Abstract
Identification of conserved proteins that act to establish the neuronal phenotype has relied predominantly on structural homologies of the underlying genes. In the case of the repressor element 1 silencing transcription factor (REST), a central player in blocking the neuronal phenotype in vertebrate non-neural tissue, the invertebrate homolog is absent, raising the possibility that distinct strategies are used to establish the CNS of invertebrates. Using a yeast two-hybrid screen designed specifically to identify functional analogs of REST, we show that Drosophila melanogaster uses a strategy that is functionally similar to, but appears to have evolved independently of, REST. The gene at the center of the strategy in flies encodes the repressor Tramtrack88 (Ttk88), a protein with no discernable homology to REST but that nonetheless is able to interact with the same transcriptional partners. Ttk88 uses the REST corepressor Drosophila CoREST to coordinately regulate a set of genes encoding the same neuronal hallmarks that are regulated by REST in vertebrates. Our findings indicate that repression is an important mechanism for regulating neuronal phenotype across phyla and suggest that co-option of a similar corepressor complex occurred to restrict expression of genes critical for neuronal function to a compartmentalized nervous system.
Keywords: CoREST, Tramtrack, REST, repression, CNS, corepressor
Introduction
Formation of the nervous system involves both intrinsic and extrinsic regulatory pathways (Edlund and Jessell, 1999). A family of Hox genes expressed in differentiating neurons exemplifies an intrinsic pathway that patterns the anteroposterior axis of the developing nervous system (Burke et al., 1995). In a complementary manner, an extrinsic silencing mechanism, targeted to the DNA by the vertebrate transcription factor repressor element 1 silencing transcription factor (REST) (Chong et al., 1995; Schoenherr and Anderson, 1995), helps restrict gene expression to neurons by blocking expression of neuronal-specific genes in non-neural cells of the embryo. REST recruits two corepressor complexes: a Sin3-histone deacetylase complex (HDAC) (Huang et al., 1999; Grimes et al., 2000; Roopra et al., 2000) and a REST corepressor (CoREST)-flavoprotein-histone deacetylase complex (Andres et al., 1999). In non-neuronal cells, this complex occupies and represses genes responsible for key neuronal functions, such as action potential propagation and neurotransmitter release and reception (Schoenherr et al., 1996; Ballas et al., 2001).
Genes involved in nervous system patterning and cell-fate decisions are highly conserved across phyla (Lowe et al., 2003). In contrast, no invertebrate studies have provided support for conservation of the REST-CoREST pathway. This may be in part because no obvious REST orthologs are present in invertebrate genomes. Recently, however, genetic studies in Caenorhabditis elegans revealed a role for CoREST in development, suggesting that a similar repressive or silencing mechanism might be operating in invertebrates (Eimer et al., 2002; Jarriault and Greenwald, 2002).
We have exploited the existence of Drosophila CoREST (dCoREST) to link mammalian and fly repressive mechanisms that regulate neuronal identity. By yeast two-hybrid screens, we isolated a single DNA-binding partner for dCoREST, Tramtrack88 (Ttk88), a known repressor that harbors no homology to REST and recognizes a distinct genetic element. The Ttk88 complex contains, in addition to dCoREST, a Drosophila flavoprotein homolog (dflavoprotein) and histone deacetylases present in the REST complex. Furthermore, Ttk88 occupies several of the same hallmark neuronal-specific genes in vivo, and knockdown of Ttk or dCoREST in S2 nonneuronal cells results in derepression of some of these genes. Our results suggest that recruitment of CoREST to neuronal-specific genes has been a successful strategy for restricting nervous systems to specialized compartments in the animal.
Materials and Methods
Yeast two-hybrid screening. A LexA-dCoREST [amino acids 64-273 dCoREST short splice variant (dCRS)] construct was used as bait to screen a Drosophila embryo Matchmaker cDNA library (Clontech, Palo Alto, CA). Screening was performed as described previously (Andres et al., 1999). Positive clones, of 107 screened, interacted specifically with CoREST as evidenced by mating assay.
Synthesis of double-stranded RNA. DNA templates were made by PCR as described by Kennerdell and Carthew (1998) with a T7 polymerase binding site at the 5′ end of each primer. The gene-specific primers minus their T7 binding sites were as follows: CoREST, forward (f)CATTCGCTCAGTTTTCTGACG and reverse (r)-CCACCGAAATGTACTCCTCC; green fluorescent protein (GFP), f-CACCATGGTGAGCAAGGGCG and r-GCGGCCGCTTTACTTGTACA; Ttk, f-AGTACAGCTTCCACCGTCACG and r-TGTCTTCATGATGGCTCACC.
Cell culture and transfections. S2 cells (obtained from the tissue-culture facility at Stony Brook; only this strain of S2s showed the dramatic knockdown of dCoREST) were grown in Schneider's Drosophila media (SDM) (Invitrogen, San Diego, CA). S2 cells were transfected using CellFectin (Invitrogen) according to manufacturer's recommendations. Transfection of double-stranded RNA (dsRNA) was adapted from Caplen et al. (2000). Cells were split to ∼70% confluency before transfection. Nucleic acids and lipid reagent were complexed as follows: 5 μg of dsRNA to 15 μl of CellFectin per 35 mm dish. Nucleic acid-lipid complexes diluted in SDM minus serum and antibiotics were added to cells and incubated overnight. An equal volume of SDM plus serum and antibiotic was added the following day.
Repression assays. S2 cells were cotransfected with 0.5 μg of an upstream activating sequence (UAS)-actin-β-galactosidase (LacZ) reporter gene and Gal4-fusion constructs at a 1:2 molar ratio of reporter to Gal4-fusion (2 μg of total DNA per 35 mm well was kept constant with pBluescript). A promoterless Renilla luciferase vector (pRL-null) (Promega, Madison, WI) was used as a transfection control. In 293 cells, the reporter was a thymidine kinase (Tk)-luciferase gene. Tk Renilla served as the transfection control plasmid (Chen et al., 1998). For assays to assess the effect of depleting CoREST on Ttk88 repression, a LexA reporter with five LexA binding sites interspersed with heat shock protein binding sites was cotransfected with a LexAttk88 fusion protein. Final DNA amounts were achieved by adjusting with pBluescript. Transfection efficiency was monitored using pRL-null (see above). Luciferase assays were performed using the Promega dual luciferase assay kit. LacZ assays were performed according to Promega with o-nitrophenyl β-d-galactopyranoside as the substrate.
Antibodies. Rabbits were immunized (Pocono Rabbit Farm, Canadensis, PA) with a glutathione S-transferase-dCoREST (amino acids 634-820) fusion protein, and serum was affinity purified. Polyclonal antibodies to Ttk69 and Ttk88 were prepared as described previously (Lehembre et al., 2000).
Coimmunoprecipitation and Western blot analysis. S2 whole-cell lysates were obtained by washing cell pellets twice in PBS and lysing for 30 min at 4°C in buffer [150 mm NaCl, 50 mm Tris, pH 8.5, 0.5 mm EDTA, 10% glycerol, 10 mm imidazole, 0.5% Triton X-100 supplemented with a protease inhibitor mixture (Sigma, St. Louis, MO), 50 μm DTT, and 500 μm PMSF]. After centrifugation (14,000 rpm, 15 min, 4°C), the supernatant [2 mg of total protein per immunoprecipitation (IP)] was precleared by incubating with IgG for 15 min followed by protein G agarose beads (60 min; Invitrogen) and incubated with antibody overnight at 4°C followed by a 2 hr incubation with protein G agarose beads. Bead-bound complexes were washed twice in IP buffer (50 mm Tris, pH 8, 150 mm NaCl, 0.1% Triton X-100, 5% glycerol), once in PBS and 0.1% NP-40, and once in PBS. Bead-bound complexes were resuspended in 10 μl of PBS and 30 μl of 2× sample buffer, boiled for 5 min, and resolved on an SDS-PAGE acrylamide gel for Western blotting analysis.
CoREST immunocytochemistry. Tissues from third instar larvae were fixed for 2 hr on ice in 4% formaldehyde, washed three times in PBS, and incubated in 100 μg/ml RNase A-PBS plus 0.01% Triton X-100 (PBT) at 37°C for 30 min. After washing three times in PBT, samples were incubated in a 1:5000 dilution of dCoREST antibody in BLOTTO (bovine lacto transfer optimizer; 5% milk powder in PBT) overnight at 4°C. Samples were then washed three times for 10 min and incubated for 1 hr at room temperature in the dark with a fluorescent secondary antibody (Alexa Fluor 488; Molecular Probes, Eugene, OR). After washing three times for 10 min in PBT and once in PBS, samples were incubated in a 1:100 dilution of phalloidin 594 (Molecular Probes), washed, and mounted in Vectashield (Vector Laboratories, Burlingame, CA) with propidium iodide. Images were collected on a Zeiss (Oberkochen, Germany) LSM510 laser-scanning microscope.
Immunostaining of polytene chromosomes. Immunostaining was performed essentially as described by Zink and Paro (1995). Briefly, salivary glands dissected from third instar larvae in 0.1% Triton X-100 in PBS were fixed in 1% Triton X-100 and 1.85% formaldehyde in PBS for 10 sec, followed by 1.85% formaldehyde and 50% acetic acid in PBS for 2 min. Squashed chromosomes were blocked (10% fetal calf serum and 0.1% Triton X-100 in PBS) and incubated with polyclonal antibodies (rat anti-TTK69, 1:200; rat anti-TTK88, 1:150; rabbit anti-dCoREST, 1:500) for 2 hr. Slides were washed twice in 200-300 mm NaCl and 0.1% Triton X-100 in PBS and once in 1% Triton X-100 in PBS and were then incubated with anti-rabbit 488 and anti-rat 594 secondary antibodies (Alexa Fluor; Molecular Probes) at a 1:250 dilution in blocking buffer for 2 hr. The wash steps were repeated, and DNA was stained in a 1 μg/ml solution of 4′,6-diamidino-2-phenylindole for 5 min before mounting in Fluoro-mount G (Southern Biotechnology, Birmingham, AL). Images were captured using a Nikon (Tokyo, Japan) Eclipse E800 microscope fitted with a Bio-Rad (Hercules, CA) Radiance Plus confocal unit.
Chromatin immunoprecipitation. Chromatin immunoprecipitation (ChIP) assays were performed as described previously (Battaglioli et al., 2002). Chromatin was cross-linked by adding 1% formaldehyde to cells for 10 min and then washed in cold PBS. DNA-protein complexes extracted using a whole-cell lysis buffer were sonicated in 600 μl of nuclear lysis buffer to obtain sheared chromatin in the range of 200-600 bp. For IP, 100 μl of sheared chromatin per IP was precleared and diluted ten times in ChIP dilution buffer for incubation overnight at 4°C with 5 μg of antibody. The next day, DNA-protein complexes collected using protein G beads (Invitrogen) were washed in a series of buffers (protocol from Upstate Biotechnology, Lake Placid, NY). Cross-linking was reversed by heating overnight at 65°C, and DNA was purified using Qiagen (Hilden, Germany) columns. Two microliters of purified chromatin were used as template for touchdown PCR. Primer sequences were as follows: dflavoprotein, f-GTGTGCACCCACAACAGATG and r-TGTGTACCTTTCTAGTGTCTTTGC; engrailed, f-AAGCAGCCACAGCAACAATA and r-CTCACTCTGCCCTGCTCTCT; embryonic lethal abnormal vision (ELAV), f-GGGAGGGTCTGGTCTTTTTC and r-GCTGGATCTGGTTCTGGAAG; β-tubulin, f-TTCTCTTTCGGCCATTATGTG and r-ACTTGGGCACAACAATTCTG; synapsin, f-AAAGTTTTCGAGCCAAGCTG and r-TTCTGTTTCTATTGTTTGGCATAAAG; nicotinic ACh receptor (nAChR), f-TTGGCAACGTGTAACCAAAA and r-GGACCATCGCGATACTGTCT; SCG10-like, f-TCTCTCTCCGCTCTCTTTGC and r-AGCGACTGACGACCACTTTT.
Quantitative PCR. RNA harvested from cells using Trizol (Invitrogen) was treated with DNase (RNase-free Turbo DNase; Ambion, Austin, TX). RNAs were reverse transcribed using either random hexamers or gene-specific primers with Superscript III (Invitrogen) at 50°C. cDNA was used as template in quantitative PCR (qPCR) using the Applied Biosystems (Foster City, CA) 7700 Sequence Detector and SYBR Green PCR master mix. To control for specific amplification, products from each primer set were visualized on agarose gels. Standard curves were generated for each primer set, and calculations were performed accordingly (see Applied Biosystems User Bulletin #2). The primer sets were as follows: glyceraldehyde-3-phosphate dehydrogenase (internal control), f AGGTGGTCTCCAACGCCTC and r-CATTGATGACCTTGGCCAGG; dflavoprotein, f-TCCGGTGTTGCTCGCTCTA and r-CATCCGTCACGCTCTCCAC; CoREST, f-CCAGTTCCACGGGAAGAGC and r-CTGGCTATGGATTTGTCTGGC; engrailed, f-TACAGCGATCGTCCCAGCTC and r-TCGTCGTTGGTCTTGTCCTTT; para, f-ACACGCAATCAATCAGTGGG and r-TCGAGCTTGTGATTGGTGTCC; nAChRβ, f-GGTGCTGGACCGCTTCTTT and r-ATTATGGCCAAGGTCCCGA; ELAV, f-CAACAAAGCCAAGTAGAGCGG and r-TGGGTTGTGGTTGCTGGTG; β-tubulin, f-TCGTTCACATCCAAGCTGGTC and r-TGCCATGCTCATCGGAGAT; SCG10, f-GAACGATCAATTGGAATGTCAAAA and r-TCGGCGGCGAACAATTT.
In situ hybridization. In situ hybridization was performed as by Klingler and Gergen (1993). Digoxigenin-labeled probes were synthesized from purified PCR product with a T7 binding site at the 5′ end for sense probe and at the 3′ end for antisense probe using T7 polymerase (Roche Products, Hertfordshire, UK). The following primers were used: dCoREST-common probe, f-CATTCGCTCAGTTTTCTGACG and r-CAACCGAAATGTACTCCTCC; dCRS-specific, f-AGATAATCGCCGTGCCTGC and r-AGATTATCGTCTGTGGCAGG. In situ hybridizations were developed for 2 hr using the dCoREST-common probe and for 2.5 hr using the dCRS-specific probe.
Results
Drosophila melanogaster has a conserved CoREST ortholog
The distinguishing motifs of CoREST proteins are an ELM2 (Egl-27 and MTA 1 homology 2) domain followed by two SANT (SWI3-ADA2-NCOR-TFIIIB) domains. Whereas sequenced mammalian genomes contain three genes coding for proteins with CoREST features, sequenced invertebrate genomes have only one. By DNA sequence, we identified a putative mammalian CoREST ortholog in Drosophila melanogaster. By sequencing expressed sequence tags, three splice variants of Drosophila CoREST were identified (Fig. 1A,C). One of these is a truncated protein, dCRS, containing only the ELM2 and SANT1 domain. The other two Drosophila splice variants contain the ELM2 and both SANT domains but differ in their inter-SANT spacing. An additional exon accounts for the increased inter-SANT spacing in the long form (dCRL), but otherwise these two proteins are identical (Fig. 1A,C). dCoREST and human CoREST (hCoREST) are most similar in the ELM2-SANT1 domains which are 66% identical and 89% similar (amino acids 92-234, dCoREST; amino acids 102-246, human CoREST). The SANT2 domain is also conserved with 43% identity and 84% similarity (amino acids 538-589, dCoREST; amino acids 380-431, hCoREST).
Figure 1.
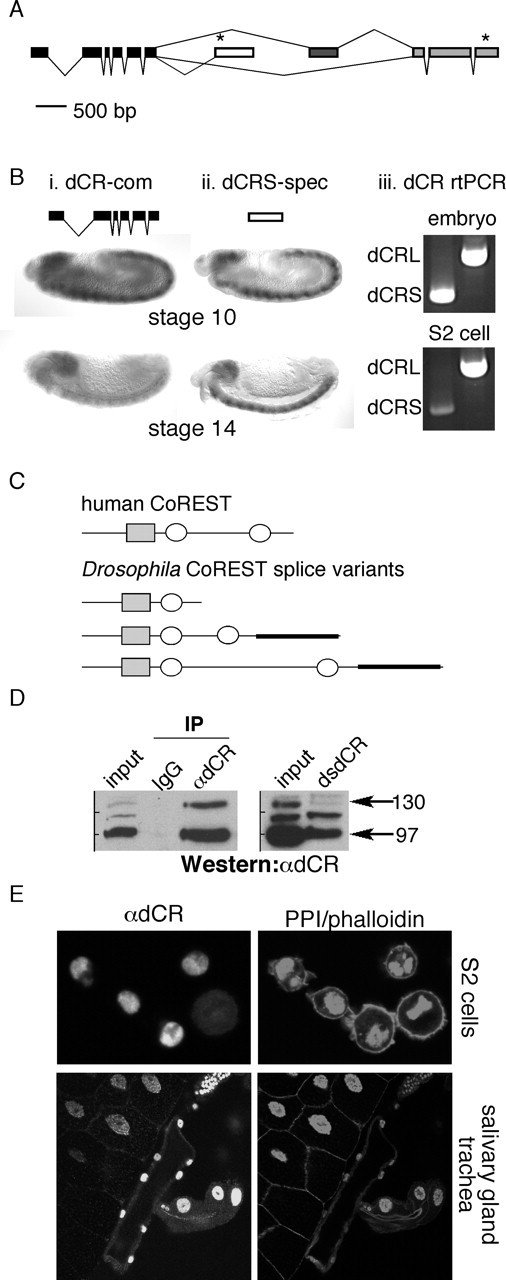
dCoREST is a functionally conserved corepressor protein. A, Intron-exon structure of dCoREST. Black indicates exons that are shared among all three splice variants. Exons unique to short and long forms of dCoREST are depicted in white and dark gray, respectively. Exons shared between the medium and long splice forms are depicted in light gray. Asterisks indicate stop codons. B, In situ hybridization and RT-PCR analysis of dCoREST splice variants. i, In situ hybridization with a common dCoREST probe (dCR-com) at indicated developmental stages. ii, In situ hybridization with a probe specific for dCRS (dCRS-spec) at indicated developmental stages. iii, RT-PCR (rtPCR) of dCRS and dCRL from entire embryos and S2 cells (non-neuronal). C, Schematic diagrams comparing deduced primary structures of human and dCoREST. Conserved ELM2 domains are represented by gray rectangles. White ovals depict SANT domains. The thick black line represents the epitope used to generate dCoREST antibody. The short, medium, and long forms of dCoREST are 273, 590, and 824 aa, respectively. D, Left, Immunoprecipitation analysis of dCoREST antibody (αdCR). S2 cells were immunoprecipitated with control antibody (IgG) or αdCR, and the Western blot was probed with αdCR. Right, S2 cells were transfected with dsdCR, and the Western blot was probed with αdCR. Arrows indicate medium (97 kDa) and long (130 kDa) forms of dCoREST protein. E, Immunohistochemical analysis of dCoREST expression in S2 cells and salivary gland and trachea of third instar larvae. Cells and tissues are counterstained with propidium iodide (PPI) (nuclei) and rhodamin-phalloidin (cellular periphery).
Drosophila CoREST is expressed ubiquitously in the embryo (Fig. 1Bi). In contrast, expression of dCRS is highly enriched in the nervous system (Fig. 1Bii). Reverse transcriptase (RT)-PCR from late-stage embryos (with nervous system) and S2 cells (nonneuronal) using primers specific for the long and short forms of dCoREST supports the more restricted expression of dCRS compared with dCRL (Fig. 1Biii).
To study dCoREST biochemically, an antibody was generated that recognizes both the medium (∼97 kDa) and long (∼130 kDa) forms of dCoREST (Fig. 1D). In immunoprecipitation, the medium and long forms of dCoREST are specifically enriched (Fig. 1D). Furthermore, dsRNA for CoREST specifically abolishes these two bands (Fig. 1D). The band at 116 kDa is a background band. The dCoREST antibody was used to examine the distribution of dCoREST in tissue. Like hCoREST, dCoREST has a nuclear localization and is expressed in all of the tissues examined (Fig. 1E) including both glia and neurons of the CNS (data not shown).
Drosophila CoREST interacts with Tramtrack, a repressor of neuronal phenotype
To identify a DNA-binding partner for dCoREST, a yeast two-hybrid screen of an embryonic cDNA fusion protein library was performed using the ELM2-SANT1 domain of dCoREST as bait. This portion of dCoREST corresponds to the REST interaction domain of human CoREST (Ballas et al., 2001). From 119 interacting clones, we isolated four classes of proteins. Only one of these, represented by five clones, was a transcription factor, the transcriptional repressor Tramtrack88. The Ttk88-dCoREST interaction was confirmed by reciprocal coimmunoprecipitations (Fig. 2A).
Figure 2.
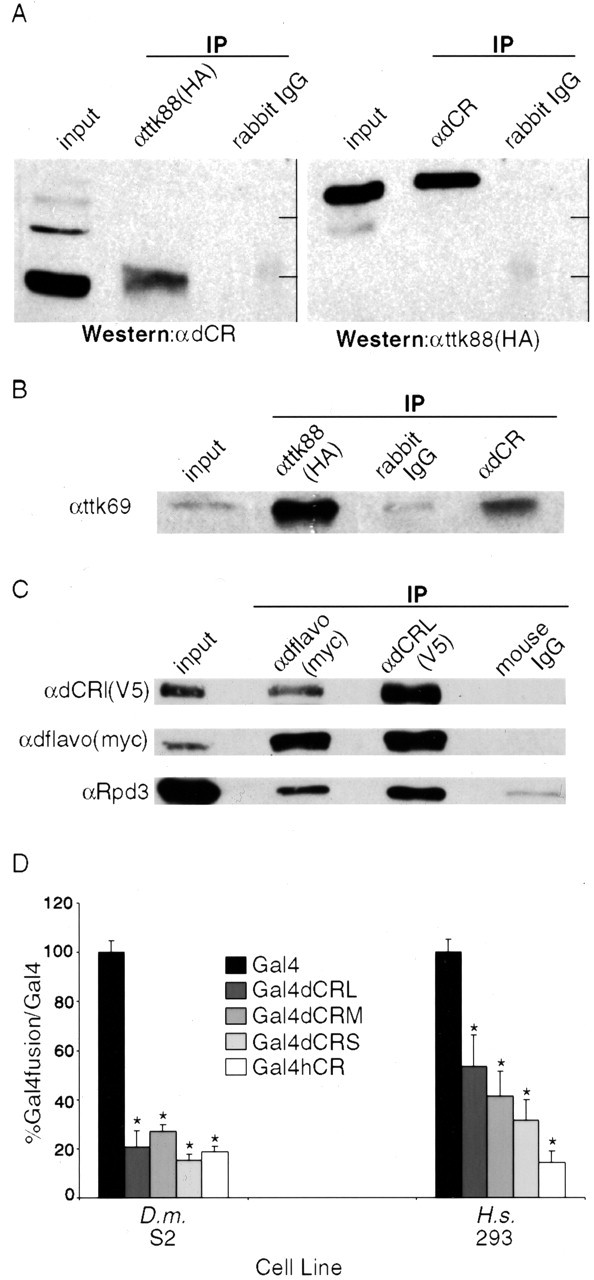
dCoREST is part of a repression complex and interacts with Tramtrack. A, Endogenous dCoREST interacts with Ttk88. Protein extracts from S2 cells transfected with HA-tagged ttk88 [ttk88(HA)] were immunoprecipitated with control antibody (IgG), αHA, and αdCR. The Western blot was cut in half; one side (left) was probed with αdCR, and the other side (right) was probed with αHA. B, dCoREST and Ttk88 are in complexes with Ttk69. Protein extracts from S2 cells transfected with HA-tagged ttk88 were immunoprecipitated with the indicated anti-bodies, and the blot was probed with Ttk69 antibody. C, dCoREST is in complexes with two proteins that also associate with human CoREST. Protein extracts from S2 cells transfected with dflavoprotein [dflavo(myc)] and dCoREST [dCRL(V5)] were immunoprecipitated with the indicated antibodies, and Western blots were probed with αV5 (top), αmyc (middle), and αRpd3 (bottom). D, dCoREST is a repressor in non-neuronal cells. Repressor assays were conducted on cell lysates from D. melanogaster (D.m.) S2 cells (left) and human [Homo sapiens (H.s.)] 293 cells (right) transfected with Gal4-CoREST fusion cDNAs (Gal4dCRL, long form of dCoREST; Gal4dCRM, medium form of dCoREST; Gal4dCRS, short form of dCoREST; hCR, human CoREST). The reporter gene contained tandem UAS sites in a promotor driving LacZ expression in S2 cells and luciferase expression in 293 cells. Reporter gene activity is expressed as a ratio of Gal4-fusion to Gal4 (S2, n = 4; 293, n = 8; *p < 0.005).
The ttk gene has two splice variants that share an N-terminal bric-a-brac-Tramtrack-Broad complex (BTB) domain but differ in their C-terminal DNA binding domains (Harrison and Travers, 1990; Read and Manley, 1992). By yeast two-hybrid assay, all three splice variants of dCoREST interacted directly with the unique C terminus of the Ttk88 splice variant (amino acids 432-811) (Fig. 3A). No Ttk69 clones were recovered in our screens, and the full-length Ttk69 clone did not interact with any of the splice variants of dCoREST in yeast (Fig. 3A). However, additional experiments demonstrate that the Ttk69 splice variant can form a complex with dCoREST and Ttk88 (Fig. 2B).
Figure 3.
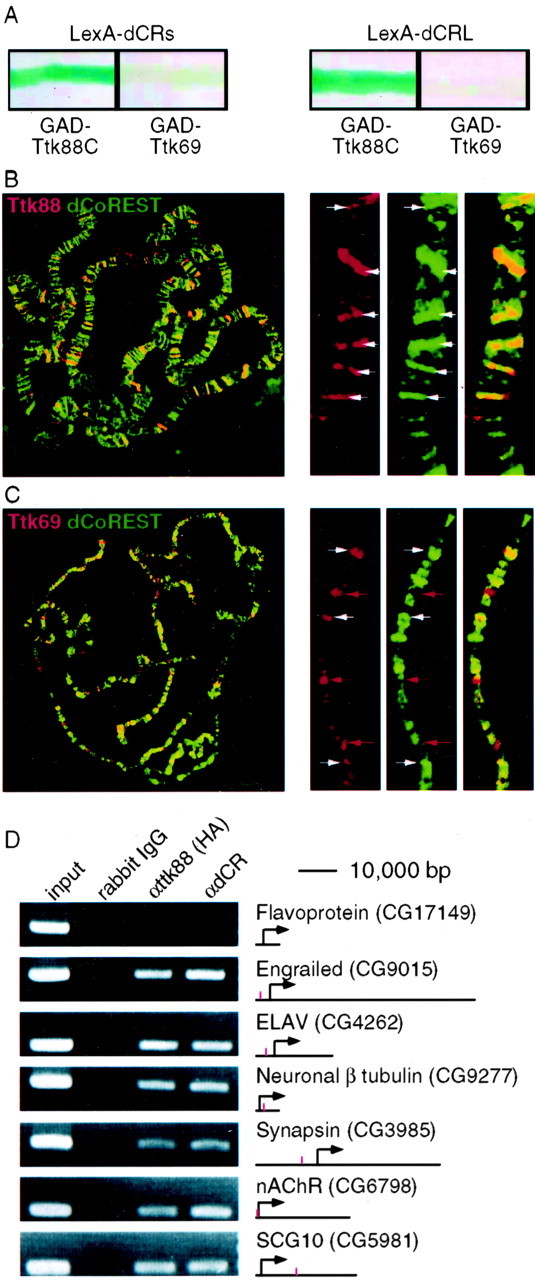
dCoREST and Ttk88 colocalize in vivo on chromatin. A, Yeast two-hybrid assay showing direct interaction between dCoREST and Ttk88 but not Ttk69. The LexA DNA binding domain was fused to either the short (LexA-dCRS, left) or the long (LexA-dCRL, right) forms of dCoREST. These fusion cDNAs were cotransformed into yeast with the Gal4 activation domain (GAD), fused to either the C terminus (amino acids 432-811) of Ttk88 (GAD-Ttk88C) or to the full-length Ttk69 (GAD-Ttk69). Blue indicates LacZ activity. B, C, Immunohistochemical analysis of polytene chromosomes stained with either Ttk88 (B, red) or Ttk69 (C, red) and dCoREST (green) antibodies. The left panel displays the staining pattern in the entire genome. The three panels to the right show αTtk88 (B) or Ttk69 (C), αdCR (middle), and the overlay of the two staining patterns (far right). White arrows mark bands that are labeled by both antibodies, and red arrows mark bands recognized by Ttk69 but not CoREST antibodies. D, Chromatin immunoprecipitation analysis of six neuronal genes and one control using αHA and αdCR antibodies.
Drosophila CoREST occurs in a corepressor complex similar to that of vertebrate CoREST, containing both a Drosophila flavoprotein and dRpd3, the Drosophila class 1 and 2 HDACs (Fig. 2C). In transient repressor assays in both Drosophila S2 cells and human 293 cells (Fig. 2D), all three dCoREST proteins expressed as Gal4-fusion proteins effectively repressed the UAS-reporter gene, demonstrating that dCoREST can function as a corepressor.
Polytene chromosomes labeled with Ttk and CoREST antibodies revealed many more CoREST sites than Ttk88 sites (Fig. 3B,C). Moreover, whereas most Ttk88 sites were also bound by CoREST (Fig. 3B), only approximately half of the Ttk69 sites also bound CoREST (Fig. 3C). The general pattern of dCoREST localization relative to bands and interbands is similar to that demonstrated previously for dRpd3 (Pile and Wassarman, 2002), suggesting that it participates in many different complexes.
Tramtrack88 targets dCoREST to neuronal-specific genes
To determine whether Tramtrack regulates neuronal phenotype in a manner similar to REST in mammals, putative Drosophila homologs of genes that are regulated by REST were tested for regulation by Ttk. Potential Ttk88 binding sites AGGGC/TGG (Read and Manley, 1992) were identified in neuronal β-tubulin (CG9277), synapsin (CG3985), nAChRβ (CG6798), SCG10-like (CG5981), and para (CG9907) genes (Fig. 3C). Synapsin, nAChRβ, and para also contained a Ttk69 recognition sequence (ttk69-GGTCCTGC).
We compared D. melanogaster and Drosophila pseudoobscura Ttk88 recognition sequences for conservation in the same set of neuronal genes. These two fly species are sufficiently divergent that conservation of promoter sequences implies functional significance. D. melanogaster engrailed, a known Ttk88 target, has 14 Ttk88 sites upstream of the transcription start site compared with 16 in D. pseudoobscura. The exact location of these sites is distinct as is their orientation (sense or antisense) relative to the gene, but the number and distribution are essentially conserved. This was also true for the para and β-tubulin genes. D. melanogaster ELAV has six Ttk88 sites 5′ of the transcription start site compared with three in D. pseudoobscura, and D. pseudoobscura synapsin has more sites than D. melanogaster synapsin, but the distribution of a subset of the sites in the two genes is similar. D. melanogaster nAChRβ is not accessible in the current assembly of D. pseudoobscura, and SCG10-like has a completely different genomic organization in D. pseudoobscura, so comparisons could not be drawn for these two genes. These findings suggest that for many of the neuronal genes, the presence of Ttk88 sites proximal to the gene is not a result of random chance but has regulatory significance (supplemental Fig. 1, available at www.jneurosci.org/cgi/content/full/24/32/7186/DC1).
Chromatin immunoprecipitations from S2 cells transfected with hemagglutinin (HA)-tagged Ttk88 were performed (Fig. 3D). Engrailed served as a positive control for the experiment, because it is regulated directly by Ttk88 (Read and Manley, 1992; Xiong and Montell, 1993). The upstream sequence of the flavoprotein served as a negative control. HA-tagged Ttk88 occupied the ELAV, β-tubulin, synapsin, nAChRβ, and SCG10 genes (Fig. 3C). The para gene was not included in this data set, because we could not amplify PCR product from this region even in control (nonprecipitated) chromatin.
Tramtrack and dCoREST are necessary to repress neuronal-specific genes in S2 cells
Double-stranded RNA targeting both Ttk69 and Ttk88 was injected into embryos. Microinjection of the dsRNA recapitulated the Ttk phenotype of extra neurons in the peripheral nervous system lineage (Kennerdell and Carthew, 1998) (data not shown). When introduced into S2 cells, the Ttk dsRNA resulted in a rapid (within 24 hr) knockdown of Ttk69 and Ttk88 protein to undetectable levels (Fig. 4A). The consequences of knocking down Ttk were examined by qPCR on Ttk88 targets. Both nAChRβ and SCG10-like were significantly derepressed (p < 0.005) in cells treated with ttk69 and ttk88 dsRNA (n = 4) versus GFP dsRNA (dsGFP) (n = 4) for 22 hr.
Figure 4.
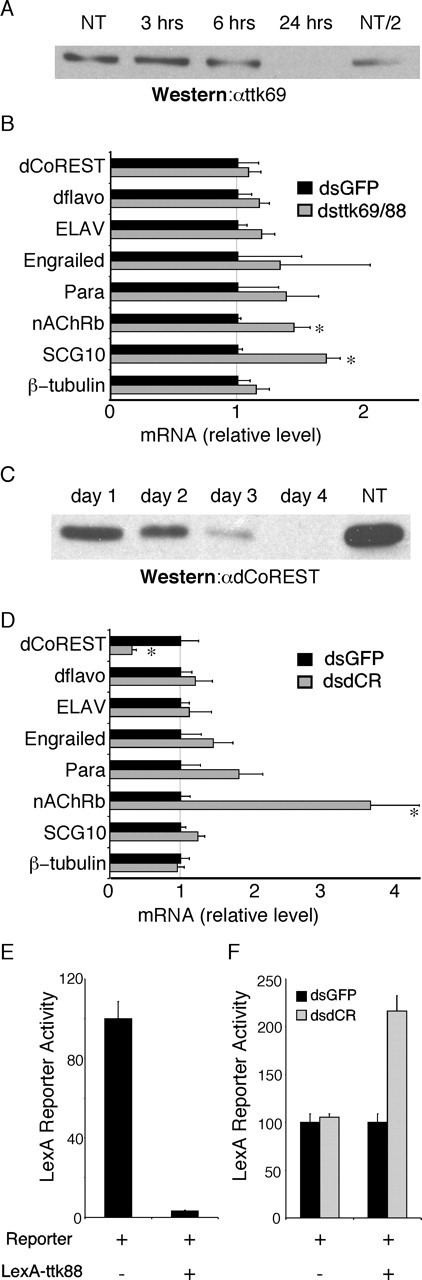
dCoREST and Ttk regulate a suite of neuronal genes. A, A Western blot analysis of the time course of ttk69/88 dsRNA knockdown of Ttk69 using an αTtk69 antibody. NT indicates not transfected and NT/2 represents one-half of the protein that was loaded in the NT lane. B, Quantitative PCR analysis of six neuronal genes, a control, and dCoREST in S2 cells treated for 24 hr with either Ttk dsRNA (dsttk69/88; n = 4) or dsGFP (n = 4). Values are the relative gene expression in dsRNA-transfected S2 cells compared with dsGFP-transfected S2 cells (data are presented as mean ± SD; *p < 0.005). C, A Western blot analysis of the time course of dsRNA knockdown of dCoREST probed with dCoREST antibody. D, Quantitative PCR analysis of six neuronal genes, a control, and dCoREST in S2 cells 4 d after transfection with either dsdCR (n = 4) or dsGFP (n = 4). Values are the relative gene expression in dsRNA-transfected S2 cells compared with dsGFP-transfected S2 cells (data are presented as mean ± SD; *p < 0.005). E, LacZ reporter assay of S2 cells cotransfected with either LexA-reporter-pBluescript or LexA-reporter-LexAttk88 fusion protein. Values are normalized to LexA-reporter-pBluescript. F, S2 cells were transfected with either dsGFP or dsdCR and cultured for 4 d. These cells were then retransfected with either LexA-reporter-pBluescript or LexA-reporter-LexAttk88 fusion protein, and LacZ assays were performed the following day. Values are normalized to GFP dsRNA treatment.
To confirm Ttk88 target genes in the embryo, Ttk88 was expressed in differentiating neurons using an ELAVGal4 driver. The neuronal driver was chosen to rule out effects of Ttk88 on cell-fate transformations that occur when Ttk is expressed in neuronal precursors. No effect was observed on neuronal morphology or on the expression levels of Ttk target genes, although expression of Ttk88 was greatly elevated (data not shown).
Double-stranded RNA was also generated against the CoREST gene. The time course for knockdown of endogenous dCoREST was surprisingly slow, taking 4 d after transfection for disappearance of the protein (Fig. 4C). In contrast, when measured by qPCR, dCoREST RNA levels reached steady state by 24 hr after transfection, suggesting that the slow time course for protein knockdown is attributable to the slow turnover of the dCoREST protein.
The slow turnover of dCoREST precluded dsRNA studies in embryos, but target gene expression levels were measured by qPCR in S2 cells. Only nAChRβ was significantly derepressed (p < 0.005) in cells transfected with dCoREST dsRNA (dsdCR) (n = 4) versus GFP dsRNA (n = 4) for 4 d (Fig. 4D). nAChRβ was also derepressed by ttk dsRNA, consistent with dCoREST functioning as an integral component of a Ttk88 repressor complex.
To further establish a direct functional link between Ttk88 and CoREST, we used a LexA reporter construct that was effectively repressed by a LexAttk88 fusion protein (Fig. 4E). Reporter activity in cells that had been depleted of CoREST using dsdCR was compared with that in cells transfected with dsGFP for the same period of time. Although reporter activity was the same in dsdCR- and dsGFP-treated cells, LexAttk88 repression was twofold less in dsdCR cells compared with dsGFP cells (Fig. 4F).
Discussion
Since its initial discovery as a REST corepressor (Andres et al., 1999), hCoREST has been isolated multiple times biochemically as a core complex consisting of CoREST, a flavoprotein that resembles polyamine oxidases, and HDACs 1 and 2 (Humphry et al., 2001; You et al., 2001; Hakimi et al., 2002; Shi et al., 2003). Unlike the case for REST, the components of the core CoREST-flavoprotein-HDAC complex have been structurally conserved across phyla, including C. elegans and D. melanogaster. In worms, a screen for genes involved in the presenilin-Notch pathway recovered CoREST and the flavoprotein (Eimer et al., 2002; Jarriault and Greenwald, 2002). The involvement of CoREST in the Drosophila Notch pathway is also supported by genetic studies (J. E. Dallman and G. Mandel, unpublished observations), suggesting a role for CoREST in nervous system development.
Drosophila has a single CoREST gene that functions as a repressor in transient assays and is in complexes with histone deacetylases in a non-neuronal cell line. Additionally, dCoREST contains a domain that only differs from the REST-interaction domain in hCoREST by 4 aa. This suggested the existence of a REST-like protein in Drosophila. The suggestion was borne out by identification of the repressor Ttk88 in two-hybrid screens with dCoREST. Like REST, Ttk88 was known, through genetic experiments, to be a repressor of neuronal cell fate (Xiong and Montell, 1993; Guo et al., 1995).
The results detailed herein indicate that Ttk88 interacts with dCoREST to repress a set of neuronal-specific genes in nonneuronal cells. Chromatin immunoprecipitation studies showed an interaction of Ttk88 at the previously identified consensus site AGGGC/TGG on several neuronal-specific genes, including ELAV, synapsin, and nAChRβ. Many of these Ttk88 sites are conserved in a distantly related fly species, D. pseudoobscura, implying that the Ttk88 sites are functionally relevant. One of the neuronal genes, nAChRβ, was derepressed by dsRNA for both Tramtrack and CoREST, a result consistent with Tramtrack and CoREST functioning as partners. There was also a coincidence of Ttk88 and CoREST on polytene chromosomes, suggesting that CoREST is a mediator of Ttk88 repression in the animal. Finally, data showing that depletion of CoREST lessens the repressor activity of Ttk88 on a Ttk88 reporter gene establish a functional link between Tramtrack88 and CoREST.
The ttk gene encodes two splice variants, Ttk69 and Ttk88. Neither protein is expressed in neurons (Harrison and Travers, 1990), but both proteins can block neuronal development (Guo et al., 1995; Giesen et al., 1997; Baonza et al., 2002). In addition, Ttk69 but not Ttk88 expression can arrest cells in the G2 stage of the cell cycle (Baonza et al., 2002). Ttk69 can recruit both the NuRD (nucleosome remodeling deacetylase) and the CtBP (C-terminus-binding protein) corepressors (Wen et al., 2000; Murawsky et al., 2001) and has the capacity to form homotetramers through the N-terminal BTB domain it shares with Ttk88 (Badenhorst et al., 2002). Despite the fact that dCoREST did not interact with Ttk69 in yeast, both were detected in coimmunoprecipitation assays in the non-neuronal S2 cells, and there was overlap between Ttk88 and Ttk69 sites on polytene chromosomes (data not shown). The pattern was not completely overlapping, however, indicating that Ttk88 and Ttk69 may also be part of distinct complexes. CoREST was associated with genes on the polytene chromosome independently of either Ttk88 or Ttk69. These data, together with the in situ hybridization results indicating that dCoREST and its splice variants are expressed in neurons whereas Ttk is not, suggest that CoREST orchestrates repression within the nervous system. Similar conclusions have been reached regarding hCoREST, although specific targets have not yet been identified.
CoREST complexes have been associated with a spreading of silencing that, in certain mammalian cell lines, appears to encompass a large cluster of neuronal-specific genes (Lunyak et al., 2002). In Drosophila, both para and nAChRβ are each in a cluster of at least five and three neuronal genes, respectively (Sawruk et al., 1990; Hong and Ganetzky, 1996). We were unable to test whether removal of Ttk resulted in coordinate derepression of the neuronal clusters in flies because most of the embryos homozygous for severe ttk alleles (Giesen et al., 1997) died before nervous system formation, similar to the situation in REST knock-out mice. With respect to gain-of-function experiments, overexpression of Ttk early in eye disc development and in sensory organ precursors suppresses the formation of photoreceptors and peripheral sensory neurons, respectively (Guo et al., 1995; Baonza et al., 2002). However, our results, taken together with previous studies (Ramaekers et al., 1997; Paquette et al., 2000; Badenhorst, 2001; Ballas et al., 2001), suggest that developmental context is a critical factor in determining the outcome of gain-of-function or loss-of-function manipulations of REST or Ttk88 in vivo (Armisen et al., 2002).
Both Ttk and REST are critical to the creation of a viable organism. However, orthologs for Ttk are present only in the fly lineage, whereas REST is identified only in the vertebrate lineage (supplemental Fig. 2, available at www.jneurosci.org/cgi/content/full/24/32/7186/DC1). Although we cannot exclude the possibility that Rest and Ttk88 diverged from a common zinc finger protein, an alternate explanation is that Ttk88 in flies has functionally replaced REST in vertebrates to limit the extent of the neural domain, as suggested by Giesen et al. (1997). The replacement during evolution of proteins of similar function but with different structures (nonorthologous gene displacement) is well documented in bacteria (Koonin et al., 1996; Galperin et al., 1998) and is thought to occur in as many as 10% of bacterial genes (Galperin et al., 1998). There are relatively few additional examples of nonorthologous gene replacement in metazoans. These findings suggest that reliance on genomic searches will underestimate conservation across species for some important developmental mechanisms.
CoREST is conserved in all animals with a nervous system, including protochordates. However, the flavoprotein and HDAC predate CoREST. Thus, we propose that CoREST co-opted the pre-existing repressor complex to regulate newly evolving repressor proteins targeting neuronal-specific genes. Tests of these ideas will require a better understanding of CoREST function in other developmental pathways and organisms.
Footnotes
This work was supported by National Institutes of Health Grants NS22518 (G.M.) and NS10700 (J.E.D.) and by a Medical Research Council Scholarship in Training Methods (A.B.). G.M. is an investigator at the Howard Hughes Medical Institute. We thank Drs. James Baker, Hilde Janssens, Paul Brehm, Jason Hodin, and Russell Doolittle for insightful discussions during the preparation of this manuscript. We also thank Dr. Wheeler for pGAD-Ttk69, Dr. Carthew for HA-tagged ttk88, Dr. Coury for luciferase constructs, and the Kernan laboratory for fly expertise.
Correspondence should be addressed to Gail Mandel, Howard Hughes Medical Institute, Department of Neurobiology and Behavior, State University of New York, Stony Brook, Stony Brook, NY 11794. E-mail: gmandel@notes.cc.sunysb.edu.
Copyright © 2004 Society for Neuroscience 0270-6474/04/247186-08$15.00/0
References
- Andres ME, Burger C, Peral-Rubio MJ, Battaglioli E, Anderson ME, Grimes J, Dallman J, Ballas N, Mandel G (1999) CoREST: a functional corepressor required for regulation of neural-specific gene expression. Proc Natl Acad Sci USA 96: 9873-9878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armisen R, Fuentes R, Olguin P, Cabrejos ME, Kukuljan M (2002) Repressor element-1 silencing transcription/neuron-restrictive silencer factor is required for neural sodium channel expression during development of Xenopus J Neurosci 22: 8347-8351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badenhorst P (2001) Tramtrack controls glial number and identity in the Drosophila embryonic CNS. Development 128: 4093-4101. [DOI] [PubMed] [Google Scholar]
- Badenhorst P, Finch JT, Travers AA (2002) Tramtrack co-operates to prevent inappropriate neural development in Drosophila Mech Dev 117: 87-101. [DOI] [PubMed] [Google Scholar]
- Ballas N, Battaglioli E, Atouf F, Andres ME, Chenoweth J, Anderson ME, Burger C, Moniwa M, Davie JR, Bowers WJ, Federoff HJ, Rose DW, Rosenfeld MG, Brehm P, Mandel G (2001) Regulation of neuronal traits by a novel transcriptional complex. Neuron 31: 353-365. [DOI] [PubMed] [Google Scholar]
- Baonza A, Murawsky CM, Travers AA, Freeman M (2002) Pointed and Tramtrack69 establish an EGFR-dependent transcriptional switch to regulate mitosis. Nat Cell Biol 4: 976-980. [DOI] [PubMed] [Google Scholar]
- Battaglioli E, Andres ME, Rose DW, Chenoweth JG, Rosenfeld MG, Anderson ME, Mandel G (2002) REST repression of neuronal genes requires components of the hSWI-SNF complex. J Biol Chem 277: 41038-41045. [DOI] [PubMed] [Google Scholar]
- Burke AC, Nelson CE, Morgan BA, Tabin C (1995) Hox genes and the evolution of vertebrate axial morphology. Development 121: 333-346. [DOI] [PubMed] [Google Scholar]
- Caplen NJ, Fleenor J, Fire A, Morgan RA (2000) dsRNA-mediated gene silencing in cultured Drosophila cells: a tissue culture model for the analysis of RNA interference. Gene 252: 95-105. [DOI] [PubMed] [Google Scholar]
- Chen G, Nguyen PH, Courey AJ (1998) A role for Groucho tetramerization in transcriptional repression. Mol Cell Biol 18: 7259-7268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong JA, Tapia-Ramirez J, Kim S, Toledo-Aral JJ, Zheng Y, Boutros MC, Altshuller YM, Frohman MA, Kraner SD, Mandel G (1995) REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell 80: 949-957. [DOI] [PubMed] [Google Scholar]
- Edlund T, Jessell TM (1999) Progression from extrinsic to intrinsic signaling in cell fate specification: a view from the nervous system. Cell 96: 211-224. [DOI] [PubMed] [Google Scholar]
- Eimer S, Lakowski B, Donhauser R, Baumeister R (2002) Loss of spr-5 bypasses the requirement for the C. elegans presenilin sel-12 by derepressing hop-1. EMBO J 21: 5787-5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY, Walker DR, Koonin EV (1998) Analogous enzymes: independent inventions in enzyme evolution. Genome Res 8: 779-790. [DOI] [PubMed] [Google Scholar]
- Giesen K, Hummel T, Stollewerk A, Harrison S, Travers A, Klambt C (1997) Glial development in the Drosophila CNS requires concomitant activation of glial and repression of neuronal differentiation genes. Development 124: 2307-2316. [DOI] [PubMed] [Google Scholar]
- Grimes JA, Nielsen SJ, Battaglioli E, Miska EA, Speh JC, Berry DL, Atouf F, Holdener BC, Mandel G, Kouzarides T (2000) The co-repressor mSin3A is a functional component of the REST-CoREST repressor complex. J Biol Chem 275: 9461-9467. [DOI] [PubMed] [Google Scholar]
- Guo M, Bier E, Jan LY, Jan YN (1995) Tramtrack acts downstream of numb to specify distinct daughter cell fates during asymmetric cell divisions in the Drosophila PNS. Neuron 14: 913-925. [DOI] [PubMed] [Google Scholar]
- Hakimi MA, Bochar DA, Chenowith J, Lane WS, Mandel G, Sheikhattar R (2002) A core-BRAF35 complex containing histone deacetylase mediates repression of neuronal-specific genes. Proc Natl Acad Sci USA 99: 7420-7425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison SD, Travers AA (1990) The tramtrack gene encodes a Drosophila finger protein that interacts with the ftz transcriptional regulatory region and shows a novel embryonic expression pattern. EMBO J 9: 207-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong CS, Ganetzky B (1996) Molecular characterization of neurally expressing genes in the para sodium channel gene cluster of Drosophila Genetics 142: 879-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Myers SJ, Dingledine R (1999) Transcriptional repression by REST: recruitment of Sin3A and histone deacetylase to neuronal genes. Nat Neurosci 2: 867-872. [DOI] [PubMed] [Google Scholar]
- Humphry GW, Wang Y, Russanova VR, Hirai T, Qin J, Nakatami Y, Howard BH (2001) Stable histone deacetylase complexes distinguished by the presence of SANT domain proteins CoREST/kiaa0071 and Mta-L1. J Biol Chem 276: 6817-6824. [DOI] [PubMed] [Google Scholar]
- Jarriault S, Greenwald I (2002) Suppressors of the egg-laying defective phenotype of sel-12 presinilin mutants implicate the CoREST corepressor complex in LIN-12/Notch signaling in C. elegans Genes Dev 16: 2713-2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennerdell JR, Carthew RW (1998) Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell 95: 1017-1026. [DOI] [PubMed] [Google Scholar]
- Klingler M, Gergen JP (1993) Regulation of runt transcription by Drosophila segmentation genes. Mech Dev 43: 3-19. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Mushegian AR, Bork P (1996) Non-orthologous gene displacement. Trends Genet 12: 334-336. [PubMed] [Google Scholar]
- Lehembre F, Badenhorst P, Muller S, Travers A, Schweisguth F, Dejean A (2000) Covalent modification of the transcriptional repressor tramtrack by the ubiquitin-related protein Smt3 in Drosophila flies. Mol Cell Biol 20: 1072-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe CJ, Wu M, Salic A, Evans L, Lander E, Stange-Thomann N, Gruber CE, Gerhart J, Kirschner M (2003) Anteroposterior patterning in hemichordates and the origins of the chordate nervous system. Cell 113: 853-865. [DOI] [PubMed] [Google Scholar]
- Lunyak VV, Burgess R, Prefontaine GG, Nelson C, Sze SH, Chenoweth J, Schwartz P, Pevzner PA, Glass C, Mandel G, Rosenfeld MG (2002) Corepressor-dependent silencing of chromosomal regions encoding neuronal genes. Science 298: 1747-1752. [DOI] [PubMed] [Google Scholar]
- Murawsky CM, Brehm A, Badenhorst P, Lowe N, Becker PB, Travers AA (2001) Tramtrack69 interacts with the dMi-2 subunit of the Drosophila NuRD chromatin remodelling complex. EMBO Rep 2: 1089-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquette AJ, Perez SE, Anderson DJ (2000) Constitutive expression of the neuron-restrictive silencer factor (NRSF)/REST in differentiating neurons disrupts neuronal gene expression and causes axon pathfinding errors in vivo Proc Natl Acad Sci USA 97: 12318-12323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pile LA, Wassarman DA (2002) Localizing transcription factors on chromatin by immunofluorescence. Methods 26: 3-9. [DOI] [PubMed] [Google Scholar]
- Ramaekers G, Usui K, Usui-Ishihara A, Ramaekers A, Ledent V, Ghysen A, Dambly-Chaudiere C (1997) Lineage and fate in Drosophila: role of the gene tramtrack in sense organ development. Dev Genes Evol 207: 97-106. [DOI] [PubMed] [Google Scholar]
- Read D, Manley JL (1992) Alternatively spliced transcripts of the Drosophila tramtrack gene encode zinc finger proteins with distinct DNA binding specificities. EMBO J 11: 1035-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roopra A, Sharling L, Wood IC, Briggs T, Bachfischer U, Paquette AJ, Buckley NJ (2000) Transcriptional repression by neuron-restrictive silencer factor is mediated via the Sin3-histone deacetylase complex. Mol Cell Biol 20: 2147-2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawruk E, Udri C, Betz H, Schmitt B (1990) SBD, a novel structural subunit of the Drosophila nicotinic acetylcholine receptor, shares its genomic localization with two alpha-subunits. FEBS Lett 273: 177-181. [DOI] [PubMed] [Google Scholar]
- Schoenherr CJ, Anderson DJ (1995) The neuron-restrictive silencer factor (NRSF): a coordinate repressor of multiple neuron-specific genes. Science 267: 1360-1363. [DOI] [PubMed] [Google Scholar]
- Schoenherr CJ, Paquette AJ, Anderson DJ (1996) Identification of potential target genes for the neuron-restrictive silencer factor. Proc Natl Acad Sci USA 93: 9881-9886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Sawada J, Sui G, Affar el B, Whetstine JR, Lan F, Ogawa H, Luke MP, Nakatani Y, Shi Y (2003) Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature 422: 735-738. [DOI] [PubMed] [Google Scholar]
- Wen Y, Nguyen D, Li Y, Lai ZC (2000) The N-terminal BTB/POZ domain and C-terminal sequences are essential for Tramtrack69 to specify cell fate in the developing Drosophila eye. Genetics 156: 195-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong WC, Montell C (1993) Tramtrack is a transcriptional repressor required for cell fate determination in the Drosophila eye. Genes Dev 7: 1085-1096. [DOI] [PubMed] [Google Scholar]
- You A, Tong JK, Grozinger CM, Schreiber SL (2001) CoREST is an integral component of the CoREST-human histone deacetylase complex. Proc Natl Acad Sci USA 98: 1454-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zink D, Paro R (1995) Drosophila Polycomb-group regulated chromatin inhibits the accessibility of a trans-activator to its target DNA. EMBO J 14: 5660-5671. [DOI] [PMC free article] [PubMed] [Google Scholar]


