Abstract
Objective
Non-alcoholic fatty liver disease (NAFLD) is a complex pathology in which several dysfunctions, including alterations in metabolic pathways, mitochondrial functionality and unbalanced lipid import/export, lead to lipid accumulation and progression to inflammation and fibrosis. The enzyme glycine N-methyltransferase (GNMT), the most important enzyme implicated in S-adenosylmethionine catabolism in the liver, is downregulated during NAFLD progression. We have studied the mechanism involved in GNMT downregulation by its repressor microRNA miR-873-5p and the metabolic pathways affected in NAFLD as well as the benefit of recovery GNMT expression.
Methods
miR-873-5p and GNMT expression were evaluated in liver biopsies of NAFLD/NASH patients. Different in vitro and in vivo NAFLD murine models were used to assess miR-873-5p/GNMT involvement in fatty liver progression through targeting of the miR-873-5p as NAFLD therapy.
Results
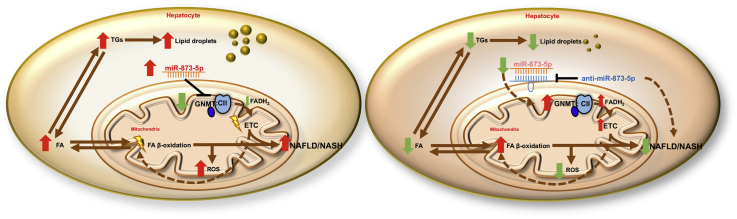
We describe a new function of GNMT as an essential regulator of Complex II activity in the electron transport chain in the mitochondria. In NAFLD, GNMT expression is controlled by miR-873-5p in the hepatocytes, leading to disruptions in mitochondrial functionality in a preclinical murine non-alcoholic steatohepatitis (NASH) model. Upregulation of miR-873-5p is shown in the liver of NAFLD/NASH patients, correlating with hepatic GNMT depletion. Importantly, NASH therapies based on anti-miR-873-5p resolve lipid accumulation, inflammation and fibrosis by enhancing fatty acid β-oxidation in the mitochondria. Therefore, miR-873-5p inhibitor emerges as a potential tool for NASH treatment.
Conclusion
GNMT participates in the regulation of metabolic pathways and mitochondrial functionality through the regulation of Complex II activity in the electron transport chain. In NAFLD, GNMT is repressed by miR-873-5p and its targeting arises as a valuable therapeutic option for treatment.
Keywords: NASH, GNMT, Mitochondria, β-oxidation, Metabolism, microRNA
Graphical abstract
Highlights
-
•
The microRNA miR-873-5p is upregulated in human and murine NAFLD/NASH livers.
-
•
miR-873-5p upregulation downregulates GNMT in the liver.
-
•
miR-873-5p inhibition reduces liver steatosis, inflammation and fibrosis in in vivo NAFLD mouse models.
-
•
GNMT is a hepatic metabolic hub with mitochondria activity through the regulation of Complex II of the ETC.
-
•
Mitochondrial GNMT deficiency compromises ETC functionality and metabolism.
List of abbreviations
- NAFLD
non-alcoholic fatty liver disease
- NASH
non-alcoholic steatohepatitis
- GNMT
Glycine N-methyltransferase
- FAO
fatty acid β-oxidation
- DNL
de novo lipogenesis
- VLDL
very-low-density lipoprotein
- miRNA/miR
microRNAs
- SAMe
S-adenosylmethionine
- SAH
S−adenosylhomocysteine
- TCA
tricarboxylic acid
- OXPHOS
oxidative phosphorylation
- ETC
electron transport chain
- ROS
reactive oxygen species
- MCD
methionine choline deficient
- MCDD
methionine choline deficient diet
- HFD
high fat diet
- HCD
high cholesterol diet
- PL
phospholipids
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- FA
fatty acid
- Ch
cholesterol
- TG
triglycerides
- SDH
succinate dehydrogenase
- OA
oleic acid
- MDMC
medium deficient in methionine and choline
- ASM
acid-soluble metabolites
- OCR
oxygen consumption rate
- OXPHOS
oxidative phosphorylation
- CII
complex II
- DMG
dimethylglycine
- SARDH
sarcosine dehydrogenase
- DMGDH
dimethylglycine dehydrogenase
- ETF
electron transfer flavoprotein
- FAD/FADH
flavin adenine dinucleotide.
1. Introduction
Non-alcoholic fatty liver disease (NAFLD) is the most common cause of chronic liver disease. There are 7.5 billion people in the world, among which it is estimated about 1.8 billion people (25%) suffer from NAFLD, making it an emerging global health problem [1]. NAFLD progresses from simple lipid accumulation (steatosis), which is considered a benign disease, to non-alcoholic steatohepatitis (NASH) with inflammation and fibrosis, representing a major risk factor for the development of cirrhosis and liver cancer [2]. Hepatic steatosis can result from different non-excluding metabolic dysregulation, including i) impaired lipid uptake; ii) alterations in mitochondrial fatty acid β-oxidation (FAO); iii) increased de novo lipogenesis (DNL); iv) and/or inefficient very-low-density lipoprotein (VLDL) assembly and secretion.
Currently, there are no approved therapies for NAFLD treatment. Current treatments aim at managing different clinical conditions such as obesity and hypertriglyceridemia. However, new therapies targeting the underlying mechanisms that trigger NAFLD are under development. Due to the expected increase both in NAFLD and NASH, the NASH market is estimated to rise about 34% in the next decade, reaching a global worth of $15 billion only in the United States [3]. Nevertheless, the development of multitarget therapies remains an important goal for the treatment of NAFLD.
Glycine N-methyltransferase (GNMT) is the most important and abundant S-adenosylmethionine (SAMe)-dependent methyltransferase in the liver. The downregulation of GNMT has been described in several manifestations of chronic liver disease, including NAFLD [4], [5], cholestasis [6], [7], cirrhosis, and liver cancer [8], [9]. Particularly, in NAFLD patients, GNMT has been described as one of the top downregulated proteins [4], [5], highlighting the importance of its downregulation as a driver of disease progression during the initial stages of the disease. In a recent study, we described the microRNA miR-873-5p as a negative posttranscriptional regulator of GNMT expression in the liver that is involved in the progression of cholestasis and fibrosis [7]. Moreover, targeting miR-873-5p recovered GNMT expression and protected from liver injury [7]. GNMT is predominantly expressed in hepatocytes, where it accounts for about 1–3% of total cytosolic proteins and it is responsible for SAMe catabolism [8]. Although it is mainly localized in the cytosol, GNMT is known to be present in other cell compartments, such as mitochondria [10] and nuclei [11], although it is unknown what roles the protein exerts in these organelles.
In recent years, several studies have described the role of mitochondria in NAFLD [12]. Mitochondria are one of the most important metabolic organelles and responsible for most ATP production in hepatocytes. Several important metabolic pathways are operative in the mitochondria, including FAO, the tricarboxylic acid (TCA) cycle, and oxidative phosphorylation (OXPHOS) in the electron transport chain (ETC). In NAFLD, a number of metabolic adaptations occur to counteract fat accumulation in the liver [12]. Increased mitochondrial FAO has been reported in NAFLD patients and murine models. However, this can result in reactive oxygen species (ROS) overproduction, leading to mitochondrial dysfunction and inflammation. Alterations in the activity of the ETC complexes, ATP generation, and oxidative metabolism have been reported in NAFLD pathogenesis [12]. Thus, a number of studies indicate that targeting mitochondrial function during NAFLD is a promising approach for the treatment of liver disease patients [13].
In this work, we have evaluated GNMT repression by miR-873-5p in the liver and its involvement in NAFLD. We describe increased hepatic miR-873-5p levels and its correlation with GNMT downregulation in NAFLD/NASH patients. MiR-873-5p upregulation was also observed in different murine NAFLD models in which the effect of targeting its expression was evaluated. We demonstrate that the efficient repression of miR-873-5p in the liver of mice fed with a methionine choline deficient diet (MCDD) results in the recovery of GNMT levels in the hepatocytes and, specifically, in the mitochondria, with protective effects against NAFLD. Mechanistically, we describe that the role of GNMT mitochondrial is mediated through the interaction and regulation of Complex II (CII) activity of the ETC. This process increased mitochondrial functionality and FAO while decreasing oxidative stress in the liver, protecting from fatty liver progression.
In summary, we describe a new essential role of GNMT in the mitochondria and demonstrate that the recovery of hepatic GNMT levels by targeting miR-873-5p emerges as a new therapeutic approach for a broad spectrum NAFLD therapies.
2. Experimental procedures
2.1. Human subjects
Liver biospecimens were obtained from obese patients diagnosed in the Marqués de Valdecilla University Hospital (MVUH, Santander, Spain). Patients were evaluated for NAFLD using multiple markers of disease after exclusion of alcoholic disease and viral hepatitis infection. The characteristics of these patients are described in Table 1. The studies were performed in agreement with the Declaration of Helsinki and national regulations. The hospital Ethics Committees approved the study procedures and written informed consent was obtained from all patients before inclusion in the study.
Table 1.
Characteristics of obese patients included in NAFLD analysis. Abbreviations: BMI (body mass index), TG (triglycerides), LDL/HDL (low/high density lipoprotein).
| Variable | Healthy | Steatosis | NASH |
|---|---|---|---|
| N | 6 | 20 | 16 |
| Age (years, mean ± SD) | 31.6 ± 5.5 | 44.6 ± 11.1 | 49 ± 11 |
| Gender (F/M) | 7/0 | 11/9 | 9/7 |
| BMI | 48.6 ± 5.4 | 47.9 ± 5.6 | 47 ± 5.7 |
| TG | 134.8 ± 93.4 | 170.3 ± 100.4 | 181.6 ± 84.7* |
| Cholesterol | 190 ± 36.6 | 172.2 ± 40.3 | 187.5 ± 32.8 |
| LDL | 110.8 ± 35 | 91.6 ± 29.6 | 106.8 ± 28 |
| HDL | 55.3 ± 12.9 | 41 ± 10 | 38.3 ± 9 |
| AST | 18.4 ± 4.1 | 27.6 ± 13 | 33 ± 20.5 |
| ALT | 18.5 ± 7.5 | 34.1 ± 19.6 | 40.5 ± 22.5 |
| Glucose | 85.8 ± 6.5 | 107.3 ± 38.9 | 115.7 ± 42* |
| NAS score | 0.6 ± 0.1 | 2.63 ± 0.25 | 4.56 ± 0.27* |
| Ballooning | 0.2 ± 0.1 | 0.42 ± 0.2 | 1.44 ± 0.13* |
| Inflammation | 0.4 ± 0.13 | 0.58 ± 0.2 | 1 ± 0.6* |
* p < 0.05 compared to healthy subjects.
2.2. Animal experimentation
2.2.1. miR-873-5p inhibition
Male 3-month old C57BL/6 mice were fed a methionine (0.1%) and choline (0%) deficient diet (MCD diet) (Research Diets, USA) or regular chow diet as a control group. After 1 week of feeding on the 0.1%MCD diet, mice were treated with anti-miR-873-5p (60μg/mouse) (Dharmacon, USA) or an unrelated miR-Ctrl using Invivofectamine® 3.0 Reagent (Thermo Fisher Scientific, Massachusetts, USA) by tail vein injection every 3–4 days until 4 weeks. At the end of the experiment, mice were sacrificed, and livers were snap frozen or fixed in formalin for subsequent analysis or subjected to different ex vivo analysis of β-oxidation and de novo lipogenesis. Male 4-month old Gnmt-KO mice fed a regular chow diet were treated with a single dose of the anti-miR-873-5p (60ug/mouse) by tail vein injection. Mice were sacrificed 4 days later, and livers were snap frozen, fixed in formalin, or used for ex vivo analysis of β-oxidation. At least five (n ≥ 5) animals were used per group. Animal procedures were approved by CIC bioGUNE's Animal Care and Use Committee and the competent authority (Diputación de Bizkaia).
2.2.2. Liver lipid quantification
30 mg of frozen livers were homogenized with ice-cold PBS. Fatty acids were measured in the homogenates using the Wako Chemicals kit (Richmond, VA, USA), and lipids were extracted and quantified as described [14]. Phosphatidilcholine (PC), phosphatidylethanolamine (PE), fatty acids (FAs), and cholesterol (Ch) were separated by thin layer chromatography (TLC) and quantified as described [15]. Triglycerides (TGs) were measured in the lipid extract with the A. Menarini Diagnostics (Italy) kit.
2.2.3. β-oxidation
Fatty acid β-oxidation was assessed as described before [16], [17], [18]. Fresh liver slices were homogenized in cold buffer (Tris–HCl 25 mM, sucrose 500 nM, EDTA Na2 pH 7.4 1 mM), sonicated (10″), and centrifuged (500 g, 10′, 4 °C). 500 μg of protein were diluted in 200 μl. The assay was performed as follows: 400 μl of assay mixture containing 0.5 μCi/ml (1–14C) palmitate was added to the samples and incubated 1 h at 37 °C in Eppendorf tubes with a Whatman paper in the cap. The reaction was stopped by adding 400 μl of perchloric acid 3M, and NaOH 1M was added to impregnate the Whatman cap. After 2 h, the Whatman cap was retired, and the radioactivity associated was measured in a scintillation counter. Eppendorf tubes were centrifuged (2100 g, 10', 4 °C). 400 μl of the supernatant were collected and the radioactivity was counted in a scintillation counter.
2.2.4. De novo lipogenesis
DNL was performed as previously described [19] with slight modifications. Briefly, freshly isolated liver tissue slices (40 mg) were incubated in high glucose DMEM with insulin (150 nM) and [3H]- Acetic acid 20 μCi/ml for 4 h. Tissue slices were washed and homogenized in PBS. Then, lipids were extracted [14] and separated [15] as previously described. Each lipid was scraped and the radioactivity was measured in a scintillation counter.
2.2.5. Succinate dehydrogenase (SDH) activity
SDH activity was measured in mitochondrial frozen liver extracts with Succinate Dehydrogenase Activity Colorimetric Assay Kit (MAK197, Sigma Aldrich) following the manufacturer's procedure. SDH activity was calculated and represented as nmol of succinate converted to fumarate/(volume/minute).
2.2.6. Lipid peroxidation
Lipid peroxidation Assay kit: Malondialdehyde (MDA) content in liver samples was quantified by using a commercially available kit from Sigma–Aldrich (St. Quentin Fallavier, France) and quantified colorimetrically (OD = 532 nm).
2.2.7. Mitochondrial isolation
Liver mitochondria were freshly isolated by differential centrifugation followed by rapid centrifugation through Percoll density gradient as previously described [20]. Alternatively, mitochondria from frozen livers were obtained using the Mitochondrial/Cytosol Fractionation Kit, Abcam (London, UK).
2.2.8. Blue native-PAGE
Purified mitochondria were solubilized in native PAGE loading buffer (Invitrogen, USA) containing 2% digitonin (Sigma, USA). Complexes were resolved by electrophoresis in 3–12% NativePAGE Novex Bis-Tris gels (Invitrogen) followed by transfer to a polyvinylidene difluoride (PVDF) membrane and western blot analysis.
2.2.9. Protein immunoprecipitation assay
Protein–Protein complexes were immunoprecipitated as described before [21] using Complex II Immunocapture Kit (ab109799) (Abcam).
2.2.10. Metabolomic analysis
Metabolites implicated in the methionine cycle and reduced/oxidized glutathione were determined by LC/MS using a Waters ACQUITY-UPLC system coupled to a Waters Micromass LCT Premier Mass Spectrometer equipped with a Lockspray ionization source as described previously [22].
2.2.11. Protein isolation and western blotting
Total protein extracts from primary hepatocytes and hepatic tissue were resolved in sodium dodecyl sulfate-polyacrylamide gels and transferred to nitrocellulose membranes. As secondary antibodies, we used anti-rabbit-IgG-HRP-linked (Cell Signaling) and anti-mouse-IgG-HRP-linked (Cell Signaling).
2.2.12. Proteomic analysis
Proteomic analysis in freshly isolated liver mitochondria was performed following the label free (LF) analysis. Protein was extracted using 7M urea, 2M thiourea, 4% CHAPS. Samples were incubated for 30 min at RT under agitation and digested following the filter-aided FASP protocol [23]. Approximately 500 ng of each sample was submitted to liquid chromatography-mass spectrometry (LC-MS) label-free analysis. Peptide separation was performed on a nanoACQUITY UPLC System (Waters) on-line connected to an LTQ Orbitrap XL mass spectrometer (Thermo Electron). An aliquot of each sample was loaded onto a Symmetry 300 C18 UPLC Trap column (180 μm × 20 mm, 5 μm (Waters)). The precolumn was connected to a BEH130 C18 column (75 μm × 200 mm, 1.7 μm (Waters), and equilibrated in 3% acetonitrile and 0.1% FA. Peptides were eluted directly into an LTQ Orbitrap XL mass spectrometer (Thermo Finnigan) through a nanoelectrospray capillary source (Proxeon Biosystems), at 300 nl/min and using a 120 min linear gradient of 3–50% acetonitrile. The mass spectrometer automatically switched between MS and MS/MS acquisition in DDA mode. Full MS scan survey spectra (m/z 400–2000) were acquired in the orbitrap with mass resolution of 30000 at m/z 400. After each survey scan, the six most intense ions above 1000 counts were sequentially subjected to collision-induced dissociation (CID) in the linear ion trap. Precursors with charge states of 2 and 3 were specifically selected for CID. Peptides were excluded from further analysis during 60 s using the dynamic exclusion feature.
Progenesis LC-MS (version 2.0.5556.29015, Nonlinear Dynamics) was used for the label-free differential protein expression analysis. One of the runs was used as the reference to which the precursor masses in all other samples were aligned to. Only features comprising charges of 2 + and 3 + were selected. The raw abundances of each feature were automatically normalized and logarithmized against the reference run. Samples were grouped in accordance to the comparison being performed, and an ANOVA analysis was performed. A peak list containing the information of all the features was generated and exported to the Mascot search engine (Matrix Science Ltd.). This file was searched against a Uniprot/Swissprot database, and the list of identified peptides was imported back to Progenesis LC-MS. Protein quantitation was performed based on the three most intense non-conflicting peptides (peptides occurring in only one protein), except for proteins with only two non-conflicting peptides. The significance of expression changes was tested at protein level, and proteins with an ANOVA p-value ≤ 0.05 were selected for further analyses.
Subsequent analysis and clustering of differentially expressed proteins was performed using Perseus software platform (http://www.perseus-framework.org) in order to group proteins according to changes in their amount. Finally, GO enrichment analysis was carried out using the STRING online tool (https://string-db.org/) in order to infer pathways and processes altered in each group and cluster.
2.2.13. RNA isolation and quantitative real-time polymerase chain reaction
Total RNA was isolated with Trizol (Invitrogen). 1–2 μg of total RNA was treated with DNAse (Invitrogen) and reverse transcribed into cDNA using M-MLV Reverse Transcriptase (Invitrogen). Quantitative real-time PCR (RT-PCR) was performed using SYBR® Select Master Mix (Applied Biosystems) and the ViiA 7 Real-Time PCR System (Applied Biosystems) by the ΔΔCt method using Arp as reference gene.
2.2.14. MicroRNA quantitative real-time PCR
RT-PCR was performed for miR-873-5p following a TaqMan® MicroRNA Reverse Transcription Kit (Life Technologies, USA) procedure using 50 ng of total RNA. qPCR was performed with the TaqMan Universal PCR Master Mix No AmpErase UNG kit following manufacturer's procedure. miR-873-5p expression levels were normalized to the U6 snRNA.
2.2.15. Immunohistochemistry
Paraffin embedded liver samples were sectioned, dewaxed, and hydrated. Immunohistochemistry was performed as previously described [24]. H&E and Sirius red staining for collagen was performed in paraffin embedded liver samples. F4/80 a membrane macrophage marker and αSMA, a marker for activated stellate cells were analyzed by immunohistochemical staining. Liver Sudan Red staining for the histological quantification of hepatic lipids was used. Stained area percentage of each sample were calculated using FRIDA software (FRamework for Image Dataset Analysis) http://bui3.win.ad.jhu.edu/frida/.
2.3. Cellular experiments
Mouse primary hepatocytes, Kupffer cells, and hepatic stellate cells were isolated as previously described [25], [26]. Briefly, mouse livers were perfused with collagenase (Worthington Biochemical Company, Freehold, USA) and hepatocytes isolated following a standard centrifugation. KC ad HSC were isolated after Percoll Plus (GE Healthcare, Little Chalfont, United Kingdom) gradient centrifugation and selective adherence. Primary hepatocytes were isolated from C57BL/6 WT and Gnmt-KO mice via collagenase as above mentioned and transfected with anti-miR-873-5p or miR-Control using dharmaFECT1 transfection reagent (Dharmacon) as previously described [7]. Hepatocytes were cultured with oleic acid (OA, 400 μM 6 h) (Sigma Aldrich) or maintained in medium deficient in methionine and choline (MDMC) (48 h). Cellular experiments were performed at least three times.
2.3.1. BODIPY staining
Hepatocytes in culture were collected in covers and incubated with BODIPY 493/503 (Molecular Probes, Thermo Fisher Scientific) at a concentration of 10 μg/ml during 45 min prior to fixation (4% paraformaldehyde). Quantification of lipid bodies was performed using the Frida Software (FRamework for Image Dataset Analysis) http://bui3.win.ad.jhu.edu/frida/.
2.3.2. Respiration studies
The cellular metabolic profile was determined using a Seahorse XF24 Extracellular Flux Analyzer (Seahorse Biosciences, USA), providing real-time measurements of the oxygen consumption rate (OCR) as previously described [27].
2.3.3. Reactive oxygen species (ROS)
ROS production in primary hepatocytes was assessed using CellROX Deep Green Reagent (Thermo Fisher Scientific). The hepatocytes were loaded with 1.5 μM CellROX in 10% FBS-MEM (10', 37 °C). The hepatocytes were then carefully washed 3 times with phosphate-buffered saline, collected, and analyzed by flow cytometry FACS Canto II (BD Biosciences, USA).
2.4. Statistical analysis
Data are represented as average ± SEM. For in vivo studies at least five (n ≥ 5) animals were used per group. For in vitro cellular studies, experiments were performed at least 3 times. mRNA/RNA data is normalized vs. control. Statistical significance was determined with Prism 5 (GraphPad Software). One-way analysis of variance (ANOVA) test was used when 3 groups were compared, while the Student's t-test was used for 2 group comparisons. A p < 0.05 was considered significant for all the comparisons except for proteomic analysis where p-adj. (q value) q < 0.05 was considered.
3. Results
3.1. GNMT and miR-873-5p expression in NAFLD
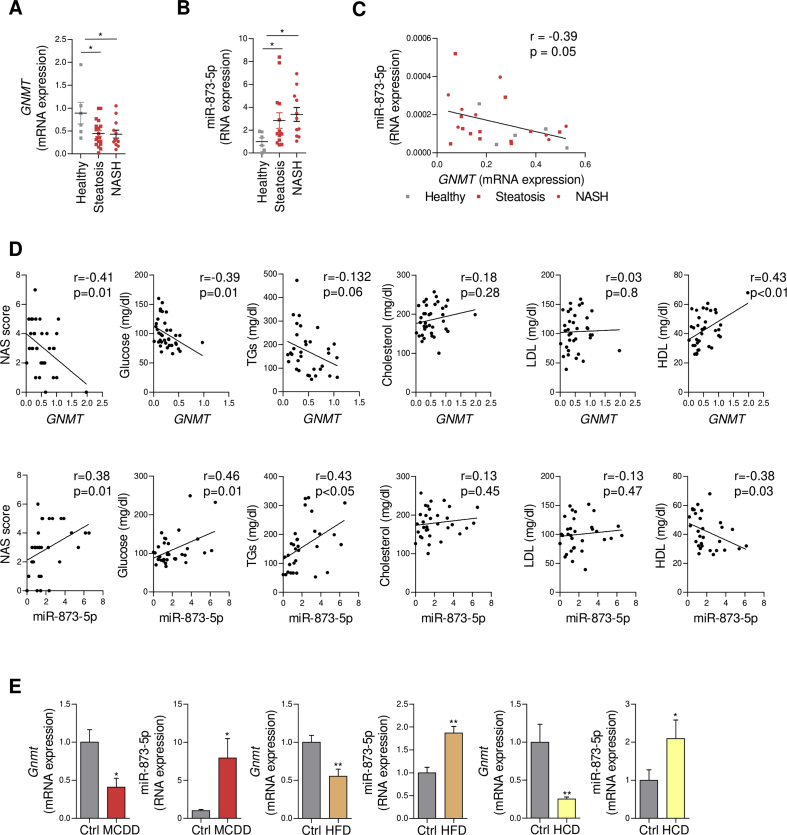
GNMT expression has been identified as part of an anti-steatotic mechanism, and its reduction has been well documented in NAFLD patients [4], [5]. The mechanisms driving GNMT downregulation in NAFLD and the potential benefit of recovering normal GNMT levels in the liver in NAFLD situations have not been previously addressed. Recently, we have described the implication of miR-873-5p in GNMT repression during fibrosis and cholestasis [7]; thus, we have analyzed the implication of this microRNA in early stages of NAFLD in association with GNMT. Consistent with previous results [4], [5], we found GNMT was downregulated in the liver of NAFLD (N = 36) compared to healthy obese (N = 6) subjects (Figure 1A). NAFLD patients were divided as simple steatosis (20) or NASH (16) stages based on clinical and histopathological features (Table 1). Conversely, miR-873-5p was progressively increased in steatosis and mainly in NASH patients (Figure 1B), correlating with GNMT reduction (Figure 1C). Moreover, we found a correlation between GNMT and miR-873-5p expression in the liver and the grade of fibrosis progression (NAS score) and metabolic parameters such as circulating glucose, TGs, and HDL levels in these patients (Figure 1D). These results indicate a matched dysregulation of GNMT, miR-873, and metabolism that correlates with NAFLD progression to more severe stages.
Figure 1.
miR-873-5p inversely correlates GNMT downregulation in the liver in NASH. (A–C) miR-873-5p and GNMT expression and correlation in the liver of a cohort of steatotic (n = 20) and NASH (n = 16) patients. (D) GNMT and miR-873-5p expression levels in the liver and correlation with (left to right) NAS score, and circulating levels of glucose, triglycerides (TGs), total cholesterol, and low- and high-density lipoprotein (LDL, and HDL). (E) miR-873-5p and Gnmt expression in indicated murine models of NAFLD, from left to right: methionine choline deficient diet (MCDD); high fat diet (HFD) and high cholesterol diet (HCD). Data shown as average ± SEM. p < 0.05 *; p < 0.01 **; p < 0.001 ***.
Gnmt and miR-873-5p regulation was also assessed in several murine models of NAFLD. Gnmt reduction in association with miR-873-5p upregulation was observed in mice fed for 4 weeks with a choline deficient and methionine-0.1% diet (MCDD), in those under a high fat diet (HFD) for 20 weeks and in mice under high cholesterol diet (HCD) for 4 weeks (Figure 1E). Even though these models differ from each other in the mechanism underlying fatty liver progression and the global metabolic effect, GNMT/miR-873-5p alterations observed in all of them suggest this axis may be involved in the regulation of a common metabolic pathway in the liver.
These data highlight the association between miR-873-5p and GNMT expression in human and mice during NAFLD.
3.2. The rescue of GNMT expression by anti-miR-873-5p reduces lipid accumulation in vitro in hepatocytes
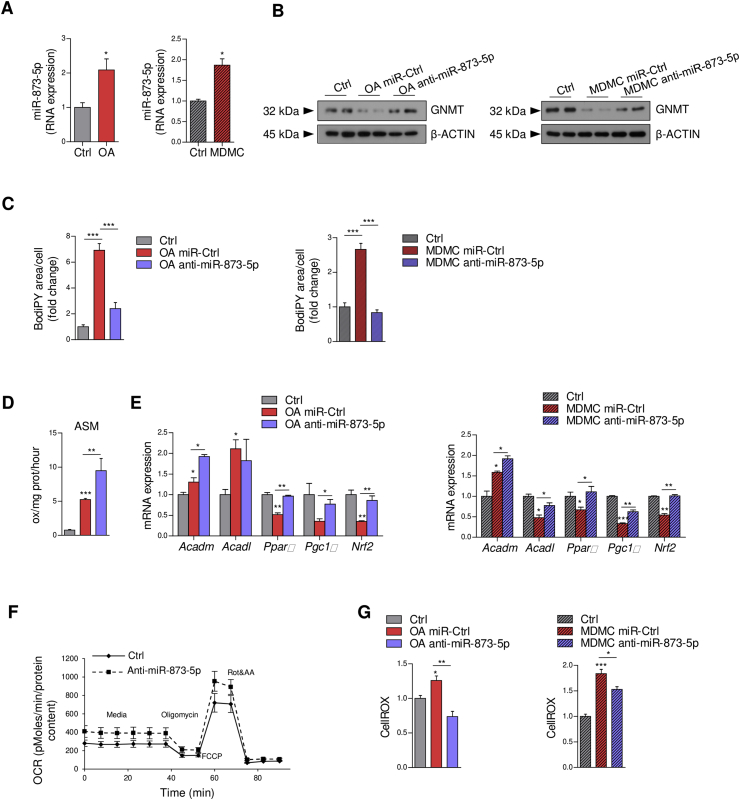
As a chronic model of GNMT depletion, the Gnmt-KO mouse develops steatosis and NASH spontaneously [28]. In this murine model, different mechanisms associated with GNMT deficiency have been described that contribute to the development of the disease [25], [28], [29], [30]. Here, we assessed the role of miR-873-5p in a steatotic in vitro model. As GNMT is known to be mainly expressed in hepatocytes [8], we first studied the role of GNMT/miR-873-5p regulation in these cells under steatotic conditions. Primary WT hepatocytes were incubated with oleic acid (OA) (400 μM, 6 h) or with a medium deficient in methionine and choline (MDMC) for 48 h. miR-873-5p was upregulated in both conditions (Figure 2A). Anti-miR-873-5p transfection (Suppl. Fig. 1A,B) resulted in GNMT recovery (Figure 2B) and in the reduction in lipid content, measured by the BODIPY staining (Figure 2C and Suppl. Fig. 1A,B). Importantly, secretion of acid-soluble metabolites (ASM) (Krebs cycle metabolites and ketones), as a measure of FA β-oxidation revealed a significant increase in cells treated with OA and transfected with anti-miR-873-5p (Figure 2D). Consistent with these results, the expression of genes associated with FA β-oxidation such as Acadm, Acadl (acyl-CoA dehydrogenase, medium and long chain), peroxisome-proliferator activated receptor alpha (Ppara), peroxisome proliferator-activated receptor gamma coactivator 1 (Pgc1α) (implicated in mitochondrial biogenesis and remodelling in most cells) and NF-E2-related factor 2 (Nrf2), a transcription factor involved in antioxidant defence, were upregulated in both models after blockade of miR-873-5p (Figure 2E).
Figure 2.
In vitro targeting of miR-873-5p decreases lipid accumulation in hepatocytes. (A,B) miR-873-5p expression and GNMT levels in primary hepatocytes cultured with oleic acid (OA) or medium deficient in methionine and choline (MDMC). (C) Quantification of BODIPY staining in primary mouse hepatocytes under indicated condition. (D) Analysis of β-oxidation and (E) qPCR analysis of β-oxidation related genes in primary hepatocytes. (F) OCR measurement of primary mouse hepatocytes with anti-miR-873-5p or miR-Ctrl by seahorse analysis. (G) ROS levels in primary hepatocytes treated with miR-Ctrl anti-miR-873-5p under indicated conditions. Data shown as average ± SEM. p < 0.05 *; p < 0.01 **; p < 0.001 ***.
The oxygen consumption rate (OCR) revealed an increase of OXPHOS by seahorse analysis in primary hepatocytes transfected with anti-miR-873-5p (Figure 2F), as a surrogate of mitochondrial functionality. Importantly, a reduction of oxidative stress (ROS) was detected in both in vitro steatotic models under anti-miR-873-5p treatment (Figure 2G).
Finally, to prove the specific targeting of miR-873-5p over Gnmt, Gnmt-KO hepatocytes were transfected with anti-miR-873-5p and incubated with oleic acid (OA) (400 μM, 6 h). Importantly, anti-miR-873-5p (Suppl. Fig. 1C) did not reduce lipid content in Gnmt-KO hepatocytes, indicating the major role of Gnmt targeting by miR-873-5p under steatotic condition in the hepatocytes (Suppl. Fig. 1C).
These results suggest a new effect of GNMT re-expression mediated by anti-miR-873-5p on fatty acid β-oxidation concomitant with a better mitochondrial functionality and a reduction in oxidative stress leading to a lower accumulation of lipids in hepatocytes.
3.3. Anti-miR-873-5p treatment reduces steatohepatitis in vivo
Previous studies have shown GNMT is mainly found in hepatocytes; however it is also expressed in other cells, such as macrophages (Kupffer Cells and hepatic stellate cells) [7], [8], [31]. Thus, to better characterize GNMT/miR-873-5p regulation in NAFLD progression, we measured their expressionGnmt levels in different hepatic cell populations freshly isolated from WT mice fed a chow or MCD diet during 4 weeks. According to previous studies Gnmt was found to be mainly expressed in hepatocytes (Suppl. Figure 2A). Moreover, MCD diet only resulted in Gnmt downregulation and miR-873-5p upregulation in hepatocytes, suggesting the importance of this miRNA-protein in these cells during NAFLD progression (Suppl. Figure 2A). These results are also in accordance with our in vitro evidence regarding the benefit of recovering GNMT levels in hepatocytes (section 3.2).
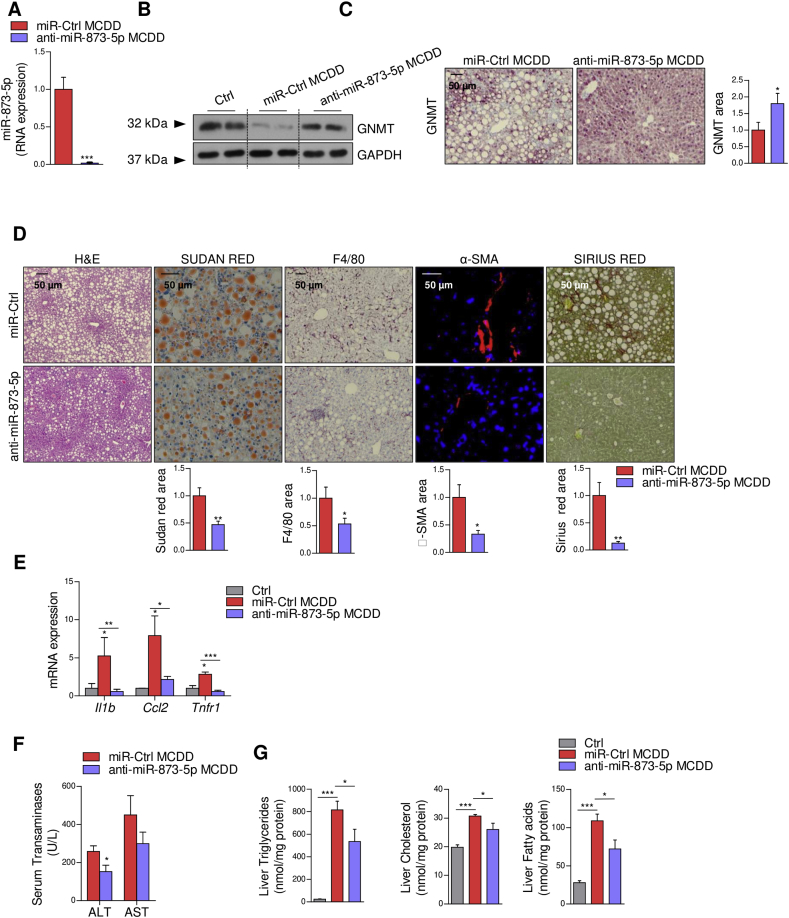
In order to analyze the role of GNMT/miR-873-5p in a preclinical model of NASH, mice were fed with a MCD diet for one week, when lipid accumulation and inflammation is initiated in the liver and GNMT/miR-873-5p misregulation is produced (Suppl. Figure B,C). The mice were then injected with a miR-Control or an anti-miR-873-5p twice a week for three more weeks under the MCD diet (Suppl. Figure 2D). Anti-miR-873-5p treatment resulted in GNMT recovery in the liver (Figure 3A–C). Histological evaluation of H&E-stained liver tissue revealed a decreased vacuolization of hepatocytes in anti-miR-873-5p-treated mice (Figure 3D). The activation of hepatic stellate cells, assayed by smooth muscle actin (SMA) staining, collagen deposition (Sirius red), inflammation (F4/80) and inflammatory gene expression (Interleukin 1b (Il1b), Chemokine (C–C motif) ligand 2 (Ccl2), and Tumor necrosis factor receptor 1 (Tnfr1)); ALT aminotransferases levels were also diminished in the livers of anti-miR873-5p-treated animals (Figure 3D–F).
Figure 3.
miR-873-5p inhibition in vivo recovers GNMT expression and reduces lipid content, inflammation and fibrosis in the liver of MCDD mice. (A) miR-873-5p levels in MCDD mice after anti-miR-873-5p administration. (B) GNMT levels determined by WB and (C) IHC in MCDD mice. (D) Liver characterization by IHC with indicated staining in MCDD mice showing H&E, lipid content (Sudan Red), inflammation (F4/80) and fibrosis (αSMA and Sirius Red). (E) mRNA analysis of indicated genes related to inflammatory response in liver extracts. (F) Serum biochemical analysis of transaminases (ALT and AST). (G) Quantification of liver lipid content showing fatty acids, triglycerides, and free cholesterol in the indicated MCDD mice. Data shown as average ± SEM. p < 0.05 *; p < 0.01 **; p < 0.001 ***.
Moreover, Sudan red, which stains triglycerides and lipids, identified a reduction of positively stained droplets in anti-miR873-5p mice (Figure 3D). Furthermore, the biochemical determination of lipid content revealed reduced levels of triglycerides, FAs and cholesterol in the livers of these mice (Figure 3G), while the levels of different types of phospholipids (PLs) remained stable or slightly decreased (Suppl. Fig. 3). Altogether, our results indicate that miR-873-5p inhibition decreases lipid content alleviating liver inflammation and fibrogenesis, globally blunting NASH progression, which appears to be mediated by the recovery of GNMT in the hepatocytes.
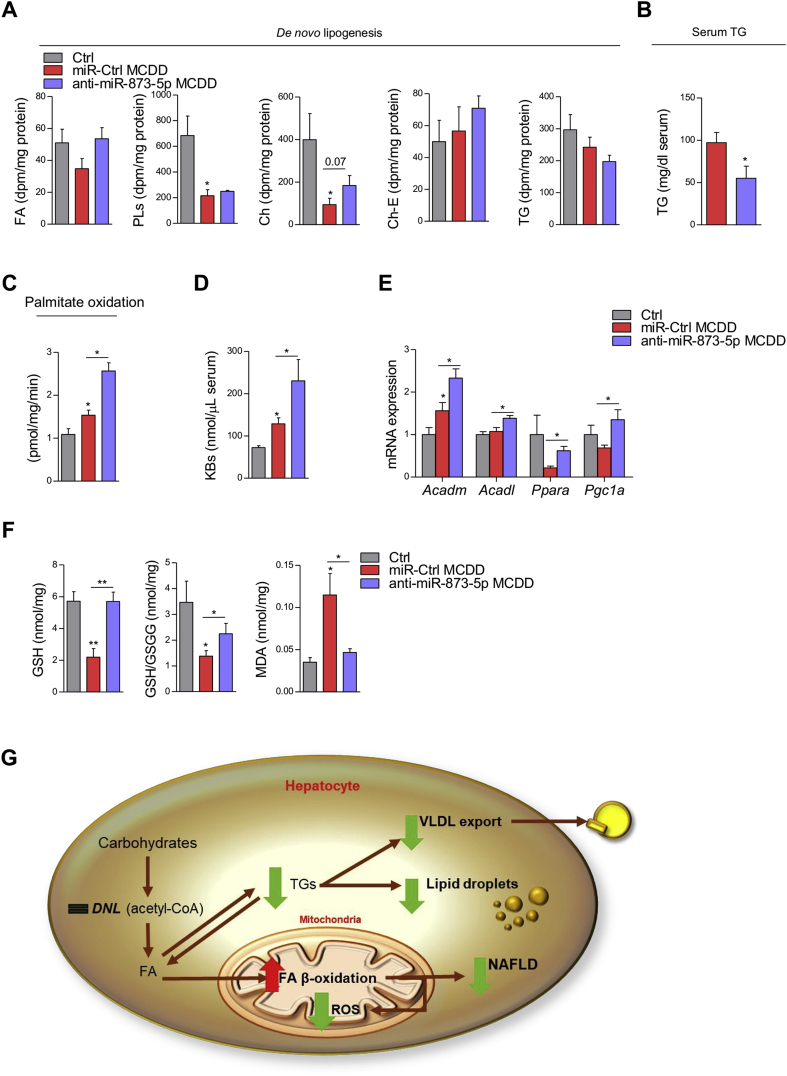
3.4. GNMT re-expression activates β-oxidation in vivo reducing steatosis
Hepatic fat can be reduced through the activation of different pathways, including triglycerides secretion into VLDLs, enhanced fatty acid β-oxidation (FAO) and/or decreased de novo lipogenesis (DNL). Therefore, we further characterized the response observed under miR-873-5p inhibition in the MCD diet mice. DNL measured by ex vivo [H3] acetate incorporation revealed no differences in the hepatic formation of FAs, PLs, free cholesterol, cholesterol ester, and TGs, after blocking miR-873-5p (Figure 4A). Moreover, reduced TG in the serum suggested decreased TG export into VLDLs in anti-miR-873-5p treated mice (Figure 4B). These data indicate that miR-873-5p regulates lipid metabolism through mechanisms different from DNL or VLDL secretion, as previously suggested in our in vitro model, in which increased FAO was shown in anti-miR-873-5p hepatocytes (section 3.2).
Figure 4.
Targeting miR-873-5p induces mitochondrial fatty acid β-oxidation. (A) Serum TG levels, (B) ex vivo determination of DNL and (C) mitochondrial FA β-oxidation measured by CO2 oxidation, in control and MCDD livers. (D) Analysis of serum ketone bodies (KBs). (E) mRNA analysis of indicated genes related to FA β-oxidation in liver extracts. (F) Analysis of oxidative stress in the MCDD livers measured by GSH and GSH/GSSG content and lipid peroxidation (MDA). (G) Schematic representation of the anti-miR-873-5p effect in the liver of MCD diet mice. Data shown as average ± SEM. p < 0.05 *; p < 0.01 **; p < 0.001 ***.
We therefore explored FAO as the main mechanism implicated in FA degradation. [C1−14] palmitate labeling revealed an increase of the CO2 released as the index FAO in anti-miR-873-5p mice (Figure 4C). Consistently, an increase in serum ketone bodies was detected (Figure 4D). These results were further supported by the induction of genes implicated in FAO, such as Acadm, Acadl, Ppara, and Pgc1a (Figure 4E).
Fatty acid β-oxidation is directly linked to the ETC. In NAFLD, mitochondrial dysfunction impairs the oxidation of FAs as a consequence of ROS production, leading to lipid accumulation within the liver. In vitro results show that improved FAO in anti-miR-873-5p-hepatocytes do not increase ROS and mitochondrial stress. Thus, we measured the oxidative stress in MCD mice by two different approaches. Decreased lipid peroxidation measured by malondialdehyde (MDA), together with the increase in reduced glutathione levels, the most important antioxidant defense system (Figure 4F), confirmed that anti-miR-873-5p treatment reduces oxidative stress in the liver in the NASH model.
The role of GNMT in mitochondrial FAO capacity mediated by anti-miR-873-5p treatment was further supported by the lack of effect of anti-miR-873-5p in the absence of GNMT in the liver. In order to prove it, 4-months old Gnmt-KO mice, when they have already developed liver steatosis and inflammation [28], were treated with the anti-miR-873-5p by tail vein injection four days, a time point in which miR-873-5p is already efficiently inhibited in the liver (Suppl. Figure 4A). According to in vitro results in Gnmt-KO hepatocytes (Section 3.2), anti-miR-873-5p treatment did not have any effect in lipid accumulation and NASH development in the livers of Gnmt-KO mice (Suppl. Figure 4B). Moreover, FAO measured by [C1−14] palmitate labeling revealed no changes in CO2 released in anti-miR-873-5p Gnmt-KO mice (Suppl. Figure 4C), further indicating the specific role of GNMT in the regulation of mitochondrial FAO and its targeting by miR-873-5p in NAFLD/NASH.
Altogether, these results confirm the implication of anti-miR-873-5p and GNMT in the induction of FAO as the main mechanism to reduce lipid accumulation in NAFLD/NASH (Figure 4G).
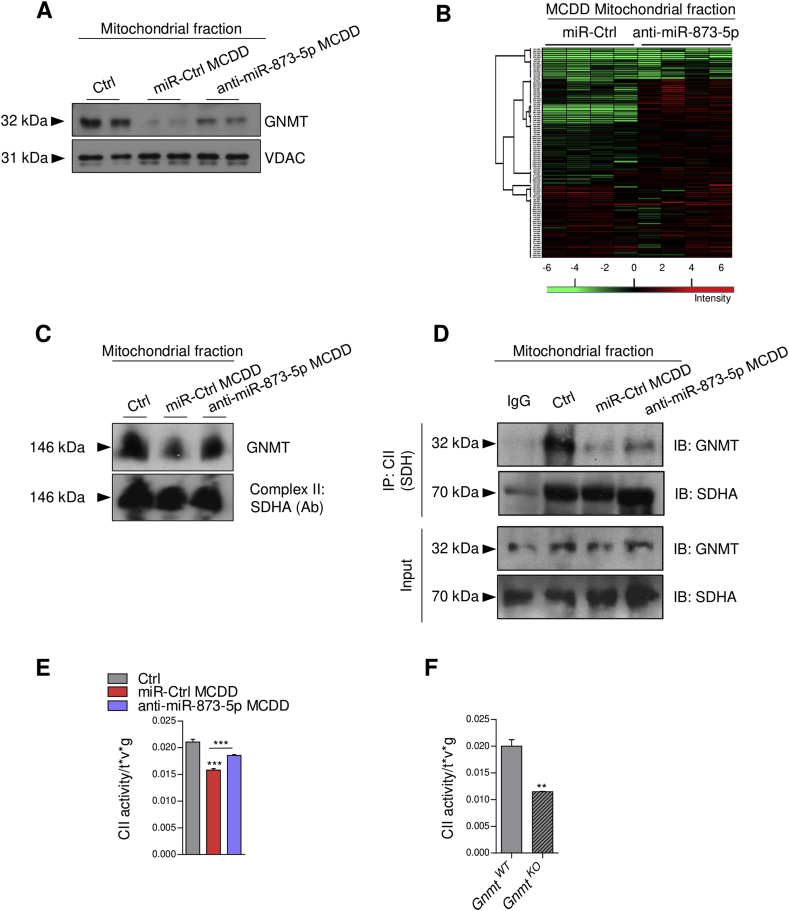
3.5. MiR-873-5p regulates mitochondrial GNMT and complex II activity in NAFLD
GNMT is an abundant cytosolic enzyme that is also present in other cellular compartments [10], [11]. To understand the impact of anti-miR-873-5p/GNMT in lipid metabolism related to FAO we evaluated the mitochondrial localization of GNMT in the MCDD fed mice. GNMT was found in the mitochondrial fraction of healthy livers, while steatotic feeding resulted in reduced mitochondrial GNMT. Remarkably, anti-miR-873-5p counteracted GNMT decrease in the mitochondria (Figure 5A and Suppl. Fig. 5A).
Figure 5.
Mitochondrial GNMT decrease drives disruptions in Complex II activity. (A) WB analysis of GNMT and (B) global proteomic profile of freshly isolated liver mitochondria in the MCDD and MCDD-anti-miR-873-5p mice compared to control diet mice, changes are presented as ratios by intensity (colour scale bar). (C) BN-PAGE of 2% digitonin solubilized mitochondrial liver extracts, transferred to a membrane and immunoblotted for GNMT and SDHA. (D) Immunoprecipitation assay of mitochondrial liver extracts immunocaptured with CII and blotted for GNMT. (E) CII activity assay performed in the liver of the MCDD and (F) in Gnmt-KO mice. Data shown as average ± SEM. p < 0.05 *; p < 0.01 **.
At present, no mitochondrial function of GNMT has been described. To characterize mitochondria in MCD diet mice, we performed proteomic analysis of freshly isolated liver mitochondria, to determine potential processes regulated by miR-873-5p. The proteomic analysis showed global proteomic alterations as a consequence of the MCD diet, as well as a result of the treatment with anti-miR-873-5p (Figure 5B). A complete list of the identified proteins significantly regulated in the analysis is provided in Supplementary Table 1. Hierarchical clustering of this proteins with PERSEUS software and gene ontology (GO) analysis using the STRING database were performed. Top regulated proteins revealed an upregulation of GO-biological processes related to oxidation-reduction processes (GO:0055114) and to metabolic and oxidative phosphorylation (OXPHOS) pathways, classified according to the KEGG database (Suppl. Table2). Moreover, other identified proteins were implicated in fatty acid degradation, antioxidation and oxidation-reduction processes (Suppl. Tables 3 and 4).
Overall, the results obtained from the proteomic analysis suggest that the most important processes altered in the mitochondria of MCDD fed mice are related to oxidation-reduction and oxidative/phosphorylation, function relying on ETC. These functions are recovered in the anti-miR-873-5p treated mice in association with GNMT recovery.
To further explore the functionality and localization of GNMT in the ETC, we performed BN-PAGE of 2% digitonin-solubilized mitochondrial liver extracts. GNMT localized with Complex II (CII) (detected by succinate dehydrogenase (SDH) subunit A) in the ETC suggesting a potential role of GNMT in CII functionality (Figure 5C).
Complex II or SDH is an important enzyme in mitochondrial metabolism since it represents the hub where ETC and TCA converge. CII was immunoprecipitated using liver mitochondrial extracts from the different MCD mice and the interaction with GNMT was examined by western blotting. GNMT co-precipitated with the CII from control diet mice. A reduction in GNMT-CII interaction was detected in liver mitochondria derived from MCDD fed mice, which was recovered in the presence of anti-miR-873-5p (Figure 5D). These results suggest that the induction of miR-873-5p during NAFLD regulates mitochondrial GNMT levels and its interaction with the CII, further indicating a mitochondrial role of GNMT.
3.6. Mitochondrial GNMT regulates complex II activity in the ETC
In order to address the functionality of GNMT in CII, SDH activity was measured in mitochondrial extracts derived from control and MCD mice. A reduction of SDH activity in MCD liver was reverted after blocking miR-873-5p (Figure 5E), suggesting that the presence of GNMT in the mitochondria and its interaction with CII enhances its activity. Interestingly, the role of GNMT in CII activity was further confirmed in Gnmt-KO mitochondrial liver extracts, where a strong reduction of SDH activity compared with WT animals was detected (Figure 5F).
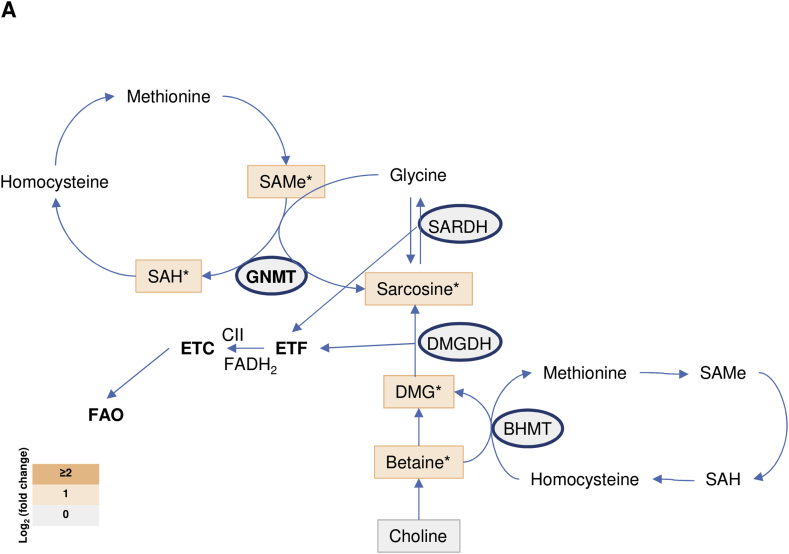
Finally, we investigated the mechanism by which GNMT levels in the mitochondria affect CII activity. GNMT is well known for participating in the methionine cycle of one-carbon metabolism, where it metabolizes SAMe to SAH, transferring the N-methyl group of SAMe to glycine to generate sarcosine. Different metabolites of one-carbon metabolism, including sarcosine and dimethylglycine (DMG) can be metabolized by sarcosine dehydrogenase (SARDH) and dimethylglycine dehydrogenase (DMGDH), respectively, in reactions producing FADH2, in which two e− are transferred to ubiquinone within the ETC. These reactions are known as the electron transfer flavoprotein: ubiquinone reductase system (ETF:QO) and occur in the mitochondria, connecting FAO, ETC, and one-carbon metabolism [32], [33]. On the other hand, mitochondrial CII transfers electrons to ubiquinone using FAD. Thus, we have analyzed whether improvement in one-carbon metabolism in the mitochondria due to GNMT rescue could be associated with the transport of e− in the ETC mediated by FAD and the increase of CII activity. Based on this hypothesis, we analyzed one-carbon metabolism in the liver of MCD mice. Mass spectrometry analysis revealed increased abundance of metabolites of one-carbon metabolites after anti-miR-873-5p treatment and GNMT recovery, including SAMe and SAH, sarcosine and other intermediates such as betaine and DMG (Figure 6). Interestingly, expression analysis of genes participating in methionine cycle metabolism, such as Mat1a and CBS did not reveal significant changes in mice fed with the MCDD or treated with the anti-miR-873-5p (data not shown), indicating that GNMT recovery by anti-miR-873-5p therapy is sufficient to restore methionine and one-carbon metabolism in the liver. Globally, these results indicate an enhanced flux within the one-carbon metabolism particularly in those steps occurring in the mitochondria, after mitochondrial GNMT recovery.
Figure 6.
Mitochondrial induction of GNMT is associated with methionine cycle activation. Log2 (fold-change) levels of metabolites and enzymes involved in methionine cycle and one-carbon metabolism and its relation in mice fed on the MCD diet and treated with the anti-miR-873-5p versus miR-Ctrl silencing. Data shown as average. p < 0.05 *; p < 0.01 **.
Overall, these results together with the localization of GNMT in the CII suggest a new association between mitochondrial one-carbon metabolism and ETC functionality regulated by GNMT.
4. Discussion
With an estimated 1.8 billion people affected by NAFLD, this chronic liver disease has turned into a global health problem with a worrying tendency [1]. Hence, there is an ample pharmacological market aiming to treat NASH-derived complications and to identify non-invasive biomarkers for the disease [3], [34]. GNMT is the most important enzyme regulating SAMe metabolism and is considered a marker of healthy liver that is frequently downregulated in chronic liver disease, including NAFLD [4], [5], cirrhosis, and hepatocellular carcinoma [7], [8], [9]. GNMT downregulation occurs during early stages of chronic liver disease; thus, it is likely that its decrease is the cause - rather than the consequence - of these pathologies. In this work, we show that miR-873-5p upregulation (recently reported to target GNMT [7]) correlates with GNMT downregulation in the liver of NAFLD/NASH patients and in different murine models, identifying a relevant mechanism mediating GNMT downregulation in NAFLD/NASH. Moreover, GNMT/miR-873-5p levels in the liver correlate with the severity of disease in human NAFLD patients, determined as NAS score, which is in accordance with our previous study [7] and further suggests GNMT downregulation is an early occurring event in liver disease that determines its progression. Previous studies have shown that miR-873-5p is regulated by the inflammatory mediator interleukin (IL)-17 [35], which is highly induced and participates in the development of NAFLD/NASH, correlating with the progression of the disease [36], [37]. Accordingly, (IL)-17 could explain the progressive increase in miR-873-5p we observe from healthy to steatosis and NASH stages.
We evaluated the therapeutic potential of targeting miR-873-5p during NAFLD development. MCDD represents an ideal model to study NAFLD progression and its complications in the liver as it leads to more severe NASH development than other models. MCDD mice show lipid accumulation, necro-inflammation, and fibrosis, resembling human pathology. MiR-873-5p inhibition restored GNMT expression reducing steatosis and NASH progression by decreasing different lipids. Even though NAFLD is a multiple-hit disease, lipid overload is considered a key driving event for its initiation and progression to more severe stages. In addition to reduced lipid accumulation, anti-miR-873-5p attenuated inflammatory and fibrogenic processes in these mice. Moreover, we provide evidence showing the global effect of miR-873-5p on lipid metabolism is mediated by the regulation of mitochondrial FAO in the hepatocytes, thereby suggesting an implication of GNMT in mitochondrial functionality. Of note, although other targets of miR-873-5p could be regulated in hepatocytes by anti-miR-873-5p, the lack of effect observed in Gnmt-KO hepatocytes in vitro and in the Gnmt-KO mice indicates Gnmt is the main target gene regulated by this microRNA under steatotic situation. Unfortunately, only oleic acid incubation could be used as in vitro model for miR-873-5p inhibition in Gnmt-KO hepatocytes, as far as the use of the MDMC in these hepatocytes would not produce similar effect due to de intrinsic high methionine levels of Gnmt-KO hepatocytes [28]. More importantly, the lack of effect in FAO oxidation upon miR-873-5p inhibition in Gnmt-KO livers strengths our hypothesis of the role of GNMT in mitochondrial functionality. Even though the Gnmt-KO may differ from our working MCDD model in terms of the mechanisms driving fatty liver development and mainly due to the high amount of SAMe levels in this mouse, we have described that these mice develop steatosis and liver inflammation by this time [28]; thus, other potential miR-873-5p targets involved in FAO should arise in this model upon anti-miR-873-5p treatment. Altogether, our results, show that GNMT is necessary for appropriate mitochondrial function and β-oxidation, and this process is altered by miR-873-5p targeting of Gnmt.
While increased mitochondrial FAO is frequently observed in steatosis, probably as a compensatory mechanism to reduce the lipid burden [12], the progression of the disease leads to decreased oxidative capacity due to an uncoupling between β-oxidation, TCA cycle, and ETC, frequently resulting in inefficient lipid metabolism and ROS overproduction in the liver [12]. Interestingly, our results suggest that miR-873-5p inhibition increases mitochondrial β-oxidation without increasing oxidative stress, which may implicate improved mitochondrial functionality and efficient e− flow in the ETC. We also show that GNMT is regulated in the mitochondria, where no function has been previously reported. GNMT interacts and regulates the activity of CII, which plays a crucial role in metabolism participating both in the ETC and the TCA cycle and contributing to ROS formation [38]. Decreased CII activity observed in MCDD-fed mice is in accordance with previous results [39], [40], [41], [42] and may implicate impaired e− flow through the ETC and decreased TCA cycle activity. On the contrary, targeting miR-873-5p and recovery of GNMT levels induces CII activity, which enhances ETC and global mitochondrial functionality, allowing a continuous lipid burning and restraining NASH progression.
As the main catabolic enzyme for SAMe, misbalance in GNMT levels may be regulating methionine metabolism in the mitochondria in our experimental model. Here we show that GNMT recovery in the mitochondria is associated with an increased amount of methionine cycle metabolites, which suggests improved methionine and one-carbon metabolism. Surprisingly, despite these global changes in methionine and one-carbon metabolism produced in mice fed the MCDD and also in those treated with the anti-miR-873-5p, lack of gene expression regulation was observed in other enzymes that participate in the cycle besides GNMT, which highlights the importance of GNMT in the regulation of these processes. Moreover, sarcosine (the product of GNMT enzymatic activity) and other metabolites increased after miR-873-5p inhibition. Besides participating in the methionine cycle, sarcosine and DMG are used as substrates by the electron transfer flavoprotein (ETF) system in reactions that produce FADH2 and transfer 2e− to ubiquinone, linking mitochondrial FAO, one-carbon metabolism, and the ETC [32], [33]. Our results indicate that the recovery of mitochondrial GNMT enhances one-carbon metabolism and suggest that interaction of GNMT with CII of the ETC, drives a metabolic adaptation in mitochondria, linking β-oxidation, ETF, and ETC. Based on our results, we propose that these adaptations may lead to improved mitochondrial functionality and OXPHOS processes, thereby reducing NASH progression.
In conclusion, the present study demonstrates that enhancing mitochondrial β-oxidation by relieving miR-873-5p repression on GNMT may be an effective mechanism to reduce lipid burden in the liver. Thus, targeting miR-873-5p may be of wide applicability for NAFLD therapies. Moreover, given the importance of GNMT in liver health and its downregulation in chronic liver disease, targeting its expression by anti-miR-873-5p may represent a novel strategy to efficiently treat liver diseases in early and advanced stages.
5. Conclusion
Our study describes the contribution of GNMT to mitochondrial functionality in hepatocytes through the binding and regulation of Complex II in the ETC, promoting fatty acid β-oxidation. Furthermore, we provide evidence for the steatotic role of miR-873-5p, a specific repressor of GNMT expression which is upregulated in the liver of NAFLD and NASH patients and in murine models. Regulation of GNMT by targeting miR-873-5p ameliorates fatty liver disease by improving mitochondrial functionality.
Financial support
This work was supported by grants from NIH (US Department of Health and Human services)- R01AT001576 (to S.C.L., J.M.M., and M.L.M.-C.), Ministerio de Economía, Industria y Competitividad: SAF2017-87301-R (to M.L.M.-C.), SAF2015-64352-R (to P.A.), Gobierno Vasco-Departamento de Salud 2013111114 (to M.L.M.-C.), Gobierno Vasco-Departamento de Educación IT-336-10 (to PA), BIOEF (Basque Foundation for Innovation and Health Research: EiTB Maratoia BIO15/CA/016/BD (M.L.M.-C.), ELKARTEK 2016, Departamento de Industria del Gobierno Vasco (to M.L.M.-C), Asociación Española contra el Cáncer (to T.C.D., P.F.-T. and M.L.M.-C.), Mitotherapeutix (to M.L.M.-C.), Daniel Alagille award from EASL (to T.C.D), Fundación Científica de la Asociación Española Contra el Cancer (AECC Scientific Foundation) Rare Tumor Calls 2017 (to M.L.M.-C.), La Caixa Foundation Program (to M.L.M.-C.), Ayudas Fundacion BBVA a Equipos de Investigación Científica 2019 (to M.L.M.-C.). Ciberehd_ISCIII_MINECO is funded by the Instituto de Salud Carlos III. We thank this work produced with the support of a 2017 Leonardo Grant for Researchers and Cultural Creators, BBVA Foundation (to M.V.R.).
This work was supported by Fonds National de la Recherche Luxembourg and the Deutsche Forschungsgemeinschaft (C12/BM/3975937, FL/997/7-1, Inter “HepmiRSTAT”, to I.B. and F.L.).
We thank MINECO for the Severo Ochoa Excellence Accreditation (SEV-2016-0644).
Author contribution
Pablo Fernández-Tussy: Acquisition of data; analysis and interpretation of data; statistical analysis; critical revision of the manuscript.
David Fernández-Ramos: Acquisition of data; analysis and interpretation of data; statistical analysis; critical revision of the manuscript.
Fernando Lopitz-Otsoa: Acquisition of data; analysis and interpretation of data; statistical analysis; critical revision of the manuscript.
Jorge Simón: Acquisition of data; analysis and interpretation of data; statistical analysis; critical revision of the manuscript.
Lucía Barbier-Torres: Acquisition of data; analysis and interpretation of data; statistical analysis; critical revision of the manuscript.
Beatriz Gomez-Santos: Acquisition of data; analysis and interpretation of data; statistical analysis.
Maitane Nuñez-Garcia: Acquisition of data; analysis and interpretation of data; statistical analysis.
Mikel Azkargorta: Acquisition of data; analysis and interpretation of data; statistical analysis.
Virginia Gutiérrez-de Juan: Acquisition of data; analysis and interpretation of data; statistical analysis; critical revision of the manuscript.
Marina Serrano-Macia: Acquisition of data; analysis and interpretation of data; statistical analysis; critical revision of the manuscript.
Rubén Rodríguez-Agudo: Acquisition of data; analysis and interpretation of data; statistical analysis; critical revision of the manuscript.
Paula Iruzibieta: Material support.
Juan Anguita: Critical revision of the manuscript.
Rui Eduarco Castro: Critical revision of the manuscript.
Devin Champagne: Material support; acquisition of data.
Mercedes Rincón: Material support.
Felix Elortza: Acquisition of data; analysis and interpretation of data; statistical analysis; critical revision of the manuscript.
Anita Arslanow: Material support; acquisition of data; analysis and interpretation of data; statistical analysis; critical revision of the manuscript.
Marcin Krawczyk: Material support; acquisition of data; analysis and interpretation of data; statistical analysis; critical revision of the manuscript.
Frank Lammert: Material support; critical revision of the manuscript
Mélanie Kirchmeyer: Material support; acquisition of data; analysis and interpretation of data; statistical analysis; critical revision of the manuscript. Obtained funding.
Iris Behrmann: Material support; acquisition of data; analysis and interpretation of data; statistical analysis; critical revision of the manuscript.
Javier Crespo: Material support.
Lu SC: Critical revision of the manuscript. Obtained funding.
José María Mato: Critical revision of the manuscript. Obtained funding.
Marta Varela-Rey: Analysis and interpretation of data. critical revision of the manuscript. Obtained funding.
Patricia Aspichueta: Acquisition of data; analysis and interpretation of data. critical revision of the manuscript.
Teresa Cardoso: Acquisition of data; analysis and interpretation of data. critical revision of the manuscript. Obtained funding.
María L. Martínez-Chantar: Study concept and design; analysis and interpretation of data; study supervision; drafting of the manuscript; obtained funding.
Conflict of interest
Dr. Mato consults for, advises for, and owns stock in Owl. He consults for and advises for Abbott. He consults for Galmed. Dr. Martínez-Chantar advises for Mitotherapeutix LLC. For the rest of the authors there is nothing to declare.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2019.08.008.
Contributor Information
Pablo Fernández-Tussy, Email: pablo.fernandeztussy@yale.edu.
David Fernández-Ramos, Email: dfernandez@cicbiogune.es.
Fernando Lopitz-Otsoa, Email: flopitz@cicbiogune.es.
Jorge Simón, Email: jsimon@cicbiogune.es.
Lucía Barbier-Torres, Email: Lucia.BarbierTorres@cshs.org.
Beatriz Gomez-Santos, Email: bgomezsantos@gmail.com.
Maitane Nuñez-Garcia, Email: maitane.nunez@gmail.com.
Mikel Azkargorta, Email: mazkargorta@cicbiogune.es.
Virginia Gutiérrez-de Juan, Email: vgutierrez@cicbiogune.es.
Marina Serrano-Macia, Email: mserrano@cicbiogune.es.
Rubén Rodríguez-Agudo, Email: rrodriguez@cicbiogune.es.
Paula Iruzubieta, Email: p.iruzubieta@gmail.com.
Juan Anguita, Email: janguita@cicbiogune.es.
Rui E. Castro, Email: ruieduardocastro@ff.ulisboa.pt.
Devin Champagne, Email: Devin.Champagne@uvm.edu.
Mercedes Rincón, Email: Mercedes.Rincon@med.uvm.edu.
Felix Elortza, Email: felortza@cicbiogune.es.
Anita Arslanow, Email: anita.arslanow@uks.eu.
Marcin Krawczyk, Email: marcin.krawczyk@uks.eu.
Frank Lammert, Email: frank.lammert@uks.eu.
Mélanie Kirchmeyer, Email: kirchmeyer.melanie@gmail.com.
Iris Behrmann, Email: iris.behrmann@uni.lu.
Javier Crespo, Email: javiercrespo1991@gmail.com.
Shelly C. Lu, Email: Shelly.Lu@cshs.org.
José M. Mato, Email: director@cicbiogune.es.
Marta Varela-Rey, Email: mvarela@cicbiogune.es.
Patricia Aspichueta, Email: patricia.aspichueta@ehu.es.
Teresa C. Delgado, Email: tcardoso@cicbiogune.es.
María L. Martínez-Chantar, Email: mlmartinez@cicbiogune.es.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Younossi Z.M., Koenig A.B., Abdelatif D., Fazel Y., Henry L., Wymer M. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology (Baltimore, Md.) 2016;64(1):73–84. doi: 10.1002/hep.28431. [DOI] [PubMed] [Google Scholar]
- 2.Ascha M.S., Hanouneh I.A., Lopez R., Tamimi T.A.-R., Feldstein A.F., Zein N.N. The incidence and risk factors of hepatocellular carcinoma in patients with nonalcoholic steatohepatitis. Hepatology (Baltimore, Md.) 2010;51(6):1972–1978. doi: 10.1002/hep.23527. [DOI] [PubMed] [Google Scholar]
- 3.Cassidy S., Syed B.A. Nonalcoholic steatohepatitis (NASH) drugs market. Nature Reviews Drug Discovery. 2016 doi: 10.1038/nrd.2016.188. [DOI] [PubMed] [Google Scholar]
- 4.Moylan C.A., Pang H., Dellinger A., Suzuki A., Garrett M.E., Guy C.D. Hepatic gene expression profiles differentiate pre-symptomatic patients with mild versus severe nonalcoholic fatty liver disease. Hepatology (Baltimore, Md.) 2014;59(2):471–482. doi: 10.1002/hep.26661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murphy S.K., Yang H., Moylan C.A., Pang H., Dellinger A., Abdelmalek M.F. Relationship between methylome and transcriptome in patients with nonalcoholic fatty liver disease. Gastroenterology. 2013;145(5):1076–1087. doi: 10.1053/j.gastro.2013.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fernández-Álvarez S., Juan V.G., Zubiete-Franco I., Barbier-Torres L., Lahoz A., Parés A. TRAIL-producing NK cells contribute to liver injury and related fibrogenesis in the context of GNMT deficiency. Laboratory Investigation; a Journal of Technical Methods and Pathology. 2015;95(2):223–236. doi: 10.1038/labinvest.2014.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fernández-Ramos D., Fernández-Tussy P., Lopitz-Otsoa F., Gutiérrez-de-Juan V., Navasa N., Barbier-Torres L. MiR-873-5p acts as an epigenetic regulator in early stages of liver fibrosis and cirrhosis. Cell Death & Disease. 2018;9(10):958. doi: 10.1038/s41419-018-1014-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Avila M.A., Berasain C., Torres L., Martín-Duce A., Corrales F.J., Yang H. Reduced mRNA abundance of the main enzymes involved in methionine metabolism in human liver cirrhosis and hepatocellular carcinoma. Journal of Hepatology. 2000;33(6):907–914. doi: 10.1016/s0168-8278(00)80122-1. [DOI] [PubMed] [Google Scholar]
- 9.Chen Y.M., Shiu J.Y., Tzeng S.J., Shih L.S., Chen Y.J., Lui W.Y. Characterization of glycine-N-methyltransferase-gene expression in human hepatocellular carcinoma. International Journal of Cancer. 1998;75(5):787–793. doi: 10.1002/(sici)1097-0215(19980302)75:5<787::aid-ijc20>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 10.Stachowicz A., Suski M., Olszanecki R., Madej J., Okoń K., Korbut R. Proteomic analysis of liver mitochondria of apolipoprotein E knockout mice treated with metformin. Journal of Proteomics. 2012;77:167–175. doi: 10.1016/j.jprot.2012.08.015. [DOI] [PubMed] [Google Scholar]
- 11.DebRoy S., Kramarenko I.I., Ghose S., Oleinik N.V., Krupenko S.A., Krupenko N.I. A novel tumor suppressor function of Glycine N-methyltransferase is independent of its catalytic activity but requires nuclear localization. PLoS One. 2013;8(7) doi: 10.1371/journal.pone.0070062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Begriche K., Massart J., Robin M.-A., Bonnet F., Fromenty B. Mitochondrial adaptations and dysfunctions in nonalcoholic fatty liver disease. Hepatology (Baltimore, Md.) 2013;58(4):1497–1507. doi: 10.1002/hep.26226. [DOI] [PubMed] [Google Scholar]
- 13.Mansouri A., Gattolliat C.-H., Asselah T. Mitochondrial dysfunction and signaling in chronic liver diseases. Gastroenterology. 2018;155(3):629–647. doi: 10.1053/j.gastro.2018.06.083. [DOI] [PubMed] [Google Scholar]
- 14.Folch J., Lees M., Sloane Stanley G.H. A simple method for the isolation and purification of total lipides from animal tissues. Journal of Biological Chemistry. 1957;226(1):497–509. [PubMed] [Google Scholar]
- 15.Ruiz J.I., Ochoa B. Quantification in the subnanomolar range of phospholipids and neutral lipids by monodimensional thin-layer chromatography and image analysis. Journal of Lipid Research. 1997;38(7):1482–1489. [PubMed] [Google Scholar]
- 16.Gao X., van der Veen J.N., Hermansson M., Ordoñez M., Gomez-Muñoz A., Vance D.E. Decreased lipogenesis in white adipose tissue contributes to the resistance to high fat diet-induced obesity in phosphatidylethanolamine N-methyltransferase-deficient mice. Biochimica et Biophysica Acta. 2015;1851(2):152–162. doi: 10.1016/j.bbalip.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 17.Hirschey M.D., Shimazu T., Goetzman E., Jing E., Schwer B., Lombard D.B. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010;464(7285):121–125. doi: 10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Porteiro B., Fondevila M.F., Delgado T.C., Iglesias C., Imbernon M., Iruzubieta P. Hepatic p63 regulates steatosis via IKKβ/ER stress. Nature Communications. 2017;8:15111. doi: 10.1038/ncomms15111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nassir F., Adewole O.L., Brunt E.M., Abumrad N.A. CD36 deletion reduces VLDL secretion, modulates liver prostaglandins, and exacerbates hepatic steatosis in ob/ob mice. Journal of Lipid Research. 2013;54(11):2988–2997. doi: 10.1194/jlr.M037812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Colell A., García-Ruiz C., Lluis J.M., Coll O., Mari M., Fernández-Checa J.C. Cholesterol impairs the adenine nucleotide translocator-mediated mitochondrial permeability transition through altered membrane fluidity. Journal of Biological Chemistry. 2003;278(36):33928–33935. doi: 10.1074/jbc.M210943200. [DOI] [PubMed] [Google Scholar]
- 21.Barbier-Torres L., Beraza N., Fernández-Tussy P., Lopitz-Otsoa F., Fernández-Ramos D., Zubiete-Franco I. Histone deacetylase 4 promotes cholestatic liver injury in the absence of prohibitin-1. Hepatology (Baltimore, Md.) 2015;62(4):1237–1248. doi: 10.1002/hep.27959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martínez-López N., García-Rodríguez J.L., Varela-Rey M., Gutiérrez V., Fernández-Ramos D., Beraza N. Hepatoma cells from mice deficient in Glycine N-methyltransferase have increased RAS signaling and activation of liver Kinase B1. Gastroenterology. 2012;143(3):787–798. doi: 10.1053/j.gastro.2012.05.050. e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wiśniewski J.R., Zougman A., Nagaraj N., Mann M. Universal sample preparation method for proteome analysis. Nature Methods. 2009;6(5):359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 24.Barbier-Torres L., Delgado T.C., García-Rodríguez J.L., Zubiete-Franco I., Fernández-Ramos D., Buqué X. Stabilization of LKB1 and Akt by neddylation regulates energy metabolism in liver cancer. Oncotarget. 2015;6(4):2509–2523. doi: 10.18632/oncotarget.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zubiete-Franco I., Fernández-Tussy P., Barbier-Torres L., Simon J., Fernández-Ramos D., Lopitz-Otsoa F. Deregulated neddylation in liver fibrosis. Hepatology (Baltimore, Md.) 2017;65(2):694–709. doi: 10.1002/hep.28933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hansen B., Arteta B., Smedsrød B. The physiological scavenger receptor function of hepatic sinusoidal endothelial and Kupffer cells is independent of scavenger receptor class A type I and II. Molecular and Cellular Biochemistry. 2002;240(1–2):1–8. doi: 10.1023/a:1020660303855. [DOI] [PubMed] [Google Scholar]
- 27.Barbier-Torres L., Iruzubieta P., Fernández-Ramos D., Delgado T.C., Taibo D., Guitiérrez-de-Juan V. The mitochondrial negative regulator MCJ is a therapeutic target for acetaminophen-induced liver injury. Nature Communications. 2017;8(1):2068. doi: 10.1038/s41467-017-01970-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martínez-Chantar M.L., Vázquez-Chantada M., Ariz U., Martínez N., Varela M., Luka Z. Loss of the glycine N-methyltransferase gene leads to steatosis and hepatocellular carcinoma in mice. Hepatology (Baltimore, Md.) 2008;47(4):1191–1199. doi: 10.1002/hep.22159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinez-Una M., Varela-Rey M., Mestre D., Fernandez-Ares L., Fresnedo O., Fernandez-Ramos D. S-adenosylmethionine increases circulating very-low density lipoprotein clearance in nonalcoholic fatty liver disease. Journal of Hepatology. 2015;62(3):673–681. doi: 10.1016/j.jhep.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martínez-Uña M., Varela-Rey M., Cano A., Fernández-Ares L., Beraza N., Aurrekoetxea I. Excess S-adenosylmethionine reroutes phosphatidylethanolamine towards phosphatidylcholine and triglyceride synthesis. Hepatology (Baltimore, Md.) 2013;58(4):1296–1305. doi: 10.1002/hep.26399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen C.-Y., Ching L.-C., Liao Y.-J., Yu Y.-B., Tsou C.-Y., Shyue S.-K. Deficiency of glycine N-methyltransferase aggravates atherosclerosis in apolipoprotein E-null mice. Molecular Medicine (Cambridge, Mass.) 2012;18:744–752. doi: 10.2119/molmed.2011.00396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoskins D.D., Mackenzie C.G. Solubilization and electron transfer flavoprtein requirement of mitochondrial sarcosine dehydrogenase and dimethylglycine dehydrogenase. Journal of Biological Chemistry. 1961;236:177–183. [PubMed] [Google Scholar]
- 33.Watmough N.J., Frerman F.E. The electron transfer flavoprotein: ubiquinone oxidoreductases. Biochimica et Biophysica Acta. 2010;1797(12):1910–1916. doi: 10.1016/j.bbabio.2010.10.007. [DOI] [PubMed] [Google Scholar]
- 34.Younossi Z., Anstee Q.M., Marietti M., Hardy T., Henry L., Eslam M. Global burden of NAFLD and NASH: trends, predictions, risk factors and prevention. Nature Reviews Gastroenterology & Hepatology. 2018;15(1):11–20. doi: 10.1038/nrgastro.2017.109. [DOI] [PubMed] [Google Scholar]
- 35.Liu X., He F., Pang R., Zhao D., Qiu W., Shan K. Interleukin-17 (IL-17)-induced microRNA 873 (miR-873) contributes to the pathogenesis of experimental autoimmune encephalomyelitis by targeting A20 ubiquitin-editing enzyme. Journal of Biological Chemistry. 2014;289(42):28971–28986. doi: 10.1074/jbc.M114.577429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gomes A.L., Teijeiro A., Burén S., Tummala K.S., Yilmaz M., Waisman A. Metabolic inflammation-associated IL-17A causes non-alcoholic steatohepatitis and hepatocellular carcinoma. Cancer Cell. 2016;30(1):161–175. doi: 10.1016/j.ccell.2016.05.020. [DOI] [PubMed] [Google Scholar]
- 37.Harley I.T.W., Stankiewicz T.E., Giles D.A., Softic S., Flick L.M., Cappelletti M. IL-17 signaling accelerates the progression of nonalcoholic fatty liver disease in mice. Hepatology (Baltimore, Md.) 2014;59(5):1830–1839. doi: 10.1002/hep.26746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wojtovich A.P., Smith C.O., Haynes C.M., Nehrke K.W., Brookes P.S. Physiological consequences of complex II inhibition for aging, disease, and the mKATP channel. Biochimica et Biophysica Acta. 2013;1827(5):598–611. doi: 10.1016/j.bbabio.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bruce K.D., Cagampang F.R., Argenton M., Zhang J., Ethirajan P.L., Burdge G.C. Maternal high-fat feeding primes steatohepatitis in adult mice offspring, involving mitochondrial dysfunction and altered lipogenesis gene expression. Hepatology (Baltimore, Md.) 2009;50(6):1796–1808. doi: 10.1002/hep.23205. [DOI] [PubMed] [Google Scholar]
- 40.García-Ruiz I., Rodríguez-Juan C., Díaz-Sanjuan T., del Hoyo P., Colina F., Muñoz-Yagüe T. Uric acid and anti-TNF antibody improve mitochondrial dysfunction in ob/ob mice. Hepatology (Baltimore, Md.) 2006;44(3):581–591. doi: 10.1002/hep.21313. [DOI] [PubMed] [Google Scholar]
- 41.García-Ruiz I., Solís-Muñoz P., Fernández-Moreira D., Grau M., Colina F., Muñoz-Yagüe T. High-fat diet decreases activity of the oxidative phosphorylation complexes and causes nonalcoholic steatohepatitis in mice. Disease Models & Mechanisms. 2014;7(11):1287–1296. doi: 10.1242/dmm.016766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pérez-Carreras M., Hoyo P.D., Martín M.A., Rubio J.C., Martín A., Castellano G. Defective hepatic mitochondrial respiratory chain in patients with nonalcoholic steatohepatitis. Hepatology. 2003;38(4):999–1007. doi: 10.1053/jhep.2003.50398. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.