Abstract
Influenza-specific immunity in humans is unique because there are repeated exposures to viral strains containing genetically conserved epitopes recruiting memory CD4 T cells and novel epitopes stimulating naïve CD4 T cells, possibly resulting in competition between memory and naïve lymphocytes. In the studies described here, we evaluated the effect of this competition on CD4 T cell and B cell response specificity using a murine model of sequential influenza infection. We found striking and selective decreases in CD4 T cell reactivity to nonconserved HA epitopes following secondary influenza infection. Surprisingly, this shift in CD4 T cell specificity was associated with dramatic decreases in HA-specific antibody. These results suggest that repeated exposure to influenza viruses and vaccines containing conserved internal proteins may have unintended and negative consequences on the ability to induce HA-specific antibody to novel pandemic strains of influenza. These finding could have important implications on pandemic influenza preparedness strategies.
Keywords: Heterosubtypic influenza infection, immunodominance, epitopes, CD4 T cells
Introduction
Influenza-specific immunity offers unique challenges to the human host because of frequent changes in the immunogen over time. Antibody-mediated immune pressure leads to emergence of immunological escape variants that circulate in the population, which in turn necessitates formulation and administration of vaccines derived from new viral strains. Also, periodically, completely novel strains with pandemic potential arise due to recombination between two viruses. Thus, both natural infection and yearly vaccination result in exposure to novel epitopes that elicit a naïve T cell response and genetically conserved determinants that stimulate memory T cells. How this competition among memory and naïve T cells influences the specificity and functionality of the protective immune response is poorly understood.
Although there is substantial literature describing the specificity of the primary T cell response to influenza (1–5), there have been relatively few studies examining how the primary T cell repertoire is remodeled with even a single subsequent encounter with a strain of influenza that shares some, but not all, of its T cell epitopes with the first virus. It is known that the CD8 T cell immunodominance hierarchy can shift following a secondary infection (6), in part as a result of differential antigen presentation (7). However, the duration of antigen presentation may also be decreased following a secondary infection; possibly due to destruction of APC by cytotoxic memory CD8 T cells (8–10). Although memory and naïve CD8 T cells have been found to expand concurrently in response to influenza infection (11), how repeat encounters with heterosubtypic strains of influenza virus shape the specificity of the CD4 T cell response against this virus has not yet been evaluated.
In this study, we have used a murine model of sequential influenza infection to examine the specificity of CD4 T cell and antibody responses following heterosubtypic infection. We found that, as expected, a secondary infection boosted responses to genetically conserved epitopes. Strikingly, this boost was accompanied by a dramatic suppression of CD4 T cell responses directed against novel influenza epitopes contained in the challenge virus. Most surprisingly, the shift in CD4 T cell specificity toward conserved internal virion proteins was associated with a decrease in hemagglutinin-specific antibody. These results suggest that periodic natural infections with influenza and the yearly administration of inactivated influenza vaccines containing internal virion proteins (12–14) could have unexpected deleterious consequences on the production of neutralizing antibody following exposure to a novel influenza strain.
Materials and Methods
Mice
C57BL/10J mice (“B10;” H-2b) were purchased from Jackson Laboratory (Bar Harbor, ME). B10.S-H2S/SqMcdJ mice (“B10.S;” H-2s) were purchased from Jackson Laboratory and bred at the University of Rochester. Animals were housed in specific pathogen free facilities and maintained according to institutional guidelines. Studies were performed in compliance with the United States Department of Health and Human Services Guide for the Care and Use of Laboratory Animals. Animal use protocols were approved by the University of Rochester Committee on Animal Resources; Animal Welfare Assurance Number A3291-01.
Synthetic peptides
17-mer peptides previously identified as I-Ab or I-As restricted influenza epitopes (2) were obtained through the NIH Biodefense and Emerging Infections Research Resources Repository, NIAID, NIH or synthesized in our facility using an Apex 396 system (AAPPTec, Louisville, KY) as described previously (15). Peptides used as antigen for restimulation are shown in Supplemental Table I (I-Ab) and Supplemental Table II (I-As), with the x139 sequence above and the X-31 sequences below. Individual peptides were used at a final concentration of 10 μM.
Influenza infection
X-31 influenza virus was provided by Dr. David Topham at the University of Rochester and x139 influenza virus was provided by Dr. Doris Bucher at New York Medical College. Mice were anesthetized by intraperitoneal (IP) injection of tribromoethanol and were infected intranasally with 30 μL X-31 in PBS at a dose of 300,000 EID50 per mouse. After 8–9 weeks, mice were infected intranasally with x139 influenza at a dose of 50,000 EID50 per mouse. Post infection, spleens and mediastinal lymph nodes (MLN) were excised and used as a source of CD4 T cells for EliSpot analysis. Syngeneic splenocytes from uninfected mice were used as antigen presenting cells (APC).
EliSpot assays
IFNγ EliSpot assays were performed as previously described (2). Briefly, CD4 T cells were enriched from splenocytes either by antibody and complement (2) or by negative selection using MACS no touch CD4 purification (Miltenyi Biotec, Gladbach, Germany) per the manufacturer’s instructions. Individual MLNs were utilized undepleted. The plates were processed and analyzed as previously described (2). Data were calculated and presented as cytokine EliSpots per million CD4 T cells with background values subtracted (typically <20 spots per well), and MLN results were normalized to account for the percentage of CD4 T cells as determined by analytical flow cytometry.
Microneutralization assay
Sera were treated with receptor-destroying enzyme per the manufacturer’s protocol (RDE; Denka Seiken, Tokyo, Japan) and heat inactivated prior to testing as previously described (15). The microneutralization titers were defined as the reciprocal of the highest serum dilution at which all of the culture wells were negative for cytopathic effect.
CD8 depletion
CD8 antibody (Clone 2.43, BioXCell, West Lebanon, NH) was used for in vivo depletion of CD8 T cells. Two hundred mg of anti-CD8 or isotype control IgG2b (BioXCell) antibodies were injected intraperitoneally every other day beginning 2 days prior to infection. At 8 days post infection, the mice were euthanized and tissues and blood were harvested for Elispot assay.
Flow cytometry
Analytical flow cytometry was performed by staining with CD4-fluorescein isothiocyanate (CD4-FITC clone RM4–4, BD Biosciences) and CD8a-FITC (Ly-2 clone 53–6.7, eBiosciences, San Diego, CA) or CD8b-FITC (H35–17.2, eBiosciences). Data were analyzed using Cell Quest software (Becton Dickinson).
ELISA assays
Blood was collected from individual mice and the presence of HA- and NP-specific antibodies in serum was determined using ELISA assays as previously described (15) using either 250 ng/100 μL of recombinant A/New Caledonia/20/99 HA protein (Protein Sciences, Meriden, CT) or 200 ng/μL recombinant A/New Caledonia/20/99 NP protein produced in house using an E. coli expression system (15). After incubation with diluted serum, the plates were washed and developed as previously described (15).
Results and Discussion
It is known that naïve and memory CD4 T cells differ with regard to their gene expression patterns and their sensitivity to antigen (16–17), but how these differences influence competitive immune responses as occur following heterosubtypic influenza infection has not been explored. To rigorously address this issue, we used an animal model of sequential infection. Mice were initially infected with “X-31”, a recombinant influenza virus containing the hemagglutinin (HA) and neuraminidase (NA) proteins of A/Aichi/2/68 (H3N2), with all other proteins derived from A/Puerto Rico/8/34 (H1N1). After waiting 8–9 weeks to establish memory, mice were infected with a reassortant virus (“x139”) composed of the HA, NA, nuclear protein (NP) and polymerase basic 1 (PB1) proteins of A/New Caledonia/20/99 virus (H1N1) with all other proteins derived from X-31. This combination of viruses thus has unrelated HA and NA proteins while most internal viral proteins remain conserved. At various time points post-infection, CD4 T cell responses were directly compared between secondary and primary x139 infections using IFNγ EliSpot assays. CD4 T cell specificity was assessed using known I-Ab and I-As restricted influenza peptides from the HA, NA, NP, M1 and PB1 proteins (2). Mice infected with X-31 eight to 9 weeks prior served as a control for waning CD4 T cell immunity.
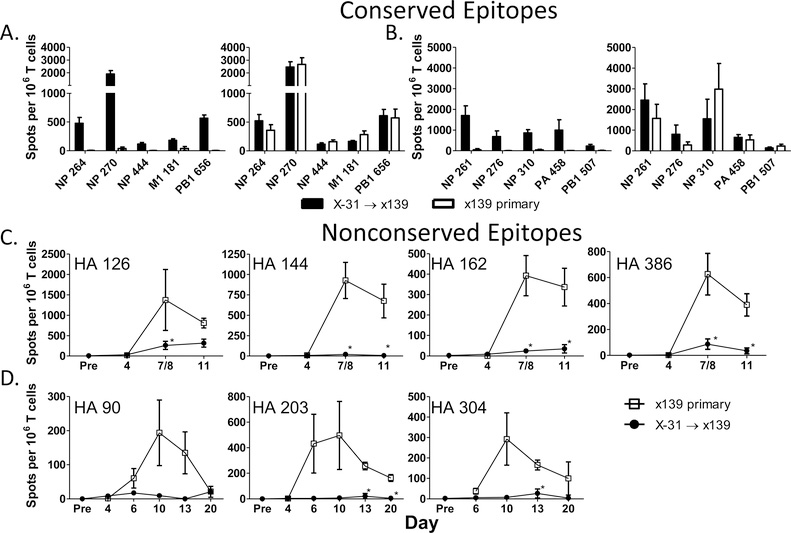
Our initial experiments revealed that CD4 T cell responses directed against conserved epitopes were maintained or boosted following a secondary heterosubtypic influenza infection (Figure 1A and B). Additionally and quite unexpectedly, responses to specificities unique to the new challenge virus were greatly diminished throughout the duration of the response compared to responses following a primary infection (Figure 1C and D). The suppression affected multiple HA epitopes in both the B10.S (Figure 1C) and B10 (Figure 1D) mouse strains, persisted through all time points tested, and was present in both the spleen and the draining mediastinal lymph node (data not shown). Collectively, these data suggest that following secondary infection with a viral strain containing both conserved and highly divergent epitopes, new specificities contributed by naïve cells are at a significant disadvantage compared to responding memory CD4 T cells devoted to the more conserved peptide epitopes.
Figure 1: CD4 T cell reactivity to nonconserved epitopes is selectively diminished following a secondary heterosubtypic influenza infection.
B10.S and B10 mice were infected sequentially with X-31 (H3N2) followed by x139 influenza (H1N1) or were only infected with x139 influenza. CD4 T cells were isolated from the spleen and EliSpot assays were performed using I-As and I-Ab restricted peptide-epitopes as restimulation antigens. The top panels depict reactivity to conserved epitopes in B10 .S (A) and B10 (B) mice at day 4 (leftward panels) and day 7 or 8 (rightward panels) following infection. Panel C demonstrates CD4 T cell reactivity to nonconserved epitopes in B10.S mice. Each time point represents data from 5–9 individual mice. Panel D shows reactivity to nonconserved epitopes in B10 mice over time; each time point shows the average of 3 individual mice. Results with a p-value of <0.05 using Welch’s T test when comparing the secondary to the primary infection are depicted with an asterisk.
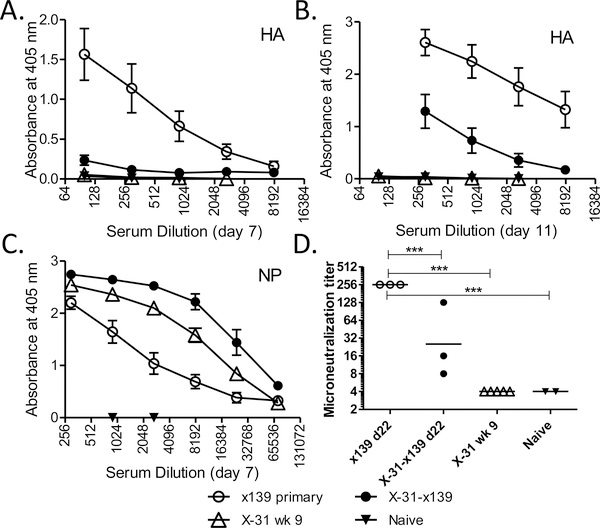
We next sought to determine if this change in CD4 T cell specificity is associated with any changes in antibody specificity or kinetics. The issue of a linkage between CD4 T cell and B cell specificities is somewhat controversial (14, 18–20). Such a linkage could occur because under physiologic conditions antigen-specific B cells predominantly internalize antigen through their B cell receptor (21) and thus, depending on the source of antigen available, not all virion proteins may be equally internalized by virus-specific B cells. This restricted antigen uptake could favor the presentation of HA-derived epitopes by HA-specific B cells and result in the selective recruitment of help from HA-specific CD4 T cells. ELISA assays were used to compare reactivity against the A/New Caledonia HA and NP proteins expressed in the challenge strain following a primary or secondary heterosubtypic infection. Unexpectedly, we found markedly diminished HA-specific antibody titers at days 7 and 11 following secondary infection (Figure 2A and B) that contrasted with accelerated kinetics of NP-specific antibody production (Figure 2C). This pattern was maintained when neutralizing antibody titers to HA were tested using a microneutralization assay (Figure 2D). These data reveal a linkage between CD4 T cell and B cell specificities and suggest that HA-specific CD4 T cells may be better able to provide help in the production of high affinity neutralizing antibody. Together, these studies suggest that CD4 T cell specificity following primary infection is substantially remodeled following secondary infection and that this change is associated with a dramatic shift in antibody specificity.
Figure 2: Heterosubtypic infection is associated with a decrease in HA-specific antibody.
HA- (Panels A and B) and NP-specific (Panel C) ELISA assays were performed on serum obtained from B10.S mice following primary and secondary influenza infection (open and closed circles, respectively). Serum from mice infected with X-31 8 to 9 weeks prior and from naïve mice served as controls (open and closed triangles, respectively). Each point represents the average of sera from 4 to 9 individual mice (experimental groups) or 2 to 5 individual mice (control groups). Error bars depict the standard error of the mean. Panel D demonstrates the results of a microneutralization assay performed on sera obtained from individual B10.S mice at 22 days post infection. Horizontal lines show the geometric mean of each group. A one-way ANOVA with Bonferroni’s Multiple Comparison Test was used to compare groups, with asterisks denoting statistical significance.
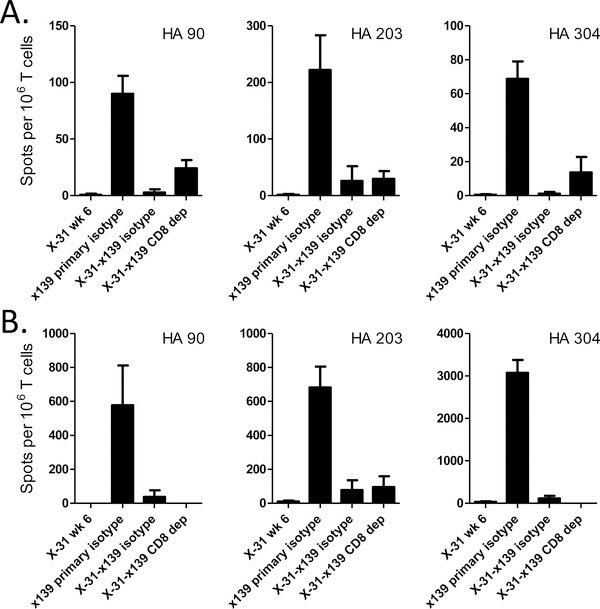
There are several possible mechanisms that may act alone or together during a heterosubtypic viral infection to account for these findings. One possibility to explain the shifts in CD4 T cell specificity is that the cytolytic activity of memory CD8 T cells decreases the duration of antigen presentation (8, 10, 22), selectively inhibiting the expansion of naïve CD4 T cells. This possibility was investigated by selective depletion of CD8 T cells prior to and throughout the duration of the secondary infection. This procedure had only minor effects on the HA-specific suppression of CD4 T cells (Figure 3), suggesting that CD8 T cell activity does not have a major role in the suppression of responses seen.
Figure 3: CD8 depletion has a modest effect on CD4 T cell reactivity to nonconserved HA epitopes.
B10 CD8 T cells were depleted in vivo throughout the duration of secondary infection (days −2, 0, 2, 4, and 6). Eight days post infection, CD4 T cell reactivity to various epitopes was examined by IFNγ Elispot assay. Panel A depicts data obtained from MACS purified CD4 T cells isolated from the spleen. Panel B shows IFNγ Elispots obtained from unpurified MLN cells. Data are presented as spots per 106 CD4 T cells, with MLN data normalized for the percent of CD4 T cells present. Flow cytometric analysis of both spleen and MLN cells indicated complete depletion of CD8 T cells.
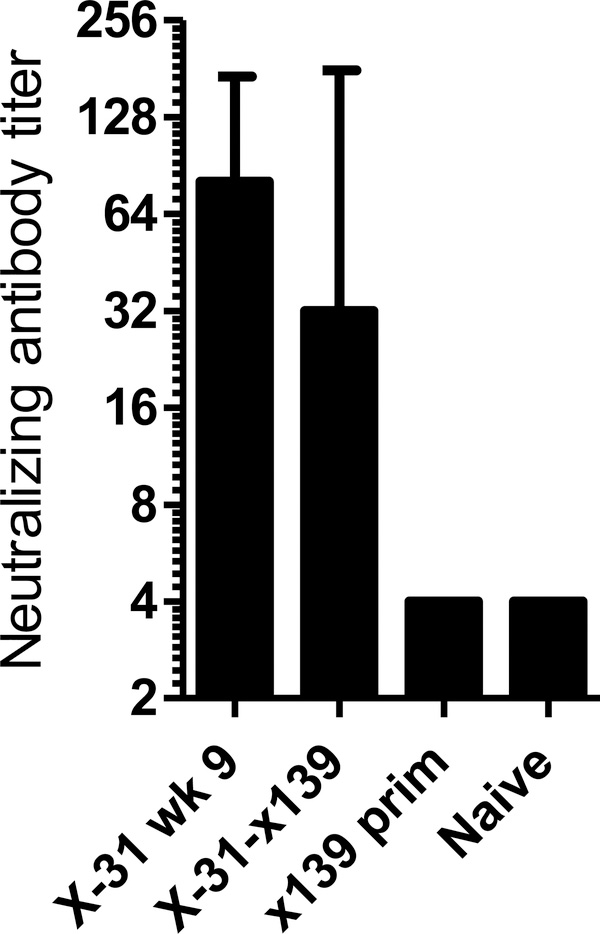
Another possibility is that the B cells primed during the primary infection are directly perturbing the secondary infection, either through cross reactive neutralizing antibody or via phenomenon of original antigenic sin. Original antigenic sin has been described to occur following sequential infection with closely related influenza variants. In this situation, infection with second, related strain induces a preferential antibody response to the first viral strain encountered, with a resulting decrease in the antibody response to the newly infecting strain (23–24). In our model of heterosubtypic infection, the infecting strains contain highly divergent hemagglutinin molecules from different HA groups (“X-31” H3; “x139” H1) that did not induce detectable levels of cross reactive antibody by either ELISA (Figure 2A and B) or microneutralization assay (Figure 2D). This argues against a role for cross reactive neutralizing antibody in either decreasing the viral load or causing original antigenic sin. Additionally, boosting of the antibody response to X-31 was not seen when X-31 neutralizing antibody titers were compared by microneutralization assay in mice infected with X-31 alone or following boosting with x139 (Figure 4). Thus, our findings do not appear to be consistent with original antigenic sin.
Figure 4: Neutralizing antibody against X-31 influenza (H3N2) is not boosted following secondary infection with x139 (H1N1).
At 22 days post infection, sera from 3 to 6 individual mice per experimental group were assayed for antibody against X-31 influenza virus by an in vitro microneutralization assay, as described in Materials and Methods. Data are depicted as the geometric mean, with error bars representing the 95% confidence interval.
Collectively these data suggest that CD4 T cells were responsible for the shifts in specificity following confrontation of the host with a novel influenza strain. This could either be the result of regulatory CD4 T cells (25) or through independent effector mechanism that result in faster viral clearance (26–27). Current experiments are designed to evaluate these possibilities more fully. Independent of the mechanisms responsible for the loss in HA-specific CD4 T cell responses, the coordinate shift in antibody response could reflect a linkage between CD4 T cell and B cell specificities. Future experiments will be able to further address the mechanism behind the shift away from HA in both CD4 T cells and B cells following rechallenge with heterosubtypic strains of influenza.
It is interesting to consider these results with respect to pandemic influenza preparedness strategies. Most clinically available inactivated influenza vaccines, including vaccines directed against pandemic strains, contain conserved internal proteins that are able to stimulate an immune response (12–14 and unpublished data). Our data suggests that simultaneous boosting of CD4 T cells specific for conserved internal proteins will result in less robust HA-specific CD4 T cell responses and concurrent lower titers of neutralizing antibodies on challenge with vaccines containing highly divergent HA proteins with few conserved HA epitopes. This could in part explain the modest responses seen following a single dose of many pandemic vaccines, including against the avian H5 (28) and H7 (29–30) strains. To promote pandemic preparedness, vaccination with highly purified avian HA molecules that promote the development of memory CD4 T cells specific for HA-specific epitopes may allow more rapid and robust antibody responses to the next pandemic strain, regardless of whether or not serological cross-reactivity is present.
Supplementary Material
Acknowledgements
The authors would like to thank Dr. Doris Bucher at New York Medical College for the x139 virus and Dr. David Topham at University of Rochester for the X-31 virus.
This work was supported by the National Institutes of Health contract number HHSN266200700008C and grant number 1K12HD068373-01.
References
- 1.Richards KA, Chaves FA, and Sant AJ 2009. Infection of HLA-DR1 transgenic mice with a human isolate of influenza a virus (H1N1) primes a diverse CD4 T-cell repertoire that includes CD4 T cells with heterosubtypic cross-reactivity to avian (H5N1) influenza virus. J. Virol 83:6566–6577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nayak JL, Richards KA, Chaves FA, and Sant AJ 2010. Analyses of the specificity of CD4 T cells during the primary immune response to influenza virus reveals dramatic MHC-linked asymmetries in reactivity to individual viral proteins. Viral Immunol. 23:169–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crowe SR, Miller SC, Brown DM, Adams PS, Dutton RW, Harmsen AG, Lund FE, Randall TD, Swain SL, and Woodland DL 2006. Uneven distribution of MHC class II epitopes within the influenza virus. Vaccine 24:457–467. [DOI] [PubMed] [Google Scholar]
- 4.Belz GT, Stevenson PG, and Doherty PC 2000. Contemporary analysis of MHC-related immunodominance hierarchies in the CD8 T cell response to influenza A viruses. J. Immunol 165:2404–2409. [DOI] [PubMed] [Google Scholar]
- 5.Zhong W, Reche PA, Lai C-C, Reinhold B, and Reinherz EL 2003. Genome-wide characterization of a viral cytotoxic T lymphocyte epitope repertoire. The Journal of Biological Chemistry 278:45135–45144. [DOI] [PubMed] [Google Scholar]
- 6.Belz GT, Xie W, Altman JD, and Doherty PC 2000. A previously unrecognized H-2D(b)-restricted peptide prominent in the primary influenza A virus-specific CD8(+) T-cell response is much less apparent following secondary challenge. J. Virol 74:3486–3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crowe SR, Turner SJ, Miller SC, Roberts AD, Rappolo RA, Doherty PC, Ely KH, and Woodland DL 2003. Differential antigen presentation regulates the changing patterns of CD8+ T cell immunodominance in primary and secondary influenza virus infections. J. Exp. Med 198:399–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang J, Huck SP, McHugh RS, Hermans IF, and Ronchese F 2006. Perforin-dependent elimination of dendritic cells regulates the expansion of antigen-specific CD8+ T cells in vivo. Proc. Natl. Acad. Sci. U. S. A 103:147–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mintern JD, Bedoui S, Davey GM, Moffat JM, Doherty PC, and Turner SJ 2009. Transience of MHC Class I-restricted antigen presentation after influenza A virus infection. Proc. Natl. Acad. Sci. U. S. A 106:6724–6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Belz GT, Zhang L, Lay MD, Kupresanin F, and Davenport MP 2007. Killer T cells regulate antigen presentation for early expansion of memory, but not naive, CD8+ T cell. Proc. Natl. Acad. Sci. U. S. A 104:6341–6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turner SJ, Cross R, Xie W, and Doherty PC 2001. Concurrent naive and memory CD8(+) T cell responses to an influenza A virus. J. Immunol 167:2753–2758. [DOI] [PubMed] [Google Scholar]
- 12.Co MD, Orphin L, Cruz J, Pazoles P, Green KM, Potts J, Leporati AM, Babon JA, Evans JE, Ennis FA, and Terajima M 2009. In vitro evidence that commercial influenza vaccines are not similar in their ability to activate human T cell responses. Vaccine 27:319–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richards KA, Chaves FA, Alam S, and Sant AJ 2012. Trivalent inactivated influenza vaccines induce broad immunological reactivity to both internal virion components and influenza surface proteins. Vaccine. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nayak JL, Fitzgerald TF, Richards KA, Yang H, Treanor JJ, and Sant AJ 2012. CD4 T-cell expansion predicts neutralizing antibody responses to monovalent inactivated pandemic H1N1 influenza vaccine. J. Infect. Dis [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alam S, and Sant AJ 2011. Infection with seasonal influenza virus elicits CD4 T cells specific for genetically conserved epitopes that can be rapidly mobilized for protective immunity to pandemic H1N1 influenza virus. J. Virol 85:13310–13321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.MacLeod MK, Kappler JW, and Marrack P 2010. Memory CD4 T cells: generation, reactivation and re-assignment. Immunology 130:10–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McKinstry KK, Strutt TM, and Swain SL 2010. The potential of CD4 T-cell memory. Immunology 130:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sette A, Moutaftsi M, Moyron-Quiroz J, McCausland MM, Davies DH, Johnston RJ, Peters B, Rafii-El-Idrissi Benhnia M, Hoffmann J, Su HP, Singh K, Garboczi DN, Head S, Grey H, Felgner PL, and Crotty S 2008. Selective CD4+ T cell help for antibody responses to a large viral pathogen: deterministic linkage of specificities. Immunity 28:847–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scherle PA, and Gerhard W 1988. Differential ability of B cells specific for external vs. internal influenza virus proteins to respond to help from influenza virus-specific T-cell clones in vivo. Proc. Natl. Acad. Sci. U. S. A 85:4446–4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johansson BE, Moran TM, and Kilbourne ED 1987. Antigen-presenting B cells and helper T cells cooperatively mediate intravirionic antigenic competition between influenza A virus surface glycoproteins. Proc. Natl. Acad. Sci. U. S. A 84:6869–6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lanzavecchia A 1990. Receptor-mediated antigen uptake and its effect on antigen presentation to class II-restricted T lymphocytes. Annu. Rev. Immunol 8:773–793. [DOI] [PubMed] [Google Scholar]
- 22.Ravkov EV, and Williams MA 2009. The magnitude of CD4+ T cell recall responses is controlled by the duration of the secondary stimulus. J. Immunol 183:2382–2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fazekas de St G, and Webster RG 1966. Disquisitions of Original Antigenic Sin. I. Evidence in man. J. Exp. Med 124:331–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim JH, Skountzou I, Compans R, and Jacob J 2009. Original antigenic sin responses to influenza viruses. J. Immunol 183:3294–3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanchez AM, Zhu J, Huang X, and Yang Y 2012. The development and function of memory regulatory T cells after acute viral infections. J. Immunol 189:2805–2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Teijaro JR, Verhoeven D, Page CA, Turner D, and Farber DL 2010. Memory CD4 T cells direct protective responses to influenza virus in the lungs through helper-independent mechanisms. J. Virol 84:9217–9226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Strutt TM, McKinstry KK, Dibble JP, Winchell C, Kuang Y, Curtis JD, Huston G, Dutton RW, and Swain SL 2010. Memory CD4+ T cells induce innate responses independently of pathogen. Nat. Med 16:558–564, 551p following 564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Treanor JJ, Campbell JD, Zangwill KM, Rowe T, and Wolff M 2006. Safety and immunogenicity of an inactivated subvirion influenza A (H5N1) vaccine. N. Engl. J. Med 354:1343–1351. [DOI] [PubMed] [Google Scholar]
- 29.Cox RJ, Madhun AS, Hauge S, Sjursen H, Major D, Kuhne M, Hoschler K, Saville M, Vogel FR, Barclay W, Donatelli I, Zambon M, Wood J, and Haaheim LR 2009. A phase I clinical trial of a PER.C6 cell grown influenza H7 virus vaccine. Vaccine 27:1889–1897. [DOI] [PubMed] [Google Scholar]
- 30.Couch RB, Patel SM, Wade-Bowers CL, and Nino D 2012. A randomized clinical trial of an inactivated avian influenza A (H7N7) vaccine. PLoS One 7:e49704. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.