Abstract
Adaptive convergent evolution, which refers to the same or similar phenotypes produced by species from independent lineages under similar selective pressures, has been widely examined for a long time. Accumulating studies on the adaptive convergent evolution have been reported from many different perspectives (cellular, anatomical, morphological, physiological, biochemical, and behavioral). Recent advances in the genomic technologies have demonstrated that adaptive convergence can arise from specific genetic mechanisms in different hierarchies, ranging from the same nucleotide or amino acid substitutions to the biological functions or pathways. Among these genetic mechanisms, the same amino acid changes in protein-coding genes play an important role in adaptive phenotypic convergence. Methods for detecting adaptive convergence at the protein sequence level have been constantly debated and developed. Here, we review recent progress on using genomic approaches to evaluate the genetic mechanisms of adaptive convergent evolution, summarize the research methods for identifying adaptive amino acid convergence, and discuss the future perspectives for researching adaptive convergent evolu-tion.
Keywords: Convergent evolution, Phenotype, Genomics, Genetic mechanism, Amino acid convergence, Adaptive evolution
1. INTRODUCTION
Convergent evolution is the independent evolution of the same or similar phenotypic traits produced by species from different evolutionary lineages [1]. Different species experience similar selective pressures from environmental conditions, and thus often produce convergent solutions to cope with similar evolutionary problems; studying convergence enables us to understand repeatability and predictability in evolution [2]. Adaptive convergent evolution can be affected by a variety of genetic and demographic factors, such as population size, gene flow, sources of adaptive alleles, and ancestral starting point [2, 3]. The adaptive phenotypic convergence mainly presents cellular, anatomical, morphological, physiological, biochemical, and behavioral similarity, which displays clearly hierarchical levels [2]. The adaptive phenotypic convergence is common in many animal and plant species [4-15]. For example, birds, bats, and insects possess analogous structures wings to independently evolve the capacity of flying [6, 7]. Other classic examples are the echolocating system of bats and marine mammals [8-11], insect and mammalian audition [12], red and green color vision in vertebrates [13], and C4 photosynthesis [14]. With technological breakthroughs in molecular biology, particularly the development and cost reduction of next-generation sequencing technology, studies on the genetic mechanisms of adaptive convergence have expanded and increased rapidly [16]. Additionally, the launch of a series of genome projects, such as EBP (the Earth BioGenome Project), 10KP (10,000 Plant Genomes Project, https://db.cngb.org/10kp/), VGP (Vertebrate Genomes Project, https://vertebrategenomesproject.org/), B10K (Bird 10,000 Genomes Project, https://b10k.genomics.cn/), Bat1K (Bat1K Project, http://bat1k.ucd.ie/), GAGA (Global Ant Genomics Alliance, http://antgenomics.dk/), i5k (Sequencing Five Thousand Arthropod Genomes, http://i5k.github.io/) and 1KITE (1K Insect Transcriptome Evolution project, http://1kite.org/), has resulted in large-scale genome-wide data, providing rich genomic resources for addressing fundamental questions about adaptive convergent evolution. These data also enable studies of adaptive convergent evolution to transition from hypothesis-driven analyses of candidate genes to discovery-driven examination on a genomic scale. The former method is only used to infer adaptive evolution for a few genes which have been assumed to be potentially related to adaptive convergence, while the latter method can allow the discovery of more candidate convergent genes, as well as further classify these genes according to their functions or detect gene-gene interactions in specific pathways.
The diversity of beneficial genetic changes fuels convergent adaptation [17]. The research on convergent molecular mechanisms has increasingly elucidated that adaptive convergence is hierarchical and may occur at different hierarchical levels: amino acid substitutions, protein-coding genes, non-coding regions, gene families, and biological functions or pathways. Among these, numerous studies have demonstrated that the same amino acid changes in protein-coding genes play crucial roles in adaptive phenotypic convergence [8-11, 13, 18, 19]. Zhang (2006) proposed four criteria to shed light on the adaptive molecular convergence [20]. Furthermore, methods for detecting adaptive amino acid convergence have been constantly debated, developed, and updated at the genome-wide level [21-27]. Additionally, genomic studies of many mammals and plants have detected this type of adaptive convergent signal [26-33]. However, studies have indicated that adaptive convergence is more general and pervasive at higher hierarchical levels, such as occurring in the same proteins, non-coding regions, gene families and even biological functions or pathways, other than the same changes at the same amino acid sites [3]. The extent of adaptive convergence largely reflected in the hierarchical level may be related to the phylogenetic level. Closely related species at an appropriate scale tend to share more similar environments with more similar genetic backgrounds, enabling them to use similar solutions (e.g. the same amino acid substitutions) to achieve adaptive convergent phenotypes. In contrast, relatively distant relatives may not show adaptive convergent changes at the protein sequence level but are more likely to show adaptive convergence at functions or pathways [2, 3].
The meaning and usage of two important terms (convergence and parallelism) in convergent evolution have been debated for many years. Several distinct criteria have been proposed to distinguish between them, considering whether homoplastic phenotypes are structurally corresponding, occur in closely related species, have the same ancestral characteristics, and are caused by similar genetics and, if so, can be identified as parallelism [34]. Additionally, it has also been suggested that convergence is used to describe a phenotypic pattern, while parallelism describes a shared molecular explanation [2, 34]. Here, we do not distinguish between convergence and parallelism, but rather consider both phenotypic convergence and molecular convergence. Molecular convergence may result in phenotypic convergence [35]. In this review, we focus on the molecular mechanisms of adaptive phenotypic convergence independently produced by species from different lineages. Furthermore, we summarize some methods for evaluating adaptive amino acid convergence at the protein sequence level and their improvements. Finally, we provide an outlook for future research of adaptive convergent evolution.
2. GENETIC MECHANISMS OF ADAPTIVE CONVERGENT EVOLUTION
Here, we present several mechanisms of the adaptive molecular convergence, research cases, as well as research advances.
2.1. Adaptive Convergence at Specific Sites
Site-specific variations include nucleotide changes in coding and non-coding sequences. Nonsynonymous mutations, which result in amino acid substitutions in protein-coding regions, are more likely to bring about spatial structure changes in proteins, in turn causing functional changes in the protein, such as higher or lower stability or affinity for substrates. Thus, the same amino acid substitutions may provide important opportunities for producing adaptive convergent phenotypes.
Hemoglobin proteins (Hb), as the most classic example, played a major role in the convergent adaptive evolution of vertebrate species faced with high-altitude hypoxic conditions. The convergent amino acid changes at some specific sites of hemoglobin contribute to convergent increases in Hb-O2 affinity in multiple high-altitude taxa. In a study of the hemoglobin evolution of Andean hummingbirds in high- and low-altitude environments, Projecto-Garcia et al. (2013) revealed two repeated amino acid replacements at β13 and β83 (both Gly→Ser), which may have directed convergent evolutionary changes in Hb-O2 affinity increases in high-altitude species with maximum elevational ranges of >3,000 m from different phylogenetical clades [36]. A similar study of the molecular evolution of hemoglobin for tit and long-tailed tit species from high-altitude Qinghai-Tibet Plateau (QTP) and the mountains of Southwest China, as well as low-altitude areas of eastern China, showed the same amino acid substitution at αA34 (Ala→Thr) in Parus humilis and Lophophanes dichrous and the same amino acid substitution at αA119 (Pro→Ala) in Aegithalos bonvaloti and Anser indicus. Both substitutions were predicted to increase Hb-O2 affinity through opposite allosteric regulation mechanisms [37].
In addition to the hemoglobin, dozens of the non-neutral convergent amino acid changes in 13 mitochondrial genes of snakes and agamid lizards [21], as well as several convergent amino acid substitutions in ATPα1 of herbivorous insect taxa [38] and Prestin of echolocating mammals [8-11] were also detected. Except for studies of adaptive convergent evolution of these few functional genes, with the development and progress in next-generation sequencing technology and its cost reduction, comparative genomic and transcriptomic analyses have been widely used in studies of adaptive convergent evolution for many species on a large scale. For example, studies employing these omics approaches, see studies of echolocation in bats and whales [33], marine mammals adapted to aquatic environments [28], bamboo-eating giant and red pandas with specialized bamboo diets and pseudothumbs [29], snub-nosed monkeys adapted to high-altitude habitats [31], mangroves adapted to aquatic environments [27], and pitcher plants adapted to carnivorous digestion [30]. All of these studies independently detected convergent amino acid substitutions in specific genes. Some substitutions were found by the population genetics studies to be fixed and were inferred to play a role in adaptive convergent adaptation according to functional assays [31].
2.2. Adaptive Convergence at Specific Genes, Non-Coding Regions and Gene Families
Although convergence at specific sites in many cases played a key role in the adaptive phenotypic convergence, other studies demonstrated that mutations at different sites in specific genes and gene families were involved in the adaptive phenotypic convergence [39]. Jawed vertebrates (gnathostomes) and jawless vertebrates (agnathans) used different convergent precursor globin proteins to transport and store O2, respectively [18]. Anser indicus and Chloephaga melanoptera employed substitutions in different subunits of hemoglobin to achieve convergent increases in Hb-O2 affinity [40]. Similarly, high-altitude parid and aegithalid species from the QTP were more likely to utilize divergent amino acid substitutions in hemoglobin to produce similar increases in Hb-O2 affinity [37]. Additionally, a comparative analysis of convergent evolution for hemoglobin function in eight pairs of high- and low-altitude waterfowl taxa indicated that convergent protein function did not require the same amino acid substitutions [41]. Candidate genes under positive selection or with similar evolutionary rate changes also suggested adaptive convergence [42-44]. Two positively selected genes (UBE2D1 and NBN) related to the response to hypoxia and DNA damage repair were commonly identified in poikilothermic animals, reflecting convergent adaptation to the high-altitude environment [42], while hundreds of genes with convergent shifts under selective pressure occurred in all three marine mammals adapted to aquatic conditions [43]. Similarly, Castiglione et al. (2017) demonstrated that the convergent visual systems of Himalayan and Andean catfishes resulted from independent but similar changes in the evolutionary rate of rhodopsin-coding sequences [44].
Most studies of adaptive convergent evolution have focused attention on mutations found in protein-coding sequences, whereas those found in non-coding regions, such as cis-regulatory mutations, have usually produced fewer pleiotropic effects with greater evolutionary flexibility, in addition to possibly regulating gene expression [45]. Thus, non-coding regions may be promising targets for the study of adaptive convergent evolution and changes in these regions should not be overlooked. Vavouri et al. (2007) observed that the highly conserved non-coding elements associated with the developmental regulatory genes evolved in parallel in worms, flies and vertebrates [46]. Parallel changes in cis-regulatory enhancers of the Shavenbaby resulted in morphological convergence between Drosophila sechellia and D. ezoana lineages [47]. Optix cis-regulatory mutations were responsible for a red wing pattern across multiple Heliconius species [48], while ebony cis-regulatory ones led to convergent male-specific pigmentation across multiple Drosophila species [49]. A recent genomic study also suggested that cis-regulatory elements may contribute to convergent phenotypic evolution [50]. Sackton et al. (2018) concluded that regulatory regions, such as conserved non-exonic elements, were highly associated with flightlessness across multiple palaeognathous birds [51].
Furthermore, gene families can be another powerful source of adaptive convergence. Specific gene families can undergo expansion and contraction when species suffer from extreme environmental stressors, such as high-altitude hypoxia [52]. The recurrent recruitment of NADP-me-IV in the NADP-me gene family caused convergence of C4 photosynthesis [14]. The repeated deployment of Zn-finger transcription factor (TF) family member, Zn-cluster TFs, has been found to facilitate the convergent evolution of yeast phenotypes in five different clades [53]. The N-methyltransferase gene family contributed to the convergent evolution of caffeine production in cacao, tea, and coffee [54].
2.3. Adaptive Convergence in Biological Functions or Pathways
An increasing number of studies have revealed a low proportion of shared specific genes or gene families, but a higher proportion of shared functions or pathways across different species. They suggested that conserved biological functions, i.e., function-level convergence of the same GO terms and KEGG pathways may underlie adaptive phenotypic convergence in species. For function-level convergence, the species were not constrained to use the same set of genes which exhibited positive selection or similar evolutionary rate shifts, but rather followed diverse trajectories with greater evolutionary flexibility. Thus, function-level convergence can play a powerful role in adaptive convergent adaptation. This type of evolutionary pattern was widely observed in many animal and plant species. Respective microbial genes related to volatile fatty acids and methane-yielding pathways were found in the high-altitude yak and Tibetan sheep [15]. Extensive overlapping functional categories related to high-altitude adaptation with accelerated evolutionary rates were identified in two highland poikilothermic animals [42]. Eusocial insects from different lineages adopted different genes but conserved metabolic pathways to evolve adaptive caste phenotypes [55]. Groundsel also recruited different genes but similar biochemical pathways to adapt to the environment [56]. Similarly, frog and lizard species groups along altitudinal gradients showed convergent and continuous adaptation to the high-altitude environment by sharing numerous similar functions, such as DNA repair and energy metabolism [57]. Convergent evolution in high-altitude environments has commonly suggested that these functions together with oxygen utilization were generally used to cope with hypoxia, hypothermia, and strong ultraviolet radiation across reptiles, amphibians, fishes, birds, mammals, and humans [52, 57-62].
3. RESEARCH METHODS OF ADAPTIVE AMINO ACID CONVERGENCE IN PROTEIN SEQUENCES
Foote et al. (2015) found that molecular convergence was relatively common for both adaptive and neutral substitutions, while molecular convergence linked to adaptive phenotypic convergence was found to be important but comparatively rare [28]. In a study of thylacine and canids having adapted extraordinary phenotypic indistinguishability, it was shown that amino acid convergence was largely in accordance with the neutral theory of molecular evolution [50]. Therefore, it is essential to distinguish adaptive convergence from non-adaptive convergence that results from stochastic processes. Methods for detecting adaptive amino acid convergence at the protein sequence level have been constantly debated, developed, and updated because of the random and false convergence unrelated to adaptation [21-27]. Here, we enumerate several methods for detecting adaptive convergence.
3.1. Data Preparation and Preprocessing
First, we must determine whether our research system is ideal to investigate adaptive convergent evolution. In an ideal system, it should at least include two focal species with convergent phenotypes or other similar traits and several control species without these characteristics. Once we confirm our study subject, we need to check whether the genomes or transcriptomes of these species are available. If they are available, we can download these data from NCBI, Ensembl or other databases; otherwise, we need to perform whole-genome sequencing or RNA sequencing. After obtaining all of the required data, we can conduct a series of data preprocessing to obtain well-aligned nucleotide and amino acid sequences across all species in the research system. Details of data preprocessing can be found in Fig. (1) [63-73].
Fig. (1).
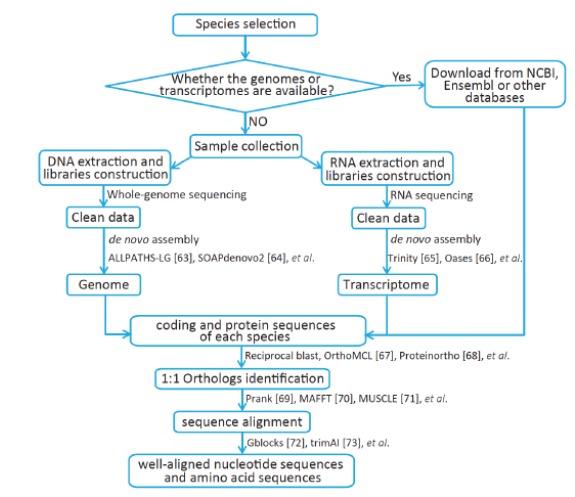
Pipeline of data preparation and preprocessing for identifying amino acid convergence.
3.2. Comparison of Sitewise Log-likelihood Support (ΔSSLS)
In this method, Castoe et al. (2009) applied Maximum Likelihood (ML) phylogenetic reconstruction to obtain site-wise log-likelihood support (SSLS) [21]. First, amino acid sequences of orthologous alignments from all species are obtained. Second, the null hypothesis (H0, phylogeny supports the known and commonly accepted species tree topology) and alternative hypothesis [H1, phylogeny supports the clustering of species that have target traits (shown in red), i.e., falsely monophyletic clustering, indicating convergence] are posited to detect genome-wide convergence (Fig. 2a). The goodness-of-fit (SSLS) of these two phylogenetic hypotheses is obtained for each amino acid site of all orthologous genes using ML. The difference in SSLS between H0 and H1 is calculated at each site [ΔSSLS (H1) = SSLS (H0) – SSLS (H1)]. Third, the ratio of convergent/divergent substitutions is used to detect whether the pair of focal lineages exhibits more convergence than other pairs in the “background” lineages [21, 24]. Non-neutral convergent amino acid substitutions are determined by comparing with empirical and model-based expectations of convergence. This method was used to study convergent evolution between snakes and agamid lizards at the mitochondrial genome level [21], Parker et al. (2013) extended it to the whole-genome level [22]. But unlike in the former example, the average difference in SSLS between H0 and H1 is calculated for each orthologous gene. An average ΔSSLS (H1) value of the orthologous gene below 0 suggests a signature of molecular convergence. In particular, the expected ΔSSLS distributions are produced by simulations, and the difference between the observed and expected mean ΔSSLS is compared to confirm the significance of convergent signals by randomization analysis. However, after Parker and his colleagues used their method to study convergent evolution in echolocating bats and dolphins, two subsequent studies criticized this work because of the many random and false convergences determined from their inadequate SSLS method [23, 24]. Zou et al. (2015) and Thomas et al. (2015) considered the ΔSSLS method unsuitable for convergence inference because it is an indirect and informal test with the inappropriate null distributions [23, 24].
3.3. Target Species Specific Amino Acid Substitution (TAAS)
In this method, amino acid sequences of orthologous alignments from all species with and without the target trait are first obtained. Amino acid substitution sites between species with target trait (i.e. target species) and species without target trait (i.e. non-target species) are examined to investigate convergent evolution of target species at the amino acid sequence level. If these amino acid types for target species (i.e., species A and B in Fig. 2b) are different from those of non-target species (i.e., species C−G in Fig. 2b) in the same site for the orthologous gene, it will be considered as convergent gene [25]. This type of site has been termed a “Target Species-specific Amino Acid Substitution” (TAAS) by Zhang et al. (2014) [25].
Fig. (2).
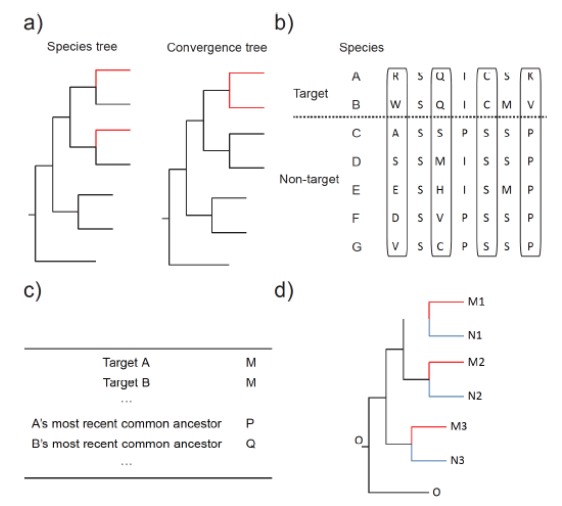
(a) Methods for evaluating amino acid convergence in protein sequences. Comparison of site-wise log-likelihood support (ΔSSLS) [21, 22] (b); Target Species-specific Amino Acid Substitution (TAAS) [25] (c); Adaptively Convergent Genes (ACGs) [26] (d); Convergence at Conservative Sites (CCS) [27].
3.4. Adaptively Convergent Genes (ACGs)
In this method, Adaptively Convergent Genes (ACGs) are defined as those that not only undergo positive selection but also include nonrandom convergent amino acid substitutions. First, positive selection is analyzed. Based on known or reconstructed phylogeny using ML or Bayesian inference which includes all species as a guide tree, paired branch-site likelihood ratio tests are applied to identify positively selected genes within target lineages (here, target lineages are species A and B) specified as the foreground branches (Fig. 2c). Paired branch-site models (Null model: model = 2, NSsites = 2, ω = 1; alternative model: model = 2, NSsites = 2, ω ≠ 1), combined with branch-site likelihood ratio tests, are run for all tests as implemented in CODEML of the PAML package [74]. Chi-square test with one degree of freedom is carried out to evaluate the significance of the compared likelihood ratio tests. The resulting P values are adjusted by the false discovery rate method for multiple testing [75, 76]. Second, genomic convergence is detected. Amino acid alignments of all orthologous genes coupled with the tree topology are used to reconstruct ancestral amino acid states using CODEML with Empirical + F model, the Jones, Taylor, and Thorton (JTT) matrix, and a discrete gamma model. Three criteria are used to identify the observed convergent sites: (i) the amino acid residues of the extant two target lineages are the same (here, target lineages are species A and B, the amino acid of both species A and B in one site is M); (ii) amino acid substitutions occur from the most recent common ancestor between the extant target lineage and its sister non-target lineage to the extant target lineage, respectively (here, the amino acids of species A and A’s most recent common ancestor in this site are different, M ≠ P; the amino acids of species B and B’s most recent common ancestor in this site are different, M ≠ Q); and (iii) if the amino acid residues of the two most recent common ancestors are identical, the site is interpreted to be “parallel” (P = Q) and if they are non-identical, it can be considered to be “convergent” (P ≠ Q) (Fig. 2c). Following the definition from Zhang and Kumar (1997), both of them are collectively considered to be convergent [77]. To reduce the impact of random convergence, JTT-fgene, JTT-fsite, or JTT-CAT models of amino acid substitution are used to further estimate the expected number of molecular convergences in each protein alignment [26]. JTT-fgene model uses the average of the equilibrium amino acid frequencies across all sites. JTT-fsite model uses the equilibrium frequencies at each site. JTT-CAT model considers the among-site heterogeneities in the equilibrium frequencies using a Bayesian mixture model. According to the method described by Zou and Zhang (2015), a Poisson test is performed to compare the difference between the observed and expected numbers of molecular convergences for each protein [26]. The false discovery rate method is also used to correct for multiple tests [75, 76]. If the number of observed convergent sites is significantly higher than that of the expected convergent sites for a gene, it will be deemed as a convergent gene. Finally, ACGs are obtained by examining the overlap between the positively selected genes and the convergent genes.
3.5. Convergence at Conservative Sites (CCS)
In this method, Xu et al. (2017) considered that convergent sites may include various types of noise arising from multiple sources, including random substitutions without being subjected to selective pressures, inaccurate inference of the ancestral characters, and inaccurate nucleotide substitution rates and patterns [26, 27, 77]. Based on these questions and problems, the method of convergence at conservative sites (CCS) was proposed to avoid the identification of random and false convergence across all sites [27]. In the method, three target species (shown in red) and their respective controlled non-target species (shown in blue) with an outgroup species (shown in black) are considered (Fig. 2d). O, M1−M3 and N1−N3 represent the observed character state of the outgroup species as the ancestral character state (shown in black), three target species (shown in red), and three non-target species (shown in blue), respectively (Fig. 2d). M1 = M2 (= M3) ≠ O and N1 = N2 = N3 = O indicates convergence at conservative sites in the target species (Fig. 2d). Three target species share the same derived states belonging to the most conservative convergence. Two of three target species share the same derived states belonging to relatively conservative convergence. For the CCS method, only conservative sites that meet N1 = N2 = N3 = O are used to infer convergence. Although this may leave out some true convergent signals, it can more efficiently exclude convergent noise caused by too many random mutations at the same site, in addition to falsely inferred ancestral states.
4. OUTLOOK FOR FUTURE RESEARCH ON ADAPTIVE CONVERGENT EVOLUTION
With the genome-wide data growth in the last decade, mechanisms of adaptive phenotypic convergence have been largely investigated at the molecular level. Recent genomic studies provide many examples of adaptive convergent evolution, which detect more candidate protein-coding genes, non-coding regions, gene families, and biological functions or pathways. These results greatly improve our understanding of convergent adaptation. However, some factors should be taken into consideration in future studies.
For the identified mutations, it is difficult to distinguish adaptive mutations from random or neutral ones. Experimental evidence can be significantly useful to address this problem. Functional assays including ancestral protein reconstruction and site-directed mutagenesis have been carried out to confirm adaptive amino acid convergence of RNases [20], Prestin [11, 32], and hemoglobin [36, 37, 41]. Turbidity assays have been performed to verify the function of fast-twitch muscle protein of calsequestrin 1 in echolocating mammals [33].
Although several studies identified specific convergence in non-coding regulatory regions [46-51], these researches more usually focused on some model species, such as worms and fruit flies or a few species with well-annotated genomes. Studies for non-coding sequences should be launched in more species.
Only a few studies have examined convergence at the gene expression level on a genome-wide scale for wild species. For example, Gallant et al. (2014) found that electric fishes convergently evolved a common gene expression pattern in TFs and developmental and cellular pathways [78]. Pfenning et al. (2014) demonstrated that a convergent gene expression pattern in specific brain regions may contribute to vocal-learning traits in both birds and humans [79]. Changes at the gene expression level are considered important for directing variances of phenotypes among species [80]. Additionally, alternative splicing may play important roles in phenotypic differences [81, 82]. Gene expression level can be changed by developmental stages, behaviors and environments [79, 83, 84]. Therefore, studies of gene expression convergence should attempt to exclude these confounding factors. Reciprocal transplant experiments or common garden experiments might be designed to exclude expression plasticity when studying convergent local adaptation [85, 86].
With advances in the high-throughput sequencing technologies, the improvement of analytical tools, and the reductions in cost, convergent changes in gene expression, alternative splicing, and regulatory regions will receive increased attention. The future studies of adaptive convergent evolution can involve greater numbers of phenotypic features and more comprehensively include a variety of factors such as sequence structure (coding sequences and non-coding regulatory sequences), gene expression, alternative splicing. Experimental studies of adaptive molecular convergence should be also conducted globally to provide direct and precise evidence to support those hypotheses.
ACKNOWLEDGEMENTS
We thank David Gabriel for his assistance. We also thank the editor for the manuscript invitation and two anonymous reviewers for the very valuable comments and suggestions.
CONSENT FOR PUBLICATION
Not applicable.
FUNDING
This work was funded by grants from the Strategic Priority Research Program, Chinese Academy of Sciences (XDA19050202 [F.L.] and XDB13020300 [F.L.]), and the State Key Program of NSFC (31630069 [F.L.] and 31572249 [F.L.]).
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCEs
- 1.Losos J.B. Convergence, adaptation, and constraint. Evolution. 2011;65(7):1827–1840. doi: 10.1111/j.1558-5646.2011.01289.x. [DOI] [PubMed] [Google Scholar]
- 2.Rosenblum E.B., Parent C.E., Brandt E.E. The molecular basis of phenotypic convergence. Annu. Rev. Ecol. Evol. Syst. 2014;45(1):203–226. [Google Scholar]
- 3.Bridgham J.T. Predicting the basis of convergent evolution. Science. 2016;354(6310):289–289. doi: 10.1126/science.aai7394. [DOI] [PubMed] [Google Scholar]
- 4.Emery N.J., Clayton N.S. The mentality of crows: Convergent evolution of intelligence in corvids and apes. Science. 2004;306(5703):1903–1907. doi: 10.1126/science.1098410. [DOI] [PubMed] [Google Scholar]
- 5.Dalziel A.C., Laporte M., Rougeux C., Guderley H., Bernatchez L. Convergence in organ size but not energy metabolism enzyme activities among wild lake whitefish (Coregonus clupeaformis) species pairs. Mol. Ecol. 2017;26(1):225–244. doi: 10.1111/mec.13847. [DOI] [PubMed] [Google Scholar]
- 6.Mitterboeck T.F., Liu S., Adamowicz S.J., Fu J., Zhang R., Song W., Meusemann K., Zhou X. Positive and relaxed selection associated with flight evolution and loss in insect transcriptomes. Gigascience. 2017;6(10):1–14. doi: 10.1093/gigascience/gix073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ben-Hamo M., Munoz-Garcia A., Larrain P., Pinshow B., Korine C., Williams J.B. The cutaneous lipid composition of bat wing and tail membranes: A case of convergent evolution with birds. Proc. Biol. Sci. 1833;2016(283):20160636. doi: 10.1098/rspb.2016.0636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Y., Cotton J.A., Shen B., Han X., Rossiter S.J., Zhang S. Convergent sequence evolution between echolocating bats and dolphins. Curr. Biol. 2010;20(2):53–54. doi: 10.1016/j.cub.2009.11.058. [DOI] [PubMed] [Google Scholar]
- 9.Liu Y., Rossiter S.J., Han X.Q., Cotton J.A., Zhang S.Y. Cetaceans on a molecular fast track to ultrasonic hearing. Curr. Biol. 2010;20(20):1834–1839. doi: 10.1016/j.cub.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 10.Rossiter S.J., Zhang S., Liu Y. Prestin and high frequency hearing in mammals. Commun. Integr. Biol. 2011;4(2):236–239. doi: 10.4161/cib.4.2.14647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Z., Qi F.Y., Zhou X., Ren H.Q., Shi P. Parallel sites implicate functional convergence of the hearing gene Prestin among echolocating mammals. Mol. Biol. Evol. 2014;31(9):2415–2424. doi: 10.1093/molbev/msu194. [DOI] [PubMed] [Google Scholar]
- 12.Montealegre Z.F., Jonsson T., Robson-Brown K.A., Postles M., Robert D. Convergent evolution between insect and mammalian audition. Science. 2012;338(6109):968–971. doi: 10.1126/science.1225271. [DOI] [PubMed] [Google Scholar]
- 13.Yokoyama S., Radlwimmer F.B. The molecular genetics and evolution of red and green color vision in vertebrates. Genetics. 2001;158(4):1697–1710. doi: 10.1093/genetics/158.4.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christin P.A., Samaritani E., Petitpierre B., Salamin N., Besnard G. Evolutionary insights on C4 photosynthetic subtypes in grasses from genomics and phylogenetics. Genome Biol. Evol. 2009;1(0):221–230. doi: 10.1093/gbe/evp020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Z., Xu D., Wang L., Hao J., Wang J., Zhou X., Wang W., Qiu Q., Huang X., Zhou J., Long R., Zhao F., Shi P. Convergent evolution of rumen microbiomes in high-altitude mammals. Curr. Biol. 2016;26(14):1873–1879. doi: 10.1016/j.cub.2016.05.012. [DOI] [PubMed] [Google Scholar]
- 16.Christin P.A., Weinreich D.M., Besnard G. Causes and evolutionary significance of genetic convergence. Trends Genet. 2010;26(9):400–405. doi: 10.1016/j.tig.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 17.Tenaillon O., Rodríguez-Verdugo A., Gaut R.L., McDonald P., Bennett A.F., Long A.D., Gaut B.S. The molecular diversity of adaptive convergence. Science. 2012;335(6067):457–461. doi: 10.1126/science.1212986. [DOI] [PubMed] [Google Scholar]
- 18.Schwarze K., Campbell K.L., Hankeln T., Storz J.F., Hoffmann F.G., Burmester T. The globin gene repertoire of lampreys: Convergent evolution of hemoglobin and myoglobin in jawed and jawless vertebrates. Mol. Biol. Evol. 2014;31(10):2708–2721. doi: 10.1093/molbev/msu216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strasser B., Mlitz V., Hermann M., Tschachler E., Eckhart L. Convergent evolution of cysteine-rich proteins in feathers and hair. BMC Evol. Biol. 2015;15(1) doi: 10.1186/s12862-015-0360-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang J.Z. Parallel adaptive origins of digestive RNases in Asian and African leaf monkeys. Nat. Genet. 2006;38(7):819–823. doi: 10.1038/ng1812. [DOI] [PubMed] [Google Scholar]
- 21.Castoe T.A., de Koning A.P.J., Kim H.M., Gu W., Noonan B.P., Naylor G., Jiang Z.J., Parkinson C.L., Pollock D.D. Evidence for an ancient adaptive episode of convergent molecular evolution. Proc. Natl. Acad. Sci. USA. 2009;106(22):8986–8991. doi: 10.1073/pnas.0900233106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parker J., Tsagkogeorga G., Cotton J.A., Liu Y., Provero P., Stupka E., Rossiter S.J. Genome-wide signatures of convergent evolution in echolocating mammals. Nature. 2013;502(7470):228–231. doi: 10.1038/nature12511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zou Z., Zhang J. No genome-wide protein sequence convergence for echolocation. Mol. Biol. Evol. 2015;32(5):1237–1241. doi: 10.1093/molbev/msv014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomas G.W.C., Hahn M.W. Determining the null model for detecting adaptive convergence from genomic data: A case study using echolocating mammals. Mol. Biol. Evol. 2015;32(5):1232–1236. doi: 10.1093/molbev/msv013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang G., Li C., Li Q., Li B., Larkin D.M., Lee C., Storz J.F., Antunes A., Greenwold M.J., Meredith R.W. Comparative genomics reveals insights into avian genome evolution and adaptation. Science. 2014;346(6215):1311–1320. doi: 10.1126/science.1251385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zou Z., Zhang J. Are convergent and parallel amino acid substitutions in protein evolution more prevalent than neutral expectations? Mol. Biol. Evol. 2015;32(8):2085–2096. doi: 10.1093/molbev/msv091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu S., He Z., Guo Z., Zhang Z., Wyckoff G.J., Greenberg A., Wu C-I., Shi S. Genome-wide convergence during evolution of mangroves from woody plants. Mol. Biol. Evol. 2017;34(4):1008–1015. doi: 10.1093/molbev/msw277. [DOI] [PubMed] [Google Scholar]
- 28.Foote A.D., Liu Y., Thomas G.W.C., Vinař T., Alföldi J., Deng J., Dugan S., van Elk C.E., Hunter M.E., Joshi V., Khan Z., Kovar C., Lee S.L., Lindblad-Toh K., Mancia A., Nielsen R., Qin X., Qu J., Raney B.J., Vijay N., Wolf J.B.W., Hahn M.W., Muzny D.M., Worley K.C., Gilbert M.T.P., Gibbs R.A. Convergent evolution of the genomes of marine mammals. Nat. Genet. 2015;47(3):272–275. doi: 10.1038/ng.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu Y., Wu Q., Ma S., Ma T., Shan L., Wang X., Nie Y., Ning Z., Yan L., Xiu Y., Wei F. Comparative genomics reveals convergent evolution between the bamboo-eating giant and red pandas. Proc. Natl. Acad. Sci. USA. 2017;114(5):1081–1086. doi: 10.1073/pnas.1613870114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fukushima K., Fang X., Alvarez-Ponce D., Cai H., Carretero-Paulet L., Chen C., Chang T.-H., Farr K.M., Fujita T., Hiwatashi Y., Hoshi Y., Imai T., Kasahara M., Librado P., Mao L., Mori H., Nishiyama T., Nozawa M., Pálfalvi G., Pollard S.T., Rozas J., Sánchez-Gracia A., Sankoff D., Shibata T.F., Shigenobu S., Sumikawa N., Uzawa T., Xie M., Zheng C., Pollock D.D., Albert V.A., Li S., Hasebe M. Genome of the pitcher plant Cephalotus reveals genetic changes associated with carnivory. Nat. Ecol. Evol. 2017;1(3):00-59. doi: 10.1038/s41559-016-0059. [DOI] [PubMed] [Google Scholar]
- 31.Yu L., Wang G.D., Ruan J., Chen Y.B., Yang C.P., Cao X., Wu H., Liu Y.H., Du Z.L., Wang X.P., Yang J., Cheng S.C., Zhong L., Wang L., Wang X., Hu J.Y., Fang L., Bai B., Wang K.L., Yuan N., Wu S.F., Li B.G., Zhang J.G., Yang Y.Q., Zhang C.L., Long Y.C., Li H.S., Yang J.Y., Irwin D.M., Ryder O.A., Li Y., Wu C.I., Zhang Y.P. Genomic analysis of snub-nosed monkeys (Rhinopithecus) identifies genes and processes related to high-altitude adaptation. Nat. Genet. 2016;48(8):947–952. doi: 10.1038/ng.3615. [DOI] [PubMed] [Google Scholar]
- 32.Liu Z., Qi F.Y., Xu D.M., Zhou X., Shi P. Genomic and functional evidence reveals molecular insights into the origin of echolocation in whales. Sci. Adv. 2018;4(10) doi: 10.1126/sciadv.aat8821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee J.H., Lewis K.M., Moural T.W., Kirilenko B., Borgonovo B., Prange G., Koessl M., Huggenberger S., Kang C., Hiller M. Molecular parallelism in fast-twitch muscle proteins in echolocating mammals. Sci. Adv. 2018 doi: 10.1126/sciadv.aat9660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scotland R.W. What is parallelism? Evol. Dev. 2011;13(2):214–227. doi: 10.1111/j.1525-142X.2011.00471.x. [DOI] [PubMed] [Google Scholar]
- 35.Stern D.L. The genetic causes of convergent evolution. Nat. Rev. Genet. 2013;14(11):751–764. doi: 10.1038/nrg3483. [DOI] [PubMed] [Google Scholar]
- 36.Projecto-Garcia J., Natarajan C., Moriyama H., Weber R.E., Fago A., Cheviron Z.A., Dudley R., McGuire J.A., Witt C.C., Storz J.F. Repeated elevational transitions in hemoglobin function during the evolution of Andean hummingbirds. Proc. Natl. Acad. Sci. USA. 2013;110(51):20669–20674. doi: 10.1073/pnas.1315456110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu X., Guan Y., Signore A.V., Natarajan C., DuBay S.G., Cheng Y., Han N., Song G., Qu Y., Moriyama H., Hoffmann F.G., Fago A., Lei F., Storz J.F. Divergent and parallel routes of biochemical adaptation in high-altitude passerine birds from the Qinghai-Tibet Plateau. Proc. Natl. Acad. Sci. USA. 2018;115(8):1865–1870. doi: 10.1073/pnas.1720487115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhen Y., Aardema M.L., Medina E.M., Schumer M., Andolfatto P. Parallel molecular evolution in an herbivore community. Science. 2012;337(6102):1634. doi: 10.1126/science.1226630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Natarajan C., Hoffmann F.G., Weber R.E., Fago A., Witt C.C., Storz J.F. Predictable convergence in hemoglobin function has unpredictable molecular underpinnings. Science. 2016;354(6310):336–339. doi: 10.1126/science.aaf9070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCracken K.G., Barger C.P., Sorenson M.D. Phylogenetic and structural analysis of the HbA (alphaA/betaA) and HbD (alphaD/betaA) hemoglobin genes in two high-altitude waterfowl from the Himalayas and the Andes: bar-headed goose (Anser indicus) and Andean goose (Chloephaga melanoptera). Mol. Phylogenet. Evol. 2010;56(2):649–658. doi: 10.1016/j.ympev.2010.04.034. [DOI] [PubMed] [Google Scholar]
- 41.Begun D.J., Natarajan C., Projecto-Garcia J., Moriyama H., Weber R.E., Muñoz-Fuentes V., Green A.J., Kopuchian C., Tubaro P.L., Alza L., Bulgarella M., Smith M.M., Wilson R.E., Fago A., McCracken K.G., Storz J.F. Convergent evolution of hemoglobin function in high-altitude Andean waterfowl involves limited parallelism at the molecular sequence level. PLoS Genet. 2015;11(12):e1005681. doi: 10.1371/journal.pgen.1005681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang Y., Wang L., Han J., Tang X., Ma M., Wang K., Zhang X., Ren Q., Chen Q., Qiu Q. Comparative transcriptomic analysis revealed adaptation mechanism of Phrynocephalus erythrurus, the highest altitude lizard living in the Qinghai-Tibet Plateau. BMC Evol. Biol. 2015;15:101. doi: 10.1186/s12862-015-0371-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chikina M., Robinson J.D., Clark N.L. Hundreds of genes experienced convergent shifts in selective pressure in marine mammals. Mol. Biol. Evol. 2016;33(9):2182–2192. doi: 10.1093/molbev/msw112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Castiglione G.M., Schott R.K., Hauser F.E., Chang B.S.W. Convergent selection pressures drive the evolution of rhodopsin kinetics at high altitudes via nonparallel mechanisms. Evolution. 2018;72(1):170–186. doi: 10.1111/evo.13396. [DOI] [PubMed] [Google Scholar]
- 45.Stern D.L. Perspective: evolutionary developmental biology and the problem of variation. Evolution. 2000;54(4):1079–1091. doi: 10.1111/j.0014-3820.2000.tb00544.x. [DOI] [PubMed] [Google Scholar]
- 46.Vavouri T., Walter K., Gilks W.R., Lehner B., Elgar G. Parallel evolution of conserved non-coding elements that target a common set of developmental regulatory genes from worms to humans. Genome Biol. 2007;8(2):15. doi: 10.1186/gb-2007-8-2-r15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Frankel N., Wang S., Stern D.L. Conserved regulatory architecture underlies parallel genetic changes and convergent phenotypic evolution. Proc. Natl. Acad. Sci. USA. 2012;109(51):20975–20979. doi: 10.1073/pnas.1207715109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reed R.D., Papa R., Martin A., Hines H.M., Counterman B.A., Pardo-Diaz C., Jiggins C.D., Chamberlain N.L., Kronforst M.R., Chen R., Halder G., Nijhout H.F., McMillan W.O. Optix drives the repeated convergent evolution of butterfly wing pattern mimicry. Science. 2011;333(6046):1137–1141. doi: 10.1126/science.1208227. [DOI] [PubMed] [Google Scholar]
- 49.Signor S.A., Liu Y., Rebeiz M., Kopp A. Genetic convergence in the evolution of male-specific color patterns in Drosophila. Curr. Biol. 2016;26(18):2423–2433. doi: 10.1016/j.cub.2016.07.034. [DOI] [PubMed] [Google Scholar]
- 50.Feigin C.Y., Newton A.H., Doronina L., Schmitz J., Hipsley C.A., Mitchell K.J., Gower G., Llamas B., Soubrier J., Heider T.N., Menzies B.R., Cooper A., O’Neill R.J., Pask A.J. Genome of the Tasmanian tiger provides insights into the evolution and demography of an extinct marsupial carnivore. Nat. Ecol. Evol. 2017;2(1):182–192. doi: 10.1038/s41559-017-0417-y. [DOI] [PubMed] [Google Scholar]
- 51.Sackton T.B., Grayson P., Cloutier A., Hu Z., Liu J.S., Wheeler N.E., Gardner P.P., Clarke J.A., Baker A.J., Clamp M., Edwards S.V. Convergent regulatory evolution and the origin of flightlessness in palaeognathous birds. bioRxiv. 2018;262584 doi: 10.1101/262584. [DOI] [PubMed] [Google Scholar]
- 52.Qu Y., Zhao H., Han N., Zhou G., Song G., Gao B., Tian S., Zhang J., Zhang R., Meng X., Zhang Y., Zhang Y., Zhu X., Wang W., Lambert D., Ericson P.G., Subramanian S., Yeung C., Zhu H., Jiang Z., Li R., Lei F. Ground tit genome reveals avian adaptation to living at high altitudes in the Tibetan plateau. Nat. Commun. 2013;4:2071. doi: 10.1038/ncomms3071. [DOI] [PubMed] [Google Scholar]
- 53.Nagy L.G., Ohm R.A., Kovacs G.M., Floudas D., Riley R., Gacser A., Sipiczki M., Davis J.M., Doty S.L., De Hoog G.S., Lang B.F., Spatafora J.W., Martin F.M., Grigoriev I.V., Hibbett D.S. Latent homology and convergent regulatory evolution underlies the repeated emergence of yeasts. Nat. Commun. 2014;5(4471) doi: 10.1038/ncomms5471. [DOI] [PubMed] [Google Scholar]
- 54.Denoeud F., Carretero-Paulet L., Dereeper A., Droc G., Guyot R., Pietrella M., Zheng C., Alberti A., Anthony F., Aprea G. The coffee genome provides insight into the convergent evolution of caffeine biosynthesis. Science. 2014;345(6201):1181–1184. doi: 10.1126/science.1255274. [DOI] [PubMed] [Google Scholar]
- 55.Berens A.J., Hunt J.H., Toth A.L. Comparative transcriptomics of convergent evolution: Different genes but conserved pathways underlie caste phenotypes across lineages of eusocial insects. Mol. Biol. Evol. 2015;32(3):690–703. doi: 10.1093/molbev/msu330. [DOI] [PubMed] [Google Scholar]
- 56.Roda F., Liu H., Wilkinson M.J., Walter G.M., James M.E., Bernal D.M., Melo M.C., Lowe A., Rieseberg L.H., Prentis P., Ortiz-Barrientos D. Convergence and divergence during the adaptation to similar environments by an Australian groundsel. Evolution. 2013;67(9):2515–2529. doi: 10.1111/evo.12136. [DOI] [PubMed] [Google Scholar]
- 57.Sun Y.B., Fu T.T., Jin J.Q., Murphy R.W., Hillis D.M., Zhang Y.P., Che J. Species groups distributed across elevational gradients reveal convergent and continuous genetic adaptation to high elevations. Proc. Natl. Acad. Sci. USA. 2018;115(45):10634–10641. doi: 10.1073/pnas.1813593115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bickler P.E., Buck L.T. Hypoxia tolerance in reptiles, amphibians, and fishes: life with variable oxygen availability. Annu. Rev. Physiol. 2007;69(1):145–170. doi: 10.1146/annurev.physiol.69.031905.162529. [DOI] [PubMed] [Google Scholar]
- 59.Ramirez J.M., Folkow L.P., Blix A.S. Hypoxia tolerance in mammals and birds: From the wilderness to the clinic. Annu. Rev. Physiol. 2007;69(1):113–143. doi: 10.1146/annurev.physiol.69.031905.163111. [DOI] [PubMed] [Google Scholar]
- 60.Moore L.G., Niermeyer S., Zamudio S. Human adaptation to high altitude: Regional and life-cycle perspectives. Yearb. Phys. Anthropol. 1998;(Suppl. 27):25–64. doi: 10.1002/(sici)1096-8644(1998)107:27+<25::aid-ajpa3>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 61.Zhang Q., Gou W., Wang X., Zhang Y., Ma J., Zhang H., Zhang Y., Zhang H. Genome resequencing identifies unique adaptations of tibetan chickens to hypoxia and high-dose ultraviolet radiation in high-altitude environments. Genome Biol. Evol. 2016;8(3):765–776. doi: 10.1093/gbe/evw032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Beall C.M., Jablonski N.G., Steegmann A.T. Human biology: An evolutionary and biocultural perspective. 2nd ed. Vol. 6. New York: John Wiley & Sons, Inc.; 2012. Human adaptation to climate: Temperature, ultraviolet radiation, and altitude. pp. 177–250. [Google Scholar]
- 63.Gnerre S., MacCallum I., Przybylski D., Ribeiro F.J., Burton J.N., Walker B.J., Sharpe T., Hall G., Shea T.P., Sykes S., Berlin A.M., Aird D., Costello M., Daza R., Williams L., Nicol R., Gnirke A., Nusbaum C., Lander E.S., Jaffe D.B. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc. Natl. Acad. Sci. USA. 2011;108(4):1513–1518. doi: 10.1073/pnas.1017351108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Luo R.B., Liu B.H., Xie Y.L., Li Z.Y., Huang W.H., Yuan J.Y., He G.Z., Chen Y.X., Pan Q., Liu Y.J., Tang J.B., Wu G.X., Zhang H., Shi Y.J., Liu Y., Yu C., Wang B., Lu Y., Han C.L., Cheung D.W., Yiu S.M., Peng S.L., Zhu X.Q., Liu G.M., Liao X.K., Li Y.R., Yang H.M., Wang J., Lam T.W., Wang J. SOAPdenovo2: An empirically improved memory-efficient short-read de novo assembler. Gigascience. 2012;1(1):18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Haas B.J., Papanicolaou A., Yassour M., Grabherr M., Blood P.D., Bowden J., Couger M.B., Eccles D., Li B., Lieber M., MacManes M.D., Ott M., Orvis J., Pochet N., Strozzi F., Weeks N., Westerman R., William T., Dewey C.N., Henschel R., LeDuc R.D., Friedman N., Regev A. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013;8(8):1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schulz M.H., Zerbino D.R., Vingron M., Birney E. Oases: robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics. 2012;28(8):1086–1092. doi: 10.1093/bioinformatics/bts094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li L., Stoeckert C.J., Roos D.S. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13(9):2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lechner M., Findeiß S., Steiner L., Marz M., Stadler P.F., Prohaska S.J. Proteinortho: Detection of (co-) orthologs in large-scale analysis. BMC Bioinformatics. 2011;12(1):1. doi: 10.1186/1471-2105-12-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Löytynoja A., Goldman N. Phylogeny-aware gap placement prevents errors in sequence alignment and evolutionary analysis. Science. 2008;320(5883):1632–1635. doi: 10.1126/science.1158395. [DOI] [PubMed] [Google Scholar]
- 70.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Edgar R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000;17(4):540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 73.Capella-Gutierrez S., Silla-Martinez J.M., Gabaldon T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25(15):1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24(8):1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 75.Benjamini Y., Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B. 1995:289–300. [Google Scholar]
- 76.Storey J.D., Tibshirani R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA. 2003;100(16):9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang J., Kumar S. Detection of convergent and parallel evolution at the amino acid sequence level. Mol. Biol. Evol. 1997;14(5):527–536. doi: 10.1093/oxfordjournals.molbev.a025789. [DOI] [PubMed] [Google Scholar]
- 78.Gallant J.R., Traeger L.L., Volkening J.D., Moffett H., Chen P-H., Novina C.D., Phillips G.N., Anand R., Wells G.B., Pinch M. Genomic basis for the convergent evolution of electric organs. Science. 2014;344(6191):1522–1525. doi: 10.1126/science.1254432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pfenning A.R., Hara E., Whitney O., Rivas M.V., Wang R., Roulhac P.L., Howard J.T., Wirthlin M., Lovell P.V., Ganapathy G., Mouncastle J., Moseley M.A., Thompson J.W., Soderblom E.J., Iriki A., Kato M., Gilbert M.T.P., Zhang G., Bakken T., Bongaarts A., Bernard A., Lein E., Mello C.V., Hartemink A.J., Jarvis E.D. Convergent transcriptional specializations in the brains of humans and song-learning birds. Science. 2014;346(6215):1256846. doi: 10.1126/science.1256846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Brawand D., Soumillon M., Necsulea A., Julien P., Csardi G., Harrigan P., Weier M., Liechti A., Aximu-Petri A., Kircher M., Albert F.W., Zeller U., Khaitovich P., Grutzner F., Bergmann S., Nielsen R., Paabo S., Kaessmann H. The evolution of gene expression levels in mammalian organs. Nature. 2011;478(7369):343–348. doi: 10.1038/nature10532. [DOI] [PubMed] [Google Scholar]
- 81.Barbosa-Morais N.L., Irimia M., Pan Q., Xiong H.Y., Gueroussov S., Lee L.J., Slobodeniuc V., Kutter C., Watt S., Colak R., Kim T., Misquitta-Ali C.M., Wilson M.D., Kim P.M., Odom D.T., Frey B.J., Blencowe B.J. The evolutionary landscape of alternative splicing in vertebrate species. Science. 2012;338(6114):1587–1593. doi: 10.1126/science.1230612. [DOI] [PubMed] [Google Scholar]
- 82.Merkin J., Russell C., Chen P., Burge C.B. Evolutionary dynamics of gene and isoform regulation in mammalian tissues. Science. 2012;338(6114):1593–1599. doi: 10.1126/science.1228186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Simonson T.S. Altitude adaptation: A glimpse through various lenses. High Alt. Med. Biol. 2015;16(2):125–137. doi: 10.1089/ham.2015.0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cheviron Z.A., Whitehead A., Brumfield R.T. Transcriptomic variation and plasticity in rufous-collared sparrows (Zonotrichia capensis) along an altitudinal gradient. Mol. Ecol. 2008;17(20):4556–4569. doi: 10.1111/j.1365-294X.2008.03942.x. [DOI] [PubMed] [Google Scholar]
- 85.Savolainen O., Lascoux M., Merila J. Ecological genomics of local adaptation. Nat. Rev. Genet. 2013;14(11):807–820. doi: 10.1038/nrg3522. [DOI] [PubMed] [Google Scholar]
- 86.Storz J.F., Scott G.R., Cheviron Z.A. Phenotypic plasticity and genetic adaptation to high-altitude hypoxia in vertebrates. J. Exp. Biol. 2010;213(24):4125–4136. doi: 10.1242/jeb.048181. [DOI] [PMC free article] [PubMed] [Google Scholar]


