Abstract
Anionic phospholipids are minor but prominent components of the plasma membrane and necessary for ion channel function. Their persistence in bulk membranes, in particular phosphatidylinositol 4,5-bisphosphate (PIP2), initially suggested they act as channel cofactors. However, recent technologies have established an emerging system of nanoscale signaling to ion channels based on lipid compartmentalization (clustering), direct lipid binding, and local lipid dynamics that allow cells to harness lipid heterogeneity to gate ion channels. The new tools to study lipid binding are set to transform our view of the membrane and answer important questions surrounding ion channel delimited processes, such as mechanosensation.
Keywords: signaling lipids, mass spectrometry, cryo-electron microscopy, super-resolution imaging, lipid rafts
Challenges and opportunities for lipid regulation of ion channels
Ion channels in lipid membranes can be regulated by extracellular ligands[1], post-translational modification[2], and the lipid that surround them[3–5]. Phosphatidylinositol 4,5 bisphosphate (PIP2) (see Glossary), a component of cell membranes, was the first known [6] and remains the most prominent anionic signaling lipid to affect ion channels [3,7–9]. As such, PIP2 has served as a model for studying this type of regulation. Initial studies (e.g. cardiac) suggested the concentration of PIP2 remains relatively high and unresponsive in the bulk plasma membrane during physiological changes, giving rise to a theory that PIP2 primarily functions as a cofactor for ion channels [3,4]. In contrast, PIP2 was also recognized for its potential role to dynamically regulate cell function [10], including localized signaling. For a detailed consideration of theories related to local PIP2 signaling, the reader is referred to Hilgemann 2007 [4]. Thus, it was hypothesized that the putative signaling role of PIP2 was dependent on its lateral mobility in the membrane, and its possible local sequestration and metabolism. However, the tools to confirm the role of PIP2 at nanoscale resolution were lacking [4]. Tools for directly measuring membrane lipids bound to ion channels were also lacking leaving the following questions unanswered: which lipids bind to channels, what are their binding affinities, where exactly do lipids bind, how are the lipids organized relative to the channels, and how are they regulating the channel?
This review first highlights the new tools for detecting membrane lipid-ion channel interactions and heterogeneity of cell membranes at nanometer scales, and then discusses how these tools have been used to show how cells harness heterogeneity of membranes to gate channels. The review ends by discussing how evolving views on lipid regulation will likely transform lipids from mostly cofactors and structural support to central orchestrators of biological function. The data herein spans molecular to cellular mechanisms. Many of the principles outlined here likely apply to the lipid regulation of transporters and many other classes of membrane proteins.
Advanced technologies for detecting membrane lipid-ion channel interactions
The tools described here are advancements that have occurred over the last five years. Specifically, mass spectrometry, detergent-based lipid binding assays, cryo-electron microscopy (EM), and super-resolution imaging techniques are discussed. Classical tools for describing interactions of ion channels with lipids have been described previously. Combining these biochemical technologies may prove to be a powerful strategy to define lipid binding to ion channels.
Mass spectrometry
It has been challenging to identify the type of membrane lipid bound to an ion channel with no prior knowledge of what that lipid might be. This is because both ion channels and lipids are hydrophobic, hence they non-specifically aggregate, masking the specific interaction. Absent a direct detection mechanism, the most common method has been to test many candidate lipids either immobilized [3] or in a functionally reconstituted system [11,12] in order to find one that binds and has a functional effect on the ion channel.
Mass spectrometry was initially limited by expensive instrumentation and technical expertise but the introduction of commercial platforms have alleviated these problems to some extent [13]. Advances in mass spectrometry now enable the direct detection of a lipid binding to an ion channel [13,14]. Electrospray ionization is used to propel the lipid-ion channel complex into a gaseous state from solution in a micelle but without disrupting the specific lipid interaction (Figure 1A). Nonspecific lipids are stripped away by the input of energy while the bound lipid is directly detected inside the mass spectrometer. This technique has been used to characterize diverse families of ion channels, such as mechanosensitive channel (MscL), aquaporin Z, ammonia channel (AmtB) [14], and, most recently, the technique has defined PIP2 interactions that influence downstream G-protein coupling [15]. Hence, the technique has broad utility. One of the great strengths of the approach is that the oligomeric state of the ion channel and detection of lipid binding can be deduced with the same experiment [16] and, in theory, the technique has the potential to identify a lipid species with no prior knowledge of the lipid. However, defining which lipids are regulating the function of a structure as opposed to only conferring stability typically requires electrophysiology experiments [17].
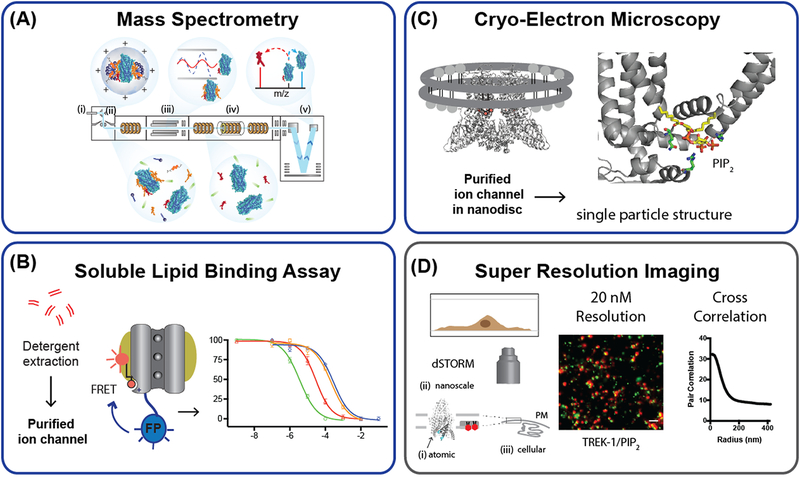
Figure 1. New tools for detecting lipid ion channel interactions.
(A) Schematic representation of the processes occurring in the mass spectrometry of membrane protein lipid interactions. (i) Ionization of the protein complex takes place while in the protective detergent micelle. (ii) Activation within the source, at high energy, is required to disrupt the micelle. (iii) Isolation of discrete m/z values is achieved in the quadrupole and is followed by activation in the collision cell (iv). (v) Following time-of-flight analysis the protein and direct protein lipid interaction is measured. (B) Detergent solubilized phosphatidylinositol 4,5-bisphosphate (PIP2) binding assay for potassium channels. The purified channel and lipids are solubilized in detergent and the binding of a fluorescent-PIP2 probe is detected by fluorescence resonance energy transfer (FRET) assay from a genetically encoded fluorescent protein (FP). (C) Cryo-electron microscopy (cryoEM), PIP2 resolved in the structure of TRPV5 channel (PDB 6DMU). A channel is purified and reconstituted into nanodiscs, i.e. a protein lipid particle, and imaged with electron microscopy. An atomic resolution structure with the lipid bound is then determined by single particle reconstruction. (D) Super resolution imaging (direct stochastic optical reconstruction microscopy (dSTORM)) of native TREK-1 and PIP2 in C2C12 myoblast cell. Cross-correlation of TREK-1 and PIP2 shows colocalization of the channel with PIP2 at specified distances in cellular membrane. The atomic scale resolution of an ion channel structure ((i.), TREK-1 PDB 4XKD) is depicted relative to the nanoscale resolution of the membrane imaged by dSTORM and the cellular resolution (iii.) imaged by diffraction limited confocal microscopy of the plasma membrane (PM). See also Figure 2A.
Soluble lipid binding assay
Knowing the binding affinity (Kd) of a lipid for an ion channel is perhaps the most important property for understanding its function in the membrane, as only the Kd can reveal the relevance of a particular lipid concentration in the membrane for that channel. Furthermore, the Kds of two distinct lipids can reveal their ability to compete for binding with an ion channel. Lastly, the Kd can help to distinguish a ligand from a cofactor. A ligand will have an affinity similar to its cellular concentration that allows it to come on and off to perform its function [5].
Recently, a proximity based assay emerged for determining the Kd of PIP2 bound to a fully assembled potassium channels [18] (Figure IB). To measure the lipid Kd, the channel was tagged with a fluorescent protein, purified in detergent, and then competition of a fluorescent lipid probe with a lipid ligand was detected by fluorescence resonance energy transfer (FRET) and bioluminescent resonance energy transfer (BRET). The soluble environment led to detecting a Kd similar to traditional ligand binding constants which are convenient for thinking about lipids as ligands and comparing their interaction within protein sites. BRET is more sensitive than FRET (~100x) but has the drawback of needing a chemical substrate to generate the signal [18]. Previous binding of PIP2 to soluble cytoplasmic fragments is unlikely to reflect the true affinity of the lipid [19].
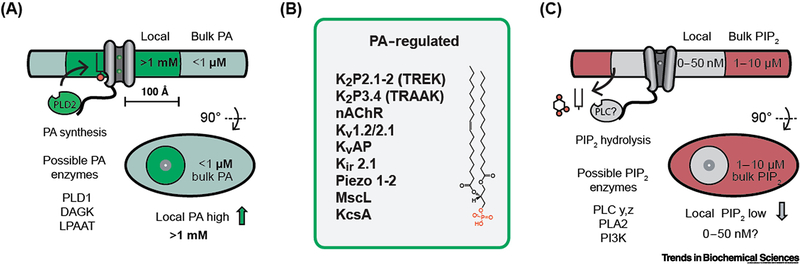
Figure 4. Lipid activation of ion channels through localized synthesis.
(A) Co-localization of TREK-1 with phosphatidic acid (PA) very convincingly established a lipid-protein based localization mechanism as a means of its activation [40]. Indirectly it also establishes the fact that local lipid gradients in a cell must exists sufficient to activate a channel. The putative concentration, local to the channel (grey cylinders), is indicated by an inset circle. PA, shown with black stick and red circle, is in concentrations low relative to a channel’s Kd. Localization of phospholipase D (PLD) and local production of PA results in a local high concentration of PA (>1 mM) and channel activation (gating). Local PA concentration is estimated from a single molecule of PA produced within 100 Å of the channel. Additional putative enzymes that could generate local PA include diacylglycerol kinase (DAGK) and lysophosphatidic acid acyltransferase (LPAAT). (B) Emerging class of PA regulated channels. At least 11 channels are known to respond directly to PA. Both nAChR and TRAAK require relatively high concentrations of PA suggestion local high concentrations may contribute to their activation [18,66]. Kv1.2/2.1 is a channel chimera [44]. TRESK is activated by a mixture of anionic lipids [71]. (C) PIP2 tonic concentration is high in the bulk membrane (~1–10 μM, red shading) [4] relative to a channels Kd (~200 nM), shown as black sticks with two red spheres. A hypothetical ion channel is shown with phospholipase C (PLC) bound to a disorder loop (black line). Local hydrolysis depletes PIP2 and decreases PIP2 binding resulting in a closed channel. Additional putative enzymes that could locally deplete PIP2 include phospholipase A2 (PLA2) and phosphoinositide 3 kinase (PI3K). Bulk PIP2 and PA concentrations are estimated from Kir and K2P lipid binding constants [18].
CryoEM
By determining the structure of lipid-ion channel complexes, lipid binding sites in a membrane protein can be identified and the lipids function can be postulated. Lipids that function as ligands or cofactors likely bind to the transmembrane domain and cause a conformational change that opens the channel. Lipids that bind to disordered loops outside the membrane likely affect channel trafficking or localization. For example, X-ray crystallography produced one of the first convincing demonstrations of a lipid bound to an ion channel—the structure of PIP2 bound to inward rectifying potassium channel subtype 2.2 (Kir2.2). PIP2 was seen bound to a specific site in the ion channel’s transmembrane domain [20], causing a conformational change consistent with the lipid gating the channel [5,20,21].
Limitations of X-ray crystallography made it difficult to study lipid-ion channel interactions more completely. Recently, cryoEM has emerged as a powerful alternative to X-ray crystallography [22]. Primarily, cryoEM has the advantage that a structure can be determined in a lipid bilayer by reconstituting the channel into nanodiscs [23], a native membrane-like state.
The heat- and capsaicin-activated transient receptor potential (TRP) vanilloid 1 (TRPV1) was the first ion channel for which a high-resolution cryoEM structure was reported in a nanodisc [23]. TRP channels remain at the forefront of the cryoEM resolution revolution, and structures of representative members of all seven sub-families have been reported [24], four of which have been determined in complex with signaling lipids [22,25–27]. The first TRP ion channel structure in lipid nanodiscs reported that the capsaicin binding site of TRPV1 under resting conditions was occupied by a phosphoinositide, another component of cell membrane. It was thus proposed that this lipid acted as competitive antagonist in these conditions [22]. This paper showed the power of the nanodisc system for determining lipid protein interactions in membrane like conditions.
Additional examples which use the power of cryoEM to determine the structure of lipid-ion channel complexes are found in the Ca2+ selective channel TRPV5 [26], the lysosomal channel TRPML1 [27], and the γ-aminobutyric acid A (GABAA) receptor [28]. TRPV5 is constitutively active in the presence of basal levels of PIP2, and a recent study used cryoEM to determine the structure of PIP2 bound to TRPV5 in lipid nanodiscs (Figure 1C). This study found that PIP2 induced a conformational rearrangement of the channel that widens the conductance pathway, allowing Ca2+ ions to pass. This structure offered the first glimpse of TRP channels in PIP2 dependent open and closed states [26]. TRPML1 is activated by PI(3,5)P2, a phosphoinositide found in the lysosomal membrane, and it is inhibited by PIP2 which is found in the plasma membrane [27]. Structures of this channel with either lipid showed that both lipids bind to the intracellular portions of the first two transmembrane segments, sites distinct from lipid binding in TRPV1, TRPV5 and TRPM8 [25,27]. Finally, using CryoEM, a PIP2 binding site was discovered in the αl subunit of GABAA receptor, which belongs to a separate super family of pentameric ligand gated ion channels that mediate fast inhibitory neurotransmission [28]. The site was speculated to contribute to localization of the channel to lipid domains within the plasma membrane, but the role of lipid regulation in GABAA was not previously known and will require further study.
Overall, CryoEM has an enormous potential to provide direct information not only on how lipids interact with ion channels but also on the conformational changes they induce in the channel protein. From the large number of recently reported ion channel structures, relatively few have been solved with their regulatory lipid bound, but given the rapid progress in the field, we will likely see more of them soon.
Super-resolution imaging
Another question that has been difficult to answer is if signaling lipids are restricted in their lateral mobility in the membrane. Determining restricted lateral mobility is critical for establishing local gradients of lipids in cell membranes. Local gradients are also critical for establishing lipids as potential central regulators of ion channel function [4]. By typical confocal microscopy, PIP2 appears relatively diffuse (i.e. there is no obvious gradient) [4,29]. But confocal microscopy diffraction is limited to ~200 nm resolution—higher resolution tells a different story [29,30].
Recently, super resolution imaging defined at least three types of lateral lipid heterogeneity (domains/clusters) in cell membranes. These include the classic lipid rafts (also called GM1 domains) [31], PIP2/protein charged clusters [29,32], and phosphatidyl inositol triphosphate (PIP3) clusters [33] (See Figure 2A). The existence of lipid heterogeneity establishes nanoscale locations within a membrane where a lipid-regulated ion channel can be activated or inhibited.
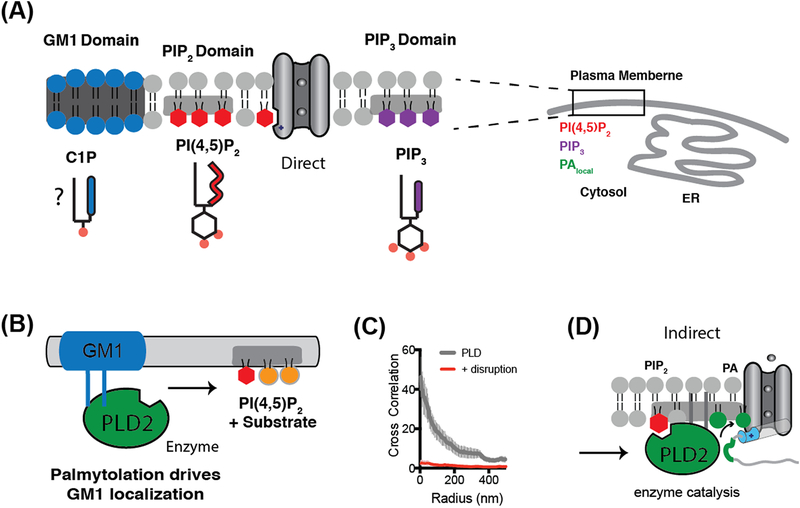
Figure 2. Lipid partitioning and clustering gives rise to lipid heterogeneity.
(A) Lipids partition into at least three domains or regions of the cell membrane. The regulation of a channel depends on its partitioning with agonizing or antagonizing anionic signaling lipids (e.g. PIP2 and PIP3). Ceramide-1-phosphated (C1P) is an anionic ceramide comprised of a phosphate head group (red dot) similar to phosphatidic acid and expected to localize to GM1 domains. The endoplasmic reticulum (ER) is shown for reference and scale. (B) PLD2 is activated by substrate presentation. GM1 domains sequester PLD2 away from its substrate phosphatidylcholine (PC, orange circles). Increases in PIP2 attract PLD2 away from the GM1 domains localizing the enzyme near PC allowing for hydrolysis. (C) Cross correlation analysis of PLD association with GM1 domains before and after disruption with methyl-beta-cyclodextrin. After disruption, PLD fails to correlate with GM1 at any distance (radius in nm, error bars shown as standard error of the mean (n=7–8). (D) PIP2 activation of PLD2 can then indirectly activate TREK-1. The enzyme localizes to the disordered C-terminus (grey strand) of TREK-1 through a PLD2 binding site (green segment). PLD2 produces anionic lipid phosphatidic acid (PA) (green lipid) which binds to lipid binding site (blue cylinder) which opens the channel. This indirect mechanism contrasts with PIP2 directly activating a channel shown in (A).
Cross-correlation analysis is a technique used to characterize colocalization of two molecules in the plasma membrane. This is particularly useful for direct stochastic optical reconstruction microscopy (dSTORM) and allows a quantitative measurement of the average distance between two populations of molecules (Figure 1D) [34]. This analysis can be important since the resolution is sufficient to observe two localized molecules as separate signals. Cross-correlation can also be used to quantitatively measure the nanoscale movement or translocation of a protein or signaling lipid between lipid domains or clusters. For example, mechanical shear causes phospholipase D2 (PLD2) to shift from GM1 domains to PIP2 clusters [32]. Due to the very small distances traveled (42 nM) this was only seen with super resolution imaging. Similar analysis should be amenable to putative nanoscale movement of channels between lipid domains.
The location of lipid regulation
Lipid based localization
Super resolution imaging has made critical advancements for answering the questions of how lipid heterogeneity emerges and how the cell harnesses heterogeneity to gate a channel. Classically, GM1 domains are thick, high in saturated lipids, and thought to utilize hydrophobicity to partition in the membrane [31]. In contrast, PIP2 is polyunsaturated [5] and has been shown to cluster with charged proteins away from GM1 domains [29] (Figure 2A). Presumably the acyl chains of PIP3 also facilitate the partitioning of PIP3 away from PIP2 [33] and GM1 domains [29,32]. Despite the advances of super resolution imaging, showing the absolute size, exact composition, and putative structures of the heterogeneity in membranes is still challenging [35]. But most agree that the membrane is not perfectly homogeneous in its distribution of lipids and proteins [36] and this alone is sufficient to postulate a mechanism for gating channels.
To harness heterogeneity for gating, a channel or a lipase that regulates the channels is targeted to lipid nanodomains [32,37]. The targeting sequesters the channel or lipase away from its ligand or substrate. Cellular mechanisms then allow the channel or lipase to translocate to its ligand or substrate over very short distances as a form of rapid regulation (Figure 2B). Best estimates from dSTORM imaging indicate translocation occurs in the 650 μsec time frame faster than what is typically detectible by electrophysiology experiments [32].
Palmitoylation can regulate the targeting of proteins in and out of nanodomains [38,39]. For example, PLD2 contains two palmitoylation sites and a PIP2 binding motif. dSTORM has shown PLD2 associates with GM1 domains, presumably using these acylations, and this inactivates it by sequestering the enzyme away from its substrate [32]. But PLD2 also binds PIP2. PIP2 is polyunsaturated hence, PIP2 binding to PLD2 opposes the palmitoylation/GM1 localization and exposes the enzyme to PC resulting in the production of PA. The generated PA then activates TWIK-related K+ channel type 1 (K2P2.1, TREK-1) (Figure 2B–D, Box 1) [18,40]. In short, the cell harnesses lipid heterogeneity to gate the channel. Disruption of the lipid domains by either anesthetics or mechanical force circumvents the endogenous regulatory system and robustly activates PLD2 [32,37]. Furthermore, any channel could theoretically be palmitoylated directly and move in and out of a GM1 domain without an enzyme intermediary.
Box 1. Lipid localization mediated by protein-protein interactions.
TREK-1, an anesthetic-sensitive inhibitory mechanosensor, was initially classified as a PIP2 activated channel [64]. Recently, phosphatidic acid (PA), a signaling lipid that facilitates activation of the nicotinic acetylcholine receptor (nAChR) [30–32], was shown to activate TREK-1 [18,40] and at least 10 other ion channels including mechanosensitive channels piezo1–2 [65], the nAChR [66], MscL [67,68], and KcsA [69,70]. TREK-1 and its homolog, TRAAK in artificial systems [18,40], are activated by the localization of PLD2 near to the channel’s disordered C-terminus [40]. The role of PLD2 and the finding that PIP2 directly antagonizes the effect of PA and phosphatidylglycerol (PG) on TREK-1 in a reconstituted system [18] argues that TREK-1 is a PA or PG-gated channel.
Presumably, the activation depends on both the Kd and local concentration of PA. The Kd of PA for TREK-1 is 9 μM [18], and it can be calculated, by the formula 1/πr2hNA, that its local concentration near TREK-1 is ~3.5 mM in the presence of PLD2. PA could diffuse away if lateral mobility is unrestricted, but experimentally this does not appear to be the case [40].
Figure IA shows a schematic of channel activation by local production of PA. Since TREK-1 is mostly inactive absent PLD2 in the membrane [40] we can estimate the effective ‘non-PLD2’ exposure to PA to be less than 500 nM—a concentration sufficiently low not to activate the channel. This mechanism is opposite the one previously considered for local PIP2 depletion [4]. In that mechanism, PIP2 remains high in the membrane and local PIP2 hydrolysis produces a localized graduation that depletes the lipid [4]. The local high concentration of PA and the potential to modulate channels with low affinity for PA suggest an emerging class of putative PA regulated channels (Figure IB).
If we consider the PIP2 scenario with an ion channel (Figure IC), PIP2 remains high and the channel binds PIP2 with a Kd ~10-fold below the concentration in the membrane such that the channel is active in the resting state. Local depletion then inactivates the channel. The model is supported by binding constants for PIP2-gated inward rectifier potassium channels (Kirs). Kirs bind with 100–250 nM affinity [18] and the estimated concentration of PIP2 in membranes is 1–10 μM [4], thus PIP2 is bound in the resting state and localization of PLC to a channel would inactivate the channel.
Numerous channels have been shown to be palmitoylated [2]. Initially palmitoylation was thought to simply increase hydrophobicity of a protein to localize the protein to the membrane [41]. This is clearly not the case with ion channels [42], as they are typically permanent integral membrane proteins usually with several transmembrane spanning segments. The role of palmitoylation localizes the channels to GM1 domains altering their membrane sorting [42] which then likely changes their exposure to lipid ligands [5].
The balance between GM1 and PIP2 signaling can be regulated by lipid composition. When arachidonic acid is added, cells rapidly take it up causing a shift in the equilibrium between GM1 domains and the disordered region, e.g. PIP2 cluster [43]. Adding cholesterol has the opposite effect, the membrane becomes thicker and favors GM1 domain formation and signaling. In theory, translocation of a channel to a GM1 domain would also expose the channel to thicker membranes or different anionic signaling lipids that could gate the channel[43]. In other words, the lipids can signal without any change in lipid concentration, simply by the channel translocation in the membrane.
Biological function of lipid signals
Lipid competition
The potentially broad applicability of the advanced tools and emerging nanoscale signaling raises the question, should lipids be more broadly viewed as central signals regulating biological function? In a biological membrane, a channel is exposed to numerous signaling lipids. Interestingly, among the PA regulated channels, almost all of them also bind PIP2 and the binding typically has an antagonizing effect. In other words, the lipid competition appears to oppose both binding and function. For example, PIP2 inhibits TREK-1 while PA activates and PA saturation inhibits Kir2 channels while PIP2 activates (Box 1) [18,20]. In Kv channels, PIP2 completely reversed the effect of PA shifting the Vmid back −40 mV toward resting membrane potentials[44].
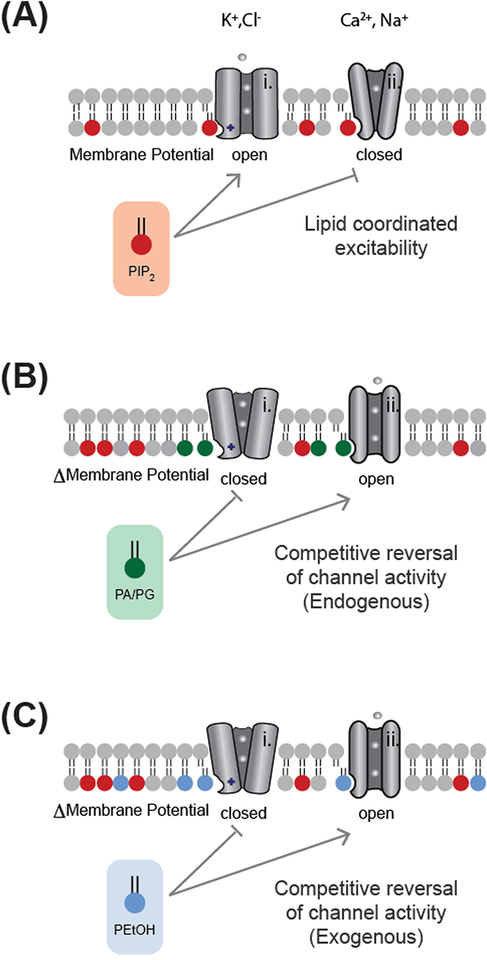
Figure 3 shows a proposed model for lipid competition. Initially, PIP2 is in high concentration and either activates or inhibits multiple classes of channels to achieve a coordinated resting membrane potential and to set cell excitability (Figure 3A). For channels regulated by both PA and PIP2 (Figure 3B), a local PA competes out PIP2 and can reverse the activity of the channel. In some sense, the PIP2 could be thought of as a tonic competitor to PA signaling, i.e. a type of cofactor as previously speculated [3,4].
Figure 3. Examples of ion channel regulation by lipid competition.
In most excitable cells, K+ channels are inhibitory and Ca2+ and Na+ channels are excitatory. (A) Lipids can regulate this excitability by increasing the opening of K+ channels (i, open) and decreasing the gating or voltage sensitivity of Ca2+ and Na+ channels (ii, closed). In general, PIP2 (red shading) increases permeability (flow of ions) of the plasma membrane, but numerous channels are exceptions. (B) In some instances, endogenous PA (green shading) can compete with PIP2 to reverse its effects (coordinated dysregulation). (C) In the presence of ethanol, PLD2 produces an unnatural lipid phosphatidylethanol (PEtOH, blue shading) which can competitively displace and reverse the effect of the endogenous lipid ligand (adapted from Chung et al. [45] under a creative commons license (CC BY-NC-ND 4.0)).
Lipid competition is also important for alcohol intoxication. PLD produces an unnatural lipid phosphatidylethanol (PEtOH) that accumulates in the membrane of cells and competes with PIP2 and PA binding to potassium channels (Figure 3C) [45]. Blocking the production of the unnatural lipid blocked an intoxicating phenotype in flies [45], suggesting a single class of toxic lipid can affect the behavior of a whole animal.
Lipids as central regulators
In light of the emerging signaling roles of PIP2 and PA, it is worth revisiting channels as downstream effectors in biological function. Of the 24 channels known to contribute to cardiac function, all 24 belong to families of channels regulated by PIP2 or PA and 17 of the 24 have identified PIP2 or PA regulation (Table 1). Their Kd’s for PIP2 and PA are not known; but experimentally, PIP2 dictates muscle (skeletal) cell excitability [46,47]. Similarly, at least 24 channels are known to contribute to pain sensation in animals (Table 1). Of those 24, 19 are known to be regulated by PIP2 or PA.
Table 1.
Lipid coordinated biological function.
| Super Family | Cardiaca | Permeabilityc | Excitab ilityd | Refe | Painb | Permeabilityc | Excitab ilityd | Refe |
|---|---|---|---|---|---|---|---|---|
| K2P | TRESKi | |||||||
| TWIK-1/2i | ? | TREK-1 | PAh↑ (PIP2h ↑↓?) | ↓↑ | [18,64] | |||
| TRAAKi | PAh↑ (PIP2↑?) |
↓ | [18,64] | TRAAK | PAh↑ (PIP2 ↑?) |
↓ | [18,64] | |
| Navi | Nav1.5 | ? | Nav1.3/1.7/1.8/1.9 | ? | ||||
| Cavi | Cav2.2 | |||||||
| Kiri | Kir2.1/2.2 | PIP2↑ (PA↓) |
↓ | [72,73] | Kir2.1 | PIP2↑ (PA↓) |
↓ | [72,73] |
| Kir6.2 | ||||||||
| Kir3.1/3.4 | PIP2↑f | ↓ | [73] | Kir3.2 | PIP2↑ | ↓ | [73] | |
| Kvi | Kv1.1/1.2 | |||||||
| Kv2.1/2.2 | ||||||||
| Kv3.4 | ||||||||
| Kv4.3 | ||||||||
| Kv7.2–3,5 | ||||||||
| hERGi | PIP2↑ | ↓ | [77] | |||||
| HCNi | HCN2/4 | PIP2↓ | ↓g | [52] | ||||
| P2Xi | P2X2/P2X3 | PIP2↑ | ↑ | [78] | ||||
| TRPi | TRPV1 | PIP2h ↓↑? | ↓↑ | [49,50] | ||||
| TRPM3 | PIP2h ↑ | ↑ | [79,80] | |||||
| TRPM8 | PIP2h ↑ | ↑ | [81] |
Channels involved in cardiac function [82].
The lipid effect the flow of ions across the membrane (permeability); up arrow (increase), or down arrow (decrease).
The lipid effect on the ability of a neuron to depolarize (excitability); up arrow (increase), or down arrow (decrease).
Reference (Ref).
Affects excitability through inactivation.
Affects excitability through voltage or cGMP dependence.
Lipid effect determined with purified reconstituted channel.
Abbreviations for ion channel names are as follows: voltage activated calcium channel (Cav), hyperpolarization-activated cyclic nucleotide-gated channel (HCN), human Ether-a-go-go-Related Gene (hERG), inward rectifying potassium channel (Kir), voltage activated potassium channel (Kv), voltage activated sodium channel (Nav), ATP-gated P2X receptor (P2X), TWIK-related acid-sensitive K+ channel (TASK), TWIK-related spinal cord potassium channel (TRESK), transient receptor potential (TRP), two-pore weak inward rectifier channel (TWIK). Additional letters and numbers after the abbreviations, indicate the channel’s subtype.
Only 5 of the channels are known to participate in both cardiac and pain processes suggesting subtype selective ligands could target pain over cardiac disease. Positive and negative allosteric modulators of lipid binding to ion channels could provide the therapeutic specificity.
In general, PIP2 appears to increase the activity of most channels and thus the permeability of the membrane (see Table 1). PIP2 also alters the excitability of the membrane. For cardiac channels, PIP2 tends to coordinate a decrease in excitability primarily by opening K+ channels. K+ channels work opposite Ca2+ and Na+ channels to decrease excitability of the cell. PIP2 can simultaneously activate a hyperpolarizing potassium channel (e.g. Kirs) and decrease depolarizing Na+ or Ca2+ channel (e.g. Cav channels) and depolarizing Cl− channels (e.g. TMEM16A) [48]. The competition of PA with PIP2 is likely to have the opposite effect, a decrease in permeability and ion channel opening.
Direct versus indirect lipid regulation
Many channels appear to be both activated and inhibited by PIP2 manipulation in cells, giving rise to a debate of PIP2’s direct effect (e.g. TREK-1 [18], TRPV1 [49,50], Kv [51], HCN [52], Cav [53], and TRP [54]). A likely contributor to the uncertainty is the fact that PIP2 has at least three independent roles in the membrane that include the following: localizing proteins to disordered regions [32], directly binding to ion channels [5], and generating second messenger signals [3]. Distinguishing direct binding of lipid versus an indirect role (e.g. through localization of the channel or an enzyme) can be challenging, especially when manipulations take place in the membrane.
TREK-1 is perhaps the best understood example of dual regulation [18,55]. When saturated with PIP2, TREK-1 is antagonized [18] (tonic inhibition). But PIP2 also localizes PLD2 near TREK-1 which indirectly activates TREK-1. Hence, PIP2 has cellular mechanisms that both activate and inhibit TREK-1. Whether these same localized and indirect mechanism contribute to the lack of apparent lipid specificity in other channels [7] is not known.
However, many ion channels have disordered loops and could bind a PA producing enzyme or PIP2 depleting enzyme. Phospholipase C (PLC) co-purifies with TRPV1 [56], glutamate receptors [57], the nAChR [58], and cation channels [59].
CONCLUDING REMARKS
Functionally reconstituting ion channels in lipid vesicles and testing ion transport of the purified channels is still the best technique to observe the putative action of a lipid with a channel. However, a reconstitution system is not without problems. Foremost, determining what lipids and proteins are needed for accurate reconstitution to recapitulate biological function is difficult. Also, there is variability in how the channel tolerates purification. Lastly, even in a purified systems, signaling molecules such as PIP2 can both activate and inhibit depending on the lipid composition or concentration [18]. Future advancements will need to address these concerns.
However, the advanced techniques described have significantly progressed the understanding of how cells harness lipid heterogeneity to gate ion channels. This system has the advantage that the concentration need not change across the entire membrane to dynamically activate or inhibit a channel [60]. The effect of heterogeneity will need to be worked out for each channel individually by identifying all the pertinent lipids bound to each channel, characterizing their direct and indirect interactions, and establishing their nanoscale function in the cells (see Outstanding Questions). The core tools are now available and set to change the role of lipids in ion channel regulation and biological function.
OUTSTANDING QUESTION.
Which channels directly bind PIP2 and which ones are indirectly modulated through PIP2 activation of regulatory proteins?
Which channels if any translocate between lipid domains as a response to changes in physiological conditions?
What are the relevant affinities for PIP2 and related anionic signaling lipids binding to ion channels? What lipids types bind to ion channels and do they compete with each other in a complex lipid environment?
How many ion channels are regulated by lipid signaling through the dynamic localization of lipid modifying enzymes?
Does a central lipid signaling-network coordinate biological function through localized lipid signals? If yes, how many of the channels participate and in what biological systems?
Highlights.
Mass spectrometry, cryoEM, and super resolution microscopy are advanced tools set to evolve the role of phosphatidylinositol 4,5 bisphosphate (PIP2) and other anionic lipids in the regulation of ion channel function.
Cells harness lipid heterogeneity to gate a channel.
Phosphatidic acid (PA) regulated channels are an emerging class of lipid regulated channels.
Nanoscale lipid gradients open up the possibility for membrane resident anionic lipids to centrally coordinate biological processes by locally regulating ion channel function.
Acknowledgements
We apologize to colleagues who work was not cited due to space limitations. Work in the authors laboratories is supported by a Director’s New Innovator Award to S.B.H. (1DP2NS087943–01) from the National Institutes of Health. We thank Nick Petersen from the laboratory of S.B.H. for helpful comments.
GLOSSARY
- AmtB
is an ammonium transport protein from Escherichia coli
- Cav
are voltage activated calcium channels that typically initiate action potentials by allowing Ca2+ ions to pass into the cytosol of a neuron
- GM1(monosialotetrahexosylganglioside) domains
is a lipid domain comprised of GM1 lipids and cholesterol. GM1 lipids are the prototypic ganglioside (glycolipid)
- KcsA
is a potassium channel from Streptomyces lividans
- Kv
are voltage gated potassium channels that typical hyperpolarize a neuron (terminate an action potential) by releasing potassium from inside the cell
- Lateral mobility
is the ability of membrane resident proteins and lipids to laterally diffuse in the two-dimensional plane of the plasma membrane
- Lipid domains
are regions within a membrane where lipids are restricted in their lateral mobility due to lipid-lipid partitioning or clustering of lipids with proteins. For example long chain saturated lipids and cholesterol (GM1 domains) partition away from polyunsaturated lipids (e.g. PIP2) The restricted movement leads to lipid heterogeneity which is harnessed by the cell to regulate ion channel gating
- MscL
is the large conductance of mechanosensation channel found in bacterial. MscL is activated by membrane stretch and helps bacteria avoid lysis due to osmotic swell
- Nanodiscs
are isolated lipid bilayers supported by a ring of scaffolding proteins. The lipid bilayer in the nanodiscs provides the most native-like environment for determining the structure, of a lipid bound to an ion channel
- Palmitoylation
is a post translational modification to a protein that places as saturated lipid (typically 14–16 carbons). Palmitoylation can localize an otherwise soluble protein to GM1 domains in the plasma membrane
- Phosphatidylinositol bisphosphate (PIP2)
is a glycol lipid (contains an inositol sugar)with two phosphorylation sites —typically located at the 4’ and 5’ positions of the inositol ring. When not in the 4,5 positions, the phosphates are indicated in parenthesis e.g., PI(3,5)P2. PIP3 contains 3 phosphates located in the 3,4, and 5’ positions. The placement of phosphates in the ring is critical for many aspects of anionic lipid signaling to ion channels. See [61] for further reading
- Phospholipase D (PLD)
is a membrane associated enzyme that hydrolyzes phosphatidylcholine (PC) into phosphatidic acid (PA) [62], an anionic signaling lipid similar to PIP2, and choline. PLD can also generate phosphatidylglycerol (PG) when glycerol is present [63]. The PLD isoform 2 (PLD2) activates TREK-1 channels
- Phospholipase C (PLC)
is a membrane associated enzyme that hydrolyzes phospholipids between the glycerol and phosphate. In the case of PIP2, PLC generates diacylglycerol and inositol triphosphate
- γ-aminobutyric acid A receptor (GABAA)
is a chloride conducting inhibitory channel activated by the neurotransmitter γ-aminobutyric acid
- Gating
is the process of a channel switching from a conducting to a non-conducting state in response to a ligand or voltage. In the conducting state, ion channels allow the passage of ions through the plasma membrane. Gating in response to voltage is ‘voltage-gated’; to extracellular neurotransmitter-ligand is ‘ligand-gated’; and to lipid is ‘lipid-gated’
- Signaling lipids
are typically low abundant anionic lipids (i.e. negative charged lipids) that signal to cells. Signaling lipids include membrane-resident lipids (e.g. PIP2 and PA) that bind to and gate ion channels. The hydrolysis of PIP2 can further signal through the generation of second messenger signals
- Second messenger signals
are intracellular signals released by the activation of an extracellular stimulus. For example, classical activation of G-protein coupled receptor (GPCR) subtype Gq activates PLC which releases intracellular inositol triphosphate from PIP2 [3]
- Transmembrane member 16A (TMEM16A)
is a calcium-activated chloride channel (CaCC)
- Transient receptor potential (TRP)
are a large family of cation channels that mediate a variety of sensations including pain, temperature, taste, pressure, and vision
- TWIK-related AA-stimulated K+ channel (TRAAK)
is a homolog of TREK-1 [40]. The similar response and affinity to PA suggests native TRAAK is likely regulated by a PA producing enzyme and responds to local high concentrations of PA [18]
- TWIK-related K+ channel subtype 1 (TREK-1
): is an inhibitory mechanosensitive K2P channel that is activated by phosphatidic acid and directly binds PLD
- Two pore domain potassium channels (K2P)
are dimeric potassium leak channels that form a pseudo-tetrameric pore
- Vmid voltage activation
is the applied voltage to a membrane that half maximally activates a voltage-gated channel. PA shifts the Vmid of Kv1.2/2.1 chimera by +40 mV (towards positive potentials). Hence, the channel requires much stronger depolarizing current to open when PA is present [44]
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hille B (2001) Ion Channels of Excitable Membranes. Sunderland, Mass Sinauer Assoc. Inc. p. 5. ISBN 0-87893-321-2. DOI: 10.1007/3-540-29623-9_5640 [DOI] [Google Scholar]
- 2.Shipston MJ (2011) Ion channel regulation by protein palmitoylation. J. Biol. Chem 286, 8709–8716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suh B-C and Hille B (2008) PIP2 is a necessary cofactor for ion channel function: how and why? Annu. Rev. Biophys 37, 175–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hilgemann DW (2007) Local PIP2 signals: when, where, and how? Pflugers Arch. 455, 55–67 [DOI] [PubMed] [Google Scholar]
- 5.Hansen SB (2015) Lipid agonism: The PIP2 paradigm of ligand-gated ion channels. Biochim. Biophys. Acta 1851, 620–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang CL et al. (1998) Direct activation of inward rectifier potassium channels by PIP2 and its stabilization by Gβγ. Nature 391, 803–6 [DOI] [PubMed] [Google Scholar]
- 7.Hilgemann DW et al. (2018) Lipid signaling to membrane proteins: From second messengers to membrane domains and adapter-free endocytosis. J. Gen. Physiol 150, jgp.201711875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gamper N and Shapiro MS (2007) Target-specific PIP2 signalling: how might it work? J. Physiol 582, 967–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Varnai P et al. (2006) Rapidly inducible changes in phosphatidylinositol 4,5-bisphosphate levels influence multiple regulatory functions of the lipid in intact living cells. J. Cell Biol 175, 377–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di Paolo G and De Camilli P (2006) Phosphoinositides in cell regulation and membrane dynamics. Nature 443, 651–7 [DOI] [PubMed] [Google Scholar]
- 11.Cheng WWL et al. (2011) Dual-mode phospholipid regulation of human inward rectifying potassium channels. Biophys. J 100, 620–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamouda AK et al. (2006) Assessing the lipid requirements of the Torpedo californica nicotinic acetylcholine receptor. Biochemistry 45, 4327–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gault J et al. (2016) High-resolution mass spectrometry of small molecules bound to membrane proteins. Nat. Methods 13, 333–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laganowsky A et al. (2014) Membrane proteins bind lipids selectively to modulate their structure and function. Nature 510, 172–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yen H-Y et al. (2018) PtdIns(4,5)P2 stabilizes active states of GPCRs and enhances selectivity of G-protein coupling. Nature 559, 423–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gupta K et al. (2017) The role of interfacial lipids in stabilizing membrane protein oligomers. Nature 541, 421–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liko I et al. (2018) Lipid binding attenuates channel closure of the outer membrane protein OmpF. Proc. Natl. Acad. Sci 115, 6691–6696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cabanos C et al. (2017) A Soluble Fluorescent Binding Assay Reveals PIP2 Antagonism of TREK-1 Channels. Cell Rep. 20, 1287–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pegan S et al. (2005) Cytoplasmic domain structures of Kir2.1 and Kir3.1 show sites for modulating gating and rectification. Nat. Neurosci 8, 279–87 [DOI] [PubMed] [Google Scholar]
- 20.Hansen SB et al. (2011) Structural basis of PIP2 activation of the classical inward rectifier K+ channel Kir2.2. Nature 477, 495–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whorton M and MacKinnon R (2011) Crystal Structure of the Mammalian GIRK2 K+ Channel and Gating Regulation by G Proteins, PIP2, and Sodium. Cell 147, 199–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao Y et al. (2016) TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action. Nature 534, 347–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cao E et al. (2013) TRPV1 structures in distinct conformations reveal activation mechanisms. Nature 504, 113–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng Y (2018) Single-particle cryo-EM—How did it get here and where will it go. Science (80-. ). 361, 876–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yin Y et al. (2019) Structural basis of cooling agent and lipid sensing by the cold-activated TRPM8 channel. Science DOI: 10.1126/science.aav9334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hughes TET et al. (2018) Structural insights on TRPV5 gating by endogenous modulators. Nat. Commun 9, 4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fine M et al. (2018) Structural basis for PIP2-mediated human TRPML1 regulation. Nat. Commun 9, 4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laverty D et al. (2019) Cryo-EM structure of the human αlβ3γ2 GABAA receptor in a lipid bilayer. Nature DOI: 10.1038/s41586-018-0833-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van den Bogaart G et al. (2011) Membrane protein sequestering by ionic protein-lipid interactions. Nature 479, 552–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chi KR (2009) Super-resolution microscopy: breaking the limits. Nat. Methods 6, 15–18 [Google Scholar]
- 31.Lingwood D and Simons K (2010) Lipid rafts as a membrane-organizing principle. Science 327, 46–50 [DOI] [PubMed] [Google Scholar]
- 32.Petersen EN et al. (2016) Kinetic disruption of lipid rafts is a mechanosensor for phospholipase D. Nat. Commun 7, 13873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang J and Richards D. a (2012) Segregation of PIP2 and PIP3 into distinct nanoscale regions within the plasma membrane. Biol. Open 1, 857–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sengupta P et al. (2013) Quantifying spatial organization in point-localization superresolution images using pair correlation analysis. Nat. Protoc 8, 345–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lyman E et al. (2018) From Dynamics to Membrane Organization: Experimental Breakthroughs Occasion a “Modeling Manifesto”. Biophys. J 115, 595–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nicolson GL (2014) The Fluid - Mosaic Model of Membrane Structure: Still relevant to understanding the structure, function and dynamics of biological membranes after more than 40 years. Biochim. Biophys. Acta - Biomembr 1838, 1451–1466 [DOI] [PubMed] [Google Scholar]
- 37.Pavel MA et al. (2018) Studies on the mechanism of general anesthesia. bioRxiv DOI: 10.1101/313973313973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zacharias DA et al. (2002) Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science 296, 913–6 [DOI] [PubMed] [Google Scholar]
- 39.Stone MB et al. (2017) Protein sorting by lipid phase-like domains supports emergent signaling function in B lymphocyte plasma membranes. Elife 6, e19891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Comoglio Y et al. (2014) Phospholipase D2 specifically regulates TREK potassium channels via direct interaction and local production of phosphatidic acid. Proc. Natl. Acad. Sci. U. S. A 111, 13547–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hancock JF et al. (1990) A polybasic domain or palmitoylation is required in addition to the CAAX motif to localize p21ras to the plasma membrane. Cell 63, 133–9 [DOI] [PubMed] [Google Scholar]
- 42.Greaves J and Chamberlain LH (2007) Palmitoylation-dependent protein sorting. J. Cell Biol 176, 249–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nayebosadri A et al. (2018) A Membrane Thickness Sensor in TREK-1 Channels Transduces Mechanical Force. SSRN Electron. J DOI: 10.2139/ssrn.3155650 [DOI] [Google Scholar]
- 44.Hite RK et al. (2014) Phosphatidic acid modulation of Kv channel voltage sensor function. Elife 3, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chung H-W et al. (2018) A Molecular Target for an Alcohol Chain Length Cutoff. J. Mol. Biol DOI: 10.1016/j.jmb.2018.11.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Flucher BE (2015) How is SR calcium release in muscle modulated by PIP(4,5)2? J. Gen. Physiol 145, 361–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Berthier C et al. (2015) Depression of voltage-activated Ca 2+ release in skeletal muscle by activation of a voltage-sensing phosphatase. J. Gen. Physiol 145, 315–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pritchard HAT et al. (2014) Inhibitory role of phosphatidylinositol 4,5-bisphosphate on TMEM16A-encoded calcium-activated chloride channels in rat pulmonary artery. Br. J. Pharmacol 171, 4311–4321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lukacs V et al. (2013) Promiscuous activation of transient receptor potential vanilloid 1 (TRPV1) channels by negatively charged intracellular lipids: the key role of endogenous phosphoinositides in maintaining channel activity. J. Biol. Chem 288, 35003–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cao E et al. (2013) TRPV1 channels are intrinsically heat sensitive and negatively regulated by phosphoinositide lipids. Neuron 77, 667–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oliver D et al. (2004) Functional conversion between A-type and delayed rectifier K+ channels by membrane lipids. Science 304, 265–70 [DOI] [PubMed] [Google Scholar]
- 52.Pian P et al. (2007) Modulation of cyclic nucleotide-regulated HCN channels by PIP2 and receptors coupled to phospholipase C. Pflugers Arch. 455, 125–45 [DOI] [PubMed] [Google Scholar]
- 53.Rodriguez-Menchaca AA et al. (2012) Dual regulation of voltage-sensitive ion channels by PIP2. Front. Pharmacol 3 SEP, 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rohacs T (2014) Phosphoinositide regulation of TRP channels. Handb. Exp. Pharmacol 223, 1143–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chemin J et al. (2007) Up- and down-regulation of the mechano-gated K2P channel TREK-1 by PIP2 and other membrane phospholipids. Pflugers Arch. Eur. J. Physiol 455, 97–103 [DOI] [PubMed] [Google Scholar]
- 56.Chuang HH (2001) Bradykinin and nerve growth factor release the capsaicin receptor from PtdIns(4, 5)P2-mediated inhibition. Nature 411, 957–962 [DOI] [PubMed] [Google Scholar]
- 57.Irino Y et al. (2005) Phospholipase Cδ4 associates with glutamate receptor interacting protein 1 in testis. J. Biochem 138, 451–6 [DOI] [PubMed] [Google Scholar]
- 58.Labriola JM et al. (2010) Phospholipase C activity affinity purifies with the Torpedo nicotinic acetylcholine receptor. J. Biol. Chem 285, 10337–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sturgeon RM and Magoski NS (2018) A closely-associated phospholipase C regulates cation channel function through phosphoinositide hydrolysis. J. Neurosci 38, 0586–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dubyak GR (2004) Cellular Homeostasis Ion homeostasis, channels, and transporters: an update on cellular mechanisms. Adv. Physiol. Educ 28, 143–154 [DOI] [PubMed] [Google Scholar]
- 61.Balla T (2013) Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol. Rev 93, 1019–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lopez I et al. (1998) Cloning and initial characterization of a human phospholipase D2 (hPLD2): ADP-ribosylation factor regulates hPLD2. J. Biol. Chem 273, 12846–12852 [DOI] [PubMed] [Google Scholar]
- 63.Yang SF et al. (1967) Transphosphatidylation by phospholipase D. J. Biol. Chem 242, 477–84 [PubMed] [Google Scholar]
- 64.Lopes CMB et al. (2005) PIP2 hydrolysis underlies agonist-induced inhibition and regulates voltage gating of two-pore domain K+ channels. J. Physiol 564, 117–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Coste B et al. (2012) Piezo proteins are pore-forming subunits of mechanically activated channels. Nature 483, 176–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Baenziger JE and DaCosta CJB (2012) Molecular mechanisms of acetylcholine receptor-lipid interactions: from model membranes to human biology. Biophys. Rev 5, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Powl AM et al. (2008) Anionic phospholipids affect the rate and extent of flux through the mechanosensitive channel of large conductance MscL. Biochemistry 47, 4317–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Powl AM et al. (2008) Importance of direct interactions with lipids for the function of the mechanosensitive channel MscL. Biochemistry 47, 12175–12184 [DOI] [PubMed] [Google Scholar]
- 69.Marius P et al. (2005) The interfacial lipid binding site on the potassium channel KcsA is specific for anionic phospholipids. Biophys. J 89, 4081–4089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Raja M et al. (2007) Phosphatidic acid plays a special role in stabilizing and folding of the tetrameric potassium channel KcsA. FEBS Lett. 581, 5715–5722 [DOI] [PubMed] [Google Scholar]
- 71.Giblin JP et al. (2018) Anionic Phospholipids Bind to and Modulate the Activity of Human TRESK Background K+ Channel. Mol. Neurobiol 1, [DOI] [PubMed] [Google Scholar]
- 72.Lee S-J et al. (2013) Secondary anionic phospholipid binding site and gating mechanism in Kir2.1 inward rectifier channels. Nat. Commun 4, 2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xie L-H et al. (2007) Activation of inwardly rectifying potassium (Kir) channels by phosphatidylinosital-4,5-bisphosphate (PIP2): interaction with other regulatory ligands. Prog. Biophys. Mol. Biol 94, 320–35 [DOI] [PubMed] [Google Scholar]
- 74.Kruse M et al. (2012) Regulation of voltage-gated potassium channels by PI(4,5)P2. J. Gen. Physiol 140, 189–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Taylor KC and Sanders CR (2016) Regulation of KCNQ/Kv7 Family Voltage-Gated K+ Channels by Lipids. Biochim. Biophys. Acta - Biomembr DOI: 10.1016/j.bbamem.2016.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wickenden AD and McNaughton-Smith G (2009) Kv7 channels as targets for the treatment of pain. Curr. Pharm. Des 15, 1773–98 [DOI] [PubMed] [Google Scholar]
- 77.Rodriguez N et al. (2010) Phosphatidylinositol-4,5-bisphosphate (PIP2) stabilizes the open pore conformation of the Kv11.1 (hERG) channel. Biophys. J 99, 1110–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bernier L-P et al. (2008) Phosphoinositides regulate P2X4 ATP-gated channels through direct interactions. J. Neurosci 28, 12938–12945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Uchida K et al. (2016) Stimulation-dependent gating of TRPM3 channel in planar lipid bilayers. FASEB J. 30, 1306–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Badheka D et al. (2015) Transient receptor potential melastatin 3 is a phosphoinositide-dependent ion channel. J. Gen. Physiol 146, 65–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zakharian E et al. (2010) Gating of transient receptor potential melastatin 8 (TRPM8) channels activated by cold and chemical agonists in planar lipid bilayers. J. Neurosci 30, 12526–12534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Grant AO (2009) Cardiac ion channels. Circ. Arrhythm. Electrophysiol 2, 185–94 [DOI] [PubMed] [Google Scholar]
- 83.Mathie A (2010) Ion channels as novel therapeutic targets in the treatment of pain. J. Pharm. Pharmacol 62, 1089–1095 [DOI] [PubMed] [Google Scholar]
- 84.Tsantoulas C and McMahon SB (2014) Opening paths to novel analgesics: The role of potassium channels in chronic pain. Trends Neurosci. 37, 146–158 [DOI] [PMC free article] [PubMed] [Google Scholar]