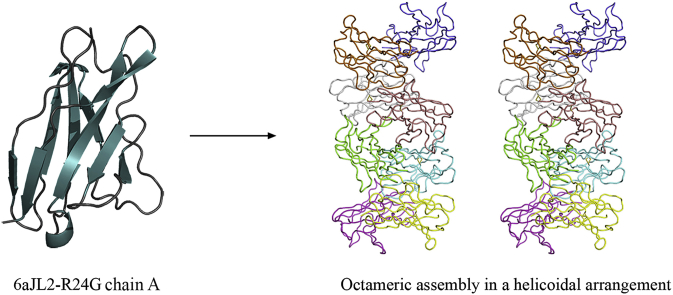
Abstract
Light chain amyloidosis is one of the most common systemic amyloidosis, characterized by the deposition of immunoglobulin light variable domain as insoluble amyloid fibrils in vital organs, leading to the death of patients. Germline λ6a is closely related with this disease and has been reported that 25% of proteins encoded by this germline have a change at position 24 where an Arg is replaced by a Gly (R24G). This germline variant reduces protein stability and increases the propensity to form amyloid fibrils. In this work, the crystal structure of 6aJL2-R24G has been determined to 2.0 Å resolution by molecular replacement. Crystal belongs to space group I212121 (PDB ID 5JPJ) and there are two molecules in the asymmetric unit. This 6aJL2-R24G structure as several related in PDB (PDB entries: 5C9K, 2W0K, 5IR3 and 1PW3) presents by crystal packing the formation of an octameric assembly in a helicoidal arrangement, which has been proposed as an important early stage in amyloid fibril aggregation. However, other structures of other protein variants in PDB (PDB entries: 3B5G, 3BDX, 2W0L, 1CD0 and 2CD0) do not make the octameric assembly, regardless their capacity to form fibers in vitro or in vivo. The analysis presented here shows that the ability to form the octameric assembly in a helicoidal arrangement in crystallized light chain immunoglobulin proteins is not required for amyloid fibril formation in vitro. In addition, the fundamental role of partially folded states in the amyloid fibril formation in vitro, is not described in any crystallographic structure published or analyzed here, being those structures, in any case examples of proteins in their native states. Those partially folded states have been recently described by cryo-EM studies, showing the necessity of structural changes in the variants before the amyloid fiber formation process starts.
Keywords: AL amyloidosis, Immunoglobulin light-chain, Crystal structure, Immune system
Graphical abstract
Highlights
-
•
The structure of 6aJL2-R24G an amyloidogenic protein has been determined at 2.0 Å.
-
•
This crystal structure and related examples presents an octameric assembly by crystal packing.
-
•
The octameric assembly in light chain proteins is not required for amyloid formation.
1. Introduction
Light chain amyloidosis (AL) is one of the most common systemic amyloidosis, characterized by the extracellular deposition of insoluble amyloid fibrils, arising from the aggregation of free monoclonal light chain immunoglobulin. These light chain proteins are secreted in excess into the bloodstream by a population of clonal plasma cells in the bone marrow [1]. The accumulation and deposition of amyloid fibrils in vital organs, without treatment, leads to progressive organ failure and the subsequent death of the patient in about 13 months [2].
AL has an estimated incidence in developed countries of 8–12 cases per million inhabitants per year. However, autopsy studies suggest that the incidence could be higher because it is often misdiagnosed [3]. The AL symptoms depend on the organ affected, although vague and nonspecific symptoms include: fatigue, weight loss, edema bleeding tendency, macroglossia, peri-orbital bruising, and hepatomegaly [4].
In almost 40% of cases, AL is diagnosed after the onset of the symptoms [5]. This is in part due to the fact that the diagnosis relies on subcutaneous fat aspiration and histopathological examination with amyloid stain, showing Congo red-positive amyloid deposits, with typical apple-green birefringence under polarized light [6].
Current treatments aim to avoid the synthesis of light chain proteins, and therefore the treatments are mostly based in chemotherapies to suppress the underlying plasma cell dyscrasia. Currently, high-dose melphalan followed by autologous peripheral blood stem cell transplantation is considered one of the most effective treatments for AL. This treatment usually increases the survival time, but rarely eliminates the disease and is not effective for rapidly progressive types [7]. To provide a basis for new treatments for AL, in vitro studies have focused on inhibiting amyloid fibril formation with small molecules such as doxycycline [8], epigallocatechin gallate [9,10], methylene blue and sulfasalazine [11].
The precursors of AL are light chain immunoglobulin proteins, mostly including only the variable region (VL). However, in some cases, the constant region (CL) has also been implicated in the disease. The VL can be from lambda (λ) or kappa (κ) germline, nevertheless, in AL patients, λ are overexpressed (λ/k = 3:1) as compared with healthy individuals [12].
It has been observed that in ~25% of the amyloidogenic λ6 proteins a change is found at position 24, where an Arg is replaced by a Gly [13]. This variant makes amyloid fibrils faster than the germline and was more thermodynamically unstable [14]. In vitro studies suggest that this mutation eliminates important interactions, including a cation-pi with Phe2, an ionic interaction with Asp93 and some H-bonds [15]. 6aJL2-R24G first crystal structure shows that in the absence of Arg24, Phe2 is reoriented to the inside of the upper hydrophobic core and a cavity is formed around the complementary determining region 1 (CDR1) [16]. Molecular dynamic simulations find differences between 6aJL2 and 6aJL2-R24G in their dynamical signatures that explain the variant increased tendency to form amyloids [17].
In this work, we determined a crystal structure by molecular replacement of recombinant 6aJL2-R24G at 2.0 Å resolution crystallized in a new condition, with two subunits in the asymmetric unit. The structure analysis shows that this structure, although crystallized in a different space group, by crystal symmetry generates octamers in a helicoidal arrangement, which is also found in structures of protein variants with PDB entries 5C9K, 2W0K, 5IR3 and 1PW3 among others [16,18,19]. This octameric assembly has been proposed as an important early stage in amyloid fibril aggregation [16]. However, other crystal structures of protein variants do not make that octameric assembly in a helicoidal arrangement (PDB entries: 3B5G, 3BDX [18], 2W0L [Unpublished], 1CD0 and 2CD0 [20] among others). The analysis presented in this work shows that the ability to form the octameric assembly in a helicoidal arrangement in light chain immunoglobulin proteins is not required for amyloid fibril formation in vitro. In addition, the critical role of partially folded states in the amyloid fibril formation in vitro, is not described in any crystallographic structure of light chain immunoglobulin proteins published to date or analyzed here, being those structures, in any case, examples of proteins in their native states. More recently, three amyloid fibril structures determined by cryo-electron microscopy (cryo-EM) showed a non-native fibril fold in mouse and human organs confirming that partially folded states are determinant in amyloid fibril aggregation [21,22].
2. Materials and methods
2.1. Protein purification and crystallization
Recombinant 6aJL2-R24G was expressed and purified following the method described [9].
6aJL2-R24G was crystallized by sitting drop vapor diffusion method at 18 °C. The drops were prepared with a robot (Mosquito LCP, TTP Labtech) in 96-well crystallization plates by mixture of protein (0.2 μl) at 7 mg/ml in 20 mM sodium phosphate buffer, pH 7.5 with the reservoir solution (0.2 μl) containing 1 M sodium/potassium tartrate, 100 mM Tris-HCl, pH 7.0 and, 200 mM lithium sulfate. Crystals appeared after one month and continued to grow for two months. Crystals were flash-cooled by immersion in liquid nitrogen using 30% glycerol in mother solution as cryoprotectant.
2.2. Data collection and crystal structure determination
A diffraction data set collected on 14-1 beamline of the Stanford Synchrotron Radiation Lightsource (SSRL, Menlo Park, USA) was indexed, integrated and scaled using XDS package [23].
The structure was determined by molecular replacement using the program PHASER [24]. The starting model was the available NMR coordinates of 6aJL2-R24G structure (PDB ID 2MKW [15]). The model was improved by rigid body refinement and geometric constraint performed in REFMAC [25]. The final model was refined alternating cycles of automatic and manual refinement with PHENIX [26] and COOT [27], respectively to a final Rcryst of 19.5% (Rfree of 24.9% calculated with 5% of the data randomly selected) at 2.0 Å (Table S1). The final structure displayed good stereochemistry, as analyzed by MOLPROBITY [28]. Structural figures were made with PyMOL Molecular Graphics, Version 1.8, Schödinger, LLC.
3. Results and discussion
The crystal structure described in this work corresponds to the 6aJL2-R24G germline variant (PDB ID 5JPJ) (using the continuous numbering as in PDB ID 2MKW) [15], crystallized in the orthorhombic space group I212121, determined by molecular replacement and refined up to 2.0 Å resolution (Table S1). The crystal asymmetric unit contains two subunits, each including residues Asn1 to Leu111 (Fig. S1A), which by crystal symmetry generates dimers formed by VL-VL domains. Native gels and NMR studies demonstrated that 6aJL2-R24G is mainly monomeric in solution. These results are in accordance with our DLS measurements of 6aJL2-R24G that show a diffusion coefficient of 1.14 × 10-7 cm2/s corresponding to an apparent radius of ~2.1 nm; this shows a good agreement with the monomer [9]. The overall structure of 6aJL2-R24G subunit has the typical immunoglobulin fold with eight β-strands, packed tightly against each other in two antiparallel β-sheets joined together by a disulfide bridge in a form of a Greek key β-barrel (Figs. S1B and S1C) [12,29]. The β-sheet one is formed by strands A, B, D and E while the β-sheet two is formed by strands: C, C′, F and G. The three CDR's are composed by residues 24–34, 50–56 and 89–95, respectively. 6aJL2-R24G comprises the conserved Trp at position 36. The r.m.s.d. between chain A and B is 0.48 Å using 111 Cα atoms. The electron density map (2Fo-Fc), countered at 1 σ around the site of Gly24 confirming the variant mutation as shown in (Fig. S1D).
The structure of the VL domain 6aJL2-R24G was superposed with the natives 6aJL2 NMR (PDB ID 2MMX, model 1) and crystal structures (PDB ID 2W0K) [15,18], with a r.m.s.d. of 2.0 Å using 106 Cα atoms and 0.52 Å using 111 Cα atoms, respectively. Also, 6aJL2-R24G was superposed with the 6aJL2-R24G NMR structure (PDB ID 2MKW, model 1) and the 6aJL2-R24G crystal structure (PDB ID 5C9K, [16]). The r.m.s.d of the structural alignments are 1.16 Å using 109 Cα residues and 0.19 Å using 111 Cα residues for entries 2 MKW and 5C9K, respectively, showing that the global structures are almost identical, particularly in the crystal structures. Besides, 6aJL2-R24G shows the Phe2 reoriented to the inside of the upper hydrophobic core similar to 5C9K structure [16].
It has been proposed that the characteristic non-helical conformation in CDR1 for germline λ6 proteins was due, to the presence of an Arg in position 24, and a Gly in this position would give rise to a different conformation. This structural rearrangement would explain, in part, the low stability (Tm (Melting temperature) in the variant is 6 °C lower than the WT) and propensity for the amyloid fibril formation [30]. However, in the 6aJL2-R24G crystallographic structures (PDB ID 5C9K and 5JPJ) there was not a helix formation in the CDR1 region, as already reported in the NMR solution structure [15].
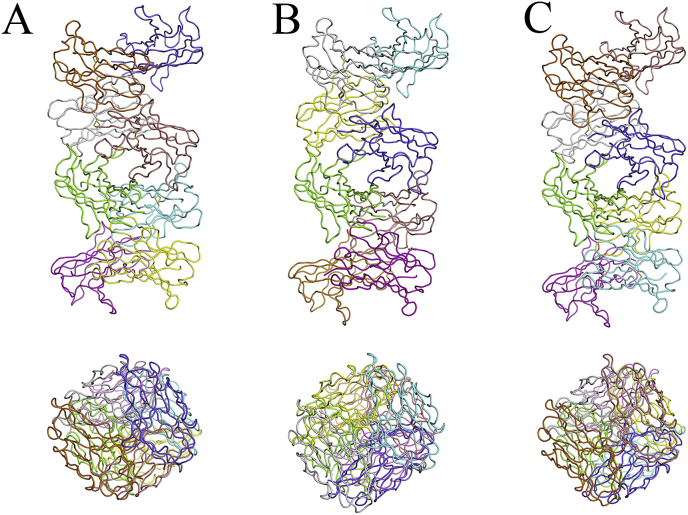
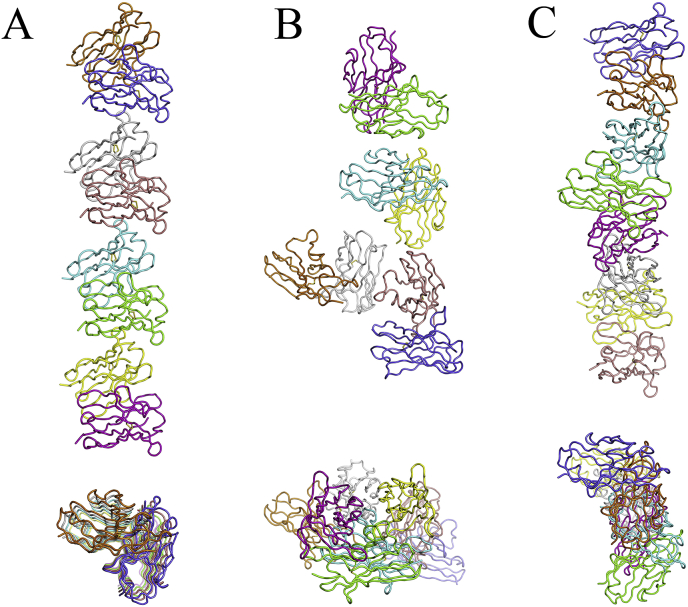
The mutational analysis conducted by a number of laboratories, points to a correlation between thermodynamic stability and amyloid fibril formation; most importantly, it appears that the population of partially folded states is critical in the initiation of the amyloid fibril reaction and certain somatic mutations appear to promote these partially folded states more than others [12]. However, more recently, it has been reported that the 6aJL2-R24G crystals (PDB ID 5C9K) revealed an octameric assembly in a helicoidal arrangement, the authors propose that direct stacking might be a plausible mechanism to understand amyloid aggregation of this protein. And this octameric assembly might represent an important early stage in amyloid aggregation [16]. We have analyzed that equivalent octameric assembly in a helicoidal arrangement are generated by crystal packing of the structures of 6aJL2-R24G: PDB ID 5JPJ (in this work), 2W0K (6aJL2 WT), 5IR3 (AR) [16,18] and 1PW3 [19] among others of 6aJL2 protein (Fig. 1). It is important to mention that the two crystal structures of 6aJL2-R24G (PDB ID 5C9K and 5JPJ) present the octameric assembly in a helicoidal arrangement, and the 6aJL2 WT (PDB ID 2W0K) crystal structure as well. In contrast, we have observed that other crystal structures of the 6aJL2 protein in PDB: PDB ID 3B5G, 3BDX [18], 2W0L [Unpublished], 1CD0 and 2CD0 [20] among others, do not form the octameric assembly in a helicoidal arrangement (Fig. 2). Even other authors have reported that a protein unstable and highly fibrillogenic as the mutant 6aJL2-P7S (Fig. 2C) (PDB ID: 3BDX), where the crystal packing analysis showed the establishment of intermolecular β-β interactions, could be relevant in the mechanism of amyloid fibril formation [18].
Fig. 1.
Octameric structures of 6aJL2 light chain variable domain generated by symmetry-related molecules in the crystal packing. A) 6aJL2-R24G of this work (PDB ID 5JPJ). B) Intrinsic octameric structure of 6aJL2-R24G (PDB ID 5C9K) [16]. C) 6aJL2 WT (PDB ID 2W0K) [18]. Views down (above) and perpendicular (below) in A), B) and C).
Fig. 2.
Structures with eight subunits of 6aJL2 generated by symmetry-related molecules in the crystal packing. A) Unstable and highly fibrillogenic P7S mutant of 6aJL2 (PDB ID 3B5G) [18]. B) Mutant H8P of 6aJL2 (PDB ID 2W0L) [Unpublished]. C) Unstable and highly fibrillogenic P7S mutant of 6aJL2 (PDB ID 3BDX) [18]. Views down (above) and perpendicular (below) in A), B) and C).
We have analyzed the four examples of 6aJL2 proteins that form the octameric assembly in a helicoidal arrangement, the other five that do not form it and the NMR structures (6aJL2 WT and mutant R24G) structurally superimposed with the structure reported in this work (PBD ID 5JPJ, subunit A). The values of R.M.S.D., space group and fibrillogenic potential are shown in Table S2. Also, in Fig. S2 all superimposed subunits are shown. The values of R.M.S.D. in general, they are very similar (0.19–0.85 Å), independently whether the 6aJL2 proteins form the octameric assembly or not, which is also observed in Fig. S2. As expected, NMR structures show the highest R.M.S.D (1.16–2.0 Å) due to their higher mobility. These 12 structures of 6aJL2 proteins analyzed generally form amyloid fibers in vitro to a greater or lesser degree (Table S2).
Furthermore, we have analyzed the 6aJL2 R24G (PDB ID 5JPJ) octameric assembly with the other four protein octameric assemblies. The artificial octamers of NMR structures were generated by superposing subunits in the 5JPJ octameric assembly. The Table S3 has the R.M.S.D values between the structures compared with 5JPJ octameric assembly. The five octameric assemblies have R.M.S.D. between 1.74 and 1.86 Å and they do not seem to present clashes between the subunits that form the octamer. However, the solution structures 2 MMX and 2 MKW clash, particularly 2 MMX (Fig. S3).
Finally, we have analyzed the crystal packing in two examples of proteins that form the octameric assembly in a helicoidal arrangement PDB ID 5JPJ and 2W0K, and two examples of variants that do not form the octamer PDB ID 3B5G and 3BDX. One example of each one is shown in Fig. S4. The crystal packing is conserved in the proteins that form the octameric assembly, but in the non-octameric proteins the packing is different. Besides, the interfaces (hydrogen bonds) between octameric proteins in general is maintained (with seven common hydrogen bonds). However, just three of the hydrogen bonds (three in each structure) found in the octameric assembly are also found in the non-octameric proteins (Table S4).
Protein crystallization and amyloid fibril formation are different processes otherwise amyloidogenic proteins would form amyloid fibrils instead of crystals. Besides, molecular dynamics simulations indicate that 6aJL2 germline variants P7S and R24G lead to a loosened native state, given their loss of interactions, and the increase in the solvent-exposed area [17]. Other experimental data agrees with these results: For AR, the most unstable and fibrillogenic λ6a protein described to date, native state fluorescence and circular dichroism characteristics were also compatible with a loosened native state [31]. In the case of 6aJL2-R24G crystal structure indicates that some interactions are loosened by the mutation, which reduces their stability and promotes the amyloid fibril formation in vitro. Considering the previous information, we conclude that the ability to form the octameric assembly in a helicoidal arrangement in light chain immunoglobulin proteins is not required for amyloid fibril aggregation in vitro. Furthermore, the existence of partially folded states crucial in amyloid fibril formation is not described in any crystal structure of light chain immunoglobulin proteins described to date. Recently, three amyloid fibril structures determined by cryo-EM showed a non-native fibril fold confirming that partially folded states are determinant in amyloid fibril aggregation [19,20]. Then, the alternative for the early stages of amyloid fibril aggregation is that the population of partially folded states in amyloidogenic proteins is determinant in the amyloid fibril reaction, and somatic mutations appear to promote these partially folded states [12]. These partially folded states have been reported in amyloidogenic proteins by molecular dynamics simulations [17], by fluorescence and circular dichroism [30] and by amyloid fibril structures determined by cryo-EM [21,22].
Acknowledgements
We thank Isabel Velázquez López (UNAM) for the generous gift of the 6aJL2-R24G plasmid. ERP acknowledge IBt Institutional budget and his economic incentive from SNI for financing his research. CA was supported by LabDP. Authors are grateful to the staff at the Stanford Synchrotron Radiation Lightsource (SSRL) beamline 14-1 for data collection facilities. Authors also thank Sonia P. Rojas Trejo for her technical assistance. Part of the research was performed at the LabDP-UAEM.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrep.2019.100682.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Dispenzieri A., Gertz M.A., Buadi F. What do I need to know about immunoglobulin light chain (AL) amyloidosis? Blood Rev. 2012;26:137–154. doi: 10.1016/j.blre.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 2.Halwani O., Delgado D.H. Cardiac amyloidosis: an approach to diagnosis and management. Expert Rev. Cardiovasc Ther. 2010;8:1007–1013. doi: 10.1586/erc.10.41. [DOI] [PubMed] [Google Scholar]
- 3.Pinney J.H., Smith C.J., Taube J.B., Lachmann H.J., Venner C.P., Gibbs S.D., Dungu J., Banypersad S.M., Wechalekar A.D., Whelan C.J., Hawkins P.N., Gillmore J.D. Systemic amyloidosis in England: an epidemiological study. Br. J. Haematol. 2013;161:525–532. doi: 10.1111/bjh.12286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yim B., Kertowidjojo E., Zhang Y., Patel P. Poor outcomes in hepatic amyloidosis: a report of 2 cases. Case Rep Oncol Med. 2016;2016:1–5. doi: 10.1155/2016/7625940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lousada I., Comenzo R.L., Landau H., Guthrie S., Merlini G. Light chain amyloidosis: patient experience survey from the amyloidosis research consortium. Adv. Ther. 2015;32:920–928. doi: 10.1007/s12325-015-0250-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gertz M.A. Immunoglobulin light chain amyloidosis: 2016 update on diagnosis, prognosis, and treatment. Am. J. Hematol. 2016;91:947–956. doi: 10.1002/ajh.24433. [DOI] [PubMed] [Google Scholar]
- 7.Skinner M., Anderson J., Simms R., Falk R., Wang M., Libbey C., Jones L.A., Cohen A.S. Treatment of 100 patients with primary amyloidosis: a randomized trial of melphalan, prednisone, and colchicine versus colchicine only. Am. J. Med. 1996;100:290–298. doi: 10.1016/s0002-9343(97)89487-9. [DOI] [PubMed] [Google Scholar]
- 8.Ward J.E., Ren R., Toraldo G., Soohoo P., Guan J., O'Hara C., Jasuja R., Trinkaus-Randall V., Liao R., Connors L.H., Seldin D.C. Doxycycline reduces fibril formation in a transgenic mouse model of AL amyloidosis. Blood. 2011;118:6610–6617. doi: 10.1182/blood-2011-04-351643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peláez-Aguilar A.E., Rivillas-Acevedo L., French-Pacheco L., Valdes-Garcia G., Maya-Martinez R., Pastor N., Amero C. Inhibition of light chain 6aJL2-R24G amyloid fiber formation associated with light chain amyloidosis. Biochemistry. 2015;54:4978–4986. doi: 10.1021/acs.biochem.5b00288. [DOI] [PubMed] [Google Scholar]
- 10.Hora M., Carballo-Pacheco M., Weber B., Morris V.K., Wittkopf A., Buchner J., Strodel B., Reif B. Epigallocatechin-3-gallate preferentially induces aggregation of amyloidogenic immunoglobulin light chains. Sci. Rep. 2017;7:1–12. doi: 10.1038/srep41515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brumshtein B., Esswein S.R., Salwinski L., Phillips M.L., Ly A.T., Cascio D., Sawaya M.R., Eisenberg D.S. Inhibition by small-molecule ligands of formation of amyloid fibrils of an immunoglobulin light chain variable domain. Elife. 2015;4:1–15. doi: 10.7554/eLife.10935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramirez-Alvarado M. Amyloid formation in light chain amyloidosis. Curr. Top. Med. Chem. 2012;12:2523–2533. doi: 10.2174/1568026611212220007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Del Pozo-Yauner L., Ortiz E., Sánchez R., Sánchez-López R., Güereca L., Murphy C.L., Allen A., Wall J.S., Fernández-Velasco D.A., Solomon A., Becerril B. Influence of the germline sequence on the thermodynamic stability and fibrillogenicity of human lambda 6 light chains. Proteins. 2008;72:684–692. doi: 10.1002/prot.21934. [DOI] [PubMed] [Google Scholar]
- 14.Mishima T., Ohkuri T., Monji A., Kanemaru T., Abe Y., Ueda T. Residual structures in the acid-unfolded states of Vlambda6 proteins affect amyloid fibrillation. J. Mol. Biol. 2009;392:1033–1043. doi: 10.1016/j.jmb.2009.07.078. [DOI] [PubMed] [Google Scholar]
- 15.Maya-Martínez R., Gil-Rodríguez P., Amero C. Solution structure of 6aJL2 and 6aJL2-R24G amyloidogenics light chain proteins. Biochem. Biophys. Res. Commun. 2015;456:695–699. doi: 10.1016/j.bbrc.2014.12.044. [DOI] [PubMed] [Google Scholar]
- 16.Luna-Martínez O.D., Hernández-Santoyo A., Villalba-Velázquez M.I., Sánchez-Alcalá R., Fernández-Velasco D.A., Becerril B. Stabilizing an amyloidogenic λ6 light chain variable domain. FEBS J. 2017;284:3702–3717. doi: 10.1111/febs.14265. [DOI] [PubMed] [Google Scholar]
- 17.Valdés-García G., Millan-Pacheco C., Pastor N. Convergent mechanisms favor fast amyloid formation in two lambda 6a Ig light chain mutants. Biopolymers. 2017;107:1–9. doi: 10.1002/bip.23027. [DOI] [PubMed] [Google Scholar]
- 18.Hernández-Santoyo A., del Pozo-Yauner L., Fuentes-Silva D., Ortiz E., Rudiño-Piñera E., Sánchez-López R., Horjales E., Becerril B., Rodríguez-Romero A. A single mutation at the sheet switch region results in conformational changes favoring λ6 light-chain fibrillogenesis. J. Mol. Biol. 2010;396:280–292. doi: 10.1016/j.jmb.2009.11.038. [DOI] [PubMed] [Google Scholar]
- 19.Wall J.S., Gupta V., Wilkerson M., Schell M., Loris R., Adams P., Solomon A., Stevens F., Dealwis C. Structural basis of light chain amyloidogenicity: comparison of the thermodynamic properties, fibrillogenic potential and tertiary structural features of four V(lambda)6 proteins. J. Mol. Recognit. 2004;17:323–331. doi: 10.1002/jmr.681. [DOI] [PubMed] [Google Scholar]
- 20.Pokkuluri P.R., Solomon A., Weiss D.T., Stevens F.J., Schiffer M. Tertiary structure of human λ6 light chains. Amyloid. 1999;6:165–171. doi: 10.3109/13506129909007322. [DOI] [PubMed] [Google Scholar]
- 21.Radamaker L., Lin Y.H., Annamalai K., Huhn S., Hegenbart U., Schönland S.O., Fritz G., Schmidt M., Fändrich M. Cryo-EM structure of a light chain-derived amyloid fibril from a patient with systemic AL amyloidosis. Nat. Commun. 2019;10:1103 1–8. doi: 10.1038/s41467-019-09032-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liberta F., Loerch S., Rennegarbe M., Schierhorn A., Westermark P., Westermark G.T., Hazenberg B.P.C., Grigorieff N., Fändrich M., Schmidt M. Cryo-EM fibril structures from systemic AA amyloidosis reveal the species complementarity of pathological amyloids. Nat. Commun. 2019;10:1104 1–110410. doi: 10.1038/s41467-019-09033-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kabsch W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr D Biol Crystallogr. 2010;66:133–144. doi: 10.1107/S0907444909047374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murshudov G.N., Vagin A.A., Dodson E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D: Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 26.Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., McCoy A.J., Moriarty N.W., Oeffner R., Read R.J., Richardson D.C., Richardson J.S., Terwilliger T.C., Zwart P.H. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of coot. Acta Crystallogr. D: Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen V.B., Arendall W.B., III, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D: Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bork P., Holm L., Sander C. The immunoglobulin fold. Structural classification, sequence patterns and common core. J. Mol. Biol. 1994;242:309–320. doi: 10.1006/jmbi.1994.1582. [DOI] [PubMed] [Google Scholar]
- 30.Del Pozo-Yauner L., Ortiz E., Becerril B. The CDR1 of the Human λVI light chains adopts a new canonical structure. Proteins. 2006;62:122–129. doi: 10.1002/prot.20779. [DOI] [PubMed] [Google Scholar]
- 31.González-Andrade M., Becerril-Luján B., Sánchez-López R., Ceceña-Álvarez H., Pérez-Carreón J.I., Ortiz E., Fernández-Velasco D.A., del Pozo-Yauner L. Mutational and genetic determinants of λ6 light chain amyloidogenesis. FEBS J. 2013;280:6173–6183. doi: 10.1111/febs.12538. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.