Abstract
Aim of the study
Chronic hepatitis C (CHC) is a viral disease with metabolic disturbances involved in its pathogenesis. Adipokines may influence the inflammatory response and contribute to development of metabolic abnormalities in CHC. Visfatin exerts immunomodulatory and insulin-mimetic effects. The aim was to measure visfatin serum concentrations and its mRNA hepatic expression in non-obese CHC patients and to assess the relationships with metabolic and histological parameters.
Material and methods
In a group of 63 non-obese CHC patients (29 M/34 F) infected with genotype 1b aged 46.6 ±14.6 years, body mass index (BMI) 24.8 ±3.0 kg/m2, serum visfatin levels and its mRNA hepatic expression were examined and the subsequent associations with metabolic and histopathological features were assessed.
Results
Serum visfatin levels were significantly higher in CHC patients compared to controls (22.7 ±5.7 vs. 17.8 ±1.5 ng/ml, p < 0.001). There was no difference in serum visfatin and its mRNA hepatic expression regardless of sex, BMI, insulin sensitivity and lipids concentrations. There was no mutual correlation between serum visfatin and visfatin mRNA hepatic expression. Hepatic visfatin mRNA levels but not visfatin serum levels were higher in patients with steatosis (1.35 ±0.75 vs. 0.98 ±0.34, p = 0.009).
Conclusions
Serum visfatin levels may reflect its involvement in chronic inflammatory processes accompanying HCV infection. Increased visfatin mRNA hepatic expression in patients with steatosis seems to be a compensatory mechanism enabling hepatocytes to survive metabolic abnormalities resulting from virus-related lipid droplet deposition prerequisite to HCV replication.
Keywords: visfatin, adipokine, chronic hepatitis C, fibrosis, insulin resistance, steatosis, liver
Introduction
Visfatin (pre-B-cell colony-enhancing factor – PBEF) was originally isolated from peripheral blood lymphocytes and identified as the product of a gene expressed in bone marrow stromal cells as well as in activated human lymphocytes [1]. Further studies revealed that this gene encodes a unique 52-kDa secreted protein with the ubiquitous nature as a nicotinamide phosphoribosyltransferase (NAmPRTase, Nampt), a cytosolic enzyme involved in nicotinamide adenine dinucleotide (NAD) biosynthesis [2]. In mammals, Nampt has two different forms: intracellular and extracellular Nampt (iNampt and eNampt, respectively). Whereas the function of iNampt as the NAD biosynthetic enzyme is established, the significance and function of eNampt have been a matter of debate [3]. Results of Revollo et al. demonstrated that eNampt/visfatin-mediated systemic NAD biosynthesis is critical for pancreatic beta cell function, suggesting a vital framework for the regulation of glucose homeostasis [4]. Together with NAD-dependent sirtuin deacetylase, visfatin plays a critical role in the regulation of glucose-stimulated insulin secretion [5]. Moreover, it was shown that visfatin is released from adipocytes [6] and plasma visfatin levels and visfatin expression in adipose tissues and liver are increased in obese subjects [7-10]. New lines of evidence show that apart from its enzymatic activity eNampt also acts as a cytokine, playing a role in immune response regulation [3], angiogenesis [11, 12] and carcinogenesis [13, 14].
Among the major causes of progressive liver diseases and reasons for liver transplantation worldwide is hepatitis C virus (HCV) infection, which leads in the majority of cases to chronic hepatitis C (CHC). This small-enveloped, single-stranded RNA(+) virus creates at least six genotypes with subtypes within genotypes [15]. HCV replication alters host lipid metabolism, leading to hepatic steatosis. Moreover, HCV-dependent insulin resistance development (by up-regulation of cytokine suppressor of cytokine signaling 3 [SOC-3]) and gluconeogenesis (induced through peroxisome proliferator-activated receptor γ coactivator Iα) are phenomena leading to complex metabolic disorders linking HCV infection with a higher risk of diabetes mellitus [16, 17]. Important liver steatosis is documented in genotype 3 HCV infection [18, 19], while growing data indicate genotype 1 to be involved in metabolic disorders [20, 21]. In genotype 3 the steatosis is strongly connected with the virus properties – so-called “viral steatosis” – whereas in genotype 1 it is defined as “metabolic steatosis”, depending on inter alia viral-induced metabolic disorders [22].
Taking into account that chronic HCV infection is associated with metabolic disorders and a specific adipocytokine profile [23, 24], we decided to evaluate visfatin serum concentrations together with visfatin gene liver tissue expression in non-obese CHC patients and assess the potential relationship with metabolic parameters and histological features.
Material and methods
The study included 63 non-obese (body mass index – BMI 24.8 ±3.0 kg/m2) CHC patients (29 M/34 F) aged 46.6 ±14.6 years.
Serum HCV-RNA was assayed with the reverse transcription polymerase chain reaction (RT-PCR) method (Amplicor Roche/Promega v.2 Diagnostic Test, Branchburg, NJ, USA) while virus genotype was assessed by a reverse-hybridization line probe assay (LiPA Versant Test, Milwaukee, WI, USA) and viral load by signal amplification nucleic acid probe assay for the quantitation of human hepatitis C viral RNA (Bayer Versant_ HCV RNA 3.0 Assay (bDNA); Bayer Diagnostics, Berkeley, CA, USA). Only HCV 1b genotype infected patients with persistently elevated alanine aminotransferase (ALT) activity for at least 6 months were included. Infection with other HCV genotypes, hepatitis B virus (HBV) and/or human immunodeficiency virus (HIV) co-infection, drug or alcohol abuse as well as neoplastic, thyroid and psychiatric diseases, diabetes mellitus and renal or heart or liver failure composed the exclusion criteria. All patients were naive according to antiviral treatment. On the day of liver biopsy a single blood sample was drawn in the morning from all patients subjected to fasting. The samples were centrifuged and serum was aliquoted and frozen at –70°C until further processing.
The control group consisted of 30 (15 M/15 F) healthy persons, with BMI ranging between 20 and 25, aged 47.9 ±14.8 years (44.7 ±14.9 and 51.1 ±14.5 years men and women, respectively). A single blood sample in the fasting state was taken in the morning. Liver biopsy was not carried out in the control group.
The study protocol was approved by the Ethical Committee of the Medical University of Silesia in Katowice, Poland, and conformed to the ethical guidelines of the Declaration of Helsinki. Informed consent was obtained for all study participants.
Biochemical and immunoenzymatic assays
Biochemical parameters were measured using routine methods. The upper limit of ALT activity was set at 38 IU/l and AST at 40 IU/l, while gamma-glutamyl-transferase (GGT) activity was set at 50 IU/l and total serum bilirubin concentration at 17 μmol/l. Insulin concentration was measured with the DiaMetra Insulin EIA Kit, Cat. No DKO076 (DiaMetra, Italy). For IR estimation the homeostasis model assessment for IR (HOMA-IR) was calculated by the formula: fasting insulin level (mUI/l) × fasting glucose level (mg/dl)/405 [25]. With respect to the HOMA-IR value, patients were divided into two subgroups – below and equal to or above 2.5. Visfatin serum concentrations were assessed in duplicate by the immunoenzymatic method with the commercially available visfatin EIA kit [Visfatin C-terminal (Human) EIA Kit, Catalogue No. EK-003-80, Phoenix Pharmaceuticals, Inc., USA; minimal detectable concentration 2.83 ng/ml].
Liver histology
Liver biopsy was performed with the Hepafix kit (B. Braun, Melsungen AG, Germany) as part of the diagnostic routine before the antiviral therapy. Tissue samples were immediately divided into a larger part for histopathological examination and a smaller one stabilized in RNAlater (Sigma-Aldrich, St. Louis, USA) and frozen at –70°C for further molecular procedures. Biopsy specimens including at least eleven portal tracts were examined independently by two experienced pathologists. Histopathological features were assessed according to Scheuer’s (necro-inflammatory activity, fibrosis), Brunt’s (steatosis) and Kleiner’s (hepatocyte ballooning) scales [26-28].
Liver tissue visfatin mRNA expression
Total RNA was isolated from liver biopsy specimens of CHC patients using the RNeasy Mini Kit (Qiagen, Hilden, Germany), followed by removing trace amounts of genomic DNA with RNase Free DNase Set (Qiagen, Hilden, Germany). RNA was quantified by measuring the absorbance at 260 and 280 nm (NanoDrop 1000 Spectrophotometer, Thermo Fisher Scientific, Wilmington, USA) and the integrity was assessed by electrophoresis in 1.2% agarose gel, ethidium bromide stained.
RNA isolates were used for cDNA synthesis by reverse transcription (RT) reaction. 2 μg of total RNA was reverse transcribed into cDNA in a total volume 20 μl using the High Capacity RNA-to-cDNA Kit (Applied Biosystems, Foster City, USA) according to the manufacturer’s instructions. Obtained cDNA was used to determine visfatin gene expression level by realtime quantitative PCR (RT-Q-PCR) assay (TaqMan system). TaqMan primers and the probe for the visfatin gene and the housekeeping gene GAPDH were bought as ready to use assays: TaqMan Gene Expression Assays (Hs 00237184_m1for visfatin) and Human GAPD Endogenous Control (FAM/MGB Probe, Non-Primer Limited) for GAPDH (Applied Biosystems, Foster City, USA). RT-Q-PCR for both genes was performed in a volume 20 μl on the ABI PRISM 7300 Real Time PCR Detection System (Applied Biosystems, Foster City, USA). For each run, a RT-Q-PCR mix was prepared on ice containing 10 μl of Applied Biosystems Universal PCR Master Mix, 1 μl of primers and probe mix and 8 μl of H2O (Qiagen, Hilden, Germany). To each well of a 96-well plate, 19 μl of QRT-PCR mix and 1 μl of cDNA samples were added. All PCRs were performed in duplicate. In all amplification reactions, a negative control was also included. Thermal cycling for both visfatin and GAPDH was initiated with an incubation step at 50°C for 2 min, followed by a first denaturation step at 95°C for 10 min, and continued with 40 cycles of 95°C for 15 s, 60°C for 1 min. The standard curves for the housekeeping gene GAPDH and the target gene were generated by serial dilutions of the control cDNA (equivalent to 1 μg of total RNA) in four 2-fold dilution steps. The expression levels of visfatin and GAPDH genes in every sample were determined from the respective standard curve and the visfatin gene expression was divided by the GAPDH gene expression to obtain a normalized target value (relative expression level).
Statistical analysis
The data were presented as mean ±SD. Differences between groups was examined through nonparametric tests (Mann-Whitney or Kruskal-Wallis), linear correlation (Spearman) and logistic regression analysis using the Statistica software version 10.0 (StatSoft Inc. USA). For all the analyses, statistical significance was determined for values of p < 0.05.
Results
Baseline characteristic of study group
The baseline clinical characteristics of the study population and control group are summarized in Table 1. There were no differences between CHC patients and the control group according to BMI and waist circumference. In CHC patients both of these parameters alongside with systolic blood pressure were significantly higher in men compared to women. Activities of ALT (p < 0.001), AST (p < 0.004), GGT (p < 0.04) and concentration of total bilirubin (p < 0.02) were higher in CHC patients. There was no significant difference with respect to the lipid profile between the two studied groups. Although there was no difference in mean fasting insulin or glucose concentration, the HOMA-IR value was significantly increased in CHC patients (p = 0.03).
Table 1.
General characteristics of CHC patients and control group
| Parameter | CHC patients n = 63 | Men n = 29 (46%) | Women n = 34 (54%) | p* | Control group n = 30 | p** |
|---|---|---|---|---|---|---|
| Age (years) | 46.6 ±14.6 | 48.2 ±16.6 | 45.3 ±12.8 | NS | 47.9 ±14.8 | NS |
| Body mass index (kg/m2) | 24.8 ±3.0 | 25.7 ±2.6 | 24.0 ±3.1 | 0.02 | 23.9 ±3.3 | NS |
| Waist circumference (cm) | 87.8 ±11.2 | 87.8 ±11.2 | 79.4 ±12.5 | 0.03 | 85.5 ±8.7 | NS |
| SBP (mmHg) | 135.0 ±18.8 | 135.0 ±18.8 | 125.0 ±12.5 | 0.02 | 115.0 ±10.5 | NS |
| DBP (mmHg) | 79.1 ±8.9 | 79.1 ±8.9 | 80.0 ±9.5 | NS | 77.1 ±6.6 | NS |
| Fasting insulin (μU/l) | 11.6 ±9.6 | 10.6 ±6.3 | 12.4 ±11.8 | NS | 10.0 ±4.5 | NS |
| Fasting glucose (mg/dl) | 93.7 ±16.0 | 95.1 ±16.4 | 92.6 ±15.8 | NS | 80.8 ±9.0 | NS |
| HOMA-IR | 2.9 ±2.7 | 2.5 ±1.3 | 3.4 ±3.5 | NS | 2.0 ±0.5 | 0.03 |
| Total bilirubin (μmol/l) | 14.3 ±8.7 | 15.1 ±4.1 | 13.6 ±11.2 | NS | 9.7 ±3.5 | 0.02 |
| GGT (U/l) | 80.7 ±86.9 | 104.8 ±119.2 | 60.2 ±34.5 | 0.04 | 26.0 ±5.5 | 0.04 |
| ALT (U/l) | 75.4 ±44.5 | 83.2 ±50.8 | 98.8 ±37.7 | NS | 25.6 ±4.3 | 0.001 |
| AST (U/l) | 51.7 ±25.3 | 54.2 ±28.5 | 59.5 ±22.5 | NS | 24.1 ±3.8 | 0.004 |
| Total cholesterol (mg/dl) | 187.8 ±117.1 | 199.5 ±170.6 | 179.4 ±53.8 | NS | 174.6 ±33.2 | NS |
| LDL cholesterol (mg/dl) | 97.6 ±45.6 | 85.0 ±38.3 | 107.6 ±49.0 | NS | 94.5 ±20.2 | NS |
| HDL cholesterol (mg/dl) | 53.7 ±24.6 | 50.3 ±27.8 | 56.0 ±22.3 | NS | 44.0 ±12.8 | NS |
| Triglycerides (mg/dl) | 131.9 ±57.8 | 136.2 ±56.3 | 129.0 ±59.6 | NS | 139.0 ±45.0 | NS |
| CRP (mg/dl) | 1.7 ±1.2 | 1.6 ±1.1 | 1.8 ±1.4 | NS | 1.5 ±1.4 | NS |
| HCV viral load (IU/l) | 2053301 ±364614 | 2781433 ±487426 | 1432246 ±1976597 | NS |
SBP – systolic blood pressure, DBP – diastolic blood pressure
p – men vs. women
p – CHC patients vs. controls
Visfatin serum concentration and hepatic mRNA expression
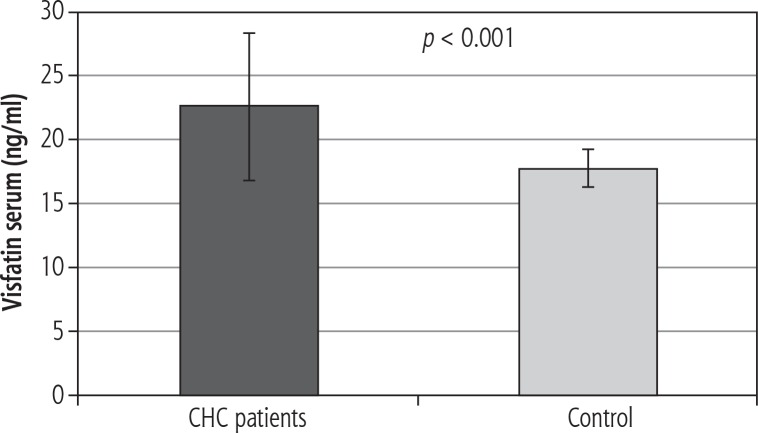
Serum visfatin levels were significantly higher in CHC patients compared to controls (22.7 ±5.7 vs. 17.8 ±1.5 ng/ml, p < 0.001) (Fig. 1). There was no difference in serum visfatin concentration between men and women with CHC (22.8 ±1.1 vs. 22.7 ±1.3 ng/ml).
Fig. 1.
Visfatin serum concentrations in CHC patients and control group
The mean visfatin mRNA expression was 1.17 ±0.09, with no difference between men and women (1.05 ±0.10 vs. 1.30 ±0.13).
Visfatin serum levels and hepatic gene expression in CHC patients according to BMI and HOMA-IR
There were no differences in serum visfatin and its hepatic mRNA expression between CHC patients with different BMI and HOMA-IR. The results are summarized in Table 2.
Table 2.
Serum visfatin concentration and visfatin mRNA in CHC patients according to sex, BMI and HOMA-IR
| Men | Women | CHC patients | ||||
|---|---|---|---|---|---|---|
| BMI < 25 kg/m2 | BMI ≥ 25 kg/m2 | BMI < 25 kg/m2 | BMI ≥ 25 kg/m2 | BMI < 25 kg/m2 | BMI ≥ 25 kg/m2 | |
| Serum visfatin (ng/ml) | 22.40 ±4.21 | 23.19 ±5.75 | 23.59 ±4.53 | 21.44 ±7.71 | 23.07 ±4.36 | 22.31 ±6.76 |
| Liver visfatin/GAPDH | 1.02 ±0.47 | 1.09 ±0.56 | 1.25 ±0.60 | 1.37 ±0.76 | 1.15 ±0.55 | 1.23 ±0.67 |
| p* | NS | NS | NS | NS | NS | NS |
| HOMA-IR < 2.5 | HOMA-IR ≥ 2.5 | HOMA-IR < 2.5 | HOMA-IR ≥ 2.5 | HOMA-IR < 2.5 | HOMA-IR ≥ 2.5 | |
| Serum visfatin (ng/ml) | 21.69 ±5.67 | 25.00 ±5.40 | 20.92 ±5.68 | 24.79 ±8.28 | 21.25 ±5.55 | 24.90 ±6.82 |
| Liver visfatin/GAPDH | 1.10 ±0.40 | 1.17 ±0.70 | 1.49 ±0.90 | 1.15 ±0.49 | 1.32 ±0.74 | 1.16 ±0.59 |
| p* | NS | NS | NS | NS | NS | NS |
p – both for serum visfatin and liver visfatin mRNA
Visfatin serum levels and hepatic gene expression in CHC patients with respect to histopathological parameters
The results of histopathological assessment are presented in Table 3. There was no difference in visfatin serum levels in CHC patients according to consecutive histological features (inflammatory activity, fibrosis and hepatocyte ballooning). Hepatic visfatin mRNA levels were significantly up-regulated in CHC patients with steatosis (p = 0.009), but did not differ between patients with various steatosis grades. The results are also shown in Table 3.
Table 3.
Serum visfatin concentration and visfatin mRNA hepatic expression in CHC patients according to liver histology
| Histopathological results | Number of patients (%) | Serum visfatin [ng/ml] | p | Liver visfatin/GAPDH | p | |
|---|---|---|---|---|---|---|
| Necro-inflammatory activity grade | 1 | 12 (18.8%) | 23.75 ±4.99 | NS | 0.93 ±0.42 | NS |
| 2 | 35 (55.1%) | 21.91 ±5.00 | 1.17 ±0.57 | |||
| 3-4 | 16 (26.1%) | 24.46 ±7.33 | 1.31 ±0.76 | |||
| Fibrosis stage | 1 | 26 (42.0%) | 22.73 ±4.40 | NS | 1.20 ±0.63 | NS |
| 2 | 26 (42.0%) | 22.57 ±6.96 | 1.10 ±0.53 | |||
| 3-4 | 11 (15.9%) | 24.39 ±5.50 | 1.24 ±0.75 | |||
| Steatosis presence | present | 31 (49.3) | 21.87 ±5.31 | NS | 1.35 ±0.75 | 0.009 |
| absent | 32 (50.7%) | 23.95 ±6.00 | 0.98 ±0.34 | |||
| Hepatocyte ballooning grade | 0-1 | 24 (38.4%) | 23.18 ±5.32 | NS | 1.20 ±0.50 | NS |
| 2 | 39 (61.6%) | 22.92 ±6.64 | 1.13 ±0.68 | |||
Relationship between serum visfatin, hepatic visfatin mRNA levels and analyzed parameters
There was no mutual correlation between serum visfatin and hepatic mRNA visfatin levels. Neither serum visfatin nor its hepatic mRNA levels were associated with any of the metabolic parameters.
Discussion
Metabolic disorders have been reported to be closely involved in CHC pathophysiology [29]. As this phenomenon is not a surprise in obese subjects, we decided to focus on non-obese CHC patients. In our study CHC patients and controls showed no differences according to BMI or any lipid profile abnormalities. In non-obese CHC patients analyzed by Chen et al. serum total cholesterol, high-density lipoprotein and low-density lipoprotein levels in HCV infected patients were significantly lower than those of healthy controls whereas serum triglycerides level was significantly elevated [23].
The HOMA-IR index was significantly higher in our CHC group, indicating glucose metabolism disturbances in patients with HCV infection even without type 2 diabetes. This phenomenon was noted in some previous studies [21, 23, 30]. Hsu et al. studying non-obese CHC patients concluded that higher IR is connected with higher HCV viral load [31]. Moreover, IR was found to be higher in those with genotype 1 infection [21, 31]. Shintani et al. in their study using transgenic mice expressing the genotype 1b HCV core protein demonstrated that this viral protein impaired insulin signaling, leading to molecular processes resulting in IR [20]. HCV core protein expression has been shown to alter postreceptor insulin signaling through its interaction with endogenous protein involved in proteasome activation and HCV core protein degradation and insulin receptor substrate-1 turnover [32].
Visfatin was shown to have phosphoribosyltransferase, cytokine and adipokine activities [33]. Its elevated serum concentrations were documented in CHC and correlated with disease severity and the presence of metabolic syndrome [23, 24, 34]. In obese and morbidly obese patients without HCV infection serum visfatin concentrations were significantly higher than in control groups, indicating its important role in metabolic syndrome [7, 9, 10]. Our present study comprised a group of non-obese CHC patients where visfatin serum concentrations were also significantly higher than in the control group. In contrast, in the group of CHC patients analyzed by Wójcik et al. no differences were noted in comparison with healthy controls, while similar to our results, in those patients without metabolic disorders HOMA-IR values were also significantly higher [30]. BMI and HOMA-IR indexes in our patients did not show any correlation with serum visfatin concentrations.
Reports on serum visfatin and histopathological examination results have also brought inconclusive data. In our previous studies the serum visfatin level was negatively associated with necro-inflammatory activity grade [34, 35], while in the current study we could not prove such a connection. Baranova et al. in their analysis of patients with HCV genotype 1 and 3 infected patients also did not observe any relationship between serum visfatin and necro-inflammatory activity [36]. Tsai et al. found higher levels of serum visfatin in cirrhotic CHC patients with hepatocellular carcinoma (HCC) compared to those without liver cancer, concluding that plasma visfatin concentration may serve as an additional tool to identify patients with more advanced liver disease [37]. In the study of Huang et al. serum visfatin levels were significantly higher in patients with advanced fibrosis [24]. However, we did not observe a similar relationship either in this CHC group or in our previously described cohort [34]. Similarly, there was no relationship between serum visfatin concentration and hepatic steatosis.
Previous observations suggested a difference of visfatin serum concentrations and even liver visfatin mRNA expression between men and women [10], but in our group of non-obese CHC patients such differences were not revealed. However, that study described obese women with fatty liver disease, so the comparison is difficult.
In our study no correlation between serum visfatin and its gene expression in liver tissue was observed regardless of inflammatory grading, fibrosis staging and ballooning hepatocyte grade. The only important difference was significantly higher visfatin mRNA expression in liver tissues with steatosis versus those without visible lipid droplets. Serum visfatin concentrations have been reported as showing a lack of genotype-specific differences [24, 35], while the mechanisms involved in hepatocyte steatosis differ between genotypes 1 and 3 [22]. HCV is proved to be an independent factor inducing a line of molecular events in infected hepatocytes. Proteomic data from cultured Huh 7.5 cells suggest that HCV induces perturbations which favor host biosynthetic activities supporting viral replication and propagation followed by a compensatory shift in metabolism aimed at maintaining energy homeostasis and cell viability during elevated viral replication and increasing cellular stress [38]. HCV relies on host lipids and exploits the lipoprotein machineries for almost all steps of its life cycle [39, 40]. The crucial role is ascribed to HCV core protein [41], whose retention and stability are regulated via proteasome activator PA28γ [32]. On the other hand, most of the intracellular functions of visfatin are due to its role as a nicotinamide phosphoribosyltransferase (iNampt) involved in nicotinamide adenine dinucleotide (NAD) biosynthesis [2, 42]. NAD biosynthesis enables cells to preserve their homeostasis in response to significant NAD-consuming events and it can be modulated by various stimuli to induce suitable NAD-mediated metabolic responses [43]. The increased visfatin mRNA hepatic level described in our study may be a compensatory mechanism in response to altered hepatocyte metabolism provoked by viral replication and steatosis.
In summary, the increased serum visfatin levels in CHC patients reflect its extracellular form produced by all cells able to respond this way to different, mostly inflammatory stimuli, induced by HCV infection. Local visfatin mRNA expression in lipid accumulated HCV-infected hepatocytes may indicate the necessity of NAD biosynthesis due to virus-induced metabolic disorders.
The findings presented in this study should be interpreted considering the limitations of using a cross-sectional study design. There were some other, unavoidable limitations. First, our study included relatively small groups of CHC patients, additionally with a small number of patients with advanced fibrosis. Second, our study did not assess patients with obesity, in whom the amount of adipose tissue can influence visfatin serum levels and interfere with the inflammatory process in the liver.
Conclusions
Serum visfatin levels may reflect its involvement in chronic inflammatory processes accompanying HCV infection. Increased visfatin mRNA hepatic expression in patients with steatosis seems to be a compensatory mechanism enabling hepatocytes to survive metabolic abnormalities resulting from virus-related lipid droplet deposition prerequisite to HCV replication.
Disclosure
Authors report no conflict of interest.
References
- 1.Samal B, Sun Y, Stearns G, et al. Cloning and characterization of the cDNA encoding a novel human pre-B-cell colony-enhancing factor. Mol Cell Biol. 1994;14:1431–1437. doi: 10.1128/mcb.14.2.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rongvaux A, Shea RJ, Mulks MH, et al. Pre-B-cell colony-enhancing factor, whose expression is up-regulated in activated lymphocytes, is a nicotinamide phosphoribosyltransferase, a cytosolic enzyme involved in NAD biosynthesis. Eur J Immunol. 2002;32:3225–3234. doi: 10.1002/1521-4141(200211)32:11<3225::AID-IMMU3225>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 3.Garten A, Petzold S, Körner A, et al. Nampt: linking NAD biology, metabolism and cancer. Trends Endocrinol Metab. 2009;20:130–138. doi: 10.1016/j.tem.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Revollo JR, Körner A, Mills KF, et al. Nampt/PBEF/Visfatin regulates insulin secretion in beta cells as a systemic NAD biosynthetic enzyme. Cell Metab. 2007;6:363–375. doi: 10.1016/j.cmet.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Imai S, Kiess W. Therapeutic potential of SIRT1 and NAMPT-mediated NAD biosynthesis in type 2 diabetes. Front Biosci (Landmark Ed) 2009;14:2983–2995. doi: 10.2741/3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tanaka M, Nozaki M, Fukuhara A, et al. Visfatin is released from 3T3-L1 adipocytes via a non-classical pathway. Biochem Biophys Res Commun. 2007;359:194–201. doi: 10.1016/j.bbrc.2007.05.096. [DOI] [PubMed] [Google Scholar]
- 7.Filippatos TD, Derdemezis CS, Kiortsis DN, et al. Increased plasma levels of visfatin/pre-B cell colony-enhancing factor in obese and overweight patients with metabolic syndrome. J Endocrinol Invest. 2007;30:323–326. doi: 10.1007/BF03346300. [DOI] [PubMed] [Google Scholar]
- 8.Varma V, Yao-Borengasser A, Rasouli N, et al. Human visfatin expression: relationship to insulin sensitivity, intramyocellular lipids, and inflammation. J Clin Endocrinol Metab. 2007;92:666–672. doi: 10.1210/jc.2006-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kukla M, Ciupińska-Kajor M, Kajor M, et al. Liver visfatin expression in morbidly obese patients with nonalcoholic fatty liver disease undergoing bariatric surgery. Pol J Pathol. 2010;6:147–153. [PubMed] [Google Scholar]
- 10.Auguet T, Terra X, Porras JA, et al. Plasma visfatin levels and gene expression in morbidly obese women with associated fatty liver disease. Clin Biochem. 2013;46:202–208. doi: 10.1016/j.clinbiochem.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 11.Bae YH, Bae MK, Kim SR, et al. Upregulation of fibroblast growth factor-2 by visfatin that promotes endothelial angiogenesis. Biochem Biophys Res Commun. 2009;379:206–211. doi: 10.1016/j.bbrc.2008.12.042. [DOI] [PubMed] [Google Scholar]
- 12.Kim SR, Bae SK, Choi KS, et al. Visfatin promotes angiogenesis by activation of extracellular signal-regulated kinase 1/2. Biochem Biophys Res Commun. 2007;357:150–156. doi: 10.1016/j.bbrc.2007.03.105. [DOI] [PubMed] [Google Scholar]
- 13.Schuster S, Penke M, Gorski T, et al. FK866-induced NAMPT inhibition activates AMPK and downregulates mTOR signaling in hepatocarcinoma cells. Biochem Biophys Res Commun. 2015;458:334–340. doi: 10.1016/j.bbrc.2015.01.111. [DOI] [PubMed] [Google Scholar]
- 14.Park HJ, Kim SR, Kim SS, et al. Visfatin promotes cell and tumor growth by upregulating Notch1 in breast cancer. Oncotarget. 2014;5:5087–5099. doi: 10.18632/oncotarget.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kupfer B. HCV virology. In: Mauss S, Berg T, Rockstroh J, Sarazzin C, Wedemeyer H, editors. Hepatology 2015. A clinical textbook. 6th ed. 2015. pp. 96–119. [Google Scholar]
- 16.Böhling A, Puchner K-P, Berg T. Hepatology 2017. A clinical textbook. 8th ed. 2017. Extrahepatic manifestations of chronic HCV; pp. 365–390. [Google Scholar]
- 17.Shlomai A, Rechtman MM, Burdelova EO, et al. The metabolic regulator PGC-Iα links hepatitis C virus infection to hepatic insulin resistance. J Hepatol. 2012;57:867–873. doi: 10.1016/j.jhep.2012.06.021. [DOI] [PubMed] [Google Scholar]
- 18.Negro F. Mechanisms and significance of liver steatosis in hepatitis C virus infection. World J Gastroenterol. 2006;12:6756–6765. doi: 10.3748/wjg.v12.i42.6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abid K, Pazienza V, de Gottardi A, et al. An in vitro model of hepatitis C virus genotype 3a-associated triglycerides accumulation. J Hepatol. 2005;42:744–751. doi: 10.1016/j.jhep.2004.12.034. [DOI] [PubMed] [Google Scholar]
- 20.Shintani Y, Fujie H, Miyoshi H, et al. Hepatitis C virus infection and diabetes: direct involvement of the virus in the development of insulin resistance. Gastroenterology. 2004;126:840–848. doi: 10.1053/j.gastro.2003.11.056. [DOI] [PubMed] [Google Scholar]
- 21.Moucari R, Asselah T, Cazals-Hatem D, et al. Insulin resistance in chronic hepatitis C: association with genotypes 1 and 4, serum HCV RNA level, and liver fibrosis. Gastroenterology. 2008;134:416–423. doi: 10.1053/j.gastro.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 22.Del Campo JA, Romero-Gómez M. Steatosis and insulin resistance in hepatitis C: a way out for the virus? World J Gastroenterol. 2009;15:5014–5019. doi: 10.3748/wjg.15.5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen L, Liu W, Lai S, et al. Insulin resistance, serum visfatin, and adiponectin levels are associated with metabolic disorders in chronic hepatitis C virus-infected patients. Eur J Gastroenterol Hepatol. 2013;25:935–941. doi: 10.1097/MEG.0b013e32835fa988. [DOI] [PubMed] [Google Scholar]
- 24.Huang JF, Huang CF, Yu ML, et al. Serum visfatin is correlated with disease severity and metabolic syndrome in chronic hepatitis C infection. J Gastroenterol Hepatol. 2011;26:530–535. doi: 10.1111/j.1440-1746.2010.06438.x. [DOI] [PubMed] [Google Scholar]
- 25.Bonora E, Targher G, Alberiche M, et al. Homeostasis model assessment closely mirrors the glucose clamp technique in the assessment of insulin sensitivity: studies in subjects with various degrees of glucose tolerance and insulin sensitivity. Diabetes Care. 2000;23:57–63. doi: 10.2337/diacare.23.1.57. [DOI] [PubMed] [Google Scholar]
- 26.Scheuer PJ. The nomenclature of chronic hepatitis: time for a change. J Hepatol. 1995;22:112–114. doi: 10.1016/0168-8278(95)80269-x. [DOI] [PubMed] [Google Scholar]
- 27.Brunt EM, Janney CG, Di Bisceglie AM, et al. Nonalcoholic steatohepatitis: a proposal for grading and staging the histological lesions. Am J Gastroenterol. 1999;94:2467–2474. doi: 10.1111/j.1572-0241.1999.01377.x. [DOI] [PubMed] [Google Scholar]
- 28.Kleiner DE, Brunt EM, van Natta M, et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41:1313–1321. doi: 10.1002/hep.20701. [DOI] [PubMed] [Google Scholar]
- 29.Kukla M, Mazur W, Bułdak RJ, Zwirska-Korczala K. Potential role of leptin, adiponectin and three novel adipokines – visfatin, chemerin and vaspin – in chronic hepatitis. Mol Med. 2011;17:1397–1410. doi: 10.2119/molmed.2010.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wójcik K, Jabłonowska E, Omulecka A, Piekarska A. Insulin resistance, adipokine profile and hepatic expression of SOCS-3 gene in chronic hepatitis C. World J Gastroenterol. 2014;20:10449–10456. doi: 10.3748/wjg.v20.i30.10449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hsu CS, Liu CJ, Liu CH, et al. High hepatitis C viral load is associated with insulin resistance in patients with chronic hepatitis C. Liver Int. 2008;28:271–277. doi: 10.1111/j.1478-3231.2007.01626.x. [DOI] [PubMed] [Google Scholar]
- 32.Moriishi K, Okabayashi T, Nakai K, et al. Proteasome activator PA28gamma-dependent nuclear retention and degradation of hepatitis C virus core protein. J Virol. 2003;77:10237–10249. doi: 10.1128/JVI.77.19.10237-10249.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim MK, Lee JH, Kim H, et al. Crystal structure of visfatin/pre-B cell colony-enhancing factor 1/nicotinamide phosphoribosyltransferase, free and in complex with the anti-cancer agent FK-866. J Mol Biol. 2006;362:66–77. doi: 10.1016/j.jmb.2006.06.082. [DOI] [PubMed] [Google Scholar]
- 34.Kukla M, Zwirska-Korczala K, Gabriel A, et al. Visfatin serum levels in chronic hepatitis C patients. J Viral Hepat. 2010;17:254–260. doi: 10.1111/j.1365-2893.2009.01174.x. [DOI] [PubMed] [Google Scholar]
- 35.Kukla M, Berdowska A, Gabriel A, et al. Association between hepatic angiogenesis and serum adipokine profile in non-obese chronic hepatitis C patients. Pol J Pathol. 2011;62:218–228. [PubMed] [Google Scholar]
- 36.Baranova A, Jarrar MH, Stepanova M, et al. Association of serum adipocytokines with hepatic steatosis and fibrosis in patients with chronic hepatitis C. Digestion. 2011;83:32–40. doi: 10.1159/000314592. [DOI] [PubMed] [Google Scholar]
- 37.Tsai IT, Wang CP, Yu TH, et al. Circulating visfatin level is associated with hepatocellular carcinoma in chronic hepatitis B or C virus infection. Cytokine. 2016;90:54–59. doi: 10.1016/j.cyto.2016.10.007. [DOI] [PubMed] [Google Scholar]
- 38.Diamond DL, Syder AJ, Jacobs JM, et al. Temporal proteome and lipidome profiles reveal hepatitis C virus-associated reprogramming of hepatocellular metabolism and bioenergetics. PLoS Pathog. 2010;6:e1000719. doi: 10.1371/journal.ppat.1000719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grassi G, Di Caprio G, Fimia GM, et al. Hepatitis C virus relies on lipoproteins for its life cycle. World J Gastroenterol. 2016;22:1953–1965. doi: 10.3748/wjg.v22.i6.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McRae S, Iqbal J, Sarkar-Dutta M, et al. The Hepatitis C Virus-induced NLRP3 inflammasome activates the sterol regulatory element-binding erotein (SREBP) and regulates lipid metabolism. J Biol Chem. 2016;291:3254–3267. doi: 10.1074/jbc.M115.694059. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41.Koike K, Tsutsumi T, Yotsuyanagi H, Moriya K. Lipid metabolism and liver disease in hepatitis C viral infection. Oncology. 2010;78(Suppl 1):24–30. doi: 10.1159/000315226. [DOI] [PubMed] [Google Scholar]
- 42.Sun Z, Lei H, Zhang Z. Pre-B cell colony enhancing factor (PBEF), a cytokine with multiple physiological functions. Cytokine Growth Factor Rev. 2013;24:433–442. doi: 10.1016/j.cytogfr.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ruggieri S, Orsomando G, Sorci L, Raffaelli N. Regulation of NAD biosynthetic enzymes modulates NAD-sensing processes to shape mammalian cell physiology under varying biological cues. Biochim Biophys Acta. 2015;1854:1138–1149. doi: 10.1016/j.bbapap.2015.02.021. [DOI] [PubMed] [Google Scholar]