Abstract
Objective
Enteroendocrine cells (EECs) of the gastro-intestinal tract sense gut luminal factors and release peptide hormones or serotonin (5-HT) to coordinate energy uptake and storage. Our goal is to decipher the gene regulatory networks controlling EECs specification from enteroendocrine progenitors. In this context, we studied the role of the transcription factor Rfx6 which had been identified as the cause of Mitchell–Riley syndrome, characterized by neonatal diabetes and congenital malabsorptive diarrhea. We previously reported that Rfx6 was essential for pancreatic beta cell development and function; however, the role of Rfx6 in EECs differentiation remained to be elucidated.
Methods
We examined the molecular, cellular, and metabolic consequences of constitutive and conditional deletion of Rfx6 in the embryonic and adult mouse intestine. We performed single cell and bulk RNA-Seq to characterize EECs diversity and identify Rfx6-regulated genes.
Results
Rfx6 is expressed in the gut endoderm; later, it is turned on in, and restricted to, enteroendocrine progenitors and persists in hormone-positive EECs. In the embryonic intestine, the constitutive lack of Rfx6 leads to gastric heterotopia, suggesting a role in the maintenance of intestinal identity. In the absence of intestinal Rfx6, EECs differentiation is severely impaired both in the embryo and adult. However, the number of serotonin-producing enterochromaffin cells and mucosal 5-HT content are increased. Concomitantly, Neurog3-positive enteroendocrine progenitors accumulate. Combined analysis of single-cell and bulk RNA-Seq data revealed that enteroendocrine progenitors differentiate in two main cell trajectories, the enterochromaffin (EC) cells and the Peptidergic Enteroendocrine (PE) cells, the differentiation programs of which are differentially regulated by Rfx6. Rfx6 operates upstream of Arx, Pax6 and Isl1 to trigger the differentiation of peptidergic EECs such as GIP-, GLP-1-, or CCK-secreting cells. On the contrary, Rfx6 represses Lmx1a and Tph1, two genes essential for serotonin biosynthesis. Finally, we identified transcriptional changes uncovering adaptive responses to the prolonged lack of enteroendocrine hormones and leading to malabsorption and lower food efficiency ratio in Rfx6-deficient mouse intestine.
Conclusion
These studies identify Rfx6 as an essential transcriptional regulator of EECs specification and shed light on the molecular mechanisms of intestinal failures in human RFX6-deficiencies such as Mitchell–Riley syndrome.
Keywords: Rfx6, Neurog3, Lmx1a, Intestine, Cell fate, Serotonin, Enteroendocrine cells, Enterochromaffin cells, Malabsorption, Mitchell–Riley syndrome
Highlights
-
•
The lack of Rfx6 impairs the differentiation of peptide-producing enteroendocrine cells.
-
•
The number of 5-HT-expressing-cells is increased in Rfx6-deficient intestine.
-
•
Intestinal inactivation of Rfx6 leads to lipid malabsorption and decreased food efficiency.
1. Introduction
Enteroendocrine cells (EECs) are one of the five main intestinal cell subtypes, which include enterocytes, goblet-, Paneth-, and tuft-cells. EECs are scattered as individual cells in the lining of the gut epithelium from the duodenum to the colon. Although they represent 1% of all intestinal epithelial cells they constitute the largest endocrine system of the body. EECs sense gut luminal factors (nutrients, microbial components) that trigger the secretion of peptide hormones or amines, regulating food intake, digestion, or glucose metabolism. Peptide hormones include Glucagon-Like Peptide 1 and 2 (GLP-1, GLP-2), Peptide YY (PYY), Gastric Inhibitory Peptide (GIP), Somatostatin (SST), Cholecystokinin (CCK), Secretin (SCT), Gastrin (GAST), Neurotensin (NTS), and Ghrelin (GHRL). The monoamine serotonin (5-hydroxytryptamine; 5-HT) is produced by enterochromaffin cells, the most abundant enteroendocrine cell type. Historically, EECs were classified using a one letter code based on their location and main secretory product. However, co-expression [1], [2] and single-cell transcriptomic studies [3], [4], [5], [6] tend to support the obsolescence of this classification [7] given that virtually all EECs express several hormones albeit not at the same level. Recent findings suggest that the hormonal repertoire of EECs might be modulated, along the crypt–villus axis, by local signals [8]. Despite these major breakthroughs, our understanding of the molecular mechanisms controlling the differentiation of EECs, both during development and renewal in the adult intestine, is still limited.
Several transcription factors have been demonstrated to regulate EECs differentiation. A master regulator is the basic helix-loop-helix (bHLH) transcription factor Neurogenin3 (Ngn3 or Neurog3), which is expressed in endocrine progenitors and determines endocrine commitment in the embryonic [9], [10], [11] and adult intestine [12]. Neurog3-expressing cells are rare hormone–negative endocrine progenitors confined to the crypts and Neurog3 expression is turned off when cells migrate towards the villi and differentiate into hormone-expressing cells. Indeed, Neurog3-deficient mice (constitutive and intestinal deletion) lack EECs. The loss of all enteroendocrine cells in mice leads to growth retardation, impaired lipid absorption, and increased lethality, underlying the importance of enteroendocrine function [12]. Neurog3-positive endocrine progenitors arise from intermediate crypt progenitors which express the bHLH transcription factor Atoh1 [13]. Downstream of Neurog3, a complex network of transcription factors controls the differentiation of subpopulations of EECs such as NeuroD1 [14], Arx [15], [16], Pax4 [15], [17], Isl1 [18], Pax6 [17], Foxa1/2 [19], or Insm1 [20].
In almost all cases, transcription factors controlling EECs differentiation also regulate pancreatic islet cell differentiation, suggesting shared genetic programs. We and others have reported the critical role of the transcription factor Regulatory Factor X 6 (Rfx6) in beta-cell development [21], [22] and function [23]. Rfx6, initially expressed broadly in the gut endoderm, becomes restricted to the endocrine lineage in the pancreas during embryogenesis and persists in adult islet cells. Rfx6 is necessary for proper islet cell development in zebrafish, mouse and Xenopus [21], [22], [24]. Rfx6-deficient mice lack alpha-, beta-, and delta-cells, are diabetic, and die shortly after birth. Rfx6 loss in adult beta cells leads to glucose intolerance, impaired beta-cell glucose sensing, and defective insulin secretion [23]. A previous study reported the expression of Rfx6 in GIP-positive enteroendocrine K-cells and the regulation of Gip promoter activity by Rfx6 [25]. Another recent study revealed that loss of Rfx6 function ex vivo, in mouse intestinal organoids, led to a reduction of EECs [5]. Still, detailed Rfx6 expression and role in mouse intestine and EECs differentiation in vivo is unknown. In humans, several mutations in RFX6 were identified as the cause of an autosomal recessive syndrome, named Mitchell–Riley syndrome, characterized by neonatal or childhood diabetes comprising hepatobiliary abnormalities and intestinal atresia [22], [26], [27], [28], [29], [30], [31], [32], [33]. Most patients present with severe congenital malabsorptive diarrhea, suggesting impaired EECs differentiation; however, this has not been studied extensively.
In this study, we investigated the function of Rfx6 in EECs differentiation in the embryonic and adult mouse. We show that EECs differentiation is severely impaired in Rfx6-deficient embryos, and only enterochromaffin cells are spared. Furthermore, we detected patches of gastric cells in the small intestine of mutant newborns indicating gastric heterotopia. Constitutive intestinal deletion of Rfx6 is found to be lethal at early post-natal stages. Deletion of Rfx6 in the adult intestine is found to induce diarrhea, impaired lipid absorption, and impaired food efficiency. Like in the embryo, adult EECs expressing peptide hormones were either lost or decreased in representation, while serotonin-positive enterochromaffin cells still developed with even slight increase in their number. Concomitantly, an increased number of Neurog3-positive enteroendocrine progenitors was also observed. Contrary to in vivo data, the removal of Rfx6 in small intestinal organoids was found to result in impaired differentiation of all EECs, including enterochromaffin cells. By comparative transcriptomic studies, we determined early Rfx6-dependent targets in the EEC lineage and identified secondary enhanced expression of neoglucogenic and nutrient absorption machinery genes reflecting adaptive response to the absence of enteroendocrine hormones. In parallel single-cell transcriptomic studies of EECs, we describe the dynamics of Rfx6 expression and expression of other known and novel intestinal transcription factors. Overall, our results show that enteroendocrine progenitors differentiate in two main cell trajectories, the enterochromaffin (EC) cells and the Peptidergic Enteroendocrine (PE) cells, the differentiation programs of which are differentially regulated by Rfx6.
2. Material and methods
2.1. Animals and animal handling
All mice were housed in an animal facility licensed by the French Ministry of Agriculture (Agreement no. B67-218-5), and all animal experiments were supervised by GG (agreement no. C67-59) and approved by the Direction des Services Vétérinaires in compliance with the European legislation on care and use of laboratory animals. Rfx6fl/+ mice have been described previously [23] and were maintained on a C57BL/6N (Taconic) background. Rfx6+/− mice have been generated by crossing Rfx6fl/+ females with CMV-Cre males. Neurog3-Cre mice are a gift from Dr. Shosei Yoshida [34], and Villin-Cre and Villin-CreERT2 were generously given by Dr. Sylvie Robine [35]. Neurog3eYFP/+ mice have been described previously [36]. Studies in adult mice were performed with males unless otherwise stated in figure legends. The proportions of mice from a given litter were kept identical between control and mutant groups. Studies in embryos were performed with combinations of males and females. Genomic tail DNA was analyzed by PCR using the primers detailed below. To achieve recombination in inducible mutants, adult mice (8–12 weeks old) were treated with tamoxifen (10 mg) (Sigma) by gavage twice per day, every second day during 5 days. Primers were as follows:
Neurog3 Forward: ctgcagtttagcagaacttcagaggga
Cre Reverse: atcaacgttttgttttcgga
Villin forward: ataggaagccagtttcccttc
ERT forward: gcattaccggtcgatgcaacgagtgatgag
ERT reverse: aggatctctagccaggcaca
Rfx6 forward: gaaggtgcacccataaaagc
Rfx6 reverse: tataagccacccagggtcag
Neurog3 forward: cggcagatttgaatgagggc
Neurog3 reverse: tctcgcctcttctggctttc
GFP forward: cctgaagttcatctgcaccac
GFP reverse: ttgtagttgtactccagcttgtgc
2.2. Histopathology and immunohistochemistry
Mouse tissues were fixed in 4% paraformaldehyde at 4 °C overnight and embedded in paraffin or Sandon Cryomatrix (Thermo Scientific). Standard histology techniques were used: for conventional histology, 7 μm paraffin sections were stained with Harris hematoxylin and eosin (H&E); for goblet cells analysis, 7 μm paraffin sections were stained with Periodic Acid-Schiff (PAS) and hematoxylin or Alcian blue (AB) (pH 2.5); for lipid and fat deposits detection, 10 μm cryo-sections were stained with Oil red O. Immunostaining on 7 (paraffin) or 10 (cryo) μm sections was performed using standard protocols. For BrdU detection assays, BrdU was injected intraperitoneally at 100 mg/kg body weight, 24 h prior sacrifice. If required, antigen retrieval was performed in 10 mM Sodium Citrate pH6 in a pressure cooker. Primary antibodies used: guinea pig anti-Neurog3 1:1000 (M. Sander, UCSD, CA, USA); goat anti-Neurog3 1:2000 (G. Gu, Vanderbilt University, TN, USA); rabbit anti-Neurog3 1:500 (2763; IGBMC); rat anti-somatostatin 1:500 (Chemicon); mouse anti-ghrelin (C. Tomasetto, IGBMC, Illkirch, France); goat anti-chromograninA 1:200 (Santa Cruz); goat anti-Cck 1:50 (Santa Cruz); guinea pig anti-glucagon 1:2000 (Linco); goat anti-glp1 1:100 (Santa Cruz); goat anti-Gip 1:100 (Santa Cruz); goat anti-gastrin 1:50 (Santa Cruz); goat anti-secretin 1:50 (Santa Cruz); goat anti-serotonin 1:2000 (Abcam); rabbit anti-serotonin 1:2000 (Diasorin Incstar); rat anti-BrdU 1:10 (AbD Serotec), rabbit anti-Rfx6 1:500 (2766, IGBMC, Illkirch, France); chicken anti-GFP 1:2000 (Abcam). Secondary antibodies conjugated to Alexa Fluor® 594, DyLight® 488, DyLight® 549 and DyLight® 649 (Jackson ImmunoResearch) were used at 1:500. For BrdU, Neurog3, and Rfx6, signal amplification was performed using biotin anti-rabbit (Jackson ImmunoResearch) coupled antibody at 1:500 and streptavidin-Cy3 (or-Dylight 488) conjugate at 1:500 (Molecular Probes). Immunofluorescent stainings on mouse intestinal organoids were performed as previously described [37].
2.3. Generation and maintenance and 4-hydroxytamoxifen treatment of mouse intestinal organoids
Mouse intestinal organoids were generated as previously described [38] from small intestine of 2–6 month-old CD1 Neurog3eYFP/+ mice, Rfx6fl/fl and Rfx6fl/fl; Villin CreERT2 mice. Briefly, crypts from duodenum or jejunum were isolated and cultured into Matrigel drops (Fisher Scientific™) to develop organoids in presence of the following medium: Advanced DMEM/F-12, HEPES 10 mM, GlutaMax 2 mM, penicillin/streptomycin 100 U/mL, supplement B27 1X (all from Gibco™), N-acetyl-l-cysteine 1 μM (Merck), human recombinant R-spondin-1 500 ng/mL (PeproTech®), murine recombinant EGF 50 ng/mL (PeproTech®) and human recombinant Noggin 100 ng/mL (R&D Systems®). Organoids were maintained at 37 °C, 5% CO2 and were split into 24-well plates every 5–7 days. To induce in vitro Cre recombinase activation and specific deletion of Rfx6, organoids derived from Rfx6fl/fl (controls) and Rfx6fl/fl; Villin-CreERT2 duodenum (n = 4 per genotype) were treated two days after seeding with 4-hydroxytamoxifen (4-OHT; Sigma–Aldrich) 1.25 μM for 15 h, after which the medium was replaced with fresh medium without 4-OHT. Organoids were harvested 8 days after 4-OHT treatment for RNA extraction.
2.4. Measurement of intestinal serotonin content
Eleven days post tamoxifen treatment, four adult males Rfx6fl/fl and three Rfx6fl/fl Villin-CreERT2 were euthanized by cervical dislocation and the entire gastrointestinal tract was excised. Using dermal biopsy punches (1.5 mm), intestinal samples were obtained from relevant sites throughout the GI tract (duodenum, jejunum, ileum, proximal colon, and distal colon). All samples were quickly homogenized and purified in ice-cold 1 M perchloric acid to precipitate proteins. Following centrifugation at 14.000G at 4°C, 15 min, after which the supernatant was collected and taken through an additional purifying step adding a similar volume of 1 M perchloric acid followed by mixing and centrifugation. The protein-free samples were diluted 2 or 10 times in MilliQ water prior HPLC injection (10 μl). 5-HT concentration was measured by HPLC-ECD (HTEC-500, Eicom) with a PP-ODS2 column (Amuza) following manufacturer's instructions regarding flow rate, mobile phase and applied potential. Retention time (3.8 s) for 5-HT was determined by a control 5-HT dilution of 1pM (Sigma–Aldrich). Results are stated as area under the curve (AUC) and presented as mean ± SD. Unpaired T-test was applied to calculate significance (*p ≤ 0.05; **p ≤ 0.01).
2.5. Morphometric analysis
Neurog3-positive cells or Neurog3/BrdU double positive cells were counted in adult mice after immunostaining on approximately 50 sections of duodenum, jejunum, ileum and colon three months after tamoxifen treatment (n = 3 per genotype). The number of Neurog3-positive cells was normalized according to the area of the sections estimated by the surface of DAPI staining. The number of Neurog3/BrdU double positive cells was normalized to the total number of Neurog3 cells, and expressed as a percentage. 5-HT positive cells were counted after immunostaining on sections of duodenum and ileum from adult animals (n = 3 per genotype), one week after tamoxifen treatment. At least 50 well-oriented crypt-villus sections per animal were analyzed and as indicated above the number of 5-HT-positive cells was normalized according to the DAPI-positive area.
2.6. Metabolic studies
Mice were fed with normal chow diet (DO3, SAFE) from weaning. Glucose tolerance tests were performed as described previously [23]. Food intake and feces production were measured over a 48 h period. Feces energy content, energy excretion, energy ingested and food efficiency were measured by the Mouse Clinical Institute (MCI) metabolic platform (http://www.ics-mci.fr). Blood analyses were performed on plasma from adult males by the MCI. The energy content of the stools was evaluated using a bomb calorimeter (C503 control, IKA). The energy excreted was calculated as feces energy content (Cal/g) X feces weight (g)/2. The energy ingested was calculated as food intake per day (g) X diet calorific value (3.5 kcal/g). The energy excreted was calculated by the difference between total calories ingested and excreted in feces. Food efficiency is the ratio of energy ingested/energy excreted per day.
2.7. Statistics
Comparisons of means and medians were performed with Graphpad Prism 7.0 or R software. Values are presented as mean ± SD or SEM. p-values were determined using the 2-tailed Student t-test with unequal variance or the 2-tailed nonparametric test of Mann–Whitney offered in Prism. p ≤ 0.05 was accepted as statistically significant.
2.8. Dissociation of mouse intestinal organoids into single cells
Organoids were dissociated to single cells as previously described, with some modifications [3]. Briefly, after medium removal and Matrigel disruption, organoids were dissociated in 500 μL of TrypLE™ Express Enzyme (1X) (Gibco™) at 37 °C for 20 min, assisted by a mechanical dissociation (up and down with a p1000 and a final passage through a syringe fitted with a 18G needle). The dissociation was stopped in a medium containing Advanced DMEM/F-12, fetal calf serum 10%, N-acetylcysteine 0.5 mM, and supplement B27 1X. The cells were pelleted, washed 2 times, and resuspended in the same medium. Finally, they were strained through a 50 μm cell strainer, and stained with DAPI (to exclude dead cells) before to be sorted by flow cytometry (FACSAria Fusion, BD).
2.9. RNA isolation and qPCR
Total RNA from whole small intestine (pups), duodenum (adults), jejunum (adults), ileum (adults), or colon (pups and adults) was extracted using TRI Reagent (Invitrogen) or RNeasy® Midi Kit (Qiagen), according to the manufacturer's instructions. Total RNA from organoids was extracted using RNeasy® Mini Kit (Qiagen) according to the manufacturer's instructions. Reverse transcription was performed using Transcriptor Reverse Transcriptase (Roche) and Random primers (Roche). Quantitative PCRs were performed using mouse-specific TaqMan primers and probes (Applied Biosystems) recognizing Neurog3 (Mm00437606_s1), ChgA (Mm00514341_m1), Arx (Mm00545903_m1), Pax4 (Mm01159036_m1), Pax6 (Mm00443081_m1), Isl1 (Mm00517585_m1), Lmx1a (Mm00473947_m1), Tac1 (Mm01166996_m1), Ucn3 (Mm00453206_s1), NeuroD1 (Mm00520715_m1), Pyy (Mm00520715_m1), Nts (Mm00481140_m1), Cck (Mm00446170_m1), Sct (Mm00441235_g1), Gip (Mm00433601_m1), Gcg/Glp1 (Mm00801712_m1), Tph1 (Mm00493794_m1), Gast (Mm00772211_g1), Sst (Mm00436671_m1), Ghrl (Mm00445450_m1), Nkx2.2 (as described in Gross S. et al., 2016; Forward primer: cctccccgagtggcagat; Reverse primer: gagttctatcctctccaaaagttcaaa; FAM-MGB Probe: ccattgactctgccccatcgctct) or UPL probe #83 for Rfx6 (Forward primer: tgcaggaaagagaaactggag; Reverse primer: ggaaatttttggcgaattgtc), with Light Cycler 480 Probes Master Mix (Roche) on Light Cycler 480 (Roche). Gene expression levels were normalized to Rplp0 (Mm01974474_gH).
2.10. Microarrays
Samples processing: Microarray analysis was performed according to Agilent protocol “One-Color Microarray-Based Gene Expression Analysis – Low Input Quick Amp Labeling” version 6.5, May 2010 (Cat #G4140-90040). Complementary RNA (cRNA) samples were linearly amplified and labeled with cyanine 3 starting from 100 ng of total RNA. Following fragmentation, labeled cRNA were hybridized on Agilent “SurePrint G3 Mouse Gene Expression 8 × 60 K Microarray” (Design ID: 028005), for 17 h, at 65 °C under 10 rpm. Following washing, the slides were scanned using an Agilent G2565CA microarray Scanner System, at a 3 μm resolution in a 20-bit scan mode, according to the “AgilentG3_GX_1Color” protocol. Data analysis: Raw.tif images were then extracted using Agilent “Feature Extraction, version 10.10.1.1” following “GE1_1010_Sep10” protocol. Median raw expression values were further normalized by quantile method [39]. Differential expression analysis between Rfx6−/− vs controls and Rfx6ΔEndo vs controls were performed using the R package limma version 3.36.5 [40]. Data have been deposited in GEO repository (GSE133038).
2.11. RNA sequencing
Ilea from adult Rfx6ΔAdInt mice was harvested at 8 days and 3 months after tamoxifen gavage. Total RNA was extracted using TRI-reagent or RNeasy® Midi Kit (Qiagen). Libraries were prepared using the total RNA-seq Ribo-zero protocol and sequenced on an Illumina HiSeq 2500 (single-end 50bp reads). Reads were identified and mapped onto the mm9 assembly of mouse genome using Tophat version 2.0.10 and the Bowtie2 version 2.1.0 aligner. Quantification of gene expression was performed using HTSeq version 0.6.1 and gene annotations from Ensembl release 67. Normalization of read counts and differential expression analysis between controls and Rfx6ΔAdInt samples were performed using the method implemented in the DESeq2 Bioconductor library version 1.0.19 [41]. Data were deposited in GEO repository (GSE133038).
2.12. Single-cell RNA-Seq; sample processing
eYFP+ cells, from dissociated Neurog3eYFP/+ intestinal organoids, were sorted directly into four 96 well plates « Precise WTA Single Cell Encoding Plate » (BD™ Precise WTA Single Cell Kit, BD Genomics) (1 cell/well) using a FACSAria Fusion (BD). DAPI was used to sort only living cells. The library was prepared according to manufacturer's recommendations (910000014 Rev. 02 11/2016, BD Genomics) and sequenced on an Illumina HiSeq 4000 using paired-end 2 × 100bp lanes sequencing. Data were deposited in GEO repository (GSE133038).
2.13. Single-cell RNA-Seq analysis
2.13.1. Demultiplexing, alignment and quantification
Fastq files were demultiplexed using the sample index encoded in reads 1. Reads 2 having a bad quality (--quality-cutoff 20,20) or corresponding to polyA sequences or to adapters were removed using Cutadapt version 1.10. Reads 2 mapping to rRNA were identified using Bowtie 2 version 2.2.8 and removed. Reads 2 were aligned to mm10 genome using STAR version 2.5.3a. After alignment, reads corresponding to technical duplicates were removed using UMI-Tools version 0.4.4 (method: “directional-adjacency”) [42]. Quantification of gene expression was performed using HTSeq version 0.6.1.post1 (parameter “--mode union”) and gene annotations from Ensembl release 90.
2.13.2. Quality filtering of cells
Cells were filtered according to several quality criteria: (i) percentage of reads uniquely aligned to genome had to be superior or equal to 40%, (ii) percentage of UMIs assigned to genes had to be superior or equal to 50%, (iii) number of assigned UMIs had to be superior or equal to 200,000, (iv) proportion of UMIs assigned to mitochondrial genes had to be inferior to 20%. Finally, 290 cells were kept for further analyses. Among these filtered cells, the number of assigned UMIs varies from 200,000 to 2.3 million. Regarding the number of expressed genes per cell, it varies from 2,777 to 10,019 with a median equal to 4,606.
2.13.3. Normalization, dimensionality reduction, and clustering
Data were then analyzed using the Seurat package version 2.3.4 [43]. Before creating the Seurat object, a gene filtering was performed. On the 29,161 genes expressed in the data, only 9,057 were retained as they met two criteria: they were coding genes or lincRNA and had at least 5 UMIs assigned in 5 samples. The normalization and scaling of the data were performed using the functions NormalizeData (method “LogNormalize”) and ScaleData implemented in Seurat.
Variable genes were identified using the FindVariableGenes function in the Seurat package: the 830 genes having a scaled dispersion superior or equal to 1 and a log-normalized average expression superior or equal to 0.2 were selected as variable genes. Then, a PCA (Principal Component Analysis) was done with the function RunPCA of Seurat, based on these 830 variable genes. The 13 first axes were used to create an UMAP, as they had a significant PCA score, according to the “Jack Straw” approach implemented in Seurat. The UMAP was performed with the function RunUMAP in Seurat (n_neighbors = 20 and min_dist = 0.3). Finally, the clustering was performed based on the two first axis of the UMAP, using the SNN (shared nearest neighbor) modularity optimization algorithm implemented in the FindClusters function of Seurat. The following parameters were used: k.param = 30, resolution = 0.9 and prune. SNN = 5/15. Eight groups of cells were found.
2.13.4. Identification of markers of the groups
Before performing the differential expression analysis, genes were filtered. On the 9,057 already filtered genes, we kept only 8,644 genes as they had a normalized expression equal or superior to 1 in at least 1% of the cells. Differential expression analysis was done using the QLF (quasi-likelihood) approach implemented in the R package edgeR version 3.20.6 [44]. The detection rate was added as a covariate to the model as it improves the performance of the analysis [45]. To compare one group of cells to the others, we used the following contrasts: 1 for said group and −1/7 for the seven other groups. One gene was considered as the marker of a group if it was significantly over-expressed in this group with a FDR inferior or equal to 10−10 and if it was expressed in at least 20% of the cells of the said group.
2.13.5. Cell trajectory
The log-transformed gene expression matrix of the 830 variable genes was used to generate diffusion map, using the function DiffusionMap (k = 15) implemented in destiny R package, version 2.12.0 [46].
3. Results
3.1. Rfx6 is expressed in endocrine progenitors in the crypts and in all nascent hormone-expressing cells at the base of villi in the adult mouse intestine
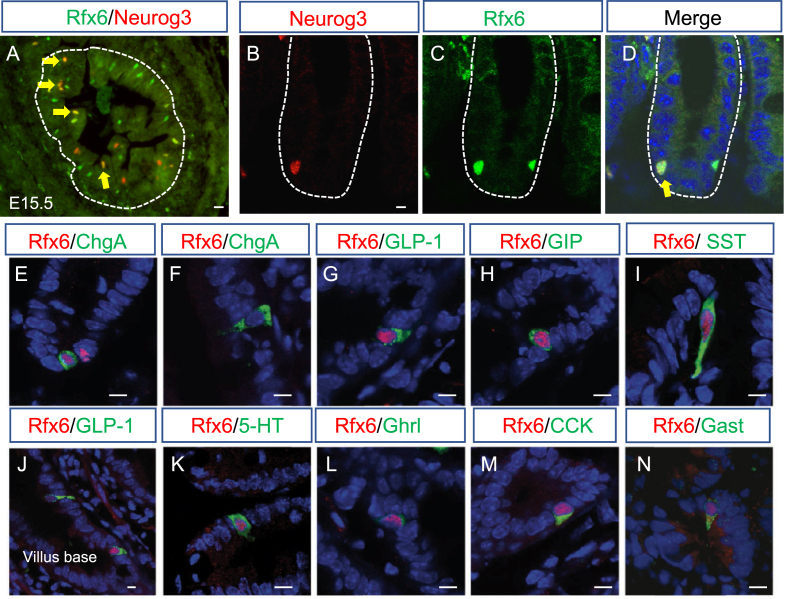
To determine Rfx6 intestinal expression, we performed a series of immunofluorescence experiments in the embryo, as well as in adult CD1 mice (Figure 1). It was previously shown that Rfx6 is expressed broadly in the gut endoderm at E9 [21], [22]. From E13.5, Rfx6 becomes restricted to a small number of scattered cells in the embryonic intestine in a pattern of developing enteroendocrine cells (Figure 1A and Suppl. Figure 1A-C). Accordingly, we found that Rfx6 is expressed in Neurog3-positive enteroendocrine progenitors during embryogenesis and in the adult small intestine. Indeed, we observed that Rfx6 is expressed in a subset of crypt based Neurog3-positive endocrine progenitor cells (40%; n = 300; Figure 1B–D). The majority of Rfx6-positive-nuclei are found in the crypts (Suppl. Figure 1D-F). Nevertheless, Rfx6 is maintained in hormone-expressing enteroendocrine cell types including incretin (GLP-1 and GIP) producing cells, as well as 5-HT enterochromaffin cells (Figure 1G–N, Suppl. Figure 1D,E). We observed that cells positive for both Rfx6 and hormones show different intensities of Rfx6 staining along the crypt-villus axis. Rfx6 staining appears to be strongest in the crypts and in the lower part of villi and becomes weaker while enteroendocrine cells differentiate and migrate up toward the villus top (Figure 1J). Indeed, Chromogranin A (ChgA)-positive or 5-HT-positive/Rfx6-negative cells can be found at the villus top indicating gradual loss of Rfx6 as enteroendocrine cells differentiate and migrate up (Figure 1F, Suppl. Figure 1D,E). In contrast, all GIP-expressing cells expressed Rfx6 (Suppl. Figure 1F).
Figure 1.
Expression of Rfx6 in the intestinal endocrine lineage of embryonic and adult mice. (A–D) Double immunofluorescence for Rfx6 (green) and Neurog3 (red) on cryosections of embryonic E15.5 (A) and adult small intestine (B–C). Yellow arrows point to Rfx6/Neurog3 double positive cells. (E–N) Double immunofluorescence for Rfx6 in red and ChromograninA (ChgA), Glucagon like peptide-1 (GLP-1), Gastric Inhibitory Polypeptide (GIP), Somatostatin (SST), Serotonin (5-HT), Ghrelin (Ghrl), Cholecystokinin (CCK), and Gastrin (Gast), in green. Nuclei are stained with DAPI (blue). Scale bar 10 μM.
3.2. Ectopic expression of gastric genes and impaired enteroendocrine cell differentiation in the embryonic intestine lacking Rfx6
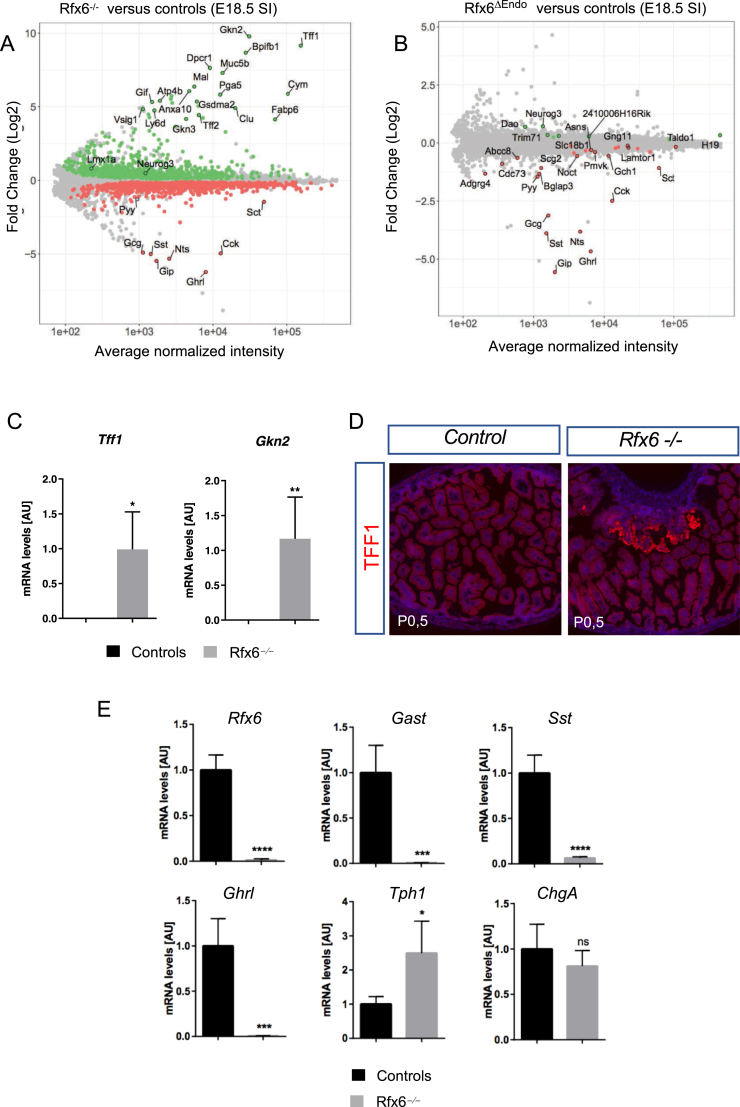
At first, Rfx6 is expressed in the embryonic gut endoderm and then becomes restricted to the enteroendocrine lineage at later stages of development suggesting different roles. To decipher Rfx6 intestinal functions, we compared the transcriptome of the embryonic small intestine of Rfx6 null mice (Rfx6−/−) as well as of mice with an endocrine specific deletion of the gene (Rfx6fl/fl; Neurog3-Cre; called thereafter Rfx6ΔEndo) to wild-type controls. We observed a strong down-regulation of the vast majority of genes encoding enteroendocrine hormones, Gcg (stands herein for preproglucagon gene), Gip, Ghrl, Sst, Nts, Cck, Pyy, Sct, both in Rfx6−/− as well as in Rfx6ΔEndo intestine (Figure 2A,B, Suppl.Tables 1 and 2). Lmx1a, a transcription factor regulating Tph1 expression and thus important for 5-HT production in EC cells, is up-regulated in Rfx6−/− intestine (Fold Change (FC) = 1.7; adjusted p-value = 0.01). Indeed, recent studies revealed the importance of Nkx2-2 transcription factor for enterochromaffin cell differentiation through control of Lmx1a expression [47]. Lmx1a in turn transactivates Tph1 encoding Tryptophan hydroxylase 1, the rate-limiting enzyme for the production of peripheral serotonin. Accordingly, we observed a trend for Tph1 to increase (FC = 1.3; p-value 0.016; adjusted p-value = 0.13).
Figure 2.
Severe impairment of enteroendocrine cell differentiation in the gastro-intestinal tract of Rfx6-deficient embryos and presence of gastric heterotopia in mutant neonates. (A, B) Gene expression profiling of the embryonic small intestine of mice with constitutive (Rfx6−/−) or endocrine-specific deletion (Rfx6ΔEndo) of Rfx6 at E18.5. (A) Rfx6−/− vs control comparison (micro-array data). MA-plot representing the log Fold-Change as a function of the mean of normalized intensity. Green and red points represent significantly (Adjusted p-value ≤ 0.1) up- and down-regulated genes, respectively. (B) Rfx6ΔEndo vs control comparison (micro-array data). MA-plot representing the log Fold-Change as a function of the mean of normalized intensity. Green and red points represent significantly (Adjusted p-value ≤ 0.1) up- and down-regulated genes, respectively. (C) RT-qPCR analysis of gastric genes in the small intestine of Rfx6−/− and wild-type mice at E18.5. Analysis was performed on n = 4 animals per genotype. (D) Immunofluorescence for TFF1 on small intestine cryosections of control and Rfx6−/− new born mice revealing gastric heterotopia. (E) RT-qPCR analysis of relevant genes in the stomach (P0.5) of Rfx6−/− and wild-type mice. Analysis was performed on n = 4 animals per genotype. Data are represented as mean ± SD; unpaired t-test, ****p ≤ 0.0001 ***, p ≤ 0.001, **p ≤ 0.01, *p ≤ 0.05, ns: not significant. AU stands for Arbitrary Unit. SI: Small Intestine. E18.5: Embryonic day 18.5. P0.5: postnatal day 0.5.
Unexpectedly, transcriptome comparison revealed that many gastric genes were strongly up-regulated in Rfx6−/− (Figure 2A), but not in Rfx6ΔEndo embryonic intestine (Figure 2B), compared with wild-type. These genes include Gkn2, Tff1, and Muc5b, which are normally expressed in pit cells of the gastric glands, as well as Atp4b, which encodes a subunit of the gastric proton pump expressed in parietal cells of the fundic gland. These data suggest that heterotopic gastric mucosa forms in the small intestine of Rfx6-deficient mice. We were able to confirm this hypothesis by RT-qPCR for Gkn2 and Tff1 and, at the protein level, for TFF1 (Figure 2C,D). In agreement with the transcriptomic data, we found many patches of TFF1-positive cells in the small intestine of Rfx6-deficient newborn mice but never in controls. Next, we investigated gastric enteroendocrine cell differentiation. Tph1 expression was, similarly to the intestine, up-regulated, while the expression of Gast, Sst, and Ghrl was severely decreased in the stomach of Rfx6-deficient newborn mice (Figure 2E). Thus, our study suggests that, in the mouse embryo, Rfx6 controls enteroendocrine cell differentiation in the gastro-intestinal tract and is required for the maintenance of intestinal cell identity.
3.3. Constitutive, but not adult-induced intestinal deletion of Rfx6, is lethal
To further study Rfx6 intestinal function and to bypass any effect related to its pancreatic function, we generated intestine-specific Rfx6-deficient mice (Rfx6fl/fl; Villin-Cre, referred as Rfx6ΔInt). Rfx6ΔInt mice died shortly after birth (between P2–P5, n = 14 litters), except one animal who survived up to 13 weeks. The presence of milk in the stomach testified that mutants were fed properly; however, they sometimes failed to thrive. Similarly to Rfx6−/− embryonic intestine, RT-qPCRs revealed that Gcg, Gip, Cck, Pyy, Nts, Sst, and Ghrl transcripts are mostly undetectable in newborns Rfx6ΔInt small intestine, Sct is decreased and Tph1 increased (Suppl. Figure 2). Because of the early lethality of Rfx6ΔInt pups, the role of Rfx6 in the adult intestine could not be studied in this model. We thus generated mice with an inducible deletion of Rfx6 (Rfx6fl/fl; Villin-CreERT2, referred as Rfx6ΔAdInt). Tamoxifen gavage efficiently deleted Rfx6, as shown by the absence of wild-type Rfx6 mRNA and protein in the small intestine and the colon of Rfx6ΔAdInt mice 8 days after treatment (Suppl. Figures 3 and 5A). Rfx6ΔAdInt mice started to lose weight one week after tamoxifen treatment, however they caught up 7 days later (Figure 3A). Only one Rfx6ΔAdInt mouse did not overcome the weight loss and died 11 days after the beginning of the treatment. Thus, except this single case, we did not observe any death upon Rfx6 intestinal deletion in the adult mouse (n ≥ 100). In summary, the lack of functional intestinal Rfx6 is lethal at early postnatal stages but is not life threatening in the adult mice.
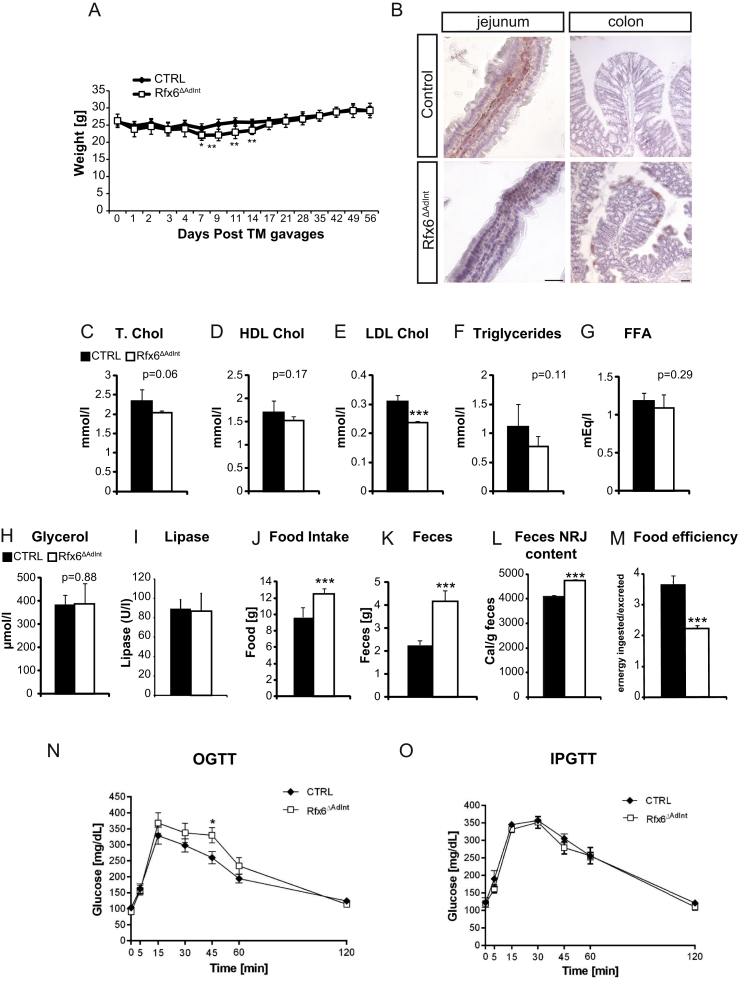
Figure 3.
Energy homeostasis in Rfx6ΔAdIntmice. (A) Weight follow-up of control and Rfx6ΔAdInt mice after tamoxifen gavages. n = 9 animals per group (6 males and 3 females) (B) Oil Red O staining on cryosections from jejunum and colon from Rfx6ΔAdInt and control animals one month after tamoxifen gavage. (C–E) Blood concentration of total (T. Chol, C), high density lipoprotein (HDL Chol, D), and low-density lipoprotein cholesterol (LDL Chol, E) in Rfx6ΔAdInt mice. (F–I) Blood concentration of triglycerides (F), free fatty acids (FFA) (G), glycerol (H) and lipase (I) in Rfx6ΔAdInt mice. (J–M) Decreased food efficiency and increased food intake and feces production in Rfx6ΔAdInt mice. Food intake (J) and feces production (K) of control and mutant mice during 48 h. (L) Energy (NRJ) content of the stools was evaluated in a bomb calorimeter and Food Efficiency (M) calculated (calories ingested/excreted in feces). (C–F) analysis performed 3 months after tamoxifen treatment. (N, O) oral (N, OGTT) and intraperitoneal (O, IPGTT) glucose tolerance tests on adult control and Rfx6ΔAdInt mice one month after tamoxifen-induced Rfx6 deletion. n = 5–6 males were analyzed per group. Data are represented as mean ± SEM; unpaired t-test, ***p ≤ 0.001, **p ≤ 0.01*, p ≤ 0.05.
3.4. Deletion of Rfx6 in the adult intestine induces diarrhea and impairs lipid absorption and food efficiency
We next investigated the metabolic consequences of induced intestinal deletion of Rfx6 in the adult mice one month after tamoxifen treatment. First, we noticed that mutant mice exhibited liquid and yellowish stools (Suppl. Figure 4A,B), suggesting diarrhea and steatorrhea. In agreement with the later hypothesis, Oil red O staining of the mutant gut revealed a clear reduction in the presence of neutral lipids content in the enterocytes and the lamina propria in the small intestine as well as accumulation of lipid droplets in the colon (Figure 3B). In addition, although total cholesterol and HDL levels are not affected, levels of LDL cholesterol were reduced in the serum of Rfx6ΔAdInt mice (Figure 3C–E). Triglycerides concentrations were also reduced, but not significantly (p = 0.11), free fatty acids and glycerol levels were not affected (Figure 3F–H). Since serum pancreatic lipase levels are unchanged (Figure 3I), these results suggest that the reduced numbers of Oil red O-positive lipid droplets and the reduced serum levels of LDL cholesterol found in Rfx6ΔAdInt mice are due to impaired lipid absorption rather than impaired lipid processing.
Glucose clearance was explored by oral (OGTT) and intraperitoneal (IPGTT) glucose tolerance tests (Figure 3N,O). After glucose gavage, blood glucose concentration raised similarly in mutant and control mice suggesting normal intestinal glucose absorption. OGTT revealed that Rfx6ΔAdInt mice are slightly glucose intolerant. This effect could result from the lack of incretin hormones. Indeed, when the incretin effect is bypassed in IPGTT, glucose clearance is unchanged. To further characterize these mice, we performed analyses of food efficiency (Figure 3J–M). Mice, in which Rfx6 was deleted from the adult intestine, showed an increased production of feces due, at least in part, to an increased food intake (Figure 3J–K). Fecal energy measurement, using bomb calorimetry (Figure 3L), revealed fecal energy loss in Rfx6ΔAdInt mice compared to the controls, suggesting malabsorption. Despite increased food intake, Rfx6ΔAdInt mice have a decreased food efficiency ratio (Figure 3M). Of note, these metabolic studies were performed with adult males and thus, results could eventually be different with females. Together, these data suggest that decreased food efficiency of Rfx6ΔAdInt mice could result from diarrhea and lipid malabsorption. This is in agreement with previous work in mice lacking all enteroendocrine cells [12] as well as with symptoms of Mitchell–Riley patients [22].
3.5. Impaired production of all enteroendocrine hormones except serotonin which is increased in Rfx6-deficient adult intestine
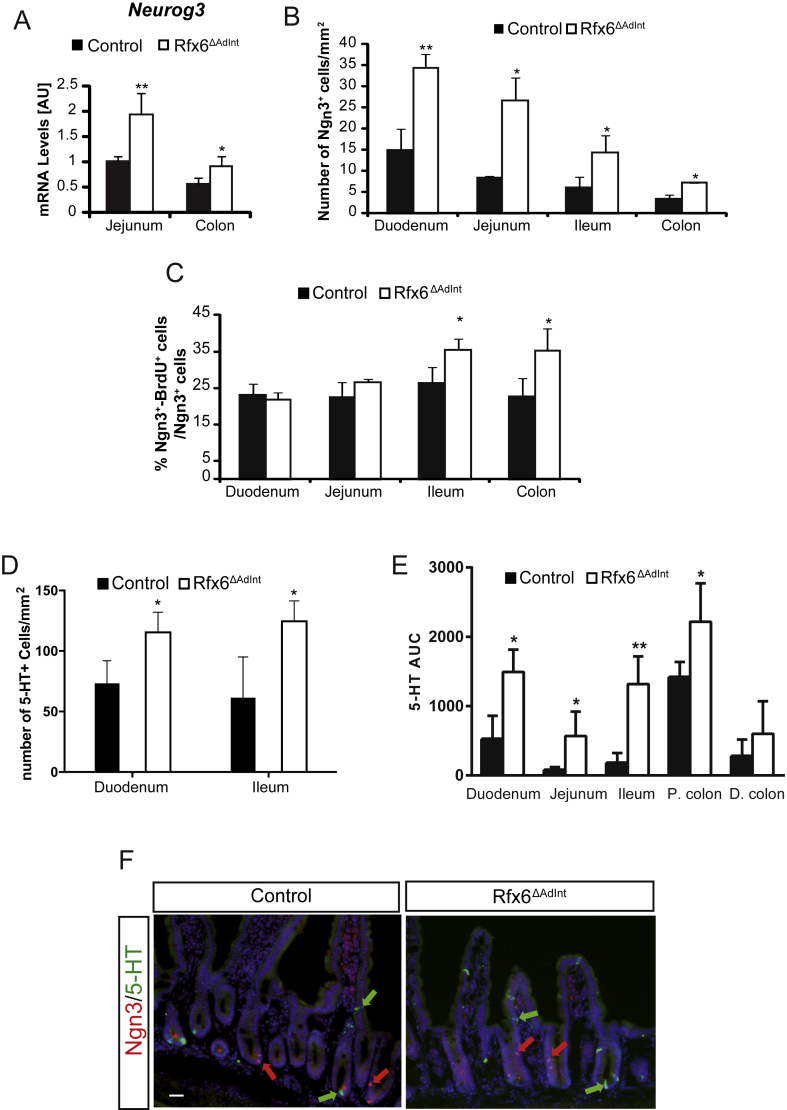
To decipher the mechanisms underlying Rfx6 function in the adult intestine, we determined the genes differentially expressed in the ileum upon Rfx6 removal one week (Figure 4A–C, Suppl. table 3) and 3 months (Figure 4D–F, Suppl. table 4) after tamoxifen treatment by RNA sequencing. RT-qPCR were also performed in other segments of the small intestine and in the colon (Suppl. Figure 5). This strategy allowed us to identify early targets and later transcriptome changes. RNA-Seq revealed that 78 and 74 genes were found up and down-regulated, respectively, 1 week after tamoxifen treatment (Adjusted p-value ≤ 0.05) while these numbers increased to 1393 and 1483 after 3 months, suggesting adaptive responses. Overall, we found that transcripts for many intestinal hormones are almost undetectable (Gcg, Pyy, Gip, Ghrl, Nts) or decreased (Cck, Sst, Nts, Sct) in the small intestine of Rfx6ΔAdInt mice compared to controls (Figure 4B,E, Suppl. Figure 5B). Gcg and Pyy transcripts are decreased as well in the mutant colon (Suppl. Figure 5B). In sharp contrast, Tph1, encoding the 5-HT/serotonin-generating enzyme in EC-cells, was up-regulated in the small and large intestine of Rfx6ΔAdInt mice. Tph1 transcripts were increased as early as one week after Rfx6 removal, in the duodenum (Suppl. Figure 5A) and ileum (Figure 4B), and after one month in the jejunum and colon of Rfx6ΔAdInt mice (Suppl. Figure 5B). Accordingly, ChgA expression, known to be enriched in EC-cells, was increased likewise upon Rfx6 deletion (Figure 4E, Suppl. Figure 5B). In agreement with the transcriptomic data, we have not been able to find any GLP1-, GIP-, CCK-, or SST-expressing cell by immunofluorescence while only few SCT-positive cells were detected (Suppl. Figure 6). To the contrary, the number of serotonin (5-HT)-positive cells was increased already 1 week after Rfx6 deletion compared to controls (Figure 5D). Consequently, mucosal 5-HT content was higher in Rfx6ΔAdInt mice (Figure 5E). These observations suggest that the differentiation of endocrine cells producing peptides is impaired in the absence of Rfx6, while the number of 5-HT cells is increased. Apart from the endocrine lineage, we did not observe any perturbation of the differentiation of other intestinal epithelial cells - including Paneth cells, goblet-cells or enterocytes - or of the crypts/villi morphogenesis (Suppl. Figure 4).
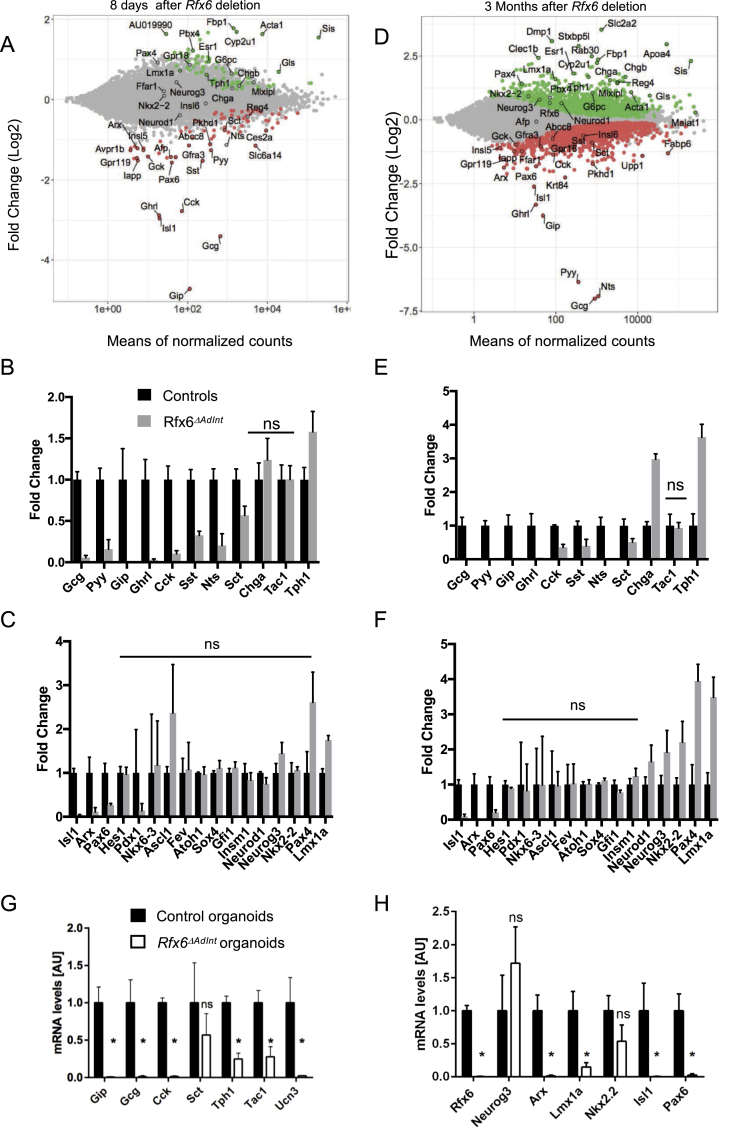
Figure 4.
Gene expression profiling of the small intestine of adult Rfx6ΔAdIntmice. (A–F) RNA-Seq analysis of the small intestine (ileum) of adult Rfx6ΔAdInt mice 8 days (A–C) and 3 months (D–F) after Rfx6 deletion. n = 3–4 animals were analyzed per group. (A, D) MA-plot representing the estimated log2 Fold-Change as a function of the mean of normalized counts. Green and red points represent significantly (adjusted p-value ≤ 0.05) up- and down-regulated genes, respectively in Rfx6ΔAdInt versus controls, 8 days (A) or 3 months (D) after Rfx6 deletion (RNA-Seq data). (B, C, E, F) Histograms showing RNA-Seq data for a selection of genes encoding enteroendocrine hormones or the rate limiting enzyme Tph1 for serotonin synthesis (B, E) or transcription factors (C, F). All controls values are adjusted to 1. ns: non-significant, adjusted p-value >0.05 between controls and mutant. For all other genes adjusted p-value is ≤ 0.05. (G, H) RT-qPCR expression analysis of relevant genes in Rfx6ΔAdint duodenal organoids, 8 days after a 15 h treatment with 1.25 μM tamoxifen (4-OHT) to delete Rfx6 gene. Data are represented as mean ± SD. Organoids were generated from n = 4 mice per group. Statistics: non-parametric Mann–Whitney test, *p ≤ 0.05, ns, non significant.
Figure 5.
Increased number of Neurog3-positive endocrine progenitors and 5-HT-positive cells upon Rfx6 deletion in the adult intestine. (A) RT-qPCR. Expression of Neurog3 mRNA in the adult jejunum and colon three months after Rfx6 deletion (n = 4 mice per group). Unpaired t-test. (B) Quantification of the number of Neurog3-positive cells per mm2 of tissue in each segment of the adult intestine (n = 3–4 mice per group) three months after Rfx6 deletion. (C) Quantification of the number of proliferating Neurog3+ cells expressed as the percentage of BrdU+/Neurog3+ cells on total Neurog3+ cells. (D) Quantification of the number of 5-HT-positive cells per mm2 of tissue in the duodenum and ileum of Rfx6ΔAdInt and control mice, 8 days after Rfx6 deletion (n = 3 mice per group). (E) Mucosal 5-HT content analysis throughout the intestinal tract from day 11 of tamoxifen-treated control Rfx6ΔAdInt mice (n = 3–4 mice per group). Biopsies were sampled from Duodenum (Duo), Jejunum (Jej), Ileum, Prox. Colon (P. colon) and Dist. Colon (D. colon). 5-HT content were analyzed on a HPLC-ECD and results stated as area under the curve (AUC). (F) Double immunofluorescence for Neurog3 and 5-HT on duodenal cryosections of control and Rfx6ΔAdInt mice (8 days post Tamoxifen treatment). Red and green arrows in D and E point to some Neurog3-and 5-HT-positive cells, respectively. Data are represented as mean ± SD; unpaired t-test was applied to calculate significance *p ≤ 0.05; **p ≤ 0.01. Scale bar represents 50 μm.
To determine the position of Rfx6 in the transcriptional network controlling enteroendocrine cell differentiation, we next examined the expression of transcription factors known to control enteroendocrine cell differentiation. We found that Arx, Isl1 and Pax6 are strongly decreased (Figure 4C,F, Suppl. Figure 5A) as early as 1 week after tamoxifen treatment, in agreement with the impaired differentiation of peptidergic enteroendocrine cells and suggesting that Rfx6 acts upstream of these transcription factors. In contrast, the expression of several transcription factors, such as Pax4, was up-regulated (Figure 4C,F, Suppl. Figure 5); Pax4 is also up-regulated in Rfx6-deficent pancreatic beta cells. Neurog3 transcripts are increased as well, suggesting that Rfx6 deletion perturbs endocrine progenitor cells development (Figure 5). Importantly, we also found that Lmx1a is increased upon Rfx6 deletion in the adult intestine, and so it is the case for Nkx2-2, an upstream regulator of Lmx1a, 3 months after tamoxifen treatment. Lmx1a upregulation likely explains the increased expression of Tph1, the rate limiting enzyme for serotonin synthesis. RNA-Seq did not reveal any changes in expression for other endocrine transcription factors, such as Insm1, Sox4, NeuroD1 or for Atoh1, which is necessary for secretory lineage specification. Unaltered gene expression reflects that the average gene expression levels are unchanged, which does not exclude variation in single cells, or that gene expression is independent of Rfx6. Taken together, our results suggest that Rfx6 controls transcriptional programs which promote the differentiation of enteroendocrine cells expressing peptide hormones (PE cells) while repressing the differentiation of enterochromaffin cells secreting serotonin (EC cells) in the digestive tract.
We next addressed whether Rfx6 similarly controls enteroendocrine cell differentiation in Rfx6ΔAdInt small intestinal organoids (Figure 4G,H). Briefly, tamoxifen was added to organoids cultures to remove Rfx6, and organoids were analyzed 8 days after treatment. As expected, RT-qPCR analysis confirmed the decreased expression of Gip, Gcg, and Cck (Sct unchanged) upon Rfx6 deletion. Unexpectedly, Tph1 expression was also decreased, as were Ucn3 and Tac1, two other markers of EC cells (see last section results). In agreement with the decreased expression of EC enriched genes, Lmx1a was down-regulated as well in this culture system, suggesting that the consequences of Rfx6 removal on EC cells differentiation programs are different in vivo and ex vivo.
3.6. Enhanced expression of neoglucogenic and nutrient absorption machinery genes in the small intestine of mice upon Rfx6 deletion
To evaluate the long-term effect of impaired enteroendocrine cell function and lipid metabolism consecutively to Rfx6 deficiency, we examined the expression of intestinal genes that are not endocrine specific. Surprisingly we observed a strong and fast upregulation of genes involved in intestinal gluconeogenesis [48]. These include Fructose 1,6 biphosphatase (Fbp1) and Glucose 6 phosphatase (G6pc) as well as Glutaminase (Gls), the enzyme which hydrolyzes glutamine, the main gluconeogenic substrate in the small intestine (Figure 4A,D, Suppl. Table 4). The expression of the gene encoding the brush border enzyme sucrase-isomaltase (Sis), which hydrolyzes dietary carbohydrates was also found to be up-regulated. A major long-term adaptation to intestinal Rfx6 deletion that was not observed at 1 week, is the strong upregulation of the glucose transporter type 2 Slc2a2 (also known as Glut2). Slc2a2 transporter is thought to facilitate the passage of dietary sugars in the blood stream [49] at the basolateral side of enterocytes. The expression of the sodium-coupled co-transporter Sglt1, which triggers glucose absorption at the apical membrane of enterocytes, was however not differentially expressed. We also found an up-regulation of Apoa4, an important chylomicron component, as well as of other apoliproteins such as Apoc3, Apoc2, and their upstream regulator, the transcription factor Creb3l3/CREB-H [50]. Together, our data suggest that Rfx6ΔAdInt mice adapt to overcome decreased food efficiency by promoting intestinal glucose and lipid absorption as well as glucose production.
3.7. Increased number of enteroendocrine progenitors in intestinal crypts of Rfx6ΔAdIntmice
Unexpectedly, when analyzing RNA-Seq data, we found increased levels of Neurog3 transcripts in the ileum of adult Rfx6ΔAdInt mice 3 months after Rfx6 deletion (Figure 4C,F). Similarly, RT-qPCR data showed that Neurog3 was up-regulated in the jejunum (2-fold) and in the colon (1.6-fold) compared to controls (Figure 5A) and in the duodenum as early as one week after Rfx6 deletion (Suppl. Figure 5A). Similar results were observed in Rfx6ΔInt neonates (Suppl. Figure 2). Morphometric analysis revealed a 1.3–5 fold increase in the number of Neurog3-cells per mm2 in all segments of the small intestine as well as in the colon of Rfx6ΔAdInt mice (Figure 5B). To determine whether the increased pool of enteroendocrine progenitors resulted from an increased proliferation of endocrine progenitors, we quantified the percentage of Neurog3+/BrdU+ double-positive cells. Neurog3 cell proliferation was not affected in the duodenum and jejunum while was increased in the ileum and colon (Figure 5C). Neurog3-positive cells did not co-stain for 5-HT (Figure 5F), suggesting that the increased number of 5-HT-positive cells does not result from premature expression of 5-HT in enteroendocrine progenitors. Taken together, our results support that the increase of Neurog3 transcripts in Rfx6-deficient intestine results from increased number of enteroendocrine progenitors.
3.8. Single cell transcriptomics revealed Rfx6 dependent genetic programs in the peptidergic enteroendocrine and enterochromaffin cell lineages
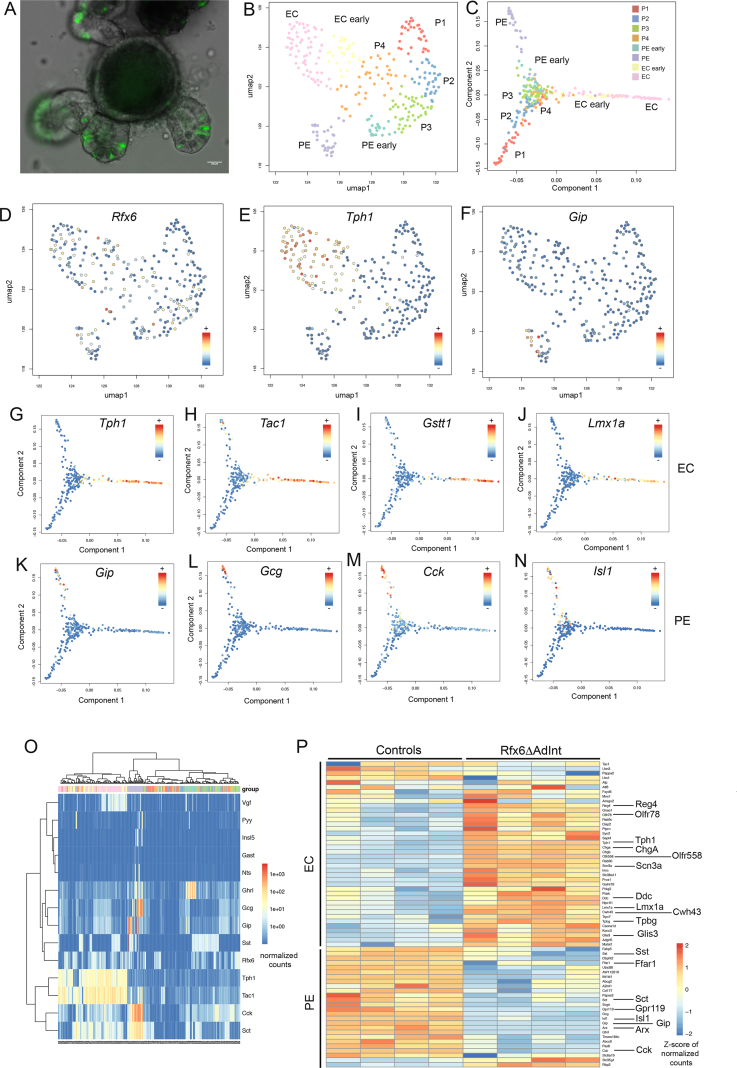
To gain knowledge into the mechanisms controlling enteroendocrine cell differentiation and diversity as well as into the role of Rfx6, we performed single-cell RNA sequencing of sorted EECs. Enteroendocrine cells were captured at various stages of their development, from endocrine progenitors to hormone expressing cells. Endocrine cells were purified from Neurog3eYFP/+ mouse intestinal organoids. In this model, due to the stability of the eYFP protein, both crypt-based endocrine progenitors as well as differentiating hormone-expressing EECs express the fluorescent reporter (Figure 6A and Suppl. Figure 7). After data filtering, 290 cells were analyzed. Shared Nearest Neighbor (SNN) clustering revealed eight groups of cells (Figure 6B and Suppl. Figure 8A) which were visualized in two dimensions using the Uniform Manifold Approximation and Projection (UMAP) algorithm. These clusters include 4 groups of progenitors (P1-4); P2–P4 expressing the endocrine progenitor marker Neurog3 (Figure 6B and Suppl. Figure 8A). Group P1 expressed markers of Paneth (Defa 17, Lyz1) and goblet (Muc2, Tff3) cells, suggesting multiple lineages priming in secretory progenitors as previously described [51]. The four other clusters represent subsets of Enterochomaffin cells (EC early and EC) expressing Tph1 (Figure 6E and Suppl. Figure 8A), and Peptidergic Enteroendocrine cells (PE early and PE) expressing Glc, Cck, Sct, Gip, Ghrl, and Sst (Figure 6F and Suppl. Figure 8A), at different stages of maturation. Clusters gene signatures included known and novel makers of EC and PE cells. Single-cell analysis confirmed that Rfx6 is expressed both in endocrine progenitors as well as in more mature enteroendocrine cells of both EC and PE lineages (Figure 6D,O) and that single EECs can co-express several hormones genes (Figure 6O). Temporal resolution of transcriptome dynamics using diffusion map algorithm predicted cell state transition trajectories. Data analysis showed that endocrine progenitors differentiate in two branches: the EC and PE lineages (Figure 6C). Examples of genes expressed along the EC and PE branches are shown in Figure 6G–N and Suppl. Figure 9. Importantly, pseudotemporal ordering of single cells also revealed dynamics of transcription factors during EC or PE cell differentiation (Figure 6 and Suppl. Figure 9). Some transcription factors were clearly enriched in EC (Lmx1a, Mnx1, Atf6, Glis3, Lhx1) or PE (Arx, Isl1, Pax6, Etv1) lineage branches. Some other transcription factors (Pax4, Neurod1, Nkx2-2, Insm1, Mlxipl), like Rfx6, were found both in progenitors and more mature enteroendocrine cells (Suppl. Figure 9 Q-X) but were not specifically enriched in EC and PE lineages according to our selection criteria.
Figure 6.
Single-cell RNA-Seq analysis of enteroendocrine cells of adult small intestine and Rfx6-dependent genetic programs. (A) Small intestinal organoid of Neurog3eYFP/+ mice (intrinsic eYFP fluorescence). eYFP labels Neurog3-positive endocrine progenitors and differentiated enteroendocrine cells. After organoid cell dissociation, fluorescence activated cell sorting (FACS) is used to isolate individual endocrine cells in 96 well plates. scRNA-Seq is then used to profile the cells. (B) Uniform Manifold Approximation and Projection (UMAP) representing the cell clusters. The 290 single cells were clustered based on the expression of 830 biologically variable genes. EC: Enterochromaffin cells; EC early: precursors of EC cells; PE: Peptidergic Enteroendocrine cells; PE early: precursors of PE cells; P1-4:groups of progenitors. (C) Cell clusters along the cell trajectory. Diffusion map, built from the log-expression of 830 biologically variables genes, was used to reconstruct the cell trajectory. (D–F) UMAP representing the expression of Rfx6, Tph1 and Gip (normalized counts) in the 290 single cells. (G–N) Expression (normalized counts) of genes enriched in the EC (G–J) and PE (K–N) branches along the cell trajectory. (O) Heatmap (normalized counts) showing expression of hormone genes and Rfx6 in individual cells. (P) Heatmap (Z-score of normalized counts) showing expression of some markers of EC and PE lineages affected by Rfx6 deletion, in Rfx6ΔAdInt (3 months after Rfx6 removal) and control conditions. Shown is the expression (row-wise Z-score), in Rfx6ΔAdInt and control conditions, of markers of EC populations (EC and/or EC-early populations) and PE populations (PE and/or PE-early populations) which are differentially expressed after Rfx6 deletion.
Our data show that Rfx6 differentially controls genetic programs regulating PE and EC cells, which appeared to be the two main EEC lineages. We thus determined, among the genes defining the signature of PE and EC lineages, which ones were actually regulated by Rfx6 in the adult small intestine (Figure 6P and Suppl. Figure 8B). As expected, most PE enriched genes, like hormone genes (e.g., Sct, Gcg, Gip, Sst, Cck), related transcription factors (e.g. Arx, Isl1, Pax6), GPCRs (e.g Gpr119, Ffar1), as well as other PE markers (Abcc8, Scgn, Fapb5) were down regulated in absence of Rfx6. Surprisingly rare PE enriched genes were either up-regulated (Rbp2) or have their expression unchanged (Gpr112, Rbp4, Scg3, data not shown). Furthermore, the vast majority of EC enriched genes were up-regulated in absence of Rfx6, including endocrine markers (Tph1, ChgA, Scn3a), transcription factors (Lmx1a, Glis3), GPCRs (Olfr558, Olfr78) or other markers (e.g Cacna1d, Disp2, Cwh43). However, the expression of highly specific markers of the EC lineage was not affected (e.g Tac1, Ucn3, Lhx1, Mnx1, Atf6) or decreased (Pappa2, Ly6a) by the loss of Rfx6. Taken together, these results suggest that Rfx6 regulates specific genetic programs in the EC and PE lineages, while others genes are independent of Rfx6.
4. Discussion
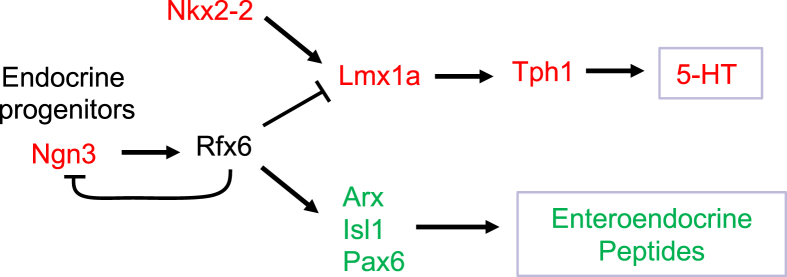
Mutations in RFX6 cause Mitchell–Riley syndrome in humans. This syndrome is characterized by neonatal diabetes and intestinal failures including malabsorption [22]. Previous studies showed that Rfx6 controls beta-cell development in the mouse embryo as well as insulin secretion in adult mouse and human beta cells [22], [23], [32]. However, the function of Rfx6 in the mouse intestine was unknown. In this study, we show that, as in the pancreas, Rfx6 is expressed in Neurog3-endocrine progenitors and persists in all hormone-expressing cells in the intestine, not only in Gip-expressing cells as previously thought [25]. In all cases, constitutive or inducible intestinal deletion of Rfx6 in mice resulted in a severe loss of enteroendocrine cells expressing peptide hormones while enterochromaffin cells were still present and genes controlling serotonin-production were increased. Unlike newborns that die, adult mice can cope with the induction of this massive reduction of peptidergic EECs. However, intestinal absorption is perturbed, leading to impaired food efficiency. RNA-Seq revealed enhanced expression of neoglucogenic and nutrient absorption machinery genes that we hypothesize to be a compensatory mechanism to cope with the malabsorption. Importantly, using single-cell transcriptomics, we found that EECs differentiate from endocrine progenitors according to two cell differentiation trajectories: the Enterochromaffin cells (EC) and Peptidergic Enteroendocrine (PE) cells. Similar conclusions were drawn recently in a single-cell profiling of EECs [5]. Identification of specific gene signatures and dynamics in the EC and PE lineages allowed us to determine their respective dependence on Rfx6. We propose that Rfx6 positively regulates genetics programs leading to PE cells differentiation, and represses 5-HT production, as well as other, but not all, EC cell specific programs (Figure 7).
Figure 7.
Proposed model for the role of Rfx6 in enteroendocrine lineages specification in the adult intestine. Neurog3 triggers the endocrine fate. Rfx6 is then activated in endocrine progenitors downstream of Neurog3 and represses Neurog3 transcription allowing differentiation to proceed. Rfx6 promotes peptidergic endocrine (PE) cell differentiation upstream of Arx, Isl1 and Pax6. Rfx6 represses, upstream of Lmx1a, a genetic program leading to Tph1 expression and 5-HT production. Green and Red: genes respectively down or up-regulated in the Rfx6ΔAdInt compared to wild-type controls.
In addition to the severe alteration in Peptidergic Enteroendocrine cell differentiation, our studies demonstrated that constitutive inactivation of Rfx6 leads to gastric heterotopia in the embryonic mouse intestine. Heterotopic gastric mucosa was reported in only 3 patients carrying compound heterozygote mutations in RFX6 [30], [31]. These patches of gastric heterotopia were revealed after histological analysis of intestinal resection to treat hemorrhagic ulcers. It has been proposed that mucosa alteration could be caused by acid secretion of the ectopic gastric tissue [31]. It is likely that gastric heterotopia is a general feature of patients with RFX6 mutations, since anemia and melena were reported in many cases and could result from intestinal bleeding [31]. Importantly, we observed gastric tissue only in the intestine of mice with a constitutive deletion of Rfx6 and never when Rfx6 was deleted in the endocrine lineage or when Rfx6 was inactivated in the adult intestine. These observations suggest that gastric heterotopia is unrelated to the impairment of the endocrine lineage but rather results from an early function of Rfx6 in mouse endoderm patterning, similarly to what has been suggested in Xenopus [24]. Nevertheless, we did not observe any gross defect of intestinal or gastric organogenesis, suggesting functional redundancy. The patchy pattern of gastric tissue in Rfx6-deficient intestine might thus be explained by the lack of functional compensation in some pluripotent endodermal stem cells that consequently lost or did not acquire the proper regional identity. In one case, ectopic pancreatic tissue was also reported in the jejunum [31], a feature that we did not observe in Rfx6-deficient mice. Of note, it has been suggested that in the human gut the loss of Foxo1 expression, particularly in EC cells, results in ectopic insulin expression [52]. We did not observe any changes in FOXO1 expression in Rfx6ΔAdInt intestine and did not detect any expression of Ins1 or Ins2 in the embryonic or adult intestine lacking Rfx6, suggesting that expanded EC cells do not express insulin.
Rfx6 inactivation led to an increased expression of Tph1, which is critical for 5-HT biosynthesis in enterochromaffin cells, at all stages and conditions studied in vivo throughout the intestine. Furthermore, we measured an increased number of 5-HT-positive cells and mucosal 5-HT content in the adult small intestine upon Rfx6 removal. We thus propose that Rfx6 represses 5-HT biosynthesis. Of note, we found that the expression of Lmx1a, which is downstream of Nkx2-2 and has been shown to control the expression of Tph1 [53], was up-regulated. This suggests that increased Tph1 results from increased Lmx1a. By contrast, Nkx2-2 transcripts were unaffected one week after Rfx6 deletion, suggesting that Rfx6 does thus not repress Lmx1a via Nkx2-2. However, 3 months after the tamoxifen treatment, Nkx2-2 expression was increased; therefore, we cannot exclude a contribution of Nkx2-2 in Lmx1a and Tph1 up-regulation at a later stage. Importantly, the unchanged expression of several specific markers of the EC lineage, identified in our single cell transcriptome study of enteroendocrine cells, such as Ucn3, Tac1, Atf6, Lmx1a, and Mnx1, suggests that Rfx6 does not act as a genetic switch promoting PE and inhibiting EC destiny. Rather, Rfx6 represses the genetic program leading to 5-HT biosynthesis and possibly others specific features of EC cells. However, as some of these genes are also expressed outside of enterochromaffin cells, such as Ucn3 and Tac1, which are found in enteric neurons [54], it is possible that we could not detect variation in their mucosal expression when performing bulk RNA-Seq on the entire intestinal tissue. Of note, the cellular origin of the increased expression of Lmx1a and Tph1 remains unclear. One hypothesis is that the loss of Rfx6 results into the ectopic induction of the serotonergic program in the PE lineage. Another possibility would be that Rfx6 modulates the levels of 5-HT in EC cells by controlling the expression of Lmx1a and Tph1. The absence of cells co-expressing Neurog3 and 5-HT in Rfx6ΔAdInt mice suggests that Rfx6 does not represses the serotonergic program in enteroendocrine progenitors but rather at later stages of EECs differentiation.
Our study also sheds light on the position of Rfx6 in the regulatory network of transcription factors controlling EECs differentiation. We found that Pax6, Isl1, and Arx are down-regulated, while Pax4 is up-regulated by the inactivation of Rfx6 suggesting that Rfx6 operates upstream of these transcription factors. In Arx-deficient mice, the expression of Gcg, Gip, Cck, and Nts was reduced [15], [16], suggesting that Rfx6 regulates the expression of these hormone-encoding genes through the regulation of Arx. Pax4 and Arx are known to repress each other in the developing pancreas [55]. However, this does not seem to be the case in the gut. Indeed, Pax4 expression is unchanged in Arx-deficient intestine [15]. Thus, in the absence of Rfx6, increased Pax4 expression is probably not a consequence of Arx down-regulation. Nevertheless, Pax4 was shown to be required for EC cell differentiation [15], in agreement with the maintenance of EC cells in Rfx6-deficient mice. Interestingly, like for Rfx6, intestinal ablation of Isl1 resulted in the loss of GLP-1, GIP, CCK, and SST expressing cells and an increase in 5-HT producing cells [18]. It is thus likely that Isl1 contributes to the regulation of PE cells differentiation, downstream of Rfx6 and also to the repression of 5-HT production through a mechanism that remains to be elucidated. We previously reported that constitutive intestinal inactivation of Neurog3 (using Villin-Cre) led to a total loss of EECs, including 5-HT expressing cells [12]. Despite this, 50% of the Neurog3ΔInt mice, which display a more severe enteroendocrine phenotype than Rfx6ΔInt mice, survived to adulthood, probably because they were kept on a more robust CD1 genetic background. Thus, Rfx6 is turned on in endocrine progenitors and operates downstream of Neurog3 to regulate a network of transcription factors including Lmx1a, Pax4, Arx, Pax6, and Isl1 which will control later steps of EEC differentiation. Our study revealed that Neurog3 expression and the number of Neurog3-positive endocrine progenitor cells increased upon induction of Rfx6 deletion in both the small intestine and colon. One explanation for this increase would be that Rfx6 normally represses Neurog3 expression in late endocrine progenitors. Alternatively, endocrine progenitors accumulate because they cannot differentiate further into peptidergic enteroendocrine cells. Interestingly, we found that Arx, Pax4, Pax6, Isl1, and Neurog3 are similarly regulated by Rfx6 in the adult gut and in the embryonic pancreas [23, and our unpublished data)] further confirming the conservation of gene regulatory pathways during the differentiation of enteroendocrine and islet cells. Of note, the use of Rfx6fl/fl mice as controls is a possible limitation of our study, given that impaired intestinal regeneration as recently been reported in Villin-CreERT2 mice [56]. Nevertheless, we believe that the lack of Rfx6 is solely causative of the enteroendocrine failure in adult mice since a very similar phenotype was observed in Rfx6−/− newborn mice which lack any CreERT2 transgene.
Unexpectedly, when we studied the consequences of the induction of Rfx6 deletion in small intestine organoid cultures, we found that the development of all EECs was impaired, including EC cells. In line with this observation, the expression of Lmx1a and Tph1 were down-regulated ex vivo upon Rfx6 deletion. A similar conclusion was reported recently in a study in which Rfx6 was inactivated directly, ex vivo, in mouse intestinal organoids [5]. We propose that these in vivo/ex vivo differences on the role of Rfx6 in EC cell development could reflect limitations of the organoid culture system to faithfully mimic alternative cell differentiation pathways when genetic networks are perturbed. We cannot exclude that the promotion of serotonergic programs in the absence of Rfx6 results from an adaptation due to a systemic effect or an effect of the microbiota that would not occur ex vivo. However, we do not favor this hypothesis as we saw the induction of Lmx1a and/or Tph1 when Rfx6 is lacking, not only in the adult but also in the embryonic intestine and in the developing stomach suggesting a general function of Rfx6 in repressing enterochromaffin cell differentiation. Nevertheless, these findings highlight the necessity of generating more sophisticated intestinal organoid models in the future, models which are innervated, vascularized, and contain a microbiome, smooth muscle or immune cells since all can eventually impact intestinal epithelial cell differentiation.
In conclusion, this study revealed that Rfx6 is essential, downstream of Neurog3, for the differentiation of peptidergic enteroendocrine cells during development and in the adult. Rfx6-dependent impairment of enteroendocrine cells perturbs food efficiency. In addition, we show that Rfx6 also represses genetic programs leading to the biosynthesis of 5-HT. These findings shed new light on the molecular mechanisms underlying intestinal malabsorption and energy metabolism in RFX6 deficiency in humans.
Author contribution
J.P. designed and performed most of the mouse experiments, analyzed the data, and wrote the paper, C.V. performed bioinformatic analysis, interpreted results, and wrote the paper. F.B. and A.D.A. performed single cell experiments, interpreted results, and wrote the paper. F.B., A.D.A., and N.P performed all organoids experiments and interpreted results. S.G., L.N., M.L.L. M.E., and C.T.C. helped initially with the design and realization of single cell experiments. C.K., S.J.C., and N.M. contributed to the bioinformatic analysis. M.L.L performed serotonin measurements. J.P. and A.D.A. performed morphometric analysis. A.M., A.B., P.S., and V.S. performed experiments and contributed to data analysis. C.L assisted with the immunostaining experiments. T.W.S. provided conceptual input. G.G. oversaw the entire project, designed experiments, analyzed data, wrote the paper, and obtained financial support.
Acknowledgements
The authors thank the members of the Genomeast platform for RNA-Seq and Doulaye Dembele for initial analyses of the data, of the Cell sorting facility, of the Mouse Clinical Institute Metabolic Exploration Plateform (MCI, Strasbourg, France) for their help with the metabolic analyses. We thank N. Messaddeq (IGBMC), for Electron microscopy and Martine Poulet for technical assistance and Tao Ye (IGBMC) for submission of data to GEO. We thank Miriam Ejarque (IGBMC), Catherine Tomasetto (IGBMC), Natalie Terry (Division of GI, Hepatology, and Nutrition Children's Hospital of Philadelphia) and Klaus Kaestner for helpful discussions. This study was funded by the Novo Nordisk Foundation (Challenge Grant NNF14OC0013655) and by Grants from the Agence Nationale pour la Recherche (ANR Rfx-PancInt) and from the Fondation pour la Recherche Médicale (FRM). IGBMC is supported by the grant ANR-10-LABX-0030-INRT, a French State fund managed by the Agence Nationale de la Recherche under the frame program Investissements d’Avenir ANR-10-IDEX-0002-02. Sara Jimenez Correa is an IGBMC international PhD programme fellow supported by the LaxEx INRT.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2019.08.007.
Contributor Information
Adèle De Arcangelis, Email: adele@igbmc.fr.
Gérard Gradwohl, Email: gradwohl@igbmc.fr.
Conflict of interest
The authors have declared no competing interest
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Egerod K.L., Engelstoft M.S., Grunddal K.V., Nøhr M.K., Secher A., Sakata I. A major lineage of enteroendocrine cells coexpress CCK, secretin, GIP, GLP-1, PYY, and Neurotensin but not somatostatin. Endocrinology. 2012;153(12):5782–5795. doi: 10.1210/en.2012-1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Habib A.M., Richards P., Cairns L.S., Rogers G.J., Bannon C.A.M., Parker H.E. Overlap of endocrine hormone expression in the mouse intestine revealed by transcriptional profiling and flow cytometry. Endocrinology. 2012;153(7):3054–3065. doi: 10.1210/en.2011-2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grün D., Lyubimova A., Kester L., Wiebrands K., Basak O., Sasaki N. Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature. 2015;525(7568):251–255. doi: 10.1038/nature14966. [DOI] [PubMed] [Google Scholar]
- 4.Haber A.L., Biton M., Rogel N., Herbst R.H., Shekhar K., Smillie C. A single-cell survey of the small intestinal epithelium. Nature. 2017;551(7680):1–28. doi: 10.1038/nature24489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gehart H., van Es J.H., Hamer K., Beumer J., Kretzschmar K., Dekkers J.F. Identification of enteroendocrine regulators by real-time single-cell differentiation mapping. Cell. 2019;176(5) doi: 10.1016/j.cell.2018.12.029. 1158–1173 e1116. [DOI] [PubMed] [Google Scholar]
- 6.Glass L.L., Calero-Nieto F.J., Jawaid W., Larraufie P., Kay R.G., Gottgens B. Single-cell RNA-sequencing reveals a distinct population of proglucagon-expressing cells specific to the mouse upper small intestine. Molecular Metabolism. 2017;6(10):1296–1303. doi: 10.1016/j.molmet.2017.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drucker D.J. Evolving concepts and translational relevance of enteroendocrine cell biology. Journal of Clinical Endocrinology & Metabolism. 2016;101(3):778–786. doi: 10.1210/jc.2015-3449. [DOI] [PubMed] [Google Scholar]
- 8.Beumer J., Artegiani B., Post Y., Reimann F., Gribble F., Nguyen T.N. Enteroendocrine cells switch hormone expression along the crypt-to-villus BMP signalling gradient. Nature Cell Biology. 2018;20(8):909–916. doi: 10.1038/s41556-018-0143-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jenny M. Neurogenin3 is differentially required for endocrine cell fate specification in the intestinal and gastric epithelium. The EMBO Journal. 2002;21(23):6338–6347. doi: 10.1093/emboj/cdf649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee C.S., Perreault N., Brestelli J.E., Kaestner K.H. Neurogenin 3 is essential for the proper specification of gastric enteroendocrine cells and the maintenance of gastric epithelial cell identity. Genes & Development. 2002;16(12):1488–1497. doi: 10.1101/gad.985002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schonhoff S.E., Giel-Moloney M., Leiter A.B. Neurogenin 3-expressing progenitor cells in the gastrointestinal tract differentiate into both endocrine and non-endocrine cell types. Developmental Biology. 2004;270(2):443–454. doi: 10.1016/j.ydbio.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 12.Mellitzer G., Beucher A., Lobstein V., Michel P., Robine S., Kedinger M. Loss of enteroendocrine cells in mice alters lipid absorption and glucose homeostasis and impairs postnatal survival. Journal of Clinical Investigation. 2010;120(5):1708–1721. doi: 10.1172/JCI40794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shroyer N.F., Helmrath M.A., Wang V.Y.C., Antalffy B., Henning S.J., Zoghbi H.Y. Intestine-specific ablation of mouse atonal homolog 1 (Math1) reveals a role in cellular homeostasis. Gastroenterology. 2007;132(7):2478–2488. doi: 10.1053/j.gastro.2007.03.047. [DOI] [PubMed] [Google Scholar]
- 14.Naya F.J., Huang H.P., Qiu Y., Mutoh H., DeMayo F.J., Leiter A.B. Diabetes, defective pancreatic morphogenesis, and abnormal enteroendocrine differentiation in BETA2/neuroD-deficient mice. Genes & Development. 1997;11(18):2323–2334. doi: 10.1101/gad.11.18.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beucher A., Gjernes E., Collin C., Courtney M., Meunier A., Collombat P. The homeodomain-containing transcription factors Arx and Pax4 control enteroendocrine subtype specification in mice. PLoS One. 2012;7(5):e36449. doi: 10.1371/journal.pone.0036449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Du A., McCracken K.W., Walp E.R., Terry N.A., Klein T.J., Han A. Arx is required for normal enteroendocrine cell development in mice and humans. Developmental Biology. 2012;365(1):175–188. doi: 10.1016/j.ydbio.2012.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Larsson L.I., St-Onge L., Hougaard D.M., Sosa-Pineda B., Gruss P. Pax 4 and 6 regulate gastrointestinal endocrine cell development. Mechanisms of Development. 1998;79(1–2):153–159. doi: 10.1016/s0925-4773(98)00182-8. [DOI] [PubMed] [Google Scholar]
- 18.Terry N.A., Walp E.R., Lee R.A., Kaestner K.H., May C.L. Impaired enteroendocrine development in intestinal-specific Islet1 mouse mutants causes impaired glucose homeostasis. The Australian Journal of Pharmacy: Gastrointestinal and Liver Physiology. 2014;307(10):G979–G991. doi: 10.1152/ajpgi.00390.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ye D.Z., Kaestner K.H. Foxa1 and Foxa2 control the differentiation of goblet and enteroendocrine L- and D-cells in mice. Gastroenterology. 2009;137(6):2052–2062. doi: 10.1053/j.gastro.2009.08.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gierl M.S., Karoulias N., Wende H., Strehle M., Birchmeier C. The zinc-finger factor Insm1 (IA-1) is essential for the development of pancreatic beta cells and intestinal endocrine cells. Genes & Development. 2006;20(17):2465–2478. doi: 10.1101/gad.381806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Soyer J., Flasse L., Raffelsberger W., Beucher A., Orvain C., Peers B. Rfx6 is an Ngn3-dependent winged helix transcription factor required for pancreatic islet cell development. Development (Cambridge, England) 2010;137(2):203–212. doi: 10.1242/dev.041673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith S.B., Qu H.-Q., Taleb N., Kishimoto N.Y., Scheel D.W., Lu Y. Rfx6 directs islet formation and insulin production in mice and humans. Nature. 2010;463(7282):775–780. doi: 10.1038/nature08748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Piccand J., Strasser P., Hodson D.J., Meunier A., Ye T., Keime C. Rfx6 maintains the functional identity of adult pancreatic β cells. Cell Reports. 2014;9(6):2219–2232. doi: 10.1016/j.celrep.2014.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pearl E.J., Jarikji Z., Horb M.E. Functional analysis of Rfx6 and mutant variants associated with neonatal diabetes. Developmental Biology. 2011;351(1):135–145. doi: 10.1016/j.ydbio.2010.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suzuki K., Harada N., Yamane S., Nakamura Y., Sasaki K., Nasteska D. Transcriptional regulatory factor X6 (Rfx6) increases gastric inhibitory Polypeptide (GIP) expression in enteroendocrine K-cells and is involved in GIP hypersecretion in high fat diet-induced obesity. Journal of Biological Chemistry. 2013;288(3):1929–1938. doi: 10.1074/jbc.M112.423137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spiegel R., Dobbie A., Hartman C., de Vries L., Ellard S., Shalev S.A. Clinical characterization of a newly described neonatal diabetes syndrome caused by RFX6 mutations. American Journal of Medical Genetics. Part A. 2011;155A(11):2821–2825. doi: 10.1002/ajmg.a.34251. [DOI] [PubMed] [Google Scholar]
- 27.Concepcion J.P., Reh C.S., Daniels M., Liu X., Paz V.P., Ye H. Neonatal diabetes, gallbladder agenesis, duodenal atresia, and intestinal malrotation caused by a novel homozygous mutation in RFX6. Pediatric Diabetes. 2014;15(1):67–72. doi: 10.1111/pedi.12063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Artuso R., Provenzano A., Mazzinghi B., Giunti L., Palazzo V., Andreucci E. Therapeutic implications of novel mutations of the RFX6 gene associated with early-onset diabetes. The Pharmacogenomics Journal. 2014;15(1):49–54. doi: 10.1038/tpj.2014.37. [DOI] [PubMed] [Google Scholar]
- 29.Zegre Amorim M., Houghton J.A.L., Carmo S., Salva I., Pita A., Pereira-da-Silva L. Mitchell-riley syndrome: a novel mutation in RFX6 gene. Case Reports in Genetics. 2015;2015:937201. doi: 10.1155/2015/937201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sansbury F.H., Kirel B., Caswell R., Lango Allen H., Flanagan S.E., Hattersley A.T. Biallelic RFX6 mutations can cause childhood as well as neonatal onset diabetes mellitus. European Journal of Human Genetics: European Journal of Human Genetics. 2015;23(12):1744–1748. doi: 10.1038/ejhg.2015.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skopkova M., Ciljakova M., Havlicekova Z., Vojtkova J., Valentinova L., Danis D. Two novel RFX6 variants in siblings with Mitchell-Riley syndrome with later diabetes onset and heterotopic gastric mucosa. European Journal of Medical Genetics. 2016;59(9):429–435. doi: 10.1016/j.ejmg.2016.08.005. [DOI] [PubMed] [Google Scholar]
- 32.Chandra V., Albagli-Curiel O., Hastoy B., Piccand J., Randriamampita C., Vaillant E. RFX6 regulates insulin secretion by modulating Ca2+ homeostasis in human β cells. Cell Reports. 2014;9(6):2206–2218. doi: 10.1016/j.celrep.2014.11.010. [DOI] [PubMed] [Google Scholar]
- 33.Khan N., Dandan W., Al Hassani N., Hadi S. A newly-discovered mutation in the RFX6 gene of the rare Mitchell-Riley syndrome. Journal of Clinical Research Pediatric Endocrinology. 2016;8(2):246–249. doi: 10.4274/jcrpe.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoshida S., Takakura A., Ohbo K., Abe K., Wakabayashi J., Yamamoto M. Neurogenin3 delineates the earliest stages of spermatogenesis in the mouse testis. Developmental Biology. 2004:447–458. doi: 10.1016/j.ydbio.2004.01.036. [DOI] [PubMed] [Google Scholar]
- 35.El Marjou F., Janssen K.-P., Chang B.H.-J., Li M., Hindie V., Chan L. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis. 2004:186–193. doi: 10.1002/gene.20042. [DOI] [PubMed] [Google Scholar]
- 36.Mellitzer G., Martin M., Sidhoum-Jenny M., Orvain C., Barths J., Seymour P.A. Pancreatic islet progenitor cells in neurogenin 3-yellow fluorescent protein knock-add-on mice. Molecular Endocrinology. 2004;18(11):2765–2776. doi: 10.1210/me.2004-0243. [DOI] [PubMed] [Google Scholar]
- 37.O'Rourke K.P., Dow L.E., Lowe S.W. Immunofluorescent staining of mouse intestinal stem cells. Bio Protocol. 2016;6(4) doi: 10.21769/bioprotoc.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sato T., Vries R.G., Snippert H.J., van de Wetering M., Barker N., Stange D.E. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459(7244):262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 39.Bolstad B.M., Irizarry R.A., Astrand M., Speed T.P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19(2):185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 40.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Research. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith T., Heger A., Sudbery I. UMI-tools: modeling sequencing errors in Unique Molecular Identifiers to improve quantification accuracy. Genome Research. 2017;27(3):491–499. doi: 10.1101/gr.209601.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Satija R., Farrell J.A., Gennert D., Schier A.F., Regev A. Spatial reconstruction of single-cell gene expression data. Nature Biotechnology. 2015;33(5):495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soneson C., Robinson M.D. Bias, robustness and scalability in single-cell differential expression analysis. Nature Methods. 2018;15(4):255–261. doi: 10.1038/nmeth.4612. [DOI] [PubMed] [Google Scholar]
- 46.Haghverdi L., Buettner F., Theis F.J. Diffusion maps for high-dimensional single-cell analysis of differentiation data. Bioinformatics. 2015;31(18):2989–2998. doi: 10.1093/bioinformatics/btv325. [DOI] [PubMed] [Google Scholar]
- 47.Gross S., Garofalo D.C., Balderes D.A., Mastracci T.L., Dias J.M., Perlmann T. The novel enterochromaffin marker Lmx1a regulates serotonin biosynthesis in enteroendocrine cell lineages downstream of Nkx2.2. Development (Cambridge, England) 2016;143(14):2616–2628. doi: 10.1242/dev.130682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Croset M., Rajas F., Zitoun C., Hurot J.M., Montano S., Mithieux G. Rat small intestine is an insulin-sensitive gluconeogenic organ. Diabetes. 2001;50(4):740–746. doi: 10.2337/diabetes.50.4.740. [DOI] [PubMed] [Google Scholar]
- 49.Kellett G.L., Brot-Laroche E., Mace O.J., Leturque A. Sugar absorption in the intestine: the role of GLUT2. Annual Review of Nutrition. 2008;28:35–54. doi: 10.1146/annurev.nutr.28.061807.155518. [DOI] [PubMed] [Google Scholar]
- 50.Lee A.-H. The role of CREB-H transcription factor in triglyceride metabolism. Current Opinion in Lipidology. 2012;23(2):141–146. doi: 10.1097/MOL.0b013e3283508fed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim T.-H., Saadatpour A., Guo G., Saxena M., Cavazza A., Desai N. Single-cell transcript profiles reveal multilineage priming in early progenitors derived from Lgr5+ intestinal stem cells. Cell Reports. 2016;16(8):2053–2060. doi: 10.1016/j.celrep.2016.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bouchi R., Foo K.S., Hua H., Tsuchiya K., Ohmura Y., Sandoval P.R. FOXO1 inhibition yields functional insulin-producing cells in human gut organoid cultures. Nature Communications. 2014;5:4242. doi: 10.1038/ncomms5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Desai S., Loomis Z., Pugh-Bernard A., Schrunk J., Doyle M.J., Minic A. Nkx2.2 regulates cell fate choice in the enteroendocrine cell lineages of the intestine. Developmental Biology. 2008;313(1):58–66. doi: 10.1016/j.ydbio.2007.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zeisel A., Hochgerner H., Lonnerberg P., Johnsson A., Memic F., van der Zwan J. Molecular architecture of the mouse nervous system. Cell. 2018;174(4) doi: 10.1016/j.cell.2018.06.021. 999-1014 e1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Collombat P. Opposing actions of Arx and Pax4 in endocrine pancreas development. Genes & Development. 2003;17(20):2591–2603. doi: 10.1101/gad.269003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bohin N., Carlson E.A., Samuelson L.C. Genome toxicity and impaired stem cell function after conditional activation of CreER(T2) in the intestine. Stem Cell Reports. 2018;11(6):1337–1346. doi: 10.1016/j.stemcr.2018.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.