripts of CaZHD6, CaZHD7, CaZHD3, and CaZHD8 have induced in response to abiotic stresses including heat, drought, salt, and cold, indicating their possible involvement in response to abiotic stresses. Our research work systemically presents the different roles of ZHD genes. We believe that this study will provide a platform for future functional characterization of ZHD genes and to deeply unfold their roles in the regulation of flowering induction in plants.
Keywords: ZHD, apple, synteny, expression analysis
Introduction
Transcription factors are playing an important role in regulating various developmental phases of the plant.1 Here, we present zinc finger-homeodomain (ZF-HD) transcription factors. The zinc finger motif is widely found in various regulatory proteins.2 It contains 2 pairs of conserved cysteine and/or histidine residues that stabilize the motif into a finger-shaped loop by coordinating with single zinc ion.3 A single protein can possess 1 or more zinc finger motifs, which are mainly involved in DNA-binding protein-protein interaction.4 In addition, the zinc finger can be divided on the bases of their nature, zinc-binding protein residues, and numbers. C2H2, C2C2, and C3H zinc fingers interact with 1 zinc ion, whereas the LIM, PHD zinc finger, and zinc finger domain coordinate 2 zinc ions.5
The zinc finger-homeodomain (ZF-HD) gene family members were first identified in C4 plant species known as Flaveria.6 Since then, the ZF-HD proteins were found to play various important roles in plant growth and development, such as the Arabidopsis ZF-HD1 was characterized to find out its functional properties. It was found that the ZF-HD1 binds to the promoter of EARLY RESPONSE TO DEHYDRATION STRESS 1 (ERD1). Its expression was induced by salt, high transpiration rate, and also by abscisic acid (ABA).7 The overexpression of ZF-HD1 along with NAC genes conferred tolerance to drought stress.7 In another report, 2 soybean ZF-HD proteins were confirmed by Park et al8 that they bind with the promoter of the gene encoding calmodulin isoform 4 (GmCaM4) and express highly under pathogen inoculation.8 These findings clarified that the ZF-HDs are mainly transcriptional regulators involved in different biological and biochemical processes by simultaneously acting with other transcriptional factors.
Flower induction is a critical developmental process but often affected by environmental and internal factors.9,10 For instance, sugar and hormones are involved in the flower developmental process.11,12 Although in hormones gibberellin (GA) was proved important for regulating flower induction by many researchers, the GA pathway is a key factor in this flower induction.12,13 The other hormones such as 6-benzylaminopurine (6-BA) and sucrose also help in promoting bud and flowers as described in the early research.11 Moreover, some gene families were also found to be crucial in flower induction. Taking the example of SPB (SQUAMOSA PROMOTER BINDING PROTEIN),14 B-Box,15 and MADS-box (DNA-binding MADS domain)16 gene families, all of them were reported to be involved in the flower developmental process.
It has been reported before that the ZF-HD proteins are crucial for flower induction. For instance, the Arabidopsis (ZF) HD transcription factors were reported for their role in flower induction.17 Furthermore, flower induction is affected by different environmental stresses but the overexpression of ZF-HD1 was reported to cop drought stress by making a bond with the promoter of ERD1.7 Therefore, it is important to study the role and expression pattern of this gene family during flower induction in 3 species: apple, peach, and Arabidopsis, under different abiotic stresses and hormonal treatments.
To our knowledge, no comprehensive study of ZHD gene family has been reported so far in apple and peach. Although the ZHD gene family has already been reported in Arabidopsis,18 tomato,19 grapes,1 and rice,20 their possible role in flowering induction is still unknown. Therefore, we have performed a systematic study to investigate the ZHD gene family member in important fruit plants and their role in flowering induction in apple. In this study, the ZHD gene family members in apple, peach, and Arabidopsis and their expression profiles under various flowering-related exogenous hormones and treatments in apple were systematically investigated. The results obtained here provide a solid foundation for further clarification of ZHD gene family roles in the plant growth and development and their contribution in flowering induction.
Materials and Methods
Identification of ZHD gene family member
In this study, all the ZHD protein sequences have been retrieved from the Arabidopsis Genome database (TAIR) database (http://www.arabidopsis.org). We retrieved the homologues with statistical expectation values (e-values) based on amino acid similarity to score sensitivity than percent identity. The similarity scoring matrices and information of explicit or implicit evolutionary model to ensure the search sensitivity and alignment accuracy of distantly related protein sequences were investigated by using the BLOSUM50 matrix program. HMM (hidden Markov model) profile of the zinc finger-homeodomain (ZHD) (pfam04770) was retrieved from the Pfam version 31.0 database.21 The heuristically BLASTp (E-value = 1e−3) identity searches were used in peach, Arabidopsis, and apple genomes. All the identified ZHD sequences were confirmed by InterPro (IPR006455) Pfam (pfam04770) and SMART for conserved domain structure and the incomplete or the absence of core domain sequences were removed.
Chromosomal localization and gene duplication
The chromosomal locations of all ZHD genes were obtained from the Genomic Database for Rosaceae (http://www.Rosaceae.org/) and for Arabidopsis (http://www.arabidopsis.org). The MapDraw was then used to find out the exact location of genes on chromosomes. The online version 4.9.1 of the Multiple Expectation for Motif Elicitation (MEME) tool (http://meme-suite.org/) was used to create the conserved and shared domains for all ZHD protein sequences.22,23 The online Gene Structure Display Server (http://gsds.cbi.pku.edu.cn) was used to construct ZHD exon-intron structure consisting of exon positions and gene length.23
Sequence alignment and polygenetic analysis
For designing the multiple alignments of ZHD sequences, DNAMAN software (Version 5.2.2; Lynnon Biosoft, CA, USA) was used. The Weblogo platform was used to generate the sequence logo (http://weblogo.berkeley.edu/logo.cgi). The phylogenetic tree was constructed with a maximum-likelihood method in MEGA 6.06 with high bootstrap (1000) repetition.24
Tandem duplication and synteny analysis
The Plant Genome Duplication Database was used to design the syntenic block (http://chibba.agtec.uga.edu/duplication/). To construct the Circos diagram, the Circos version 0.63 (http://circos.ca/) was used. In addition, to find out the Tandem duplication of ZHD genes in apple, peach, and Arabidopsis, the physical location of a gene on the chromosome was used. Adjacent homologous ZHD genes on the same chromosome having 1 intervening gene were classified as tandemly duplicated.
Plant materials and treatments
A total of 72 uniform 6-year-old ‘Fuji’/T337/Malus robusta Red apple trees were used in this study. The studied materials were grown at the experimental orchard of Northwest Agriculture and Forestry University in Yangling (108°04′ E, 34°16′ N), China. The materials were further distributed into 4 groups: 18 were treated with 6-BA, 18 were treated with sugar, 18 cultivars were sprayed with GA, whereas 18 were sprayed with water and used as a control. Three blocks with 3 replications were designed for each group. The plants were treated with hormones by following the study of Zhang et al12 with slight modification. GA3 of 700 mg L−1 (Sigma, Deisenhofen, Germany) was sprayed once on a clear morning at the rate of 30 DAFB (days after full bloom) (9 May). In addition, trees were sprayed with 300 mg L−1 of 6-BA (Sigma) on a clear morning at the rate of 30 DAFB (9 May). Moreover, the sugar treatment composed of spraying trees 2 times with 15 000 and 20 000 mg L−1 sucrose on clear mornings at 30 and 37 DAFB (9 May and 16 May). Furthermore, the hand wand sprayer with low pressure was used for all the treatments. The terminal buds on current-year spurs (<5 cm) were collected at 30, 50, and 70 DAFB and preserved in liquid nitrogen. Pepper seedlings were exposed to low, high temperature, and drought conditions in the chamber for abiotic stresses, whereas some pepper seedlings were treated with 300 mM NaCl. The samples were collected at 0, 3, 6, and 12 h after post-treatment.25 After that, the collected samples were stored at −80°C to extract RNA and study gene expression.
Sample collection
Tissue from each collected organ was used for the investigation of gene expression in 6-year-old ‘Fuji’/T337/Malus robusta Red at the end of April 2017. Among them, bud samples were collected along with mature leaves. Three independent experimental blocks and 3 biological replications from each experimental block were collected. Total RNA was extracted by Extraction Kit (Invitrogen, NY, USA), and the RNA samples were then purified using an RNeasyMinElute TM Cleanup Kit (Qiagen, Germany). The quality and quantity of RNA were checked by nanodrop and finally adjusted to 2000 ng/µL. cDNA was synthesized by SuperMix kit, with DNA remover according to the manufactured protocol and the final volume of cDNA was obtained up to 300 ng/µL, which was further diluted with TE (Tris-EDTA) buffer up to 50 ng/µL and used for quantitative real time polymerase chain reaction (qRT-PCR) analysis. For qRT-PCR analysis, cDNA was diluted up to 100 ng/µL. All the primers were designed from apple, Arabidopsis, and peach ZHD sequences for RT-PCR using primer 6.0 (Supplemental Table S4). Moreover, the primer pair was tested through standard RT-PCR to check the size specificity of the amplified product by 1% agarose gel electrophoresis.
qRT-PCR analysis
The RT-PCR was performed in an iCycleriQ Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). Each reaction consisted of 5 μL SYBR Premix ExTaq (Takara, Kyoto, Japan), 2 μL cDNA samples, 0.5 μL of each primer (10 μM), and 2 μL ddH2O in a reaction system of 10 μL. The thermal cycle was as follows: 95°C for 3 min, followed by 40 cycles at 94°C for 15 s, 62°C for 20 s, and 72°C for 20 s. Melting-curve analysis was performed directly after RT-PCR to verify the presence of gene-specific PCR products. This analysis was done at 94°C for 15 s, followed by a constant increase from 60 to 95°C at a 2% ramp rate. The EF-1α gene (Gen Bank accession no. DQ341381) and 18S rRNA were used as an internal control and served as a standard gene for normalizing all mRNA expression levels. The relative amount of template present in each PCR amplification mixture was evaluated by using the 2−ΔΔCtmethod, where the mean values were obtained from 3 independent PCR amplifications. The expression data of each gene were converted into log2 ratio and fold changes. For fold change (FC = log2 final value – log2 initial value), FC = 2 greater was considered upregulated and FC = 2 or less than 2 was considered downregulated.
Statistical analysis
Statistical analysis was performed using SPSS software (version 25.0; SPSS Inc., Chicago, IL, USA) and 95% confidence intervals (P ⩽ .05) were considered statistically significant. The data were plotted by GraphPad Prism 7.0 (GraphPad Software, Inc., LA Jolla, CA, USA).
Results
Identification and classification of zinc finger-homeodomain gene family in apple and peach
The Arabidopsis zinc finger-homeodomain genes were used as quarry sequences against the HMM algorithm21 to identify, classify, and characterize the zinc finger-homeodomain gene family members in apple and peach. A total of 31 and 10 zinc finger-homeodomain gene family members were investigated in apple and peach, respectively. The number of ZHD gene members was diverse in the plant such as 31 ZHD in apple, 17 ZHD genes were found in Arabidopsis, and 10 ZHD members were found in peach in this study (Table 1). The entire assumed zinc finger-homeodomain family members lack transmembrane segment except for MdZHD11 (Supplemental Figure S1). Furthermore, the physiochemical appearances of zinc finger-homeodomain members were calculated through the EXPASY PROTOPARAM (http://www.expasy.org/tools/protparam.html) online tool (Supplemental Table S1). The supposed length and molecular weights of the zinc finger-homeodomain proteins vary widely, ranging from 9.53 (MdZHD28) to 78.98 kDa (MdZHD11). A total of 39 out of 55 zinc finger-homeodomain proteins were basic in nature based on their isoelectric point, which was greater than 7; however, the rest of the proteins were categorized into acidic proteins. All the zinc finger-homeodomain genes were grouped into unstable proteins excluding MdZHD3 and MdZHD30 genes because the instability index of most of the zinc finger-homeodomain family members was greater than 40. All the zinc finger-homeodomain members were identified to be hydrophilic based on their grand average of hydropathicity (GRAVY) value. The GC content of the ZHD gene family was between 34.6% (MdZH11) and 63.7% (PpZHD9) in the studied plants. We further found that 29 zinc finger-homeodomain proteins were found on the antisense strand, whereas the rest of the zinc finger-homeodomain proteins were located on the sense strand. Furthermore, the range of aliphatic index values was 41.41 (lowest) (PpZHD6) to 81.69 (highest) (PpZHD3). Gly (G), Ser (S), and Pro (P) are the major amino acids of the zinc finger-homeodomain proteins, whereas other most abundant amino acids are Glu (E), Lys (K) Asn (A), and Arg (R), depending on the particular zinc finger-homeodomain protein (Supplemental Table S1). Moreover, we studied the predicted location of the zinc finger-homeodomain proteins in the cells.26 The results showed that the maximum number of ZHD proteins is located in the nucleus portion of the cells; however, some members are distributed in mitochondria, cytoplasm, extracellular, and plasma membrane of the cells (Supplemental Table S1).
Table 1.
Nomenclature identification, chromosomal location, coding sequence (CDS), and peptide length and weight of ZHD genes in Arabidopsis, apple, and peach.
| Name | ID | Location | Genomic | CDS | Protein | MW |
|---|---|---|---|---|---|---|
| AtZHD1 | At5g65410 | Chr5: 26136002..26137554 | 1553 | 840 | 279 | 31.10 |
| AtZHD2 | At4g24660 | Chr4: 12722912..12725835 | 1284 | 663 | 220 | 24.02 |
| AtZHD3 | At2g02540 | Chr2: 683625..685496 | 1685 | 933 | 310 | 34.72 |
| AtZHD4 | At1g14440 | Chr1: 4938754..4941332 | 2579 | 939 | 312 | 35.48 |
| AtZHD5 | At1g75240 | Chr1: 28241011..28242795 | 1785 | 930 | 309 | 33.89 |
| AtZHD6 | At2g18350 | Chr2: 7971017..7972396 | 1065 | 789 | 262 | 29.95 |
| AtZHD7 | At3g50890 | Chr3: 18916014..18917386 | 1373 | 750 | 249 | 28.66 |
| AtZHD8 | At5g15210 | Chr3: 18916014..18917386 | 1743 | 816 | 271 | 29.38 |
| AtZHD9 | At3g28920 | Chr3: 10940305..10941833 | 1529 | 939 | 312 | 33.56 |
| AtZHD10 | At5g39760 | Chr5: 15911350..15912860 | 1511 | 1005 | 334 | 36.39 |
| AtZHD11 | At1g69600 | Chr1: 26182156..26183546 | 1391 | 729 | 242 | 26.28 |
| AtZHD12 | At5g60480 | Chr5: 24323371..24324335 | 965 | 673 | 223 | 25.24 |
| AtZHD13 | At5g42780 | Chr5: 17154718..17155757 | 1040 | 729 | 242 | 27.97 |
| AtZHD14 | At1g14687 | Chr1: 5047782..5048753 | 972 | 507 | 168 | 19.92 |
| AtZHD15 | AT1G74660 | Chr1:28045472..28048289 | 935 | 309 | 102 | 11.21 |
| AtZHD16 | AT3G28917 | Chr3:10924708..10925813 | 1106 | 303 | 101 | 10.11 |
| AtZHD17 | AT1G18835 | Chr1:6495844..6496491 | 648 | 267 | 22 | 9.75 |
| PpZHD1 | ppa024080m | scaffold_1:27083790..27084839 | 1050 | 1050 | 350 | 38.08 |
| PpZHD2 | ppa023369m | scaffold_1:33211050..33212142 | 1093 | 933 | 311 | 34.18 |
| PpZHD3 | ppa015411m | scaffold_1:42239945..42240970 | 1026 | 1026 | 342 | 37.46 |
| PpZHD4 | ppa009679m | scaffold_1:46661094..46661942 | 849 | 849 | 283 | 30.43 |
| PpZHD5 | ppa008908m | scaffold_3:19674589..19675917 | 1329 | 948 | 316 | 33.93 |
| PpZHD6 | ppa013998m | scaffold_3:19683618..19684281 | 1064 | 279 | 93 | 9.88 |
| PpZHD7 | ppa024260m | scaffold_5:15780700..15780984 | 285 | 285 | 95 | 10.41 |
| PpZHD8 | ppa011508m | scaffold_5:18046719..18047505 | 787 | 624 | 208 | 22.76 |
| PpZHD9 | ppa009712m | scaffold_6:18513271..18514119 | 849 | 846 | 282 | 30.16 |
| PpZHD10 | ppa020272m | scaffold_7:16871070..16872065 | 996 | 996 | 332 | 36.49 |
| MdZHD1 | MDP0000804580 | chr2:9073140..9074075 | 936 | 936 | 312 | 34.12 |
| MdZHD2 | MDP0000208739 | chr2:12795643..12796605 | 963 | 960 | 320 | 35.28 |
| MdZHD3 | MDP0000640510 | chr3:21086230..21086857 | 628 | 627 | 207 | 13.73 |
| MdZHD4 | MDP0000318292 | chr6:3322762..3323531 | 710 | 666 | 222 | 24.84 |
| MdZHD5 | MDP0000123799 | chr6:13926570..13927400 | 831 | 828 | 276 | 30.77 |
| MdZHD6 | MDP0000181626 | chr6:17681637..17682467 | 831 | 828 | 276 | 30.71 |
| MdZHD7 | MDP0000253357 | chr6:25989027..25989323 | 297 | 294 | 98 | 11.04 |
| MdZHD8 | MDP0000790705 | chr6:30114168..30114776 | 609 | 609 | 203 | 22.05 |
| MdZHD9 | MDP0000163471 | chr8:4149654..4150691 | 1038 | 1035 | 345 | 37.32 |
| MdZHD10 | MDP0000287735 | chr8:10761185..10761505 | 321 | 321 | 107 | 11.34 |
| MdZHD11 | MDP0000259786 | chr8:23251460..23260953 | 9494 | 2109 | 703 | 78.98 |
| MdZHD12 | MDP0000372694 | chr8:35492826..35493914 | 1089 | 1089 | 363 | 39.30 |
| MdZHD13 | MDP0000934043 | chr9:3127906..3128178 | 273 | 273 | 91 | 9.62 |
| MdZHD14 | MDP0000821959 | chr9:3140644..3141705 | 1062 | 1062 | 354 | 38.45 |
| MdZHD15 | MDP0000787720 | chr11:20708937..20709761 | 825 | 825 | 275 | 29.30 |
| MdZHD16 | MDP0000614531 | chr12:26103667..26104719 | 1053 | 1053 | 351 | 37.57 |
| MdZHD17 | MDP0000314479 | chr13:2921413..2929343 | 7931 | 645 | 215 | 23.22 |
| MdZHD18 | MDP0000286456 | chr13:4549358..4550359 | 1002 | 1002 | 334 | 36.31 |
| MdZHD19 | MDP0000322140 | chr14:25816559..25816843 | 285 | 282 | 94 | 10.46 |
| MdZHD20 | MDP0000438181 | chr14:25820956..25821240 | 285 | 285 | 95 | 10.46 |
| MdZHD21 | MDP0000277477 | chr14:33594885..33600655 | 5771 | 660 | 220 | 24.18 |
| MdZHD22 | MDP0000199202 | chr15:31938023..31939068 | 1046 | 837 | 279 | 29.72 |
| MdZHD23 | MDP0000462091 | chr15:43935418..43936239 | 822 | 822 | 274 | 29.58 |
| MdZHD24 | MDP0000249735 | chr15:54534805..54554864 | 20060 | 1602 | 534 | 58.45 |
| MdZHD25 | MDP0000200799 | chr16:3106786..3107699 | 914 | 747 | 249 | 32.08 |
| MdZHD26 | MDP0000532986 | chr16:3126785..3127753 | 969 | 969 | 323 | 35.05 |
| MdZHD27 | MDP0000199110 | chr16:3128389..3129357 | 969 | 966 | 322 | 35.05 |
| MdZHD28 | MDP0000847771 | chr17:3466484..3466756 | 273 | 273 | 91 | 9.53 |
| MdZHD29 | MDP0000801659 | chr17:3502796..3503806 | 1011 | 1011 | 337 | 37.01 |
| MdZHD30 | MDP0000380874 | chr17:10645267..10645992 | 726 | 318 | 106 | 11.60 |
| MdZHD31 | MDP0000023799 | unanchored:6165680..6166480 | 801 | 594 | 198 | 22.08 |
Systematic evolutionary relationship, gene structural diversity, and motif analysis
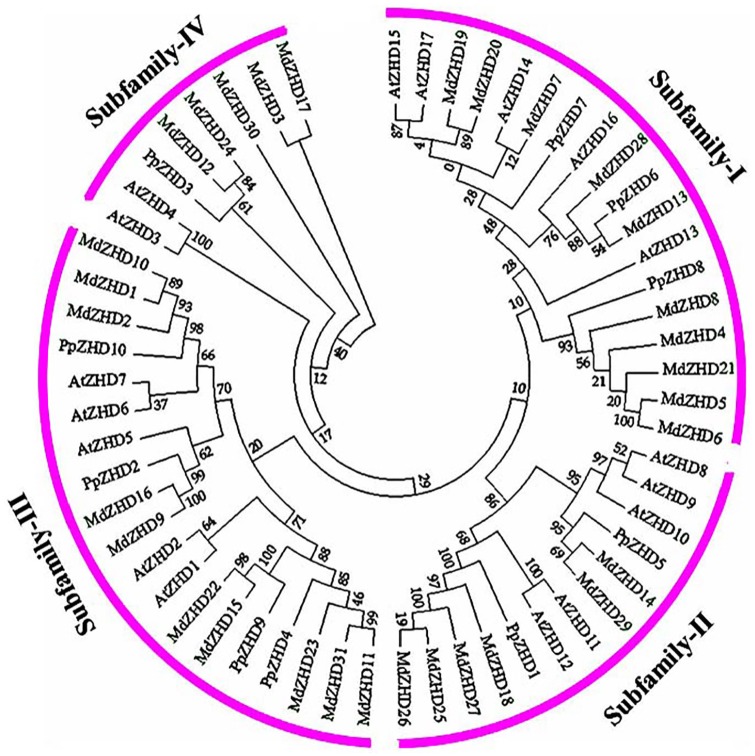
The multiple sequence alignments and phylogenetic relationship analysis of apple, peach, and Arabidopsis zinc finger-homeodomain gene family members were performed to study the evolutionary phylogenetic relationships and functional divergence among zinc finger-homeodomain genes in plants (Figure 1). The MEGA 6.06 Software was used to build an unrooted maximum-likelihood phylogenetic tree to investigate the evolutionary relationship. This study divided the zinc finger-homeodomain genes into 4 well-conserved subfamilies (Figure 1). In subfamily I, we found 18 ZHD members, and all the members of subfamily I belonged to MIF class of ZHD gene family apart from PpZHD8, MdZHD4, MdZHD5, MDZHD6, MdZHD8, and MdZHD21 genes, whereas the remaining MIF genes were detected in subfamily II (MdZHD18, MdZHD27, and MdZHD29), subfamily IV (MdZHD3, MdZHD12, and MdZHD17), and subfamily III (MdZHD10). A total of 13 ZHD members were found in subfamily II, and all the ZHD genes showed quite similar intron-exon and motif distribution in this subfamily (Figure 1). In subfamily III, we found 20 genes, and their gene structure and motif arrangement were also similar to each other. In subfamily IV, we noted a few number of ZHD genes. The motif 8 was only detected in the apple and peach ZHD members of subfamily III, whereas motif 9 was only detected in some members of subfamilies I and III, respectively. Similarly, we also noted that motif 6 was present in the ZHD members of subfamily II. Overall, we found quite alike gene structure, motif distribution, and physiochemical properties within every subfamily.
Figure 1.
Systematic evolutionary relationships of ZHD gene family in apple, peach, and Arabidopsis among 4 lineages within the subgroup. The 4 conserved subfamilies are marked by different numbers and represented as subfamilies I, II, III, and IV.
Exon-intron structural diagram of the zing finger-homeodomain gene members was constructed according to their genomic and coding sequences to study the structural diversity of ZHD genes in plants (Supplemental Figure S2). We found the range of intron numbers from 1 (MdZHD21, MdZHD29, MdZHD30, and MdZHD31) to 5 (MdZHD11) in the present investigation. However, we observed that a maximum number of ZHD family members possessed no introns in apple. We also noted that all the ZHD members were found without intron in Arabidopsis and peach, and they possessed only single exon. The MEME online server found a total of 10 motifs and was named motif 1-10 (Supplemental Figure S3). The results revealed that motifs 1 and 2 were the highly repeated motifs found in 52 and 50 ZHD gene members, respectively. The next most frequently occurred motif was motif 3 (42), followed by motifs 4 (39) and 5 (38), respectively (Supplemental Table S2). Similarly, motif 7 was the longest motif based on width, composed of 41 sequences, followed by motif 3, motif 2, and motif 6, respectively. The longest motif, namely motif-7, was only found in 4 ZHD members (MdZHD18, MdZHD25, MdZHD26, and MdZHD27).
Chromosomal location, multiple alignments, gene duplication, and gene ontology of zinc finger-homeodomain genes
For better understanding of ZHD members, we classified the apple, peach, and Arabidopsis ZHD members based on their location. The annotation information and chromosome position of the zinc finger-homeodomain genes indicated that zinc finger-homeodomain genes are randomly scattered on the chromosomes in the genome of the apple, peach, and Arabidopsis (Supplemental Figure S4). We found that all the MdZHD genes were found on all the chromosomes in the apple genome excluding chromosome numbers 1, 4, 5, 7, 9, and 10 (Supplemental Figure S4A). The highest number of ZHD members (MdZHD4-8) was observed on chromosome number 6, followed by chromosome no 8. Three MdZHD gene members were located on each chromosome number 14, 15, 16, and 17, whereas 1 MdZHD gene member was localized on each chromosome number 3, 11, 12, and 13, respectively. Moreover, chromosome number 2 possessed 2 ZHD members in apple genome. In the Arabidopsis genome, ZHD members were found on all chromosomes. Four ZHD members were distributed on each chromosome number 1 and 5 in the Arabidopsis genome (Supplemental Figure S4B). The rest of the 6 ZHD members were observed on chromosome numbers 1, 2, and 3. In peach, 4 PpZHD members were found on scaffold 1. Two PpZHD members were found on each scaffold-3 and 5, respectively. Similarly, 1 PpZHD member was localized on each chromosome number 2 and 4 in the peach genome (Supplemental Figure S4C).
The conserved domains of the zinc finger-homeodomain gene family were predicted through Pfam, SMART, Inter Pro Scan, Conserved Domain Database (CDD), National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/cdd/), and Scan Prosite databases (Supplemental Table S3). We noted that all the ZHD proteins possessed ZF domain; however, HD domain was not detected for some ZHD members (Supplemental Table S3). The proteins lacking HD domain were classified as MIF (MINI ZINC FINGER) proteins in this study. The family-specific domains including HD and ZF domains were aligned by DNAMAN software (Supplemental Figure S5), and the logo was constructed via Web logo (http://weblogo.berkeley.edu/logo.cgi) online server (Supplemental Figure S6).
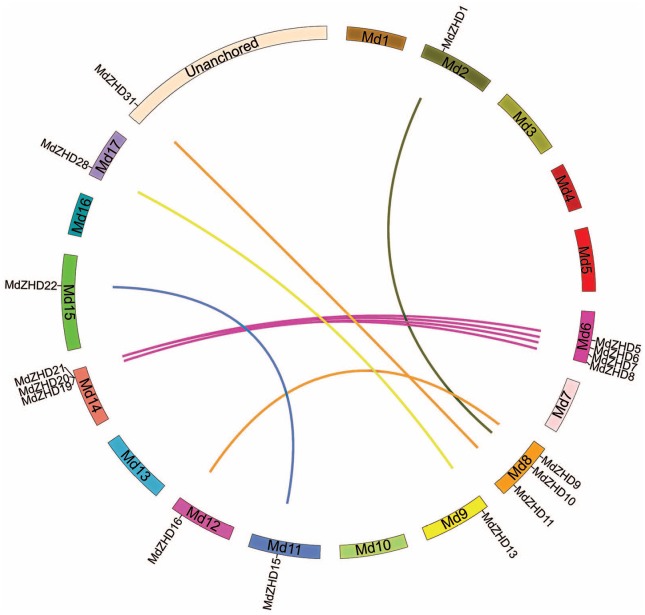
The mechanism of gene duplication including tandem duplication, segmental duplication, and transposition (retro and replicate transposition) is vital for the evolution of plants.27 Therefore, we also investigated the possibility of gene duplication in the ZHD gene family in apple and peach. Both the segmental and tandem duplications were studied in this finding. We found only segmental duplication in apple genome (Figure 2). Circos program was used to construct a diagram to draw the duplicated blocks in the apple genome. We identified that a total of 14 MdZHD pairs were located in the segmentally duplicated regions on different chromosomes in the apple genome. However, no tandem duplication region was detected among the ZHD family members in the apple and peach. The findings showed that only segmental duplication may contribute to the evolution process of the ZHD gene family in plants.
Figure 2.
Synteny analysis of ZHD gene family members in apple. Chromosomes of apple are shown in different colors and in circular form. The approximate positions of the ZHD genes are marked with a short black line on the circle. Colored curves denote the syntenic relationships between duplicated pairs.
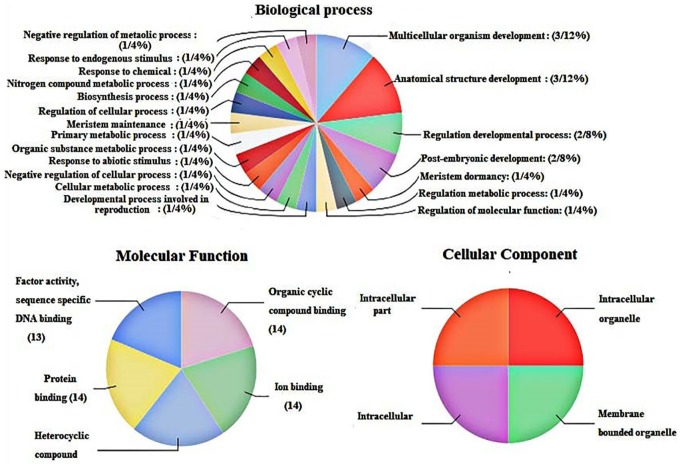
Furthermore, we performed gene ontology (GO) enrichment analysis of ZHD gene family members to predict the possible function of ZHD proteins in the biological process, molecular level, and cellular compartments (Figure 3). At cellular component, we found that ZHD gene may play a vital role in the intracellular membrane, intracellular organelles, and other membrane-bound organelles (Figure 3). Gene ontology analysis revealed that ZHD proteins may work as ion binding, heterocyclic, and organic cyclic compounds at the molecular level. Moreover, we further noted that ZHD protein could be a potential marker for certain biological process including regulation of RNA biosynthesis process, DNA transcriptional regulation, negative regulation of cellular metabolism, positive transcriptional regulation, and cellular macromolecule biosynthesis activities (Figure 3). The results of GO analysis suggest that ZHD may be involved in the metabolic network and regulate various biological processes and pathways in plants.
Figure 3.
The GO Ontology analysis of ZHD gene family members in apple, Arabidopsis, and peach.
Developmental and tissue-specific expression profile of ZHD gene family members
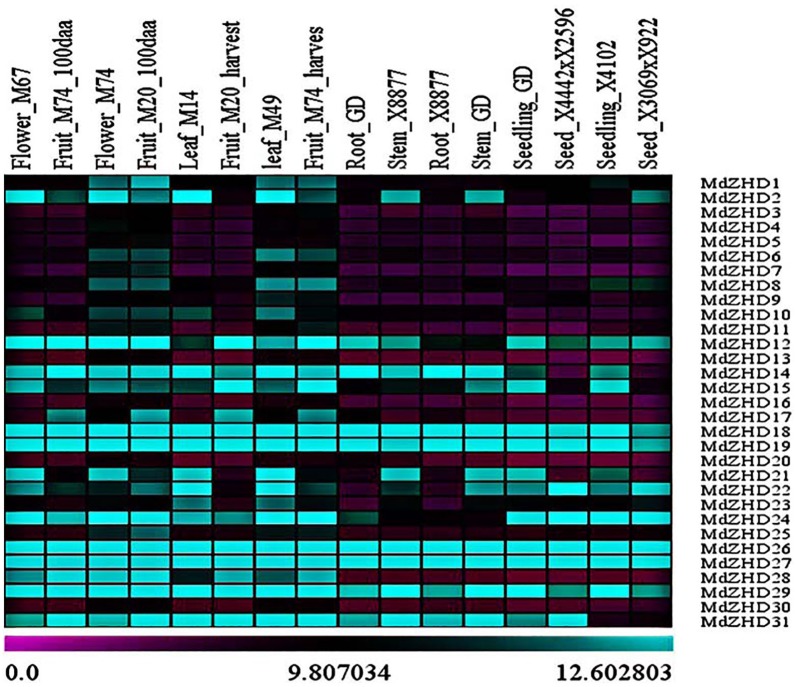
The transcript profile obtained from ArrayExpress of a zinc finger-homeodomain gene family is presented in the form of a heat map, from purple-black-blue, evaluating the transcript profile percentage (Figure 4). The transcript of ZHD gene family members in 7 different tissues comprising leaves, flowers, fruits, seeds, stems, roots, and seedlings from 10 apple varieties (M67, M74, M20, M14, M49, M74, GD, and X8877) and 2 hybrids was studied. The ZHD gene family members showed a distinct transcript level among the studied tissues. We observed that a high number of ZHD gene family members were greatly expressed in the flower_M74, fruit_M20_100daa, leaf_M49, and fruit_M74_hurvest. MdZHD12, MdZHD15, MdZHD18, MdZHD19, MdZHD24, MdZHD26, MdZHD27, and MdZHD29 genes showed high expression level in all the studied tissues, whereas MdZHD3, MdZHD4, MdZHD5, MdZHD13, MdZHD16, MdZHD20, and MdZHD30 were detected with less transcript level.
Figure 4.
The expression profiles obtained from the ArrayExpress data, displaying diverse expression levels of apple ZHD genes in different tissues and organs. The relative transcript level of ZHD gene members based on ArrayExpress data was presented as heat maps from purple to blue reflecting relative signal values, where dark purple boxes represent stronger downregulated expression and dark blue boxes represent stronger upregulation.
Organ-specific expression of zinc finger-homeodomain gene family
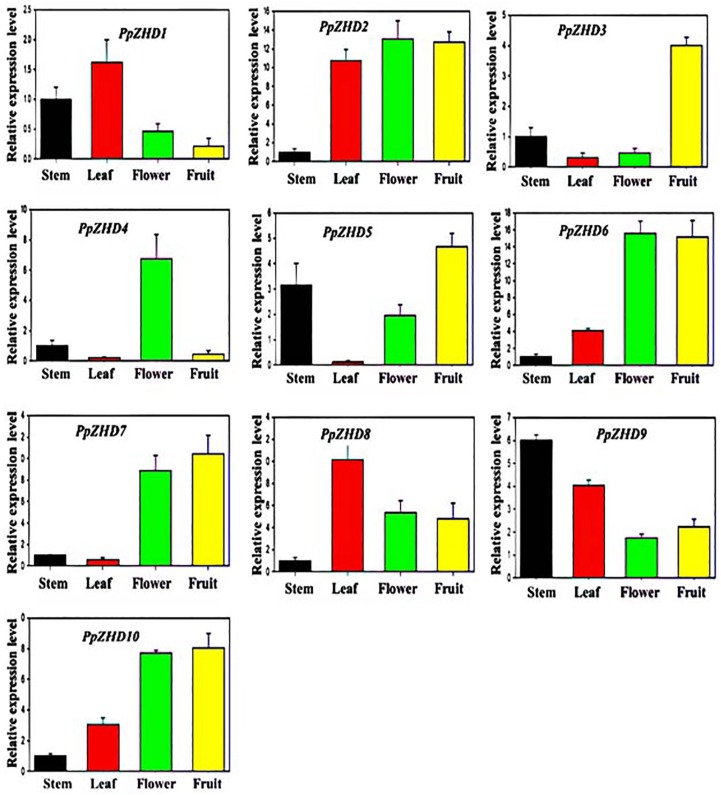
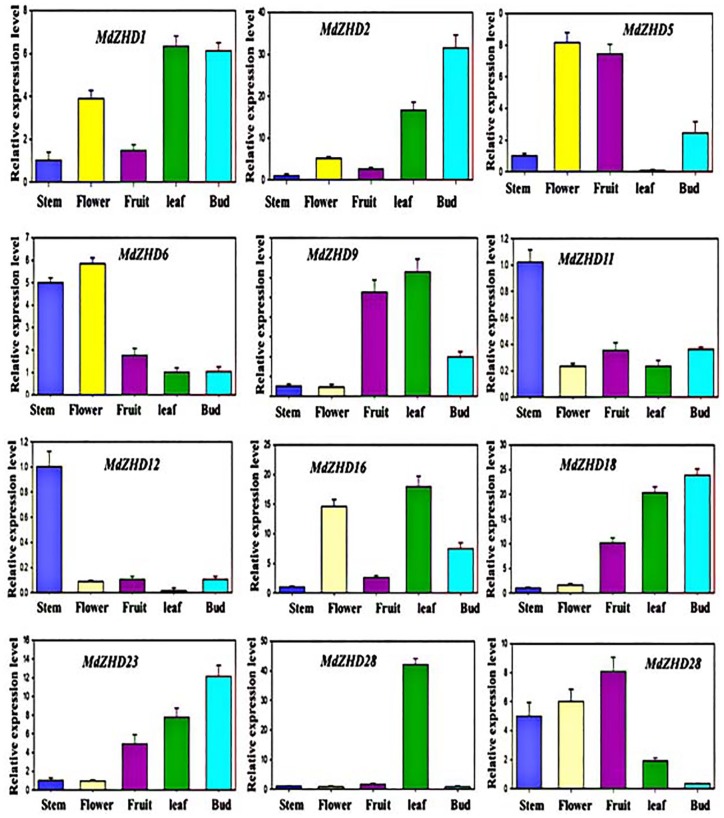
In this study, Arabidopsis ZHD gene family showed significant expression among the tested tissues (Supplemental Figure S7). We observed that the expression profile of AtZHD1, AtZHD3, AtZHD5, AtZHD6, AtZHD7, AtZHD8, and AtZHD10 was high in leaf, whereas the remaining Arabidopsis ZHD genes were found with less transcript level in leaf. The expression profiles of the entire Arabidopsis ZHD gene were less in stem excluding AtZHD4 and AtZHD9 genes. Similarly, the transcripts of AtZHD1, AtZHD3, and AtZHD7 were high in seedling; however, the remaining genes displayed less expression. Less transcript level was also detected for all the AtZHD gene family members in flower except AtZHD1 and AtZHD3. The expression level of ZHD gene family members was not significantly high in the root. Furthermore, we also investigated the expression profile of the ZHD gene family member in peach (Figure 5). We found that all the peach ZHD gene family members showed high expression level in fruit apart from PpZHD1 and PpZHD4. In leaf, PpZHD1, PpZHD2, PpZHD6, PpZHD8, PpZHD9, and PpZHD10 showed significant expression, whereas the remaining genes including PpZHD3, PpZHD4, PpZHD5, and PpZHD7 were found with less expression in leaf. A high transcript level was detected for PpZHD5 and PpZHD9 in the stem of peach. In flower, the transcript levels of PpZHD2, PpZHD4, PpZHD6, PpZHD7, PpZHD8, and PpZHD10 were high, whereas other PpZHD genes showed lesser expression level. Moreover, we also evaluated the transcript level of the ZHD gene family member in different tissues of apple including stem, leaf, fruit, flower, and bud. This study found high expression level for ZHD genes among the tested tissues (Figure 6). In flowering-related bud, the transcript level of the MdZHD1, MdZHD2, MdZHD16, MdZHD18, and MdZHD23 was high compared with other apple ZHD genes. Similarly, some apple ZHD genes, named MdZHD1, MdZHD2, MdZHD9, MdZHD16, MdZHD18, MdZHD23, and MdZHD26, were highly expressed in apple leaf. Low transcript level was detected for the entire apple ZHD gene family member excluding MdZHD6 and MdZHD28. Similarly, MdZHD1, MdZHD2, MdZHD5, MdZHD6, MdZHD16, and MdZHD29 were greatly transcribed in flower, whereas the remaining genes were less transcribed. Low expression level was observed for the entire MdZHD gene family members except MdZHD5, MdZHD9, MdZHD18, MdZHD23, and MdZHD29 genes in fruit. Altogether, we observed the diverse expression pattern for each gene in every tissue. A maximum number of ZHD family members were expressed in leaf, signifying their role in leaf development. Some genes were also expressed in flower and fruit, indicating their involvement in these important tissues of the plant. Moreover, the transcribed rate of a few ZHD gene family members was also significantly high in other studied tissues. All these results shed light on the contribution of ZHD family members in plant growth and development; however, further experiment is required to unravel the exact function of the ZHD gene family.
Figure 5.
Expression profile of the PpZHD genes in tested tissues. The graphs indicate the tissue-specific expression level in the peach plant. The samples were collected in different developmental stages and were analyzed through qRT-PCR. The x-axis indicates the tissues. The y-axis shows the relative expression level of each tissue. The error bars indicate the standard deviations of the three independent qRT-PCR biological replicates.
Figure 6.
Expression profile of the MdZHD genes in tested tissues. The graphs indicate the tissue-specific expression level in apple plant. The samples were collected in different developmental stages and were analyzed through qRT-PCR. The x-axis indicates the tissues. The y-axis shows the relative expression level of each tissue. The error bars indicate the standard deviations of the three independent qRT-PCR biological replicates.
The response of ZHD gene family members to exogenous flowering-related treatments in apple
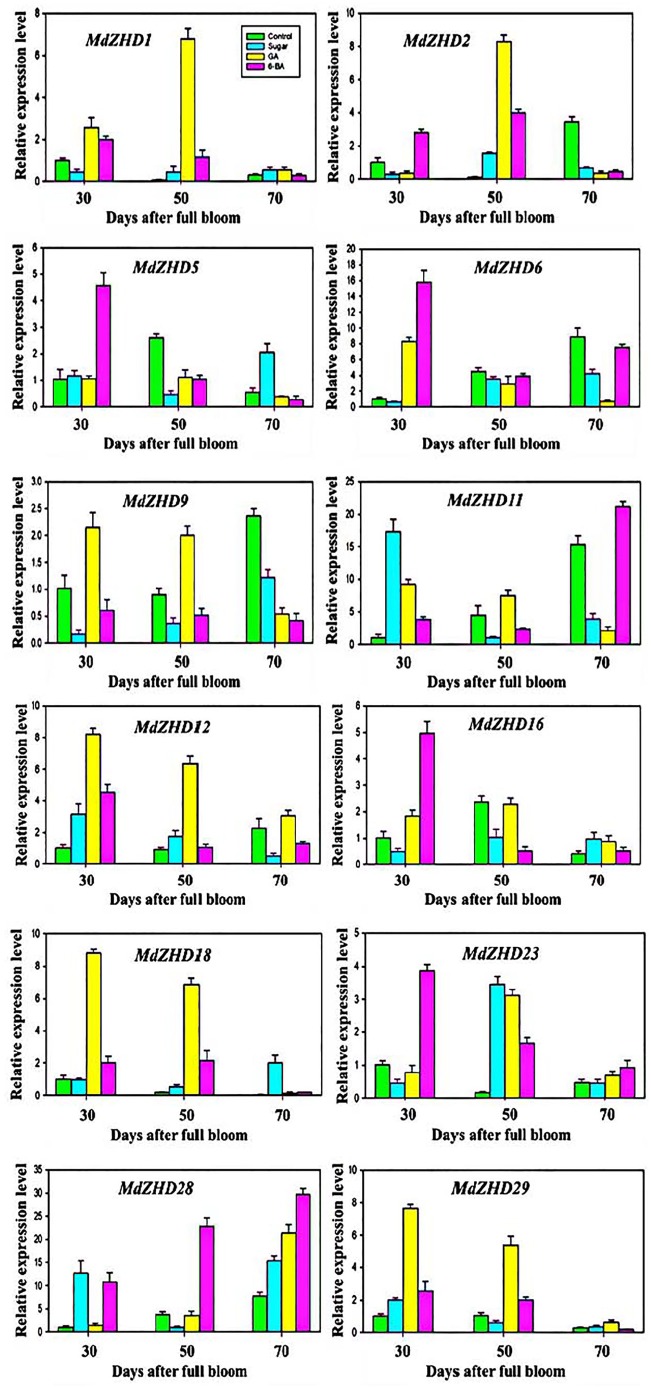
Previously, it has been reported that exogenous GA, ABA, and sugar alter the flowering rate in plants. Gibberellin reduced the flowering rate, whereas 6-BA and sugar triggered the flowering rate among these exogenous treatments.15,28-30 Based on this evidence, here we also used GA, 6-BA, and sugar to investigate the transcript level of the ZHD gene family and their possible role in flowering induction in apple (Figure 7). We noticed the distinct gene expression profile for each ZHD gene family member under flowering-related treatments. The transcript level of the entire ZHD gene family members was low in response to exogenous sugar at all the three time-points; however, the expression levels of MdZHD11, MdZHD23, and MdZHD28 were upregulated at one and two time-points, respectively (Figure 6). MdZHD11 gene was found with high expression level at 30 DAFB, MdZHD23 was upregulated at 50 DAFB, whereas MdZHD28 gene was detected at 30 and 50 DAFB. Under exogenous GA treatment, we noticed that the expression profiles of MdZHD1, MdZHD2, MdZHD6, MdZHD9, MdZHD11, MdZHD12, MdZHD18, MdZHD23, MdZHD28, and MdZHD29 were promoted at least 1 or 2 time points, whereas the transcribed rate of the remaining ZHD members was low at all the 3 time points (Figure 7). In response to exogenous GA, we observed high expression pattern at 30 and 50 DAFB time points compared with 70 DAFB time point. Moreover, the expression level of MdZHD1, MdZHD12, and MdZHD29 were high at all the three time-points (30, 50, and 70 DAFB) compared with control samples. Furthermore, we also used the exogenous 6-BA to identify the expression pattern of the apple ZHD gene family members (Figure 7). The ZHD gene family members displayed diverse expression pattern under exogenous 6-BA treatment. The entire ZHD gene family member showed the high transcribed rate at least at one or two time points under exogenous 6-BA application apart from MdZHD1, MdZHD9, MdZHD18, and MdZHD29 genes. The transcript of MdZHD23 and MdZHD28 genes was high at all the studied time points than the transcript of ZHD gene family members in control. The transcript of a maximum number of genes was promoted at 30 DAFB, followed by 50 and 70 DAFB, respectively. Overall, we observed variations in the transcript level of the ZHD gene family under applied exogenous treatment, and the expression of every gene was greatly affected in response to at least a single treatment. The differences in the transcript level under each treatment indicate the response of ZHD gene family to exogenous flowering-related treatments and their contribution to the flowering induction; however, further deep molecular experiment will reveal the role of ZHD gene family in the flowering induction and the development of plant architecture.
Figure 7.
Inducible expression profile of apple ZHD gene family members in response to flowering-related treatments (sugar, GA, and 6-BA). The x-axis indicates the treatment. The y-axis shows the relative expression level of each treatment compared to control. The error bars indicate the standard deviations of the three independent qRT-PCR biological replicates.
The transcript level of ZHD gene family members under exogenous abiotic stresses in pepper
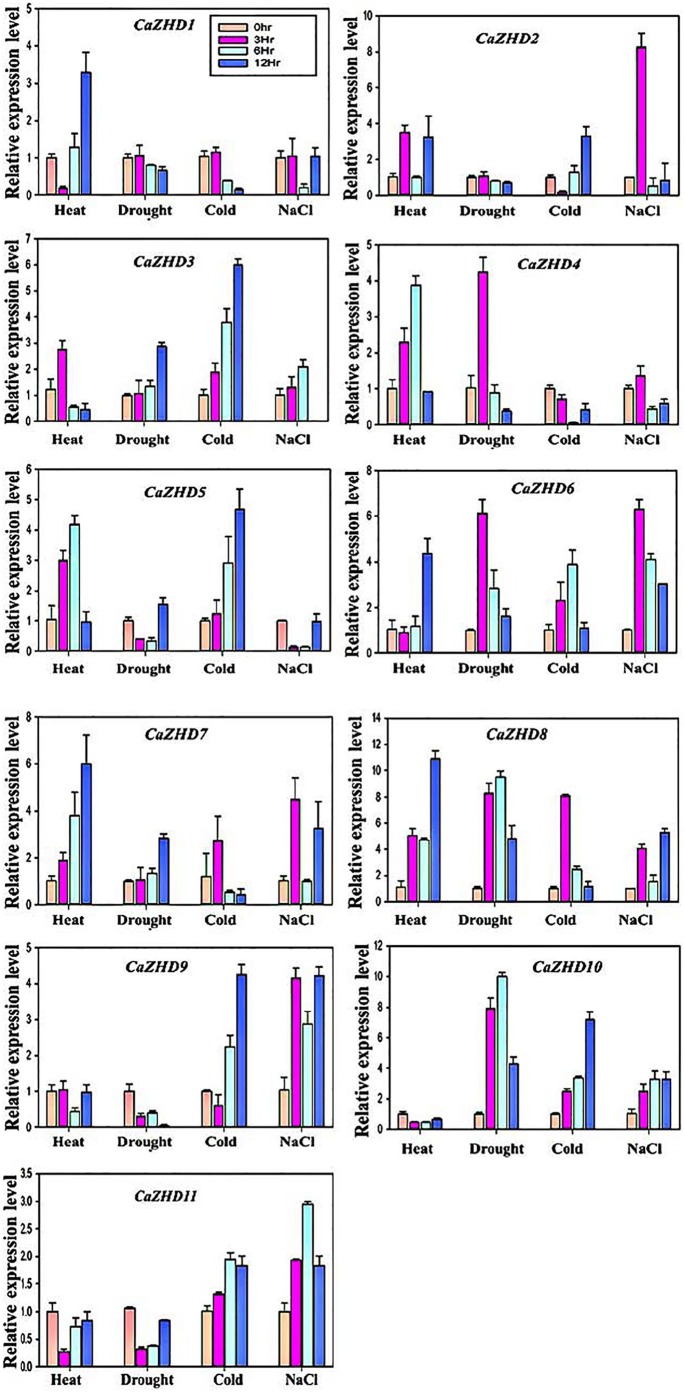
The transcript analysis of gene families provides essential evidence for gene function. Thus, the qRT-PCR was used to explore the expression profile of the ZHD genes under different abiotic stresses, including heat, drought, cold, and salt in pepper. The results showed that the pepper ZHD gene family shared distinct transcript level under these abiotic studies (Figure 8). More than 2-fold variance in the transcript level was considered to be the true differences for all the pepper ZHD genes. In this study, we found that CaZHD3, CaZHD6, CaZHD7, and CaZHD8 genes were upregulated at least 1 or more time points under all the 4 abiotic stresses, whereas CaZHD1 gene showed a low level of the transcript under these abiotic stresses apart from 12 h time point under heat stress (Figure 8). The expression profiles of CaZHD2 and CaZHD10 were high in response to all abiotic stresses at different time points excluding CaZHD2 expression in response to drought, whereas CaZHD10 under heat treatment. Moreover, the remaining 3 ZHD members including CaZHD4, CaZHD5, and CaZHD9 were induced under 2 abiotic stresses, whereas these genes showed a low level of the transcript under 2 abiotic stresses. For instance, the transcript level of CaZHD4 was high in response to heat and drought; however, lesser under cold and salt, CaZHD5 displayed high expression level under heat and cold, whereas low expression profile in response to drought and salt, whereas the transcript profiles of CaZHD9 and CaZHD11 were the opposite of CaZHD4 gene, high expression under cold and salt, but low level in response to heat and drought stresses. Overall, each pepper ZHD gene family member showed a different expression pattern under the multivariate abiotic stresses. The distinctive inducible transcript level of the pepper ZHD genes family member in response to multivariate abiotic stresses may signify the possible role of ZD gene family in response to different abiotic stresses. However, further studies are required to evaluate intensely the specific performance of the ZHD gene family in plant multivariate stresses.
Figure 8.
Inducible expression profile of pepper ZHD gene family members in response to abiotic treatments (heat, drought, cold and salt). The x-axis indicates the treatment. The y-axis shows the relative expression level of each treatment compared to control. The error bars indicate the standard deviations of the three independent qRT-PCR biological replicates.
Discussion
Identification and evolution of ZHD gene family in plants
The plant-specific ZHD gene family influences various biological processes in plants, including growth and development and responses to abiotic stress and phytohormones.31 Previously, it was postulated that plant-specific ZHD gene family has been found in land plants, but not in algae,18 signifying that ZHD gene family may have evolved before the divergence of land plants but after the separation of land plant lineages from single-celled algae. Previous studies reported the plant-specific ZHD gene family in Arabidopsis (17), rice (15), tomato (22), grapes (13), and Brassica rapa (31). In this study, we found a total of 10, 17, and 31 ZHD members in peach, Arabidopsis, and apple, respectively (Table 1). Among them, 2, 3, and 11 genes were found to be MIF (MINI ZINC FINGER) class of the ZHD gene family in peach, Arabidopsis, and apple, respectively. We noticed that MIF proteins share high levels of sequence similarity with the ZF domain of the identified ZHD proteins, suggesting that MIF proteins might interfere with the functions of ZHD proteins through their ZF domain. This study found similar number of ZHD gene family member in Arabidopsis; however, the number of ZHD gene family members was higher in apple and lesser in peach compared with the reported ZHD members in other plants. The number of ZHD genes in apple (750 Mb) is relatively small compared with the smaller-genome plants including Arabidopsis (164 Mb), rice (441 Mb), B. rapa (283.8 Mb), and grapes (466 Mb), whereas similar to B. rapa ZHD gene family, indicating that the duplication events might lead to the expansion of the ZHD gene families in smaller-genome size plants during evolution. For example, it was reported that 4 different types of large-scale duplications occurred in the genome of Arabidopsis, and most of the Arabidopsis genes ascended as a result of that duplication in the genome.18,32,33 Furthermore, the conserved ZF-HD dimer domains and protein parameters of the ZHD gene family members in apple, peach, and Arabidopsis were similar suggesting that ZHD proteins shared common gene structure and domain architecture during evolutions. The Arabidopsis, apple, and peach proteins were divided into 4 subfamilies (Figure 1), and we observed quite a common feature for ZHD gene family members within each subfamily. Previous studies reported that MIF class of ZHD family members of various crops phylogenetically clustered into a distinct clade from ZHD proteins. Similar clustering was observed for MIF proteins in this study, where most of the MIF proteins were clustered into a separate subfamily (subfamily I). This attitude of MIF proteins implying that MIF protein may lose the HD domain after deriving from ZHD genes.
Multiple Expectation for Motif Elicitation analysis found various conserved motifs in the studied ZHD proteins (Supplemental Figure S3), and we observed similar motif distribution within subfamilies, suggesting their functional similarity within the same subfamily. The previous studies found that most of the ZHD genes were intronless; however, this study found 1 to 5 introns for some ZHD genes (Supplemental Figure S2) signifying that exon/intron gain/loss exonization/pseudo-exonization, and insertion/deletion are responsible for the structural divergence of genes. Moreover, the similar exon-intron distribution within subfamilies proposes that ZHD shared highly conserved architecture during evolution. Segmental duplication, tandem duplication, and transposition are the major contributors to gene duplication in evolution.23 In this study, we found that 14 pairs of genes arose from segmental duplication in apple (Figure 2). Moreover, no tandem duplicated gene was investigated suggesting that segmental duplication may play a vital role in the expansion of the ZHD gene family in plants.
Expression profiling ZHD genes in various organs indicating their role in plant growth and development
The members of the same gene family have a common transcript level in plants. This may coordinate and/or differ in the functional interaction of the family members. Previous studies found that ZHD gene family regulates various biological processes in plants, including growth and development and responses to abiotic stress and phytohormones.1,18,31 In tomato, high expression was observed for the SlZHD members, indicating their role in flower bud development.19 Similarly, 3 SlZHD genes, namely, SlZHD7, SlZHD8, and SlZHD9, were more expressed in leaves signifying their role in leaf development. In Arabidopsis, ZHD gene family members were found with high transcript level in floral tissues, revealing their vital regulatory role in floral tissue development.17 In grapes, the VvZHD genes exhibit varied transcript level in 6 organs including root, stem, leaves, flower, fruit, and tendrils, signifying widespread roles in plant growth and development.1 The ZHD proteins work as transcriptional controllers with unique biochemical properties that control the regulation of floral development in Arabidopsis.6 Similarly, in this study, the ArrayExpress data were used to investigate the expression pattern of the zinc finger-homeodomain gene family members in different tissues and organs, and their possible task in apple growth and development (Figure 4). The ArrayExpress data confirmed that the expression of ZHD family members was more in flowering and leaf-related samples. Some members were also expressed highly in other tissues like stem, root, and seedling. Moreover, qRT-PCR found that most of the ZHD members were highly expressed of leaf and seedling in Arabidopsis (Supplemental Figure S6). Some ZHD members showed significant expression in other tissues of Arabidopsis, including flower, stem, and root. Furthermore, most of the ZHD gene family members showed a high transcript level in flower, leaf, and fruit in peach (Figure 5). On the contrary, a high number of ZHD was greatly expressed in leaf, bud, flower, and fruit, whereas only MdZHD6 gene was detected with high transcript activities in the stem (Figure 6). Previously, it was reported that leaves and buds were greatly affected by the flower induction in Arabidopsis and other plants.13,34 In this study, we also found some ZHD members that were greatly expressed in the leaves and bud samples, suggesting their possible contribution to the flower induction in the plant. The results further revealed that the differences in the transcript level in the tissues may play important role in plant development and some ZHD members have a specific function in different developmental stages.
Inducible expression patterns under a number of flowering-related treatments and abiotic stresses infer the vital contributions of ZHD gene members to flowering induction and abiotic stresses
Several studies have proposed that ZHD genes are vital for the regulation of various biological processes in plants, including growth and development and responses to abiotic stress and phytohormones.1,18,31 In Arabidopsis, AtZHD1 works as a transcription regulator that sticks to the promoter region of ERD1 (EARLY RESPONSE TO DEHYDRATION STRESS 1), and its transcript was greatly affected under drought, salinity, and ABA.7 Moreover, ZFHD1 combines with some NAC proteins, and the overexpression of ZFHD1 and NAC genes enhanced tolerance under drought stress in Arabidopsis.17 Recently, some studies revealed the function of ZHD genes in other plants. For example, GmZF-HD1 and GmZF-HD2 directly triggered calmodulin isoform 4 (GmCaM4) coding gene on pathogen stimulation in soybean.8 It was also reported that 4 ZHD genes have also been involved in gene regulation in rice.35 In grapes, VvZHD genes showed substantial increase in their transcript level under dehydration and/or high salinity.1 Similarly, they also found that the differences in transcript level of VvZHD genes under different plant hormones proposed potentially important targets for promoting the resistance of grape to stress conditions,1 as previously it was found that ZHD gene family control flower development in plants.
Previous studies investigated that a number of environmental factors including temperatures, hormones, and photosynthesis control the flower induction in plants.24,36,37 It was also found that hormones and sugar change the flowering rate in apple; however, the association of apple ZHD genes with hormones and sugar remains unknown. Till now, GA, 6-BA, and sugar are considered the major factors linked with flower induction. Previously, it was also found that exogenous treatment (GA3, 6-BA, and sugar) can affect the flowering rates.28-30 Although some studies investigated that the ZHD genes influence the regulation of plant growth and development. Nevertheless, the possible contribution of ZHD genes in the flower induction in fruit species is still unknown.
Based on these results, in this study, we found that ZHD gene family members exhibited different transcript levels in response to flowering-related hormones and treatments at a different time point in flowering buds (Figure 7). In response to exogenous GA, we noted that the transcript of some ZHD genes was markedly affected at 50 DAFB. Similarly, the transcripts of some apple ZHD genes were triggered at different time points in flowering-related buds in response to exogenous sugar and 6-BA, suggesting the potential role of ZHD gene family in flowering induction. Furthermore, we also found some pepper ZHD gene families, namely, CaZHD3, CaZHD6, CaZHD7, and CaZHD8, which were highly expressed under abiotic stresses including heat, drought, cold, and salt (Figure 8). Some ZHD gene family members displayed high transcript level under all the applied abiotic stresses, whereas some were highly expressed under 2 different abiotic stresses at certain time points.
Altogether, most ZHD genes were expressed at significantly higher levels under exogenous flowering-related treatments and abiotic stresses at different time points in apple and pepper, signifying their possible contribution in the flowering induction and abiotic stresses in apple and pepper. Moreover, we further suggested that ZHD gene family may accomplish numerous tasks in plant growth and development and in response to flowering-related hormonal applications and abiotic treatments. But their exact role remains unknown, and further research must be done to reveal the particular role of the ZHD gene family in plant growth and development.
Conclusion
This context identified a total of 17, 10, and 31 ZHD genes in the Arabidopsis, peach, and apple genome, which were distributed unevenly in the genome. We classified these ZHD gene family members and performed their phylogenetic, structural, synteny, and functional analyses. Furthermore, we explored their transcript level in different tissues including root, leaf, stem, flower, fruit, and seedling in Arabidopsis, peach, and apple. Moreover, the expression pattern of ZHD members was evaluated in response to various exogenous flowering-related treatments and abiotic stresses. The findings of this study indicate that ZHD gene family may play a vital role in the plant growth and development and some of the members may participate in the flower induction in fruit plants. Also, it could be worth studying the expression of these genes in different phytohormones and metals such as melatonin,38 brassinosteroids,39,40 strigolactones,41 and iron stress.42 These results may be valuable for the betterment of flower induction in plants.
Supplemental Material
Supplemental material, Suply_xyz20395669ea5ba for Zinc Finger-Homeodomain Genes: Evolution, Functional Differentiation, and Expression Profiling Under Flowering-Related Treatments and Abiotic Stresses in Plants by Abdullah Shalmani, Izhar Muhammad, Rahat Sharif, CaiPing Zhao, Uzair Ullah, Dong Zhang, Xiu-Qing Jing, Bakht Amin, Peng Jia, Muhammad Mobeen Tahir, Ze Xu, Kun-Ming Chen and Na An in Evolutionary Bioinformatics
Footnotes
Funding:The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was financially supported by the National Science and Technology Supporting Project (2013BAD20B03), China Apple Research System (CARS-28), National Spark Plan Program (2014GA850002), Science and Technology Innovative Engineering Project in Shaanxi province, China (2015NY114), China Postdoctoral Science Foundation (2014M56806), Yangling Subsidiary Center Project of the National Apple Improvement Center, and Collaborative Innovation of the Center for Shaanxi Fruit Industry Development.
Declaration of Conflicting Interests:The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: AS, IM and RS conceived the manuscript. CZ, UU, DZ, XQJ, BA, PJ, MMT and ZX helped in performing the experiment and bioinformatics work. KMC and NA supervised and financed the research work.
Supplemental material: Supplemental material for this article is available online.
References
- 1. Wang H, Yin X, Li X, et al. Genome-wide identification, evolution and expression analysis of the grape (Vitis vinifera L.) zinc finger-homeodomain gene family. Int J Mol Sci. 2014;15:5730-5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Krishna SS, Majumdar I, Grishin NV. Structural classification of zinc fingers: survey and summary. Nucleic Acids Res. 2003;31:532-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Klug A, Schwabe J. Protein motifs 5. Zinc fingers. FASEB J. 1995;9:597-604. [PubMed] [Google Scholar]
- 4. Mackay JP, Crossley M. Zinc fingers are sticking together. Trends Biochem Sci. 1998;23:1-4. [DOI] [PubMed] [Google Scholar]
- 5. Yanagisawa S. Dof domain proteins: plant-specific transcription factors associated with diverse phenomena unique to plants. Plant Cell Physiol. 2004;45:386-391. [DOI] [PubMed] [Google Scholar]
- 6. Windhovel A, Hein I, Dabrowa R, Stockhaus J. Characterization of a novel class of plant homeodomain proteins that bind to the C4 phosphoenolpyruvate carboxylase gene of Flaveria trinervia. Plant Mol Biol. 2001;45:201-214. [DOI] [PubMed] [Google Scholar]
- 7. Tran LSP, Nakashima K, Sakuma Y, et al. Co-expression of the stress-inducible zinc finger homeodomain ZFHD1 and NAC transcription factors enhances expression of the ERD1 gene in Arabidopsis. Plant J. 2007;49:46-63. [DOI] [PubMed] [Google Scholar]
- 8. Park HC, Kim ML, Lee SM, et al. Pathogen-induced binding of the soybean zinc finger homeodomain proteins GmZF-HD1 and GmZF-HD2 to two repeats of ATTA homeodomain binding site in the calmodulin isoform 4 (GmCaM4) promoter. Nucleic Acids Res. 2007;35:3612-3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Guitton B, Kelner JJ, Velasco R, Gardiner SE, Chagne D, Costes E. Genetic control of biennial bearing in apple. J Exp Bot. 2011;63:131-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xing L-B, Zhang D, Li Y-M, et al. Transcription profiles reveal sugar and hormone signaling pathways mediating flower induction in apple (Malus domestica Borkh.). Plant Cell Physiol. 2015;56:2052-2068. [DOI] [PubMed] [Google Scholar]
- 11. Li Y, Zhang D, Xing L, Zhang S, Zhao C, Han M. Effect of exogenous 6-benzylaminopurine (6-BA) on branch type, floral induction and initiation, and related gene expression in ‘Fuji’apple (Malus domestica Borkh). Plant Growth Regul. 2016;79:65-70. [Google Scholar]
- 12. Zhang S, Zhang D, Fan S, et al. Effect of exogenous GA3 and its inhibitor paclobutrazol on floral formation, endogenous hormones, and flowering-associated genes in ‘Fuji’apple (Malus domestica Borkh.). Plant Physiol Biochem. 2016;107:178-186. [DOI] [PubMed] [Google Scholar]
- 13. Porri A, Torti S, Romera-Branchat M, Coupland G. Spatially distinct regulatory roles for gibberellins in the promotion of flowering of Arabidopsis under long photoperiods. Development. 2012;139:2198-2290. [DOI] [PubMed] [Google Scholar]
- 14. Li J, Hou H, Li X, et al. Genome-wide identification and analysis of the SBP-box family genes in apple (Malus domestica Borkh.). Plant Physiol Biochem. 2013;70:100-114. [DOI] [PubMed] [Google Scholar]
- 15. Shalmani A, Fan S, Jia P, et al. Genome identification of B-BOX gene family members in seven rosacea species and their expression analysis in response to flower induction in Malus domestica. Molecules. 2018;23:1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kumar G, Arya P, Gupta K, Randhawa V, Acharya V, Singh AK. Comparative phylogenetic analysis and transcriptional profiling of MADS-box gene family identified DAM and FLC-like genes in apple (Malusx domestica). Sci Rep. 2016;6:20695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tan Irish VF. The Arabidopsis zinc finger-homeodomain genes encode proteins with unique biochemical properties that are coordinately expressed during floral development. Plant Physiol. 2006;140:1095-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hu W, dePamphilis CW, Ma H. Phylogenetic analysis of the plant-specific zinc finger-homeobox and mini zinc finger gene families. J Integr Plant Biol. 2008;50:1031-1045. [DOI] [PubMed] [Google Scholar]
- 19. Khatun K, Nath UK, Robin AHK, et al. Genome-wide analysis and expression profiling of zinc finger homeodomain (ZHD) family genes reveal likely roles in organ development and stress responses in tomato. BMC Genomics. 2017;18:695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jain M, Tyagi AK, Khurana JP. Genome-wide identification, classification, evolutionary expansion and expression analyses of homeobox genes in rice. FEBS J. 2008;275:2845-2861. [DOI] [PubMed] [Google Scholar]
- 21. Finn RD, Bateman A, Clements J, et al. Pfam: the protein families database. Nucl Acid Res. 2013;42:D222-D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bailey TL, Williams N, Misleh C, Li WW. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369-W373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hu B, Jin J, Guo A-Y, Zhang H, Luo J, Gao G. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2014;31:1296-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725-2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Guo W-L, Chen R-G, Gong Z-H, Yin Y-X, Li D-W. Suppression subtractive hybridization analysis of genes regulated by application of exogenous abscisic acid in pepper plant (Capsicum annuum L.) leaves under chilling stress. PLoS ONE. 2013;8:e66667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yu C-S, Cheng C-W, Su W-C, et al. CELLO2GO: a web server for protein subCELlular LOcalization prediction with functional gene ontology annotation. PLoS ONE. 2014;9:e99368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kong H, Landherr LL, Frohlich MW, Leebens-Mack J, Ma H, dePamphilis CW. Patterns of gene duplication in the plant SKP1 gene family in angiosperms: evidence for multiple mechanisms of rapid gene birth. Plant J. 2007;50:873-885. [DOI] [PubMed] [Google Scholar]
- 28. Gangappa SN, Botto JF. The BBX family of plant transcription factors. Trends Plant Sci. 2014;19:460-470. [DOI] [PubMed] [Google Scholar]
- 29. Chia T, Muller A, Jung C, Mutasa-Gottgens ES. Sugar beet contains a large CONSTANS-LIKE gene family including a CO homologue that is independent of the early-bolting (B) gene locus. J Exp Bot. 2008;59:2735-2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chu Z, Wang X, Li Y, et al. Genomic organization, phylogenetic and expression analysis of the B-BOX gene family in tomato. Front Plant Sci. 2016;7:1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang W, Wu P, Li Y, Hou X. Genome-wide analysis and expression patterns of ZF-HD transcription factors under different developmental tissues and abiotic stresses in Chinese cabbage. Mol Genetic Genom. 2016;291:1451-1464. [DOI] [PubMed] [Google Scholar]
- 32. Vision TJ, Brown DG, Tanksley SD. The origins of genomic duplications in Arabidopsis. Science. 2000;290:2114-2117. [DOI] [PubMed] [Google Scholar]
- 33. Blanc G, Hokamp K, Wolfe KH. A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res. 2003;13:137-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Galvao VC, Horrer D, Kuttner F, Schmid M. Spatial control of flowering by DELLA proteins in Arabidopsis thaliana. Development. 2012:4072. [DOI] [PubMed] [Google Scholar]
- 35. Figueiredo DD, Barros PM, Cordeiro AM, et al. Seven zinc-finger transcription factors are novel regulators of the stress responsive gene OsDREB1B. J Exp Bot. 2012;63:3643-3656. [DOI] [PubMed] [Google Scholar]
- 36. Griffiths S, Dunford RP, Coupland G, Laurie DA. The evolution of CONSTANS-like gene families in barley, rice, and arabidopsis. Plant Physiol. 2003;131:1855-1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ledger S, Strayer C, Ashton F, Kay SA, Putterill J. Analysis of the function of two circadian-regulated CONSTANS-LIKE genes. Plant J. 2001;26:15-22. [DOI] [PubMed] [Google Scholar]
- 38. Sharif R, Xie C, Zhang H, et al. Melatonin and its effects on plant systems. Molecules. 2018;23:E2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Clouse SD, Sasse JM. Brassinosteroids: essential regulators of plant growth and development. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:427-451. [DOI] [PubMed] [Google Scholar]
- 40. Gudesblat GE, Russinova E. Plants grow on brassinosteroids. Curr Opin Plant Biol. 2011;14:530-537. [DOI] [PubMed] [Google Scholar]
- 41. Brewer PB, Koltai H, Beveridge CA. Diverse roles of strigolactones in plant development. Mol Plant. 2013;6:18-28. [DOI] [PubMed] [Google Scholar]
- 42. Muhammad I, Jing X-Q, Shalmani A, et al. Comparative in silico analysis of Ferric Reduction Oxidase (FRO) genes expression patterns in response to abiotic stresses, metal and hormone applications. Molecules. 2018;23:1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, Suply_xyz20395669ea5ba for Zinc Finger-Homeodomain Genes: Evolution, Functional Differentiation, and Expression Profiling Under Flowering-Related Treatments and Abiotic Stresses in Plants by Abdullah Shalmani, Izhar Muhammad, Rahat Sharif, CaiPing Zhao, Uzair Ullah, Dong Zhang, Xiu-Qing Jing, Bakht Amin, Peng Jia, Muhammad Mobeen Tahir, Ze Xu, Kun-Ming Chen and Na An in Evolutionary Bioinformatics