Abstract
In biology, there are an abundant number of self-assembled structures organized according to hierarchical levels of complexity. In some examples, the assemblies formed at each level exhibit unique properties and behaviors not present in individual components. Viruses are an example of such where first individual subunits come together to form a capsid structure, some utilizing a scaffolding protein to template or catalyze the capsid formation. Increasing the level of complexity, the viral capsids can then be used as building blocks of higher-level assemblies. This has inspired scientists to design and construct virus capsid-based functional nanomaterials. This review provides some insight into the assembly of virus capsids across several length scales, and certain properties that arise at different levels, providing examples found in naturally occurring systems and those that are synthetically designed.
Introduction
Virus capsids are diverse and elegant examples of biomolecular nanostructures, which are hierarchically self-assembled from a limited number of macromolecular subunits. Self-assembly is a common mechanism for the construction of complex biological systems whereby smaller building blocks spontaneously come together to form larger, hierarchically organized, structures. Biology has evolved to assemble hierarchically ordered structures, which exhibit properties that arise from the collective interaction of the individual building blocks [1,2]. Bacterial microcompartments, for example, are cellular containers that self-assemble from protein subunits and encapsulate a series of enzymes [3]. These containers act as organelle-like compartments that segregate chemical reactions within prokaryotic cells leading to specialized catalytic function. Inspired by nature, materials with multiple levels of hierarchy have been synthetically constructed using modular approaches [4,5]; that is, the formation of individual virus-like particles (VLPs) through self-assembly of capsid subunits, and the assembly of these VLP building blocks into ordered three-dimensional arrays [6•]. The assembly behavior and resultant lattice structure are a consequence of inter-particle interactions mediated by the particle exterior and because this construction approach is modular, it holds the promise that a large diversity of functional cargos could be encapsulated inside of VLPs and assembled into superlattices using the same assembly approach. Thus, the construction of virus-based assemblies at multiple length scales presented here could be widely applicable for the formation of a range of complex materials.
Scaffold protein-mediated assembly
Both viral and non-viral assemblies often require macro-molecular scaffolds to template formation of the proper structure. Tubulin, for example, is essential for cell proliferation and its polymerization requires regulatory proteins that mediate the assembly and disassembly of the microtubule network [7–9]. Similarly, chaperone proteins behave as scaffolds to promote proper intracellular protein-folding, and prevent aggregation in the cytosol due to the high macromolecular concentrations found within cellular environments [10]. However, there are many more examples of scaffold-mediated assembly in the realm of viral capsid self-assembly.
The role of scaffold proteins (SP) in viral capsid assembly can be to prevent the formation of aberrant assemblies, act as a catalyst during nucleation, promote proper inter-subunit interactions between coat protein (or major capsid protein) subunits, and prevent formation of particles that may otherwise form due to the spontaneous radius of curvature of the CP subunits [11–13,14••]. In systems where SP is necessary, it is typically required for the proper formation of an initial assembly intermediate, the procapsid or prohead [12,15]. This assembly is accomplished via both long-range and short-range non-covalent interactions between the coat protein (CP) and SP, some of which can subsequently be disrupted during the maturation of the procapsid [5,11,15,16,17••]. Considering the critical role of SP and the environment in which viral particles assemble, the interactions between the CP and SP are often highly specific. The presence of a SP can also lower the threshold or critical CP concentration required for capsid formation making the overall assembly process more favorable [17••].
In general, large icosahedral viruses require scaffolding protein [14••]. A few bacteriophage examples such as P22 [18–21], λ [22–24], T7 [25–27], T4 [28–30], and ϕ29 [31–33] all require scaffolding proteins to template the assembly of the initial procapsid structure. Viruses from the herpesviridae also require an internal scaffolding protein, and assemble via a similar mechanism to these bacteriophages, where first a prohead structure is formed before the packaging of DNA [34,35]. Oligomeric forms of SPs (monomers, dimers, and tetramers) template CP assembly by either first forming a SP ‘core’ around which the CPs can accumulate (e.g.: T4) or the SP remains largely dissociated and co-assembles with the CP (e.g.: P22 (Figure 1a), λ, T7, and ϕ29). The conformational flexibility of many of these scaffolding proteins allows for structural adaptability, which is critical for driving interactions between non-equivalent capsid proteins.
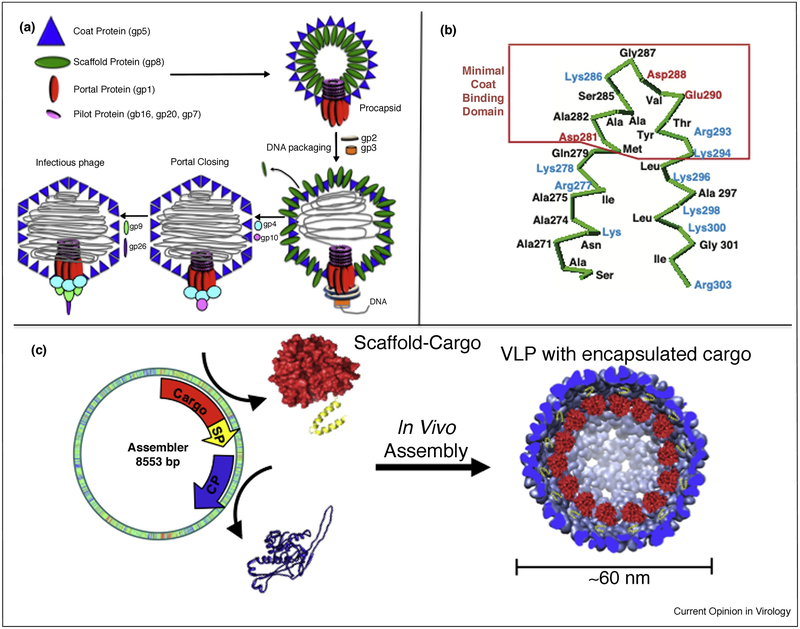
Figure 1.
Roles of bacteriophage P22 scaffolding protein (SP). (a) The assembly pathway of bacteriophage P22. A variety of gene products come together to form the capsid including coat protein (CP) gp5 and SP gp8. SP is responsible for co-assembling with CP and templating the procapsid structure. As DNA is packaged, the binding domain is lost due to a CP conformation shift and SP is ejected from the capsid. (b) The C-terminus of gp8 SP, including the 35 amino acids necessary for interacting with CP for assembly of the procapsid. The N-terminus is not required for templating capsid assembly [20]. (c) Truncating the N-terminus of SP and fusing a cargo to it allowed for the programmed encapsulation of cargo non-native to P22, including a variety of different enzymes [36].
In the case of bacteriophage T4, the essential scaffolding proteins form a core structure with at least four other proteins, even in the absence of the major capsid protein [28], before capsid assembly. In P22, the flexible nature of the internal scaffolding protein has allowed materials chemists to repurpose these viral capsids and drive the encapsulation of cargo molecules, not native to the P22 structure, into the capsid [36–40,41•,42•]. The SP binding domain is a structured helix-turn-helix (hth) located near the C-terminus, while the N-terminus of the SP is largely disordered and can be truncated without losing the ability to template P22 assembly (Figure 1b) [20]. Fusing protein cargos, such as fluorescent proteins or catalytically active enzymes, to the truncated N-terminus or C-terminus, while preserving the integrity of the SP hth binding domain, has provided a general approach for encapsulating a wide variety of functional proteins for use in synthetic materials design and construction (Figure 1c).
Capsid–capsid interactions and collective behavior
The functional complexity achieved by biological systems often arises from global behavior that is the result of collective properties not present in the behavior of the independent components before assembly [1,2,43]. This can be observed in entire ecosystems as well as on a cellular level, where the individual sub-cellular compartments act together (i.e. the cell) to carry out reactions beyond the capabilities of each individual compartment [44••]. Furthermore, when individual cells come together to form specific tissues, additional functionality is achieved as a result of the collective cellular behavior [45]. In the case of viral capsids, emergent properties are evident upon assembly of a single capsid from individual protein subunits, including the ability to package cargo such as DNA and other macromolecules, provide a protective barrier for the interior components, and in infectious viruses the ability to infect host organisms [46•].
Adding complexity to the hierarchical organization, such as in the interactions between individual viral capsids, can lead to collective behavior not present in the individual capsid. When considering the processes of viral infection, the intracellular environment undergoes dramatic rearrangement due to the increasing concentration of capsids and other viral components [47–50]. For both bacterial and eukaryotic organisms parts of the cellular machinery migrate to the periphery of the cell altering cellular organization [51], and the cytoskeleton shifts in order to accommodate the viral replication sites, which often include electron-dense aggregates referred to as viral factories (Figure 2d) [49,52,53•]. Viral factories assist in the synthesis of viral components, fabrication of capsids, packaging of genomic material, and most importantly for this discussion, are associated with the high local concentration of viral particles within the intracellular environment [51,54].
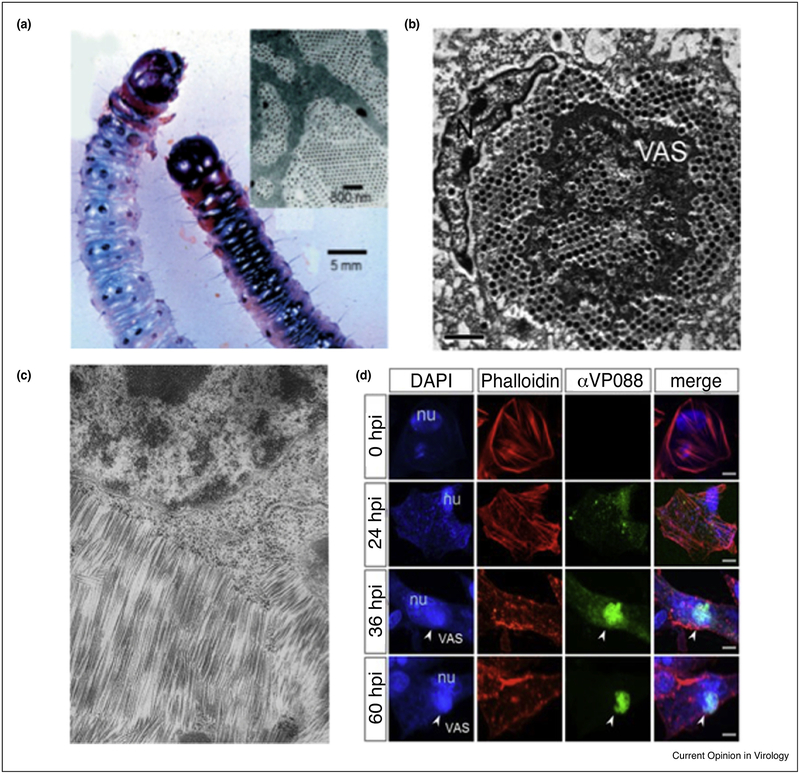
Figure 2.
Capsid–capsid interactions can induce structural changes. (a) A larvae infected (left) and not infected (right) with Wiseana Iridovirus. The infected larvae display iridescence and discoloration due to the crystalline packing arrangement of the virus capsid in the infected cells [57]. (b) The paracrystalline array formed by the Singapore Grouper Iridovirus in an infected cell [48]. (c) Hexagonally packed TMV crystals embedded in the cytoplasm [47]. (d) Confocal microscope fluorescence imaging of a cell upon formation of a virus factory, monitored after hours post infection (hpi). The nucleus (blue-DAPI stained) and the F-actin (red-phalloidin stained) both go through large conformation changes upon formation of a virus factory (green) [63•].
Not only does the high viral-particle concentration result in cytopathic effects, but also there are several examples where the inter-capsid interactions give rise to new properties, something that has been exploited by material scientists. In one example, viral factories are associated with the paracrystalline arrays of iridovirus formed inside the cytoplasm of infected cells (Figure 2a and b) [48]. Though the mechanism of this assembly remains largely unknown, the result is the crystalline close packing of the iridovirus particles giving rise to the iridescent properties associated with high levels of infection [55,56]. The iridescence is indicative of infection; however, this feature has prompted the study and use of this virus toward the design and construction of bio-photonic crystals [57–59]. The rod-shaped Tobacco Mosaic Virus (TMV) also adopts similar crystalline packing densities, where hexagonal crystals have been observed in the cytoplasm shortly after infection (Figure 2c) [47]. These nematic liquid-crystal domains formed by TMV, exhibit intrinsic magnetic properties [60–62], show Bragg diffraction, and form iridescent gels [50].
Overall, the formation of these crystalline arrays is driven in part by shape complementarity, where the shape of the individual components determines the orientation, global entropic gain due to displacement of water molecules, the local concentration of individual particles, excluded volume effects, and factors dependent on ionic strength and other environmental conditions.
Synthetic higher-order assembly
Materials scientists increasingly draw inspiration for the design of new functional materials [5,64–67] from the multi-length scale assemblies found in nature. The study of these materials is important for understanding self-assembly in biological systems, which are often dynamic, complex, and difficult to study in their native forms. The synthetic use of directed self-assembly via non-covalent interactions, has highlighted the possibilities for nano-scale and micro-scale structures, unattainable via covalent bond formation. Designing synthetic systems provides an opportunity to limit and control the study to a few parameters. Exploiting some of the characteristics of self-assembled structures can lead to the synthesis of multicomponent materials designed for applications in the fields of biomedicine, energetics, and catalysis [68,69••,70–74,75••]. For example, similar to the photonic properties observed in the ordered lattices of iridoviruses, crystalline assemblies of individual synthetic nanoparticles have been shown to have unique magnetic, electronic, photonic, and plasmonic properties [76,77].
Virus and virus-like particles (VLPs) are attractive building blocks for the synthetic construction of higher order assemblies due to their monodispersity in size and shape [69••,78,79]. Both isotropic and anisotropic viral nanoparticles have been studied for the design and synthesis of self-assembled nano-arrays. Tobacco mosaic virus (TMV) and M13 phage are two examples of anisotropic rod-shaped nanoparticles with a propensity to orient along their long axis and form liquid crystal-like domains, and therefore are appealing candidates for higher order assemblies. TMV has been used as a building block for the construction of superlattices [80–82], as a template for ordered silica mesophases [83], and the formation of 2-D/3-D assemblies [84–86] through non-covalent interactions including electrostatics [86], depletion forces [80,81], and metal coordination [82,84]. Similarly, the high aspect ratio and helical nanofibrous shape of phage M13 make it ideal for the development of both covalently and non-covalently assembled liquid-crystals [87,88], supramolecular structures [89], nanoporous networks [90], with very long range (cm length scale) order [91••], and hierarchically organized M13 phage-based nano-fiber materials with piezoelectric properties [92].
Symmetrical icosahedral virus particles such as those derived from bacteriophage P22 [6•,93,94], Cowpea chlorotic mottle virus (CCMV) [95], Cowpea mosaic virus (CPMV) [96], and bacteriophage Qβ [97] have also been used for directed construction of synthetic higher-order assemblies via non-covalent interactions. CCMV and Qβ have been assembled into binary 3D superlattices using tunable electrostatic interactions with Au nanoparticles and poly-peptide functionalized bio-macromolecules (Figure 3a) [98], and complementary oligonucleotide interactions [95,97]. Complementary oligonucleotide-functionalized CPMV particles have been used to assemble 3D arrays with tunable melting temperatures by varying the degree of complementarity between the nucleotides [96]. The icosahedral capsids additionally contain hollow interiors, capable of encapsulating functional cargo, highlighting the potential for user-defined functionalities of the resulting higher ordered assemblies. Functional CCMV VLPs have been used as building blocks in the formation of non-lattice higher-order 2D thin films and 3D hydrogels [99,100]. CPMV and Qβ have been utilized to encapsulate non-native cargo but lattice formation of these functional VLPs remains to be demonstrated [101–103].
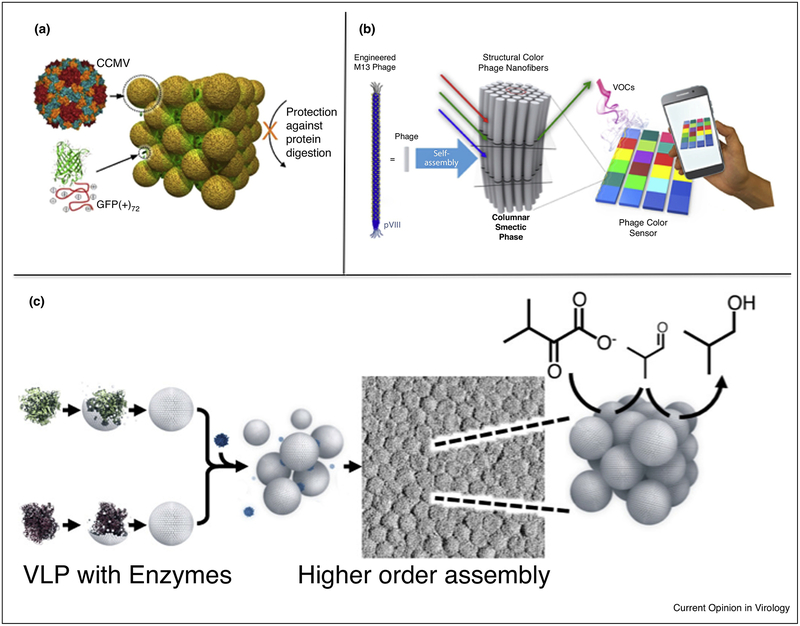
Figure 3.
Virus capsid-based synthetic high-order assemblies. (a) Electrostatically driven co-crystals are formed through the interactions between the negatively charged CCMV capsid and super-charged cationic polypeptide fused to a fluorescent protein [98]. (b) A sensor was constructed using M13 Phage where environmental stimuli triggered a change in the spacing between phages resulting in an optical response [105]. (c) Coupling the ability to encapsulate enzyme cargo and tune inter-particle interactions, bacteriophage P22-based fcc lattices were formed capable of multi-step catalysis toward the enzymatic synthesis of isobutanol [6•].
However, studies using the VLP derived from the bacteriophage P22 have combined the capabilities of encapsulating active enzyme cargos, and the ability to design and construct protein cage superlattices. Utilizing the functionally active P22 VLPs as building blocks, the construction of catalytically active arrays using protein linkers and polymer-mediated assembly has been demonstrated [6•,93]. The exterior of the P22 VLPs was modified with a range of small peptides, to tune the surface charge. When allowed to interact with oppositely charged branched polymers (PAMAM dendrimers), the P22 variants assembled into ordered lattices with very pronounced ionic strength dependence (Figure 3c) [6•]. The resulting lattices were investigated by small angle X-ray scattering (SAXS) and electron microscopy that revealed ordered face centered cubic arrangements of VLPs. These higher order assemblies were driven by the nature of the exterior capsid surface and were independent of the encapsulated cargo, thus catalytic enzymes encapsulated within the P22 remained active when assembled in the higher order structures. This afforded the design and construction of extended materials with demonstrated multistep enzyme catalysis for the formation of isobutanol. Since the VLP-based lattice materials required less solvation than the individual capsids, this allowed for very high enzyme and substrate concentrations in the reactions thereby increasing catalytic efficiency considerably [6•]. To further enhance the robustness of this design and construction, a ditopic ‘cementing’ protein was used to lock the P22 lattice into place forming a Protein Macromolecular Framework (PMF) from which the dendrimer could be removed at high ionic strength without disrupting the lattice [93]. Higher order assemblies constructed from P22 VLPs demonstrated properties observed in the extended lattice that were different from those of the individual particles. This was shown when the assembled lattice incorporated and bound a charged macromolecule under ionic strength conditions where there was no interaction with individual P22 particles [94]. Collective behavior has also been observed in ordered domains assembled from M13 phage that displayed colors in the visible range due to light refraction (Figure 3b). Upon changes in humidity, or through specifically designed interactions with small molecules, the assemblies underwent a structural change that caused the inter-particle distances to shift and change color in an easily measurable way [104,105]. These properties are a result of the extended lattice and are not present in the individual virus particles.
Conclusion
The field of VLP-based materials assembly to create higher-order structures is an emerging area, even though the hierarchical organizations of assemblies such as TMV or iridovirus have been known for decades. However, the collective properties and the potential applications for synthetic higher order assemblies have not been fully realized for many of these systems. Virology has provided scientists with inspiration and a framework for using virus-based building blocks to design and synthesize a new generation of functional biomaterials. Understanding self-assembly across multiple length scales has guided the development of these systems with the ability to tune the properties of individual particles, as well as new approaches for higher order assembly of functional materials.
Acknowledgements
The authors would like to acknowledge funding support from the U.S. Department of Energy, Office of Basic Energy Sciences, Division of Materials Sciences and Engineering (DESC0016155), the National Institutes of Health (R01 AI104905), and the National Science Foundation (NSF-BMAT DMR-1507282 and EEC-720625). E.S. was supported by the Graduate Training Program in Quantitative and Chemical Biology under Award Number T32 GM109825 and Indiana University. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Novikoff AB: The concept of integrative levels and biology. Science 1945, 101:209–215. [DOI] [PubMed] [Google Scholar]
- 2.Vicsek T, Zafeiris A: Collective motion. Phys Rep 2012, 517:71–140. [Google Scholar]
- 3.Chowdhury C, Sinha S, Chun S, Yeates TO, Bobik TA: Diverse bacterial microcompartment organelles. Microbiol Mol Biol Rev 2014, 78:438–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mendes AC, Baran ET, Reis RL, Azevedo HS: Self-assembly in nature: using the principles of nature to create complex nanobiomaterials. Wiley Interdiscip Rev: Nanomed Nanobiotechnol 2013, 5:582–612. [DOI] [PubMed] [Google Scholar]
- 5.Stupp SI, Zha RH, Palmer LC, Cui H, Bitton R: Self-assembly of biomolecular soft matter. Faraday Discuss 2013, 166:9–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uchida M, McCoy K, Fukuto M, Yang L, Yoshimura H, Miettinen HM, LaFrance B, Patterson DP, Schwarz B, Karty JA et al. : Modular self-assembly of protein cage lattices for multistep catalysis. ACS Nano 2017, 12:942–953. [DOI] [PMC free article] [PubMed] [Google Scholar]; • This paper illustrates how bacteriophage P22 can be used to construct a superlattice structure that is capable of multistep catalysis towards the synthesis of iso-butanol. Utilizing the scaffolding protein as means by which non-native cargo can be encapsulated, two different populations of enzymes are encapsulated. The exterior of the capsids was modified to tune the inter-particle interactions, without affecting the catalytic activity of the enzymes within.
- 7.Hlavanda E, Kovács J, Oláh J, Orosz F, Medzihradszky KF, Ovádi J: Brain-specific p25 protein binds to tubulin and microtubules and induces aberrant microtubule assemblies at substoichiometric concentrations. Biochemistry 2002, 41:8657–8664. [DOI] [PubMed] [Google Scholar]
- 8.Schofield AV, Gamell C, Suryadinata R, Sarcevic B, Bernard O: Tubulin polymerizing protein 1 (TPPP1) phosphorylation by Rho-associated coiled-coil kinase (ROCK) and Cyclin dependent kinase 1 (Cdk1) inhibits microtubule dynamics to increase cell proliferation. J Biol Chem 2013, 288:7907–7917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tokesi N, Lehotzky A, Horváth I, Szabó B, Oláh J, Lau P, Ovádi J: TPPP/p25 promotes tubulin acetylation by inhibiting histone deacetylase 6. J Biol Chem 2010, 285:17896–17906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frydman J: Folding of newly translated proteins in vivo: the role of molecular chaperones. Ann Rev Biochem 2001, 70:603–647. [DOI] [PubMed] [Google Scholar]
- 11.Chen D-H, Baker ML, Hryc CF, DiMaio F, Jakana J, Wu W, Dougherty M, Haase-Pettingell C, Schmid MF, Jiang W et al. : Structural basis for scaffolding-mediated assembly and maturation of a dsDNA virus. Proc Natl Acad Sci U S A 2011, 108:1355–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aksyuk AA, Rossmann MG: Bacteriophage assembly. Viruses 2011, 3:172–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parent KN, Doyle SM, Anderson E, Teschke CM: Electrostatic interactions govern both nucleation and elongation during phage P22 procapsid assembly. Virology 2005, 340:33–45. [DOI] [PubMed] [Google Scholar]
- 14.Li S, Roy P, Travesset A, Zandi R: Why large icosahedral viruses need scaffolding proteins. Proc Natl Acad Sci U S A 2018, 115:10971–10976. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Using a computational approach, this article provides a rationale for the presence of scaffolding proteins in large icosahedral viruses.
- 15.Dokland T: Scaffolding proteins and their role in viral assembly. Cell Mol Life Sci 1999, 56:580–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parker MH, Prevelige PE Jr: Electrostatic interactions drive scaffolding/coat protein binding and procapsid maturation in bacteriophage P22. Virology 1998, 250:337–349. [DOI] [PubMed] [Google Scholar]
- 17.Hagan MF, Zandi R: Recent advances in coarse-grained modeling of virus assembly. Curr Opin Virol 2016, 18:36. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Great source for gaining insight into the computational approach for studying virus-particle assembly and the driving forces associated with the process.
- 18.Lander GC, Tang L, Casjens SR, Gilcrease EB, Prevelige P, Poliakov A, Potter CS, Carragher B, Johnson JE: The structure of an infectious P22 virion shows the signal for headful DNA packaging. Science 2006, 312:1791–1795. [DOI] [PubMed] [Google Scholar]
- 19.King J, Botstein D, Casjens S, Earnshaw W, Harrison S, Lenk E: Structure and assembly of the capsid of bacteriophage P22. Phil Trans R Soc Lond B 1976, 276:37–49. [DOI] [PubMed] [Google Scholar]
- 20.Weigele PR, Sampson L, Winn-Stapley D, Casjens SR: Molecular genetics of bacteriophage P22 scaffolding protein’s functional domains. J Mol Biol 2005, 348:831–844. [DOI] [PubMed] [Google Scholar]
- 21.Casjens S, King J: P22 morphogenesis I: catalytic scaffolding protein in capsid assembly. J Supramol Struct 1974, 2:202–224. [DOI] [PubMed] [Google Scholar]
- 22.Georgopoulos C, Tilly K, Casjens S : Lambdoid phage head assembly. Lambda II 1983:279–304. [Google Scholar]
- 23.Ziegelhoffer T, Yau P, Chandrasekhar GN, Kochan J, Georgopoulos C, Murialdo H: The purification and properties of the scaffolding protein of bacteriophage lambda. J Biol Chem 1992, 267:455–461. [PubMed] [Google Scholar]
- 24.Murialdo H, Becker A: Head morphogenesis of complex double-stranded deoxyribonucleic acid bacteriophages. Microbiol Rev 1978, 42:529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Serwer P: Internal proteins of bacteriophage T7. J Mol Biol 1976, 107:271–291. [DOI] [PubMed] [Google Scholar]
- 26.Cerritelli ME, Studier WF: Assembly of T7 capsids from independently expressed and purified head protein and scaffolding protein. J Mol Biol 1996, 258:286–298. [DOI] [PubMed] [Google Scholar]
- 27.Cerritelli ME, Conway JF, Cheng N, Trus BL, Steven AC: Molecular mechanisms in bacteriophage T7 procapsid assembly, maturation, and DNA containment. Adv Protein Chem 2003, 64:301–324. [DOI] [PubMed] [Google Scholar]
- 28.Mesyanzhinov VV, Sobolev BN, Marusich EI, Prilipov AG, Efimov VP: A proposed structure of bacteriophage T4 gene product 22—a major prohead scaffolding core protein. J Struct Biol 1990, 104:24–31. [DOI] [PubMed] [Google Scholar]
- 29.Mathews CK: Bacteriophage T4. eLS 2001:1–11. [Google Scholar]
- 30.Yap ML, Rossmann MG: Structure and function of bacteriophage T4. Future Microbiol 2014, 9:1319–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Viñuela E, Camacho A, Jiménez F, Carrascosa JL, Ramirez G, Salas M: Structure and assembly of phage Φ29. Phil Trans R Soc Lond B 1976, 276:29–35. [DOI] [PubMed] [Google Scholar]
- 32.Morais MC, Kanamaru S, Badasso MO, Koti JS, Owen BA, McMurray CT, Anderson DL, Rossmann MG: Bacteriophage φ29 scaffolding protein gp7 before and after prohead assembly. Nat Struct Mol Biol 2003, 10:572. [DOI] [PubMed] [Google Scholar]
- 33.Fu C-y Morais MC, Battisti AJ Rossmann MG, Prevelige PE Jr: Molecular dissection of ø29 scaffolding protein function in an in vitro assembly system. J Mol Biol 2007, 366:1161–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Preston VG, McDougall IM: Regions of the herpes simplex virus scaffolding protein that are important for intermolecular self-interaction. J Virol 2002, 76:673–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rixon F: Structure and assembly of herpesviruses. Seminars in Virology. Elsevier; 1993:135–144. [Google Scholar]
- 36.Patterson DP, Prevelige PE, Douglas T: Nanoreactors by programmed enzyme encapsulation inside the capsid of the bacteriophage P22. ACS Nano 2012, 6:5000–5009. [DOI] [PubMed] [Google Scholar]
- 37.O’Neil A, Reichhardt C, Johnson B, Prevelige PE, Douglas T: Genetically programmed in vivo packaging of protein cargo and its controlled release from bacteriophage P22. Angew Chem Int Ed 2011, 50:7425–7428. [DOI] [PubMed] [Google Scholar]
- 38.O’Neil A, Prevelige PE, Basu G, Douglas T: Coconfinement of fluorescent proteins: spatially enforced communication of GFP and mCherry encapsulated within the P22 capsid. Biomacromolecules 2012, 13:3902–3907. [DOI] [PubMed] [Google Scholar]
- 39.Patterson DP, Schwarz B, El-Boubbou K, van der Oost J, Prevelige PE, Douglas T: Virus-like particle nanoreactors: programmed encapsulation of the thermostable CelB glycosidase inside the P22 capsid. Soft Matter 2012, 8:10158–10166. [Google Scholar]
- 40.Patterson DP, Schwarz B, Waters RS, Gedeon T, Douglas T: Encapsulation of an enzyme cascade within the bacteriophage P22 virus-like particle. ACS Chem Biol 2013, 9:359–365. [DOI] [PubMed] [Google Scholar]
- 41.Sharma J, Uchida M, Miettinen HM, Douglas T: Modular interior loading and exterior decoration of a virus-like particle. Nanoscale 2017, 9:10420–10430. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Describes a useful methodology for control over the stoichiometry of encapsulated macromolecules.
- 42.Jordan PC, Patterson DP, Saboda KN, Edwards EJ, Miettinen HM, Basu G, Thielges MC, Douglas T: Self-assembling biomolecular catalysts for hydrogen production. Nat Chem 2016, 8:179. [DOI] [PubMed] [Google Scholar]; • This is one of the many examples that virus-particles have been re-purposed for designing functional materials. In this study, bacteriophage P22 particles are used as hollow capsids that encapsulate enzymes involved in the production of hydrogen gas. When comparing the activity, the encapsulated enzyme shows a 100-fold increase in enzymatic activity compared to the same free enzyme.
- 43.Ponge J-F: Emergent properties from organisms to ecosystems: towards a realistic approach. Biol Rev 2005, 80:403–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kubitschke H, Morawetz EW, Käs JA, Schnauß J: Physical properties of single cells and collective behavior. Quantification of Biophysical Parameters in Medical Imaging. Springer; 2018:89–121. [Google Scholar]; •• This review shows properties and collective behavior found in cells and their surrounding environments. This includes the biophysical characterization of biopolymers, cells, and tissues and the emergent mechanical properties that arise at each increasing level of organization.
- 45.Noll N, Mani M, Heemskerk I, Streichan SJ, Shraiman BI: Active tension network model suggests an exotic mechanical state realized in epithelial tissues. Nat Phys 2017, 13:1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sanjuán R: Collective infectious units in viruses. Trends Microbiol 2017, 25:402–412. [DOI] [PMC free article] [PubMed] [Google Scholar]; • This paper offers insight into the cooperative and collective properties that arise during a viral infection. It covers topics such as the recruitment of other macromolecules in the surrounding environment, virus–virus interactions, and also non-cooperative interactions.
- 47.Kolehmainen L, Zech H, Von Wettstein D: The structure of cells during tobacco mosaic virus reproduction: mesophyll cells containing virus crystals. J Cell Biol 1965, 25:77–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu Y, Tran BN, Wang F, Ounjai P, Wu J, Hew CL: Visualization of assembly intermediates and budding vacuoles of Singapore Grouper Iridovirus in Grouper embryonic cells. Sci Rep 2016, 6:18696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Netherton C, Moffat K, Brooks E, Wileman T: A guide to viral inclusions, membrane rearrangements, factories, and viroplasm produced during virus replication. Adv Virus Res 2007, 70:101–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oster G: Two-phase formation in solutions of tobacco mosaic virus and the problem of long-range forces. J Gen Physiol 1950, 33:445–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Novoa RR, Calderita G, Arranz R, Fontana J, Granzow H, Risco C: Virus factories: associations of cell organelles for viral replication and morphogenesis. Biol Cell 2005, 97:147–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reissig M, Howes DW, Melnick JL: Sequence of morphological changes in epithelial cell cultures infected with poliovirus. J Exp Med 1956, 104:289–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chaikeeratisak V, Nguyen K, Khanna K, Brilot AF, Erb ML, Coker JK, Vavilina A, Newton GL, Buschauer R, Pogliano K et al. : Assembly of a nucleus-like structure during viral replication in bacteria. Science 2017, 355:194–197. [DOI] [PMC free article] [PubMed] [Google Scholar]; • This paper illustrates the formation of viral intracellular compartments in a bacterial cell post infection
- 54.de Castro IF, Volonté L, Risco C: Virus factories: biogenesis and structural design. Cell Microbiol 2013, 15:24–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.İnce İ,Özcan O, Ilter-Akulke A, Scully E, Özgen A: Invertebrate iridoviruses: a glance over the last decade. Viruses 2018, 10:161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Williams T, Barbosa-Solomieu V, Chinchar VG: A decade of advances in iridovirus research. Adv Virus Res 2005, 65:173–248. [DOI] [PubMed] [Google Scholar]
- 57.Juhl SB, Chan EP, Ha YH, Maldovan M, Brunton J, Ward V, Dokland T, Kalmakoff J, Farmer B, Thomas EL: Assembly of Wiseana iridovirus: viruses for colloidal photonic crystals. Adv Funct Mater 2006, 16:1086–1094. [Google Scholar]
- 58.Large M: Optical Biomimetics: Materials and Applications. Elsevier; 2012. [Google Scholar]
- 59.López C: Materials aspects of photonic crystals. Adv Mater 2003, 15:1679–1704. [Google Scholar]
- 60.Urakami N, Imai M, Sano Y, Takasu M: The isotropic–nematic transition and the phase separation of the tobacco mosaic virus particles with polysaccharide. J Chem Phys 1999, 111:2322–2328. [Google Scholar]
- 61.Wu Z, Zierold R, Mueller A, Ruff SE, Ma C, Khan AA, Geiger F, Sommer BA, Knez M, Nielsch K et al. : Preparation and magnetoviscosity of nanotube ferrofluids by viral scaffolding and ALD on porous templates. Phys Status Solidi B 2010, 247:2412–2423. [Google Scholar]
- 62.Charles SW: Alignment of biological assemblies using magnetic fluids—a review. J Magn Magn Mater 1990, 85:277–284. [Google Scholar]
- 63.Yuan Y, Wang Y, Liu Q, Zhu F, Hong Y: Singapore grouper iridovirus protein VP088 is essential for viral infectivity. Sci Rep 2016, 6:31170. [DOI] [PMC free article] [PubMed] [Google Scholar]; • This work provides an excellent visualization of the cellular conformational shifts and reorganization upon infection with an Iridovirus. Using fluorescence microscopy, the formation of a viral factory is seen at various stages after infection.
- 64.Zhang S: Fabrication of novel biomaterials through molecular self-assembly. Nat Biotechnol 2003, 21:1171. [DOI] [PubMed] [Google Scholar]
- 65.Ariga K, Lee MV, Mori T, Yu X-Y, Hill JP: Two-dimensional nanoarchitectonics based on self-assembly. Adv Colloid Interface Sci 2010, 154:20–29. [DOI] [PubMed] [Google Scholar]
- 66.Whitesides GM, Grzybowski B: Self-assembly at all scales. Science 2002, 295:2418–2421. [DOI] [PubMed] [Google Scholar]
- 67.Huang MH, Thoka S: Formation of supercrystals through self-assembly of polyhedral nanocrystals. Nano Today 2015, 10:81–92. [Google Scholar]
- 68.Chen C, Liu K, Li J, Yan X: Functional architectures based on self-assembly of bio-inspired dipeptides: structure modulation and its photoelectronic applications. Adv Colloid Interface Sci 2015, 225:177–193. [DOI] [PubMed] [Google Scholar]
- 69.Schwarz B, Uchida M, Douglas T: Biomedical and catalytic opportunities of virus-like particles in nanotechnology. Advances in Virus Research. Elsevier; 2017:1–60. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• This is an excellent review on how scientists have utilized virus-like particles for applications across a variety of different fields. After providing a general description of virus-like particles and their structures, it described machinery, in which virus particles are used as metabolic compartments for biomedical delivery and imaging, as templates for constrained polymerization reactions, and as immune-stimulatory agents.
- 70.Yoo J-W, Irvine DJ, Discher DE, Mitragotri S: Bio-inspired, bioengineered and biomimetic drug delivery carriers. Nat Rev Drug Discov 2011, 10:521. [DOI] [PubMed] [Google Scholar]
- 71.Thorkelsson K, Bai P, Xu T: Self-assembly and applications of anisotropic nanomaterials: a review. Nano Today 2015, 10:48–66. [Google Scholar]
- 72.Berezhnoy NV, Korolev N, Nordenskiöld L: Principles of electrostatic interactions and self-assembly in lipid/peptide/DNA systems: applications to gene delivery. Adv Colloid Interface Sci 2014, 205:221–229. [DOI] [PubMed] [Google Scholar]
- 73.Sanchez C, Arribart H, Guille MMG: Biomimetism and bioinspiration as tools for the design of innovative materials and systems. Nat Mater 2005, 4:277. [DOI] [PubMed] [Google Scholar]
- 74.Kim S, Park CB: Bio-inspired synthesis of minerals for energy, environment, and medicinal applications. Adv Funct Mater 2013, 23:10–25. [Google Scholar]
- 75.Mateu MG: Assembly, engineering and applications of virus-based protein nanoparticles. Protein-based Engineered Nanostructures. Springer; 2016:83-120. [DOI] [PubMed] [Google Scholar]; •• This chapter not only covers how virus particles have been repurposed, but also covers basic principles behind virus assembly and the various pathways through which assembly can take place. Additionally provides a general overview on the basic structure of virus-capsids that have been studied. This is a great source for citations on virus assembly, virus structure, and virus engineering.
- 76.Suryanarayana C: The structure and properties of nanocrystalline materials: issues and concerns. JOM J Miner Metals Mater Soc 2002, 54:24–27. [Google Scholar]
- 77.Braga D, Grepioni F: Crystal engineering: from molecules and crystals to materials. Crystal Engineering: From Molecules and Crystals to Materials. Springer; 1999:421–441. [Google Scholar]
- 78.Roldao A, Silva AC, Mellado MCM, Alves PM, Carrondo MJ: Viruses and virus-like particles in biotechnology: fundamentals and applications In Comprehensive Biotechnology. Volume 1: Scientific Fundamentals in Biotechnology, 2nd edition Edited by Moo-Young M. UK: Elsevier/Pergamon, Oxford; 2011:625–649. [Google Scholar]
- 79.Mateu MG: Virus engineering: functionalization and stabilization. Protein Eng Des Sel 2010, 24:53–63. [DOI] [PubMed] [Google Scholar]
- 80.Li T, Zan X, Sun Y, Zuo X, Li X, Senesi A, Winans RE, Wang Q, Lee B: Self-assembly of rodlike virus to superlattices. Langmuir 2013, 29:12777–12784. [DOI] [PubMed] [Google Scholar]
- 81.Li T, Zan X, Winans RE, Wang Q, Lee B: Biomolecular assembly of thermoresponsive superlattices of the tobacco mosaic virus with large tunable interparticle distances. Angew Chem Int Ed Engl 2013, 125:6770–6774. [DOI] [PubMed] [Google Scholar]
- 82.Li T, Winans RE, Lee B: Superlattice of rodlike virus particles formed in aqueous solution through like-charge attraction. Langmuir 2011, 27:10929–10937. [DOI] [PubMed] [Google Scholar]
- 83.Fowler CE, Shenton W, Stubbs G, Mann S: Tobacco mosaic virus liquid crystals as templates for the interior design of silica mesophases and nanoparticles. Adv Mater 2001, 13:1266–1269. [Google Scholar]
- 84.Nedoluzhko A, Douglas T: Ordered association of tobacco mosaic virus in the presence of divalent metal ions. J Inorg Biochem 2001, 84:233–240. [DOI] [PubMed] [Google Scholar]
- 85.Gebhardt R, Teulon J-M, Pellequer J-L, Burghammer M, Colletier J-P, Riekel C: Virus particle assembly into crystalline domains enabled by the coffee ring effect. Soft Matter 2014, 10:5458–5462. [DOI] [PubMed] [Google Scholar]
- 86.Yang L, Wang S, Fukuto M, Checco A, Niu Z, Wang Q: Structure and interaction in 2D assemblies of tobacco mosaic viruses. Soft Matter 2009, 5:4951–4961. [Google Scholar]
- 87.Lee BY, Zhang J, Zueger C, Chung W-J, Yoo SY, Wang E, Meyer J, Ramesh R, Lee S-W: Virus-based piezoelectric energy generation. Nat Nanotechnol 2012, 7:351. [DOI] [PubMed] [Google Scholar]
- 88.Lee S-W, Belcher AM: Virus-based fabrication of micro-and nanofibers using electrospinning. Nano Lett 2004, 4:387–390. [Google Scholar]
- 89.Chung W-J, Oh J-W, Kwak K, Lee BY, Meyer J, Wang E, Hexemer A, Lee S-W: Biomimetic self-templating supramolecular structures. Nature 2011, 478:364. [DOI] [PubMed] [Google Scholar]
- 90.Courchesne NMD, Klug MT, Chen PY, Kooi SE, Yun DS, Hong N, Fang NX, Belcher AM, Hammond PT: Assembly of a bacteriophage-based template for the organization of materials into nanoporous networks. Adv Mater 2014, 26:3398–3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sawada T, Serizawa T: Filamentous viruses as building blocks for hierarchical self-assembly toward functional soft materials. Bull Chem Soc Japan 2018, 91:455–466. [Google Scholar]; •• This is a great review that focuses on the recent interests in utilization M13 phage as a building block in higher order assemblies and how those assemblies have been used as functional biomaterials. The paper provides general information on the M13 structure and then provides examples of how it has been incorporated into 2D and 3D assemblies.
- 92.Heo K, Jin H-E, Kim H, Lee JH, Wang E, Lee S-W: Transient self-templating assembly of M13 bacteriophage for enhanced biopiezoelectric devices. Nano Energy 2019, 56:716–723. [Google Scholar]
- 93.McCoy K, Uchida M, Lee B, Douglas T: Templated assembly of a functional ordered protein macromolecular framework from P22 virus-like particles. ACS Nano 2018, 12:3541–3550. [DOI] [PubMed] [Google Scholar]
- 94.Aumiller WM Jr, Uchida M, Biner DW, Miettinen HM, Lee B, Douglas T: Stimuli responsive hierarchical assembly of P22 virus-like particles. Chem Mater 2018, 30:2262–2273. [Google Scholar]
- 95.Kostiainen MA, Hiekkataipale P, Laiho A, Lemieux V, Seitsonen J, Ruokolainen J, Ceci P: Electrostatic assembly of binary nanoparticle superlattices using protein cages. Nat Nanotechnol 2013, 8:52. [DOI] [PubMed] [Google Scholar]
- 96.Strable E, Johnson JE, Finn M: Natural nanochemical building blocks: icosahedral virus particles organized by attached oligonucleotides. Nano Lett 2004, 4:1385–1389. [Google Scholar]
- 97.Cigler P, Lytton-Jean AK, Anderson DG, Finn M, Park SY: DNA-controlled assembly of a NaTl lattice structure from gold nanoparticles and protein nanoparticles. Nat Mater 2010, 9:918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Korpi A, Ma C, Liu K, Nonappa, Herrmann A, Ikkala O, Kostiainen MA: Self-assembly of electrostatic cocrystals from supercharged fusion peptides and protein cages. ACS Macro Lett 2018, 7:318–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liu A, de Ruiter MV, Zhu W, Maassen SJ, Yang L, Cornelissen JJ: Compartmentalized thin films with customized functionality via interfacial cross-linking of protein cages. Adv Funct Mater 2018:1801574. [Google Scholar]
- 100.Yang L, Liu A, de Ruiter MV, Hommersom CA, Katsonis N, Jonkheijm P, Cornelissen JJ: Compartmentalized supramolecular hydrogels based on viral nanocages towards sophisticated cargo administration. Nanoscale 2018, 10:4123–4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Douglas T, Young M: Host-guest encapsulation of materials by assembled virus protein cages. Nature 1998, 393:152. [Google Scholar]
- 102.Pokorski JK, Breitenkamp K, Liepold LO, Qazi S, Finn M: Functional virus-based polymer-protein nanoparticles by atom transfer radical polymerization. J Am Chem Soc 2011, 133:9242–9245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Prasuhn DE Jr, Yeh RM, Obenaus A, Manchester M, Finn M: Viral MRI contrast agents: coordination of Gd by native virions and attachment of Gd complexes by azide-alkyne cycloaddition. Chem Commun 2007:1269–1271. [DOI] [PubMed] [Google Scholar]
- 104.Oh J-W, Chung W-J, Heo K, Jin H-E, Lee BY, Wang E, Zueger C, Wong W, Meyer J, Kim C et al. : Biomimetic virus-based colourimetric sensors. Nat Commun 2014, 5:3043. [DOI] [PubMed] [Google Scholar]
- 105.Lee JH, Fan B, Samdin TD, Monteiro DA, Desai MS, Scheideler O, Jin HE, Kim S, Lee SW: Phage-based structural color sensors and their pattern recognition sensing system. ACS Nano 2017, 11:3632–3641. [DOI] [PubMed] [Google Scholar]