Abstract
Background:
MicroRNAs participate in many molecular mechanisms and signaling trans-duction pathways that are associated with plant stress tolerance by repressing expression of their target genes. However, how microRNAs enhance tolerance to low temperature stress in plant cells remains elusive.
Objective:
In this investigation, we demonstrated that overexpression of the rice microRNA528 (Os-miR528) increases cell viability, growth rate, antioxidants content, ascorbate peroxidase (APOX) activi-ty, and superoxide dismutase (SOD) activity and decreases ion leakage rate and thiobarbituric acid reac-tive substances (TBARS) under low temperature stress in Arabidopsis (Arabidopsis thaliana), pine (Pi-nus elliottii), and rice (Oryza sativa).
Methods:
To investigate the potential mechanism of OsmiR528 in increasing cold stress tolerance, we examined expression of stress-associated MYB transcription factors OsGAMYB-like1, OsMYBS3, OsMYB4, OsMYB3R-2, OsMYB5, OsMYB59, OsMYB30, OsMYB1R, and OsMYB20 in rice cells by qRT-PCR.
Results:
Our experiments demonstrated that OsmiR528 decreases expression of transcription factor OsMYB30 by targeting a F-box domain containing protein gene (Os06g06050), which is a positive regulator of OsMYB30. In OsmiR528 transgenic rice, reduced OsMYB30 expression results in in-creased expression of BMY genes OsBMY2, OsBMY6, and OsBMY10. The transcript levels of the OsBMY2, OsBMY6, and OsBMY10 were elevated by OsMYB30 knockdown, but decreased by Os-MYB30 overexpression in OsmiR528 transgenic cell lines, suggesting that OsmiR528 increases low temperature tolerance by modulating expression of stress response-related transcription factor.
Conclusion:
Our experiments provide novel information in increasing our understanding in molecular mechanisms of microRNAs-associated low temperature tolerance and are valuable in plant molecular breeding from monocotyledonous, dicotyledonous, and gymnosperm plants.
Keywords: Cold stress, Gene expression, MicroRNAs, Molecular breeding, Pinus, Transcription factor
1. INTRODUCTION
MicroRNAs regulate expression of stress-responsive genes through direct degradation or repression of target genes that are associated with abiotic stress-related defense pathway in plants [1]. MicroRNAs have been established as important factors that regulate the response to stresses. For example, miR319 and miR393 regulate expression of cold-related genes in sugarcane (Saccharum officinarum L.) [2], and miR169 functions in freezing tolerance in alfalfa (Medicago sativa L.) [3]. MicroRNAs accumulated differentially at different developmental stages in rice (Oryza sativa L.) anthers under cool conditions [4] and in grass (Brachypodium distachyon L.) under freezing stress condition [5]. In Arabidopsis (Arabidopsis thaliana L.), overexpression of miR156 has been reported to play a role in delayed flowering at low temperature [6]. MicroRNAs negatively regulate gene expression by translational inhibition or degradation of target messenger RNAs [7]. However, miR402 has been reported to have a positive effect in regulating seed germination in stressed Arabidopsis through regulating DNA demethylation when plants are placed under stress conditions [8].
MicroRNAs and their targets have been used for modifying plant traits [9]. In tomato (Solanum lycopersicum L.), miR156a and miR397 have been reported to play roles in response to low temperature stress and miR156a and miR397 transgenic plants have practical application for a low temperature tolerance [10]. In Cassava (Manihot esculenta L.), microRNAs improve chilling resistance that could be beneficial to breeding [11]. In grapevine (Vitis vinifera L.), a diverse set of microRNAs have been reported to regulate expression of AP2, bHLH, GRAS, SBP, MYB, and bZIP under low temperature stress and these target genes are involved in cold stress response [12]. MicroRNA408 is a highly conserved miRNA in plants. Expression of miR408 is significantly affected by stress including salinity, cold, oxidative stress, drought and osmotic stress in Arabidopsis [13].
Transcription factors regulate expression of many stress response-associated genes that are related to low temperature tolerance in plants [14, 15]. MYB transcription factors are important proteins that are associated with response to different stresses in a large number of plant species [15]. In A. thaliana, MYB15 is associated with regulation of expression of CBF genes for low temperature tolerance [16]. In wheat (Triticum aestivum L.), MYB4 plays a role in response to cold and wound stress [17]. In O. sativa, MYB transcription factors may play important roles in low temperature stress signal transduction [18]. In white spruce [Picea glauca (Moench) Voss], MYB14 transcripts accumulating most strongly and rapidly under stress and MYB14 may be related to a number of defense response implicating flavonoids [19]. In Bambara groundnut (Vigna subterranean L.), low temperature stress can affect the growth and development through MYB transcription factor regulation [20]. In sweet orange (Citrus sinensis L.), MYB transcriptional activator regulates anthocyanin production under cold stress [21].
MYB transcription factors regulate multiple environmental stress responses [15]. Overexpression of GaMYB62L, a R2R3 MYB transcription factor, improves drought, salt and cold tolerance in transgenic A. thaliana [22]. MYB transcription factors are involved in complex biological responses during the process of drought and cold tolerance [23]. MYB-bHLH-WD40 (MBW) complex plays important roles in regulating the expression of the genes that are associated with the biosynthesis of anthocyanins [24]. MYB transgenic A. thaliana were found to be more tolerant to the low temperature stress [25]. MYB transgenic tomato lines increase anthocyanin synthesis under the high light condition and enhance low temperature tolerance [26]. Myb41-overexpressing lines increase transcriptional responses to cold stress [27]. Expression analysis derived from whole-genome microarray data demonstrated that MYBs function in resistance against cold stress in Chinese cabbage [Brassica rapa subsp. chinensis (L.)] [28]. Expression analysis of MYB transcription factors demonstrated that MYB transcription factors are associated with both cold stress and different mechanisms of regulation in plants [29]. Transcriptional regulation of expression of genes associated with cold stress is important for cold tolerance in plants. For example, MYB14 encodes a nuclear protein. Knock-down of MYB14 by microRNA increased the tolerance to freezing stress [30]. In watermelon (Citrullus lanatus), miR159-5p, miR858, and miR8029-3p regulate target genes involved in signal transduction under cold stress [31]. In T. aestivum, miRNAs differentially regulate expression of transcription factors that are involved in cold stress response [32]. In V. vinifera, cold stress may induce expression of several miRNAs that may contribute to cold stress response [12].
MicroRNA528 (miR528) is an important monocot-specific RNA that may contribute to multiple stress responses. Overexpression of miR528 in transgenic creeping bentgrass (Agrostis stolonifera L.) improves abiotic stress resistance [33]. In maize (Zea mays L.), the expression levels of miR528 increased upon hormone depletion and cold stress [34]. Sequence profiling data of rice leaf samples demonstrated that miR528 is being down-regulated in two drought-tolerant rice varieties [35]. In T. aestivum, miR528 is involved in the activation of signal transduction pathways and is associated with abiotic stresses [36]. In Z. mays, a total of 40 conserved miRNA families including miR528 were identified to be involved in biotic stresses [37]. However, OsmiR528 increased cold stress tolerance has not been reported in plants. In this investigation, we demonstrated OsmiR528 enhanced low temperature tolerance by decreasing expression of stress responsive-associated transcription factor OsMYB30 and regulating expression of stress-response genes in three species including Arabidopsis (Arabidopsis thaliana), pine (Pinus elliottii), and rice (Oryza sativa).
2. MATERIALS AND METHODS
2.1. Plasmid Constructs
The sequence of OsmiR528 [33] and the vector pBI121 were used to construct pBI-OsmiR528. The pBI121 plasmid and the DNA fragment of OsmiR528 were digested at 37oC, and digested and purified DNA was ligated to produce pBI-OsmiR528. Expression vector pBI-OsmiR528 was introduced into Agrobacterium tumefaciens strain LBA4404 by electroporation. Expression vector pCambia1301H-OsMYB30 was constructed by insert OsMYB30 (AK112056.1) into pCambia1301H as previously described [38]. The control cell lines are transformed with vectors that do not have the test genes or microRNA528.
2.2. Transformation of Plant Cells
Transgenic cells of Arabidopsis (Arabidopsis thaliana), pine (Pinus elliottii), and rice (Oryza sativa) were produced as previously described [39], using Agrobacterium tumefaciens strain LBA4404 carrying expression vector pBI-OsmiR528. For the overexpression of OsMYB30 in rice, the expression vector pCambia1301H-OsMYB30 was used to transform rice cells as previously described [40, 41].
2.3. RNA Isolation and Northern Blot Analysis
For northern blotting analysis, total RNA was isolated from the experimental Arabidopsis, rice, and pine cells. The RNeasy Mini Plant Kit (Germantown, MD, USA) was used. Six μg of total RNA was examined by gel electrophoresis. The northern blotting analysis was carried out as previously described [42]. The Digoxigenin (DIG)-labelling OsmiR528 DNA (Roche Diagnostics) was used as the hybridization probe. U6 RNA was used as the RNA loading control. Three cell lines of Arabidopsis (a109, a217, and a313), 3 cell lines of pine (p103, p213, and p307), and 3 cell lines of rice (r120, r203, and r368) were used for cold-induced oxidative damage experiments.
2.4. Cold Treatment, Cell Viability and Growth Rate Determination
The transgenic cells and control cells (non-transgenic cultures) were cultured for 6 weeks before the treatment of cold stress. Cold treatment of Arabidopsis (a109, a217, and a313), pine (p103, p213, and p307), and rice (r120, r203, and r368) cell lines was performed as previously described [39], by transferring the cells into chambers with different temperatures (-10, 4, 12, and 24oC). Cells were treated in the chambers with different temperatures for 16 hours. Then the transgenic cells and control cells (non-transgenic cultures) returned to the normal growth condition for 2 days, followed by the determination of cell viability. Chemical staining was carried out to determine the rate of cell viability. The chemical dyes are fluorescein diacetate for viable cells and phenosafranine for dead cells. A volume of 20 μl of the culture medium containing each dye was mixed with a 20 μl of cells on a microscope slide. Viable cells and dead cells were counted after 5 min. The cell growth rate was also measured 2 days after treatment in chambers with different temperatures using the turbidimetric measurement [43]. The rate of average growth was expressed as mg/g FW/day.
2.5. Determination of Total Antioxidant Levels, the Activity Of β–Amylase, Maltose, Sucrose, and Fructose Contents
Total antioxidant levels were determined from 2-days-old cell samples (300 mg) using an antioxidant assay kit (Sigma–Aldrich) following the product manual. Antioxidant reactions were performed in a 96-well plate by reading endpoint absorbance at 405 nm using a plate reader (BioTek, Synergy 2, Winooski, VT, USA). The activities of β–amylase, maltose, sucrose, and fructose contents were determined as previously described [38].
2.6. Thiobarbituric Acid Reactive Substances (TBARS) Determination
The amount of thiobarbituric acid reactive substances (TBARS) derived from the thiobarbituric acid (TBA) reaction was used to express the lipid peroxidation, as previously described [39]. Cell cultures (3g) of Arabidopsis (a109, a217, and a313), pine (p103, p213, and p307), and rice (r120, r203, and r368) were used in this experiment as previously described [39]. The absorbance of the sample extracts of different cell lines was determined at 532 nm. The absorption of the control was subtracted from the samples [39].
2.7. Measurement of Peroxidase (APOX) and Superoxide Dismutase (SOD) Activity
Measurements of APOX and SOD activities were performed as described before [39]. Three grams of non-transgenic control and transgenic cell cultures of Arabidopsis (a109, a217, and a313), pine (p103, p213, and p307), and rice (r120, r203, and r368) were extracted under 4oC in 3 ml of assay buffer that includes 50 mM phosphate buffer (pH 7.4), 0.5% (v/v) Triton X-100, 1 mM EDTA, and 1 g PVP at 4 oC. After centrifugation, the supernatant was collected and used for the determination of SOD activity, as described by Becana et al. [44]. APOX activity was detected, as described previously [45].
2.8. Measurement of Ion Leakage
Ion leakage was determined using cells of different lines as previously described [46, 47]. Cells of Arabidopsis (a109, a217, and a313), pine (p103, p213, and p307), and rice (r120, r203, and r368) and the control were placed in 1 ml of dH2O for one hour with a moderate shaking incubator. The conductivity of cells was measured using a conductivity meter (B-173, Horiba, Kyoto, Japan) before and after the cells derived from different species were autoclaved. The ratio of conductivity values before and after autoclaving cells derived from different species was expressed as ion leakage.
2.9. Overexpression and Knockdown of the OsMYB30 Gene in Rice Lines
Rice cell lines r120, r203, and r368 were used for the overexpression of OsMYB30 as previously described [39], using Agrobacterium-mediated transformation. Knockdown of OsMYB30 was performed using siRNA-mediated gene silencing. The siRNA (5′-UGUUAUUGCCAAGCACUU AAA-3′) was delivered into rice cell lines r120, r203, and r368 by particle bombardment.
2.10. RNA Isolation and qRT–PCR
Total RNA was isolated from cells of rice using an RNeasy Mini Plant Kit (Germantown, MD, USA) by following the manual. The cDNA was synthesized using cDNA synthesis kit (Invitrogen) by following the manufacturer’s manual. qRT–PCR was performed on the 7500 Real-Time PCR instrument (Applied Biosystems) by following the manufacturer’s manual. The internal control is the rice GAPDH gene. The primer sequences are listed in Table 1. The relative expression level of different genes was determined as described before [38]. The data obtained from the experiments were normalized to the control. The delta-delta Ct method was applied to calculate the expression values. U6 gene was used as the internal reference control.
2.11. Luciferase Reporter Assay
The promoter sequence of OsMYB30 was purified and inserted into the luciferase reporter expression vector as previously described [48, 49]. The OsmiR528 targets Os06g 06050, Os06g11310, Os08g04310, Os06g37150, and Os07g38290 were inserted into the effector. The control effector is the vector expressing the GAL4 DNA-binding domain (DB). 2 μg reporter, 2 μg effector, 0.04 μg internal control pPTRL, and the Dual-Luciferase Reporter Kit (Promega, Madison, WI, USA) were used to measure the luciferase activity as previously described [48, 49].
2.12. Statistical Analyses
Statistical analysis was performed using the software of SAS (Cary, NC, USA), using ANOVA models. The significant differences among average values of cells obtained from Arabidopsis (a109, a217, and a313), pine (p103, p213, and p307), and rice (r120, r203, and r368) were made at 5% level of probability. Each value of different cell lines derived from Arabidopsis (a109, a217, and a313), pine (p103, p213, and p307), and rice (r120, r203, and r368) was presented as means ± SD.
3. RESULTS
3.1. Overexpression of OsmiR528 Enhances Low Temperature Tolerance
To determine the function of OsmiR528 in cold stress tolerance, cells of Arabidopsis (a109, a217, and a313), pine (p103, p213, and p307), and rice (r120, r203, and r368) were generated through Agrobacterium-mediated transformation. Overexpression of OsmiR528 was confirmed in A. thaliana
3.2. Overexpression of OsmiR528 Enhances Activity of Antioxidative Enzymes in Transgenic Cells Under Cold Stress
To examine the effects of OsmiR528 on the level of antioxidants and the activity of antioxidative enzymes, the amount of antioxidants and TBARS, the activity of APOX and SOD were analyzed in transgenic cells. Compared to the control, overexpression of OsmiR528 increases the levels of antioxidants, decreases the amount of TBARS, and increases the activity of APOX and SOD in cells of Arabidopsis (Fig. 2a, 2d, 2g, and 2j), pine (Fig. 2b, 2e, 2h, and 2k), and rice (Fig. 2c, 2f, 2i, and 2l), respectively, under 4oC treatment. Under 24oC condition, there was no significant difference in the levels of antioxidants, the amount of TBARS, and the activity of APOX and SOD between cells overexpressing OsmiR528 and the control.
Fig. (2).
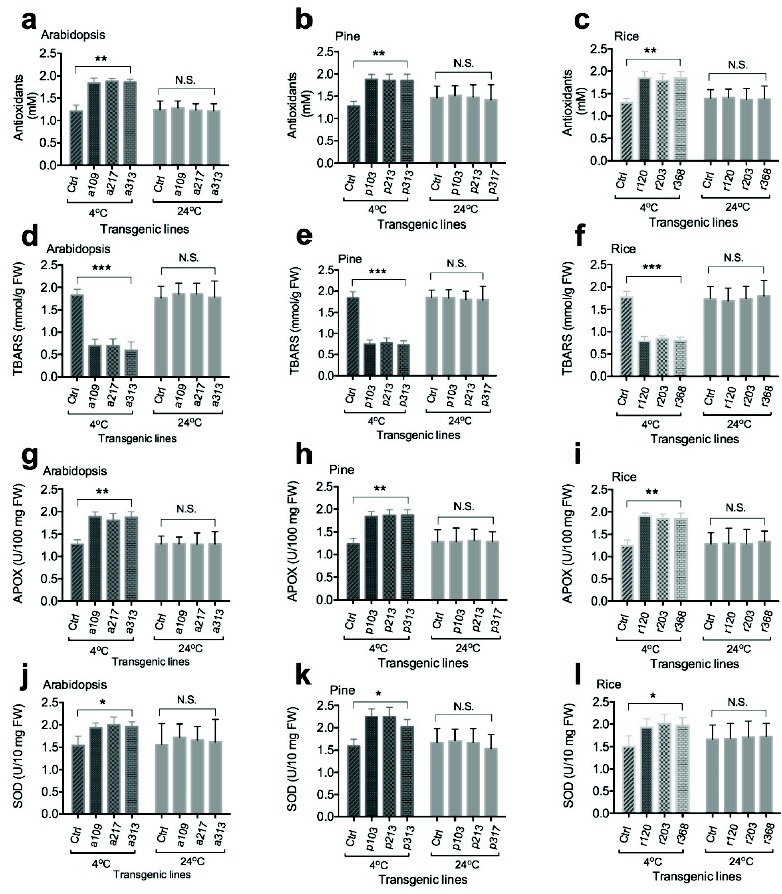
Overexpression of OsmiR528 enhances activity of antioxidative enzymes in transgenic cells under cold stress. Overexpression of OsmiR528 increases the levels of antioxidants, the amount of TBARS, the activity of APOX and SOD in cells of Arabidopsis (a, d, g, and j), pine (b, e, h, and k), and rice (c, f, i, and l), respectively. The statistically significant difference between groups was determined by one-way ANOVA. Data are presented as means of three independent experiments. Error bars represent standard error. The asterisk indicates significant differences compared to the control, as assessed by a t-test. *P<0.05, **P<0.01, ***P<0.001, significant relative to control. N.S., no statistical significance.
3.3. MYB Transcription Factors in OsmiR528 Transgenic Rice Cells
To investigate how OsmiR528 overexpression enhances low temperature stress tolerance in A. thaliana, P. elliottii, and O. sativa, expression of nine MYB transcription factors was examined in OsmiR528 transgenic rice cell lines (r120, r203, and r368). Results of qRT-PCR demonstrated that the expression of stress-related MYB transcription factors OsGAMYB-like1 (Fig. 3a), OsMYBS3 (Fig. 3b), OsMYB4 (Fig. 3c), OsMYB3R-2 (Fig. 3d), OsMYB5 (Fig. 3e), OsMYB59 (Fig. 3f), OsMYB1R (Fig. 3h), and OsMYB20
Fig. (3).
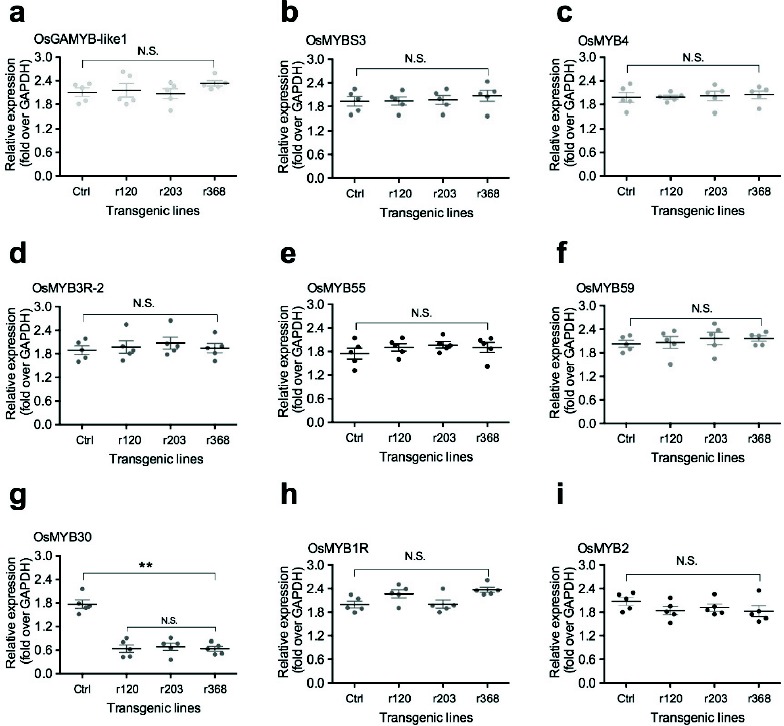
Expression of stress-related MYB transcription factors in OsmiR528 transgenic rice cells. Expression of stress-related MYB transcription factors OsGAMYB-like1 (a), OsMYBS3 (b), OsMYB4 (c), OsMYB3R-2 (d), OsMYB5 (e), OsMYB59 (f), OsMYB30 (g), OsMYB1R (h), and OsMYB20 (i) in OsmiR528 transgenic cell lines. The statistically significant difference between groups was determined by one-way ANOVA. Data are presented as means of five independent experiments. Error bars represent standard error. The asterisk indicates significant differences compared to the control, as assessed by a t-test. **P<0.01, significant relative to control. N.S., no statistical significance.
(Fig. 3i) did not alter between OsmiR528 transgenic cell lines and the control. However, expression of OsMYB30 (Fig. 3g) significantly decreased in OsmiR528 transgenic cell lines, compared to the control.
3.4. OsmiR528 Regulates Expression of OsMYB30 by Targeting F-box Domain Containing Protein (Os06g06050) in Rice Cells
To study the potential mechanism of OsmiR528 in cold stress tolerance, we searched several microRNA databases.
Based on the miRNA database (http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=tarbase/index&mirnas =MIMAT0002884) search results, there are five predicted targets for OsmiR528 in rice, including one F-box domain containing protein (Os06g06050), two plastocyanin-like domain containing proteins (Os06g11310 and Os08g04310), OsAAO (ascorbate oxidase, Os06g37150), and OsCBP1 (Cu2+binding domain-containing protein 1, Os07g38290). OsmiR528 targets Os06g06050 (Fig. 4a). Overexpression of OsmiR528 significantly decreased expression of Os06g06050 (Fig. 4b). To determine the relationship between the OsmiR528 targets and OsMYB30 expression, the luciferase reporter assay was used. The activity of luciferase was significantly increased when the OsMYB30 promoter was in the reporter and the Os06g06050 was in the effector (Fig. 4c-d). Other OsmiR528 targets did not change the luciferase activity significantly (Fig. 4e-l). These results suggested that decreased expression of OsMYB30 induced by the overexpression of OsmiR528 is not due to the degradation of transcription factor OsMYB30 mRNA, but the fact that target (Os06g06050) of OsmiR528 is a positive regulator of OsMYB30.
Fig. (4).
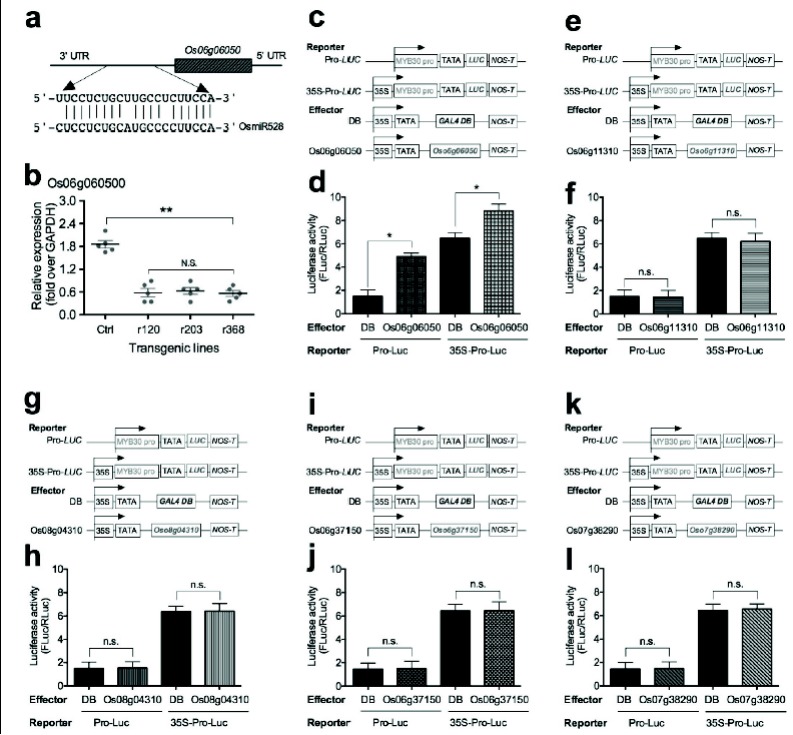
OsmiR528 regulates expression of OsMYB30 by targeting a F-box domain containing protein (Os06g06050) in rice cells. (a) Binding sequence of OsmiR528 in the 3’-UTR of Os06g06050. (b) Overexpression of OsmiR528 decreased expression of Os06g06050 in transgenic rice cell lines. (c-d) The luciferase reporter (c) using the OsMYB30 promoter as reporter and the Os06g06050 as an effector and the luciferase activity (d). (e-f) The luciferase reporter (e) with the OsMYB30 promoter as reporter and the Os06g11310 as an effector and the luciferase activity (f). (g-h) The luciferase reporter with the OsMYB30 promoter as reporter and the Os08g04310 as an effector (g) and the luciferase activity (h). (i-j) The luciferase reporter with the OsMYB30 promoter as reporter and the Os06g37150 as an effector (i) and the luciferase activity (h). (k-l) The luciferase reporter with the OsMYB30 promoter as reporter and the Os07g38290 as an effector (k) and the luciferase activity (l).
3.5. Expression of BMY Genes Regulated by Transcription Factor OsMYB30
Transcription factor OsMYB30 negatively regulates expression of BMY (β-amylase) genes in rice [38]. To examine the effect of overexpression of OsmiR528 on the expression of BMY genes, qRT-PCR was used to amplify transcripts of BMY2, BMY6, and BMY10 in OsmiR528 transgenic cells.
Results derived from the experiments demonstrated that overexpression of OsmiR528 increases expression of BMY2 (Fig. 5a), BMY6 (Fig. 5b), and BMY10 (Fig. 5c). In OsmiR528 transgenic cell, overexpression of OsMYB30 decreases expression of BMY2 (Fig. 5d), BMY6 (Fig. 5e), and BMY10 (Fig. 5f) in OsmiR528 transgenic cells. Silencing of OsMYB30 by siRNA results in increased expression of BMY2 (Fig. 5g), BMY6 (Fig. 5h), and BMY10 (Fig. 5i) in OsmiR528 transgenic cell lines.
Fig. (5).
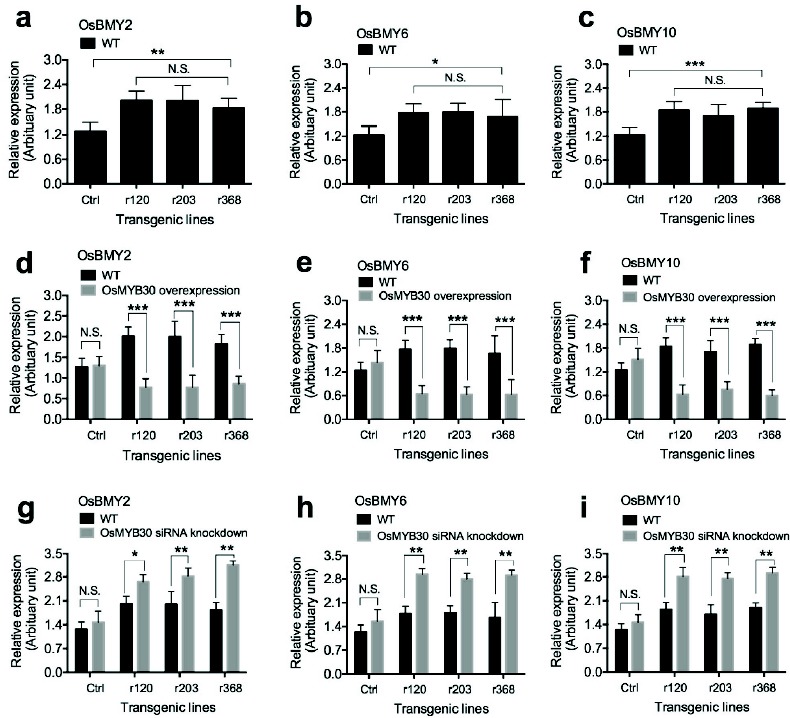
Expression of BMY genes regulated by transcription factor OsMYB30 in rice cells. Overexpression of OsmiR528 increases expression of BMY2 (a), BMY6 (b), and BMY10 (c). Overexpression of OsMYB30 decreases expression of BMY2 (d), BMY6 (e), and BMY10 (f) in OsmiR528 transgenic cell lines. Silencing of OsMYB30 by siRNA results in increased expression of BMY2 (g), BMY6 (h), and BMY10 (i) in OsmiR528 transgenic cell lines. The statistically significant difference between groups was determined by one-way ANOVA. Data are presented as means of three independent experiments. Error bars represent standard error. The asterisk indicates significant differences compared to the control, as assessed by a t-test. *P<0.05, **P<0.01, ***P<0.001, significant relative to control. N.S., no statistical significance.
3.6. Overexpression of miR528 Increases β-Amylase Activity and Sugar Content
To further confirm whether OsmiR528 alters expression of BMY genes at the protein level and whether the activity of BMY enzyme is correlated with its catalytic chemical product, β–amylase activity and maltose, sucrose, and fructose content were measured. The results revealed that the overexpression of OsmiR528 increases β–amylase activity and maltose, sucrose, and fructose content in cells of Arabidopsis (Fig. 6a, 6d, 6g, and 6j), pine (Fig. 6b, 6e, 6h, and 6k), and rice (Fig. 6c, 6f, 6i, and 6l). These experimental results demonstrated that OsmiR528 played an important role in the metabolism of starch by reducing expression of OsMYB30 that resulted in increased expression of BMY genes, which control the metabolism of starch and lead to changes of maltose, sucrose, and fructose content in OsmiR528 transgenic rice lines. Based on our experimental results, we propose a model showing OsmiR528-mediated pathways for cold stress tolerance in plant cells (Fig. 7). Under normal condition, less OsmiR528 causes little or no OsMYB30 degradation (Fig. 7a) or little or no degradation of the stress-responsive gene repressor (Fig. 7a) that results in cold stress sensitive. Overexpression of OsmiR528 causes OsMYB30 degradation (Fig. 7b) or degradation of the stress-responsive gene repressor (Fig. 7b) that leads to increased expression of BMY genes or stress-response gene, which enhances cold stress tolerance. Both (Fig. 7a) and (Fig. 7b) indicate a pathway supported by experimental evidence or reports from literature. Both (Fig. 7a) and (Fig. 7b) show a suggested pathway.
Fig. (6).
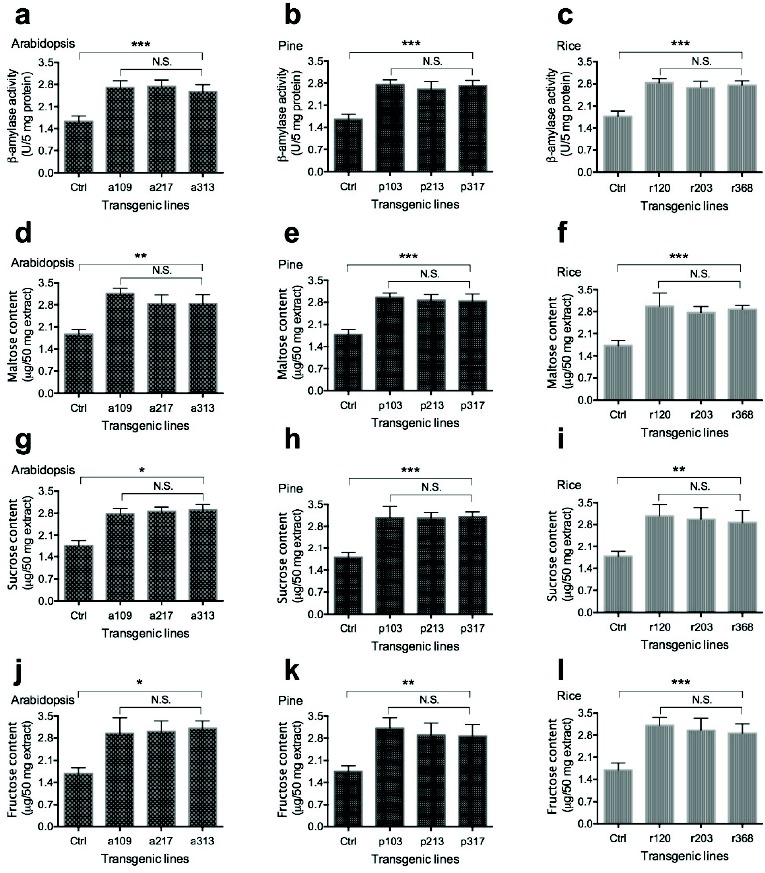
Overexpression of OsmiR528 increases β-amylase activity and sugar content in plant cells. Overexpression of OsmiR528 increases β–amylase activity and maltose, sucrose, and fructose content in cells of Arabidopsis (a, d, g, and j), pine (b, e, h, and k), and rice (c, f, i, and l). The statistically significant difference between groups was determined by one-way ANOVA. Data are presented as means of three independent experiments. Error bars represent standard error. The asterisk indicates significant differences compared to the control, as assessed by a t-test. *P<0.05, **P<0.01, ***P<0.001, significant relative to control. N.S., no statistical significance.
Fig. (7).
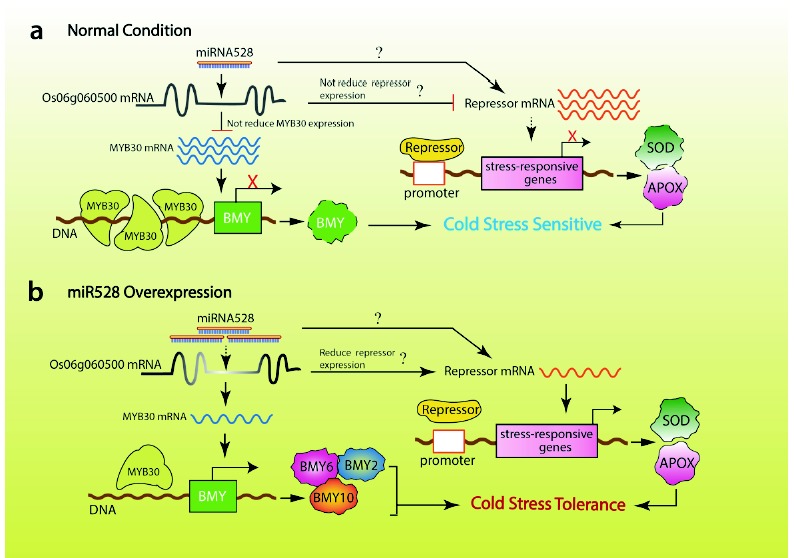
A model showing pathways for cold stress tolerance in plant cells. Under normal condition, less OsmiR528 does not reduce expression of OsMYB30 degradation or expression of the stress-response gene repressor (a) that results in cold stress sensitive. Overexpression of OsmiR528 reduces expression of OsMYB30 or expression of the stress-response gene repressor (b) that leads to increased expression of BMY genes or stress-response gene, which enhances cold stress tolerance. Both (a) and (b) indicate a pathway supported by experimental evidence or reports from literature. In both (a) and (b), arrows with question mark show a suggested pathway. Dotted arrow indicates a suggested step.
4. DISCUSSION
In this investigation, we first describe that overexpression of OsmiR528 enhances cold stress tolerance in Arabidopsis (Arabidopsis thaliana), pine (Pinus elliottii), and rice (Oryza sativa). MicroRNAs are very important in regulating stress response in plants. Different microRNAs have been documented to function in plants via modulating expression of other genes or transcription factors [15, 50-54]. In Arabis alpina, miR172 and miR156 regulate plant response to cold by targeting the APETALA2 transcription factor and SPL (SQUAMOSA PROMOTER BINDING PROTEIN LIKE) transcription factors [51]. In plant parasitic nematodes, some miRNAs negatively regulated expression of their target genes, suggesting that miRNAs might participate in ecological adaptation [55]. In Yerba mate (Ilex paraguariensis), a diverse supply of microRNA precursors was involved in heat and oxidative stress and pathogen response [56]. In A. thaliana, miR172 targeted SCHLAFMUTZE (SMZ) to regulate the flowering response [57]. Expression of miR408 regulated sensitivity to abiotic stress including cold and drought stress [13]. Overexpression of miR402 accelerated the seed germination and seedling growth of Arabidopsis under abiotic stress including cold stress [8]. In O. sativa, miR319 played regulatory roles in the development of leaf and in the tolerance of cold stress [58]. Our results demonstrated that the overexpression of OsmiR528 enhanced cold stress tolerance in Arabidopsis, pine, and rice by decreasing expression of MYB transcription factor. OsMYB30 has been reported to be involved in cold stress tolerance [38]. We identified that the target (Os06g06050) of OsmiR528 is a positive regulator of OsMYB30.
In this investigation, we find that OsmiR528 enhances low temperature tolerance by decreasing expression of OsMYB30. The target (Os06g06050) of OsmiR528 positively regulates expression of OsMYB30. MYB transcription factors play important roles in tolerance of abiotic stress [38]. In the paper mulberry (Broussonetia papyrifera L.), MYB transcription factors enhanced the tolerance of cold stress [59]. Transcription factor MYB96 induced by cold stress integrated the abscisic acid (ABA) and cold signaling pathways to activate freezing tolerance [52]. In A. thaliana, MYB transcription factor EARLY FLOWERING MYB PROTEIN (EFM) regulated plant responses to low temperature and light that are involved in the timing of reproduction [60]. MYB1 from apples improved abiotic stress tolerance by regulating expression of NtDREB1/CBF, NtERD10B and NtERD10C [61]. Genome-wide analysis of transcripts of transcription factors involved in plant abiotic stress tolerance demonstrated that Myb transcription factor protein families might be important in the control of mRNA stability [62]. In Sheep grass (Festuca ovina L.), MYB1 enhances salt stress by regulating different signaling pathways including the DREB1/CBF- and MYB2-mediated signaling pathway [52]. In O. sativa, transcription factor MYB2 plays an important role in increasing tolerance of rice to cold stress [63]. In A. thaliana, MYB15 is an important transcription factor that regulates the expression of BREB1/CBF genes under cold stress [16]. Our experimental results from Arabidopsis, pine, and rice demonstrated that OsmiR528 enhances cold stress tolerance by decreasing expression of OsMYB30 in rice and increases expression of BMY genes, which results in increased sugar content in cells.
In A. thaliana, P. elliottii, and O. sativa, our experimental results first support that OsmiR528 enhances cold stress tolerance in three plant species. MicroRNAs play important roles in regulating tolerance of salt, cold, and drought stresses by regulating expression of their target genes. In T. asetivum, miRNAs regulate expression of transcription factors that are involved in gene regulation in cold stress response [32]. Many families of transcription factor improve plant growth under normal and stress conditions through the interaction of these regulators in the plant growth and development [64]. In C. lanatus, mRNA-seq analysis revealed that miR159-5p, miR858, miR8029-3p, and miR0048-3p were important in regulating the expression of target genes associated with signal transduction under cold stress [31]. In S. lycopersicum, miR160, miR165, miR166, miR171, miR398, miR408, miR827, miR9472, miR9476 and miR9552 were very important miRNAs involved in tomato response to drought stress [65]. In A. thaliana, miRNA159 degrades the transcripts of MYB101 and MYB33 in vitro and in vivo during seedling stress responses [66]. Genome-wide expression analysis demonstrated that 109 miRNAs are involved in transcription factor and stress-related signaling pathways under abiotic stress in S. lycopersicum [67], 230 miRNAs and their targets have been identified to be involved in cold stress response by Solexa sequencing in V. vinifera. [12]. Under drought and salinity stresses, 337 cotton miRNAs were identified and miR164, miR172, miR396, miR1520, and miR6158 may contribute to cotton (Gossypium hirsutum L.) stress-resistant breeding [68]. In A. thaliana, P. elliottii, and O. sativa, our experimental results first support that OsmiR528 enhances cold stress tolerance in three plant species. These results provide new information in increasing our understanding of molecular mechanisms of microRNAs-related cold stress tolerance and are valuable in plant molecular breeding.
MicroRNAs repress expression of their target genes through different processes, including degrading the mRNAs of their target genes, destabilizing the mRNA through shortening of its poly(A) tail, and decreasing the translation efficiency of the mRNA into proteins by ribosomes. Based on the miRNA database (http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=tarbase/index&mirnas =MIMAT000 2884) search results, there are five predicted targets for miR528 in O. sativa, including one F-box domain containing protein (Os06g06050), two plastocyanin-like domain containing proteins (Os06g11310 and Os08g04310), OsAAO (ascorbate oxidase, Os06g37150), and OsCBP1 (Cu2+binding domain-containing protein 1, Os07g38290). MicroRNA528 has been reported to play a role in ROS accumulation and antiviral defense control [69]. MicroRNA528 also affects lodging resistance of maize by regulating lignin biosynthesis under nitrogen-luxury conditions [70]. We find that OsMYB30 is not a target of OsmiR528. The target (Os06g06050) of OsmiR528 is a positive regulator of OsMYB30. Overexpression of OsmiR528 decreased the expression of Os06g06050 that reduced the expression of OsMYB30. Reduced expression of OsMYB30 resulted in increased expression of BMY genes OsBMY2, OsBMY6, and OsBMY10, which enhanced cold stress tolerance.
CONCLUSION
Results in this investigation demonstrated that OsmiR528 enhances cold stress tolerance by reducing expression of OsMYB30. Under normal condition, less OsmiR528 causes little or no decrease of OsMYB30 expression or little or no decrease in the expression of the stress-response gene repressor that results in cold stress sensitive. Overexpression of OsmiR528 causes decreased expression of OsMYB30 or decreased expression of the stress-response gene repressor that leads to increased expression of BMY genes or stress-response genes, which enhances cold stress tolerance through increasing cell viability, growth rate, antioxidants content, ascorbate peroxidase (APOX) activity, and superoxide dismutase (SOD) activity and decreasing ion leakage rate and thiobarbituric acid reactive substances (TBARS) under cold stress in A. thaliana, P. elliottii, and O. sativa. Expressions of stress-related MYB transcription factors OsGAMYB-like1, OsMYBS3, OsMYB4, OsMYB3R-2, OsMYB5, OsMYB59, OsMYB30, OsMYB1R, and OsMYB20 in transgenic cells have been examined. OsmiR528 decreases expression of transcription factor OsMYB30 by targeting F-box domain containing protein gene (Os06g 06050). Reduced OsMYB30 expression results in increased expression of BMY genes OsBMY2, OsBMY6, and OsBMY 10. The expression levels of the OsBMY2, OsBMY6, and OsBMY10 were elevated by OsMYB30 knockdown, and the expression levels of the OsBMY2, OsBMY6, and OsBMY10 were reduced by OsMYB30 overexpression in OsmiR528 transgenic cells, indicating that OsmiR528 enhances low temperature tolerance by modulating low temperature stress response-associated genes. Results derived from the experiments provide novel information for increasing our understanding in molecular mechanisms of microRNAs-associated low temperature stress tolerance and are valuable in plant molecular breeding from monocotyledonous, dicotyledonous, and gymnosperm plants including A. thaliana, P. elliottii, and O. sativa.
Fig. (1).
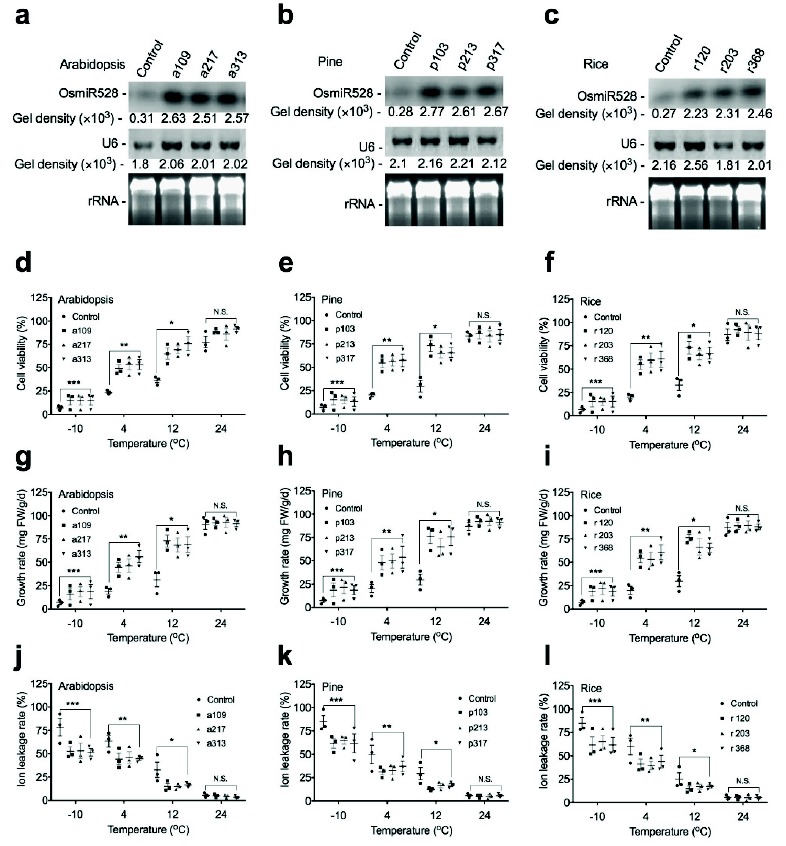
Overexpression of OsmiR528 enhances cold stress tolerance in transgenic cells. Overexpression of OsmiR528 was confirmed by northern blotting analysis in Arabidopsis (a), pine (b), and rice (c), respectively. Overexpression of OsmiR528 increases the cell viability, the cell growth rate, and decreases the ion leakage rate in cells of Arabidopsis (d, g, and j), pine (e, h, and k), and rice (f, i, and l), respectively. The statistically significant difference between groups was determined by one-way ANOVA. Data are presented as means of three independent experiments. Error bars represent standard error. The asterisk indicates significant differences compared to the control, as assessed by a t-test. *P<0.05, **P<0.01, ***P<0.001, significant relative to control. N.S., no statistical significance.
Table 1. Primers used in this study.
| Gene Name | Primers (5’ to 3’) |
|---|---|
| OsGAMYB-like1 | Forward: TCTGAGTTCTTCCTTTCCAATC Reverse: GATACATCGTGCCTCCAGAAA |
| OsMYB2 | Forward: ATGGACATGGCGCACGAGAG Reverse: CACACGGCGGCCTGGGTGG |
| OsMYBS3 | Forward: CCTTTCTGGCAAAATCAGAAAGA Reverse: ATGAACTGGAACAGGCTTGACA |
| OsMYB4 | Forward: CCGTGTGCGCCAAGGA Reverse: GAAGCTGTCGTCGATCTGGAA |
| OsMYB3R-2 | Forward: CAGGGTTTCTATCTCGTTCC Reverse: ATTTCCAAGCCCTTCCAC |
| OsMYB30 | Forward: ACTCCGGGATGGAGATGAG Reverse: GATGAACAGCTTGAGCCAGA |
| OsMYB55 | Forward: ATTACCCTCC ACTCCGTCCT CGGC Reverse: TCCAGTAGTTCTTGATCTCGTTGTC |
| OsMYB59 | Forward: GAGGCGTGCATCAGGAGCCTCG Reverse: GGCGGCGACGAACTGCCGGTG |
| OsMYB1R | Forward: GAGCAGCATGCAACAGTTTCTC Reverse: GGCTCCCGTTGTAGCTCAAC |
| OsBMY2 | Forward: AGATAAGGGAAAATCGGTTGG Reverse: TTAACAGCGACGGCAAAG |
| OsBMY6 | Forward: TTTCACCCGTAAGACTTA Reverse: AGTGGAAGAAGATTCAGTA |
| OsBMY10 | Forward: TGCCGATTGTCTGAATAAC Reverse: GAGATCCGAGCGATACTT |
(Fig. 1a), P. elliottii (Fig. 1b), and O. sativa (Fig. 1c), respectively. Results derived from the experiments of low temperature stress demonstrated that overexpression of OsmiR528 elevates the percentage of cell viability, the rate of cell growth, and reduces the rate of ion leakage in A. thaliana (Fig. 1d, 1g, 1j), P. elliottii (Fig. 1e, 1h, and 1k), and O. sativa (Fig. 1f, 1i, and 1l) at low temperature, respectively.
ACKNOWLEDGEMENTS
The authors are grateful to Bradshaw, Dr. Lischewski, and Dr. Andersen-Ranberg of Duke University for their critical reading and suggestions during the preparation of this manuscript.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
No Animals/Humans were used for studies that are the basis of this research.
CONSENT FOR PUBLICATION
Not applicable.
AUTHORS’ CONTRIBUTIONS
Wei Tang and Wells A. Thompson have done the analysis, imaging studies and contributed to the analysis of the manuscript. Wei Tang and Wells A. Thompson have contributed sample collection, experiments, and data analysis, and improvement of the manuscript. Wei Tang has conceived the experiments and contributed to the analysis. Wei Tang writes the manuscript. All authors reviewed the manuscript.
AVAILABILITY OF DATA AND MATERIALS
The data supporting the findings is available upon request.
FUNDING
This investigation was supported by a grant from the Education Committee of Hubei Providence of China and by a grant from the National Natural Science Foundation of China (31270740). We thank Dr. Prasad for support.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
SUPPLEMENTARY MATERIAL
Supplementary material is available on the publishers Web site along with the published article.
REFERENCES
- 1.Devi S.J., Madhav M.S., Kumar G.R., Goel A.K., Umakanth B., Jahnavi B., Viraktamath B.C. Identification of abiotic stress miRNA Transcription Factor Binding Motifs (TFBMs) in rice. Gene. 2013;531(1):15–22. doi: 10.1016/j.gene.2013.08.060. [DOI] [PubMed] [Google Scholar]
- 2.Yang Y., Zhang X., Chen Y., Guo J., Ling H., Gao S., Su Y., Que Y., Xu L. Selection of reference genes for normalization of microRNA expression by RT-qPCR in sugarcane buds under cold stress. Front. Plant Sci. 2016;7(1):86. doi: 10.3389/fpls.2016.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shu Y., Liu Y., Li W., Song L., Zhang J., Guo C. Genomewide investigation of microRNAs and their targets in response to freezing stress in Medicago sativa L., based on high-throughput sequencing. G3 Bethseda, 2016;6(1):755–765. doi: 10.1534/g3.115.025981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maeda S., Sakazono S., Masuko-Suzuki H., Taguchi M., Yamamura K., Nagano K., Endo T., Saeki K., Osaka M., Nabemoto M., Ito K., Kudo T., Kobayashi M., Kawagishi M., Fujita K., Nanjo H., Shindo T., Yano K., Suzuki G., Suwabe K., Watanabe M. Comparative analysis of microRNA profiles of rice anthers between cool-sensitive and cool-tolerant cultivars under cool-temperature stress. Genes Genet. Syst. 2016;91(1):97–109. doi: 10.1266/ggs.15-00056. [DOI] [PubMed] [Google Scholar]
- 5.Bredow M., Vanderbeld B., Walker V.K. Knockdown of ice-binding proteins in Brachypodium distachyon demonstrates their role in freeze protection. PLoS One. 2016;11:e0167941. doi: 10.1371/journal.pone.0167941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim J.J., Lee J.H., Kim W., Jung H.S., Huijser P., Ahn J.H. The microRNA156-squamosa promoter binding protein-like3 module regulates ambient temperature-responsive flowering via flowering locus T in Arabidopsis. Plant Physiol. 2012;159(1):461–478. doi: 10.1104/pp.111.192369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gebelin V., Argout X., Engchuan W., Pitollat B., Duan C., Montoro P., Leclercq J. Identification of novel microRNAs in Hevea brasiliensis and computational prediction of their targets. BMC Plant Biol. 2012;12(1):18. doi: 10.1186/1471-2229-12-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim J.Y., Kwak K.J., Jung H.J., Lee H.J., Kang H. MicroRNA402 affects seed germination of Arabidopsis thaliana under stress conditions via targeting DEMETER-LIKE Protein3 mRNA. Plant Cell Physiol. 2010;51(1):1079–1083. doi: 10.1093/pcp/pcq072. [DOI] [PubMed] [Google Scholar]
- 9.Kamthan A., Chaudhuri A., Kamthan M., Datta A. Small RNAs in plants: recent development and application for crop improvement. Front. Plant Sci. 2015;6(1):208. doi: 10.3389/fpls.2015.00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen H., Chen X., Chai X., Qiu Y., Gong C., Zhang Z., Wang T., Zhang Y., Li J., Wang A. Effects of low temperature on mRNA and small RNA transcriptomes in Solanum lycopersicoides leaf revealed by RNA-Seq. Biochem. Biophys. Res. Commun. 2015;464(3):768–773. doi: 10.1016/j.bbrc.2015.07.029. [DOI] [PubMed] [Google Scholar]
- 11.Zeng C., Chen Z., Xia J., Zhang K., Chen X., Zhou Y., Bo W., Song S., Deng D., Guo X., Wang B., Zhou J., Peng H., Wang W., Peng M., Zhang W. Chilling acclimation provides immunity to stress by altering regulatory networks and inducing genes with protective functions in cassava. BMC Plant Biol. 2014;14(207) doi: 10.1186/s12870-014-0207-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun X., Fan G., Su L., Wang W., Liang Z., Li S., Xin H. Identification of cold-inducible microRNAs in grapevine. Front. Plant Sci. 2015;6(1):595. doi: 10.3389/fpls.2015.00595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma C., Burd S., Lers A. miR408 is involved in abiotic stress responses in Arabidopsis. Plant J. 2015;84(1):169–187. doi: 10.1111/tpj.12999. [DOI] [PubMed] [Google Scholar]
- 14.Sosa-Valencia G., Palomar M., Covarrubias A.A. Reyes, J.L. The legume miR1514a modulates a NAC transcription factor transcript to trigger phasiRNA formation in response to drought. J. Exp. Bot. 2017;68(8):2013–2026. doi: 10.1093/jxb/erw380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen X., Guo X., Guo X., Zhao D., Zhao W., Chen J., Li T. PacMYBA, a sweet cherry R2R3-MYB transcription factor, is a positive regulator of salt stress tolerance and pathogen resistance. Plant Physiol. Biochem. 2017;112(1):302–331. doi: 10.1016/j.plaphy.2017.01.015. [DOI] [PubMed] [Google Scholar]
- 16.Agarwal M., Hao Y., Kapoor A., Dong C.H., Fujii H., Zheng X., Zhu J.K.A. R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J. Biol. Chem. 2006;281(49):37636–37645. doi: 10.1074/jbc.M605895200. [DOI] [PubMed] [Google Scholar]
- 17.Al-Attala M.N., Wang X., Abou-Attia M.A., Duan X., Kang Z. A novel TaMYB4 transcription factor involved in the defence response against Puccinia striiformis f. sp. tritici and abiotic stresses. Plant Mol. Biol. 2014;84(4-5):589–603. doi: 10.1007/s11103-013-0156-7. [DOI] [PubMed] [Google Scholar]
- 18.Bai B., Wu J., Sheng W.T., Zhou B., Zhou L.J., Zhuang W., Yao D.P., Deng Q.Y. Comparative analysis of anther transcriptome profiles of two different rice male sterile lines genotypes under cold stress. Int. J. Mol. Sci. 2015;16(5):11398–11416. doi: 10.3390/ijms160511398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bedon F., Bomal C., Caron S., Levasseur C., Boyle B., Mansfield S.D., Schmidt A., Gershenzon J., Grima-Pettenati J., Seguin A., MacKay J. Subgroup 4 R2R3-MYBs in conifer trees: gene family expansion and contribution to the isoprenoid- and flavonoid-oriented responses. J. Exp. Bot. 2010;61(14):3847–3864. doi: 10.1093/jxb/erq196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bonthala V.S., Mayes K., Moreton J., Blythe M., Wright V., May S.T., Massawe F., Mayes S., Twycross J. Identification of gene modules associated with low temperatures response in Bambara Groundnut by network-based analysis. PLoS One. 2016;11(2):e0148771. doi: 10.1371/journal.pone.0148771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Butelli E., Licciardello C., Zhang Y., Liu J., Mackay S., Bailey P., Reforgiato-Recupero G., Martin C. Retrotransposons control fruit-specific, cold-dependent accumulation of anthocyanins in blood oranges. Plant Cell. 2012;24(3):1242–1255. doi: 10.1105/tpc.111.095232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Butt H.I., Yang Z., Chen E., Zhao G., Gong Q., Yang Z., Zhang X., Li F. Functional characterization of cotton GaMYB62L, a novel R2R3 TF in transgenic Arabidopsis. PLoS One. 2017;12(1):e0170578. doi: 10.1371/journal.pone.0170578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davey M.W., Graham N.S., Vanholme B., Swennen R., May S.T., Keulemans J. Heterologous oligonucleotide microarrays for transcriptomics in a non-model species; a proof-of-concept study of drought stress in Musa. BMC Genomics. 2009;10(1):436. doi: 10.1186/1471-2164-10-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hichri I., Barrieu F., Bogs J., Kappel C., Delrot S., Lauvergeat V. Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J. Exp. Bot. 2011;62(8):2465–2483. doi: 10.1093/jxb/erq442. [DOI] [PubMed] [Google Scholar]
- 25.Imtiaz M., Yang Y., Liu R., Xu Y., Khan M.A., Wei Q., Gao J., Hong B. Identification and functional characterization of the BBX24 promoter and gene from chrysanthemum in Arabidopsis. Plant Mol. Biol. 2015;89(1):1–19. doi: 10.1007/s11103-015-0347-5. [DOI] [PubMed] [Google Scholar]
- 26.Kiferle C., Fantini E., Bassolino L., Povero G., Spelt C., Buti S., Giuliano G., Quattrocchio F., Koes R., Perata P., Gonzali S. Tomato R2R3-MYB proteins SlANT1 and SlAN2: same protein activity, different roles. PLoS One. 2015;10(8):e0136365. doi: 10.1371/journal.pone.0136365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lippold F., Sanchez D.H., Musialak M., Schlereth A., Scheible W.R., Hincha D.K., Udvardi M.K. AtMyb41 regulates transcriptional and metabolic responses to osmotic stress in Arabidopsis. Plant Physiol. 2009;149(4):1761–1772. doi: 10.1104/pp.108.134874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saha G., Park J.I., Ahmed N.U., Kayum M.A., Kang K.K., Nou I.S. Characterization and expression profiling of MYB transcription factors against stresses and during male organ development in Chinese cabbage (Brassica rapa ssp. pekinensis). Plant Physiol. Biochem. 2016;104(1):200–215. doi: 10.1016/j.plaphy.2016.03.021. [DOI] [PubMed] [Google Scholar]
- 29.Tombuloglu H., Kekec G., Sakcali M.S., Unver T. Transcriptome-wide identification of R2R3-MYB transcription factors in barley with their boron responsive expression analysis. Mol. Genet. Genomics. 2013;288(3-4):141–155. doi: 10.1007/s00438-013-0740-1. [DOI] [PubMed] [Google Scholar]
- 30.Chen Y., Chen Z., Kang J., Kang D., Gu H., Qin G. AtMYB14 regulates cold tolerance in arabidopsis. Plant Mol. Biol. Report. 2013;31(1):87–97. doi: 10.1007/s11105-012-0481-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H., Dong Y., Chang J., He J., Chen H., Liu Q., Wei C., Ma J., Zhang Y., Yang J., Zhang X. High-throughput microRNA and mRNA sequencing reveals that microRNAs may be involved in melatonin-mediated cold tolerance in Citrullus lanatus L. Front. Plant Sci. 2016;7(1):1231. doi: 10.3389/fpls.2016.01231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song G., Zhang R., Zhang S., Li Y., Gao J., Han X., Chen M., Wang J., Li W., Li G. Response of microRNAs to cold treatment in the young spikes of common wheat. BMC Genomics. 2017;18(1):212. doi: 10.1186/s12864-017-3556-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yuan S., Li Z., Li D., Yuan N., Hu Q., Luo H. Constitutive expression of rice MicroRNA528 alters plant development and enhances tolerance to salinity stress and nitrogen starvation in creeping bentgrass. Plant Physiol. 2015;169(1):576–593. doi: 10.1104/pp.15.00899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chavez-Hernandez E.C., Alejandri-Ramirez N.D., Juarez-Gonzalez V.T., Dinkova T.D. Maize miRNA and target regulation in response to hormone depletion and light exposure during somatic embryogenesis. Front. Plant Sci. 2015;6(1):555. doi: 10.3389/fpls.2015.00555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheah B.H., Nadarajah K., Divate M.D., Wickneswari R. Identification of four functionally important microRNA families with contrasting differential expression profiles between drought-tolerant and susceptible rice leaf at vegetative stage. BMC Genomics. 2015;16(1):692. doi: 10.1186/s12864-015-1851-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ragupathy R., Ravichandran S., Mahdi M.S., Huang D., Reimer E., Domaratzki M., Cloutier S. Deep sequencing of wheat sRNA transcriptome reveals distinct temporal expression pattern of miRNAs in response to heat, light and UV. Sci. Rep. 2016;6:39373. doi: 10.1038/srep39373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thiebaut F., Rojas C.A., Grativol C., Motta M.R., Vieira T., Regulski M., Martienssen R.A., Farinelli L., Hemerly A.S., Ferreira P.C. Genome-wide identification of microRNA and siRNA responsive to endophytic beneficial diazotrophic bacteria in maize. BMC Genomics. 2014;15:766. doi: 10.1186/1471-2164-15-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lv Y., Yang M., Hu D., Yang Z., Ma S., Li X., Xiong L. The OsMYB30 transcription factor suppresses cold tolerance by interacting with a jaz protein and suppressing beta-amylase expression. Plant Physiol. 2017;173(2):1475–1491. doi: 10.1104/pp.16.01725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang W., Page M. Transcription factor AtbZIP60 regulates expression of Ca2+ -dependent protein kinase genes in transgenic cells. Mol. Biol. Rep. 2013;40(1):2723–2732. doi: 10.1007/s11033-012-2362-9. [DOI] [PubMed] [Google Scholar]
- 40.Shimono M., Sugano S., Nakayama A., Jiang C.J., Ono K., Toki S., Takatsuji H. Rice WRKY45 plays a crucial role in benzothiadiazole-inducible blast resistance. Plant Cell. 2007;19(6):2064–2076. doi: 10.1105/tpc.106.046250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Toki S., Hara N., Ono K., Onodera H., Tagiri A., Oka S., Tanaka H. Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J. 2006;47(6):969–976. doi: 10.1111/j.1365-313X.2006.02836.x. [DOI] [PubMed] [Google Scholar]
- 42.Tang W., Newton R.J., Weidner D.A. Genetic transformation and gene silencing mediated by multiple copies of a transgene in eastern white pine. J. Exp. Bot. 2007;58(3):545–554. doi: 10.1093/jxb/erl228. [DOI] [PubMed] [Google Scholar]
- 43.Sung Z.R. Turbidimetric measurement of plant cell culture growth. Plant Physiol. 1976;57:460–462. doi: 10.1104/pp.57.3.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Becana M., Aparicio-Tejo P., Irigoyen J.J., Sanchez-Diaz M. Some enzymes of hydrogen peroxide metabolism in leaves and root nodules of Medicago sativa. Plant Physiol. 1986;82(4):1169–1171. doi: 10.1104/pp.82.4.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tang W., Newton R.J. Peroxidase and catalase activities are involved in direct adventitious shoot formation induced by thidiazuron in eastern white pine (Pinus strobus L.) zygotic embryos. Plant Physiol. Biochem. 2005;43(8):760–769. doi: 10.1016/j.plaphy.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 46.Tang W., Charles T.M., Newton R.J. Overexpression of the pepper transcription factor CaPF1 in transgenic Virginia pine (Pinus Virginiana Mill.) confers multiple stress tolerance and enhances organ growth. Plant Mol. Biol. 2005;59(4):603–617. doi: 10.1007/s11103-005-0451-z. [DOI] [PubMed] [Google Scholar]
- 47.Tang W., Newton R.J., Li C., Charles T.M. Enhanced stress tolerance in transgenic pine expressing the pepper CaPF1 gene is associated with the polyamine biosynthesis. Plant Cell Rep. 2007;26(1):115–124. doi: 10.1007/s00299-006-0228-0. [DOI] [PubMed] [Google Scholar]
- 48.Abe H., Yamaguchi-Shinozaki K., Urao T., Iwasaki T., Hosokawa D., Shinozaki K. Role of arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell. 1997;9(10):1859–1868. doi: 10.1105/tpc.9.10.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raffaele S., Vailleau F., Leger A., Joubes J., Miersch O., Huard C., Blee E., Mongrand S., Domergue F., Roby D. A MYB transcription factor regulates very-long-chain fatty acid biosynthesis for activation of the hypersensitive cell death response in Arabidopsis. Plant Cell. 2008;20(3):752–767. doi: 10.1105/tpc.107.054858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Almeida T., Pinto G., Correia B., Santos C., Goncalves S. QsMYB1 expression is modulated in response to heat and drought stresses and during plant recovery in Quercus suber. Plant Physiol. Biochem. 2013;73(1):274–281. doi: 10.1016/j.plaphy.2013.10.007. [DOI] [PubMed] [Google Scholar]
- 51.Bergonzi S., Albani M.C., Ver Loren Van Themaat E., Nordstrom K.J., Wang R., Schneeberger K., Moerland P.D., Coupland G. Mechanisms of age-dependent response to winter temperature in perennial flowering of Arabis alpina. Science. 2013;340(6136):1094–1097. doi: 10.1126/science.1234116. [DOI] [PubMed] [Google Scholar]
- 52.Cheng L., Li X., Huang X., Ma T., Liang Y., Ma X., Peng X., Jia J., Chen S., Chen Y., Deng B., Liu G. Overexpression of sheepgrass R1-MYB transcription factor LcMYB1 confers salt tolerance in transgenic Arabidopsis. Plant Physiol. Biochem. 2013;70(1):252–260. doi: 10.1016/j.plaphy.2013.05.025. [DOI] [PubMed] [Google Scholar]
- 53.Lee H.G., Seo P.J. The MYB96-HHP module integrates cold and abscisic acid signaling to activate the CBF-COR pathway in Arabidopsis. Plant J. 2015;82(6):962–977. doi: 10.1111/tpj.12866. [DOI] [PubMed] [Google Scholar]
- 54.He Q., Jones D.C., Li W., Xie F., Ma J., Sun R., Wang Q., Zhu S., Zhang B. Genome-wide identification of R2R3-MYB genes and expression analyses during abiotic stress in Gossypium raimondii. Sci. Rep. 2016;6(22980) doi: 10.1038/srep22980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang Q.X., Cheng X.Y., Mao Z.C., Wang Y.S., Zhao L.L., Yan X., Ferris V.R., Xu R.M., Xie B.Y. MicroRNA discovery and analysis of pinewood nematode Bursaphelenchus xylophilus by deep sequencing. PLoS One. 2010;5(10):e13271. doi: 10.1371/journal.pone.0013271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Debat H.J., Grabiele M., Aguilera P.M., Bubillo R.E., Otegui M.B., Ducasse D.A., Zapata P.D., Marti D.A. Exploring the genes of yerba mate (Ilex paraguariensis A. St.-Hil.) by NGS and de novo transcriptome assembly. PLoS One. 2014;9(10):e109835. doi: 10.1371/journal.pone.0109835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mathieu J., Yant L.J., Murdter F., Kuttner F., Schmid M. Repression of flowering by the miR172 target SMZ. PLoS Biol. 2009;7:e1000148. doi: 10.1371/journal.pbio.1000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang C., Li D., Mao D., Liu X., Ji C., Li X., Zhao X., Cheng Z., Chen C., Zhu L. Overexpression of microRNA319 impacts leaf morphogenesis and leads to enhanced cold tolerance in rice (Oryza sativa L.). Plant Cell Environ. 2013;36(12):2207–2218. doi: 10.1111/pce.12130. [DOI] [PubMed] [Google Scholar]
- 59.Peng X., Wu Q., Teng L., Tang F., Pi Z., Shen S. Transcriptional regulation of the paper mulberry under cold stress as revealed by a comprehensive analysis of transcription factors. BMC Plant Biol. 2015;15:108. doi: 10.1186/s12870-015-0489-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yan Y., Shen L., Chen Y., Bao S., Thong Z. Yu, H. A MYB-domain protein EFM mediates flowering responses to environmental cues in Arabidopsis. Dev. Cell. 2014;30(4):437–448. doi: 10.1016/j.devcel.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 61.Wang R.K., Cao Z.H., Hao Y.J. Overexpression of a R2R3 MYB gene MdSIMYB1 increases tolerance to multiple stresses in transgenic tobacco and apples. Physiol. Plant. 2014;150(1):76–87. doi: 10.1111/ppl.12069. [DOI] [PubMed] [Google Scholar]
- 62.Chiba Y., Mineta K., Hirai M.Y., Suzuki Y., Kanaya S., Takahashi H., Onouchi H., Yamaguchi J., Naito S. Changes in mRNA stability associated with cold stress in Arabidopsis cells. Plant Cell Physiol. 2013;54(2):180–194. doi: 10.1093/pcp/pcs164. [DOI] [PubMed] [Google Scholar]
- 63.Yang A., Dai X., Zhang W.H.A. R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J. Exp. Bot. 2012;63(7):2541–2556. doi: 10.1093/jxb/err431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Samad A.F.A., Sajad M., Nazaruddin N., Fauzi I.A., Murad A.M.A., Zainal Z., Ismail I. MicroRNA and transcription factor: key players in plant regulatory network. Front. Plant Sci. 2017;8:565. doi: 10.3389/fpls.2017.00565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Candar-Cakir B., Arican E., Zhang B. Small RNA and degradome deep sequencing reveals drought-and tissue-specific micrornas and their important roles in drought-sensitive and drought-tolerant tomato genotypes. Plant Biotechnol. J. 2016;14(8):1727–1746. doi: 10.1111/pbi.12533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reyes J.L., Chua N.H. ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. Plant J. 2007;49(4):592–606. doi: 10.1111/j.1365-313X.2006.02980.x. [DOI] [PubMed] [Google Scholar]
- 67.Din M., Barozai M.Y. Profiling microRNAs and their targets in an important fleshy fruit: Tomato (Solanum lycopersicum). Gene. 2014;535(2):198–203. doi: 10.1016/j.gene.2013.11.034. [DOI] [PubMed] [Google Scholar]
- 68.Xie F., Wang Q., Sun R., Zhang B. Deep sequencing reveals important roles of microRNAs in response to drought and salinity stress in cotton. J. Exp. Bot. 2015;66(3):789–804. doi: 10.1093/jxb/eru437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu J., Yang Z., Yang S., Yao S., Zhao Y., Wang P., Li X., Song L., Jin T., Zhou Y., Lan L., Xie X., Zhou C., Chu Y., Qi X., Cao Y., Li Y. ROS accumulation and antiviral defence control by microRNA528 in rice. Nat. Plants. 2017;3:16203. doi: 10.1038/nplants.2016.203. [DOI] [PubMed] [Google Scholar]
- 70.Sun Q., Liu X., Yang J., Liu W., Du Q., Wang H., Fu C., Li W-X. MicroRNA528 affects lodging resistance of maize by regulating lignin biosynthesis under nitrogen-luxury conditions. Mol. Plant. 2018;11(6):806–814. doi: 10.1016/j.molp.2018.03.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material is available on the publishers Web site along with the published article.
Data Availability Statement
The data supporting the findings is available upon request.


