Abstract
Background
Although the distribution of Giardia duodenalis genotypes in humans has been increasingly reported in recent years, data on possible differences in pathogen transmission between age groups and virulence between genotypes are scarce. The purpose of this study is to investigate the genetic diversity of G. duodenalis in humans in Spain and compare the distribution of G. duodenalis assemblages A and B between children and adults and clinical presentations between the two genotypes.
Methods
In the present study, 125 microscopy-positive fecal samples were collected from humans in Spain over a 7-year period. PCR and sequence analyses of the triosephosphate isomerase, β-giardin and glutamate dehydrogenase genes were used to identify the multilocus genotypes of G. duodenalis.
Results
Sequence analysis of three genetic loci identified both G. duodenalis assemblages A (29) and B (66), with co-infections of the two in two patients. Among the sequences obtained in this study, four multilocus genotypes (MLGs) of the sub-assemblage AII were observed within assemblage A. In contrast, 19 MLGs were detected within assemblage B due to the high sequence diversity at each locus. One MLG, however, was found in 51.9% (27/52) of assemblage B samples. Children were more commonly infected by assemblage B (44/53 or 83%) than adults (22/42 or 52.4%; χ2 = 10.371, df = 1, P = 0.001). Asymptomatic infection was more common in patients with assemblage A (4/29 or 13.8%) than in those with assemblage B (1/66 or 1.5%; χ2 = 6.091, df = 1, P = 0.029), and the frequency of abdominal pain occurrence was higher in assemblage B patients (65/66 or 98.5%) than assemblage A patients (25/29 or 86.2%; χ2 = 6.091, df = 1, P = 0.029).
Conclusions
These results illustrate the existence of differences in genotype distribution between children and adults and clinical presentations between G. duodenalis genotypes. They are useful in understanding the transmission of G. duodenalis in humans in Spain.
Keywords: Giardia duodenalis, Multilocus genotyping, Assemblage, Spain
Background
Giardia duodenalis is a common gastrointestinal pathogen in a wide range of vertebrates, including humans and domestic animals [1]. As one of the most common enteric pathogens, G. duodenalis is mainly transmitted through contact with infected persons or ingestion of contaminated food or water [2]. It colonizes the upper small intestine, causing asymptomatic, acute or chronic infections [3]. Worldwide, it is estimated that 280 million people are infected by G. duodenalis every year [4]. Human infection rates are generally high in developing countries, with pre-school children having the highest infection rates [1, 5]. Infection rates are also higher in children in industrialized nations, although older people can also have symptomatic G. duodenalis infections [6]. Symptoms of giardiasis include diarrhea, nausea, vomiting, abdominal pain, bloating, epigastric pain and weight loss [2, 7], which can develop 6–15 days after infection [8]. In developing countries, giardiasis has been further associated with growth retardation and poor cognitive performance in children [9].
Giardia duodenalis is a multispecies complex with at least eight recognized assemblages or genotypes (A-H) based on the genetic characterization of pathogens in clinical specimens [10, 11]. Among them, assemblages A and B are major human pathogens [1]. Assemblage A is further detected in livestock and companion animals, which are more often infected with their own host-adapted genotypes (assemblages C-F). In contrast, assemblage B is commonly reported in only a small number of animal species as the dominant G. duodenalis genotype [1]. Evidence for the occurrence of zoonotic transmission has come from the finding of the same subtypes or multilocus genotypes (MLGs) in humans and animals in the same area. For example, some human isolates of sub-assemblage AI clustered together with animal isolates in a recent study in Sweden [12]. Thus, both assemblages A and B are considered zoonotic pathogens, although the significance of zoonotic infection in giardiasis epidemiology remains unclear [1]. In addition, a small number of infections with assemblage E have been reported in humans in Australia, Brazil and Egypt [13].
Molecular analysis of more than 2800 G. duodenalis-positive samples from humans indicates that assemblage B (accounting for ~ 58% giardiasis cases) has a higher prevalence than assemblage A (accounting for ~ 37% giardiasis cases) worldwide [14]. The distribution of the two assemblages, however, differs among areas. For example, only assemblage A was detected in some studies in Uganda, USA, Canada and Korea [15], only assemblage B was detected in a study in India [16], while similar occurrence of assemblages A and B was identified studies of giardiasis in Slovenia, Netherlands and Albania [15, 17, 18]. Some previous studies carried out in Spain demonstrated a different distribution of G. duodenalis assemblages among some provinces. For instance, assemblages A and B were detected in 27.4 and 72.6%, respectively, of G. duodenalis-positive patients in Madrid [19], while an equal distribution of assemblages A and B was identified in La Rioja [20].
Although G. duodenalis is a common human pathogen worldwide, few data are available on differences in pathogen transmission between children and adults and clinical presentations between genotypes. One study in Australia indicated that adults were more commonly infected with G. duodenalis assemblage A compared with children [21]. Differences in virulence between assemblages A and B have been reported, with assemblage B being more virulent in most recent reports and more common in outbreaks [13]. In Spain, the few recent molecular epidemiological surveys indicate that assemblage B is the dominant G. duodenalis in humans [19, 20, 22]. The infection rates varied from 3.1% to 17.8% among these Spanish studies [20, 23, 24]. However, only one or two of the three commonly used genotyping loci, namely β-giardin (bg), glutamate dehydrogenase (gdh) and triosephosphate isomerase (tpi), were used in these studies. In addition, all samples in each survey were collected from the same city.
To fill some of the knowledge gaps described above, in the present study, multilocus genotyping targeting the three classical genetic loci (bg, gdh and tpi) was used to characterize G. duodenalis in clinical samples collected from humans in multiple locations in Spain between 2012 and 2018. The distribution of G. duodenalis genotypes was compared between children and adults and clinical presentations compared between assemblages A and B to assess the possibility of differences in the transmission of G. duodenalis between the two populations and virulence between the two assemblages.
Methods
Sample collection
A total of 125 fecal samples positive for G. duodenalis by microscopy were used in this study. They were from out-patients in hospitals in 10 Spanish provinces during 2012 (n = 9), 2015 (n = 27), 2016 (n = 33), 2017 (n = 25) and 2018 (n = 31) and submitted to a commercial laboratory, Synlab Diagnosticos Globales (Barcelona, Spain), for the detection of enteric pathogens, including common viruses, bacteria and parasites; samples positive for these bacteria and viruses were not further tested for enteric parasites. Each patient was supplied with one or more sterile plastic containers (1–3) with 5 ml of merthiolate-iodine-formaldehyde (MIF) solution to preserve the fecal samples; these were kept at room temperature until they were examined by microscopy. Among them, 118 were from patients suffering from diarrhea or other intestinal symptoms, while 7 were from asymptomatic persons who requested testing specifically for G. duodenalis because of unknown reasons. The patients were 1–75 years of age, with a median age of 10 years, and comprised 67 males and 58 females.
Microscopy analysis of G. duodenalis
The fecal samples were analyzed for G. duodenalis by direct microscopic examination of wet mount of fecal materials fixed with MIF solution as previously described [25]. Briefly, wet mounts were made using 20 μl of the upper suspension of the MIF-fixed sample. The entire area of the wet mounts was examined under a BX50 light microscope (Olympus, Tokyo, Japan) using 20× and 40× objectives. The samples positive for G. duodenalis were washed by centrifugation and stored in 70% ethanol at 4 °C before being shipped to the laboratory at the Centers for Disease Control and Prevention for molecular analysis.
DNA extraction and PCR amplification
Approximately 500 μl of suspension of the ethanol-preserved fecal sample was washed twice with distilled water by centrifugation at 2000× g for 10 min to remove ethanol. Genomic DNA was extracted from the washed fecal material using a FastDNA SPIN Kit for Soil (MP Biomedicals, Santa Ana, CA, USA) as described [26] and stored at − 80 °C until being analyzed by PCR within six months.
To identify MLGs of G. duodenalis, a 532-bp fragment of the tpi gene, a 511-bp fragment of the bg gene and a 530-bp fragment of the gdh gene were amplified using nested PCR [1]. To reduce the effect of residual PCR inhibitors in the extracted DNA, 400 ng/μl of non-acetylated bovine serum albumin (Sigma Aldrich, St. Louis, MO, USA) was used in primary PCR. Each DNA sample was analyzed in duplicate, using both positive (assemblage E) and negative (reagent-grade water) controls in each analysis. The secondary PCR products were visualized by 1.5% agarose gel electrophoresis.
Sequence analysis
All positive secondary PCR products from the three genetic loci generated in the study were sequenced in both directions using the secondary primers and the BigDye Terminator v.3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA) on an ABI 3730 Genetic Analyzer (Applied Biosystems). The nucleotide sequences of each gene were assembled using ChromasPro v.1.32 (http://technelysium.com.au/ChromasPro.html) and aligned with reference sequences (U57897, AY072723, AY072724, AY178737 and EF507651 for assemblage A, and KX468986, L02116, EU272153, FJ560565, HM140723, AB781124, KX960128, EU637581, KX469029, KJ888980, AY072728, HM165218, AY072727, EF507671, EU834843, KY696790 and KM977636 for assemblage B) downloaded from the GenBank database using the software ClustalX (http://www.clustal.org/) to identify the genotypes and subtypes of G. duodenalis. The evolution of assemblage B MLGs was assessed by using eBURST v.3.0 (http://eBURST.mlst.net/). Samples with apparent presence of double peaks at any of the loci during DNA sequencing were excluded in the MLG analysis of sequence data.
Data analysis
The Chi-square test was used to compare differences in infection rates of assemblages among children and adult and clinical symptoms. Differences were considered significant at the level of P < 0.05. The probability for the occurrence of asymptomatic infection or abdominal pain between assemblages A and B was measured by the odds ratio (OR) together with its 95% confidence intervals (95% CI). Statistical analysis was performed using SPSS Statistics v.21.0 for Windows (IBM Corp., Armonk, NY, USA).
Results
Detection and identification of G. duodenalis genotypes
Altogether, 125 G. duodenalis-positive fecal samples were included in this study. They were mostly diagnosed by microscopy detection of cysts in fecal samples (Fig. 1a). One fecal sample, however, was collected from a patient who was initially diagnosed as positive for G. duodenalis based on the detection of trophozoites in duodenum biopsies (Fig. 1b). Twelve samples collected during the period that had co-infections with other enteric protozoa, including Blastocystis hominis (n = 11), Endolimax nana (n = 2), Entamoeba coli (n = 2), Cryptosporidium sp. (n = 1) and Iodamoeba butschlii (n = 1). They were not included in the analysis of the associations between symptoms and assemblages of G. duodenalis.
Fig. 1.
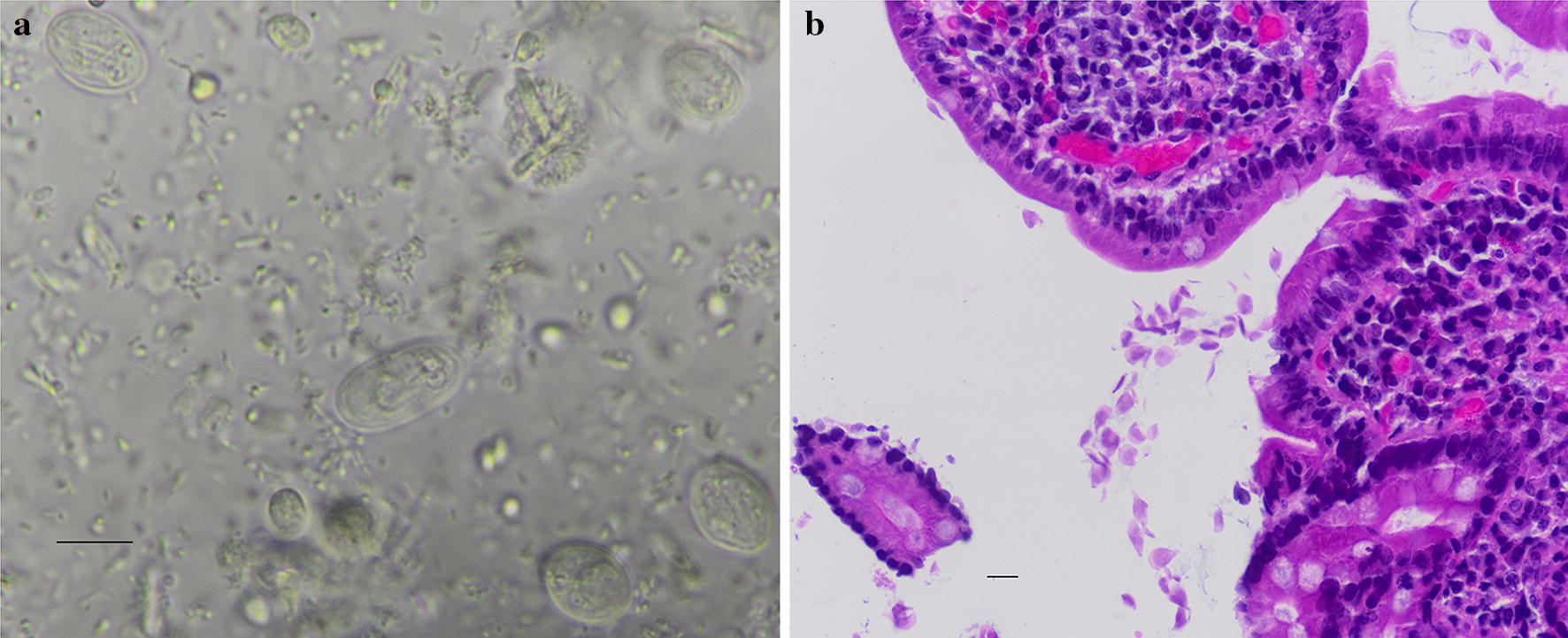
Cysts (a) and trophozoites (b) of Giardia duodenalis in a fecal sample and a duodenum biopsy of patients in Spain, respectively. The cysts were detected in microscopic analysis of wet mount of fecal material fixed with the merthiolate-iodine-formaldehyde solution under an Olympus BX43 using a 100× objective, while the trophozoites were detected by microscopic analysis of hematoxylin-eosin stained tissue section under an Olympus BX43 using a 60× objective. Scale-bars: 10 µm
Among the 125 fecal samples, 97 were positive in PCR analyses of the three genetic loci. The number of PCR-positive samples were 89, 84 and 87 at the tpi, bg and gdh loci, respectively. The amplification rates were 8/9 (88.9%), 23/27 (85.2%), 25/33 (75.8%), 20/25 (80%) and 21/31 (67.7%) for samples collected during 2012, 2015, 2016, 2017 and 2018, respectively. Among the 97 genotyped samples, 78 were successfully sequenced at all three genetic loci; the remaining 19 samples were successfully genotyped at only one (n = 12) or two (n = 7) genetic loci. Two genotypes of G. duodenalis were detected among the 97 genotyped samples, namely assemblages A (29, 30%) and B (66, 68%), with two samples having both (Table 1). Between the two samples with mixed genotypes, one was identified as assemblage B at the tpi locus but assemblage A at bg and gdh loci, while the other was identified as assemblage B at the gdh locus but assemblage A at tpi and bg loci.
Table 1.
Occurrence of Giardia duodenalis genotypes (assemblages A and B) by age, gender, geographical source and symptom of the 125 microscopy-positive patients enrolled in the study in Spain during 2012–2018
| Group | Case count | No. PCR-positive | Assemblage | Clinical symptom | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | A + B | Abdominal pain | Bloating | Diarrhea | Nausea | Dehydration | Weight loss | Epigastric pain | Normal | |||
| Age (years) | |||||||||||||
| 0–5 | 46 | 38 | 6 | 31 | 1 | 37 (44) | 19 (24) | 7 (8) | 3 (3) | 2 (2) | 2 (2) | 0 | 1 (2) |
| 6–12 | 22 | 16 | 3 | 13 | 0 | 16 (21) | 10 (13) | 2 (2) | 1 (1) | 0 | 1 (1) | 0 | 0 (1) |
| 23–75 | 57 | 43 | 20 | 22 | 1 | 39 (53) | 11 (16) | 10 (14) | 0 | 0 | 1 (1) | 5 (6) | 4 (4) |
| Gender | |||||||||||||
| Male | 67 | 55 | 15 | 39 | 1 | 52 (62) | 23 (29) | 9 (13) | 3 (3) | 1 (1) | 1 (1) | 3 (3) | 3 (5) |
| Female | 58 | 42 | 14 | 27 | 1 | 40 (56) | 17 (24) | 10 (11) | 1 (1) | 1 (1) | 3 (3) | 2 (3) | 2 (2) |
| Geographical source | |||||||||||||
| Barcelona | 87 | 72 | 24 | 46 | 2 | 69 (83) | 28 (33) | 12 (13) | 3 (3) | 1 (1) | 2 (2) | 3 (3) | 3 (4) |
| Málaga | 13 | 8 | 3 | 5 | 0 | 7 (12) | 5 (9) | 1 (2) | 0 | 0 | 1 (1) | 1 (1) | 1 (1) |
| Mallorca | 6 | 5 | 1 | 4 | 0 | 5 (6) | 2 (2) | 0 | 0 | 0 | 0 | 1 (2) | 0 |
| Madrid | 6 | 4 | 0 | 4 | 0 | 4 (6) | 3 (5) | 3 (5) | 0 | 0 | 0 | 0 | 0 |
| Tarragona | 5 | 4 | 1 | 3 | 0 | 4 (4) | 1 (1) | 1 (1) | 0 | 0 | 1 (1) | 0 | 0 (1) |
| Sevilla | 3 | 2 | 0 | 2 | 0 | 2 (3) | 1 (2) | 1 (1) | 0 | 0 | 0 | 0 | 0 |
| Girona | 2 | 1 | 0 | 1 | 0 | 0 (1) | 0 | 0 | 0 | 0 | 0 | 0 | 1 (1) |
| Cadiz | 1 | 0 | 0 | 0 | 0 | 0 (1) | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Valladolid | 1 | 1 | 0 | 1 | 0 | 1 (1) | 0 | 1 (1) | 1 (1) | 1 (1) | 0 | 0 | 0 |
| Badajoz | 1 | 0 | 0 | 0 | 0 | 0 (1) | 0 (1) | 0 (1) | 0 | 0 | 0 | 0 | 0 |
| Subtotal | 125 | 97 | 29 | 66 | 2 | 92 (118) | 40 (53) | 19 (24) | 4 (4) | 2 (2) | 4 (4) | 5 (6) | 5 (7) |
Note: Numbers in parentheses are the total numbers of samples examined, while those without parentheses are numbers of PCR-positive samples
Distribution of G. duodenalis genotypes by age, gender, location and clinical symptom
Among the 125 microscopy-positive samples, 46 (36.8%), 22 (17.6%) and 57 (45.6%) were from the age groups of 0–5, 6–12 and 23–75 years, respectively (Table 1). The numbers of PCR-positive samples from the three age groups were 38, 16 and 43, respectively. There was a significant difference in the distribution of G. duodenalis assemblages between children and adults (χ2 = 10.371, df = 1, P = 0.001). Children under 12 years-old were more commonly infected by assemblage B (44/53 or 83.0%) than assemblage A (9/53 or 17.0%). In contrast, adults had similar distribution of assemblages A (20/42 or 47.6%) and B (22/42 or 52.4%). Among the 97 PCR-positive samples, 55 and 42 were from male and female patients, respectively. There was no significant difference in the distribution of G. duodenalis assemblages A and B between male (15/54 or 27.8% and 39/54 or 72.2%, respectively) and female (14/41 or 34.1% and 27/41 or 65.9%, respectively) patients (χ2 = 0.446, df = 1, P = 0.328).
Eighty-seven (69.6%) of the 125 microscopy-positive samples were collected from Barcelona. Among them, 72 samples were PCR-positive for G. duodenalis, including 24 and 46 samples identified as assemblages A and B, respectively (two samples had both). The remaining 38 samples (30.4%) were collected from 9 other provinces, with 25 of them being PCR-positive. Among the latter, 5 and 20 samples were identified as assemblages A and B, respectively.
Abdominal pain was the most common clinical symptom of giardiasis: 118 (94.4%) of the 125 patients had the symptom (Table 1). The other two main clinical manifestations included bloating and diarrhea: 53 and 24 of the study patients, respectively, had these symptoms. Other clinical manifestations such as nausea, dehydration, weight loss and epigastric pain were observed in 4, 2, 4 and 6 patients, respectively. In addition, 7 patients were asymptomatic at the time of sampling.
The distribution of G. duodenalis genotypes by clinical symptom is shown in Table 2. Patients infected with assemblages A and B had a different frequency of asymptomatic infection (χ2 = 6.091, df = 1, P = 0.029). Therefore, assemblage A-infected patients (4/29 or 13.8%) were more likely to have asymptomatic infection than assemblage B-infected patients (1/66 or 1.5%) (OR: 10.4, 95% CI: 1.108–97.625). In addition, the frequency (% occurrence) of abdominal pain was significantly different in patients infected by assemblages A and B (χ2 = 6.091, df = 1, P = 0.029). Among the 29 assemblage A-infected patients, 25 (86.2%) had abdominal pain. In contrast, among the assemblage B-infected patients, 65 of 66 (98.5%) had this symptom. Other clinical symptoms such as bloating, diarrhea, nausea, dehydration, weight loss and epigastric pain were equally distributed between patients infected with the two assemblages (Table 2).
Table 2.
Distribution of clinical symptoms in 97 PCR-positive patients enrolled in the study by assemblage (A or B) of Giardia duodenalis
| Group | Case count | Clinical symptom | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Abdominal pain | Bloating | Diarrhea | Nausea | Dehydration | Weight loss | Epigastric pain | Normal | ||
| Assemblage A | 29 | 25 | 9 | 5 | 1 | 0 | 1 | 3 | 4 |
| Assemblage B | 66 | 65 | 30 | 13 | 3 | 1 | 3 | 2 | 1 |
| Assemblage A + B | 2 | 2 | 1 | 1 | 0 | 1 | 0 | 0 | 0 |
| Subtotal | 97 | 92 | 40 | 19 | 4 | 2 | 4 | 5 | 5 |
Distribution of assemblage A subtypes
Within assemblage A, only the A2 subtype was detected in 27 (100%) assemblage A-positive samples at the tpi locus, with the sequence being identical to U57897 in GenBank. In contrast, three subtypes were observed among the 28 assemblage A-positive samples at the bg locus, with sequences being identical to AY072723 (A2) in 10 (35.7%) samples, AY072724 (A3) in 17 (60.7%) samples and AB469365 (A5) in one (3.6%) sample. Two subtypes were observed at the gdh locus among the 27 assemblage A-positive samples, with the sequences being identical to AY178737 (A2) in 9 (33.3%) samples and EF507651 (A4) in 18 (66.7%) samples (Table 3).
Table 3.
Distribution of multilocus genotypes of Giardia duodenalis in 76 patient samples with complete data at all three genetic loci in the study conducted in Spain during 2012–2018
| Sample ID | Genotype (GenBank ID) | MLG type (n) | ||
|---|---|---|---|---|
| tpi | bg | gdh | ||
| 38808, 42849, 42853, 42855, 44333, 44945, 45105, 45598, 45599, 45606 | A2 (U57897) | A3 (AY072724) | A2 (AY178737) | AII-9 (10) |
| 42838, 42844, 44335, 44347, 44944, 45610, 45751 | A2 (U57897) | A2 (AY072723) | A2 (AY178737) | AII-1 (7) |
| 44943, 44948, 44949, 45608, 45611, 45745 | A2 (U57897) | A3 (AY072724) | A4 (EF507651) | AII-4 (6) |
| 45612 | A2 (U57897) | A2 (AY072723) | A4 (EF507651) | AII-8 (1) |
| 38810, 38811, 42835, 42837, 42845, 42848, 42857, 42858, 42863, 44329, 44330, 44332, 44334, 44338, 44343, 44357, 44358, 44359, 44361, 44947, 45099, 45100, 45102, 45103, 45602, 45609, 45756 | B (KX468986) | B2 (KX960128) | BIV (EF507671) | MLG B1 (27) |
| 42860, 44939, 44940, 45107, 45600 | B (KX468986) | B1 (EU637581) | B3 (EU834843) | MLG B2 (5) |
| 38807, 44354, 44355 | B (KX468986) | B2 (KX960128) | B3 (EU834843) | MLG B3 (3) |
| 38814, 45607 | B (L02116) | B (MG754396) | B3 (EU834843) | MLG B4 (2) |
| 45613 | B (KX468986) | B2 (KX960128) | KY696790-B | MLG B5 (1) |
| 42846 | B (KX468986) | B4 (KX469029) | BIV (EF507671) | MLG B6 (1) |
| 44356 | B (KX468986) | B (KJ888980) | BIV (EF507671) | MLG B7 (1) |
| 45106 | B (KX468986) | B (AY072728) | B3 (EU834843) | MLG B8 (1) |
| 45101 | B (KX468986) | B (MG754396) | B (KY696790) | MLG B9 (1) |
| 45104 | B (KX468986) | B (MG754396) | B3 (EU834843) | MLG B10 (1) |
| 38815 | B (L02116) | B1 (EU637581) | BIV (EF507671) | MLG B11 (1) |
| 45604 | B (L02116) | B1 (EU637581) | B3 (EU834843) | MLG B12 (1) |
| 44352 | B (L02116) | B2 (KX960128) | B3 (EU834843) | MLG B13 (1) |
| 42834 | B (L02116) | B2 (KX960128) | BIV (EF507671) | MLG B14 (1) |
| 38813 | B (L02116) | B (MG754395) | B (MG754398) | MLG B15 (1) |
| 45755 | B (L02116) | B (AY072727) | BIV (EF507671) | MLG B16 (1) |
| 42841 | B (EU272153) | B (AY072728) | B2 (KM977636) | MLG B17 (1) |
| 42862 | B (FJ560565) | B6 (HM165218) | BIV (EF507671) | MLG B18 (1) |
| 44351 | B (HM140723) | B (MG754396) | B3 (EU834843) | MLG B19 (1) |
Distribution of assemblage B subtypes
More subtypes were detected among assemblage B samples at each of the three genetic loci (Table 3 and Additional file 1: Table S1). Compared with the reference sequence KX468986, 1–5 single nucleotide substitutions (SNPs) were detected at eight positions at the tpi locus among assemblage B samples in this study. Altogether, seven subtypes of assemblage B were observed at the tpi locus, 6 of which were identical to KX468986, L02116, EU272153, FJ560565, HM140723 and AB781124 in the GenBank. The remaining one had 99% sequence identity to KX469015, was named as TB1, and deposited in GenBank under the accession number MG754394. Among them, the KX468986 sequence type was the dominant subtype, being detected in 47 of 66 assemblage B-positive samples. The second most common sequence type was L02116, which was detected in 9 samples. The remaining 5 sequence types were only detected in one or two samples during the five years of study.
Similarly, compared with the reference KX960128, 10 subtypes were observed at the bg locus among the 66 assemblage B samples, including three novel subtypes (Table 3 and Additional file 1: Table S1). The latter were named as BB1, BB2 and BB3, with sequences being deposited in GenBank under accession numbers MG754395–MG754397. Among the assemblage B samples, KX960128-B2 was the dominant subtype at the bg locus, being detected in 36 samples. This was followed by EU637581-B1, which was detected in 7 samples. The remaining subtypes were detected in 1–5 samples.
At the gdh locus, seven subtypes were observed among 66 assemblage B samples (Table 3 and Additional file 1: Table S1). Compared with the reference EF507671, 1–11 SNPs were detected at 19 positions, with three novel subtypes being identified. The latter were named as GB1, GB2 and GB3, with sequences being deposited in GenBank under the accession numbers MG754398, MG754399 and MG767308. Among the assemblage B samples, EF507671-BIV was the dominant subtype, being detected in 33 samples. This was followed by EU834843-B3, which was detected in 21 samples. The remaining subtypes were each observed in only one or two samples.
Distribution of G. duodenalis MLGs
Altogether, four MLGs were presented among the 24 assemblage A samples that were successfully subtyped at all three genetic loci (Table 3). Among them, AII-9 was the dominant MLG, being detected in 10 samples. The remaining MLGs, including AII-1, AII-4 and AII-8, were detected in 7, 6 and 1 samples, respectively.
In contrast, 19 MLGs were detected among the 52 assemblage B samples that were successfully subtyped at all three genetic loci (Table 3). Among them, MLG-B1 was the dominant type, being detected in 27 (51.9%) samples. MLG-B2, MLG-B3 and MLG-B4 were less frequent, being detected in 5, 3 and 2 samples, respectively. The remaining 15 MLGs (MLG-B5 to MLG-B19) were each detected in only one samples.
In the eBURST analysis of the MLG data from assemblage B, one cluster and three singletons of MLGs were observed. In the cluster, MLG-B3 was the primary founder and other common MLGs, such as MLG-B1, MLG-B10 and MLG-B14, were subgroup founders. The most commonly detected MLG, MLG-B1, appeared to be a single locus variant of the primary founder MLG-B3 (Fig. 2).
Fig. 2.
Genetic relationship of multilocus genotypes (MLGs) of Giardia duodenalis assemblage B based on eBURST analysis of sequences of the triosephosphate isomerase, β-giardin, and glutamate dehydrogenase genes. Each MLG is represented by a dot. The size of each dot is proportional to the number of samples with the MLG. The primary founder of the group is colored blue, while the subgroup founder is colored yellow. Single-locus variants are connected by lines. MLG-B15, MLG-B17 and MLG-B18 form singletons outside the main cluster
Discussion
In the present study, we assessed the genetic diversity of G. duodenalis in humans in Spain during 2012–2018. Among the 125 microscopy-positive samples collected from out-patients in ten provinces, 97 (77.6%) generated the expected products in PCR. The genotyping results obtained suggest the occurrence of both assemblages A and B in Spain, with assemblage B (66/95 or 69.5%) being twice as common as assemblage A (29/95 or 30.5%). No assemblage E infection was detected in this study. This is consistent with observations in three previous studies in Spain, which had also shown a more frequent occurrence of assemblage B than assemblage A in humans in Zaragoza, Madrid and La Rioja provinces [19, 20, 22]. The failure in PCR amplification of DNA from some microscopy-positive samples could be due to the initial short storage of the fecal material in MIF, which contains PCR-unfriendly preservative formalin; PCR is known to be more sensitive than conventional microscopy in detecting G. duodenalis [27, 28].
The distribution of G. duodenalis genotypes appears to be different between children and adults in Spain. In this study, 83% (44/53) of G. duodenalis-positive children were infected with assemblage B, compared with 52.4% (22/42) in adults (χ2 = 10.371, df = 1, P = 0.001). Similar results were obtained in one previous study in Egypt, where children had a higher frequency of assemblage B (24/34 or 70.6%) than assemblage A (10/34 or 29.4%), while adults had a similar frequency of both assemblages (12/26 or 46.2% and 14/26 or 53.8%, respectively) [29]. In contrast, the opposite was observed in a study conducted in the UK in which assemblages A and B were equally distributed in children of 0–9 years, assemblage B was more common in adults of 30–49 years, while assemblage A was more common in adults over 50 years [30]. These results indicate that the dominant genotypes of G. duodenalis could change in humans over age, perhaps as a reflection of the development of acquired immunity and/or differences in exposures.
The clinical symptoms caused by assemblages A and B appear to be different in the Spanish population. In the present study, abdominal pain was the most common clinical symptom, but patients infected by G. duodenalis assemblage B were more likely to develop it than patients infected by assemblage A (OR: 10.4, 95% CI: 1.108–97.625). This is different from data from the UK, in which the frequency of abdominal pain occurrence was similar between patients infected with the two assemblages [31]. In addition, assemblage A-infected patients were more likely to have asymptomatic infections in our study (OR: 10.4, 95% CI: 1.108–97.625), which is similar to observations in previous studies in Ethiopia, Netherlands and Argentina [32–35]. However, in another study of giardiasis in children under five years in Spain, symptoms were more common in patients infected by assemblage A than by assemblage B [22]. Similarly, assemblage B-infected patients were more likely to have vomiting than assemblage A-infected patients in a study in England [30], while there was no significant difference in the occurrence of vomiting between infections with assemblages A and B in the present study. Therefore, multiple factors could affect the clinical presentations of G. duodenalis genotypes.
Anthroponotic transmission appears to play an important role in giardiasis epidemiology in Spain. In the present study, almost all assemblage A-positive samples were identified as having sub-assemblage AII, which is known to preferentially infect humans [1]. This is similar to previous observations in South America [36, 37]. In addition, assemblage B was the dominant genotype in this study, which is consistent with data from South America and Africa [5, 35, 38, 39]. Assemblage B detected in these studies has only occasionally been detected in farm animals or companion animals. Therefore, both observations above suggest that G. duodenalis in the Spanish population could be mostly anthroponotic in origin.
In the present study, a higher genetic heterogeneity was observed in assemblage B than in assemblage A. Among the latter, most of the cases except one belonged to three MLGs within the sub-assemblage AII. In contrast, 19 MLGs were detected among assemblage B isolates, which is consistent with previous observations [30, 40, 41]. The identification of MLGs within assemblage B is complicated by the possible presence of sequence heterozygosity at some of the genetic loci, which may lead to the over-estimation of MLGs [19, 40]. Therefore, meiotic recombination, the presence of sequence heterozygosity and concurrence of mixed subtypes could all contribute to the high MLG numbers within assemblage B [42, 43]. Nevertheless, in agreement with the observation with assemblage A in this study, over half (27/52 or 51.9%) of the assemblage B infections in the Spanish population were caused by one MLG, indicating the possible circulation of one subtype in the community, and the occurrence of clonal expansion of limited number of subtypes, a finding in agreement with the result of the eBURST analysis (Fig. 2).
Conclusions
Results of the MLG analysis demonstrate a common occurrence of both assemblages A and B of G. duodenalis in the Spanish population, with a different distribution of the two genotypes between children and adults and some genotype-associated differences in clinical presentations or virulence. Despite the observation of high genetic diversity in G. duodenalis at the subtype level, several MLGs appear to have high occurrence in the human populations examined. The genetic identity of G. duodenalis at both the genotype and subtype levels suggests that anthroponotic transmission is important giardiasis epidemiology in the study area. Further studies using a better epidemiological design and more advanced molecular biologic tools are, however, needed to confirm these hypotheses.
Supplementary information
Additional file 1: Table S1. Distribution of Giardia duodenalis subtypes in 21 human samples with partial or discrepant data at three genetic loci in Spain.
Acknowledgements
We thank Dr Adriana Casado of Synlab for the picture of G. duodenalis-infected intestinal issue. The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the US Centers for Disease Control and Prevention.
Abbreviations
- PCR
polymerase chain reaction
- MLGs
multilocus genotypes
- tpi
triosephosphate isomerase
- bg
β-giardin
- gdh
glutamate dehydrogenase
- SNPs
single nucleotide substitutions
Authors’ contributions
YYF and LHX designed the study. YFW and OGM performed the experiments. OGM, DMR, LO, JH and YQG collected and analyzed the data. YFW, OGM and LHX interpreted the results and prepared the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Key R&D Programme of China (2017YFD0501305 to YYF). YFW was partially supported by the China Scholarship Council. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
The data supporting the conclusions of this article are included within the article and its additional file. Representative nucleotide sequences generated in this study were submitted to the GenBank database under the accession numbers MG754394–MG754399 and MG767308.
Ethics approval and consent to participate
Samples used in the study were submitted to a diagnostic laboratory by hospitals as part of the routine diagnosis of causes for diarrhea and enteric illnesses. No formal consent was needed for the collection and submission of clinical diagnostic samples. Residual samples were used in the study after the removal of identifiable personal information other than the demographic and clinical data collected at the time of sample submission. The work was covered by Human Subjects Protocol no. 990115, which was reviewed and approved by the Institutional Reviewing Board of the Centers for Disease Control and Prevention (CDC), Atlanta, USA. The research was further approved by the ethics committee of East China University of Science and Technology, Shanghai, China.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yuanfei Wang, Email: zhizunbao19@163.com.
Olga Gonzalez-Moreno, Email: olga.gonzalez@synlab.es.
Dawn M. Roellig, Email: iyd4@cdc.gov
Laura Oliver, Email: laura.oliver@synlab.es.
Jordi Huguet, Email: jordi.huguet@synlab.es.
Yaqiong Guo, Email: guoyq@scau.edu.cn.
Yaoyu Feng, Email: yyfeng@ecust.edu.cn.
Lihua Xiao, Email: lxiao1961@gmail.com.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13071-019-3692-4.
References
- 1.Feng Y, Xiao L. Zoonotic potential and molecular epidemiology of Giardia species and giardiasis. Clin Microbiol Rev. 2011;24:110–140. doi: 10.1128/CMR.00033-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ryan U, Hijjawi N, Feng Y, Xiao L. Giardia: an under-reported foodborne parasite. Int J Parasitol. 2019;49:1–11. doi: 10.1016/j.ijpara.2018.07.003. [DOI] [PubMed] [Google Scholar]
- 3.Einarsson E, Ma’ayeh S, Svard SG. An up-date on Giardia and giardiasis. Curr Opin Microbiol. 2016;34:47–52. doi: 10.1016/j.mib.2016.07.019. [DOI] [PubMed] [Google Scholar]
- 4.Lane S, Lloyd D. Current trends in research into the waterborne parasite Giardia. Crit Rev Microbiol. 2002;28:123–147. doi: 10.1080/1040-840291046713. [DOI] [PubMed] [Google Scholar]
- 5.Squire SA, Ryan U. Cryptosporidium and Giardia in Africa: current and future challenges. Parasit Vectors. 2017;10:195. doi: 10.1186/s13071-017-2111-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Painter JE, Gargano JW, Collier SA, Yoder JS. Giardiasis surveillance—United States, 2011–2012. MMWR Suppl. 2015;64:15–25. [PubMed] [Google Scholar]
- 7.Ankarklev J, Jerlstrom-Hultqvist J, Ringqvist E, Troell K, Svard SG. Behind the smile: cell biology and disease mechanisms of Giardia species. Nat Rev Microbiol. 2010;8:413–422. doi: 10.1038/nrmicro2317. [DOI] [PubMed] [Google Scholar]
- 8.Watkins RR, Eckmann L. Treatment of giardiasis: current status and future directions. Curr Infect Dis Rep. 2014;16:396. doi: 10.1007/s11908-014-0396-y. [DOI] [PubMed] [Google Scholar]
- 9.Berkman DS, Lescano AG, Gilman RH, Lopez SL, Black MM. Effects of stunting, diarrhoeal disease, and parasitic infection during infancy on cognition in late childhood: a follow-up study. Lancet. 2002;359:564–571. doi: 10.1016/S0140-6736(02)07744-9. [DOI] [PubMed] [Google Scholar]
- 10.Caccio SM, Lalle M, Svard SG. Host specificity in the Giardia duodenalis species complex. Infect Genet Evol. 2018;66:335–345. doi: 10.1016/j.meegid.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 11.Heyworth MF. Giardia duodenalis genetic assemblages and hosts. Parasite. 2016;23:13. doi: 10.1051/parasite/2016013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ankarklev J, Lebbad M, Einarsson E, Franzen O, Ahola H, Troell K, et al. A novel high-resolution multilocus sequence typing of Giardia intestinalis assemblage A isolates reveals zoonotic transmission, clonal outbreaks and recombination. Infect Genet Evol. 2018;60:7–16. doi: 10.1016/j.meegid.2018.02.012. [DOI] [PubMed] [Google Scholar]
- 13.Xiao L, Feng Y. Molecular epidemiologic tools for waterborne pathogens Cryptosporidium spp. and Giardia duodenalis. Food Waterborne Parasitol. 2017;8–9:14–32. doi: 10.1016/j.fawpar.2017.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ryan U, Caccio SM. Zoonotic potential of Giardia. Int J Parasitol. 2013;43:943–956. doi: 10.1016/j.ijpara.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 15.Caccio SM, Thompson RC, McLauchlin J, Smith HV. Unravelling Cryptosporidium and Giardia epidemiology. Trends Parasitol. 2005;21:430–437. doi: 10.1016/j.pt.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 16.Tak V, Mirdha BR, Yadav P, Vyas P, Makharia GK, Bhatnagar S. Molecular characterisation of Giardia intestinalis assemblages from human isolates at a tertiary care centre of India. Indian J Med Microbiol. 2014;32:19–25. doi: 10.4103/0255-0857.142236. [DOI] [PubMed] [Google Scholar]
- 17.Soba B, Islamovic S, Skvarc M, Caccio SM. Multilocus genotyping of Giardia duodenalis (Lambl, 1859) from symptomatic human infections in Slovenia. Folia Parasitol (Praha). 2015;62:062. doi: 10.14411/fp.2015.062. [DOI] [PubMed] [Google Scholar]
- 18.Berrilli F, Di Cave D, D’Orazi C, Orecchia P, Xhelilaj L, Bejko D, et al. Prevalence and genotyping of human isolates of Giardia duodenalis from Albania. Parasitol Int. 2006;55:295–297. doi: 10.1016/j.parint.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 19.de Lucio A, Martinez-Ruiz R, Merino FJ, Bailo B, Aguilera M, Fuentes I, et al. Molecular genotyping of Giardia duodenalis isolates from symptomatic individuals attending two major public hospitals in Madrid, Spain. PLoS ONE. 2015;10:e0143981. doi: 10.1371/journal.pone.0143981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Azcona-Gutierrez JM, de Lucio A, Hernandez-de-Mingo M, Garcia-Garcia C, Soria-Blanco LM, Morales L, et al. Molecular diversity and frequency of the diarrheagenic enteric protozoan Giardia duodenalis and Cryptosporidium spp. in a hospital setting in Northern Spain. PLoS ONE. 2017;12:e0178575. doi: 10.1371/journal.pone.0178575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Asher AJ, Hose G, Power ML. Giardiasis in NSW: identification of Giardia duodenalis assemblages contributing to human and cattle cases, and an epidemiological assessment of sporadic human giardiasis. Infect Genet Evol. 2016;44:157–161. doi: 10.1016/j.meegid.2016.06.051. [DOI] [PubMed] [Google Scholar]
- 22.Sahagun J, Clavel A, Goni P, Seral C, Llorente MT, Castillo FJ, et al. Correlation between the presence of symptoms and the Giardia duodenalis genotype. Eur J Clin Microbiol Infect Dis. 2008;27:81–83. doi: 10.1007/s10096-007-0404-3. [DOI] [PubMed] [Google Scholar]
- 23.Mateo M, Mateo M, Montoya A, Bailo B, Saugar JM, Aguilera M, et al. Detection and molecular characterization of Giardia duodenalis in children attending day care centers in Majadahonda, Madrid, Central Spain. Medicine (Baltimore). 2014;93:e75. doi: 10.1097/MD.0000000000000075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cardona GA, Carabin H, Goni P, Arriola L, Robinson G, Fernandez-Crespo JC, et al. Identification and molecular characterization of Cryptosporidium and Giardia in children and cattle populations from the province of Alava, North of Spain. Sci Total Environ. 2011;412–413:101–108. doi: 10.1016/j.scitotenv.2011.09.076. [DOI] [PubMed] [Google Scholar]
- 25.Golvan YJ, Drouhet E, Segrerain G, Mariat F. Laboratory examinations. Techniques in parasitology and mycology. II. Med Mycol. 1972;353:499. [Google Scholar]
- 26.Jiang J, Alderisio KA, Singh A, Xiao L. Development of procedures for direct extraction of Cryptosporidium DNA from water concentrates and for relief of PCR inhibitors. Appl Environ Microbiol. 2005;71:1135–1141. doi: 10.1128/AEM.71.3.1135-1141.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gotfred-Rasmussen H, Lund M, Enemark HL, Erlandsen M, Petersen E. Comparison of sensitivity and specificity of 4 methods for detection of Giardia duodenalis in feces: immunofluorescence and PCR are superior to microscopy of concentrated iodine-stained samples. Diagn Microbiol Infect Dis. 2016;84:187–190. doi: 10.1016/j.diagmicrobio.2015.11.005. [DOI] [PubMed] [Google Scholar]
- 28.Adeyemo FE, Singh G, Reddy P, Stenstrom TA. Methods for the detection of Cryptosporidium and Giardia: from microscopy to nucleic acid based tools in clinical and environmental regimes. Acta Trop. 2018;184:15–28. doi: 10.1016/j.actatropica.2018.01.011. [DOI] [PubMed] [Google Scholar]
- 29.El Basha NR, Zaki MM, Hassanin OM, Rehan MK, Omran D. Giardia assemblages A and B in diarrheic patients: a comparative study in Egyptian children and adults. J Parasitol. 2016;102:69–74. doi: 10.1645/14-676. [DOI] [PubMed] [Google Scholar]
- 30.Minetti C, Lamden K, Durband C, Cheesbrough J, Fox A, Wastling JM. Determination of Giardia duodenalis assemblages and multi-locus genotypes in patients with sporadic giardiasis from England. Parasit Vectors. 2015;8:444. doi: 10.1186/s13071-015-1059-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Breathnach AS, McHugh TD, Butcher PD. Prevalence and clinical correlations of genetic subtypes of Giardia lamblia in an urban setting. Epidemiol Infect. 2010;138:1459–1467. doi: 10.1017/S0950268810000208. [DOI] [PubMed] [Google Scholar]
- 32.Gelanew T, Lalle M, Hailu A, Pozio E, Caccio SM. Molecular characterization of human isolates of Giardia duodenalis from Ethiopia. Acta Trop. 2007;102:92–99. doi: 10.1016/j.actatropica.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 33.Flecha MJ, Benavides CM, Tissiano G, Tesfamariam A, Cuadros J, de Lucio A, et al. Detection and molecular characterisation of Giardia duodenalis, Cryptosporidium spp. and Entamoeba spp. among patients with gastrointestinal symptoms in Gambo Hospital, Oromia Region, southern Ethiopia. Trop Med Int Health. 2015;20:1213–1222. doi: 10.1111/tmi.12535. [DOI] [PubMed] [Google Scholar]
- 34.Homan WL, Mank TG. Human giardiasis: genotype linked differences in clinical symptomatology. Int J Parasitol. 2001;31:822–826. doi: 10.1016/S0020-7519(01)00183-7. [DOI] [PubMed] [Google Scholar]
- 35.Molina N, Minvielle M, Grenovero S, Salomon C, Basualdo J. High prevalences of infection with Giardia intestinalis genotype B among children in urban and rural areas of Argentina. Ann Trop Med Parasitol. 2011;105:299–309. doi: 10.1179/136485911X12987676649665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oliveira-Arbex AP, David EB, Oliveira-Sequeira TC, Bittencourt GN, Guimaraes S. Genotyping of Giardia duodenalis isolates in asymptomatic children attending daycare centre: evidence of high risk for anthroponotic transmission. Epidemiol Infect. 2016;144:1418–1428. doi: 10.1017/S0950268815002514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramirez JD, Heredia RD, Hernandez C, Leon CM, Moncada LI, Reyes P, et al. Molecular diagnosis and genotype analysis of Giardia duodenalis in asymptomatic children from a rural area in central Colombia. Infect Genet Evol. 2015;32:208–213. doi: 10.1016/j.meegid.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 38.Atherton R, Bhavnani D, Calvopina M, Vicuna Y, Cevallos W, Eisenberg J. Molecular identification of Giardia duodenalis in Ecuador by polymerase chain reaction–restriction fragment length polymorphism. Mem Inst Oswaldo Cruz. 2013;108:512–515. doi: 10.1590/S0074-02762013000400019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lebbad M, Ankarklev J, Tellez A, Leiva B, Andersson JO, Svard S. Dominance of Giardia assemblage B in Leon, Nicaragua. Acta Trop. 2008;106:44–53. doi: 10.1016/j.actatropica.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 40.Caccio SM, Beck R, Lalle M, Marinculic A, Pozio E. Multilocus genotyping of Giardia duodenalis reveals striking differences between assemblages A and B. Int J Parasitol. 2008;38:1523–1531. doi: 10.1016/j.ijpara.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 41.Wegayehu T, Karim MR, Li J, Adamu H, Erko B, Zhang L, et al. Multilocus genotyping of Giardia duodenalis isolates from children in Oromia Special Zone, central Ethiopia. BMC Microbiol. 2016;16:89. doi: 10.1186/s12866-016-0706-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cooper MA, Adam RD, Worobey M, Sterling CR. Population genetics provides evidence for recombination in Giardia. Curr Biol. 2007;17:1984–1988. doi: 10.1016/j.cub.2007.10.020. [DOI] [PubMed] [Google Scholar]
- 43.Hussein AI, Yamaguchi T, Nakamoto K, Iseki M, Tokoro M. Multiple-subgenotype infections of Giardia intestinalis detected in Palestinian clinical cases using a subcloning approach. Parasitol Int. 2009;58:258–262. doi: 10.1016/j.parint.2009.04.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Distribution of Giardia duodenalis subtypes in 21 human samples with partial or discrepant data at three genetic loci in Spain.
Data Availability Statement
The data supporting the conclusions of this article are included within the article and its additional file. Representative nucleotide sequences generated in this study were submitted to the GenBank database under the accession numbers MG754394–MG754399 and MG767308.



