Abstract
Objective
The aim of this study was to use whole genome sequencing (WGS) help detect de novo mutations or pathogenic genes of Mayer-Rokitansky-Küster-Hauser syndrome type 1(MRKH syndrome type 1).
Study design
This was a case-parent trios study. Nine unrelated probands, with MRKH syndrome type 1 and their parents were enrolled. The enrollment, sequencing process, establishment of the de novo mutations detecting procedure and experiment part were performed over a 2-year period.
Results
we detected 632 de novo single nucleotide variants (SNVs), 267 de novo small insertions/deletions (indels), 39 de novo structural variations (SVs) and 28 de novo copy number alterations (CNAs). Three novel damaging coding de novo SNVs with three damaging coding de novo genes (PIK3CD, SLC4A10 and TNK2) were revealed. Two SNVs were annotated of the promoter region of gene NBPF10 and 3'UTR of NOTCH2NL, potentially contributing to the pathogenesis of MRKH.
Conclusion
We identified five de novo mutations in BAZ2B, KLHL18, PIK3CD, SLC4A10 and TNK2 by performing WGS, the functional involvement of all deleterious mutations in MRKH candidate genes of the trios warrant further study. WGS may complement conventional array to capture the complete landscape of the genome in MRKH.
Keywords: Next-generation sequencing, Whole-genome sequencing, Mayer–Rokitansky–Küster–Hauser syndrome, De novo mutations
Introduction
Mayer–Rokitansky–Küster–Hauser (MRKH) syndrome (OMIM # 277000) is characterized by congenital hypoplasia of the uterus, cervix and upper two-thirds of the vagina during fetal development [1]. MRKH syndrome is a rare birth defect that affects approximately 1 in 4,500–5,000 newborn girls [2]. Typically, individuals with MRKH syndrome have normal ovarian function and a normal female karyotype (46,XX) [3]. MRKH syndrome can be classified into type 1 (isolated; OMIM 277000), and type 2 (associated with renal, skeletal, cardiovascular and other malformations). MRKH Type 2 is also known as MURCS (Mullerian Duct Aplasia, Unilateral Renal Agenesis, And Cervicothoracic Somite Anomalies (OMIM 601076).
MRKH has been reported to be a sporadic malformation with characteristic familial clustering and a high risk of associated malformations in trios of affected women. The etiology of MRKH syndrome remains unknown. Several genes have been linked to MRKH, such as HOXA7 [4], HOXA13, PBX1 [5], AMH [6], RBM8A andTBX6 [6]. These studies have failed to provide sufficient insight into MRKH syndrome and the primary developmental anomalies. De novo mutations play an important role in human disease [7]. Furthermore there are few genetic studies exploring the possible genetic etiology of MRKH syndrome [8]. Whole exome and whole genome studies are two approaches to identify de novo mutations in candidate genes for particular developmental conditions. This approach has been used extensively in uncovering potential new genes involved in autism spectrum disorder [9]. A recent study using whole-exome sequencing (WES) combined with SNP arrays identified five novel mutated genes among seven patients with MRKH type 1 [10]. These mutations were recurrent in more than two individuals, but the study does not include analysis of the corresponding parent samples.
In this study, we hypothesize that the MRKH type 1 may be due to the occurrence of de novo mutations. We used the first whole genome sequencing (WGS) in full parent-child MRKH type 1 trio families to unequivocally identify these events.
Material and methods
Samples from MRKH syndrome-affected families
Nine unrelated Chinese patients with MRKH Type 1 and their parents were recruited from March 2014 to November 2014 in the Department of Obstetrics and Gynecology of Luohu Hospital. Blood samples and phenotypic data were collected from probands and their parents (Table S1). All participants provided written consent. Approval for the study was obtained from the Ethics Committees of Luohu Hospital.
Sequencing, alignment and variant discovery
Genomic DNA of probands and their parents (trio families) were extracted from blood and sequenced using HiSeq X Ten sequencing system (500 bps library, 150 bps paired reads). The genome mean depth was at least 50× each sample (probands: ˜57×, maternal: ˜56× and paternal: ˜53×), and the coverage of each sample was > 99% (Fig. S2 and Table S2). Reading were aligned with the reference genome (hg19, GRCh37) by the Burrows-Wheeler Aligner, BWA, version 0.59 [11]. Sequence Alignment/Map package (SAM tools [12]: Picard tools (version 1.52)) was used to remove duplications. The single nucleotide variants (SNVs) and small insertions/deletions (Indels) were identified using the Genome Analysis Toolkit, GATK version 3.4 [13]. All SNV/Indels mutation events were presented in Table S3. The effects (e.g., missense, nonsense, or frameshift mutations) and classifications (e.g., in exonic, intronic, or intergenic regions) of variants across the genome were annotated by ANNODB (unpublished software) and Oncotator [14]. The flowchart of the analysis protocol is shown in Fig. S1.
De novo SNV detection
A potential de novo SNV mutation (DNM) was identified following both of the two conditions: (1) one site is heterozygous (novel allele frequency >30%) or homozygous with non-reference SNV in the proband. (2) This site is homozygous (novel allele frequency <5%) with reference nucleotide in both parents. In addition, other filters: (1) the read depth (DP) at the variant position of each member of the trio families should be more than 10×. (2) a phred-scaled quality score(PSQ) more than 30 [15]. (3) a QualByDepth (QD: variant confidence from the QUAL field/unfiltered depth) of more than 10. (4) a HomopolymerRun (Hrun: Largest contiguous homopolymer run of the variant allele in either direction on the reference) less than 5. (5) a MappingQualityZero (MQ0: total count across all samples that had reads with a mapping quality of zero) less than 4.
De novo indel detection
A similar filtering protocol was applied to detect de novo indels: the proband should be heterozygous or homozygous at the indel site, whereas the both parents should be homozygous without indels at the same position [15]. Furthermore, other filters: DP > 10 at the variant position of each member of the trio families; PSQ more than 30 [15]; QD > 10; Hrun less than 5 and MQ0 less than 4.
Variant experimental validation
To validate genomic loci genotypes in offspring and parents was using the Sanger sequencing. A total of eight of nine in exon region putative DNMs sites were validated. In subsequent exonic analyses, we combine sites with complete validation data of de novo Mutations (Table S7).
Variant characterization
When assessing inherited variants, we removed those that are common in the population (i.e., found in dbSNP135). Deleterious variants obtained from nonsense, splice site and frameshift. We defined damaging missense SNVs as those that both Sorting Intolerant From Tolerant (SIFT) [16] and PolyPhen-2 [17] predicted to be damaging via the variant effect predictor(Table S8).
De novo CNA detection
The SegSeq [18] was used to detect copy number alterations (CNAs). Proband-specific CNAs, that did satisfy the following criteria would be discarded: (1) q0 (the fraction of reads with zero mapping quality in the called CNA regions) < 50%. (2) Corrected P-value < 0.05. (3) Overlap rate of repeat database (STR: simple tandem repeat, DGV, genomic SuperDups) < 50%. Putative de novo CNAs for each trio family were detected by the following filters: (1) the normalized average RD signal in the child was < 1.4 (for deletions) or >2.6 (for duplications); and (2) the normalized average RD signal in each parent was between1.6 and 2.4.
To infer recurrently amplified or deleted genomic regions, we re-implemented the GISTIC 2.0 algorithm [19], using copy numbers in 100-kb windows as markers. G-scores were calculated for genomic regions on the basis of the frequency and amplitude of amplifications or deletions affecting each gene. A significant CNA region was defined as having amplification/deletion with |G-score| > 0.1, corresponding to a P-value threshold of 0.01 from the permutation-derived null distribution.
De novo structural variations (SVs) detection
Based on the reliability of the algorithm, the Meerkat 0.185 [20] was used with suggested parameters for de novo SVs and breakpoints detection of 9 trio families. In summary, we mapped whole reads against the human reference genome (hg19) to find soft-clipped and unmapped reads. Then re-mapped these reads to identify discordant read pairs and searched for reads that were covered the candidate breakpoints, and refined precise breakpoints by local alignments. De novo SVs were generated by filtering out SVs events in parents, and appear only in the offspring. Only high confidence events were used for downstream analysis.
Network and pathway analysis
Projecting data onto network contexts is a powerful way to unravel patterns embedded in seemingly, scattered large data sets to amplify knowledge discovery related to complex diseases [21]. We conducted a comprehensive literature review of all variant genes with de novo SNVs variants and searched for the connections between the genes.
The Functional Interaction (FI) network of these genes was generated with the use of the application ReactomeFIViz [21] on the Cystoscope platform [22]. The genes focus of the FI Network by an spectral partition based network clustering algorithm [23]. Nodes in different network modules show in different colors. All white node genes of this Network were supported by at least one reference from the literature or in our study, other gray genes were through calculation of the algorithm, all of these genes were associated with a relevant pathway in the Pathway Database (Fig. S3).
Result
Genetic variants in WGSV
We performed whole genome next generation sequencing (WGS) on 9 trio families. The mean sequencing depth was around 50× (46.48× - 71.22×; Fig. S2) and the mean coverage was more than 99% (Table S2). Totally, we detected an average of 3,909,478 SNPs and 797,951 indels (Table S3) per sample. The mutation rate was approximately 1% [24], which is similar with the results of 1000 Genome Project [25], UK10 K project [26] and ASD project [7].
De novo mutations
We detected 632 de novo SNVs (70.2 per individual), 267 de novo indels (29.7 per individual), 28 de novo CNAs (3.1 per individual; Table S9) and 39 de novo SVs (4.4 per individual; Table S12). 9 de novo SNVs were located in exonic regions (1 mutation per individual; Table S13), and 7 of these were an amino acid change mutations (one was in Dbsnp135) and the other 2 were synonymous. Functional analysis with both PolyPhen-2 and SIFT predicted that 4 of the 6 missense mutations would be deleterious (Table S8).
267 highly confident de novo INDELs were retained for further analysis (Tables S4 and S5).
MRKH 6 had a missense mutation (G1535A; R512Q) in PIK3CD (OMIM: 602839; RefSeq mRNAs: NM_005026). MRKH 4 had a The c.1706C > G mutation in TNK2. MRKH 9 had a missense de novo mutation (C709 T; R237C) in SLC4A10 (OMIM: 605556; RefSeq mRNAs: NM_001178015).
De novo SNVs in non-coding regions
In non-coding regions, we identified 623 de novo SNVs, most of which were singletons and newly discovered based on the ENCODE database (Tables S4 and S5).We found 168 variants that mapped to DNase1 hypersensitive sites (DHS: 99), transcription factor binding sites (TFP: 52), enhancer elements [14], and Pseudogene [3]. 29 of 168 mutations were detected as recurrent mutations (Table S6). One mutation (chr1: 145291364A > T) presented in MRKH 1 was located in 3'UTR of NOTCH2NL. The region is also the promoter of NBPF10 (Table 2). A de novo mutations (chr1: 145291369 G > A) was identified in the 3'UTR of NOTCH2NL in MRKH 8.
Table 2.
The noncoding. recurrent NBPF10 by whole-genome sequencing (WGS) analysis.
| No. | Target gene | Noncoding. recurrent | Family ID | ||
|---|---|---|---|---|---|
| 1 | NBPF10 (Promoter) | TFP(BATF|chr1:145291202-145292202) | Trio01(chr1:145291364) | Trio08(chr1:145291369) | Trio09(chr1:145292171) |
| TFP(NFKB1|chr1:145290201-145291965) | Trio01(chr1:145291364) | Trio08(chr1:145291369) | Trio10(chr1:145290279) | ||
*TFP -transcription factor binding peak.
De novo SVs
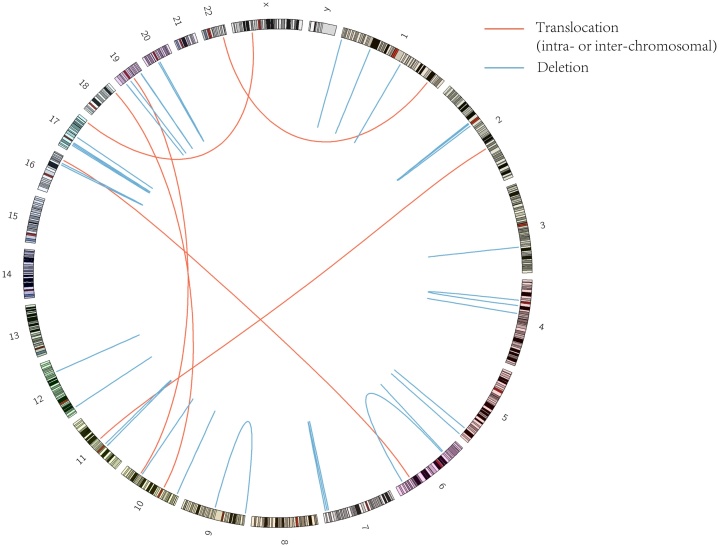
A total of 39 de novo SVs were identified in all 9 trio families. Statistic data and a full list of de novo SVs are presented in Tables S10, S11 and S12. Six types of SVs were observed, including deletions insertions(del_ins), tandem duplications (tandem_dups), inversions_reverse(invers_r), inter-chromosomal translocations(transl_inter), deletions, insertions, and inversions_follow (invers_f). Of these SVs, 31% were deletions, about 15% were inter-chromosomal translocations(transl_inter). For these 39 SVs, fork stalling and template switching (FoSTeS), non-homologous end joining (NHEJ) and alternative end joining(alt-EJ) were the dominant mechanisms.
In 9 trio families’ genomes, we found that 15 SVs occurred in type of gene-gene and were predicted as a direct link to a gene such as DUSP16 (dual specificity phosphatase 16), KIF6(kinesin family member 6), PRKCZ(protein kinase C zeta), and ZNF665(zinc finger protein 665), and 19 genes contained a breakpoint in one of these trio families. Specifically, one breakpoint in chr17 is effect to the target gene: TBC1D16, (Intron &Promoter) (Table S10) (Fig. 1, inter-chromosomal translocations (chr17-chrX)).
Fig. 1.
De novo structural variations.
Copy-number alterations (CNAs)
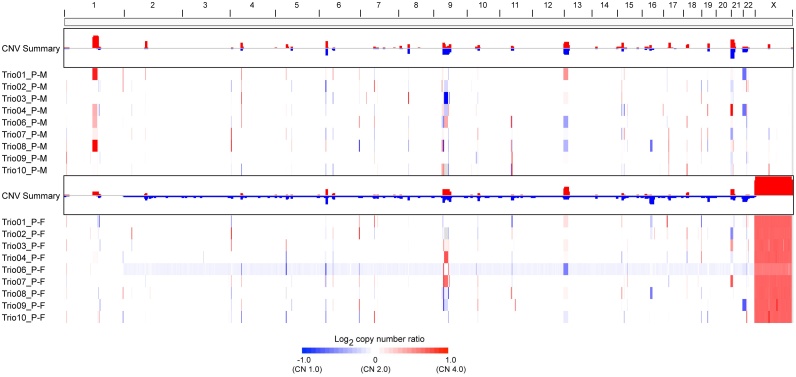
We profiled the 9 trio families for CNAs using WGS and found abnormalities of chromosomal regions that identified a total of 28 de novo CNAs. A full list of CNAs is presented in Table S9. These CNAs occurs on different chromosomes, including chr1, chr2, chr3, chr7, chr8, chr10, chr11, chr13, chr14, chr15, chr16, chr17, chr20, chr22 and the region of CNA in exonic is ranging from 4k to 200k.
Profiling of CNAs identified many putative disease related genes, that may be implicated in MRKH. We applied GSITIC analysis to WGS data to identify recurrent focal CNAs, and this analysis yielded 23 regions of focal amplification in proband-Mother and 18 regions of focal amplification in proband-Father (Table S14), several of which included CNAs previously detected in MRKH type 1, which encompassed genes such as OR2T2, LOC727924, OR4N4, OR4M2, PSG9.
A significant discovery, was the frequent amplification of PSG9, pregnancy specific beta-1 glycoprotein 9, which is gained in proband-mother (4/9) and lost in proband-father (5/9). (Figs. S4 and S5).
We identified 38 focal deletion regions in proband-Mother and 48 regions in proband-Father, which were also frequently truncated in MRKH. One of the most common focal deletions, detected in 9 trio familiess (90%), was a deleted region at 15q11.2 containing LOC727924, OR4N4 and OR4M2 (Fig. 2) and is a previously reported genomic alteration in MRKH.
Fig. 2.
Landscape of CNV.
Inherited dominant mutations in autosomal genes
With analysis of Autosomal Genes with inherited dominant mutations, one gene in exon was yielded, and the gene named KRTAP5-10, Keratin Associated Protein 5-10. In non-coding regions, We found 3 recurrent variants that mapped to enhancer elements, with target gene that DUOX1(Dual Oxidase 1), MLLT4(Myeloid/Lymphoid Or Mixed-Lineage Leukemia; Translocated To, 4) and RPS6KA2(Ribosomal Protein S6 Kinase A2).These functional data indicated that these 3 genes might plays an important role in MRKH syndrome (Tables S15 and S16).
Network and pathway analysis
In this study, only genes interacting with one pathway gene show to minimize a diagram of clutter (Fig. S2). Newly added genes from pathway database color in grey, while original genes color in white. FIs extracted from pathways show as solid lines (for example, EGFR-PIK3CD, while those predicted based on NBC show as dashed lines (for example, TNK2-EGFR). These Extracted FIs involved in activation might play an important role possibly in MRKH syndrome.
Discussion
We detected eight de novo SNVs in nine families, in which three deleterious variants were validated. Our study also identified a number of previously unreported functional genomic regions, some of which include SV and CNV regions. Previous studies on MRKH patients using low resolution microarrays or gene panels alone identified very few potentially causative gene variants [27]. Therefore, WGS may complement conventional array and WES to capture the complete landscape of the genome in MRKH.
Multiple lines of evidences have reported that complex genetic factors are implicated in MRKH (CNAs, indels and SNVs) [2,10,[28], [29], [30], [31], [32], [33], [34]]. The clinical characterizations of MRKH syndrome are varied and complicated. As previously reported, the genes mutations linked with MRKH syndrome are believed to be associated with early embryonic developmental and might be located in several chromosomal loci [35]. Many individuals with MRKH have potential defects on several chromosomes, including 1, 4, 7, 8, 10, 11, 16, 17, 22 and X [33,[36], [37], [38], [39], [40]].
In these 9 trio families’ genomes, 15 SVs occurred in type of gene-gene and were predicted as a direct link to a gene. These results suggested that, different mutational mechanisms could act on the patients with MRKH syndrome, and the results indicated that MRKH syndrome have alterations affecting the short length of chromosome, such as chr1, chr13 and chr19. (Fig. 1 landscape of SV).
Among the significant MRKHS-associated genes identified in our study, BAZ2B, KLHL18, PIK3CD, SLC4A10 and TNK2 showed extensive genetic pleiotropy:
PIK3CD (OMIM: 602839; RefSeq mRNAs: NM_005026) generates phosphatidylinositol 3,4, 5-trisphosphate (PIP3). PIP3 plays a key role by recruiting PH domain-containing proteins to the membrane, including AKT1 and PDPK1, and activating signaling cascades involved in cell growth, survival, proliferation, motility and morphology.
The c.1706C > G mutation in TNK2 indicated that it should be correlated with Ack1 tyrosine kinase activation occurring on chromosome 3. Specifically, TNK2 was reported to be related to tumors, such as non-small cell lung cancer (PMID: 24461128) and bronchopulmonary dysplasia (PMID: 23897914). Diseases associated with this gene include infantile-onset mesial temporal lobe epilepsy with severe cognitive regression.
SLC4A10 is a membrane-bound transporter that facilitates is transfer of glucose and other sugars, bile salts, organic acids, metal ions and amine compounds into the cell. The role of these genes in the development of disease needs to be studied in model organism (e.g. mouse) (Table 1).
Table 1.
Summary of the three damaged de novo SNVs by whole-genome sequencing (WGS) analysis.
| No. | Genes | Cytoband | Genome_Change | cDNA_Change | Codon_Change | Protein_Change | Family ID |
|---|---|---|---|---|---|---|---|
| 1 | TNK2 | 3q29 | g.chr3:195597011G > C | c.1517C > G | c.(1516-1518)cCc > cGc | p.P506R | trio04 |
| 2 | PIK3CD | 1p36.2 | g.chr1:9780813G > A | c.1535 G > A | c.(1534-1536)cGg > cAg | p.R512Q | trio06 |
| 3 | SLC4A10 | 2q24.2 | g.chr2:162719515C > T | c.709C > T | c.(709-711)Cgt > Tgt | p.R237C | trio09 |
The de novo mutations (chr1: 145291369 G > A) was identified in the 3'UTR of NOTCH2NL, indicating the important influence of NOTCH2NL on MRKH processing. This gene is typically associated with Hajdu-Cheney syndrome and the region containing these genes involves the copy number alterations and chromosomal structural variation based on the literature.
However, this compound inheritance model gene dosage model is less likely to underlie the etiology of MRKH, Therefore, trait-specific genetic modifiers of the identified MRKHS-associated genes, second locus genomic modifiers in enhancers/promoters, and potential gene-environment interactions still demand further investigation.
In summary, the genetic diversity identified within our population further highlights the clinical heterogeneity of MRKH. BAZ2B, KLHL18, PIK3CD, SLC4A10 and TNK2 were identified as novel disease-associated genes. The ethnicity upon our rare-inheritance model of disease is unknown. The functions and mechanisms of all deleterious mutations in MRKH candidate genes of the trio families warrant further study.
Conclusion
We identified five de novo mutations in BAZ2B, KLHL18, PIK3CD, SLC4A10 and TNK2 by performing WGS, the functional involvement of all deleterious mutations in MRKH candidate genes of the trios warrant further study. WGS may complement conventional array to capture the complete landscape of the genome in MRKH.
Declaration of Competing Interest
A signed declaration of no conflict of interest has been provided by all authors involved in the study.
Acknowledgements
We wish to thank all subjects for participating in the present study. Personal acknowledgements should precede those of institutions or agencies. We would like to thank Xizhuo Sun and Song Wu for providing computational resources and storage resources. The study was supported by Health and Family Planning Commission of Guangdong Province (grant number A2018431), Shenzhen Healthcare Research Project (grant numbers 201601055 & SZLY2017020) and Sanming Project of Medicine in Shenzhen (grant numbers SZSM201601044). PHX thanks the inimitable care and support of FJL over the last 10 years and I love you and our two lovely daughters. Will you spend the rest of your life with me?
References
- 1.Golan A.L.R., Bukovsky I., Caspi E. Congenital anomalies of the Mullerian system. Fertil Steril. 1989;51(51):55. doi: 10.1016/s0015-0282(16)60660-x. [DOI] [PubMed] [Google Scholar]
- 2.Folch M., Pigem I., Konje J.C. Mullerian agenesis: etiology, diagnosis, and management. Obstet Gynecol Surv. 2000;55(10):644–649. doi: 10.1097/00006254-200010000-00023. [DOI] [PubMed] [Google Scholar]
- 3.Pan H.X., Luo G.N. Phenotypic and clinical aspects of Mayer-Rokitansky-Kuster-Hauser syndrome in a Chinese population: an analysis of 594 patients. Fertil Steril. 2016;106(5):1190–1194. doi: 10.1016/j.fertnstert.2016.06.007. [DOI] [PubMed] [Google Scholar]
- 4.Chen X., Mu Y., Li C., Li G., Zhao H., Qin Y. Mutation screening of HOXA7 and HOXA9 genes in Chinese women with Mullerian duct abnormalities. Reprod Biomed Online. 2014;29(5):595–599. doi: 10.1016/j.rbmo.2014.07.012. [DOI] [PubMed] [Google Scholar]
- 5.Burel A., Mouchel T., Odent S., Tiker F., Knebelmann B., Pellerin I. Role of HOXA7 to HOXA13 and PBX1 genes in various forms of MRKH syndrome (congenital absence of uterus and vagina) J Negat Results Biomed. 2006;5:4. doi: 10.1186/1477-5751-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oppelt P., Strissel P.L., Kellermann A., Seeber S., Humeny A., Beckmann M.W. DNA sequence variations of the entire anti-Mullerian hormone (AMH) gene promoter and AMH protein expression in patients with the Mayer-Rokitanski-Kuster-Hauser syndrome. Hum Reprod. 2005;20(1):149–157. doi: 10.1093/humrep/deh547. [DOI] [PubMed] [Google Scholar]
- 7.Michaelson J.J., Shi Y., Gujral M., Zheng H., Malhotra D., Jin X. Whole-genome sequencing in autism identifies hot spots for de novo germline mutation. Cell. 2012;151(7):1431–1442. doi: 10.1016/j.cell.2012.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palumbo P., Antona V., Palumbo O., Piccione M., Nardello R., Fontana A. Variable phenotype in 17q12 microdeletions: clinical and molecular characterization of a new case. Gene. 2014;538(2):373–378. doi: 10.1016/j.gene.2014.01.050. [DOI] [PubMed] [Google Scholar]
- 9.Jiang Y.H., Yuen R.K., Jin X., Wang M., Chen N., Wu X. Detection of clinically relevant genetic variants in autism spectrum disorder by whole-genome sequencing. Am J Hum Genet. 2013;93(2):249–263. doi: 10.1016/j.ajhg.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen M.J., Wei S.Y., Yang W.S., Wu T.T., Li H.Y., Ho H.N. Concurrent exome-targeted next-generation sequencing and single nucleotide polymorphism array to identify the causative genetic aberrations of isolated Mayer-Rokitansky-Kuster-Hauser syndrome. Hum Reprod. 2015;30(7):1732–1742. doi: 10.1093/humrep/dev095. [DOI] [PubMed] [Google Scholar]
- 11.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramos A.H., Lichtenstein L., Gupta M., Lawrence M.S., Pugh T.J., Saksena G. Oncotator: cancer variant annotation tool. Hum Mutat. 2015;36(4):E2423–9. doi: 10.1002/humu.22771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murai K.K., Nguyen L.N., Irie F., Yamaguchi Y., Pasquale E.B. Control of hippocampal dendritic spine morphology through ephrin-A3/EphA4 signaling. Nat Neurosci. 2003;6(2):153–160. doi: 10.1038/nn994. [DOI] [PubMed] [Google Scholar]
- 16.Ng P.C., Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31(13):3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adzhubei I., Jordan D.M., Sunyaev S.R. 2013. Predicting functional effect of human missense mutations using PolyPhen-2. Current protocols in human genetics / editorial board, Jonathan L Haines [et al.] Chapter 7:Unit7 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chiang D.Y., Getz G., Jaffe D.B., O’Kelly M.J., Zhao X., Carter S.L. High-resolution mapping of copy-number alterations with massively parallel sequencing. Nat Methods. 2009;6(1):99–103. doi: 10.1038/nmeth.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mermel C.H., Schumacher S.E., Hill B., Meyerson M.L., Beroukhim R., Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12(4):R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang L., Luquette L.J., Gehlenborg N., Xi R., Haseley P.S., Hsieh C.H. Diverse mechanisms of somatic structural variations in human cancer genomes. Cell. 2013;153(4):919–929. doi: 10.1016/j.cell.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu G., Dawson E., Duong A., Haw R., Stein L. Reactome FIViz: a Cytoscape app for pathway and network-based data analysis. F1000Research. 2014;3:146. doi: 10.12688/f1000research.4431.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newman M.E. Modularity and community structure in networks. Proc Natl Acad Sci U S A. 2006;103(23):8577–8582. doi: 10.1073/pnas.0601602103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barreiro L.B., Laval G., Quach H., Patin E., Quintana-Murci L. Natural selection has driven population differentiation in modern humans. Nat Genet. 2008;40(3):340–345. doi: 10.1038/ng.78. [DOI] [PubMed] [Google Scholar]
- 25.Genomes Project C, Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Consortium U.K., Walter K., Min J.L., Huang J., Crooks L., Memari Y. The UK10K project identifies rare variants in health and disease. Nature. 2015;526(7571):82–90. doi: 10.1038/nature14962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takahashi K., Hayano T., Sugimoto R., Kashiwagi H., Shinoda M., Nishijima Y. Exome and copy number variation analyses of Mayer–Rokitansky–Küster– Hauser syndrome. Hum Genome Var. 2018;5(1) doi: 10.1038/s41439-018-0028-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oppelt P., Renner S.P., Kellermann A., Brucker S., Hauser G.A., Ludwig K.S. Clinical aspects of Mayer-Rokitansky-Kuester-Hauser syndrome: recommendations for clinical diagnosis and staging. Hum Reprod. 2006;21(3):792–797. doi: 10.1093/humrep/dei381. [DOI] [PubMed] [Google Scholar]
- 29.McGowan R., Tydeman G., Shapiro D., Craig T., Morrison N., Logan S. DNA copy number variations are important in the complex genetic architecture of mullerian disorders. Fertil Steril. 2015;103(4) doi: 10.1016/j.fertnstert.2015.01.008. 1021–30 e1. [DOI] [PubMed] [Google Scholar]
- 30.Rall K., Eisenbeis S., Barresi G., Ruckner D., Walter M., Poths S. Mayer-Rokitansky-Kuster-Hauser syndrome discordance in monozygotic twins: matrix metalloproteinase 14, low-density lipoprotein receptor-related protein 10, extracellular matrix, and neoangiogenesis genes identified as candidate genes in a tissue-specific mosaicism. Fertil Steril. 2015;103(2) doi: 10.1016/j.fertnstert.2014.10.053. 494-502.e3. [DOI] [PubMed] [Google Scholar]
- 31.Sun X., Su Y., He Y., Zhang J., Liu W., Zhang H. New strategy for in vitro activation of primordial follicles with mTOR and PI3K stimulators. Cell Cycle. 2015;14(5):721–731. doi: 10.1080/15384101.2014.995496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tewes A.C., Rall K.K., Romer T., Hucke J., Kapczuk K., Brucker S. Variations in RBM8A and TBX6 are associated with disorders of the mullerian ducts. Fertil Steril. 2015;103(5):1313–1318. doi: 10.1016/j.fertnstert.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 33.Williams L.S., Demir Eksi D., Shen Y., Lossie A.C., Chorich L.P., Sullivan M.E. Genetic analysis of Mayer-Rokitansky-Kuster-Hauser syndrome in a large cohort of families. Fertil Steril. 2017;108(1) doi: 10.1016/j.fertnstert.2017.05.017. 145–51.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watanabe K., Kobayashi Y., Banno K., Matoba Y., Kunitomi H., Nakamura K. Recent advances in the molecular mechanisms of Mayer-Rokitansky-Kuster-Hauser syndrome. Biomed Rep. 2017;7(2):123–127. doi: 10.3892/br.2017.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Connell M., Owen C., Segars J. Genetic syndromes and genes involved in the development of the female reproductive tract: a possible role for gene therapy. J Genetic Syndr Gene Ther. 2013:4. doi: 10.4172/2157-7412.1000127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pizzo A., Lagana A.S., Sturlese E., Retto G., Retto A., De Dominici R. Mayer-Rokitansky-Kuster-Hauser syndrome: embryology, genetics and clinical and surgical treatment. ISRN Obstet Gynecol. 2013;2013 doi: 10.1155/2013/628717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ledig S., Schippert C., Strick R., Beckmann M.W., Oppelt P.G., Wieacker P. Recurrent aberrations identified by array-CGH in patients with Mayer-Rokitansky-Kuster-Hauser syndrome. Fertil Steril. 2011;95(5):1589–1594. doi: 10.1016/j.fertnstert.2010.07.1062. [DOI] [PubMed] [Google Scholar]
- 38.Nik-Zainal S., Strick R., Storer M., Huang N., Rad R., Willatt L. High incidence of recurrent copy number variants in patients with isolated and syndromic Mullerian aplasia. J Med Genet. 2011;48(3):197–204. doi: 10.1136/jmg.2010.082412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morcel K., Watrin T., Jaffre F., Deschamps S., Omilli F., Pellerin I. Involvement of ITIH5, a candidate gene for congenital uterovaginal aplasia (Mayer-Rokitansky-Kuster-Hauser syndrome), in female genital tract development. Gene Expr. 2012;15(5–6):207–214. doi: 10.3727/105221613x13571653093169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morcel K., Dallapiccola B., Pasquier L., Watrin T., Bernardini L., Guerrier D. Clinical utility gene card for: Mayer-Rokitansky-Kuster-Hauser syndrome. Eur J Hum Genetics: EJHG. 2012;20(2) doi: 10.1038/ejhg.2011.158. [DOI] [PMC free article] [PubMed] [Google Scholar]