Abstract
After human genome sequencing and rapid changes in genome sequencing methods, we have entered into the era of rapidly accumulating genome-sequencing data. This has derived the development of several types of methods for representing results of genome sequencing data. Circular genome visual-ization tools are also critical in this area as they provide rapid interpretation and simple visualization of overall data. In the last 15 years, we have seen rapid changes in circular visualization tools after the de-velopment of the circos tool with 1-2 tools published per year. Herein we have summarized and revisited all these tools until the third quarter of 2018.
Keywords: Circular visualization, Circos, Genomics, Next-generation sequencing, Data modeling, BLAST alignment
1. INTRODUCTION
Genomic data visualization is the hallmark of genetics and genomic studies. With the rapid amplification of genomic data after the year 1995, both prokaryotic and eukaryotic genomic visualization has become the center stage of genome research. This has rapidly faced challenges with the great leap in the next-generation DNA sequencing technologies [1]. With the rapid advancements in whole genome sequencing technologies, there is a massive leap into comparative genomics analyses for various purposes [2]. Hence, there is a need of extensive genomic data visualization methods resolving various biological implications [1]. Visualization approaches are essential method for data modeling, analysis and data representation. It plays a crucial role in the statistical analysis of multi-dimensional genomic data that describes their relationships [3]. However, still, there is a lack of rearrangement of visualization of genome annotation in sequencing. To challenge this problem, genomic circular visualization is a virtual paradigm for comparative genomics dataset in order to view the correlations between the amount of sequencing data and its annotations from high-throughput sequencing technology [3]. It displays a map that shows relationships between genomic intervals. In the last 2 decades or so, there have been rapid improvements in circular data visualization methods (Fig. 1) and tools. Herein, this study reviews the circular visualization of genomic tools in multidimensional genomic big data.
Fig. (1).
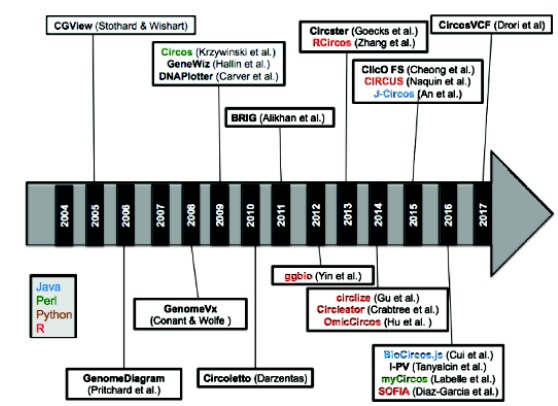
Timeline of genome visualization tools using circular methods or circos-based methods.
2. SUMMARY OF CIRCULAR VISUALIZATION TOOLS
We have summarized all circular visualization tools in Fig. 1 and Table 1.
Table 1. Summary of bioinformatics tools for circular visualization of genomic data.
| Tool | Input | Programming Language* | Website | Refs. |
|---|---|---|---|---|
| Circular Genome Viewer (CGView) | FASTA protein sequences, protein GI, UniProt | J | http://wishart.biology.ualberta.ca/cgview/ | [12] |
| CGView server | Various formats | J | http://stothard.afns.ualberta.ca/cgview_server/ | [28] |
| GView | Various formats | J | https://www.gview.ca/ | [29] |
| GView server | Various formats | J | https://server.gview.ca/ | |
| CGView Comparison Tool (CCT) | Various formats | J | http://stothard.afns.ualberta.ca/downloads/CCT/ | |
| BLAST Ring Image Generator (BRIG) | Various formats | http://sourceforge.net/projects/brig/. | [22] | |
| GenomeDiagram | Various formats | PY | http://bioinf.scri.ac.uk/lp/programs.html | [13] |
| GenomeVx | Flat file of GeneBank | C++ | http://wolfe.ucd.ie/GenomeVx/ | [14] |
| Circos | GFF-style data | PL | http://circos.ca/ | [5] |
| DNAPlotter | Sequence formats | J | https://www.sanger.ac.uk/science/tools/dnaplotter | [15] |
| Circoletto | Fasta format | PL | http://tools.bat.infspire.org/circoletto/ | [4] |
| Circleator | GenBank Sequence Alignment/SAM or BAM format | PL | http://jonathancrabtree.github.io/Circleator/ | [18] |
| RCircos | Specific data frame | R | https://bitbucket.org/henryhzhang/rcircos | [16] |
| Circlize | Specific data frame | R | https://github.com/jokergoo/circlize | [17] |
| OmicCircos | Specified matrix data | R | https://www.bioconductor.org/packages/release/bioc/html/OmicCircos.html | [19] |
| Circular Interactive Layout Converter Free Services (clicO FS) | Three type of file: Karyotype, Data, Configuration file | RU | http://clicofs.codoncloud.com | [10] |
| CIRCUS | SAM, BAM, Annotation, CNV and, variant files. | R | -- | [20] |
| CircosVCF | VCF file | WT | http://www.ariel.ac.il/research/fbl/software | [8] |
| J-Circos | Flat file format | JS | https://sourceforge.net/projects/jcircos/ | |
| BioCircos.js | Various formats | JS | http://bioinfo.ibp.ac.cn/biocircos/ | [11] |
| Interactive Protein Sequence Visualization (I-PV) | Protein sequence, conservation and SNV data | PL | http://i-pv.org/ | [7] |
| SOFIA | Various formats | R | https://cggl.horticulture.wisc.edu/ | [21] |
| Circos for Genomics and Transcriptomics Data Visualization (CGDV) | Various formats | WT | https://cgdv-upload.persistent.co.in/cgdv/ | [24] |
| CiVi | Various formats | PY, JS | http://www.cbs.dtu.dk/services/gwBrowser | [25] |
*Programming Language: J – Java; JS – Javascript; PL – Perl; PY – Python; R – R-programming language; RU – Ruby, WT - Webtool
2.1. Circoletto
Circoletto is a flexible, compatible suite in the rearrangement of genomic sequence for visualization in a comparative analysis written in Perl. It works in close combination with BLAST output and circos. Circoletto acquires either BLAST alignment output or query sequence and database sequence to predict e-value for best local alignment. Circoletto can use cusmtomized annotation files provided by the users with more information of the sequence relations and annotations Circoletto displays the different functional annotations from these files using customizable color codes. A ribbon-like structure is generated that represents sequence alignment calculated by BLAST. Width of the ribbon represents alignment length and bit score is generated in four quartiles represented in a different color pattern of ribbon such as blue represents the worst with below 25% bitscore, green is lower medium bandwidth after 25% bitscore, orange stands for the third position, and finally red represents the best bandwidth between 75%-100% bitscore followed by the black band which placed on the top that represents the best alignment between query sequence and their corresponding database sequence [4].
2.2. Circos
Circos is a command line software written on perl programming language for visualization of genomic sequence and features [5] and this tool is now available in various formats and it is the most commonly used tool for genomic visualization (http://circos.ca/).
This tool helps in constructing the circular plots of establishing a genomic relationship of various kinds and one of such relationship is shown in Fig. 2. Circos have an effective feature that displays genomic variations, sequence alignment, a genomic assembly that focuses on the difference and similarities between genomes [5].
Fig. (2).
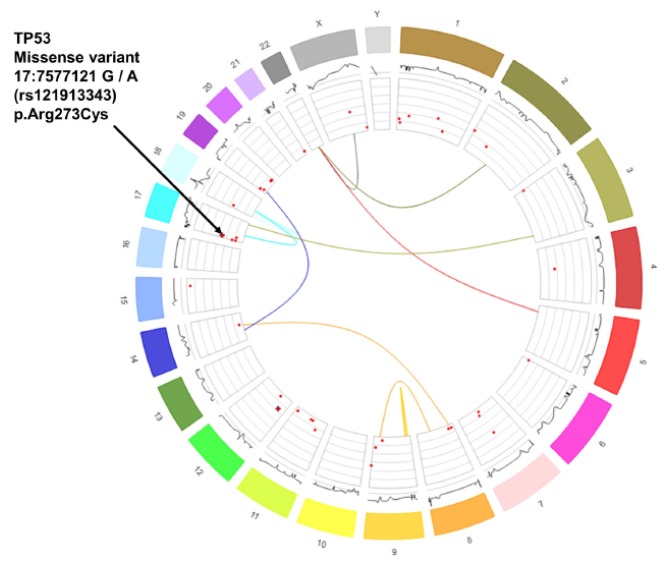
Visualization of whole genome by using Circos [5]. Data taken from the ICGC, specifically pancreatic adenocarcinoma tumor samples from various donors. It shows a number of layers, starting from the outer layer representing color and size distinguished chromosomes, moving clockwise from chromosome 1 to 22, then X and Y. Inside the chromosomes ring, you can see a line representation of copy number mutations. When expanded (by clicking a chromosome), more details on the precise copy number at specific locations are revealed. Hover the bars for further details.
2.3. J-Circos
J-Circos is a javascript-based genomic visualization and is robust in nature that increases the sensitivity of packages in an operating system [6]. Each circle indicates gene expression, gene fusions, and change in chromosomal rearrangement [6]. The observation of arranged in a) color, which is selected by three integer number and are separated by commas, b) chromosomes concatenated one by one according to their size file, c) Beginning of nucleotide position, and d) length that represents UCSC-bigWig/bedgraph/Circos-wiggle/Circos-bridge lines to construct complex interactions [6]. For Translocation and fusion, J-circos generate circos bridge for visualization. In the final output, the outer ring represents the chromosomes, which are bind one to one according to the size of the chromosome [6].
2.4. Interactive Protein Sequence Visualization (i-PV)
Interactive Protein Sequence Visualization (i-PV) is an interactive genomic data visualizer [7]. I-PV uses J-circos for automation ofchecking errors and duplicates and matched against provided input file [6]. i-PV provides information about sequence conservation, amino acid properties and mutational profiles [7]. i-PV assists users interactively in feature extraction with tracking and extraction functions and final output is available as an interactive output HTML window open automatically [7].
2.5. CircosVCF
CircosVCF has an interactive GUI that provides variation information of the genomic using genetic variant data generated from variant call format (vcf) as depicted in Fig. (3) [8].
Fig. (3).
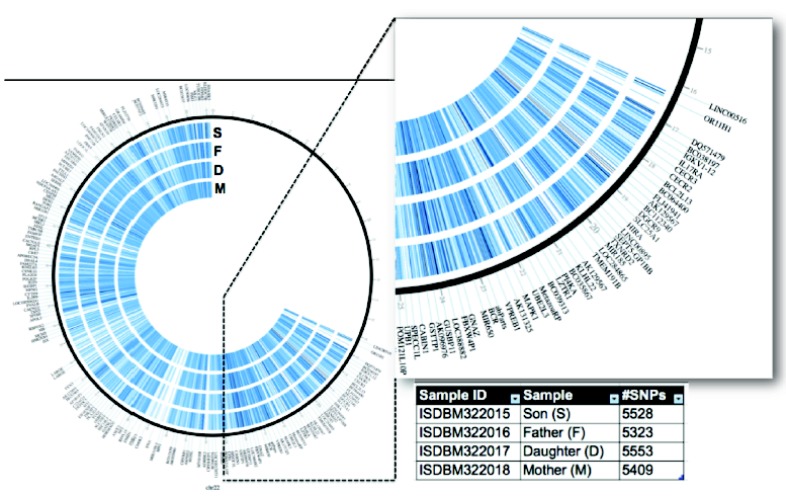
Genetic variant visualization on circus plot using circosVCF [8]. All genetic variants of human chromosome 21 were visualized based on the familial whole genome data from Corpas et al. [27], which included data from a couple and their two children. The region between 14 Mb to 25 Mb of the chromosome was zoomed.
It identifies the SNPs regions as well as calculates the SNPs density in genomic location. c. The darker color on the circos plot represents denser regions based on its genotype where yellow represents the homozygosity to the parent genome, red stands for the homozygosity to the alternative genome, whereas blue stand is indicativelevels of heterozygosity [8].
2.6. Circular Interactive Layout Converter Free Services (clicO FS)
ClicO FS is implemented in Ruby program is a user-friendly web-based service, that allows users to generate a circular ring of genomic data easily [9]. Unlike circos, clicO FS required two input files, namely karyotype file that defines the axes of chromosomes, and data file. There are benefits for the registered users as they can work and store multiple projects in ClicO FS [9]. Currently, Improvised version of ClicO FS supports plugin-based some specific feature such as applying BLAST to comparative genomics data, genetic linkage map data and transcriptome analysis data and generation of circus-like images [9].
2.7. BioCircos.js
BioCircos.js is a flexible and powerful web-based application for the circular visualization. It is implemented on JavaScript is running at the backend of the application. Biocircos.js generates high graphics quality based on D3 (Data-Driven-Documents). This tool provides output of several types like covering mutational hotspots (CNVs, Indel and SNPs), outputs in several formats like heatmap, scatter, and histogram) and depicting several patterns like expression and interactions [10].
2.8. Circular Genome Viewer (CGView)
Circular Genome Viewer (CGView) is a comparative genomics server for circular visualization [11]. It is based on the perl program and heavily added by BLAST-based homology searches. The input genomic sequence file, its BLAST results and GFF files are processed by another perl program using user-defined criteria. It generates an XML file for the CGView map-drawing program and genetic features are mapped into different colors [11]. The maps generated by the CGView Server consist of concentric feature rings as shown in Fig. 4. These rings are used to display gene information read from the primary sequence file, features or analysis results from the GFF files, base composition plots, ORFs, start and stop codons, and BLAST results. CGView colors features according to genetic information types, and in some cases the height of the feature is adjusted to reflect their properties [11].
Fig. (4).
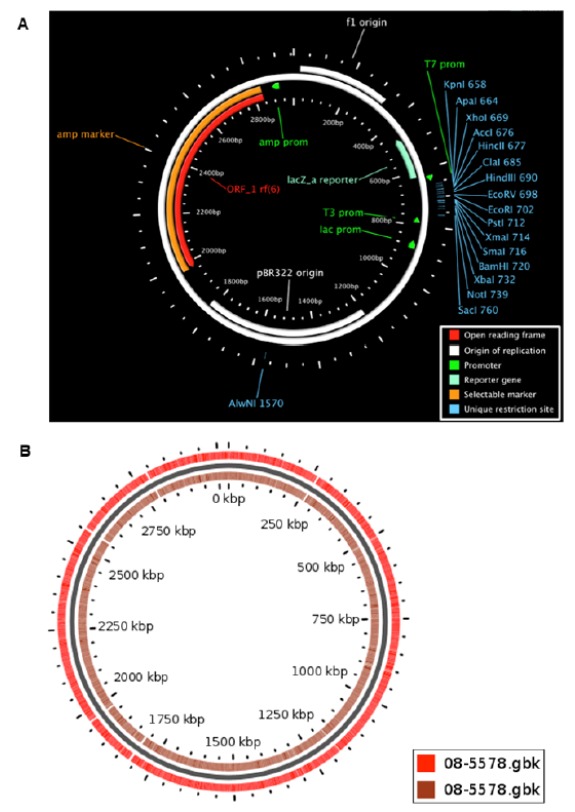
GCview generates images showing the genomic regions that contain the genes of interest. A) The same map rendered at an expanded size and centered on a region of interest, B) A CGView map of the Escherichia coli genome, with genes marked as arrows along with GC content.
2.9. GenomeDiagram
GenomeDiagram is a python-based application for the visualization and comparative analysis of large-scale genomic sequence [12]. This application creates a series of concentric rings of genomic information and these rings possesses genomic features or graphs about the genomic fragments/locations from the reference genome. This application provides different genomic features in different colors to visualization and it uses scalable vector for creating graphs of different types [12]. From this SVG file, the Genome Diagram can convert images to either a static or stream image [12].
2.10. GenomeVx
GenomeVx is a web-based tool implemented in C++ program and the user can itself make changes in circular maps, high-quality graphics for publication, mapping of mitochondrial and chloroplast genomes and of large plasmids as given in Fig. (5).
Fig. (5).
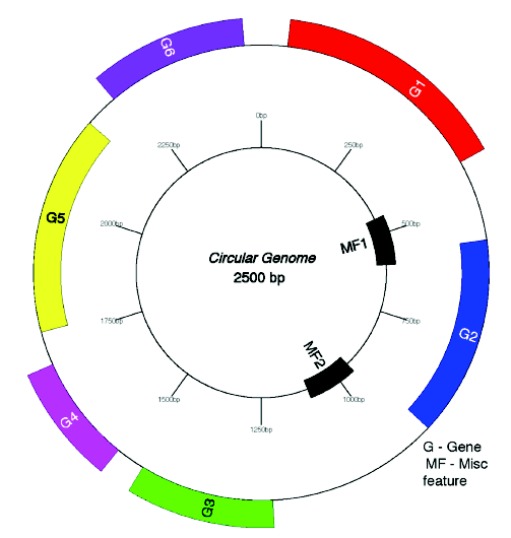
Visualization of different genes by GenomeVX. Genes are colored in groups based on the first two letters of the gene name. A user specified number of evenly spaced scale indices is produced after rounding the genome size to the nearest two digits (thus, the distance between the last label and 0 kb may be greater than between the other markers). GenomeVx can orient the map either anticlockwise from 3 O’clock (the convention for chloroplast genomes) or clockwise from 12 O’clock (the convention for mitochondrial genomes).
The user can import flat file of GenBank and generate output in PDF format. After uploading input file, it generates the list of editable functions, so that users can use the options for editing/changes according to their need. GenomeVx is the simplest tool for the generation genomic visualization without resorting to ad-hoc solutions. It can easily access without the installation of location software [13].
2.11. DNAPlotter
DNAPlotter is a Java based standalone application, with an interactive interface with customized features and module, changes can occur immediately to the circular visualized figure [14]. Several types of data formats can be read by the DNAPlotter aided by the Artemis library [14]. It shows GC contents in the input file by counting the number of GC content and calculates GC screw [(G − C)/(G + C)] value in the form of linear graphs. Genomic sequence use as an input to DNAPlotter and in resultant it shows comparative analysis [14].
2.12. RCircos
RCircos is a flexible Circos-based R-library [15], which generates circular genomic structures with providing several information such as such as chromosomal name and genomic locations [15]. Graphical implementation of RCircos is based on tracking 2D-plot using standard R-plot functionality [15].
2.13. Circlize
Circlize is R-based implementation for generating simplified circular map of genomic data. This is enhanced version of circos visualization use basic graphics of constructing circular map like lines, points. This tool can be easily customized to new types of graphics. Apart from that, data construction or visualization has coherent correlation [16].
2.14. Circleator
Circleator visualization tool implemented in standalone Perl application and produce different figure format for publication and ready circular figures of genomics data. Circleator is highly configured and incorporated into CloVR. It includes predefined composition files and an implementation of the library for well-defined circular visualization that allows creating complex figures without any expertise in programming. Bioperl supported file format that includes GenBank sequence alignment/map and BGZF-compressed (SAM/BAM) alignment files. The generated output consists of genomic variations, gene expression in tab-delimited data and is scored on the basis of e-value [17].
2.15. OmicCircos
OmicCircos is a comprehensive and highly useful bioconductor package for circular genomic data visualization and it produces output of high-quality figures and statistical overviews from wide arrays of data types [18]. OmicCircos generates output of several genomic features, which include point and copy number mutations, expression data, and DNA-methylation patterns [18]. OmicCircos has three main functions as segAnglePo, circos and simcircos, for generation of genomic segmental information, circular graphics and simulation data, respectively [18]. OmicCircos is a user-friendly package with options for track drawing and zooming in and out [18].
2.16. CIRCUS
CIRCUS is a biocondutor package, which is used to analyze genomic sequence for structural variations in high throughput sequencing [19]. The CIRCUS output contains several types of rings for dedicated for chromosomal numbers, genomic fragment information, genomic annotations, read coverage and mutational profiles [19].
2.17. SOFIA
Recently developed tool called SOFIA is a highly flexible in analysis and representations of data generated in different types of studies like linkage mapping, quantitative trait loci (QTL) mapping, association studies, and comparative genomics. This tool can generate different types high-resolution plots [20], which are suitable for publication. This tool runs on Perl native-circos in the R environment. Additionally SOFIA is user-friendly and it only needs basic understanding of programming [20].
2.18. BLAST Ring Image Generator (BRIG)
BLAST-Ring Image Generator (BRIG) is a creates circular data using the BLAST-based alignment process [21]. BRIG computes output using BLAST-based homology detection of the genome of prokaryotes with other published genomes and the simulated draft genomes [21]. The BLAST score matches coloured concentric rings by indicating a defined percentage identity in sequence comparison. BRIG also generates genome assembly information such as read coverage, assembly breakpoints and collapsed repeats In mapping process of sequencing technique, unassembled sequencing reads against more than one parent reference genome which increases ist versatility. BRIG is useful tool, which is easy to learn and use [21].
2.19. ggbio
ggbio is extension of the grammar of graphics approach used by ggplot2 (https://ggplot2.tidyverse.org). It is based on bioconductor library for genomic plots of high throughput sequencing data as given in Fig. (6). It can generate genomic maps in various formats including circular version. In output, it generates detailed description of genomic location information followed by genomic variations [22].
Fig. (6).
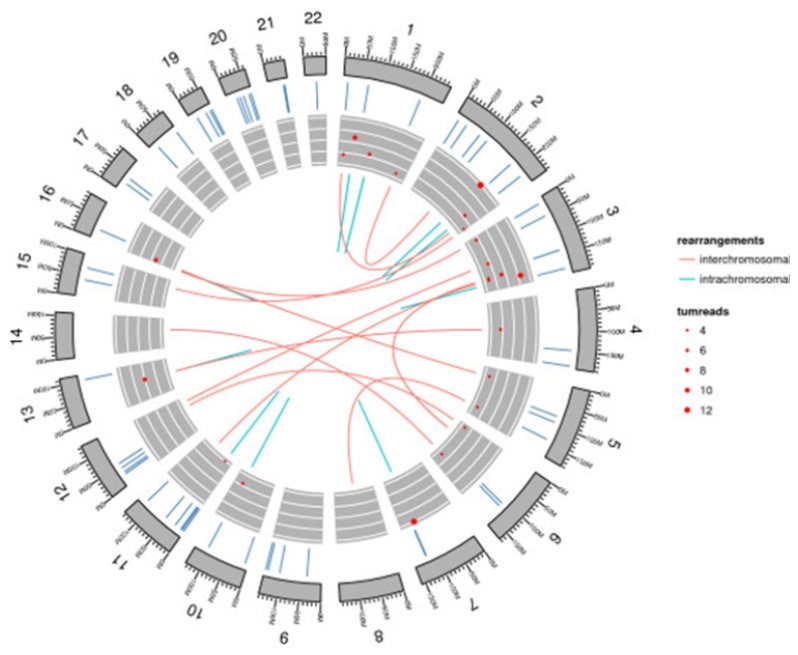
Chromosomal visualization by ggbio. This circular map shows chromosomal segments, which indicates somatic mutations. It also visualizes intermolecular and intramolecular interactions rearrangements.
2.20. Circos for Genomics and Transcriptomics Data Visualization (CGDV)
Circos-based CGDV is a webtool, which is capable of automatic and seamless circular visualization of various large sequencing data including genomics and transcriptomics [23]. CGDV takes inputs as several types of genomic data formats and plots circular results [23]. All intermediate files for generating circos are handled by CGDV in automatically.
2.21. CiVi
CiVi is a simple to use web service tool for generation of circular graph to analyze microbial genomes and annotations of sequence [24]. The generated output comprises of several observational features such as gene name, COG class, PFAM domain, GC content, and subcellular localization can be comprehensively viewed [24]. CiVi depicts several genomic results focusing on three major types as (i) genome-wide distribution (ii) provided experimental data, and (iii) the local orientation and location with respect to neighboring genes as given in Fig. 7. CiVi is a highly useful tool for publication-ready images with minimal training for beginners [24].
Fig. (7).
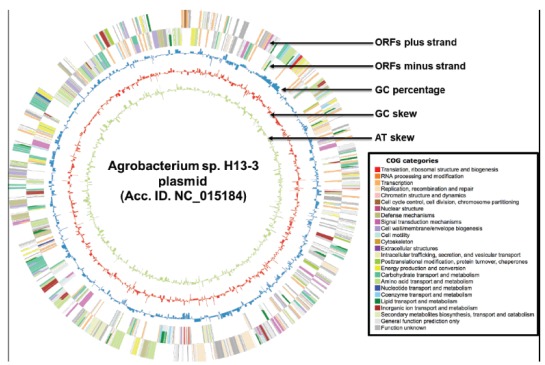
CiVi based circular map of Agrobacterium sp. H13-3 plasmid. There are five rings in generated, which indicates ORFs plus strand (outermost ring), ORFs minus strand, GC content, GC skew, AT skew (innermost ring).
2.22. GeneWiz
GeneWiz is an interactive web-based genomic application for circular depiction of genomic data using genomic alignments of homologous segments [25]. Furthermore, it can easily calculate phenotypic features of DNA such as curvature or stacking energy along the chromosome. GeneWiz is user-friendly application with providing users to select various genomic data, ranges and features and also options to change color and zooming settings [25].
2.23. Circster
Circster is a web-based interactive Circos-style genome visualizer [26]. It is user-friendly, GUI based and requires no programming language skills [27]. It is developed by Galaxy-team and it implemented into Galaxy genomics workbench (http://galaxyproject.org). Hence, it is able to use Galaxy framework and its features in visualization process.
2.24. myCircos.js
myCircos is a web-based application written in perl for visualization or generation of circos plots and only formatted data files are required as an input. Mycircos have many features of database, which is repository of previously generated plots and generates into interactive SVG format.
CONCLUSION
All in all, we have described here genomic visualization tools for both prokaryotic genomes as well as eukaryotic genomes. In the last 15 years, we have seen revolutionary changes in genomic generation with advancement in genomic technologies. This has led into rapid development of circular visualizations tools with average 1-2 new tools per year developed for this purpose. This review provided a comprehensive summary of these tools and it will be useful for others to choose circular visualization tools for their purposes. These genomic visualization tools help a biologist with various genomic information, necessary for inferring conclusions related with genetics, molecular biology and biotechnology.
ACKNOWLEDGEMENTS
Declared none.
CONSENT FOR PUBLICATION
Not applicable.
AUTHORS’ CONTRIBUTIONS
Conceptualization, Abhishek Kumar; writing—original draft preparation, Alisha Parveen, Abhishek Kumar and Sukant Khurana; writing—review and editing, Alisha Parveen, Abhishek Kumar and Sukant Khurana; visualization, Alisha Parveen, Abhishek Kumar; supervision, Abhishek Kumar and Sukant Khurana.
FUNDING
None.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Koboldt D.C., Steinberg K.M., Larson D.E., Wilson R.K., Mardis E.R. The next-generation sequencing revolution and its impact on genomics. Cell. 2013;155(1):27–38. doi: 10.1016/j.cell.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dark M.J. Whole-genome sequencing in bacteriology: State of the art. Infect. Drug Resist. 2013;6(1):115–123. doi: 10.2147/IDR.S35710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stothard P., Grant J.R., Van Domselaar G. Visualizing and comparing circular genomes using the CG View family of tools. Brief. Bioinform. 2017;346(6215) doi: 10.1126/science.1256846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Darzentas N. Circoletto: Visualizing sequence similarity with Circos. Bioinformatics. 2010;26(20):2620–2621. doi: 10.1093/bioinformatics/btq484. [DOI] [PubMed] [Google Scholar]
- 5.Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009;19(9):1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.An J., Lai J., Sajjanhar A., Batra J., Wang C., Nelson C.C. J-Circos: An interactive Circos plotter. Bioinformatics. 2015;31(9):1463–1465. doi: 10.1093/bioinformatics/btu842. [DOI] [PubMed] [Google Scholar]
- 7.Tanyalcin I., Al Assaf C., Gheldof A., Stouffs K., Lissens W., Jansen A.C. I-PV: A CIRCOS module for interactive protein sequence visualization. Bioinformatics. 2016;32(3):447–449. doi: 10.1093/bioinformatics/btv579. [DOI] [PubMed] [Google Scholar]
- 8.Drori E., Levy D., Smirin-Yosef P., Rahimi O., Salmon-Divon M. CircosVCF: Circos visualization of whole-genome sequence variations stored in VCF files. Bioinformatics. 2017;33(9):1392–1393. doi: 10.1093/bioinformatics/btw834. [DOI] [PubMed] [Google Scholar]
- 9.Cheong W.H., Tan Y.C., Yap S.J., Ng K.P. ClicO FS: An interactive web-based service of Circos. Bioinformatics. 2015;31(22):3685–3687. doi: 10.1093/bioinformatics/btv433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cui Y., Chen X., Luo H., Fan Z., Luo J., He S., Yue H., Zhang P., Chen R. BioCircos.js: An interactive Circos JavaScript library for biological data visualization on web applications. Bioinformatics. 2016;32(11):1740–1742. doi: 10.1093/bioinformatics/btw041. [DOI] [PubMed] [Google Scholar]
- 11.Grin I., Linke D. GCView: the genomic context viewer for protein homology searches. Nucleic Acids Res. 2011;39(1):353–356. doi: 10.1093/nar/gkr364. (Web Server issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pritchard L., White J.A., Birch P.R., Toth I.K. GenomeDiagram: a python package for the visualization of large-scale genomic data. Bioinformatics. 2006;22(5):616–617. doi: 10.1093/bioinformatics/btk021. [DOI] [PubMed] [Google Scholar]
- 13.Conant G.C., Wolfe K.H. GenomeVx: Simple web-based creation of editable circular chromosome maps. Bioinformatics. 2008;24(6):861–862. doi: 10.1093/bioinformatics/btm598. [DOI] [PubMed] [Google Scholar]
- 14.Carver T., Thomson N., Bleasby A., Berriman M., Parkhill J. DNAPlotter: Circular and linear interactive genome visualization. Bioinformatics. 2009;25(1):119–120. doi: 10.1093/bioinformatics/btn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang H., Meltzer P., Davis S. RCircos: An R package for Circos 2D track plots. BMC Bioinformatics. 2013;14(1):244. doi: 10.1186/1471-2105-14-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gu Z., Gu L., Eils R., Schlesner M., Brors B. Circlize Implements and enhances circular visualization in R. Bioinformatics. 2014;30(19):2811–2812. doi: 10.1093/bioinformatics/btu393. [DOI] [PubMed] [Google Scholar]
- 17.Crabtree J., Agrawal S., Mahurkar A., Myers G.S., Rasko D.A., White O. Circleator: Flexible circular visualization of genome-associated data with BioPerl and SVG. Bioinformatics. 2014;30(21):3125–3127. doi: 10.1093/bioinformatics/btu505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu Y., Yan C., Hsu C.H., Chen Q.R., Niu K., Komatsoulis G.A., Meerzaman D. OmicCircos: A simple-to-use R package for the circular visualization of multidimensional omics data. Cancer Inform. 2014;13(1):13–20. doi: 10.4137/CIN.S13495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Naquin D., d’Aubenton-Carafa Y., Thermes C., Silvain M. CIRCUS: A package for Circos display of structural genome variations from paired-end and mate-pair sequencing data. BMC Bioinformatics. 2014;15(1):198. doi: 10.1186/1471-2105-15-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diaz-Garcia L., Covarrubias-Pazaran G., Schlautman B., Zalapa J. SOFIA: An R package for enhancing genetic visualization with circos. J. Hered. 2017;108(4):443–448. [Google Scholar]
- 21.Alikhan N.F., Petty N.K., Ben Zakour N.L., Beatson S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genomics. 2011;12(1):402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yin T., Cook D., Lawrence M. ggbio: An R package for extending the grammar of graphics for genomic data. Genome Biol. 2012;13(8):77. doi: 10.1186/gb-2012-13-8-r77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jha V., Singh G., Kumar S., Sonawane A., Jere A., Anamika K. CGDV: A webtool for circular visualization of genomics and transcriptomics data. BMC Genomics. 2017;18(1):823. doi: 10.1186/s12864-017-4169-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Overmars L., van Hijum S.A., Siezen R.J., Francke C. CiVi: circular genome visualization with unique features to analyze sequence elements. Bioinformatics. 2015;31(17):2867–2869. doi: 10.1093/bioinformatics/btv249. [DOI] [PubMed] [Google Scholar]
- 25.Hallin P.F., Staerfeldt H.H., Rotenberg E., Binnewies T.T., Benham C.J., Ussery D.W. GeneWiz browser: An interactive tool for visualizing sequenced chromosomes. Stand. Genomic Sci. 2009;1(2):204–215. doi: 10.4056/sigs.28177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goecks J., Eberhard C., Too T., Galaxy T., Nekrutenko A., Taylor J. Web-based visual analysis for high-throughput genomics. BMC Genomics. 2013;14(1):397. doi: 10.1186/1471-2164-14-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Corpas M., Valdivia-Granda W., Torres N., Greshake B., Coletta A., Knaus A., Harrison A.P., Cariaso M., Moran F., Nielsen F., Swan D., Weiss Solis D.Y., Krawitz P., Schacherer F., Schols P., Yang H., Borry P., Glusman G., Robinson P.N. Crowdsourced direct-to-consumer genomic analysis of a family quartet. BMC Genomics. 2015;16(1):910. doi: 10.1186/s12864-015-1973-7. [DOI] [PMC free article] [PubMed] [Google Scholar]


