Abstract
Alzheimer’s disease (AD) is a type of neurodegenerative disorder and the most common form of dementia. MicroRNA (miRNA) has been shown to play a role in various diseases, including AD. It also has been reported to regulate autophagy. We extracted miRNA from blood samples and constructed an miRNA-101a lentivirus vector. In this study we found the level of miRNA-101a was significantly reduced in the plasma of patients with AD and APPswe/PS1ΔE9 transgenic mice. The relative expression of miRNA-101a exhibited a relatively high diagnostic performance (area under receiver operating characteristic curve: 0.8725) in the prediction of AD with a sensitivity of 0.913 and a specificity of 0.733 at the threshold of 0.6463. Under electron microscopy, autophagic vacuoles in AD-related cells numbered more than the cells up-regulating miRNA-101a in the in vitro experiments. Dual-luciferase reporter assay and Western blot results proved that the MAPK1 pathway plays a role in the formation of autophagic vacuoles in AD. This study found that the autophagy phenomenon regulated by miRNA-101a via the MAPK pathway might be a new mechanism in AD. This could provide new insights into AD formation and treatment.
Keywords: MicroRNA-101a, Alzheimer’s disease, autophagy, MAPK1
Introduction
Alzheimer’s disease (AD) is a type of neurodegenerative disorder and the most common form of dementia1. A number of complex neuropathologic factors play a role in AD, such as neurofibrillary tangles, neuritic plaques, neuroinflammation, and neuronal shrinkage2–4. Although these hallmarks have been suspected to be the cause of AD, a clear understanding of AD mechanisms has not yet been reached5.
Autophagy is a cellular process that involves self-degradation and recycling of macromolecules and cellular organelles6,7. It is a degradation pathway for the turnover of dysfunctional organelles or aggregated proteins in the cell. Recent reports found that autophagy plays a role in AD8. The accumulation of lysosomes and their hydrolases within neurons is a well-established neuropathologic feature of AD. Lysosomal pathology in the AD brain involves extensive alterations of macroautophagy. Autophagic vacuoles were uncommon in brains devoid of AD pathology but were abundant in AD brains, particularly within neurotic processes, including synaptic terminals9.
It has been reported that the MAPK1 signaling pathway plays a crucial role in the regulation of many cellular biological processes, including autophagy10. Interestingly, MAPK has special functions as both a positive and a negative regulator of autophagy11,12. The alternative process for autophagy is regulated by several autophagic proteins, including beclin-1. MAPK can promote autophagy through its phosphorylation of BCL2, which releases beclin-1 from its association with BCL2 to function in autophagosome formation13. Autophagic vesicles have recently been shown to contain AβPP, as well as the secretase activities required to generate Aβ, and are particularly highly enriched in β-secretase enzymatic activity and γ-secretase complex components14. All of these data demonstrate that autophagy plays an important role in the formation of AD.
A variety of biological processes are modulated by microRNAs (miRNAs). Several in vitro and in vivo studies have explored the functional roles of miRNAs in the pathogenesis of AD15. Previous studies have documented that a number of miRNAs, including the miRNA-146, miRNA-101, miRNA-9, miRNA-29, miRNA-107, miRNA-106 and miRNA-153 families, are deregulated in AD brains and play vital roles in the pathogenesis of AD. Dysregulation of miRNAs has been reported to contribute to AD via modulation of autophagy. Such findings are not surprising considering the fact that miRNAs are key regulators of autophagy. The possible mechanisms of how miRNA-autophagy regulates the formation of AD and whether MAPK plays a role in this process are not clear.
The importance of the miRNA–autophagy interconnection is only beginning to be elucidated. It will be intriguing to further understand these interactions in the coming years. Consequently, many of these findings may provide promising possibilities for future treatment strategies in AD.
Materials and Methods
Sample Collection
The study population was recruited at The First Affiliated Hospital of Zhengzhou University, China, following a standardized protocol in compliance with the National Institute of Neurological and Communicative Disorders and Stroke-Alzheimer’s Disease and Related Disorders Association criteria for probable AD. The severity of dementia was assessed by the Mini-Mental Status Examination (MMSE) and the Clinical Dementia Rating scale. Computed tomography and magnetic resonance imaging were also performed to validate the diagnosis of AD.
Control participants, without dementia, matched for age, sex and education were recruited from the same hospital. Control participants had no evidence of clinical dementia, MMSE scores >28, and no evidence of any significant abnormalities (e.g. cerebrovascular disease) on structural neuroimaging. Recruited participants underwent a complete clinical and laboratory investigation. Exclusion criteria were based on history of depression or psychosis, alcohol or substance abuse or use of psychoactive medications. After a signed informed consent by patients and parents, experiments with human samples were performed in accordance with the Declaration of Helsinki, as part of a protocol approved by the institutional review board.
Sample Preparation and miRNA Extraction
The blood samples were collected in NaF/KOx tubes. At 4°C, the blood samples were centrifuged at 3000 r/min for 10 min and the supernatants were transferred into new tubes. The plasma samples were stored at −80°C until use. The miRNA was extracted from 400 µl plasma samples using a miRcute miRNA isolation kit (TIANGEN Biotech, Beijing, China). The samples were eluted in a final volume of 30 µl. A NanoDrop 2000 Spectrophotometer (Thermo Scientific, Middlesex, MA, USA) was used to measure the total RNA concentrations and purities.
MiRNA Microarray Assay and Data Analysis
miRNA was sent to LC Sciences (delivered by the Lianchuan Biotechnology Company, Hangzhou, China) for miRNA expression profiling using their proprietary μParaflo microfluidic chip, which contains 2042 human mature miRNA probes (Sanger miRBase 19.0). By using poly (A) polymerase, the assay began with the extension of the miRNA samples at the 3′ end with a poly (A) tail. For subsequent fluorescent dye staining, an oligonucleotide tag was ligated to the poly (A) tail. Hybridization was performed overnight on a μParaflo microfluidic chip by using a micro-circulation pump. Tag-conjugated Cy3 dye was circulated through the microfluidic chip for dye staining after RNA hybridization. Fluorescent images were collected by a laser scanner (GenePix 4000B, Microarray Scanner) and digitized (Array-Pro image analysis software, Media Cybernetics Rockville, MD, USA). The data were analyzed by first subtracting the background and then normalizing the signals with a locally weighted regression (Lowess) filter. The miRNA gene targets were determined with the TargetScan Human online miRNA database (http://mirfocus.org/index.php).
Quantitative Real-time Polymerase Chain Reaction Verification
Complementary DNAs (cDNAs) were generated for the miRNA samples from the ‘screening’ sets via reverse transcription with the Reverse Transcription System (Promega, A3500, Madison, WI, USA), which consists of stem-looped reverse transcription primers. By using the LightCycler 1.5 Real-time Polymerase Chain Reaction (PCR) system (Roche Applied Science, Penzberg, Germany), triplicate quantitative real-time (qRT)-PCRs of the cDNA sample were performed. The U6 small RNA and β-actin mRNA were used as internal controls. All reactions were run in triplicate and the primers were followed: F 5′GGCAGTTATCACAGTGCTGATGCT3′, R: 5′GCGCGTACAGTACTGTGATAACTGAA3 ′ for miRNA101a; F: 5′CGCTTCGGCAGCACATATAC3′ and R: 5′TTCACGAATTTGCGTGTCAT3′ for U6; F: 5′GTCACCAACTGGGACGACAT3′ and R: 5′GAGGCGTACAGGGATAGCAC3′ for β actin mRNA.
Animals and Tissues
The APPswe/PS1ΔE9 transgenic mice used in the present study were provided by the Department of Laboratory Animal Science, Peking University Health Science Centre (Beijing, China). The APPswe/PS1ΔE9 transgenic mice were produced via co-injections of APPswe and PS1ΔE9 plasmids on a C57BL/6 J genetic background. APPswe/PS1ΔE9 double transgenic mice develop behavioral phenotypic and pathological features that make them useful as an AD model. These mice exhibit spatial memory deficits at 3 months of age and senile plaques in the brain tissue at 4.5 months of age16.
In Situ Hybridization
In situ hybridization was performed with the MicroRNA ISH Buffer and Controls Kit according to manufacturer’s protocol (Boster Biological Technology Ltd., WuHan, China) with some modifications. Briefly, the brain tissues of 0- and 9-month-old APPswe/PS1ΔE9 double transgenic mice and age-matched C57BL/6 J mice were fixed with 4% paraformaldehyde solution with 1/1000 diethylpyrocarbonate, and embedded in paraffin. The slides were deparaffinized and incubated with proteinase-K for 10 min at 37°C and washed with phosphate-buffered saline (PBS). After washing in PBS, the sections were prehybridized for 2–4 h at 38–42°C in prehybridization buffer. Hybridization with DIG-labelled riboprobes was performed overnight at 38–42°C in hybridization buffer. The hybridization buffer contained 5 nM double-DIG LNA™ microRNA probes for miRNA-101a-3p or scramble-miRNA as the negative control (Boster, WuHan, China). After hybridization, the sections were washed in 5 × SSC (sodium solution citrate) for 10 min, 0.5 × SSC for 15 min and 0.2 × SSC for 15 min at room temperature. Blocking was performed for 2 h at 37°C with blocking buffer and alkaline phosphatase-conjugated Fab anti-DIG antibody. Staining was performed using 3,3-diaminobenzidine (DAB).
miRNA-101a Lentivirus Vector Construction
The miRNA-101a (MI0000148) sequence was obtained from the miRBase database. The pFU-GW-RNAi vector was linearized by double-enzyme (XbaI and HpaI) restriction (New England BioLabs), and the target gene was introduced into the AQ1 vector to obtain the pGC-LV recombinant vector, which was transformed into competent cells. After being identified by PCR and sequenced, the competent cells, connected with the lentiviral packaging vectors, pHelper1.0 and pHelper2.0 (Genechem Co. Ltd., Shanghai, China), were concurrently transfected into 293 T packaging cells by lipofectamine 2000 (Invitrogen, Waltham, MA, USA) to create the virus. The virus titer was detected throughout the entire dilution method. A lentiviral vector containing only green fluorescent protein was prepared by the same method to act as the control vector. Lentiviral vectors were stored at −80°C17.
Cell Culture and Treatment
Human neuroblastoma SH-SY5Y cells were maintained and cultured as previously described. Briefly, SH-SY5Y cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM)-GlutaMAX (Invitrogen) supplemented with 10% fetal bovine serum (FBS). For all experiments, cells were differentiated 24 h after plating by treatment with 10 mM all-trans retinoic acid (Sigma Aldrich, St. Louis, MO, USA) in complete growth medium for 7 days. Media were changed every 2 days. For oxidative stress studies, differentiated SH-SY5Y cells were treated with H2O2 (500 mM final concentration in medium) for 30 min as previously described18. After 24 h, these cells were infected with 0.03 μg miRNA-101a-LV or negative control RNA (FU-RNAi-NC-LV), with optimal infection efficiency occurring at 30–50% confluence. The cultured SH-SY5Y were divided into four groups: control group: SH-SY5Y cells; AD cell model group: SH-SY5Y cells treated with treated with H2O2; transfected group: (the AD model cells transfected with miRNA-101a); and negative control group: (AD model cells transfected with FU-RNAi-NC-LV). Cells were prepared for transmission electron microscopy (TEM) and Western blot.
TEM
The three groups were fixed with 2.5% glutaraldehyde in 0.1 M PBS for 90 min at room temperature, and then fixed in 1% osmium tetroxide for 30 min. The cells were progressively dehydrated in a 10% graded series of 50–100% ethanol and propylene oxide, and then embedded in Epon 812 resin after being washed with PBS. The blocks were cut into ultrathin sections by using a microtome, which were then stained with saturated uranyl acetate and lead citrate. The cellular ultrastructure was examined by using a transmission electron microscope (Olympus CKX41, U-CTR30-2).
Dual-Luciferase Reporter Assay
The 5′-flanking regions of the mmu-miRNA-101 were synthesized. According to the Miranda system, segments in the 3′-UTR of the mouse MAPK1 were amplified by genomic PCR and cloned between the Xho1-Not1 sites of psiCHECK-2 (Promega).
The 293 T-cells were seeded at a density of 2 × 105 cells/well in 24-well plates and co-transfected with 500 ng plasmid DNA in psiCHECK2 (Promega), and 1 μl lipofectamine 2000 (Invitrogen). A luciferase activity assay was performed 48 h after transfection using the dual-luciferase reporter assay system (Promega). All transfection experiments were performed in triplicate and repeated at least in triplicate. The following primer sequences were used: MAPK1SallF:5′ACGCGTCGACATTGGTCAGGACAAGGGCTCAGAG 3′, MAPK1NotIR:5′ATAAGAATGCGGCCGCGTAATTCTTTTTACAAGTCAGGTGCCATAAAC 3′; mutMAPK1F:5′TCTGACATGGCTCTGTATCTGCTCTACAGTACGGATGCCTTTTTGGTGTTGTATCCC 3′, mutMAPK1 R:5′GGGATACAACACCAAAAAGGCATCCGTACTGTAGAGCAGATACAGAGCCATGTCAGA 3′.
Western Blot
Primary antibodies used in these experiments were anti-MAPK1 (1:1000, Santa Cruz Biotechnology), anti-LC3 (1: 1000, Santa Cruz Biotechnology), and β-actin protein (1: 1000; sc-81178; Santa Cruz Biotechnology). Cells from each group were digested (50 mM Tris–HCl (pH 6.8), 10 mM ethylenediaminetetraacetic acid (EDTA), 2% sodium dodecyl sulfate (SDS), 5 mM dithiothreitol, 0.5 mM phenylmethanesulfonyl fluoride) and then the supernatants were collected after centrifugation of 15,000 × g for 1 h. Cell lysate (100 ml) was collected to perform protein quantification using the Bradford method. Equal amounts of protein (20 mg) from each cell sample were separated by SDS- polyacrylamide gel electrophoresis (PAGE) on an 8% polyacrylamide gel and transferred to polyvinylidene fluoride membrane (Millipore Corp., Bedford, MA, USA). This membrane was incubated overnight with a primary antibody (dilution 1:1000) at 41°C, then incubated with a horse radish peroxidase-conjugated secondary antibody (Zymed Laboratory, San Francisco, CA, USA) for 1 h at room temperature. Detection of reactive antigens was performed using an enhanced chemiluminescence (ECL) kit (Santa Cruz Biotechnology). The resulting image was analyzed with ChemiImager 4000 (Alpha Innotech, San Leandro, CA, USA) for protein band densitometry.
Statistical Analysis
The data were analyzed by one-tailed unpaired Student’s t test for two groups in the experiments using Prism 5 software. For three or more groups in the experiment, the data were analyzed by one-way analysis of variance followed by Bonferroni’s multiple comparison tests by using Prism 5 software. A probability of p < 0.05 was considered significant in all comparisons.
Results
Patients
MiRNA array assays were used to profile the miRNAs of plasma samples from five patients with AD and five healthy volunteers. The results were confirmed in the plasma of a validation cohort of 46 patients with AD and 60 healthy volunteers using real-time PCR. The baseline characteristics are listed in Table 1. There were no significant differences in age or sex between the patients with AD and control groups in the microarray data. There were no significant differences in age or sex between the patients with AD and control groups in the qRT-PCR data.
Table 1.
Baseline Characteristics. Overall and by Group.
| Group | AD | Normal |
|---|---|---|
| Sex (M/F) | 15/36 | 30/35 |
| Age (y) | 69.2±3.5 | 70.2±2.8 |
| MMSE score | 29±1 | 22±1 |
AD: Alzheimer’s disease; MMSE: Mini-Mental Status Examination.
miRNA-101a is Significantly Decreased in Patients with AD and APPswe/PS1ΔE9 Mice with Increased Age
miRNA Expression in the Peripheral Blood Plasma of Patients with AD
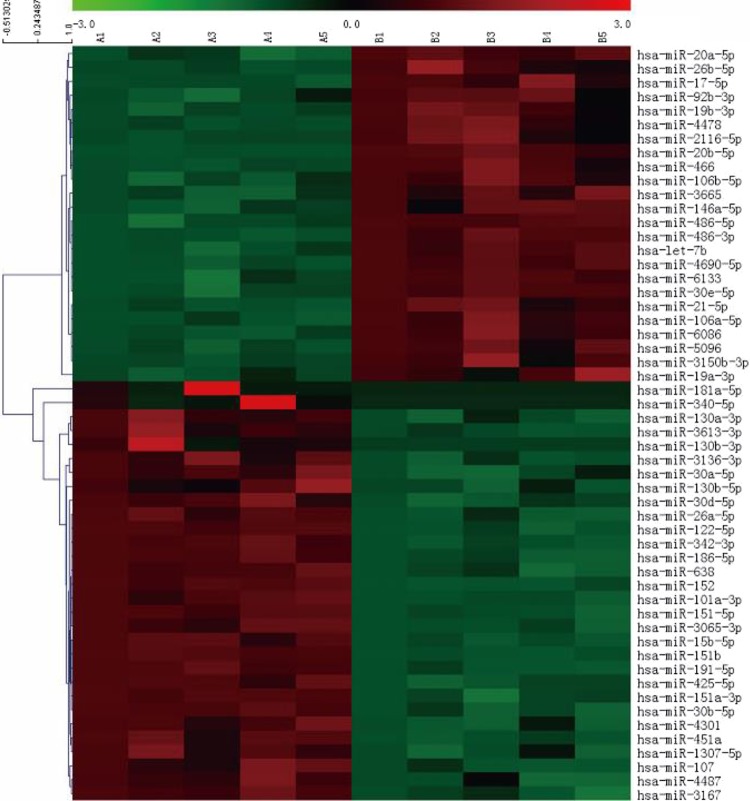
The miRNA array assays were used to profile the miRNAs of plasma samples from five patients with AD and five healthy volunteers. A total f 54 miRNAs exhibited differences in expressions that were exceeded two-fold between the AD and control plasma samples based on the microarray analyses (p < 0.05). Overall, 30 miRNAs were downregulated, including miRNA-186-5p, miRNA -36b-5p, miRNA -15b-5p, miRNA -151a-5p, miRNA -181a-5p, miRNA-101a-3p and miRNA -3167, and 24 miRNAs were upregulated, including miRNA -106a-5p, miRNA -6133, miRNA -146a-5p, let-7b and miRNA -30e-5p. K-means clustering analysis identified 14 miRNAs with distinct temporal expression patterns that were in the same cluster according to the microarray results, including miRNA -3167, miRNA -342-3p, miRNA -151-5p, miRNA -451a, miRNA -122-5p, miRNA -186-5p, miRNA -638, miRNA -4487, miRNA-101a-3p, miRNA -30d-5p, miRNA -107, miRNA -3065-3p, miRNA -30b-5p and miRNA -26a-5p. Of the miRNAs that were predicted to regulate AD genes, only miRNA-101a-3p exhibited a reciprocal pattern of expression. Gene ontology analysis revealed that miRNA-101a-3p potentially targets 621 genes, 65 of which are associated with AD. Thus, miRNA-101a-3p might play an important role in the mechanism of AD (Figure 1).
Fig. 1.
Heatmap of miRNA microarray. Hierarchical clustering of differentially expressed miRNAs was shown in paired AD-control samples. A total of 54 miRNAs exhibited differences in expression between the patient with AD and control plasma samples that exceeded two-fold based on the microarray assay. Red indicates overexpression; green represents downregulation.
AD: Alzheimer’s disease; miRNA: microRNA.
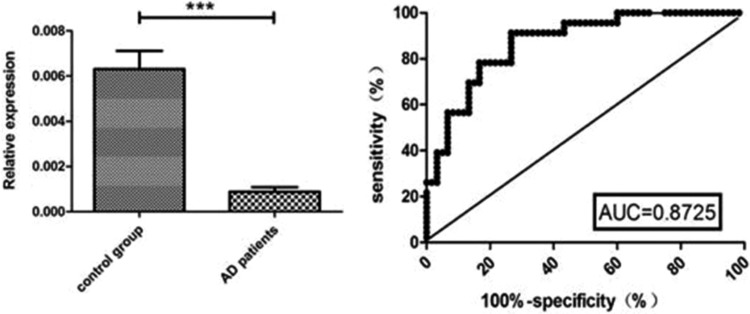
The results were confirmed in the plasma of a validation cohort of 46 patients with AD and 60 healthy volunteers using real-time PCR. The qRT-PCR results indicated that the relative expressions of miRNA-101a were significantly more downregulated in the AD patient group than in the control group; the expression in the AD group was significantly more downregulated compared with that of the control group (p < 0.01). The relative expressions of miRNA-101a exhibited a relatively high diagnostic performance (area under receiver operating characteristic curve: 0.8725) in the prediction of AD with a sensitivity of 0.913 and a specificity of 0.733 at the threshold of 0.6463 (Figure 2).
Fig. 2.
The concentration of miRNA-101a in AD and control groups. The left panel shows the relative expressions of miRNA-101a. The relative expression of miRNA-101a was significantly downregulated in the AD group compared with the control group (p < 0.01). The right panel shows the ROC curve.
AD: Alzheimer’s disease; miRNA: microRNA; ROC: receiver operating characteristic.
miRNA Expression in APPswe/PS1ΔE9 Mice with Increased Age
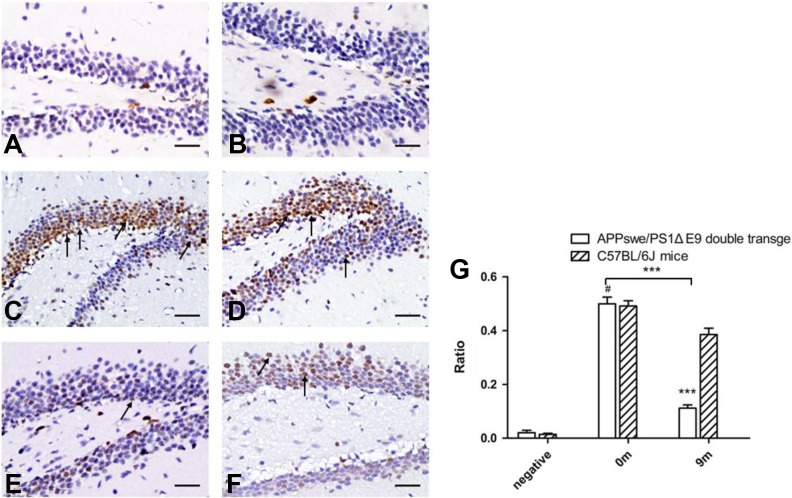
Meanwhile, we tested the expression of miRNA-101a in the hippocampus of APPswe/PS1ΔE9 mice. The expression of miRNA-101a in the brain tissue declined gradually with increasing age (Figure 3).
Fig. 3.
In situ hybridization of brain tissue. For each pair of images, the APPswe/PS1ΔE9 double transgenic mice (A, C, and E) are shown on the left, and the age-matched C57BL/6 J mice (B, D, and F) are shown on the right. A and B show the negative control. C and D are from 0-month-old mice, and in situ hybridization revealed no difference in miRNA-101a-3p expression between C and D. E and F are from 9-month-old mice, and in situ hybridization revealed that the level of miRNA-101a-3p expression in E was lower than that in F. (Original magnification ×200.)
miRNA: microRNA.
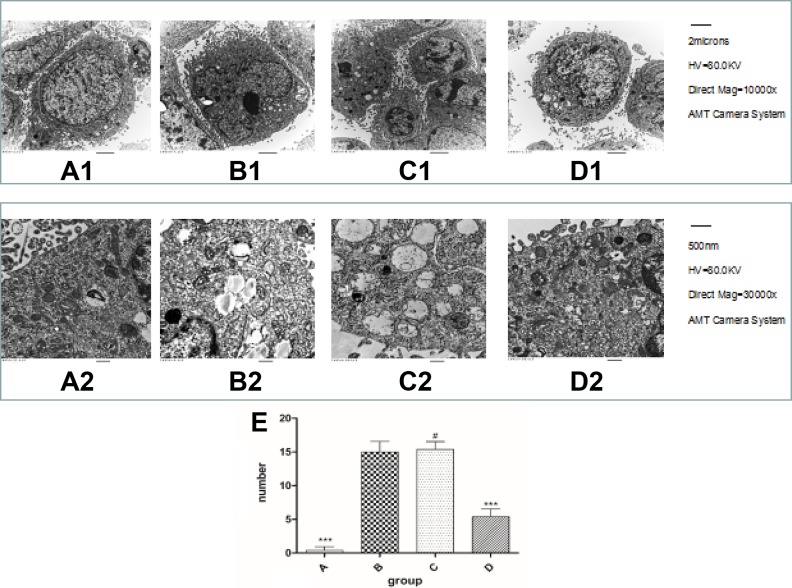
Changes in Autophagy Revealed by TEM in AD Model Cells
TEM is the standard method to detect autophagy. To further investigate whether miRNA-101a regulated the autophagy phenomenon in the modulated Alzheimer’s-associated pathogenesis, we used TEM to detect the four groups described earlier. The SH-SY5Y cells exhibited the normal ultrastructural morphology of cytoplasm, organelles and nuclei (Figure 4A1, A2). The AD model cells had abundant autophagic vacuoles and lysosomes (Figure 4B1, B2). After transfection with miRNA-101a, the autophagic vacuoles and lysosomes numbered less than the AD model cells (Figure 4D1, D2). However, the morphological change in the negative control group (Figure 4C1, C2) was similar to AD model cells.
Fig. 4.
The formation of autophagy bubbles revealed by transmission electron microscopy. A1–D1 ×10,000; A2–D2 ×30,000. A1 and A2 are SH-SY5Y cells. They showed abundant and normal morphology of cytoplasm, cell organelles and nuclei. B1 and B2 were from the AD model cells. There were abundant autophagic vacuoles and lysosomes. C1 and C2 were from the negative control group; the autophagic vacuoles are similar to the AD cell model. After transfection with miRNA-101a, the autophagic vacuoles and lysosomes numbered fewer than the AD model cells, D1 and D2.
AD: Alzheimer’s disease.
miRNA-101a Might Regulate Autophagy Through the MAPK Pathway in AD Model Cells
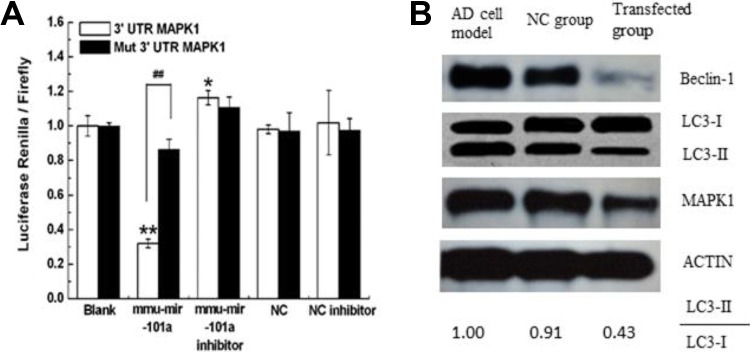
To explore the possible mechanism by which miRNA-101a regulates autophagy in Alzheimer’s-associated pathogenesis, we performed a search for genes regulated by miRNA-101a using the Miranda program. After the preliminary screening and experiment, we chose to investigate MAPK1 as a possible target of miRNA-101a. We constructed a luciferase reporter plasmid. Then HEK-293 T-cells were co-transfected with the vector and mmu-miRNA-101a or control and the relative luciferase activity was determined. The result showed that when compared with the control, the relative luciferase activity was significantly decreased by miRNA-101a, while the luciferase activity was not altered by the vector containing the mutant 3′-UTR (p < 0.01; Figure 5A). The suppression depended on the presence of miRNA-101a targeting sequences. It indicated that MAPK1 was targeted by miRNA-101a.
Fig. 5.
MiRNA-101a regulated autophagy might through MAPK pathway. A shows miRNA-101a targeted MAPK1 by dual-luciferase assay. Relative luciferase activity assays of luciferase reporters with MAPK1 or mut-MAPK1 3’-UTR were performed after co-transfection with miRNA-101a mimics, inhibitor or control. Transfected with psiCHECK2-MAPK1 (blank), co-transfected with miRNA-101a, co-transfected with miRNA-101a inhibitor, miRNA-101a negative control (NC), and miRNA-101a- (NC inhibitor) were established. The relative luciferase activity was significantly reduced when MAPK1-UTR reporter vectors containing the MAPK1 binding site were co-transfected together with mmu-miRNA-101a compared with the blank and NC (n = 6; *p < 0.01). This reduction was not observed when mut-MAPK1 or control expression vectors were used (n = 6; # p > 0.05). B shows the expression of protein MAPK1, beclin-1, and LC3 in the three groups. In the AD cell model, the key proteins for autophagy were highly regulated. After transfection with miRNA-101a, the expressions of beclin-1 and LC3-II were downregulated. In the NC group, the expressions of MAPK1, beclin-1, and LC3-II were similar to the AD model cells.
AD: Alzheimer’s disease; miRNA: microRNA; NC: negative control.
Then we checked the expression of MAPK1 protein and the autophagy index protein LC3 and beclin-1 in the three groups by Western blot (Figure 5B).
Discussion
MicroRNAs are abundantly expressed in the brain where they play important roles in neural development and function. They are also involved in many aspects of neuronal biology, including proliferation, apoptosis, synaptic plasticity, and neuroprotection19,20. Recently, it has been reported that miRNAs are detectable in plasma and that circulating miRNAs have a potential as new biomarkers for various cancers, diabetes and neurodegenerative diseases.
Changes in microRNA expression have been observed in the brains of patients affected by various neurological diseases, including AD. Indeed, several in vitro and in vivo studies have explored the functional roles of microRNAs in AD pathogenesis15. In the present study, 54 miRNAs exhibited differences in expression that exceeded two-fold between the AD and control plasma samples based on microarray analysis (p < 0.05). Gene ontology analysis revealed that miRNA-101a-3p potentially targets 621 genes, and 65 of these genes are associated with AD. Thus, miRNA-101a-3p might play an important role in the mechanism of AD. After verification of the results by real-time PCR, miRNA-101a was selected for further analyses in AD (Figure 1).
Little research has examined the applications of miRNA in AD, particularly plasma miRNA-101a. We validated the expression of miRNA-101a in another AD group and in the APPswe/PS1ΔE9 transgenic mouse brain tissues. Two independent studies have demonstrated reduced expression of miRNA-101a in AD brain samples relative to controls21,22. In our study, the expression of miRNA-101a in the brain tissues of APPswe/PS1ΔE9 double transgenic mice was found to be significantly reduced compared with the control group of age-matched C57 BL/6 J mice (Figure 3). These results pointed out that miRNA-101a might play a role in AD.
Recent studies have found that autophagy plays an important role in the formation of AD. Moreover, miRNAs might regulate this process. In the future, stem cells therapies may be helpful in this23–26. In order to discuss the possible mechanism, we built the AD cell model and constructed a lentiviral vector mouse miRNA-101a to study its role in the formation of AD. Our results demonstrated that autophagy was activated in SH-SY5Y cells treated with H2O2 (Figure 4B1, B2). Interestingly after transfection with miRNA-101a, the autophagy bubbles decreased (Figure 4D1, D2). These results confirmed that autophagy plays a role in AD, and miR-101a had a negative regulation in this phenomenon.
Previous studies have suggested that the MAPK1 pathway can regulate autophagy. In addition, it also plays a role in AD. In this study, the data from the dual-luciferase assay found that MAPK1 could be regulated by miRNA-101a (Figure 5A). We found the expression of MAPK1 and beclin-1 was decreased after overexpression of miRNA-101a in AD model cells, suggesting that miRNA-101a can regulate the formation of autophagy in AD and this process might be modulated via MAPK1.
Meanwhile, other unknown factors might also be involved in differentiation, which require exploration in future studies. In conclusion, our data suggest that miRNA-101a is significantly decreased in patients with AD and AD animal models with increased age and it can regulate the autophagy phenomenon by targeting the MAPK pathway. Therefore, our results may prompt further investigation into this possible mechanism in AD.
Acknowledgements
The authors are grateful for the assistance from The First Affiliated Hospital of Zhengzhou University, China, for assistance with neuronal culture techniques.
Footnotes
Ethical Approval: This study was approved by The First Affiliated Hospital of Zhengzhou University ethics committee. The research plan submitted by this project conforms to the ethical principles of experimental animal research and the Declaration of Helsinki. The feeding and death methods of animals conformed to the requirements of scientific research ethics, and gave the maximum protection to the safety of animals. The researching method of patients’ blood sample accords with the requirements of scientific research ethics. We did not bring unnecessary injury to the animals and patients, and had a scientific basis for the research and design.
Statement of Human and Animal Rights: This article does not contain any studies with human or animal subjects.
Statement of Informed Consent: There are no human subjects in this article and informed consent is not applicable.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This research was supported by the National Natural Science Foundation of China (grant nos. 81371385 and 81071114.) and a grant from The First Affiliated Hospital of Zhengzhou University, China.
ORCID iD: John H. Zhang  https://orcid.org/0000-0002-4319-4285
https://orcid.org/0000-0002-4319-4285
References
- 1. Turner RS. Alzheimer’s disease. Semin Neurol. 2006;26(5):499–506. [DOI] [PubMed] [Google Scholar]
- 2. Selkoe DJ. Cell biology of protein misfolding: the examples of Alzheimer’s and Parkinson’s diseases. Nat Cell Biol. 2004;6(11):1054–1061. [DOI] [PubMed] [Google Scholar]
- 3. Li X, Li TQ, Andreasen N, Wiberg MK, Westman E, Wahlund LO. Ratio of Abeta42/P-tau181p in CSF is associated with aberrant default mode network in AD. Sci Rep. 2013;3:1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rowe CC, Ackerman U, Browne W, Mulligan R, Pike KL, O’Keefe G, Tochon-Danguy H, Chan G, Berlangieri SU, Jones G, Dickinson-Rowe KL, et al. Imaging of amyloid beta in Alzheimer’s disease with 18F-BAY94-9172, a novel PET tracer: proof of mechanism. Lancet Neurol. 2008;7(2):129–135. [DOI] [PubMed] [Google Scholar]
- 5. Huang Y, Mucke L. Alzheimer mechanisms and therapeutic strategies. Cell. 2012;148(6):1204–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451(7182):1069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6(4):463–477. [DOI] [PubMed] [Google Scholar]
- 8. Lee S, Sato Y, Nixon RA. Primary lysosomal dysfunction causes cargo-specific deficits of axonal transport leading to Alzheimer-like neuritic dystrophy. Autophagy. 2011;7(12):1562–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nixon RA, Wegiel J, Kumar A, Yu WH, Peterhoff C, Cataldo A, Cuervo AM. Extensive involvement of autophagy in Alzheimer disease: an immuno-electron microscopy study. J Neuropathol Exp Neurol. 2005;64(2):113–122. [DOI] [PubMed] [Google Scholar]
- 10. Zhao Y, Li X, Ma K, Yang J, Zhou J, Fu W, Wei F, Wang L, Zhu WG. The axis of MAPK1/3-XBP1u-FOXO1 controls autophagic dynamics in cancer cells. Autophagy. 2013;9(5):794–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Xu P, Das M, Reilly J, Davis RJ. JNK regulates FoxO-dependent autophagy in neurons. Genes Dev. 2011;25(4):310–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Waetzig V, Herdegen T. Context-specific inhibition of JNKs: overcoming the dilemma of protection and damage. Trends Pharmacol Sci. 2005;26(9):455–461. [DOI] [PubMed] [Google Scholar]
- 13. Wei Y, Pattingre S, Sinha S, Bassik M, Levine B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell. 2008;30(6):678–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Moreira PI, Duarte AI, Santos MS, Rego AC, Oliveira CR. An integrative view of the role of oxidative stress, mitochondria and insulin in Alzheimer’s disease. J Alzheimers Dis. 2009;16(4):741–761. [DOI] [PubMed] [Google Scholar]
- 15. Delay C, Mandemakers W, Hebert SS. MicroRNAs in Alzheimer’s disease. Neurobiol Dis. 2012;46(2):285–290. [DOI] [PubMed] [Google Scholar]
- 16. Yuan SM, Gao K, Wang DM, Quan XZ, Liu JN, Ma CM, Qin C, Zhang LF. Evodiamine improves congnitive abilities in SAMP8 and APP(swe)/PS1(DeltaE9) transgenic mouse models of Alzheimer’s disease. Acta Pharmacol Sin. 2011;32(3):295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408. [DOI] [PubMed] [Google Scholar]
- 18. Shen C, Chen Y, Liu H, Zhang K, Zhang T, Lin A, Jing N. Hydrogen peroxide promotes Abeta production through JNK-dependent activation of gamma-secretase. J Biol Chem. 2008;283(25):17721–17730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, Lin DW, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105(30):10513–10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bell JD, Cho JE, Giffard RG. MicroRNA changes in preconditioning-induced neuroprotection. Transl Stroke Res. 2017;8(6):585–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Long JM, Lahiri DK. MicroRNA-101 downregulates Alzheimer’s amyloid-beta precursor protein levels in human cell cultures and is differentially expressed. Biochem Biophys Res Commun. 2011;404(4):889–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hebert SS, Horre K, Nicolai L, Papadopoulou AS, Mandemakers W, Silahtaroglu AN, Kauppinen S, Delacourte A, De Strooper B. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer’s disease correlates with increased BACE1/beta-secretase expression. Proc Natl Acad Sci USA. 2008;105(17):6415–6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liska MG, Crowley MG, Borlongan CV. Regulated and unregulated clinical trials of stem cell therapies for stroke. Transl Stroke Res. 2017;8(2):93–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Delavaran H, Aked J, Sjunnesson H, Lindvall O, Norrving B, Kokaia Z, Lindgren A. Spontaneous recovery of upper extremity motor impairment after ischemic stroke: implications for stem cell-based therapeutic approaches. Transl Stroke Res. 2017;8(4):351–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bang OY, Moon GJ, Kim DH, Lee JH, Kim S, Son JP, Cho YH, Chang WH, Kim YH; investigators S-t. Stroke induces mesenchymal stem cell migration to infarcted brain areas via CXCR4 and C-Met signaling. Transl Stroke Res. 2017;8(5):449–460. [DOI] [PubMed] [Google Scholar]
- 26. Takagi T, Yoshimura S, Sakuma R, Nakano-Doi A, Matsuyama T, Nakagomi T. Novel regenerative therapies based on regionally induced multipotent stem cells in post-stroke brains: their origin, characterization, and perspective. Transl Stroke Res. 2017;8(6):515–528. [DOI] [PubMed] [Google Scholar]