Abstract
Identification of the protein targets of bioactive small molecules is a routine challenge in chemical biology and phenotype-based drug discovery. Recent years have seen an explosion of approaches to meeting this challenge, but the traditional method of affinity pulldowns remains a practical choice in many contexts. This technique can be used as long as an affinity probe can be synthesized, usually with a crosslinking moiety to enable photo-affinity pulldowns. It can be applied to varied tissue types and can be performed with minimal specialized equipment. Here, we provide our protocol for photo-affinity pulldown experiments, with notes on making this method generally applicable to varied target identification challenges.
Keywords: Target identification, off-target, mass spectrometry, synthetic chemistry, photoreactive group, biotin, small molecule, natural product, click chemistry, inhibitor
1. Introduction
1.1. The need for target identification
A crucial task in many chemical biology contexts is finding the protein(s) with which a small molecule interacts. A classic case is the identification of targets and thus mechanism-of-action for bioactive natural products (Corson & Crews, 2007). But target identification is also part of many other research scenarios. For instance, with the growing popularity of cell-based and other phenotypic small molecule screens for drug or probe discovery (Moffat, Vincent, Lee, Eder, & Prunotto, 2017), hit compounds that induce a phenotype of interest also require target identification. An equally important task is confirming that target-directed compounds do indeed bind the expected proteins, and identifying off-targets of such compounds as well (Van Vleet, Liguori, Lynch, Rao, & Warder, 2018). This is a crucial aspect of validating chemical probes (Blagg & Workman, 2017; Drewes & Knapp, 2018). Therefore, target identification experiments are important tools in discovering new biology, validating hits, and developing leads.
1.2. Choice of methods
There is a growing array of methods for small molecule target identification. Several recent reviews cover this field admirably, so we will be brief here (Chang, Kim, & Kwon, 2016; Drewes & Knapp, 2018; Schurmann, Janning, Ziegler, & Waldmann, 2016; Tulloch et al., 2018; Ziegler, Pries, Hedberg, & Waldmann, 2013). The least definitive methods are cell-based readouts, for instance assessment of phosphorylation of select targets. Unfortunately, many “target” proteins are claimed for small molecules based solely on these kinds of phenotypes. The flawed logic used is, for example “compound X is a MEK inhibitor since MEK’s downstream target ERK is dephosphorylated in compound X-treated cells”. Lacking any biochemical validation that “compound X” interacts with MEK, these kinds of claims are misleading, but all too common, especially for promiscuous natural products such as curcumin (Nelson et al., 2017). Similarly, assessment of gene expression in response to compound treatment is also correlative, even when done at a genome-wide scale. But a powerful variant is the use of comparative gene expression profiling, where the genomic effects of a compound are compared with gene expression profiling of compounds of known mechanism or of gene knockouts (Hughes et al., 2000). Similarly, screening for suppressors or enhancers of compound function with other small molecules or gene knockdown/knockout approaches can yield useful results (synthetic lethality experiments are a subset of this approach) (Schenone, Dancik, Wagner, & Clemons, 2013). Although historically done in genetically tractable model organisms or with siRNA or shRNA libraries, the advent of genome editing technologies such as CRISPR/Cas9 make these “big data” approaches more tractable. Similar approaches can be used with proteomic, metabolomic, or even high-content imaging profiles (Kapoor, Waldmann, & Ziegler, 2016; Tulloch et al., 2018).
Also gaining popularity is the cellular thermal shift assay (CETSA; known as thermal proteome profiling [TPP] when performed proteome-wide), which takes advantage of the thermal stabilization usually offered to a protein by a small molecule ligand (Mateus, Maatta, & Savitski, 2016). Thermal denaturation across a temperature range followed by proteomic analysis of proteins soluble at each temperature with and without compound can yield specific binding proteins. Similar approaches include drug affinity response target stability (DARTS), which exploits the relative protease-resistance of compound-bound proteins (Pai et al., 2015), and stability of proteins from rates of oxidation (SPROX), which uses differential sensitivity to hydrogen peroxide (Strickland et al., 2013). The helpful reviews cited above cover the strengths and weaknesses plus successful applications of all these techniques, and others. Importantly, all of these require availability of advanced genomics or proteomics capabilities. Techniques that can be performed in the absence of these resource-intensive methodologies remain valuable.
1.3. Photo-affinity chromatography: background
Affinity chromatography, using a modified compound “bait” to identify target proteins, is a tried-and-true approach to target identification, in use for half a century (Cuatrecasas, 1970). Some key examples of compound-target pairs identified by affinity chromatography are indicated in Table 1. Photo-crosslinking adds an extra dimension to the pulldown approach, enabling purification of weak and low-abundance interactors by engendering a covalent interaction between the affinity reagent and its target(s) by ultraviolet (UV) light treatment. The most widely-used photo-crosslinking moieties are benzophenone, aryl azide, and diazirines (Murale, Hong, Haque, & Lee, 2016).
Table 1.
Some classic compound-target pairs identified by affinity chromatography
| Compound | Target | Reference |
|---|---|---|
| norepinephrine | β-adrenergic receptor | (Lefkowitz, Haber, & O’Hara, 1972) |
| FK506 | FKBP12 | (Harding, Galat, Uehling, & Schreiber, 1989) |
| geldanamycin | HSP90 | (Whitesell, Mimnaugh, De Costa, Myers, & Neckers, 1994) |
| fumagillin | methionine aminopeptidase 2 | (Sin et al., 1997) |
| parthenolide | IκB kinase β | (Kwok, Koh, Ndubuisi, Elofsson, & Crews, 2001) |
| pateamine A | eukaryotic initiation factor 4A | (Low et al., 2007) |
| thalidomide | cereblon | (Ito et al., 2010) |
This approach is conceptually appealing, especially to researchers used to immunoprecipitation or other pulldown strategies, with which it shares methodological parallels (DeCaprio & Kohl, 2017). It can also be performed with minimal specialized equipment, although does require peptide mass fingerprinting (Thiede et al., 2005) or shotgun proteomics (Nogueira & Domont, 2014) for final identification, but this routine proteomic technique is readily available at universities and through various commercial vendors for minimal cost.
Other advantages of this approach are its flexibility in terms of input tissue, types of small molecule and protein targets that can be identified, and low cost. The major shortcoming is the need for synthetic chemistry to generate the photo-affinity probe reagent, with concomitant required knowledge of compound structure-activity relationships (SAR). A secondary drawback is the potential for finding interactions that are biophysically “real” but not biologically relevant. However, these are usually easily ruled out in subsequent validation experiments. There is much room for modification of this general approach, including variants such as activity-based protein profiling, in which the affinity reagent becomes covalently attached to the target via the target’s enzymatic activity (Wang et al., 2017).
1.4. Photo-affinity chromatography: overview
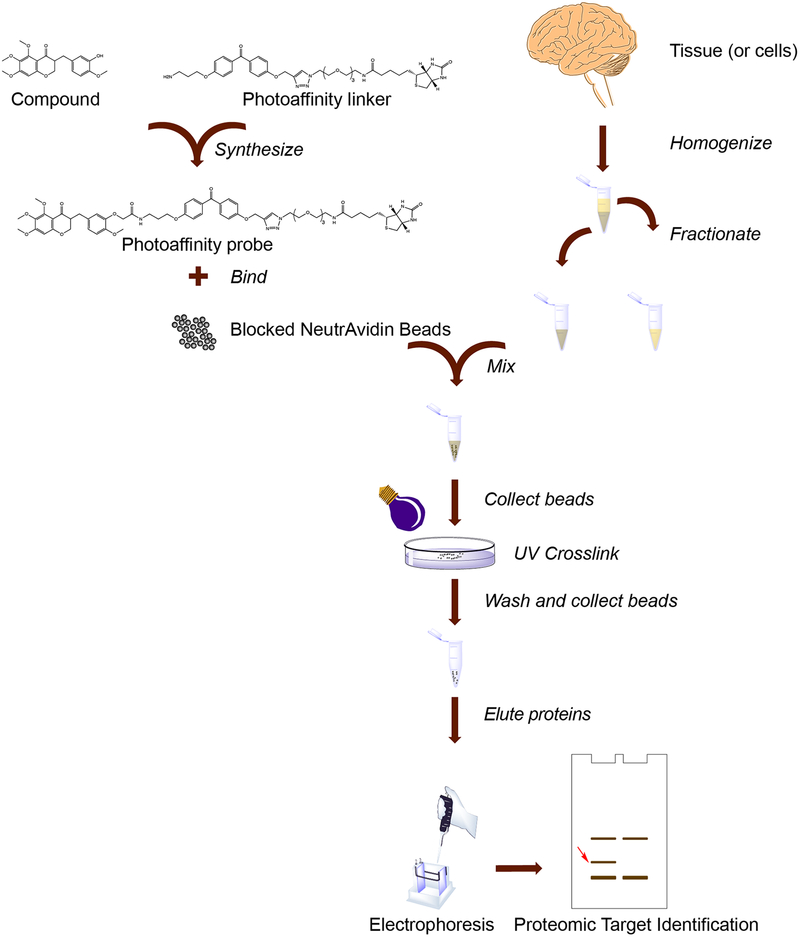
A typical photo-affinity pulldown procedure includes several steps (Fig. 1). First, a photo-affinity probe is synthesized, consisting of an active derivative of the compound of interest linked to a photoreactive moiety, linked to a “handle,” usually biotin due to its ease of use. This probe compound can either be exposed directly to a lysate, or it can be bound to beads first, with subsequent blocking of the beads. Next, a protein lysate is prepared, with fractionation as necessary. The lysate is exposed to photo-affinity probe compound followed by binding to beads, or to compound-bound beads directly. Photo-crosslinking is performed, then extensive washing of the beads removes non-specific binding proteins. Probe-bound proteins are eluted from the beads and separated by gel electrophoresis, with detection by silver staining. Unique bands are identified by mass spectrometry, followed by validation experiments.
Fig. 1.
Overview of the photoaffinity pulldown procedure. As indicated, major steps include biotin-containing photo-affinity reagent synthesis and binding to beads, along with tissue or cell lysis, homogenization, and fractionation. Cell lysates are mixed with beads and crosslinked with ultraviolet light, then beads are washed, proteins eluted, and electrophoresis performed. Unique gel bands are identified by a proteomic approach such as peptide mass fingerprinting. Validation experiments subsequent to target identification are not shown.
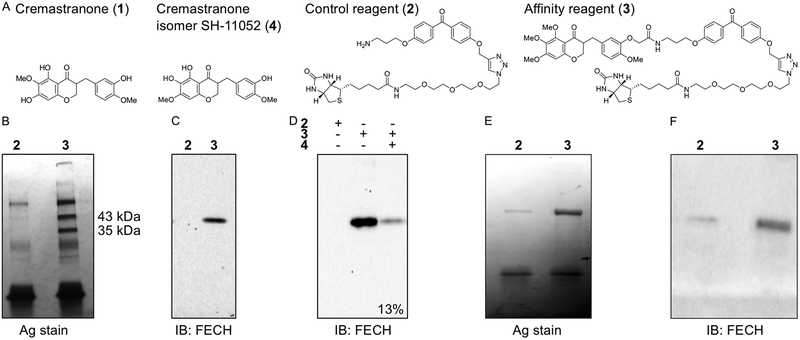
We describe here our general protocol for photo-affinity pulldown experiments. This protocol, with slight variations, has successfully identified the following: dCTP pyrophosphatase as a target of the anticancer natural product triptolide (Corson, Cavga, Aberle, & Crews, 2011), ferrochelatase as a target of the antiangiogenic natural product cremastranone (Basavarajappa et al., 2017), and soluble epoxide hydrolase as a target of the novel antiangiogenic compound SH-11037 (Sulaiman et al., 2018). A similar approach has been used to confirm targeting binding and rule out off-targets of a PI3K-targeting PROTAC (Hines, Gough, Corson, & Crews, 2013), and a TEAD4·Yap1 protein-protein interaction inhibitor (Bum-Erdene et al., in press). As shown in Fig. 2, using the identification of ferrochelatase as cremastranone’s target as an example, a successful pulldown starts with affinity reagents (Fig. 2A), and yields a small number of clear protein bands present in the photo-affinity pulldown condition but not with a control compound (Fig. 2B). After proteomic identification, immunoblot can confirm the target protein (Fig. 2C), with subsequent validation in a competition experiment (Fig. 2D). Finally, another useful validation experiment is isolation of recombinant target protein with the affinity reagent (Fig. 2E,F); see Section 2.9 below for details.
Fig. 2.
Example photo-affinity pulldown outcomes: identification of ferrochelatase (FECH) as a target of the antiangiogenic natural product, cremastranone. (A) Chemical structures of cremastranone (1) (Lee et al., 2014), control and active photo-affinity reagents (2, 3) (Lee et al., 2016), and active isomer used for competition experiments (4) (Basavarajappa et al., 2014). (B) Proteins pulled down with indicated reagents in photoaffinity chromatography were separated on SDS-PAGE and silver stained. Note specific bands at 43 and 35 kDa, which were excised and subjected to proteomic identification, leading to identification of the upper band as FECH. (C) Immunoblot of pulled down proteins using antibody against FECH. (D) Immunoblot of pulled down proteins from competition assay with excess active cremastranone isomer (4); relative quantification of band intensity shown. (E) Silver stained SDS-PAGE gel of recombinant human FECH protein pulled down using photoaffinity chromatography. (F) Anti-FECH immunoblot of a similar pulldown experiment. Adapted with permission from (Basavarajappa et al., 2017), © The Authors.
2. Experimental Considerations
2.1. Probe design
Typically, photo-affinity probes for chemical proteomics techniques contain three functional groups, a ligand (a bioactive small molecule), a photoreactive group, and an affinity tag (biotin), that are connected by linkers. A photo-reactive group can be irradiated with UV light upon reversible or irreversible complexation with a target protein. The formation of a highly reactive intermediate, such as a nitrene, carbene, or diradical by photo-irradiation leads to covalent binding between the ligand and the target protein (Murale et al., 2016). In particular, three kinds of photo-reactive groups consisting of aryl azides (i.e., tetrafluorophenyl azide), aliphatic and aromatic diazirines (i.e., trifluoromethylphenyl diazirine), and benzophenones have been used in photo-affinity labeling (Fleming, 1995; Smith & Collins, 2015; Sumranjit & Chung, 2013). We have chosen to use benzophenone due to its advantages of chemical stability and facile modifications over other photo-reactive groups. Benzophenone is reversibly activated by UV light (long wavelength, ~330–360 nm) and generates a triplet diradical to react with protein functional groups via a sequential abstraction-recombination mechanism (Dorman & Prestwich, 1994; Vodovozova, 2007). The building blocks of a substituted-benzophenone are commercially available, in contrast to aryl azides and diaziridines. However, the disadvantages are the long irradiation time needed to bind the targets and benzophenone’s bulkiness, potentially affecting binding capability and specific labeling.
Benzophenone can be connected between a ligand and biotin through a linker that will not disturb the interaction with the binding pocket of a putative protein of interest. An appropriate linker between a ligand, a photoreactive group and biotin is necessary to provide enough space to minimize steric hindrance, which would hamper the interactions of the ligand moiety with the target protein(s) in cells (Kapoor et al., 2016). Aliphatic alkyl chains, polyethylene glycol (PEG) groups, peptides and so on have been used as linkers (spacers), connecting the three functional groups. Generally, a hydrophilic PEG such as di- or triethylene glycol ether is used rather than the hydrophobic alkyl linkers which may bind proteins non-specifically (Ziegler et al., 2013). PEG linkers can contribute improved physicochemical properties, such as solubility, to photoaffinity probes. Another hydrophilic linker, a polyproline peptide with a rod-like helix structure was reported to increase the success rates of finding a low-abundance protein from cell lysates (Sato et al., 2007). Linker length must be determined empirically, but the probe design we use finds a useful balance between flexibility and retention of target binding.
The photo-affinity probes can be utilized for the preparative purification of target protein(s) from relevant biological extracts, after verifying retention of their biological activity, which should ideally be done in the same assay used for identifying the bioactivity of the parent compound. However, sometimes this may not be possible: by their nature, photoaffinity probe compounds are quite large, and therefore may not have solubility or cellular uptake amenable to all assays. One way to avoid these issues is to perform in-cell or in-lysate copper(I)-catalyzed azide alkyne cycloaddition (CuAAC), the so-called “click” reaction. This facile, bio-orthogonal reaction makes possible treatment with a minimally-modified (and thus more likely active) ligand compound, with subsequent covalent attachment of the biotin linker (Hong, Steinmetz, Manchester, & Finn, 2010). It also allows modular attachment of other moieties such as fluorophores for determination of subcellular localization. Our “clickable” probe design (see Section 2.2) enables this possibility if desired.
In cases where affinity reagent activity cannot be adequately tested directly, an excellent understanding of the SAR prior to probe synthesis is particularly important. Suitable sites for immobilization should be predicted by SAR study with a variety of analogs modified on different positions. For example, our trifunctional photo-affinity probe 3 of antiangiogenic homoisoflavonoids (Lee et al., 2016) is designed based on previous SAR studies (Basavarajappa et al., 2015). Otherwise, multiple different kinds of photo-affinity probes can be produced to find the best immobilization of a ligand without dramatic loss of activity or conferring steric hindrance with respect to target binding.
Multiple “handles” can be used in these probe compounds, including HaloTag™ (Ohana et al., 2015), or functional groups designed to react with functionalized beads, e.g., amines that react with NHS-ester beads. However, we employ biotin due to its ease of use and very strong yet non-covalent interaction with its substrate, avidin. Several variants of avidin are available linked to beads for affinity purifications (Thermo Fisher Scientific, 2010). The natural avidin is glycosylated and therefore positively charged, resulting in background binding of negatively charged cellular components. The widely-used streptavidin has a near-neutral isoelectric point, but contains an integrin-binding motif that can increase background binding. Neutravidin™, a deglycosylated avidin, avoids this issue as well, and is our first choice of binding matrix.
2.2. Probe synthesis
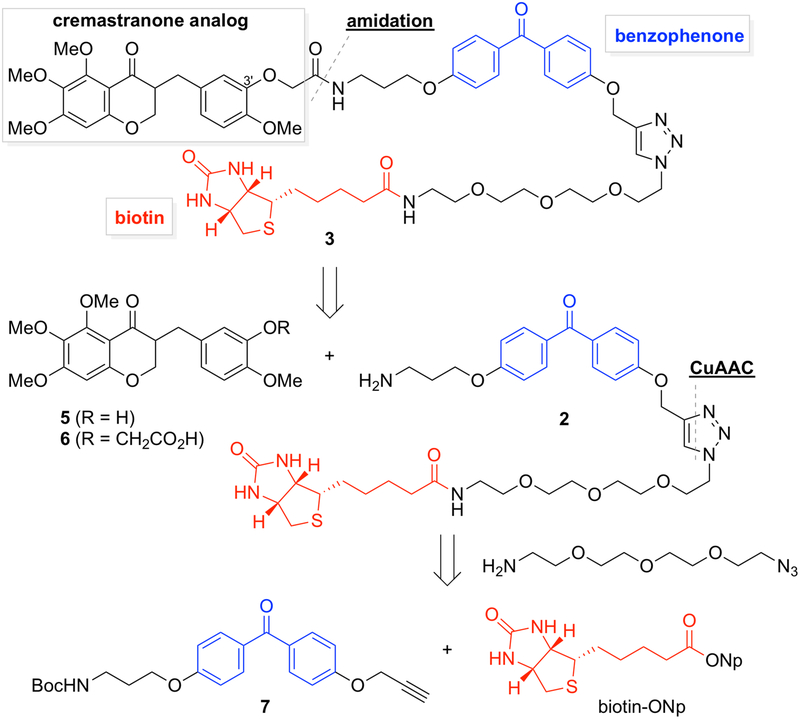
Our synthetic plan for photo-affinity probes for cremastranone is outlined in Fig. 3 (Lee et al., 2016). This general strategy can be applied to other ligand compounds of interest with a free hydroxyl or amine. The phenol group on the C-3ʹ position of the homoisoflavonoid scaffold is attached with a moiety of benzophenone and biotin linked with a triethylene glycol ether. Importantly, two kinds of photoaffinity probes should be utilized simultaneously. Along with a homoisoflavonoid-containing photo-affinity probe 3, a negative control probe 2 which has just benzophenone and biotin without the homoisoflavonoid scaffold is required. This approach can differentiate ligand-specific binding protein(s) from those bound nonspecifically to both matrices. An even better negative control, if available, is a probe containing an inactive analog of the compound of interest (Ziegler et al., 2013). In the course of retrosynthetic analysis of photo-affinity probe 3 (Fig. 3), the linkage between homoisoflavonoid 6 and benzophenone-biotin 2 can be formed by an amidation reaction. CuAAC can be used to bind the 4,4ʹ-disubstituted benzophenone 7 having a propargyl group and a functionalized biotin moiety from the treatment of 11-azido-3,6,9-trioxaundecan-1-amine with biotin-ONp (Meldal & Tornoe, 2008).
Fig. 3.
Retrosynthetic analysis of cremastranone-based photoaffinity probe via amidation and CuAAC (Copper-catalyzed Azide-Alkyne Cycloaddition).
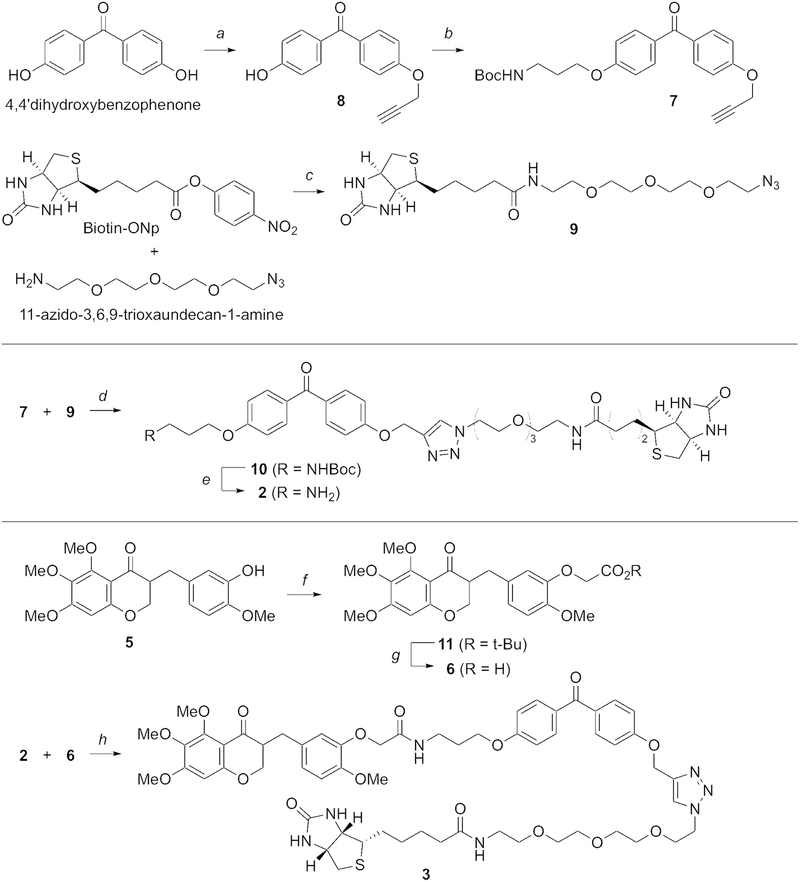
Our synthesis of photo-affinity probe 3 (Fig. 4) commences with O-dialkylation of 4,4ʹ-dihydroxybenzophenone as a starting material by the treatment with propargylic bromide and potassium carbonate, followed by the introduction of 3-(Boc-amino)-1-propanol by using a diisopropyl azodicarboxylate-mediated Mitsunobu reaction. Propargyl bromide is used at only half the amount of 4,4ʹ-dihydroxy-benzophenone in order to increase the yield of 8 and to minimize the formation of the side product having two propargyl moieties. The commercially available Biotin-ONp is coupled with 11-azido-3,6,9-trioxaundecan-1-amine, followed by the CuAAC reaction between a propargyl group of 7 and an azido group of 9 to afford the 1,2,3-triazole-containing benzophenone-biotin 10. TFA-promoted Boc deprotection of 10 gives rise to the photo-affinity probe 2 as a negative control. The phenol group of homoisoflavonoid 5 is alkylated with t-butyl bromoacetate and potassium carbonate, followed by the Boc-deprotection to afford the resulting aryloxyacetic acid 6 in good yield. Finally, the HBTU-mediated amidation of the acid 6 and the amine 2 completes the synthesis of our desired photo-affinity probe 3 (Fig. 4) (Lee et al., 2016).
Fig. 4.
Synthesis of negative control photoaffinity probe (2) and ligand-containing photo-affinity probe (3). Reagents and conditions: a) K2CO3, propargylic bromide, acetone, 90%; b) diisopropyl azodicarboxylate, 3-(tert-butoxycarbonylamino)-1-propanol, PPh3, THF, 82%; c) 11-azido-3,6,9-trioxaundecan-1-amine, iPr2NEt, CH2Cl2, 79%; d) CuSO4·5H2O, sodium ascorbate, t-BuOH/H2O, 80%; e) TFA, CH2Cl2, 90%; f) t-butyl bromoacetate, K2CO3, acetone, reflux, 85%; g) TFA, CH2Cl2, 99%; h) 2, HBTU, iPr2NEt, DMF, 38%. For further details, see Lee et al., 2016.
2.3. Bead preparation
As noted, photo-affinity reagent can be exposed to protein source and then linked to beads (this is the only possibility if using whole cells), or photo-affinity probe can be bound to beads first, and then the beads can be exposed to a cell or tissue lysate. Either method can work, but we have had most success with the latter, which is what we describe here. One advantage of pre-labeling the beads is the ability to give the affinity reagent a long incubation to bind to the beads without fear of target protein degradation. Another advantage of pre-labeling is the opportunity to block the beads with excess biotin after compound binding to reduce background from endogenously biotinylated proteins in the target protein lysate. However, for either method it is prudent to pre-bind the beads with a blocking protein to avoid non-specific protein binding. We use cytochrome c for this blocking as it is a very small (~12 kDa) protein, generally outside the size range of most proteins of interest and thus unlikely to interfere with gel-based pulldown analysis. However, other proteins could be used if necessary.
2.4. Protein source
The choice of tissue or cell type is largely dependent on where the compound’s target is likely to be expressed. We routinely use brain tissue as a rich, readily available source of diverse proteins (estimates of protein expression indicate the brain as a tissue with a large variety of proteins expressed (Uhlen et al., 2015)). Bovine brain (readily available from abattoirs) has been used by some investigators in the past (Crews, Collins, Lane, Snapper, & Schreiber, 1994), but we prefer porcine brain due to reduced risk of potential prion biohazards (Biosafety in Microbiological and Biomedical Laboratories, 2009). For a more homogeneous protein source, cultured cells can be used. We have had success using HeLa S3 cells, a subclone of this cervical cancer cell line adapted to suspension culture (Puck, Marcus, & Cieciura, 1956). These can be grown in large amounts; we have obtained these from the U.S. National Cell Culture Center (https://oldsite.cellculturecompany.com/nc3/) to avoid the logistical challenges of growing large amounts of cells ourselves.
The amount of tissue used is also a key question, and can dictate what cell sources are feasible. We prefer to use large amounts (20 g of brain tissue or cell pellet), to maximize the likelihood of detecting potential low abundance proteins. However, such amounts of tissue require significant workup and fractionation (see Section 2.5, below), and may not be practical for many protein sources. It is possible to perform this protocol on a micro scale with just a few million cells, especially for a high-abundance target, strong binding, and/or detection via immunoblot rather than proteomics. In all cases, tissue or cell pellets should be flash frozen immediately after harvest to minimize protein degradation. Even if the tissue is to be used immediately, flash freezing is still recommended as it aids cell lysis.
2.5. Fractionation
Although photo-affinity reagent labeling of whole live cells is possible, cell lysis increases exposure of cellular contents to the photo-affinity reagent molecule, especially if the photo-affinity reagent is poorly cell-penetrant. Lysis should be done under isotonic conditions and with as little detergent as possible, to minimize denaturation of proteins. We include cell fractionation as part of our protocol. If a compound target is known or suspected to be found in a specific organelle, then organelle purification (Castle, 2003) will increase likelihood of finding the target and enriching it. However, even if the target subcellular location is unknown, we find that fractionation into soluble and insoluble fractions helps simplify analysis by reducing the number of distinct proteins present in each fraction. This makes identification of unique bands on silver-stained gels more straightforward. The tradeoff is an overall dilution of protein during fractionation, potentially leading to undetectable levels of target protein. Doing pulldowns on fractions also requires more photo-affinity probe compound than a single pulldown on a whole-cell lysate. Note that the extensive fractionation described in the two protocols below is optimized for large amounts of brain tissue or cells as starting material. Starting from smaller amounts of material can be considerably simpler, for instance simply lysing a small cell pellet by trituration followed by removal of debris by centrifugation at 14,000×g, 10 min. Even the protocols listed here can be adjusted to improve interactions with target proteins, for instance by varying the NaCl concentration and/or detergents used for membrane protein solubilization, for instance 0.1–1% Triton X-100, 0.1–5.8% octyl glucoside, 0.2–2% dodecyl maltoside (Schimerlik, 2001).
2.6. Binding and crosslinking
The time allowed for photo-affinity probe to find and interact with its target and come to a binding equilibrium is dependent on the affinity between the two partners. Generally, longer binding times are better to maximize signal, with the key caveat that protein or compound degradation must be considered. We routinely use a 75-minute binding interval (performed at 4°C to minimize protein degradation), but this can readily be optimized. Similarly, UV crosslinking time can be varied to find a balance between ensuring a near-complete reaction, and avoiding over-heating or drying out of samples; we usually use 30 minutes. We use a long-wave UV lamp (Blak-Ray™ from UVP) for crosslinking, but other options are possible, including DNA crosslinkers (such as a Stratalinker™) with the default short-wave UV fluorescent bulbs replaced with long-wave UV bulbs, or even fluorescent desk lamps with appropriate bulbs, such as the F15T8/ BLB 15 W fluorescent tubes (Corson et al., 2011), available from online retailers. To maximize intensity, the light source should be positioned as close as possible to the sample. Output at sample distance should be measured with a light meter and recorded. For the crosslinking step, samples should be in shallow uncovered dishes to avoid UV absorption by plastic lids and/or buffer. Maintaining one sample unexposed to crosslinking can be informative, as it can reveal if the compound of interest binds to the protein target covalently without the need for crosslinking (Corson et al., 2011).
2.7. Washing
Optimization of wash parameters is also useful. Our standard protocol (Section 3.8) includes five washes, two with isotonic NaCl, two with high NaCl, and one with no NaCl, all in the presence of 1% Triton X-100 detergent. In our hands, this yields a fairly clean pulldown for multiple targets. However, fewer washes may be required for some targets to maximize signal-to-noise. And choice of detergent and concentration can also both be varied to increase signal and/or reduce background. We perform the whole protocol in “batch mode”, i.e., adding each wash to beads in a tube, mixing, then centrifuging and removing the supernatant from the agarose beads. However, it is also feasible to run the entire protocol on an FPLC system, passing lysate over a small column packed with compound-bound beads, then flowing each wash through the column. This allows more extensive washing, but usually requires a larger bead volume than we work with routinely. It is also possible to use magnetic beads (such as streptavidin Dynabeads™ from Thermo Fisher) in batch mode, saving time and effort on centrifugation steps, but increasing costs relative to agarose beads.
2.8. Analysis
There are multiple possibilities for analyzing pulldown results. If a target protein is known or expected, perhaps based on informatic data (McMasters, 2018), immunoblot using standard methods (Kurien & Scofield, 2015) offers a simple, sensitive, and definitive method of detecting the pulled down protein. However, in most cases, target proteins will be unknown, at least initially. Shotgun proteomics of the entire pulled-down proteome may be used to directly seek differentially abundant proteins in photo-affinity reagent versus control pulldown eluates (Nogueira & Domont, 2014). It is even possible to design a cell-based pulldown experiment using isotope-enriched vs. normal medium to distinguish photo-affinity reagent and control conditions in an experiment using stable-isotope labeling by amino acids in cell culture (SILAC) (Chen, Wei, Ji, Guo, & Yang, 2015) or other labeled experimental design (Wang et al., 2017). But we prefer visualization on a silver stained gel first, followed by excision of differentially-abundant protein bands and then proteomic identification by either peptide mass fingerprinting or shotgun proteomics, given the wide availability of this latter technique (Kim & Cho, 2019). This approach enables visual confirmation of a target band, which is helpful for publication. It also gives information on identified protein size to make positive protein identification easier, and provides a highly enriched input into the proteomics workflow, making data analysis simpler by reducing noise. Disadvantages of gel-based analysis include the possibility of missing low-abundance targets that are not visible on a silver-stained gel, or those that co-migrate with a non-specific band. This approach can also result in increased cost if many bands are selected, and increased risk of contamination due to added handling of samples.
2.9. Validation
Validation of target proteins identified by proteomics is essential. Fortunately, there are multiple options for this. A straightforward and necessary first step is an immunoblot of the pulldown eluates confirming that the identified protein of interest is present in the pulldown but not control lanes (Fig. 2C) and at the right size. It can also be helpful to probe the blot with a horseradish peroxidase-labeled streptavidin followed by enhanced chemiluminescence detection, which will identify all biotinylated proteins on the blot, helping to confirm or identify bands of interest.
A second key step is a competition assay. This can be included in the initial study design if desired, but can also be performed in a follow-up experiment, potentially on a smaller scale since less protein is often required for a pulldown followed by immunoblot detection than in an initial proteomic experiment (Fig. 2D). In competition experiments, the cell lysate is incubated with an excess (10–100-fold) of the untagged compound of interest or an active analog prior to exposure to the probe. If an interaction is specific, the free compound should saturate binding sites on the target protein, reducing or eliminating interaction with the probe.
Another useful validation experiment is pulldown of the identified target protein from an orthogonal system such as bacterial cells expressing recombinant protein (Fig. 2E, F). This ensures that the interaction is direct, and not through a protein complex. In parallel with this, if recombinant protein is available, biophysical techniques such as surface plasmon resonance (SPR) (Piliarik, Vaisocherova, & Homola, 2009) and isothermal titration calorimetry (ITC) (Callies & Hernandez Daranas, 2016) can be used to confirm and quantify the interaction. Likewise, if the target protein is an enzyme, appropriate enzymatic assays can be used to assess if the small molecule is an inhibitor of its target.
If recombinant protein is not available, CETSA (Jafari et al., 2014) can be used to confirm binding to the endogenous protein, and functional experiments such as siRNA knockdown in cells can validate if the target has the expected function. From there, more extensive analyses can be explored: mutagenesis or structure elucidation to determine binding site, gene editing to produce resistant cells, and more (Moustakim et al., 2018). A well-validated pulldown target identification experiment can offer a strong basis for further experimentation.
3. Pulldown of cremastranone target proteins
3.1. Equipment & supplies
Rotor-stator generator probe homogenizer (e.g. Fisher Scientific PowerGen™ Model 500) with disposable plastic or reusable metal probe
Disposable 1.5 mL microcentrifuge tubes
Disposable 50 mL conical tubes
Ultracentrifuge tubes
3.5 cm diameter plastic petri dishes
Probe sonicator (e.g. QSonica Q125)
Tube rotator
Benchtop centrifuge
Ultracentrifuge
Microcentrifuge
Dounce homogenizer
Long-wave UV lamp (e.g. Mercury bulb H44GS100 [Sylvania] in a Blak-Ray™ 100A lamp [UVP])
Hamilton syringe (50 μL volume) with blunt needle
Heating block
SDS-PAGE unit and power supply
Dishes for silver staining
Lightbox
Rotary evaporator system
Hot plate stirrer
600 MHz NMR
LC-MS spectrometer
3.2. Reagents
Note: where suppliers and catalog numbers are suggested, other reagents may also be appropriate
NeutrAvidin™ agarose beads (Thermo Fisher Cat. # 29200)
Cytochrome c (Sigma Cat. # C7752)
SDS-PAGE gels (4–20% precast gradient gels are ideal for maximizing separation)
d-(+)-biotin (Santa Cruz Cat. # sc-204706)
Porcine brain, flash-frozen
Cell pellets, flash-frozen
Methanol
Acetic acid
HPLC-grade water
Ethanol
Acetone
Dichloromethane (CH2Cl2)
Tetrahydrofuran (THF)
N,N-Dimethylformamide (DMF)
Sodium thiosulfate
Silver nitrate
Potassium carbonate (K2CO3)
Magnesium sulfate (MgSO4)
Triphenylphosphine
N,N-Diisopropylethylamine (Sigma Cat. # D125806)
4,4ʹ-Dihydroxybenzophenone (Sigma Cat. # D110507)
Propargyl bromide (TCI Cat. # P0484)
3-(tert-Butoxycarbonylamino)-1-propanol (TCI Cat. # H0900)
11-Azido-3,6,9-trioxaundecan-1-amine (Sigma Cat. # 17758)
Biotin p-nitrophenol ester (Biotin-ONp) (FutureChem Cat. # 2476601)
tert-Butyl bromoacetate (TCI Cat. # B1473)
Diisopropyl azodicarboxylate (TCI Cat. # A1246)
CuSO4·5H2O (Sigma Cat. # 209198)
Trifluoroacetic acid (TFA) (Sigma Cat. # T6508)
Hexafluorophosphate Benzotriazole Tetramethyl Uronium (HBTU) (TCI Cat. # B1657)
Silica gel 60 (230–400 mesh, Merck)
3.3. Buffers
50 mM biotin solution: Dissolve the powder in 1M Tris·Cl pH 7.4 buffer. The powder will dissolve only when the pH is ~ 7.4.
Isotonic Lysis Buffer (Buffer A):
25 mM Tris-HCl, pH 7.4
150 mM NaCl
2.5 mM sodium pyrophosphate
0.1 mM sodium orthovanadate
1 mM PMSF
10 μg/mL aprotinin
10 μg/ mL leupeptin
Dissolution Buffer/Wash Buffer 1 (Buffer B): Buffer A + 1% Triton X-100
Wash Buffer 2: Buffer B with a total of 350 mM NaCl
Wash Buffer 3: Buffer B without NaCl
10× SDS-PAGE running buffer:
60.6 g Tris base
288 g glycine
1% sodium dodecyl sulfate (SDS)
Make to 2 L with ultrapure water, adjust pH to 8.3
2× SDS-PAGE loading dye:
1.248 mL 2 M Tris-HCl, pH 6.8
2 mL 2-mercaptoethanol
12.752 mL ultrapure water
4 mL glycerol
0.8 g SDS
A few grains of bromophenol blue
Developing Solution:
30 g Na2CO3·H2O
1.4 mL 37% formaldehyde solution
50 mL of 0.2 g/L sodium thiosulfate solution
Make to 1L with HPLC-grade water
3.4. Probe synthesis (Fig. 4)
Preparation of (4-hydroxyphenyl)(4-(prop-2-yn-1-yloxy)phenyl)methanone (8). Add K2CO3 (0.16 g) and propargyl bromide (0.11 mL) to an acetone solution (5 mL) of 4,4ʹ-dihydroxybenzophenone (0.5 g) at 0 °C. Stir at 50 °C for 3 h, and add H2O (10 mL) at ambient temperature. The solution is extracted three times with ethyl acetate and washed with brine, dried over MgSO4, filtered, and concentrated. The residue is purified by flash column chromatography on silica gel (ethyl acetate / n-hexane = 1 : 2) to afford 8 (0.26 g, 90%).
Preparation of tert-butyl (3-(4-(4-(prop-2-yn-l-yloxy)benzoyl)phenoxy)propyl)carbamate (7). Add 8 (0.14 g), PPh3 (0.15 g), and diisopropyl azodicarboxylate (0.13 mL) to a THF solution (2 mL) of 3-(tert-butoxycarbonylamino)-1-propanol (0.12 g) at ambient temperature. Stir for 3 h, and the solution is diluted with ethyl acetate and washed with brine, dried over MgSO4, filtered and concentrated. The residue is purified by flash column chromatography on silica gel (ethyl acetate / n-hexane = 1 : 3) to afford 7 (0.19 g, 82%).
Preparation of N-(2-(2-(2-(2-Azidoethoxy)ethoxy)ethoxy)ethyl)-5-((3aS,4S,6aR)-2-oxohexahydro-lH-thieno[3,4-d]imidazol-4-yl)pentanamide (9). Add 11-azido-3,6,9-trioxaundecan-1-amine (1.6 mL) and N,N-diisopropylethylamine (4.3 mL) to a DMF solution (30 mL) of biotin-ONp (1.0 g) at ambient temperature. Stir for 12 h, and add H2O (1 mL). The solution is extracted three times with ethyl acetate and washed with brine, dried over MgSO4, filtered and concentrated. The residue is purified by flash column chromatography on silica gel (CH3OH / CH2Cl2 = 1 : 10) to afford 9 (1.5 g, 79%).
Preparation of Boc-protected benzophenone-biotin (10). Add 9 (0.11 g), CuSO4·5H2O (60 mg) and sodium ascorbate (1.0 M in H2O, 8 drops) to a t-BuOH/H2O solution (6 mL, 1 : 1) of 7 (0.1 g) at ambient temperature. Stir for 24 h, and add H2O (1 mL). The solution is extracted with ethyl acetate and washed with brine, dried over MgSO4, filtered and concentrated. The residue is purified by flash column chromatography on silica gel (CH3OH / CH2Cl2 = 1 : 10) to afford 10 (0.17 g, 80%).
Preparation of ammonium salt of benzophenone-biotin (2). To a CH2Cl2 solution (2 mL) of 10 (20 mg, 0.028 mmol), add TFA (0.4 mL) at 0 °C. Stir for 2.5 h at ambient temperature, and the solution is concentrated under reduced pressure to afford 2 (19 mg, 90%).
Preparation of tert-butyl 2-(2-methoxy-5-((5,6,7-trimethoxy-4-oxochroman-3-yl)methyl)phenoxy)acetate (11). Add tert-butyl bromoacetate (48 μL) and K2CO3 (0.1 g) to an acetone solution (5 mL) of 3-(3-hydroxy-4-methoxybenzyl)-5,6,7-trimethoxychroman-4-one (5) (0.1 g) at ambient temperature and reflux for 3 h. The solution is diluted with ethyl acetate and washed with brine, dried over MgSO4, filtered and concentrated. The residue is purified by flash column chromatography on silica gel (ethyl acetate / n-hexane = 1 : 2) to afford 11 (0.12 g, 85%).
Preparation of 2-(2-methoxy-5-((5,6,7-trimethoxy-4-oxochroman-3-yl)methyl)phenoxy)acetic acid (6). Add TFA (0.7 mL) to a CH2Cl2 solution (5 mL) of 11 (0.12 g). Stir for 2 h and the solution is concentrated to afford 6 (0.12 g, 99%).
Preparation of cremastranone-benzophenone-biotin probe (3). Add HBTU (20 mg) and N,N-diisopropylethylamine (22 μL) to a DMF solution (1 mL) of 6 (24 mg). Stir for 30 min, and add a DMF solution (0.5 mL) of 2 (38 mg). Stir for 24 h, and the reaction mixture is diluted with ethyl acetate, dried over MgSO4, filtered and concentrated. The residue is purified by flash column chromatography on silica gel (CH3OH / CH2Cl2 = 1 : 10) to afford 3 (20 mg, 38%).
Structures of final compounds should be confirmed by 1H and 13C NMR, and purity >95% ascertained by LC-MS.
3.5. Bead preparation
Take 1 mL NeutrAvidin™ beads, 50% slurry per treatment condition
Wash 3× in 500 μL isotonic lysis buffer (Buffer A)
Add 100 μM affinity reagent or linker control in an appropriate volume of Buffer A (200–300 μL), bind with rotation overnight, 4°C
Add biotin in Buffer A to a final concentration of 1 mM (i.e., 1 in 50 dilution), bind with rotation 1h, 4°C
To block, add cytochrome c in Buffer A to a final concentration of 1 mg/mL, bind with rotation 1h, 4°C
Wash 3× in 500 μL Buffer A, centrifuging at 500×g after each wash
Resuspend in Buffer A to a total volume of 1 mL
3.6. Cell lysis and fractionation: brain tissue
Suspend a 20 g tissue slice in 50 mL cold lysis buffer (Buffer A)
Homogenize on ice using probe homogenizer, power setting 6, 2×15 s
Centrifuge 2000×g, 5 minutes, 4°C
Reserve pelleted large tissue fragments (P1), transfer supernatant (S1) to Dounce homogenizer
Homogenize with 50 strokes on ice
Sonicate 10 minutes, with 60% amplitude, 10 s on and 40 s off
Centrifuge 11,000×g, 30 minutes, 4°C
Collect pellet (P2) and supernatant (S2)
Resuspend pellet P2 in Buffer B
Homogenize P2 with 25 strokes of a Dounce homogenizer
Centrifuge 11,000×g, 30 minutes, 4°C
Collect supernatant (S3)
Use P2, S2, and S3 fractions for pulldown
3.7. Cell lysis and fractionation: cell pellets
Thaw cell pellet on ice (e.g., 5L culture equivalent of HeLa S3 cells)
Suspend in 3–10 mL cold lysis buffer (Buffer A)
Homogenize with 50 strokes of a Dounce homogenizer on ice
Centrifuge 2000×g, 2 minutes, 4°C
Save pelleted nuclei, transfer supernatant to ultracentrifuge tubes
Ultracentrifuge 100,000×g, 45 minutes, 4°C
Collect supernatant (=S100 fraction)
Wash pellet with 100 μL Buffer A
Resuspend pellet in Buffer B
Homogenize with 25 strokes of a Dounce homogenizer on ice
Centrifuge 10,000×g, 10 minutes
Save pelleted insoluble material, keep supernatant (=P100 fraction)
Use both S100 and P100 fractions for pulldown
3.8. Binding, crosslinking, and pulldown
Divide each lysate fraction evenly between experimental conditions (at a minimum, control and photo-affinity reagent conditions)
If performing a competition experiment, add 1–10 mM competitor compound to appropriate lysates 15–60 minutes prior to adding beads
Add prepared beads to fractionated protein lysates, rotate 1–2 hours, 4°C
Centrifuge 500×g, 2 minutes
Resuspend in 500 μL Buffer A or Buffer B, matching input buffer
Transfer to 6 cm tissue culture plate (or 6-well plate for smaller volumes), uncovered
Expose to 365 nm UV light, 30–45 minutes, 4°C
Collect liquid and beads into 1.5 mL tube
Wash plate with 500 μL Buffer B and add to tube, centrifuge 500×g, 2 minutes
Wash 2× 5 minutes with 500 μL Buffer B, centrifuge 500×g, 2 minutes after each
Wash 2× 5 minutes with Wash Buffer 2, centrifuge 500×g, 2 minutes after each
Wash 5 minutes with Wash Buffer 3, then centrifuge 500×g, 2 minutes
Remove all liquid with Hamilton or other narrow-gauge syringe
Add 300 μL 2× SDS-PAGE loading dye
Heat tubes at 70°C for 10 minutes (beads may be stored at −20°C for several weeks after this step)
Centrifuge briefly, then load 3 μL on gel for streptavidin-HRP or immunoblot, or 35–75 μL on gel for silver staining
Also load molecular weight marker, but use ≤1 μL to avoid an overly strong signal, and leave a lane between this marker and pulldown samples to avoid signal bleed
Run SDS-PAGE under standard conditions until dye front reaches bottom of gel
3.9. Silver staining
Pre-warm 2 g/L silver nitrate to 37°C
Wash gel 2×20 minutes (or up to overnight) in 50% methanol/10% acetic acid/40% HPLC-grade water (all water used in this process should be HPLC-grade), using 50–100 mL for each wash, in a clean, dedicated gel tray
Wash 10 minutes in 20% ethanol
Wash 10 minutes in water
Reduce with 0.2 g/L sodium thiosulfate, 1 minute
Wash 2×20 s with water
Incubate with pre-warmed 2 g/L silver nitrate, 37°C, 30 minutes
Wash 20 s with water
Incubate with developing solution, rocking for <30 s; discard
Add fresh developing solution and incubate just until desired signal is reached: bands should be mid-brown but background should still be only faintly yellow
Immediately transfer to 1% acetic acid stop bath
Gels may be kept in the stop bath for many days, transferred to water, or dried
3.10. Analysis
Examine gel on lightbox, but avoid keratin contamination by wearing gloves, mask, and hairnet; photograph
Seek bands present with the photo-affinity reagent but absent or faint with the control compound and competition condition; there will always be some non-specific, endogenously biotinylated protein bands
If bands are faint or overlapping, consider running more material or a different percentage gel for the molecular weight of interest
Excise bands of interest, plus cognate region of control lanes, using a sterile, disposable scalpel blade; photograph gel after excisions, and annotate band sizes
Send bands for proteomic analysis at a core or commercial facility, being sure to specify the correct species for database analysis
In assessing the resulting data, look for peptide and protein matches unique or highly enriched in the size-matched pulldown vs. control samples, with an appropriate protein size based on the migration of the band
4. Summary
Although a wealth of methods are possible for small molecule target identification, a carefully designed photo-affinity pulldown approach can yield rich data. As described in this chapter, optimal results are obtained through thorough photoaffinity reagent design; appropriate choice and fractionation of protein source; optimization of binding, crosslinking and washing; and appropriate analysis and validation experiments. By judiciously considering these factors, it is possible to find novel targets and/or off-targets, leading to new biology and validation of drug leads or chemical probes.
Acknowledgements
We thank members of the Corson laboratory for comments on the manuscript. Related work in the Corson laboratory was supported by NIH/NEI R01EY025641, the Retina Research Foundation, the International Retinal Research Foundation, and the Showalter Research Trust. SY Seo was supported by the Bio & Medical Technology Development Program of the National Research Foundation funded by the Korean government, MSIP (NRF-2017M3A9C8027781)
Contributor Information
Seung-Yong Seo, College of Pharmacy, Gachon University, Incheon, Republic of Korea.
Timothy W. Corson, Indiana University School of Medicine, Indianapolis, IN, United States.
References
- Basavarajappa HD, Lee B, Fei X, Lim D, Callaghan B, Mund JA, … Corson TW (2014). Synthesis and mechanistic studies of a novel homoisoflavanone inhibitor of endothelial cell growth. PloS One, 9(4), e95694. doi: 10.1371/journal.pone.0095694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basavarajappa HD, Lee B, Lee H, Sulaiman RS, An H, Magana C, … Corson TW (2015). Synthesis and biological evaluation of novel homoisoflavonoids for retinal neovascularization. Journal of Medicinal Chemistry, 58(12), 5015–5027. doi: 10.1021/acs.jmedchem.5b00449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basavarajappa HD, Sulaiman RS, Qi X, Shetty T, Sheik Pran Babu S, Sishtla KL, … Corson TW (2017). Ferrochelatase is a therapeutic target for ocular neovascularization. EMBO Molecular Medicine, 9(6), 786–801. doi: 10.15252/emmm.201606561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biosafety in Microbiological and Biomedical Laboratories. (2009). (Chosewood LC & Wilson DE Eds. 5th ed.). Washington, DC: U.S. Department of Health and Human Services. [Google Scholar]
- Blagg J, & Workman P (2017). Choose and use your chemical probe wisely to explore cancer biology. Cancer Cell, 32(1), 9–25. doi: 10.1016/j.ccell.2017.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bum-Erdene K, Zhou D, Gonzalez-Gutierrez G, Ghozayel MK, Si Y, Xu D, … Meroueh SO (in press). Small-molecule covalent modification of conserved cysteine leads to allosteric inhibition of the TEAD4•Yap1 protein-protein interaction. Cell Chemical Biology. [DOI] [PubMed] [Google Scholar]
- Callies O, & Hernandez Daranas A (2016). Application of isothermal titration calorimetry as a tool to study natural product interactions. Natural Product Reports, 33(7), 881–904. doi: 10.1039/c5np00094g [DOI] [PubMed] [Google Scholar]
- Castle JD (2003). Purification of organelles from mammalian cells. Current Protocols in Immunology, Chapter 8, Unit 8 1B. doi: 10.1002/0471142735.im0801bs56 [DOI] [PubMed] [Google Scholar]
- Chang J, Kim Y, & Kwon HJ (2016). Advances in identification and validation of protein targets of natural products without chemical modification. Natural Product Reports, 33(5), 719–730. doi: 10.1039/c5np00107b [DOI] [PubMed] [Google Scholar]
- Chen X, Wei S, Ji Y, Guo X, & Yang F (2015). Quantitative proteomics using SILAC: Principles, applications, and developments. Proteomics, 15(18), 3175–3192. doi: 10.1002/pmic.201500108 [DOI] [PubMed] [Google Scholar]
- Corson TW, Cavga H, Aberle N, & Crews CM (2011). Triptolide directly inhibits dCTP pyrophosphatase. Chembiochem, 12(11), 1767–1773. doi: 10.1002/cbic.201100007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corson TW, & Crews CM (2007). Molecular understanding and modern application of traditional medicines: triumphs and trials. Cell, 130(5), 769–774. doi: 10.1016/j.cell.2007.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crews CM, Collins JL, Lane WS, Snapper ML, & Schreiber SL (1994). GTP-dependent binding of the antiproliferative agent didemnin to elongation factor 1 alpha. Journal of Biological Chemistry, 269(22), 15411–15414. [PubMed] [Google Scholar]
- Cuatrecasas P (1970). Protein purification by affinity chromatography. Derivatizations of agarose and polyacrylamide beads. Journal of Biological Chemistry, 245(12), 3059–3065. [PubMed] [Google Scholar]
- DeCaprio J, & Kohl TO (2017). Immunoprecipitation. Cold Spring Harbor Protocols, 2017(12), pdb prot098640. doi: 10.1101/pdb.prot098640 [DOI] [PubMed] [Google Scholar]
- Dorman G, & Prestwich GD (1994). Benzophenone photophores in biochemistry. Biochemistry, 33(19), 5661–5673. [DOI] [PubMed] [Google Scholar]
- Drewes G, & Knapp S (2018). Chemoproteomics and chemical probes for target discovery. Trends in Biotechnology. doi: 10.1016/j.tibtech.2018.06.008 [DOI] [PubMed] [Google Scholar]
- Fleming SA (1995). Chemical reagents in photoaffinity labeling. Tetrahedron, 51(46), 12479–12520. doi: 10.1016/0040-4020(95)00598-3 [DOI] [Google Scholar]
- Harding MW, Galat A, Uehling DE, & Schreiber SL (1989). A receptor for the immunosuppressant FK506 is a cis-trans peptidyl-prolyl isomerase. Nature, 341(6244), 758–760. doi: 10.1038/341758a0 [DOI] [PubMed] [Google Scholar]
- Hines J, Gough JD, Corson TW, & Crews CM (2013). Posttranslational protein knockdown coupled to receptor tyrosine kinase activation with phosphoPROTACs. Proceedings of the National Academy of Sciences of the United States of America, 110(22), 8942–8947. doi: 10.1073/pnas.1217206110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong V, Steinmetz NF, Manchester M, & Finn MG (2010). Labeling live cells by copper-catalyzed alkyne--azide click chemistry. Bioconjugate Chemistry, 21(10), 1912–1916. doi: 10.1021/bc100272z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes TR, Marton MJ, Jones AR, Roberts CJ, Stoughton R, Armour CD, … Friend SH (2000). Functional discovery via a compendium of expression profiles. Cell, 102(1), 109–126. [DOI] [PubMed] [Google Scholar]
- Ito T, Ando H, Suzuki T, Ogura T, Hotta K, Imamura Y, … Handa H (2010). Identification of a primary target of thalidomide teratogenicity. Science, 327(5971), 1345–1350. doi: 10.1126/science.1177319 [DOI] [PubMed] [Google Scholar]
- Jafari R, Almqvist H, Axelsson H, Ignatushchenko M, Lundback T, Nordlund P, & Martinez Molina D (2014). The cellular thermal shift assay for evaluating drug target interactions in cells. Nature Protocols, 9(9), 2100–2122. doi: 10.1038/nprot.2014.138 [DOI] [PubMed] [Google Scholar]
- Kapoor S, Waldmann H, & Ziegler S (2016). Novel approaches to map small molecule-target interactions. Bioorganic and Medicinal Chemistry, 24(15), 3232–3245. doi: 10.1016/j.bmc.2016.05.020 [DOI] [PubMed] [Google Scholar]
- Kim YI, & Cho JY (2019). Gel-based proteomics in disease research: Is it still valuable? Biochimica et Biophysica Acta: Proteins and Proteomics, 1867(1), 9–16. doi: 10.1016/j.bbapap.2018.08.001 [DOI] [PubMed] [Google Scholar]
- Kurien BT, & Scofield RH (2015). Western blotting: an introduction. Methods in Molecular Biology, 1312, 17–30. doi: 10.1007/978-1-4939-2694-7_5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok BH, Koh B, Ndubuisi MI, Elofsson M, & Crews CM (2001). The anti-inflammatory natural product parthenolide from the medicinal herb Feverfew directly binds to and inhibits IkappaB kinase. Chemistry and Biology, 8(8), 759–766. [DOI] [PubMed] [Google Scholar]
- Lee B, Basavarajappa HD, Sulaiman RS, Fei X, Seo SY, & Corson TW (2014). The first synthesis of the antiangiogenic homoisoflavanone, cremastranone. Organic and Biomolecular Chemistry, 12(39), 7673–7677. doi: 10.1039/c4ob01604a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B, Sun W, Lee H, Basavarajappa H, Sulaiman RS, Sishtla K, … Seo SY (2016). Design, synthesis and biological evaluation of photoaffinity probes of antiangiogenic homoisoflavonoids. Bioorganic and Medicinal Chemistry Letters, 26(17), 4277–4281. doi: 10.1016/j.bmcl.2016.07.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefkowitz RJ, Haber E, & O’Hara D (1972). Identification of the cardiac beta-adrenergic receptor protein: solubilization and purification by affinity chromatography. Proceedings of the National Academy of Sciences of the United States of America, 69(10), 2828–2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low WK, Dang Y, Schneider-Poetsch T, Shi Z, Choi NS, Rzasa RM, … Liu JO (2007). Isolation and identification of eukaryotic initiation factor 4A as a molecular target for the marine natural product Pateamine A. Methods in Enzymology, 431, 303–324. doi: 10.1016/S0076-6879(07)31014-8 [DOI] [PubMed] [Google Scholar]
- Mateus A, Maatta TA, & Savitski MM (2016). Thermal proteome profiling: unbiased assessment of protein state through heat-induced stability changes. Proteome Science, 15, 13. doi: 10.1186/s12953-017-0122-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMasters DR (2018). Knowledge-based approaches to off-target screening. Methods in Enzymology, 610, 311–323. doi: 10.1016/bs.mie.2018.09.023 [DOI] [PubMed] [Google Scholar]
- Meldal M, & Tornoe CW (2008). Cu-catalyzed azide-alkyne cycloaddition. Chemical Reviews, 108(8), 2952–3015. doi: 10.1021/cr0783479 [DOI] [PubMed] [Google Scholar]
- Moffat JG, Vincent F, Lee JA, Eder J, & Prunotto M (2017). Opportunities and challenges in phenotypic drug discovery: an industry perspective. Nature Reviews Drug Discovery, 16(8), 531–543. doi: 10.1038/nrd.2017.111 [DOI] [PubMed] [Google Scholar]
- Moustakim M, Felce SL, Zaarour N, Farnie G, McCann FE, & Brennan PE (2018). Target identification using chemical probes. Methods in Enzymology, 610, 27–58. doi: 10.1016/bs.mie.2018.09.013 [DOI] [PubMed] [Google Scholar]
- Murale DP, Hong SC, Haque MM, & Lee JS (2016). Photo-affinity labeling (PAL) in chemical proteomics: a handy tool to investigate protein-protein interactions (PPIs). Proteome Science, 15, 14. doi: 10.1186/s12953-017-0123-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson KM, Dahlin JL, Bisson J, Graham J, Pauli GF, & Walters MA (2017). The essential medicinal chemistry of curcumin. Journal of Medicinal Chemistry, 60(5), 1620–1637. doi: 10.1021/acs.jmedchem.6b00975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogueira FC, & Domont GB (2014). Survey of shotgun proteomics. Methods in Molecular Biology, 1156, 3–23. doi: 10.1007/978-1-4939-0685-7_1 [DOI] [PubMed] [Google Scholar]
- Ohana RF, Kirkland TA, Woodroofe CC, Levin S, Uyeda HT, Otto P, … Wood KV (2015). Deciphering the cellular targets of bioactive compounds using a chloroalkane capture tag. ACS Chemical Biology, 10(10), 2316–2324. doi: 10.1021/acschembio.5b00351 [DOI] [PubMed] [Google Scholar]
- Pai MY, Lomenick B, Hwang H, Schiestl R, McBride W, Loo JA, & Huang J (2015). Drug affinity responsive target stability (DARTS) for small-molecule target identification. Methods in Molecular Biology, 1263, 287–298. doi: 10.1007/978-1-4939-2269-7_22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piliarik M, Vaisocherova H, & Homola J (2009). Surface plasmon resonance biosensing. Methods in Molecular Biology, 503, 65–88. doi: 10.1007/978-1-60327-567-5_5 [DOI] [PubMed] [Google Scholar]
- Puck TT, Marcus PI, & Cieciura SJ (1956). Clonal growth of mammalian cells in vitro; growth characteristics of colonies from single HeLa cells with and without a feeder layer. Journal of Experimental Medicine, 103(2), 273–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S, Kwon Y, Kamisuki S, Srivastava N, Mao Q, Kawazoe Y, & Uesugi M (2007). Polyproline-rod approach to isolating protein targets of bioactive small molecules: isolation of a new target of indomethacin. Journal of the American Chemical Society, 129(4), 873–880. doi: 10.1021/ja0655643 [DOI] [PubMed] [Google Scholar]
- Schenone M, Dancik V, Wagner BK, & Clemons PA (2013). Target identification and mechanism of action in chemical biology and drug discovery. Nature Chemical Biology, 9(4), 232–240. doi: 10.1038/nchembio.1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimerlik MI (2001). Overview of membrane protein solubilization. Current Protocols in Neuroscience, Chapter 5, Unit 5 9. doi: 10.1002/0471142301.ns0509s02 [DOI] [PubMed] [Google Scholar]
- Schurmann M, Janning P, Ziegler S, & Waldmann H (2016). Small-molecule target engagement in cells. Cell Chem Biol, 23(4), 435–441. doi: 10.1016/j.chembiol.2016.03.008 [DOI] [PubMed] [Google Scholar]
- Sin N, Meng L, Wang MQ, Wen JJ, Bornmann WG, & Crews CM (1997). The anti-angiogenic agent fumagillin covalently binds and inhibits the methionine aminopeptidase, MetAP-2. Proceedings of the National Academy of Sciences of the United States of America, 94(12), 6099–6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E, & Collins I (2015). Photoaffinity labeling in target- and binding-site identification. Future Medicinal Chemistry, 7(2), 159–183. doi: 10.4155/fmc.14.152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickland EC, Geer MA, Tran DT, Adhikari J, West GM, DeArmond PD, … Fitzgerald MC (2013). Thermodynamic analysis of protein-ligand binding interactions in complex biological mixtures using the stability of proteins from rates of oxidation. Nature Protocols, 8(1), 148–161. doi: 10.1038/nprot.2012.146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulaiman RS, Park B, Sheik Pran Babu SP, Si Y, Kharwadkar R, Mitter SK, … Corson TW (2018). Chemical proteomics reveals soluble epoxide hydrolase as a therapeutic target for ocular neovascularization. ACS Chemical Biology, 13(1), 45–52. doi: 10.1021/acschembio.7b00854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumranjit J, & Chung SJ (2013). Recent advances in target characterization and identification by photoaffinity probes. Molecules, 18(9), 10425–10451. doi: 10.3390/molecules180910425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thermo Fisher Scientific. (2010). Antibodies, avidins and lectins In The Molecular Probes Handbook—A Guide to Fluorescent Probes and Labeling Technologies (11 ed.). [Google Scholar]
- Thiede B, Hohenwarter W, Krah A, Mattow J, Schmid M, Schmidt F, & Jungblut PR (2005). Peptide mass fingerprinting. Methods, 35(3), 237–247. doi: 10.1016/j.ymeth.2004.08.015 [DOI] [PubMed] [Google Scholar]
- Tulloch LB, Menzies SK, Coron RP, Roberts MD, Florence GJ, & Smith TK (2018). Direct and indirect approaches to identify drug modes of action. IUBMB Life, 70(1), 9–22. doi: 10.1002/iub.1697 [DOI] [PubMed] [Google Scholar]
- Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, … Ponten F (2015). Proteomics. Tissue-based map of the human proteome. Science, 347(6220), 1260419. doi: 10.1126/science.1260419 [DOI] [PubMed] [Google Scholar]
- Van Vleet TR, Liguori MJ, Lynch JJ 3rd, Rao M, & Warder S (2018). Screening strategies and methods for better off-target liability prediction and identification of small-molecule pharmaceuticals. SLAS Discovery, 2472555218799713. doi: 10.1177/2472555218799713 [DOI] [PubMed] [Google Scholar]
- Vodovozova EL (2007). Photoaffinity labeling and its application in structural biology. Biochemistry (Mosc.), 72(1), 1–20. [DOI] [PubMed] [Google Scholar]
- Wang J, Wong YK, Zhang J, Lee YM, Hua ZC, Shen HM, & Lin Q (2017). Drug target identification using an iTRAQ-based quantitative chemical proteomics approach-based on a target profiling study of andrographolide. Methods in Enzymology, 586, 291–309. doi: 10.1016/bs.mie.2016.09.049 [DOI] [PubMed] [Google Scholar]
- Whitesell L, Mimnaugh EG, De Costa B, Myers CE, & Neckers LM (1994). Inhibition of heat shock protein HSP90-pp60v-src heteroprotein complex formation by benzoquinone ansamycins: essential role for stress proteins in oncogenic transformation. Proceedings of the National Academy of Sciences of the United States of America, 91(18), 8324–8328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler S, Pries V, Hedberg C, & Waldmann H (2013). Target identification for small bioactive molecules: finding the needle in the haystack. Angewandte Chemie. International Edition In English, 52(10), 2744–2792. doi: 10.1002/anie.201208749 [DOI] [PubMed] [Google Scholar]