Abstract
Background
Shiga toxin-producing Escherichia coli (STEC) are emerging foodborne pathogens that are public health concern. Cattle have been identified as the major STEC reservoir. In the present study, we investigated the prevalence and characteristics of STEC strains in beef cattle from a commercial farm in Sichuan province, China.
Results
Among 120 beef cattle fecal samples, stx genes were positive in 90% of samples, as assessed using TaqMan real-time PCR, and 87 (72.5%) samples were confirmed to yield at least one STEC isolate by culture using four selective agars, MacConkey, CHROMagar™ ECC, modified Rainbow® Agar O157, and CHROMagar™ STEC, from which 31, 32, 91, and 73 STEC strains were recovered, respectively. A total of 126 STEC isolates were selected and further characterized. Seventeen different O:H serotypes were identified, all of which belonged to the non-O157 serotypes. One stx1 subtype (stx1a) and three stx2 subtypes (stx2a, stx2c, and stx2d) were present among these isolates. The intimin encoding gene eae, and other adherence-associated genes (iha, saa, and paa) were present in 37, 125, 74, and 30 STEC isolates, respectively. Twenty-three isolates carried the virulence gene subA, and only one harbored both cnf1 and cnf2 genes. Three plasmid-origin virulence genes (ehxA, espP, and katP) were present in 111, 111, and 7 isolates, respectively. The 126 STEC isolates were divided into 49 pulsed-field gel electrophoresis (PFGE) patterns.
Conclusions
Our study showed that the joint use of the selective MacConkey and modified Rainbow® Agar O157 agars increased the recovery frequency of non-O157 STEC strains in animal feces, which could be applied to other samples and in regular STEC surveillance. Moreover, the results revealed high genetic diversity of non-O157 STEC strains in beef cattle, some of which might have the potential to cause human diseases.
Electronic supplementary material
The online version of this article (10.1186/s12866-019-1582-8) contains supplementary material, which is available to authorized users.
Keywords: Shiga toxin, Escherichia coli, O157:H7, Non-O157 STEC, Beef cattle, Culture medium
Background
Shiga toxin-producing Escherichia coli (STEC) strains are significant foodborne zoonotic pathogens that are associated with illnesses ranging from mild diarrhea to hemorrhagic colitis (HC) and life-threatening hemolytic uremic syndrome (HUS) complications in humans [1]. Since the 1980s, more than 400 STEC serotypes have been reported worldwide, among which O157:H7 was the top causative serotype related to foodborne illnesses [2]. In recent years, sporadic cases or outbreaks caused by non-O157 STEC strains have been increasing [3–5]. Non-O157 STEC strains have been responsible for approximately 64% of STEC infections each year, particularly strains of several serogroups (O26, O45, O103, O111, O121, and O145, termed the “big six”), which generally possess the adhesin intimin (eae gene), a pathogenic marker for enterohemorrhagic E. coli (EHEC) and accounted for the majority of non-O157 STEC illnesses [2, 6, 7].
Shiga toxin (Stx), including Shiga toxin 1 (Stx1) and Shiga toxin 2 (Stx2) (encoded by stx1 and stx2 genes), are essential virulence factors in STEC strains [8]. Several Stx1 subtypes (Stx1a, Stx1c, and Stx1d) and Stx2 subtypes (Stx2a to Stx2h) have been identified in E. coli strains [9, 10]. The presence of stx2a and/or stx2c genes is more frequently related to severe clinical diseases [11]. In addition to Stx, a number of virulence factors are associated with pathogenicity. Intimin, encoded by the eae gene in the locus of enterocyte effacement (LEE), could induce attaching and effacing (A/E) lesions [12]. Strains containing a Shiga toxin gene together with the LEE island are classified as EHEC, which are associated with more severe clinical symptoms in humans [13]. Other adhesion-related genes such as iha (IrgA homolog adhesin), efa1 (EHEC factor for adherence 1), saa (STEC autoagglutinating adhesin), and paa (porcine A/E associated protein) also play important roles in bacterial adhesion [14–16]. Cytotoxic necrotizing factors, encoded by the cnf1 and cnf2 genes, can impair the function of epithelial and immune cells. Subtilase (subAB) is an AB5 toxin that can lead to diseases including renal damage, hemolytic anemia, and HUS-related pathological features [1, 15]. The astA gene encodes a toxin that is structurally related to the heat-stable enterotoxin of enterotoxigenic E. coli [17]. In addition, the virulence factors carried on the pO157 plasmid are involved in STEC pathogenicity. For example, enterohemolysin (ehxA) can destroy mammalian cell membranes; serine protease (espP) and catalase-peroxidase (katP) contribute to the colonization of STEC strains in the human intestinal tract; and ToxB contributes to the adherence of O157:H7 to Caco-2 cells [18].
Although many domestic and wild animals can serve as a reservoir of STEC strains [19, 20], ruminants, especially cattle, have been recognized as the main reservoir and play an important role in the epidemiology of STEC infections [8, 21]. The transmission route includes ingestion of contaminated food or water, direct contact, or exposure to the environment [21]. The prevalence of STEC strains in cattle varied from 0.4 to 74.0% because of the differences in cattle categories, farm environments, and isolation methods [7, 22].
Being able to reliably detect STEC strains in different matrices could improve surveillance activities for emergent strains. To improve the recovery frequency of STEC strains, especially non-O157 STEC strains, from various samples, culture methods using selective chromogenic agars have been attempted [23–26]. Our previous investigation demonstrated that most of non-O157 STEC isolates recovered from diverse sources (animals, foodstuffs, and humans) in China were sensitive to tellurite ingredients, which resulted in poor growth on tellurite-amended agars. The recovery frequency of STEC strains from complex matrices could thus be improved by the combined use of less selective and highly selective agars [27]. In this study, we investigated the prevalence of STEC in the feces of beef cattle (collected from a commercial farm in China) using different selective chromogenic media. We further characterized these isolates by serotyping, stx subtyping, virulence gene profiling, and pulsed-field gel electrophoresis (PFGE) typing.
Results
Prevalence of stx genes in beef cattle fecal samples
Among the 120 fecal samples screened by TaqMan real-time PCR, 108 (90%) were positive for stx1 or stx2 or both (cycle threshold (Ct) values below 40), among which 80.8% of the samples were positive for both stx1 and stx2, while 9.2% (11 samples) were only stx2 positive (Additional file 1: Table S1).
STEC isolates recovered from different selective chromogenic agars
Four different selective chromogenic agars, i.e. MAC (MacConkey), CH-ECC (CHROMagar™ ECC), RBA-NT (modified Rainbow® Agar O157 and CH-STEC (CHROMagar™ STEC), were used simultaneously to isolate STEC strains from all 120 samples. RBA-NT agar gave the highest positive culture rate of 59.2% (71/120), followed by CH-STEC agar (47.5%, 57/120). MAC and CH-ECC agars gave the same positive culture rate of 24.2% (29/120). In total, 72.5% (87/120) of the samples yielded one or more STEC isolate by the combined use of the four agars (Table 1). Notably, STEC isolates from two stx-PCR negative samples were recovered from both RBA-NT and CH-STEC agars. There was no significant correlation between the Ct values of stx-real-time PCR and the positive culture (Additional file 1: Table S1).
Table 1.
STEC isolates recovered from different chromogenic agars
| Agars | No. of samples with STEC isolates (%) | No. of STEC isolates | stx1 (%) | stx2 (%) | stx1 + stx2 (%) |
|---|---|---|---|---|---|
| MAC | 29 (24.2) | 31 | 1 (3.2) | 10 (32.3) | 20 (64.5) |
| CH-ECC | 29 (24.2) | 32 | 1 (3.2) | 10 (31.3) | 21 (65.6) |
| RBA-NT | 71 (59.2) | 91 | 29 (31.9) | 34 (37.4) | 28 (30.8) |
| CH-STEC | 57 (47.5) | 73 | 22 (30.1) | 31 (42.5) | 20 (27.4) |
| Total | 87 (72.5) | 126a | 31a | 45a | 50a |
aNumber of STEC isolates selected for further analyzing. MAC, MacConkey; CH-ECC, CHROMagar™ ECC, RBA-NT, modified Rainbow® Agar O157; CH-STEC, CHROMagar™ STEC
The performance of the four chromogenic agars was evaluated. Only six samples yielded STEC isolates on all four agars. Although RBA-NT and CH-STEC agars gave higher culture positive rates, the combined use of MAC and RBA-NT agars could cover almost all strains cultured from the other two agars (except for one sample) (Fig. 1). In total, 31, 32, 91, and 73 stx-positive isolates were recovered from 120 samples on MAC, CH-ECC, RBA-NT, and CH-STEC agars, respectively. Twenty (64.5%) and 21 (65.6%) strains that were positive for the stx1 + stx2 genes were found on MAC and CH-ECC agars, respectively. Among the isolates recovered on RBA-NT agar, 29 stx1-positive and 34 stx2-positive strains were tested, and 28 tested positive for both stx1 and stx2 genes. Of the 73 isolates found on CH-STEC agar, 22 and 31 tested positive for the stx1 and stx2 genes, respectively, and 20 tested positive for both genes. With the exception of the different stx types/subtypes present in the same sample, only one representative STEC isolate from each sample was kept. Finally, one isolate carrying the stx1, stx2, or stx1 + stx2 gene was chosen from 53 samples each; two isolates per sample were chosen from 29 samples; and three isolates per sample were chosen from five samples. A total of 126 stx-positive isolates were all confirmed to be E. coli and were selected for further characterization (Table 1 and Fig. 2).
Fig. 1.
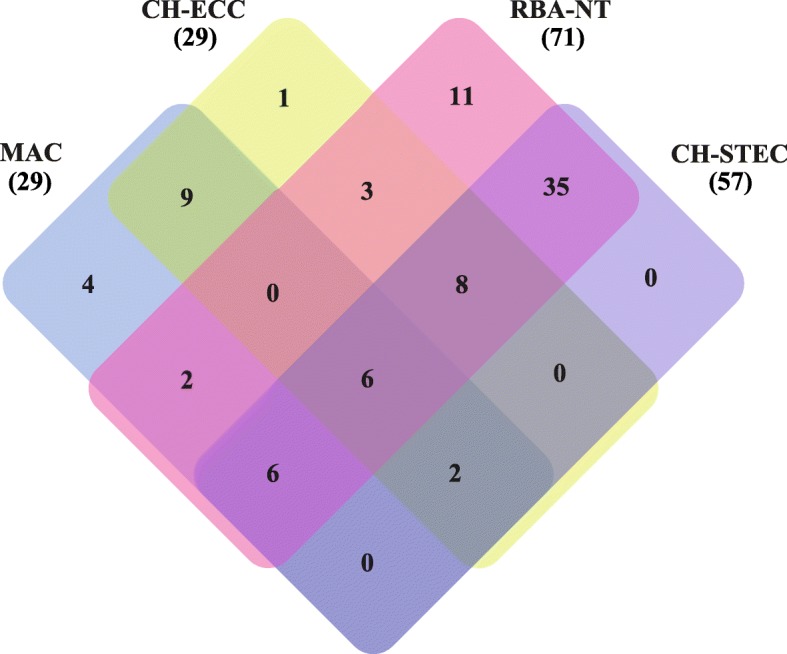
Venn diagram showing the numbers of common and unique samples that recovered STEC isolates on MAC, CH-ECC, RBA-NT, and CH-STEC agars. The number of culture-positive samples is listed in each of the diagram components. The total number of samples that recovered STEC isolates for each agar is given in parentheses
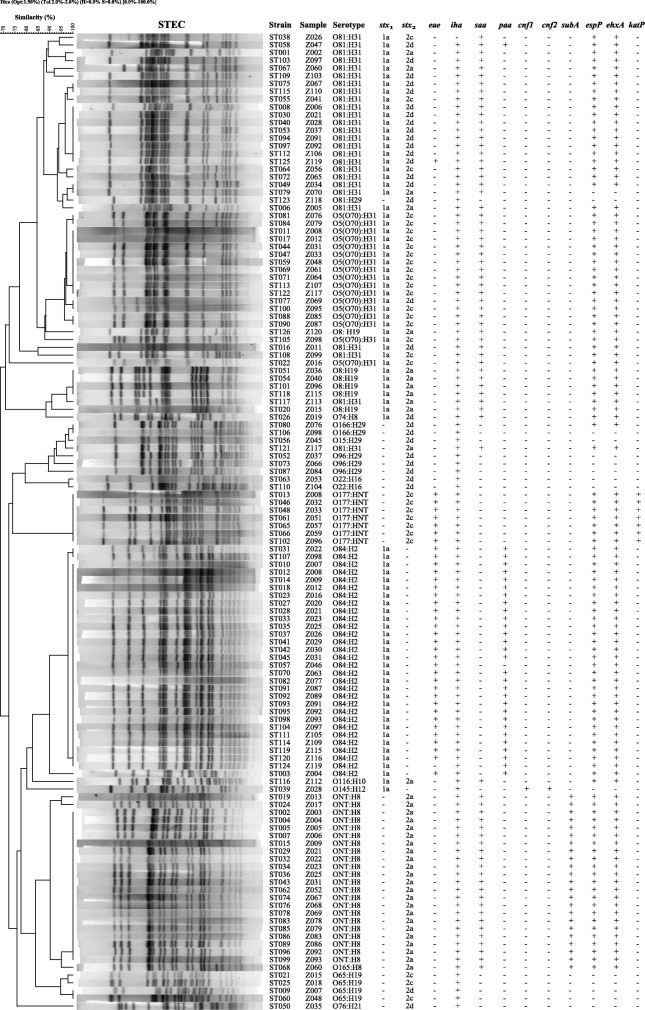
Fig. 2.
PFGE profiles of 126 non-O157 STEC isolates from beef cattle. The corresponding isolate names; samples names; serotypes; stx1 and stx2 subtypes; and the profiles of eae, iha, saa, paa, cnf1, cnf2, subA, espP, ehxA, and katP genes; are listed on the right
Serogroups and serotypes
Fifteen different O serogroups and nine different H types were identified among the 126 STEC isolates, which belonged to 17 different serotypes, including O5(O70):H31, O8:H19, O15:H29, O22:H16, O65:H19, O74:H8, O76:H21, O81:H31, O81:H29, O84:H2, O96:H29, O116:H10, O145:H12, O165:H8, O166:H29, O177:HNT, and ONT:H8. The most common serotype was O84:H2 (23.81%), followed by O81:H31 (20.63%), ONT:H8 (17.46%), and O5(O70):H31 (13.49%). Twenty-two and seven isolates were untypable for the O and H antigens, respectively (Table 2).
Table 2.
Serotypes, stx subtypes and virulence genes of 126 beef cattle STEC isolates
| Serotype | No. of isolates | stx subtypes | Intimin gene | Adherence-related genes | Other virulence-associated genes | Plasmid genes | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| stx 1 | stx 2 | eae | iha | saa | paa | cnf1 | cnf2 | subA | ehxA | espP | katP | ||
| O5(O70):H31 a | 16 | stx 1a | stx 2c | – | + | + | – | – | – | – | + | + | – |
| O5(O70):H31 a | 1 | stx 1a | stx 2d | – | + | + | – | – | – | – | + | + | – |
| O8: H19 | 6 | stx 1a | stx 2a | – | + | + | – | – | – | – | + | + | – |
| O15:H29 | 1 | stx 2d | – | + | – | – | – | – | – | – | – | – | |
| O22:H16 | 2 | stx 2d | – | + | – | – | – | – | – | – | – | – | |
| O65:H19 | 1 | stx 2d | – | + | – | – | – | – | – | – | – | – | |
| O65:H19 | 3 | stx 2c | – | + | – | – | – | – | – | – | – | – | |
| O74:H8 | 1 | stx 1a | stx 2d | – | + | + | – | – | – | – | + | + | – |
| O76:H21 | 1 | stx 2d | – | + | – | – | – | – | – | – | – | – | |
| O81:H29 | 1 | stx 2d | – | + | – | – | – | – | – | – | – | – | |
| O81:H31 | 4 | stx 1a | stx 2a | – | + | + | – | – | – | – | + | + | – |
| O81:H31 | 1 | stx 1a | stx 2a | – | + | + | – | – | – | – | – | – | – |
| O81:H31 | 1 | stx 2a | – | + | + | – | – | – | – | + | + | – | |
| O81:H31 | 3 | stx 1a | stx 2c | – | + | + | – | – | – | – | + | + | – |
| O81:H31 | 1 | stx 1a | stx 2c | – | – | + | – | – | – | – | + | + | – |
| O81:H31 | 14 | stx 1a | stx 2d | – | + | + | – | – | – | – | + | + | – |
| O81:H31 | 1 | stx 1a | stx 2d | + | + | + | – | – | – | – | + | + | – |
| O81:H31 | 1 | stx 1a | stx 2d | – | + | + | + | – | – | – | + | + | – |
| O84:H2 | 28 | stx 1a | + | + | – | + | – | – | – | + | + | – | |
| O84:H2 | 1 | stx 1a | – | + | – | + | – | – | – | + | + | – | |
| O84:H2 | 1 | stx 1a | + | + | – | – | – | – | – | + | + | – | |
| O96:H29 | 3 | stx 2d | – | + | – | – | – | – | – | – | – | – | |
| O116:H10 | 1 | stx 1a | stx 2a | – | + | + | – | – | – | – | + | + | – |
| O145:H12 | 1 | stx 1a | – | + | – | – | + | + | – | – | – | – | |
| O165:H8 | 1 | stx 2a | – | + | + | – | – | – | + | + | + | – | |
| O166:H29 | 1 | stx 2d | – | + | – | – | – | – | – | + | + | – | |
| O166:H29 | 1 | stx 2d | – | + | – | – | – | – | – | – | – | – | |
| O177: HNT | 7 | stx 2c | + | + | – | – | – | – | – | + | + | + | |
| ONT:H8 | 22 | stx 2a | – | + | + | – | – | – | + | + | + | – | |
| Total | 126 | 1 b | 3 b | 37 | 125 | 74 | 30 | 1 | 1 | 23 | 111 | 111 | 7 |
a, These isolates were agglutinated with both O5 and O70 antisera
b, Number of stx1 or stx2 subtypes identified
Subtypes of stx
Of the 126 isolates, 31 (24.6%) were positive for stx1 only, 45 (35.7%) were positive for stx2 only, and 50 (39.7%) were positive for both stx1 and stx2. Only the stx1a subtype was identified in the 81 stx1-positive STEC isolates. Among 95 stx2-positive isolates, three subtypes, i.e. stx2a, stx2c, and stx2d, were identified in 36, 30, and 29 isolates, respectively (Table 2).
Presence of virulence genes
All STEC isolates carried at least one of the virulence-related genes tested. Thirty-seven isolates were eae positive, which belonged to only three serotypes (O81:H31, O84:H2, and O177:HNT). Among the putative adhesin genes (iha, efa1, saa, and paa) screened, iha (99.2%) was the most prevalent, followed by saa (58.7%) and paa (23.8%); however, efa1 was not detected in any isolate. Among the virulence-associated genes (cnf1, cnf2, astA, and subA) tested, 23 (18.3%) isolates were positive for subA, and only one isolate contained both cnf1 and cnf2. The astA gene was absent in all STEC isolates. Three plasmid-related virulence genes (ehxA, espP, and katP) were present in 111, 111 and 7 isolates, respectively. Notably, ehxA-positive isolates also carried espP, and the seven katP-positive isolates harbored both ehxA and espP. None of the 126 isolates were positive for toxB (Table 2).
Pulsed field gel electrophoresis (PFGE)
Genomic DNA from all 126 isolates was digested using Xba I and separated using PFGE to investigate their genetic relationships. An UPGMA (unweighted pair group method with arithmetic mean) dendrogram showed that the STEC isolates were genetically diverse, with nodes linking single isolates or groups of isolates at less than 80% similarity. In total, 49 different PFGE patterns were generated among the 126 STEC isolates. There were two predominant PFGE patterns, which contained 27 and 20 isolates each. Most isolates in the same PFGE pattern tended to have the same serotype, stx subtypes, and virulence gene profiles. All isolates recovered from the same sample showed different PFGE patterns, serotypes, or virulence gene profiles to each other (Fig. 2).
Discussion
Cattle have been identified as dominant reservoirs of STEC without showing any clinical signs themselves, and the consumption of food or water contaminated with bovine feces is often linked to STEC infection [11]. The prevalence of STEC in cattle has been reported recently in the United States, France, Australia, Japan, Brazil, and other countries [1–3, 13, 28–30]. According to previous reports, the prevalence rates of O157 and non-O157 STEC strains ranged from 22 to 62.7% and 2.1 to 70.1%, respectively, in different categories of cattle [22, 31]. In recent years, molecular techniques have been developed to target STEC strains. A TaqMan real-time PCR method was adopted in this study to screen for the presence of STEC strains. A high stx-positive rate (90%) was obtained in beef cattle fecal samples after enrichment, showing the high sensitivity of the real-time PCR method [32]. The isolation rate in this study was 72.5% by culture, indicating a high prevalence of STEC in these beef cattle. The difference of STEC prevalence rates might depend on the various cattle species in certain geographic areas and different detection methods [22]. The failure to recover isolates from a fraction of stx-positive samples in this study indicated that the stx gene might have been amplified from non-E. coli bacteria, stx-phages, or free DNA molecules in the background flora. Another possibility is perturbation of background microflora, or low levels of STEC strains in the stx-positive samples. Notably, two stx-negative samples yielded STEC strains using the culture method, indicating that the presence of STEC strains in cattle fecal samples may be underestimated if only stx-positive samples are subjected to culture. The disagreement between culture- and PCR-based methods to detect STEC strains in beef fecal samples was also observed in a previous report [33]. One of the reasons for the misidentification of culture-positive samples by real-time PCR was likely explained by the inhibitors in samples inducing an increase of Ct values [34].
Currently, there is no single or combination of multiple selective agars capable of identifying all STEC serogroups [35]. As suggested by our previous study, using inclusive agars or less selective agars in combination with highly selective agars would increase the probability of recovering STEC strains from complex matrices [27]. By combining two inclusive and less selective agars (MAC and CH-ECC) and two highly selective agars (RBA-NT and CH-STEC), a culture positive rate of 78.7% (85/108) from the stx-positive samples was obtained, which was higher than those reported in our previous investigation in yak (61.6%) [36], pig (24.4%) [37], and pika (36.2%) [19]. In general, one to three STEC strains were obtained using the four agars for each sample. One strain was predominantly obtained on MAC or CH-ECC agar, which mainly carried stx1 + stx2 genes. One or two strains were predominantly obtained on RBA-NT and CH-STEC agars; however, there was no obvious stx subtype difference between the two agars (Table 1). Notably, the combination of MAC and RBA-NT could cover almost all the culture positive samples (except for one sample) (Fig. 1). Both CH-STEC and RBA-NT use potassium tellurite as a selective additive to isolate specific bacteria; however, STEC isolates display a great variation in tellurite resistance [27, 38]. Our study indicated that MAC may cover the shortage of tellurite-amended agars, and that the combined using MAC and RBA-NT agars could increase the recovery frequency of non-O157 STEC from animal fecal samples.
O:H serotyping of STEC strains has been used widely to identify a strain’s potential to cause severe diseases. O157:H7 is the most well known STEC serotype that causes infections and outbreaks worldwide; however, other serotypes might show regional variations. In particular, STEC strains that possess both the Stx toxin and intimin are the causative agents of severe clinical outcomes, and are also classified as EHEC [13]. Five O groups (O157, O26, O103, O111, and O145), are known as the “big five” EHEC in the European Union, while six O types (O26, O45, O103, O111, O121, and O145), or the “big six”, have been recognized as being responsible for most of the clinical non-O157 STEC infections in the United States [39]. The O157:H7 serotype was not identified in the present study, as was the case with our previous surveys in yak, pig, pika, and raw meat [19, 36, 37, 40], implying a low prevalence of O157 STEC in China. Another possible reason for failure to isolate an O157 STEC might be the method used. Isolation of O157 STEC strains often requires more targeted methods, and the use of immunomagnetic separation (IMS) may improve the isolation sensitivity of O157 strains. However, one of the “big six” serotypes, O145:H12, was identified in this study. Several serogroups of bovine-origin such as O8, O15, O22, O84, O165, and O116 were also detected, some of which are associated with human diseases [1, 3, 31, 41].
STEC is characterized by the production of Stx1, Stx2, or both, and several Stx1/Stx2 subtypes have been described in E. coli [10]. A novel subtype of Stx1e was identified in Enterobacter cloacae [42]. Recently, we identified a novel Stx2 subtype, Stx2h, in E. coli strains from wild marmots in the Qinghai-Tibetan plateau, China [9]. Stx subtypes differ dramatically in their pathogenic potency. The Stx1a, Stx2a, Stx2c, and Stx2d subtypes are commonly reported as associated with HC and HUS [39]. In this study, all stx1-positive strains in beef cattle carried the stx1a subtype, which was consistent with previous reports [3, 8], while the most prevalent stx2 subtypes were stx2a subtypes (53.3%). Three stx subtype combinations, i.e. stx1a + stx2a, stx1a + stx2c, and stx1a + stx2d were detected in 39.7% of the isolates, suggesting that some STEC isolates from cattle have a high pathogenic potential.
To further evaluate the potential virulence of STEC isolates from the cattle, virulence factor genes were tested. Intimin, encoded by the eae gene, plays an important role in bacterial colonization, and several studies have documented that STEC strains carrying the eae gene, which are also identified as EHEC strains, are highly associated with severe human diseases and outbreaks [43, 44]. The eae-positive rate among the 126 STEC isolates in this study was 29.4%, 29 eae-positive isolates carried stx1a, seven harbored stx2c, and one possessed both stx1a and stx2d, which was similar to previous reports [8, 30], implying their high pathogenic potential. Other adherence-related genes, iha, saa and paa, were present at varying frequencies among the isolates, which may have involved cattle colonization by eae-negative non-O157 STEC strains [44]. Additionally, co-existence of ehxA and espP, two plasmid-origin virulence factors, were observed, which contrasted with reports of non-O157 STEC isolates from other sources [45]. The subtilase toxin encoded by subA gene was described in STEC O113:H21, which was related to an outbreak of HUS [46]. In total, 23 isolates from the cattle fecal samples harbored subA genes; however, these STEC strains belonged to the O165 and untypable O serogroups.
PFGE is often used as the gold standard molecular typing method for intestinal pathogens [47]. For those two or three colonies selected from the same sample with different phenotypic or genetic properties, different PFGE patterns, serotypes, and virulence gene profiles were observed, which indicated that beef cattle were colonized by different STEC strains. Some isolates from different samples showed identical PFGE patterns, serotypes, and virulence gene profiles, suggesting that multiple isolates from different beef cattle may belong to the same STEC clone and cross contamination might have occurred on the farm.
Conclusions
A considerable number of non-O157 STEC strains were isolated from beef cattle feces in a farm in China. Based on their serotypes, stx subtypes, and the presence of virulence genes, some non-O157 STEC isolates from beef cattle may have the potential to cause human diseases. A combination of selective MAC and RBA-NT media could increase the recovery frequency of non-O157 STEC during regular animal surveillance.
Methods
Sample collection and enrichment
A total of 120 beef cattle fecal samples were collected from five different pens in a commercial beef cattle farm in Zigong city, Sichuan province, China, in May 2017. All fresh fecal samples were stored separately in 2 ml sterile tubes containing Luria-Bertani medium (LB, Land Bridge, Beijing, China) with 30% glycerol, and then transported immediately to the laboratory in the National Institute for Communicable Disease Control and Prevention, China CDC, in ice bags under cold conditions. After arrival in the laboratory, the sample was frozen at − 80 °C until culture. Each fecal sample was homogenized in 5 ml of E. coli broth (EC broth, Land Bridge, Beijing, China) after thawing, and then incubated at 37 °C for 18 to 24 h with agitation.
DNA extraction and stx screening by TaqMan real-time PCR
A portion (1.5 ml) of each enrichment culture was transferred to a new tube for DNA extraction and centrifugation. Briefly, 150 μl of rapid lysis buffer (100 mM NaCl, 1 mM EDTA [pH 9.0], 10 mM Tris-HCl [pH 8.3], 1% Triton X-100) were added to 1.5 ml of each centrifuged pellet. The mixture was boiled for 10 min, followed by centrifuged at 13,000×g for 10 min, and the supernatant containing the DNA was used as a template in real-time PCR assays. The primers and probes set for stx1/stx2 detection was prepared as previously described [32].
Isolation of STEC strains
Approximately 10 μl of each enrichment culture was streaked directly onto CHROMagar™ ECC agar (CH-ECC, CHROMagar, Paris, France), MacConkey agar (MAC, Land Bridge, Beijing, China), and Rainbow® Agar O157 (RBA, Biolog Inc., Hayward, CA, USA), supplemented with 10 μg/ml novobiocin and 0.8 μg/ml potassium tellurite (modified, RBA-NT), and CHROMagar™ STEC agar (CH-STEC, CHROMagar, Paris, France), respectively. After overnight incubation at 37 °C, ten presumptive colonies on each plate were picked and subjected to colony PCR to detect the stx1 and stx2 genes according to the method revealed in a previous study [19], including green-blue or colorless colonies on CH-ECC agar; pink or colorless colonies on MAC agar; purple, grey, or mauve colonies on RBA-NT agar; and mauve colonies on CH-STEC agar. The stx-positive colonies were then plated onto LB agar and incubated at 37 °C overnight to obtain a single colony for further identification. Only one STEC isolate from each sample was chosen for further characterization if only identical stx types/subtypes were present on the four different agars. Isolates on inclusive agars (MAC and CH-ECC agars) were selected as a priority, while other isolates with same stx types/subtypes were eliminated.
Biochemical test and serotyping of STEC isolates
All stx-positive isolates were confirmed biochemically as E. coli using API® 20E biochemical test strips (bioMérieux, Lyon, France). The O serogroup of each isolate was preliminary screened using a PCR method with O antigen specific primers [48] and confirmed by using all O1-O188 E. coli antisera (SSI Diagnostica, Hillerød, Denmark). The entire coding sequence of the fliC gene was amplified by PCR, and then sequenced to determine the H type, as described previously [19].
Identification of virulence and adherence factor genes
The presence of the intimin-encoding gene (eae), putative adhesin genes (iha, efa1, saa and paa), virulence-associated genes (cnf1, cnf2, astA, and subA), and the large heterologous virulence plasmid genes (ehxA, katP, espP, and toxB) in all STEC isolates were detected as previously described [36].
Subtyping of stx
The stx1 and stx2 subtypes were determined using a PCR-based subtyping method [10]. The complete stx1 and/or stx2 genes of certain STEC isolates were amplified and sequenced to verify the PCR-based subtyping results.
Pulsed-field gel electrophoresis (PFGE)
PFGE was performed according to the STEC subtyping protocol from PulseNet, USA (https://www.cdc.gov/pulsenet/pdf/ecoli-shigella-salmonella-pfge-protocol-508c.pdf). Briefly, the genomic DNA was digested for 2 h with 45 U of XbaI (Takara, Dalian, China) at 37 °C. The digested samples were placed on 1% SeaKem Gold agarose and electrophoresis was carried out at 6.0 V/cm for 18 h with an initial switch time value of 6.8 s and final switch time of 35.5 s. Images were captured using the Gel Doc™ XR+ System (Bio-Rad, Hercules, CA, USA). PFGE patterns were analyzed and a UPGMA dendrogram was constructed using the BioNumerics software (Applied Maths, Belgium).
Additional file
Table S1. Prevalence of stx and STEC in beef cattle feces. (PDF 39 kb)
Acknowledgements
Not applicable.
Abbreviations
- A/E
Attaching and Effacing
- CH-ECC
CHROMagar™ ECC
- CH-STEC
CHROMagar™ STEC
- EHEC
Enterohemorrhagic E. coli
- HC
Hemorrhagic colitis
- HUS
Hemolytic uremic syndrome
- IMS
Immunomagnetic separation
- LEE
Locus of enterocyte effacement
- MAC
MacConkey
- PFGE
Pulsed-Field Gel Electrophoresis
- RBA-NT
modified Rainbow® Agar O157
- STEC
Shiga toxin-producing Escherichia coli
- Stx
Shiga toxin
- Stx1
Shiga toxin 1
- Stx2
Shiga toxin 2
- UPGMA
Unweighted Pair Group Method with Arithmetic mean
Authors’ contributions
RF and YX designed the project; RF, SF, QL, and HW carried out the sampling work; RF, KS, XY, HS, YXu, and BH carried out the experiments; RF, KS, JZ, and YX analyzed the data; RF drafted the manuscript; XB and YX revised the manuscript. All authors had read and approved the manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (81772152 and 81701977), the National Basic Research Priorities Program of China (2015CB554201), and the National Science and Technology Major Project (2018ZX10201001 and 2018ZX10301407). The funding bodies had no role in the design of the study and collection, analysis, and interpretation of data and in writing this manuscript.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Ethics approval and consent to participate
This study is part of STEC active surveillance program conducted in China. The collection of feces from commercial beef cattle was approved by the owners of the farm. Feces were immediately collected after the beef cattle moved away from their stool, without any animal manipulation. This study was reviewed and officially approved by the ethics committee of the National Institute for Communicable Disease Control and Prevention, China CDC, in accordance with the medical research regulations of National Health Commission of the People’s Republic of China (approval number: ICDC-2017006).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Ruyue Fan and Kun Shao contributed equally to this work.
Contributor Information
Ruyue Fan, Email: fryforever@163.com.
Kun Shao, Email: shaokun1006@163.com.
Xi Yang, Email: 449795091@qq.com.
Xiangning Bai, Email: baixiangning@icdc.cn.
Shanshan Fu, Email: 543195534@qq.com.
Hui Sun, Email: Sunhui@icdc.cn.
Yanmei Xu, Email: xyanmei2006@126.com.
Hong Wang, Email: 460973389@qq.com.
Qun Li, Email: lqzgcdc@163.com.
Bin Hu, Email: wubz9670@126.com.
Ji Zhang, Email: j.zhang7@massey.ac.nz.
Yanwen Xiong, Phone: +8610 58900749, Email: xiongyanwen@icdc.cn.
References
- 1.Gonzalez AG, Cerqueira AM, Guth BE, Coutinho CA, Liberal MH, Souza RM, Andrade JR. Serotypes, virulence markers and cell invasion ability of Shiga toxin-producing Escherichia coli strains isolated from healthy dairy cattle. J Appl Microbiol. 2016;121(4):1130–1143. doi: 10.1111/jam.13230. [DOI] [PubMed] [Google Scholar]
- 2.Mellor GE, Fegan N, Duffy LL, Mc MK, Jordan D, Barlow RS. National survey of Shiga toxin-producing Escherichia coli serotypes O26, O45, O103, O111, O121, O145, and O157 in Australian beef cattle feces. J Food Prot. 2016;79(11):1868–1874. doi: 10.4315/0362-028X.JFP-15-507. [DOI] [PubMed] [Google Scholar]
- 3.Dong HJ, Lee S, Kim W, An JU, Kim J, Kim D, Cho S. Prevalence, virulence potential, and pulsed-field gel electrophoresis profiling of Shiga toxin-producing Escherichia coli strains from cattle. Gut Pathog. 2017;9:22. doi: 10.1186/s13099-017-0169-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luna-Gierke RE, Griffin PM, Gould LH, Herman K, Bopp CA, Strockbine N, Mody RK. Outbreaks of non-O157 Shiga toxin-producing Escherichia coli infection: USA. Epidemiol Infect. 2014;142(11):2270–2280. doi: 10.1017/S0950268813003233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Valilis E, Ramsey A, Sidiq S, DuPont HL. Non-O157 Shiga toxin-producing Escherichia coli-a poorly appreciated enteric pathogen: systematic review. Int J Infect Dis. 2018;76:82–87. doi: 10.1016/j.ijid.2018.09.002. [DOI] [PubMed] [Google Scholar]
- 6.Iweriebor BC, Iwu CJ, Obi LC, Nwodo UU, Okoh AI. Multiple antibiotic resistances among Shiga toxin producing Escherichia coli O157 in feces of dairy cattle farms in eastern cape of South Africa. BMC Microbiol. 2015;15:213. doi: 10.1186/s12866-015-0553-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim SA, Park SH, Lee SI, Ricke SC. Rapid and simple method by combining FTA card DNA extraction with two set multiplex PCR for simultaneous detection of non-O157 Shiga toxin-producing Escherichia coli strains and virulence genes in food samples. Lett Appl Microbiol. 2017;65(6):482–488. doi: 10.1111/lam.12805. [DOI] [PubMed] [Google Scholar]
- 8.Jajarmi M, Imani Fooladi AA, Badouei MA, Ahmadi A. Virulence genes, Shiga toxin subtypes, major O-serogroups, and phylogenetic background of Shiga toxin-producing Escherichia coli strains isolated from cattle in Iran. Microb Pathog. 2017;109:274–279. doi: 10.1016/j.micpath.2017.05.041. [DOI] [PubMed] [Google Scholar]
- 9.Bai X, Fu S, Zhang J, Fan R, Xu Y, Sun H, He X, Xu J, Xiong Y. Identification and pathogenomic analysis of an Escherichia coli strain producing a novel Shiga toxin 2 subtype. Sci Rep. 2018;8(1):6756. doi: 10.1038/s41598-018-25233-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scheutz F, Teel LD, Beutin L, Pierard D, Buvens G, Karch H, Mellmann A, Caprioli A, Tozzoli R, Morabito S, et al. Multicenter evaluation of a sequence-based protocol for subtyping Shiga toxins and standardizing Stx nomenclature. J Clin Microbiol. 2012;50(9):2951–2963. doi: 10.1128/JCM.00860-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mekata H, Iguchi A, Kawano K, Kirino Y, Kobayashi I, Misawa N. Identification of O serotypes, genotypes, and virulotypes of Shiga toxin-producing Escherichia coli isolates, including non-O157 from beef cattle in Japan. J Food Prot. 2014;77(8):1269–1274. doi: 10.4315/0362-028X.JFP-13-506. [DOI] [PubMed] [Google Scholar]
- 12.Smith JL, Fratamico PM, NWt G. Shiga toxin-producing Escherichia coli. Adv Appl Microbiol. 2014;86:145–197. doi: 10.1016/B978-0-12-800262-9.00003-2. [DOI] [PubMed] [Google Scholar]
- 13.Bibbal D, Loukiadis E, Kerouredan M, Ferre F, Dilasser F, Peytavin de Garam C, Cartier P, Oswald E, Gay E, Auvray F, et al. Prevalence of carriage of Shiga toxin-producing Escherichia coli serotypes O157:H7, O26:H11, O103:H2, O111:H8, and O145:H28 among slaughtered adult cattle in France. Appl Environ Microbiol. 2015;81(4):1397–1405. doi: 10.1128/AEM.03315-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bai X, Hu B, Xu Y, Sun H, Zhao A, Ba P, Fu S, Fan R, Jin Y, Wang H, et al. Molecular and phylogenetic characterization of non-O157 Shiga toxin-producing Escherichia coli strains in China. Front Cell Infect Microbiol. 2016;6:143. doi: 10.3389/fcimb.2016.00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thomas RR, Brooks HJ, O'Brien R. Prevalence of Shiga toxin-producing and enteropathogenic Escherichia coli marker genes in diarrhoeic stools in a New Zealand catchment area. J Clin Pathol. 2017;70(1):81–84. doi: 10.1136/jclinpath-2016-203882. [DOI] [PubMed] [Google Scholar]
- 16.Toma C, Martinez Espinosa E, Song T, Miliwebsky E, Chinen I, Iyoda S, Iwanaga M, Rivas M. Distribution of putative adhesins in different seropathotypes of Shiga toxin-producing Escherichia coli. J Clin Microbiol. 2004;42(11):4937–4946. doi: 10.1128/JCM.42.11.4937-4946.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Savarino SJ, Fasano A, Watson J, Martin BM, Levine MM, Guandalini S, Guerry P. Enteroaggregative Escherichia coli heat-stable enterotoxin 1 represents another subfamily of E. coli heat-stable toxin. Proc Natl Acad Sci U S A. 1993;90(7):3093–3097. doi: 10.1073/pnas.90.7.3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lim JY, Yoon J, Hovde CJ. A brief overview of Escherichia coli O157:H7 and its plasmid O157. J Microbiol Biotechnol. 2010;20(1):5–14. [PMC free article] [PubMed] [Google Scholar]
- 19.Bai X, Zhang W, Tang X, Xin Y, Xu Y, Sun H, Luo X, Pu J, Xu J, Xiong Y, et al. Shiga toxin-producing Escherichia coli in plateau pika (Ochotona curzoniae) on the Qinghai-Tibetan plateau, China. Front Microbiol. 2016;7:375. doi: 10.3389/fmicb.2016.00375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dias D, Caetano T, Torres RT, Fonseca C, Mendo S. Shiga toxin-producing Escherichia coli in wild ungulates. Sci Total Environ. 2019;651(Pt 1):203–209. doi: 10.1016/j.scitotenv.2018.09.162. [DOI] [PubMed] [Google Scholar]
- 21.Padola NL, Etcheverria AI. Shiga toxin-producing Escherichia coli in human, cattle, and foods. Strategies for detection and control. Front Cell Infect Microbiol. 2014;4:89. doi: 10.3389/fcimb.2014.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caceres ME, Etcheverria AI, Fernandez D, Rodriguez EM, Padola NL. Variation in the distribution of putative virulence and colonization factors in Shiga toxin-producing Escherichia coli isolated from different categories of cattle. Front Cell Infect Microbiol. 2017;7:147. doi: 10.3389/fcimb.2017.00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gill A, Huszczynski G, Gauthier M, Blais B. Evaluation of eight agar media for the isolation of Shiga toxin-producing Escherichia coli. J Microbiol Methods. 2014;96:6–11. doi: 10.1016/j.mimet.2013.10.022. [DOI] [PubMed] [Google Scholar]
- 24.Wang H, Chen Z, Jiang X. Improving the enrichment and plating methods for rapid detection of non-O157 Shiga toxin-producing Escherichia coli in dairy compost. J Food Prot. 2016;79(3):413–420. doi: 10.4315/0362-028X.JFP-15-249. [DOI] [PubMed] [Google Scholar]
- 25.Wylie JL, Van Caeseele P, Gilmour MW, Sitter D, Guttek C, Giercke S. Evaluation of a new chromogenic agar medium for detection of Shiga toxin-producing Escherichia coli (STEC) and relative prevalences of O157 and non-O157 STEC in Manitoba, Canada. J Clin Microbiol. 2013;51(2):466–471. doi: 10.1128/JCM.02329-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zelyas N, Poon A, Patterson-Fortin L, Johnson RP, Lee W, Chui L. Assessment of commercial chromogenic solid media for the detection of non-O157 Shiga toxin-producing Escherichia coli (STEC) Diagn Microbiol Infect Dis. 2016;85(3):302–308. doi: 10.1016/j.diagmicrobio.2016.03.013. [DOI] [PubMed] [Google Scholar]
- 27.Fan R, Bai X, Fu S, Xu Y, Sun H, Wang H, Xiong Y. Tellurite resistance profiles and performance of different chromogenic agars for detection of non-O157 Shiga toxin-producing Escherichia coli. Int J Food Microbiol. 2018;266:295–300. doi: 10.1016/j.ijfoodmicro.2017.12.016. [DOI] [PubMed] [Google Scholar]
- 28.Lee K, Kusumoto M, Iwata T, Iyoda S, Akiba M. Nationwide investigation of Shiga toxin-producing Escherichia coli among cattle in Japan revealed the risk factors and potentially virulent subgroups. Epidemiol Infect. 2017;145(8):1557–1566. doi: 10.1017/S0950268817000474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mir RA, Weppelmann TA, Kang M, Bliss TM, DiLorenzo N, Lamb GC, Ahn S, Jeong KC. Association between animal age and the prevalence of Shiga toxin-producing Escherichia coli in a cohort of beef cattle. Vet Microbiol. 2015;175(2–4):325–331. doi: 10.1016/j.vetmic.2014.12.016. [DOI] [PubMed] [Google Scholar]
- 30.Adamu MS, Ugochukwu ICI, Idoko SI, Kwabugge YA, Abubakar NS, Ameh JA. Virulent gene profile and antibiotic susceptibility pattern of Shiga toxin-producing Escherichia coli (STEC) from cattle and camels in Maiduguri, north-eastern Nigeria. Trop Anim Health Prod. 2018;50(6):1327–1341. doi: 10.1007/s11250-018-1565-z. [DOI] [PubMed] [Google Scholar]
- 31.Hussein HS, Bollinger LM. Prevalence of Shiga toxin-producing Escherichia coli in beef cattle. J Food Prot. 2005;68(10):2224–2241. doi: 10.4315/0362-028x-68.10.2224. [DOI] [PubMed] [Google Scholar]
- 32.Hara-Kudo Y, Konishi N, Ohtsuka K, Iwabuchi K, Kikuchi R, Isobe J, Yamazaki T, Suzuki F, Nagai Y, Yamada H, et al. An interlaboratory study on efficient detection of Shiga toxin-producing Escherichia coli O26, O103, O111, O121, O145, and O157 in food using real-time PCR assay and chromogenic agar. Int J Food Microbiol. 2016;230:81–88. doi: 10.1016/j.ijfoodmicro.2016.03.031. [DOI] [PubMed] [Google Scholar]
- 33.Noll LW, Shridhar PB, Dewsbury DM, Shi X, Cernicchiaro N, Renter DG, Nagaraja TG. A comparison of culture- and PCR-based methods to detect six major non-O157 serogroups of Shiga toxin-producing Escherichia coli in cattle feces. PLoS One. 2015;10(8):e0135446. doi: 10.1371/journal.pone.0135446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Verhaegen Bavo, De Reu Koen, De Zutter Lieven, Verstraete Karen, Heyndrickx Marc, Van Coillie Els. Comparison of Droplet Digital PCR and qPCR for the Quantification of Shiga Toxin-Producing Escherichia coli in Bovine Feces. Toxins. 2016;8(5):157. doi: 10.3390/toxins8050157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brusa V, Pineyro PE, Galli L, Linares LH, Ortega EE, Padola NL, Leotta GA. Isolation of Shiga toxin-producing Escherichia coli from ground beef using multiple combinations of enrichment broths and selective agars. Foodborne Pathog Dis. 2016;13(3):163–170. doi: 10.1089/fpd.2015.2034. [DOI] [PubMed] [Google Scholar]
- 36.Bai X, Zhao A, Lan R, Xin Y, Xie H, Meng Q, Jin D, Yu B, Sun H, Lu S, et al. Shiga toxin-producing Escherichia coli in yaks (Bos grunniens) from the Qinghai-Tibetan plateau, China. PLoS One. 2013;8(6):e65537. doi: 10.1371/journal.pone.0065537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meng Q, Bai X, Zhao A, Lan R, Du H, Wang T, Shi C, Yuan X, Bai X, Ji S, et al. Characterization of Shiga toxin-producing Escherichia coli isolated from healthy pigs in China. BMC Microbiol. 2014;14:5. doi: 10.1186/1471-2180-14-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lewis GL, Jorgensen QR, Loy JD, Moxley RA. Tellurite resistance in Shiga toxin-producing Escherichia coli. Curr Microbiol. 2018;75(6):752–759. doi: 10.1007/s00284-018-1444-x. [DOI] [PubMed] [Google Scholar]
- 39.Group FWSE Hazard identification and characterization: criteria for categorizing Shiga toxin-producing Escherichia coli on a risk basis (dagger) J Food Prot. 2019;82(1):7–21. doi: 10.4315/0362-028X.JFP-18-291. [DOI] [PubMed] [Google Scholar]
- 40.Bai X, Wang H, Xin Y, Wei R, Tang X, Zhao A, Sun H, Zhang W, Wang Y, Xu Y, et al. Prevalence and characteristics of Shiga toxin-producing Escherichia coli isolated from retail raw meats in China. Int J Food Microbiol. 2015;200:31–38. doi: 10.1016/j.ijfoodmicro.2015.01.018. [DOI] [PubMed] [Google Scholar]
- 41.Bonardi S, Alpigiani I, Tozzoli R, Vismarra A, Zecca V, Greppi C, Bacci C, Bruini I, Brindani F. Shiga toxin-producing Escherichia coli O157, O26 and O111 in cattle faeces and hides in Italy. Vet Rec Open. 2015;2(1):e000061. doi: 10.1136/vetreco-2014-000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Probert WS, McQuaid C, Schrader K. Isolation and identification of an Enterobacter cloacae strain producing a novel subtype of Shiga toxin type 1. J Clin Microbiol. 2014;52(7):2346–2351. doi: 10.1128/JCM.00338-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Werber D, Fruth A, Buchholz U, Prager R, Kramer MH, Ammon A, Tschape H. Strong association between Shiga toxin-producing Escherichia coli O157 and virulence genes stx2 and eae as possible explanation for predominance of serogroup O157 in patients with haemolytic uraemic syndrome. Eur J Clin Microbiol Infect Dis. 2003;22(12):726–730. doi: 10.1007/s10096-003-1025-0. [DOI] [PubMed] [Google Scholar]
- 44.Etcheverria AI, Padola NL. Shiga toxin-producing Escherichia coli: factors involved in virulence and cattle colonization. Virulence. 2013;4(5):366–372. doi: 10.4161/viru.24642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fu S, Bai X, Fan R, Sun H, Xu Y, Xiong Y. Genetic diversity of the enterohaemolysin gene (ehxA) in non-O157 Shiga toxin-producing Escherichia coli strains in China. Sci Rep. 2018;8(1):4233. doi: 10.1038/s41598-018-22699-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paton AW, Woodrow MC, Doyle RM, Lanser JA, Paton JC. Molecular characterization of a Shiga toxigenic Escherichia coli O113:H21 strain lacking eae responsible for a cluster of cases of hemolytic-uremic syndrome. J Clin Microbiol. 1999;37(10):3357–3361. doi: 10.1128/jcm.37.10.3357-3361.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kang E, Hwang SY, Kwon KH, Kim KY, Kim JH, Park YH. Prevalence and characteristics of Shiga toxin-producing Escherichia coli (STEC) from cattle in Korea between 2010 and 2011. J Vet Sci. 2014;15(3):369–379. doi: 10.4142/jvs.2014.15.3.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Iguchi A, Iyoda S, Seto K, Morita-Ishihara T, Scheutz F, Ohnishi M, Pathogenic E. coli working Group in Japan: Escherichia coli O-genotyping PCR: a comprehensive and practical platform for molecular O serogrouping. J Clin Microbiol. 2015;53(8):2427–2432. doi: 10.1128/JCM.00321-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Prevalence of stx and STEC in beef cattle feces. (PDF 39 kb)
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.