Abstract
Brain-derived neurotrophic factor (BDNF), a member of the neurotrophin family, modulates neuronal survival, differentiation, and synaptic function. Reduced BDNF expression in the cortex caused by mutation of the huntingtin gene has been suggested to play a role in the striatal degeneration observed in Huntington's disease. BDNF expression rises dramatically in the cortex during the first few weeks of postnatal life in mice. Previously, it has been impossible to study the specific long-term effects of BDNF absence on CNS structures because of the early postnatal lethality of BDNF-/- mice. Mice harboring a floxed BDNF gene were bred with Emx1IREScre/+ mice to generate Emx-BDNFKO mice that lack cortical BDNF but are viable. Adult Emx-BDNFKO mice display a hindlimb clasping phenotype similar to that observed in mouse models of Huntington's disease. The striatum of postnatal Emx-BDNFKO mice was reduced in volume compared with controls, and the most abundant neuron type of the striatum, medium spiny neurons (MSNs), had shrunken cell somas, thinner dendrites, and fewer dendritic spines at 35 d of age. Although significant striatal neuron losses were not detected at 35 or 120 d postnatal, 35% of striatal neurons were missing in Emx-BDNFKO mice aged beyond 1 year. Thus, cortical BDNF, although not required for the generation or near-term survival of MSN, is necessary for normal striatal neuron dendrite morphology during the period when BDNF expression rises in the cortex. Furthermore, a long-term in vivo requirement for cortical BDNF in supporting the survival of MSNs is revealed.
Keywords: BDNF, Huntington's disease, dendrite, striatum, Cre-lox, neurotrophin, mouse, mutant
Introduction
Huntington's disease (HD), an inherited neurodegenerative disorder, results from the expansion of a CAG trinucleotide repeat region in the huntingtin gene, which lengthens a glutamine stretch in the Huntingtin protein (Htt) (Andrew et al., 1993; Benitez et al., 1994). Mutant Htt causes the death of neurons, particularly medium spiny neurons (MSNs), which account for ∼90% of striatal neurons. The demise of MSNs causes motor, cognitive, and behavioral dysfunction (Reiner et al., 1988). One possible mechanism leading to cell loss in HD is a reduction of neurotrophic factor function (Cattaneo et al., 2001).
Mammalian neurons depend on neurotrophic factors for survival, differentiation, and proper function. An important family of neurotrophic factors, the neurotrophins, includes brain-derived neurotrophic factor (BDNF), which is believed to act in many autocrine, paracrine, retrograde, and anterograde neural signaling pathways by activating the TrkB receptor tyrosine kinase (von Bartheld et al., 1996; Bibel and Barde, 2000; Huang and Reichardt, 2001). Although BDNF and TrkB coexpress within some brain regions, they do not always overlap. Notably, in the intact striatum, TrkB mRNA is present but BDNF mRNA is not, and from cortical ablations it was concluded that most striatal BDNF is anterogradely transported from the cortex (Altar et al., 1997).
Loss of BDNF through cortical ablation or gene mutation in the mouse resulted in reduced expression of neuronal markers, including parvalbumin, calbindin, and the dopamine-regulated phosphoprotein, DARPP-32, within the striatum (Jones et al., 1994; Altar et al., 1997; Ivkovic and Ehrlich, 1999). Striatal MSNs express TrkB (Merlio et al., 1992; Costantini et al., 1999) and display BDNF immunoreactivity (Ferrer et al., 2000). In vitro and in vivo studies indicate that BDNF can influence MSN survival and differentiation (Mizuno et al., 1994; Widmer and Hefti, 1994; Nakao et al., 1995; Ventimiglia et al., 1995; Alcantara et al., 1997; Ivkovic and Ehrlich, 1999); however, these experiments do not directly examine the long-term consequences of the loss of cortical BDNF on MSNs.
Chronic depletion of BDNF likely contributes to HD pathology. Absence of wild-type Htt results in decreased expression of BDNF within the cortex, and one important function of Htt may be activation of BDNF transcription (Zuccato et al., 2001). The majority of cortical neurons expressing BDNF also express Htt, and in a rat model of HD, reduction of Htt levels precedes reduction of BDNF levels (Fusco et al., 2003). Patients with HD have reduced BDNF transcript in the cortex and decreased BDNF protein in cortex and striatum (Ferrer et al., 2000; Zuccato et al., 2001). In addition to directly affecting MSN survival and differentiation, reduced striatal BDNF could lead to increased susceptibility of MSNs to excitotoxicity and other insults (Duan et al., 2003). To elucidate specific BDNF functions in the development of the striatum and the pathology of HD, we have analyzed the consequences of an absence of cortical BDNF for MSNs using forebrain-specific BDNF mutant mice (Gorski et al., 2003a). We report here that cortical BDNF has a relatively minor role in determining the number of MSNs early in life but plays a major role in MSN dendritic structure. Moreover, the forebrain-specific BDNF mutant mice display aspects of behavioral and anatomical abnormalities seen in mouse models of HD.
Materials and Methods
Generation of forebrain-restricted BDNF mutant mice. All animal procedures were conducted in accord with Public Health Service guidelines and with the approval of the University of Colorado Institutional Animal Care and Use Committee. Emx1IRESCre/+ induces efficient forebrain-specific, Cre-mediated loxP recombination (Gorski et al., 2002). Emx1IRESCre/+;BDNFneo/+ mice, where BDNFneo is a null allele that has a neomycin cassette in place of the BDNF coding exon (Jones et al., 1994), and BDNFlox/+ mice were bred; one-eighth of the offspring contain all three mutated alleles, i.e., Emx1IRESCre/+;BDNFneo/lox, and are referred to as “Emx-BDNFKO.” The mice used here were backcrossed at least 6-10 generations onto the C57BL/6J strain. Both BDNF+/+ and Emx1IRESCre/+;BDNF+/+ were considered wild type in data collection, because no abnormalities were detected in the Emx1IRESCre/+;BDNF+/+ genotype. Heterozygous mice were BDNFneo/lox. PCR analysis was used to determine genotype. BDNF protein was quantified using the BDNF EmaxTM ImmunoAssay System (Promega, Madison, WI). Tissue was dissected and weighed. Protein was extracted and quantified as per manufacturer's protocol, except for a change in lysis buffer (to 50 mm Tris-HCl, 0.6 m NaCl, 0.2% Triton X-100, 1% BSA, 0.1 m benzothonion chloride, 1 mm benzamidine, 0.1 mm PMSF, pH 7.4). All samples from an individual animal were run in triplicate, and resulting quantities were averaged.
Immunocytochemistry and X-gal stain. Postnatal mice below the age of 8 d were anesthetized by hypothermia on wet ice. Older mice were heavily sedated by intraperitoneal injection of 2.5% avertin (20 μl/gm) in PBS, pH 7.4 (100% avertin is 10 gm tribromoethanol, 10 ml tert-amyl alcohol) and then treated in the same manner as described previously (Vigers et al., 2000). Primary antibodies included mouse monoclonal anti-NeuN (neuronal-specific nuclear protein) (1:1000; Chemicon, Temecula, CA) and a rabbit-anti ChAT antibody (Ab) (1:000; Chemicon). The secondary antibodies used were biotinylated goat anti-rabbit or biotinylated goat anti-mouse Ab (Jackson ImmunoResearch Laboratories, West Grove, PA) diluted 1:250 in blocking solution. Visualization was by diaminobenzidine reaction as described in Vigers et al. (2000). Slides were counterstained, when appropriate, with cresyl violet. β-gal expression in CNS tissues was visualized histochemically by staining with X-gal as described previously (Vigers et al., 2000).
Behavioral analysis. Wild-type, heterozygous, and Emx-BDNFKO mice at various ages were analyzed for limb clasping by suspending them from their tails at least 1 foot above a surface for 1 min. A clasping event was defined by the retraction of either or both hindlimbs into the body and toward the midline. Mice were scored on a simple “yes” or “no” basis, with the investigator being blind to genotype.
Motor tests were performed with 1-, 2-, and 4-month-old mice. The rotarod apparatus (San Diego Instruments, San Diego, CA) was used in an acceleration mode gradually increasing from 4 to 40 rpm over the course of 5 min. Mice were placed on the apparatus, and rotation was initiated. Latency to fall was recorded automatically. Mice that were still on the wheel at 5 min were considered complete responders. They were removed from the wheel, and their time was noted as 300 sec. Trials were given ∼1 hr apart, with three trials per day for 3 d. All trials were undertaken in the morning before noon. The three trials for any given mouse were averaged for that day to give the mouse a single time for that day. Experiments were run with the investigator blind to genotype.
Volumetric and morphometric analyses. Brains from the various ages were sectioned sagittally at 100 μm thickness to facilitate measurements. Strict morphological criteria were used consistently in all mice to determine the boundaries of these brain regions. Briefly, the superior boundary of the striatum was defined by the corpus callosum, the lateral boundary by the external capsule, and the medial boundary by the lateral ventricle and the corpus callosum. The ventral boundary of the striatum was delineated by the anterior commissure, excluding the nucleus accumbens. For the cortex, the primary ventral boundary was the corpus callosum. A line connecting the rhinal fissure to the corpus callosum was used in more medial sections to define the anterior ventral portion of the neocortex. Entorhinal cortex was also included in cortical volume measurements. Hippocampal outlines encompassed the dentate gyrus, the CA1-3 fields of Ammon's horn, and the subiculum, but not the presibiculum or fimbria hippocampus. Starting with one of the four most lateral sections, selected on a random basis across brains, every fourth section through the lateromedial extent of a hemisphere of the brain was analyzed. Both hemispheres of a given brain were analyzed separately; using this sampling strategy, ∼20 histological sections per brain were analyzed. All volumetric quantifications were performed with a Nikon Eclipse E600 with a 4×/0.20 objective, a motorized Biopoint XYZ axis computer-controlled stage (Ludl Electronics, Hawthorne, NY), a CCD MTI video camera (Dage-MTI, Michigan City, IN), and Stereo-Investigator, a morphometry and stereology software package (v4.37, MicroBrightField, Colchester, VT). When calculating the volume of neocortex, hippocampus, and neostriatum, the boundaries were defined and the volumes were determined with the StereoInvestigator software according to the Cavalieri principle (Regeur and Pakkenberg, 1989). The Gundersen estimated coefficient of error ranged from 0.03 to 0.05. All morphometric quantification was conducted with the investigator blind to genotype.
Cell counting. The total number of neurons in each striatum was estimated using a fractionator sampling design (Gundersen et al., 1988; West et al., 1991). Serial coronal sections (40 μm) were obtained from postnatal day (P) 35, ∼P120, and 1- to 1.5-year-old mutant and control mice using a cryostat (Leica, Bannockburn, IL), and anti-NeuN immunocytochemistry was performed to identify neurons (Mullen et al., 1992). Counterstaining with cresyl violet allowed delineation of the area of the striatum in each section, defined by the boundaries of the lateral ventricle and outer edge of the globus pallidus medially, the external capsule laterally, and the anterior commissure ventrally. In each of the sections sampled, striatal neurons were counted, using NeuN-positive nuclei as the counting unit according to optical dissector rules (Gundersen et al., 1988). Labeled profiles were counted only if the first recognizable profile of the cell came into focus within the counting frame (West et al., 1991). NeuN-positive cells were counted on every 12th coronal section, extending from the most rostral to the most caudal parts of the striatum. Counts were made at regular predetermined intervals (x = 550 μm; y = 550 μm) within an unbiased counting frame of known area (30 × 30 μm = 900 μm2) that was superimposed on the image of the tissue sections viewed under a 60×, numerical aperture 1.40 oil-immersion objective with microscope, camera, and stage the same as for the volumetric analysis. The frame was positioned randomly by StereoInvestigator software (MicroBrightField), leading to a systematic sample of the area occupied by the striatum; 20 μm was defined for the z-dimension of the counting brick with a 2 μm guard on either side. Because of the limited availability, cost, and difficulty in obtaining older mice, 1- to 1.5-year-old brains were cut at the midline, and only one hemisphere per mouse was used for the purposes of cell counts.
DiOlistics and analysis of cell morphology. Gene gun cartridges were prepared, and 200 μm coronal brain slices were blasted with DiI- and DiO-coated tungsten beads as described (Gorski et al., 2003a). Images taken at 0.5 μm intervals at 1000× magnification were obtained on a Zeiss Axioplan deconvolution microscope (Zeiss, Oberkochen, Germany) equipped with a Hamamatsu C4742-95 digital camera (Hamamatsu, Bridgewater, NJ). Adjacent images were deconvolved to nearest neighbors and analyzed using the Open Lab software suite (Improvision, Coventry, UK). Quantification was performed on digitally magnified images (2×).
Cell soma cross-sectional areas were measured as described previously (Gorski et al., 2003a). Stereotypical MSNs were chosen as cells with a roundish soma and multiple dendrites with numerous spines. Measurements were taken in the image plane where both the edge of the nucleus and plasma membrane were in sharp focus (three mice per genotype; 12 cells per mouse). Dendrite branch diameter was measured 5-10 μm from the soma for primary dendrites, or from the previous branch point for higher-order dendrites, and where the edges of the dendrite were in focus. Dendritic spines of striatal medium spiny neurons were defined as protuberances extending 0.5-5 μm from the dendrite (where measurements were from base to tip of protrusion) for the purposes of length and density measurements. Only well defined spines extending parallel to the plane of the captured image were measured for length. Spine density was defined as the number of spines along the length of the 1°, 2°, 3°, 4°, and 5° dendrite segments divided by the length of that dendrite segment. Calculations were limited to spines found to be at least 20 μm away from the origin of the 1° dendrite and only for dendritic segment lengths of ≥10 μm. Spine density measurements for a given genotype were binned into 0.5 μm groups to calculate the distribution of spine densities. Statistical analyses were done using the Prism (version 2.0; Graph-Pad, San Diego, CA) software one-way ANOVA test with p values reported as determined by the Newman-Keuls post hoc test. A total of three mice per genotype and 10-20 cells per animal were analyzed for dendritic thickness, spine length, and density; 2708 spines were quantified for wild type, 3454 for heterozygous, and 2013 for mutant.
Results
BDNF expression is not apparent in the striatum
To study the developmental timing of possible sources of BDNF relevant to the mouse striatum, we used a BDNFlacZ/+ mouse line in which lacZ is targeted to the BDNF gene (Bennett et al., 1999). No traces of blue staining were found in the striatum at any age examined, including embryonic day (E) 15.5, E17.5, and birth (see supplemental Fig. 1, available at www.jneurosci.org) or P4, P8, P15, and P35 (Fig. 1). BDNF expression, as visualized by X-gal staining, was limited to a small number of cells in the hippocampus at E15.5. By E17.5, there is expression in the piriform cortex, hippocampus, thalamus, hypothalamus, and amygdaloid nucleus, but only rare stained cells are seen in neocortex and none in striatum. This was much the same for P0 (see supplemental Fig. 1, available at www.jneurosci.org). Neocortical expression was apparent in the deep layers V-VI of the neocortex as early as P4 and became increasingly prevalent with age (Fig. 1). Expression is initially seen rostrally and then rises in a rostral to caudal manner with increasing age, mirroring developmental maturation of the cortex. At P8 there are scattered X-gal-positive cells in somatosensory cortex that become more numerous and cover a wider area caudally. By P15, there is extensive staining in both somatosensory and motor cortices and in all cortical layers, although fewer cells in layer V. Starting at ∼P2, some layer V cortical axon collaterals form connections with striatal neurons (Nisenbaum et al., 1998; Sheth et al., 1998) and thus could provide BDNF to the striatum beginning with the onset of cortical BDNF expression at ∼P4.
Figure 1.
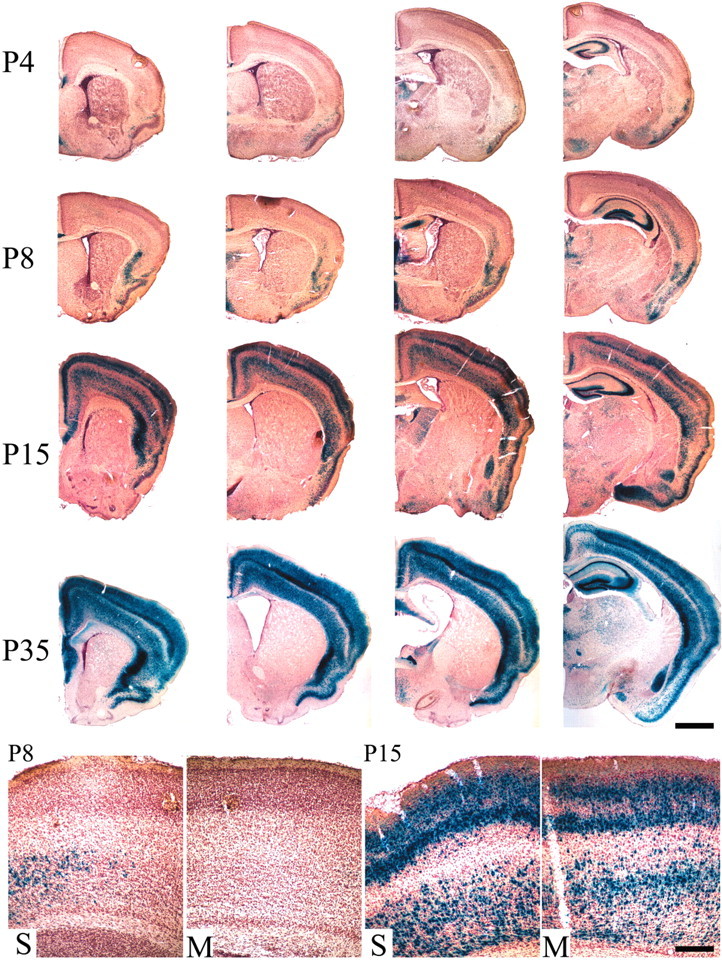
The striatum lacks apparent BDNF expression. BDNFlacZ/+ mice demonstrate substantial expression of the BDNF gene within the cortex with undetectable levels within the striatum. Shown are images (20×)of 40 μm coronal sections in anterior to posterior series from P4, P8, P15, and P35 mice stained with X-gal and counterstained with Neutral Red. Scale bar, 1 mm. At the bottom, 100× images of somatosensory (S) and motor (M) cortices of P8 and P15 mice. Scale bar, 200 μm.
Most striatal BDNF is derived from the cortex
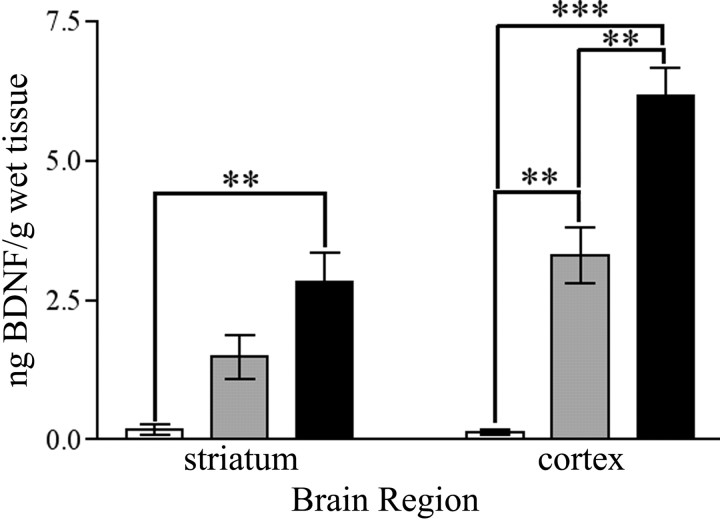
On the basis of lesion experiments, cortical neurons are considered to be the primary source of BDNF for the striatum (Altar et al., 1997). Emx-BDNFKO mice have near-complete elimination of cortical BDNF protein because of Cre-mediated recombination by ∼E10.5 (Gorski et al., 2003a). ELISA analysis demonstrated a ∼95% reduction in BDNF protein concentration in the striatum of P35 Emx-BDNFKO mice compared with wild-type littermates (Fig. 2) (p < 0.01; n = 3 mice per genotype). In addition to the cortex, the thalamus and midbrain-hindbrain (specifically substantia nigra) produce BDNF (Hofer et al., 1990; Seroogy et al., 1994) and send projections to the striatum. Little recombination of the BDNFlox allele occurs in the thalamus and midbrain-hindbrain of Emx-BDNFKO mice, however, and Emx-BDNFKO mice retain near-heterozygous levels of BDNF protein in these structures (Gorski et al., 2003a). Because striatal cells do not express detectable levels of BDNF mRNA in the intact animal (Altar et al., 1997), and there is also little recombination of the BDNFlox allele in the striatum of Emx-BDNFKO mice, we also conclude that the primary source of striatal BDNF protein is from the cortex. The small amount of residual BDNF in the striatum of Emx-BDNFKO mice is likely caused by anterograde delivery from subcortical striatal afferents, such as those from the substantia nigra.
Figure 2.
Quantification of BDNF protein in cortex and striatum at P35. BDNF protein was quantified by ELISA for striatum and dorsal cortex and expressed as nanograms of BDNF protein per gram of wet tissue. These extracts were obtained from P35 mice; n = 3 mice per genotype (*p<0.05;**p<0.01;***p<0.001; one-way ANOVA with a Tukeypost hoctest). White bars are Emx-BDNFKO, gray bars are heterozygotes, and black bars are wild type.
Emx-BDNFKO mice display phenotypic behavior indicative of motor dysfunction
Hindlimb and forelimb clasping have been observed in transgenic lines in which there is motor dysfunction or degeneration, including HD mouse models expressing mutant huntingtin, Hoxb8 knock-outs, and staggerer mutants in the RORalpha gene (Lalonde, 1987; Hamilton et al., 1996; van den Akker et al., 1999; Auerbach et al., 2001; Guidetti et al., 2001; van Dellen et al., 2001). Given the proposed role of Htt as an activator of cortical BDNF transcription (Zuccato et al., 2001), we tested the response of Emx-BDNFKO mice to tail suspension at varying ages (1-12 months). Wild-type and heterozygous mice often splay their legs out when suspended, and never more than half would exhibit clasping at any age studied (Fig. 3A). A majority of Emx-BDNFKO mice clasped at all ages during tail suspension, and all did at ∼P120 and older. As Emx-BDNFKO mice age, the clasping behavior progresses from just involving the hindlimbs to the forelimbs as well. It is interesting to note that when these mice were tested for motor function on the accelerating rotarod at three different ages (1, 2, and 4 months), they displayed no severe abnormality (Fig. 3B-D). In fact, at 1 month, Emx-BDNFKO mice were significantly better at rotarod then wild-type and heterozygous littermates, perhaps surprising in light of the anatomical studies described below.
Figure 3.
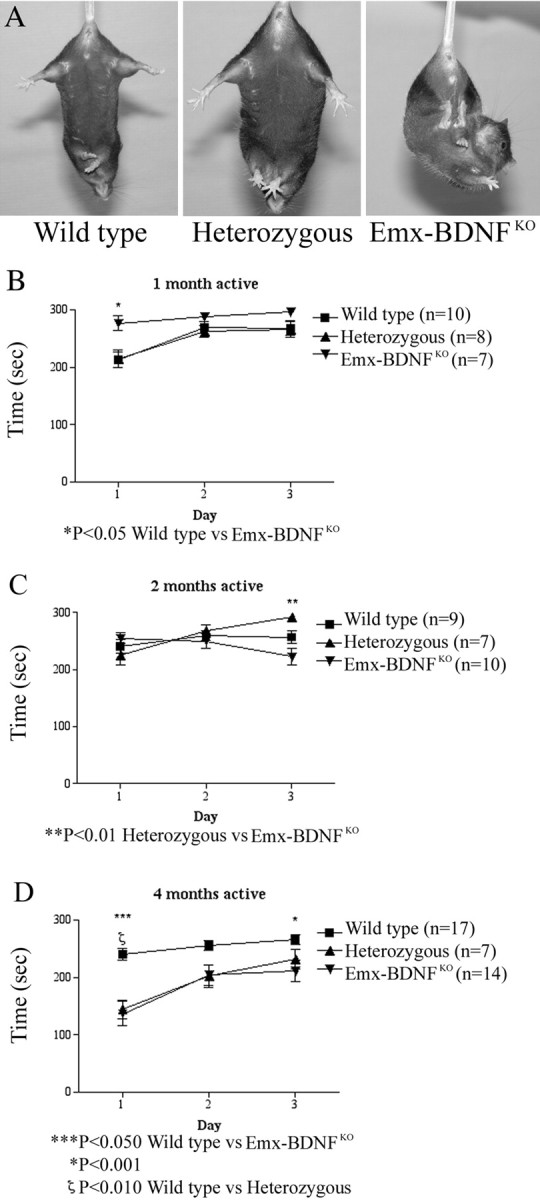
Emx-BDNFKO mice exhibit a foot-clasping phenotype. Emx-BDNFKO, heterozygous, and wild-type mice of ages 1, 2, and 4 months were suspended by their tails for 1 min. Clasping was defined as the balling up of one or both of the hindlimb paws, accompanied by pulling them into the body and movement of the limbs toward the midline. A, Four-month-old wild-type, heterozygous, and Emx-BDNFKO mice undergoing tail suspension. B, Rotarod performance of 1-month-old mice (wild type, n = 10; heterozygotes, n = 8; Emx-BDNFKO, n = 7; *p < 0.05 for Emx-BDNFKO vs wild type). C, Two-month-old mice (wild type, n = 9; heterozygotes, n = 7; Emx-BDNFKO, n = 10; **p < 0.01 for heterozygous vs Emx-BDNFKO). D, Four-month-old mice (wild type, n = 17; heterozygotes, n = 7; Emx-BDNFKO, n = 14; *p < 0.05 wild type vs Emx-BDNFKO; ζp < 0.01 wild type vs heterozygous; ***p < 0.001 wild type vs Emx-BDNFKO). Squares are wild type, triangles are heterozygotes, and circles are Emx-BDNFKO.
Emx-BDNFKO mice have reduced CNS volumes
To generally assess the morphological effects that Emx1IRESCre-mediated loss of BDNF has on the mouse CNS, the volumes of hippocampus, neocortex, and striatum of the mice were measured. All three regions express TrkB mRNA and protein, and cells in these regions are known to respond to BDNF. Three ages (P14, P35, and ∼P120) were examined to determine how the absence of BDNF affects these regions over time. At P14, a statistically significant 12% reduction in hippocampal volumes was detected in Emx-BDNFKO compared with wild-type controls. At P35, the hippocampal volumes of Emx-BDNFKO mice were 10% less than wild-type volumes. By ∼P120, average hippocampal volumes were within ∼3% of wild-type values (Fig. 4A). Neither difference (at P35 and ∼P120) was statistically significant.
Figure 4.
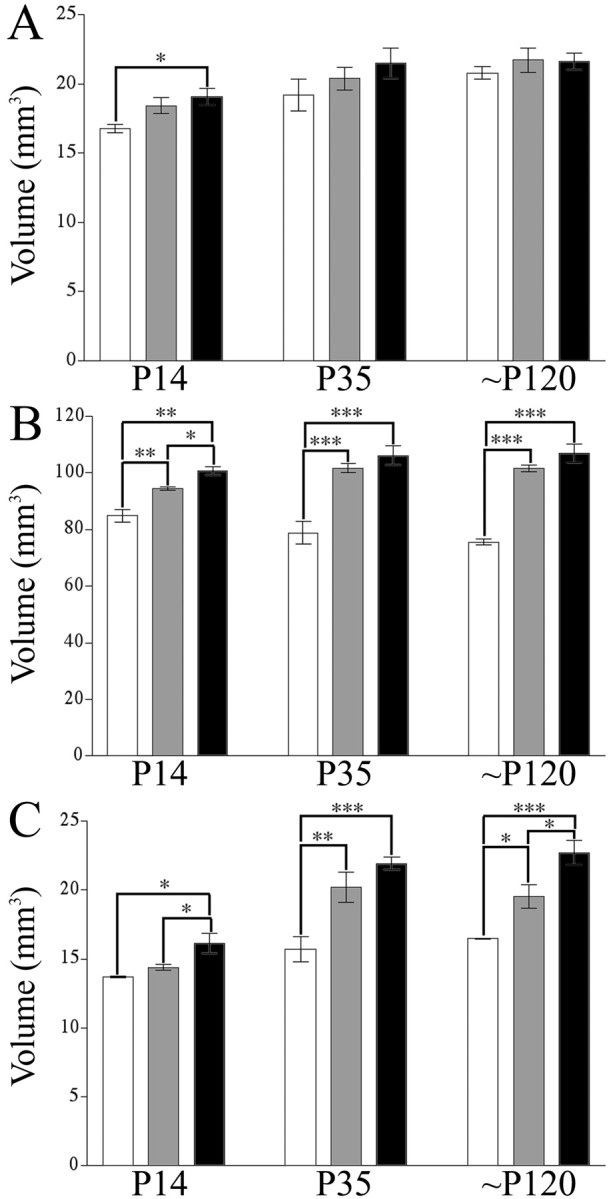
CNS volume reduction in Emx-BDNFKO mice at P14, P35, and ∼P120. Specific CNS structures are reduced in volume in the absence of BDNF as assessed by the Cavalieri estimator. A, Hippocampus. B, Neocortex. C, Striatum (*p < 0.05; **p < 0.01; ***p < 0.001; one-way ANOVA with a Tukey post hoc test). White bars are Emx-BDNFKO, gray bars are heterozygotes, and black bars are wild type.
In contrast to the hippocampus, the cortical volume of Emx-BDNFKO mice was smaller than controls at all ages examined (Fig. 4B). At P14, a statistically significant 16% reduction was observed between Emx-BDNFKO mice and wild-type controls. The magnitude of the reduction in cortical volume increased at later ages. At P35, the cortex of Emx-BDNFKO mice was ∼25% smaller than wild type, whereas at ∼P120, mutants were 30% smaller than the cortical volume of wild types. Cortical volumes of heterozygous mice were only a few percent lower than wild-type volumes, and this difference was statistically significant only at P14.
Striatal volume was significantly reduced in Emx-BDNFKO mice compared with wild type at all ages analyzed (Fig. 4C). At P14, the striatal volume of Emx-BDNFKO mice was 15% less than wild-type littermates. Similar to observations made in the cortex, the difference in striatal volumes between mutant and wild-type mice widened at P35 (33% reduction). This difference persisted to ∼P120, suggesting that the striatal phenotype of Emx-BDNFKO did not worsen with advancing age from P35 onward.
In summary, the loss of brain volume associated with an absence of cortical BDNF was partially region specific. Hippocampi of mutant mice were reduced only in early postnatal development, whereas initial reductions in cortex and striatum became larger with age.
Loss of striatal neuron number in old but not young adult Emx-BDNFKO mice
To determine whether the decrease in striatal volume was caused by cell loss or a reduction in cell size, NeuN-positive neurons in the striatum were counted using an optical fractionator probe. Total neuronal numbers within the striatum of Emx-BDNFKO mice were estimated to be 14.2% lower than that of wild-type mice at P35 but only 6% lower at ∼P120. Neither difference proved to be significant (Fig. 5B)(p = 0.26, n = 3 mice/genotype for P35; p = 0.48, n = 5 for wild type and mutants and n = 6 for heterozygotes for ∼P120). It is not until much later ages (1-1.5 years) that a significant reduction in NeuN-positive cells of 35% is seen between Emx-BDNFKO mice and wild-type controls (Fig. 5B) (p < 0.05; n = 4 for wild type and mutant and n = 5 for heterozygotes).
Figure 5.
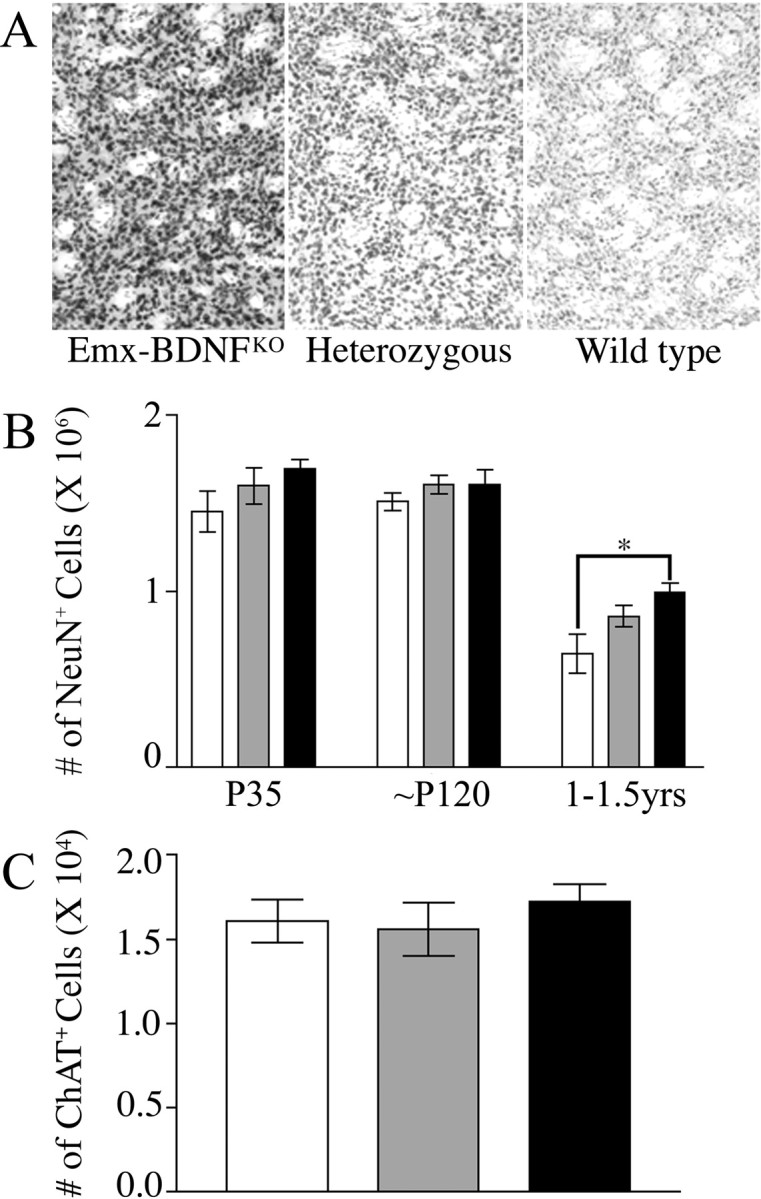
Loss of striatal neurons in old but not young adult Emx-BDNFKO mice. A, Representative images of striatum from all three genotypes immunostained for NeuN and counter-stained with cresyl violet (40 μm cryostat sections at 100× magnification). B, Optical fractionator estimate of the number of NeuN-positive cells in the striatum of P35, ∼P120, and 1- to 1.5-year-old mice. C, Estimate of the number of ChAT-positive cells in the striatum of ∼P120 mice. White bars are Emx-BDNFKO, gray bars are heterozygotes, and black bars are wild type.
Similar results at ∼P120 were obtained for specific counts of cholinergic large aspiny neurons in the neostriatum. A slight decrease of ∼7-10% in the number of ChAT cells in Emx-BDNFKO and heterozygous mice compared with wild-type controls was not statistically significant (Fig. 5C) (p = 0.86; n = 3 mice per genotype).
MSNs are smaller with thinner dendrites
To test whether the loss of cortical BDNF alters MSN morphology, we labeled neurons using DiOlistics. The cross-sectional area of MSN somas from P35 Emx-BDNFKO mice was reduced by 25% when compared with wild-type and heterozygous controls (Fig. 6A). There was no significant difference between animals of different genotypes in terms of the number of 1° dendrites (Fig. 6C). The average number of primary dendrites was 7 (range 4-11). The diameter of all portions of the dendritic tree, determined by analyzing different dendritic orders, was reduced in Emx-BDNFKO mice as compared with wild-type and heterozygous controls, by ∼40 and ∼30%, respectively (Fig. 6B) (p < 0.001 for all orders). The dendritic thickness of heterozygous mice was reduced by ∼10% compared with wild type.
Figure 6.
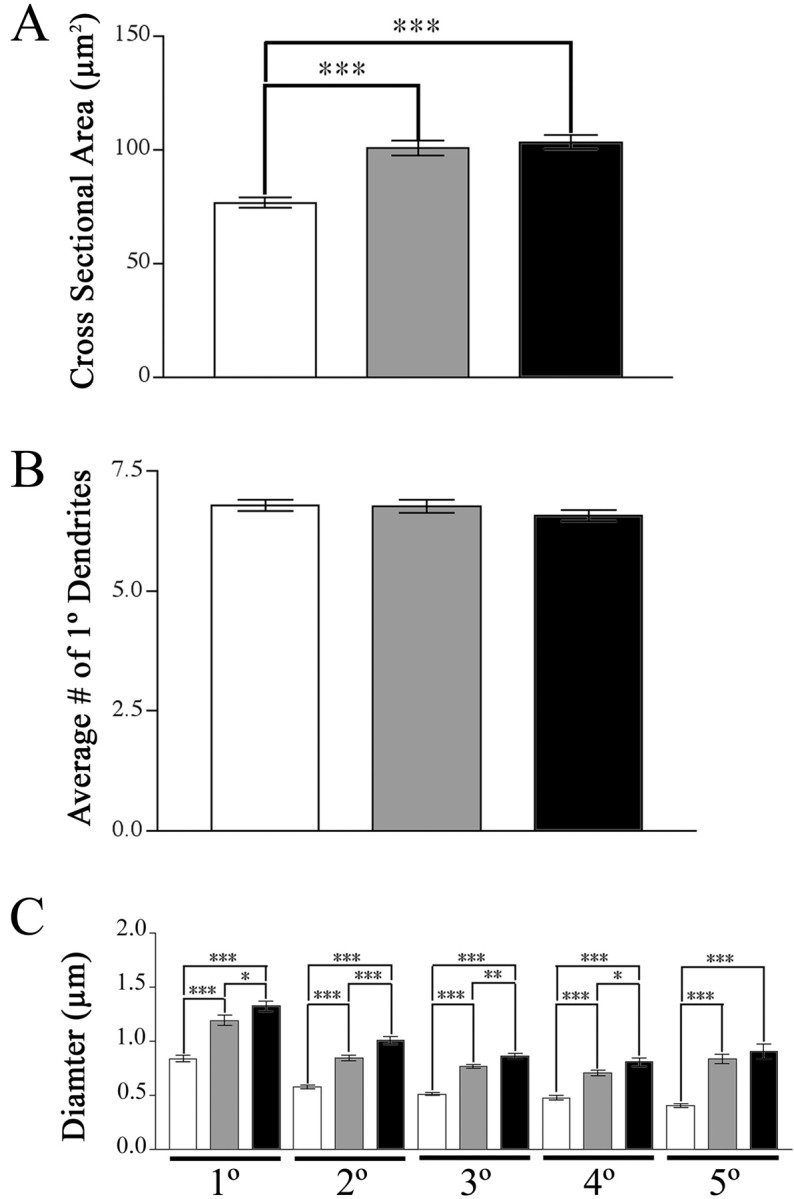
Reduced soma and dendrite size inEmx-BDNFKO mice at P35. Neurons were labeled using DiOlistics and analyzed using fluorescent deconvolution microscopy. A, Cross-sectional areas of medium spiny neurons of the striatum (Emx-BDNFKO, 76.66 ± 2.14 μm2; heterozygous, 100.7 ± 3.32 μm2; wild type, 103.3 ± 3.08 μm2; represented as mean ± SEM). B, Number of primary dendrites on MSNs from each of the genotypes. C, Diameters of primary through fifth-order dendrites (*p < 0.05; **p < 0.01; ***p < 0.001; one-way ANOVA with a Tukeypost hoctest). White bars areEmx-BDNFKO, gray bars are heterozygotes, and black bars are wild type.
Number and length of MSN dendritic spines reduced in Emx-BDNFKO mice
Most of the corticostriatal connections are excitatory and occur on dendritic spines. Spine number and morphology are good indicators of the status of corticostriatal synaptic connections (Kemp and Powell, 1971a,b; Dube et al., 1988). Because BDNF can be anterogradely transported by cortical neurons and released at synapses and its TrkB receptors are found in postsynaptic densities (von Bartheld et al., 1996; Wu et al., 1996; Aoki et al., 2000; Kohara et al., 2003), BDNF is poised to influence corticostriatal connectivity. If cortical BDNF supplied anterogradely is indeed important for establishing or maintaining synapses with MSNs or altering their strength, one might expect that loss of BDNF would result in altered dendritic spine numbers or morphology. To determine the effects of cortical BDNF on MSN dendritic spines, we analyzed spine density and length in P35 mice. Representative images of dendrites of the same order from P35 mice suggested a significant reduction in spine density in Emx-BDNFKO mice compared with controls (Fig. 7A).
Figure 7.
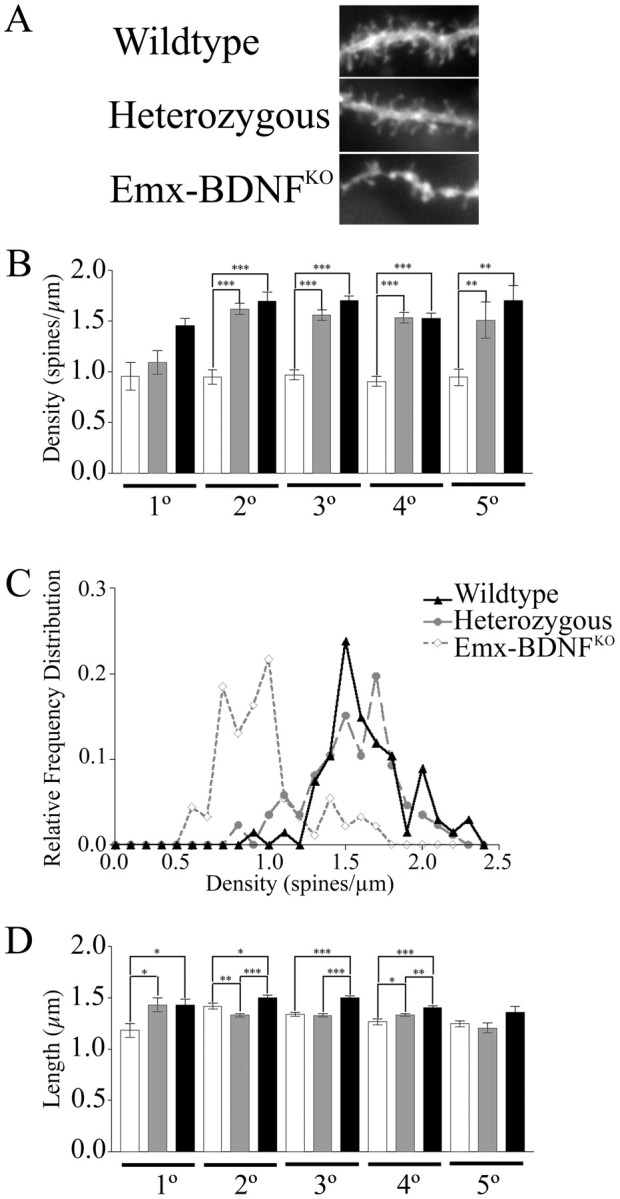
Reduced MSN spine density and length in Emx-BDNFKO mice. A, Representative images of DiI-labeled dendrite segments from the various genotypes at P35. B, Spine density per unit length of dendrite for different orders of dendrites. C, Relative frequency distributions of spine densities for each of the genotypes for pooled dendritic orders. D, Spine lengths for multiple orders of dendrites (Emx-BDNFKO as percentage of wild type: 1°, 90.1%, p < 0.05; 2°, 83.6%, p < 0.001; 3°, 89.1%, p < 0.01; 4°, 87.4, p < 0.05; 5°, 92.3%, p > 0.05; *p < 0.05; **p < 0.01; ***p < 0.001; one-way ANOVA with a Tukey post hoc test). White bars are Emx-BDNFKO, gray bars are heterozygotes, and black bars are wild type.
Quantification revealed a 30-40% reduction in spine density between Emx-BDNFKO MSN dendrites compared with heterozygous and wild-type control MSNs (Fig. 7B). The decrease in spine density was seen on every order of dendrite. Although heterozygous mice had slightly lower densities compared with wild-type mice, the difference was not significant in any of the dendrite orders measured. Spine length in both heterozygous and Emx-BDNFKO MSN was ∼10% shorter than in wild type (p < 0.03 for 1°; p < 0.001 for 2°, 3°, 4°; p = 0.07 for 5°; n = 3 mice per genotype). Spine lengths from different order dendrites were not statistically different when compared within a genotype.
Discussion
Our results demonstrate the absence of detectable striatal X-gal signal in BDNFlacZ/+ reporter mice and the near complete loss of BDNF in the striatum of Emx-BDNFKO mice. From these data we conclude that BDNF in the striatum originates primarily from the cortex via anterograde transport, as suggested previously (Altar et al., 1997). Loss of cortical BDNF early in development results in a decrease in cortical and striatal volumes (Fig. 4). Not all CNS structures are so grossly affected in the absence of BDNF. The hippocampus of Emx-BDNFKO mice as compared with wild-type mice at P14 showed only a transient and minor reduction. In seeking an explanation for the relative subtlety of these affects, we discovered an increase of ∼2× of NT-3 protein in several brain regions of Emx-BDNFKO mice (Gorski et al., 2003a). Because of the relatively high NT-3 expression levels present in the hippocampus (Friedman et al., 1991; Vigers et al., 2000), NT-3 might compensate for the reduction of BDNF in the hippocampus. Alternatively, the hippocampus could receive BDNF from another source, via either anterograde or retrograde transport. The amygdala, thalamus, hypothalamus, and ventral tegmental area all send and receive projections from the hippocampus, and each expresses BDNF in the mouse (Hofer et al., 1990).
The piriform cortex of BDNFLacZ/+ mice is the first cortical region to begin expressing BDNF at E17.5, as visualized by X-gal stain, which then increases in a lateral to medial and rostral to caudal manner with age. By P15, when decreases in striatal and cortical volume appear in Emx-BDNFKO mice, X-gal stain has been apparent in all layers of the cortex in rostral regions (Fig. 1). Because corticostriatal axons undergo significant arborization of collaterals within the striatum by P7 (Sheth et al., 1998), cortical BDNF could be influencing cells in both the cortex and (by anterograde transport) the striatum. The reduction in BDNF expression found in both Emx-BDNFKO and heterozygous mice might cause the lack of difference between these genotypes in striatal volume seen at P14. By P35 and ∼P120, striatums of Emx-BDNFKO mice exhibit smaller volumes than both heterozygous and wild-type mice. Notably, although the reduction seen in the cortex of BDNFneo/lox heterozygous mice relative to wild type at P14 ceases to be significant at P35 and ∼P120, the striatum of heterozygotes remains smaller than wild type at ∼P120. Thus, we hypothesize that the striatal sensitivity to BDNF levels may exceed that of the cortex. This could contribute to the preferential degeneration of the striatum in HD patients.
The lack of a significant loss of striatal neurons in Emx-BDNFKO mice at younger ages indicates that cortical BDNF does not play a strong proliferative-survival role for striatal MSNs up to ∼P120. Similarly, Gorski et al. (2003a) did not detect cortical neuron losses in Emx-BDNFKO mice at P35; however, examination of 1- to 1.5-year-old mice (Fig. 5B) does indicate a significant decrease in the number of striatal neurons compared with wild type. This demonstrates that BDNF is required for long-term survival of striatal neurons. Whether this neuronal loss is caused by a direct BDNF trophic survival function only apparent with aging, increased susceptibility to excitotoxic injury (Martinez-Serrano and Bjorklund, 1996; Alberch et al., 2002), or some other type of insult (Perez-Navarro et al., 2000; Galvin and Oorschot, 2003) is unclear. These possible roles for BDNF will require future examination. Importantly, our results do not exclude BDNF from some previously described roles in striatal neuron biochemical differentiation (Mizuno et al., 1994; Ventimiglia et al., 1995; Ivkovic et al., 1997).
Analysis of the morphology of MSNs revealed significant decreases in soma area, dendrite thickness, and especially dendritic spine density. Reduced soma size and dendrite thickness indicate that the cells failed to thrive in a BDNF-poor environment, although their ability to create the proper number of primary dendrites appears unaffected. A reduction in dendrite diameter would be predicted to cause a higher resistance to current flow, and thus these MSNs could require greater synaptic stimulation to reach threshold for firing action potentials. Altered Ca2+ dynamics and signaling processes might also accompany spine loss (Segal and Andersen, 2000).
Dendritic spines on MSNs likely indicate asymmetric synapses with cortical afferents; thus the ∼30-40% spine loss in Emx-BDNFKO mice suggests reduced connectivity between cortex and striatum. Curtailed connection between cortex and striatum could result in MSNs failing to receive cortically derived maturation signals and factors other than BDNF. A negative feedback loop could arise in which the loss of BDNF causes morphological changes in MSNs, such as cell shrinkage and loss of spines, which in turn further cripples the ability of MSNs to support cortical afferents, leading to additional changes in the MSNs. The MSN reduction in spines of Emx-BDNFKO mice closely parallels that seen in cortical ablation experiments (Kemp and Powell, 1971b) and HD model mice (Guidetti et al., 2001). Although we examined spines only at P35, the volume of the striatum and the total number of striatal neurons remain virtually unchanged between P35 and ∼P120 (Figs. 3C, 4B), which we hypothesize to mean that no further morphological degeneration of MSNs occurs between these ages. Thus, absence of cortical BDNF reveals that almost half of MSN spines depend directly or indirectly on BDNF, whereas the others do not or have some compensatory factor(s).
Although evidence for both retrograde and anterograde transport of BDNF within the CNS exists, the results of these genetically based experiments clearly demonstrate the case for an important role for anterograde transport in regulating dendritic morphology, as also demonstrated in studies of cortical GABAergic neurons (Kohara et al., 2003). Intriguingly, the similarities between the striatal and cortical dendritic phenotypes as defined in this study and by Gorski et al. (2003a) and S.R. Zeiler, J.A. Gorski, and K.R. Jones (unpublished observations) could suggest that much of CNS BDNF function is through anterograde delivery.
The data on spine number and morphology contain two noteworthy points. First, when the distribution of spine density on individual dendrites was graphed, ∼15% of dendrites in Emx-BDNFKO mice had wild-type densities. Johnston et al. (1990) reported that ∼15% of MSNs receive input from the substantia nigra pars compacta. Nigral neurons would still produce BDNF in Emx-BDNFKO mice (Fig. 6C). This implies that some amount of BDNF may be transported anterogradely from the nigra to a subset of MSNs in the striatum, which agrees with previous studies (Friedman et al., 1993; Altar et al., 1997; Altar and DiStefano, 1998).
Second, unlike other findings from our laboratory in which loss of cortical BDNF results in greater spine length for pyramidal neurons of Emx-BDNFKO and heterozygous mice (Zeiler et al., unpublished observations), the mean spine lengths from striatal neurons of Emx-BDNFKO and heterozygous mice were shorter than wild type at P35. This indicates that the cortical spines in the wild types lack maturity by classical definitions of spine morphology and that the mutant spines are more mature (Nimchinsky et al., 2002). This discrepancy could be the result of hypertrophy of spines in wild-type mice caused by BDNF activity (Tyler and Pozzo-Miller, 2003). The spine phenotype in mutants resembles previous cortical lesion experiments in cats (Kemp and Powell, 1971b) in which a similar shrinkage of spines and spine heads of MSNs was observed.
Loss of BDNF in the cortex (early in development) leads to a reduction in cellular size and complexity of layer II-III cortical neurons (Gorski et al., 2003a). This reduction in gross cellular morphology in the cortex could extend to the corticostriatal afferents, primarily originating in layer V, leading to reduced axonal arborization; thus fewer synapses exist between cortical neurons and MSNs, resulting in sickly, shrunken cells. The additional absence of BDNF normally transported from the cortex to the striatum might increase the severity of the phenotype. This scenario suggests combined direct and indirect effects of the loss of cortical BDNF on MSNs; however, loss of BDNF does not completely eliminate growth and differentiation of these cells, and notably, many spines are still present, suggesting that many corticostriatal synapses remain. Regardless, we conclude that cortical BDNF plays an important role in the growth and differentiation of MSNs.
Emx-BDNFKO mice exhibit clasping of hindlimbs, a stereotyped behavioral phenotype indicative of neurological dysfunction, that becomes increasingly severe as these mice age. A similar clasping phenotype has been seen in HD model mice (Carter et al., 1999; Auerbach et al., 2001; Guidetti et al., 2001). Interestingly, Emx-BDNFKO mice do not perform poorly on rotarod tests, whereas HD model mice exhibit both clasping phenotype and poor rotarod test performance (Carter et al., 1999; Guidetti et al., 2001). The similarities and subtle differences in behavioral phenotypes between HD model mice and Emx-BDNFKO mice opens the possibility that a close comparison of the two model systems could be used to identify specific neurological foundations for BDNF-dependent aspects of the HD phenotype.
Similarities to HD were of interest given that one of the putative functions of wild-type Htt is to regulate BDNF expression in the cortex, and diminution of cortical BDNF could contribute to HD (Zuccato et al., 2001). Recent experiments demonstrate that Emx-BDNFKO mice suffer from an inability to perform a horizontal-vertical discrimination task, a test for complex learning, and complex learning is also impaired in HD (Schmidtke et al., 2002; Gorski et al., 2003b). Thus, our results support a link between cortical BDNF levels and specific aspects of HD pathology attributable to failure of Htt function and subsequent decrement of BDNF (Cattaneo et al., 2001; Zuccato et al., 2001; Fusco et al., 2003). We have demonstrated here that MSNs receiving a paucity of BDNF exhibit a cellular phenotype that mirrors aspects of the HD phenotype, and therefore analysis of Emx-BDNFKO mice should help to elucidate the potential role of BDNF in this tragic disease.
Footnotes
This work was supported by a Howard Hughes Medical Institute fellowship and funding from the National Institutes of Health Minority Biomedical Research Support Initiative for Minority Student Development Grant (Z.C.B.), a Burroughs-Wellcome New Investigator in Pharmacology Award, grants from the American Cancer Society, the Colorado Council for Research and Creative Work, the Muscular Dystrophy Association, the March of Dimes Birth Defects Foundation (1-FY03-94), and a grant from the Hereditary Disease Foundation (K.R.J.). We thank Dipesh Amin, Tiffany Farnum, Susan Tamowski, Dr. Steven Zeiler, and Dr. Alison Vigers for technical support, Dr. Paula Bickford for the use of her stereology equipment, Dr. Jeanne Wehner for use of her rotarod apparatus, Dr. Min Han for use of microscope and OpenLab software, Dr. Brad Olwin for use of his cryostat, and Dr. Jeffrey Bennett, Dr. Leslie Leinwand, Dr. Mike Klymkowsky, Dr. Robert E. Boswell, and Dr. Tintin Su for assistance in the writing of this manuscript.
Correspondence should be addressed to Kevin R. Jones, 347 University Campus Box, Department of Molecular, Cellular, and Developmental Biology, University of Colorado, Boulder, CO 80309. E-mail: krjones@stripe.colorado.edu.
Copyright © 2004 Society for Neuroscience 0270-6474/04/244250-09$15.00/0
References
- Alberch J, Perez-Navarro E, Canals JM (2002) Neuroprotection by neurotrophins and GDNF family members in the excitotoxic model of Huntington's disease. Brain Res Bull 57: 817-822. [DOI] [PubMed] [Google Scholar]
- Alcantara S, Frisen J, del Rio JA, Soriano E, Barbacid M, Silos-Santiago I (1997) TrkB signaling is required for postnatal survival of CNS neurons and protects hippocampal and motor neurons from axotomy-induced cell death. J Neurosci 17: 3623-3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altar CA, DiStefano PS (1998) Neurotrophin trafficking by anterograde transport. Trends Neurosci 21: 433-437. [DOI] [PubMed] [Google Scholar]
- Altar CA, Cai N, Bliven T, Juhasz M, Conner JM, Acheson AL, Lindsay RM, Wiegand SJ (1997) Anterograde transport of brain-derived neurotrophic factor and its role in the brain. Nature 389: 856-860. [DOI] [PubMed] [Google Scholar]
- Andrew SE, Goldberg YP, Kremer B, Telenius H, Theilmann J, Adam S, Starr E, Squitieri F, Lin B, Kalchman MA, et al. (1993) The relationship between trinucleotide (CAG) repeat length and clinical features of Huntington's disease. Nat Genet 4: 398-403. [DOI] [PubMed] [Google Scholar]
- Aoki C, Wu K, Elste A, Len G, Lin S, McAuliffe G, Black IB (2000) Localization of brain-derived neurotrophic factor and TrkB receptors to postsynaptic densities of adult rat cerebral cortex. J Neurosci Res 59: 454-463. [DOI] [PubMed] [Google Scholar]
- Auerbach W, Hurlbert MS, Hilditch-Maguire P, Wadghiri YZ, Wheeler VC, Cohen SI, Joyner AL, MacDonald ME, Turnbull DH (2001) The HD mutation causes progressive lethal neurological disease in mice expressing reduced levels of huntingtin. Hum Mol Genet 10: 2515-2523. [DOI] [PubMed] [Google Scholar]
- Benitez J, Fernandez E, Garcia Ruiz P, Robledo M, Ramos C, Yebenes J (1994) Trinucleotide (CAG) repeat expansion in chromosomes of Spanish patients with Huntington's disease. Hum Genet 94: 563-564. [DOI] [PubMed] [Google Scholar]
- Bennett JL, Zeiler SR, Jones KR (1999) Patterned expression of BDNF and NT-3 in the retina and anterior segment of the developing mammalian eye. Invest Ophthalmol Vis Sci 40: 2996-3005. [PubMed] [Google Scholar]
- Bibel M, Barde YA (2000) Neurotrophins: key regulators of cell fate and cell shape in the vertebrate nervous system. Genes Dev 14: 2919-2937. [DOI] [PubMed] [Google Scholar]
- Carter RJ, Lione LA, Humby T, Mangiarini L, Mahal A, Bates GP, Dunnett SB, Morton AJ (1999) Characterization of progressive motor deficits in mice transgenic for the human Huntington's disease mutation. J Neurosci 19: 3248-3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattaneo E, Rigamonti D, Goffredo D, Zuccato C, Squitieri F, Sipione S (2001) Loss of normal huntingtin function: new developments in Huntington's disease research. Trends Neurosci 24: 182-188. [DOI] [PubMed] [Google Scholar]
- Costantini LC, Feinstein SC, Radeke MJ, Snyder-Keller A (1999) Compartmental expression of trkB receptor protein in the developing striatum. Neuroscience 89: 505-513. [DOI] [PubMed] [Google Scholar]
- Duan W, Guo Z, Jiang H, Ware M, Li XJ, Mattson MP (2003) Dietary restriction normalizes glucose metabolism and BDNF levels, slows disease progression, and increases survival in huntingtin mutant mice. Proc Natl Acad Sci USA 100: 2911-2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dube L, Smith AD, Bolam JP (1988) Identification of synaptic terminals of thalamic or cortical origin in contact with distinct medium-size spiny neurons in the rat neostriatum. J Comp Neurol 267: 455-471. [DOI] [PubMed] [Google Scholar]
- Ferrer I, Goutan E, Marin C, Rey MJ, Ribalta T (2000) Brain-derived neurotrophic factor in Huntington disease. Brain Res 866: 257-261. [DOI] [PubMed] [Google Scholar]
- Friedman WJ, Ernfors P, Persson H (1991) Transient and persistent expression of NT-3/HDNF mRNA in the rat brain during postnatal development. J Neurosci 11: 1577-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman WJ, Ibanez CF, Hallbook F, Persson H, Cain LD, Dreyfus CF, Black IB (1993) Differential actions of neurotrophins in the locus coeruleus and basal forebrain. Exp Neurol 119: 72-78. [DOI] [PubMed] [Google Scholar]
- Fusco FR, Zuccato C, Tartari M, Martorana A, De March Z, Giampa C, Cattaneo E, Bernardi G (2003) Co-localization of brain-derived neurotrophic factor (BDNF) and wild-type huntingtin in normal and quinolinic acid-lesioned rat brain. Eur J Neurosci 18: 1093-1102. [DOI] [PubMed] [Google Scholar]
- Galvin KA, Oorschot DE (2003) Continuous low-dose treatment with brain-derived neurotrophic factor or neurotrophin-3 protects striatal medium spiny neurons from mild neonatal hypoxia/ischemia: a stereological study. Neuroscience 118: 1023-1032. [DOI] [PubMed] [Google Scholar]
- Gorski JA, Talley T, Qiu M, Puelles L, Rubenstein JL, Jones KR (2002) Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. J Neurosci 22: 6309-6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski JA, Zeiler SR, Tamowski S, Jones KR (2003a) Brain-derived neurotrophic factor is required for the maintenance of cortical dendrites. J Neurosci 23: 6856-6865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski JA, Balogh SA, Wehner JM, Jones KR (2003b) Learning deficits in forebrain-restricted brain-derived neurotrophic factor mutant mice. Neuroscience 121: 341-354. [DOI] [PubMed] [Google Scholar]
- Guidetti P, Charles V, Chen EY, Reddy PH, Kordower JH, Whetsell Jr WO, Schwarcz R, Tagle DA (2001) Early degenerative changes in transgenic mice expressing mutant huntingtin involve dendritic abnormalities but no impairment of mitochondrial energy production. Exp Neurol 169: 340-350. [DOI] [PubMed] [Google Scholar]
- Gundersen HJ, Bendtsen TF, Korbo L, Marcussen N, Moller A, Nielsen K, Nyengaard JR, Pakkenberg B, Sorensen FB, Vesterby A, West MJ (1988) Some new, simple and efficient stereological methods and their use in pathological research and diagnosis. Apmis 96: 379-394. [DOI] [PubMed] [Google Scholar]
- Hamilton BA, Frankel WN, Kerrebrock AW, Hawkins TL, FitzHugh W, Kusumi K, Russell LB, Mueller KL, van Berkel V, Birren BW, Kruglyak L, Lander ES (1996) Disruption of the nuclear hormone receptor RORalpha in staggerer mice. Nature 379: 736-739. [DOI] [PubMed] [Google Scholar]
- Hofer M, Pagliusi SR, Hohn A, Leibrock J, Barde YA (1990) Regional distribution of brain-derived neurotrophic factor mRNA in the adult mouse brain. EMBO J 9: 2459-2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang EJ, Reichardt LF (2001) Neurotrophins: roles in neuronal development and function. Annu Rev Neurosci 24: 677-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivkovic S, Ehrlich ME (1999) Expression of the striatal DARPP-32/ARPP-21 phenotype in GABAergic neurons requires neurotrophins in vivo and in vitro J Neurosci 19: 5409-5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivkovic S, Polonskaia O, Farinas I, Ehrlich ME (1997) Brain-derived neurotrophic factor regulates maturation of the DARPP-32 phenotype in striatal medium spiny neurons: studies in vivo and in vitro. Neuroscience 79: 509-516. [DOI] [PubMed] [Google Scholar]
- Johnston JG, Gerfen CR, Haber SN, van der Kooy D (1990) Mechanisms of striatal pattern formation: conservation of mammalian compartmentalization. Brain Res Dev Brain Res 57: 93-102. [DOI] [PubMed] [Google Scholar]
- Jones KR, Farinas I, Backus C, Reichardt LF (1994) Targeted disruption of the BDNF gene perturbs brain and sensory neuron development but not motor neuron development. Cell 76: 989-999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp JM, Powell TP (1971a) The synaptic organization of the caudate nucleus. Philos Trans R Soc Lond B Biol Sci 262: 403-412. [DOI] [PubMed] [Google Scholar]
- Kemp JM, Powell TP (1971b) The termination of fibres from the cerebral cortex and thalamus upon dendritic spines in the caudate nucleus: a study with the Golgi method. Philos Trans R Soc Lond B Biol Sci 262: 429-439. [DOI] [PubMed] [Google Scholar]
- Kohara K, Kitamura A, Adachi N, Nishida M, Itami C, Nakamura S, Tsumoto T (2003) Inhibitory but not excitatory cortical neurons require presynaptic brain-derived neurotrophic factor for dendritic development, as revealed by chimera cell culture. J Neurosci 23: 6123-6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalonde R (1987) Motor abnormalities in staggerer mutant mice. Exp Brain Res 68: 417-420. [DOI] [PubMed] [Google Scholar]
- Martinez-Serrano A, Bjorklund A (1996) Protection of the neostriatum against excitotoxic damage by neurotrophin-producing, genetically modified neural stem cells. J Neurosci 16: 4604-4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlio JP, Ernfors P, Jaber M, Persson H (1992) Molecular cloning of rat trkC and distribution of cells expressing messenger RNAs for members of the trk family in the rat central nervous system. Neuroscience 51: 513-532. [DOI] [PubMed] [Google Scholar]
- Mizuno K, Carnahan J, Nawa H (1994) Brain-derived neurotrophic factor promotes differentiation of striatal GABAergic neurons. Dev Biol 165: 243-256. [DOI] [PubMed] [Google Scholar]
- Mullen RJ, Buck CR, Smith AM (1992) NeuN, a neuronal specific nuclear protein in vertebrates. Development 116: 201-211. [DOI] [PubMed] [Google Scholar]
- Nakao N, Brundin P, Funa K, Lindvall O, Odin P (1995) Trophic and protective actions of brain-derived neurotrophic factor on striatal DARPP-32-containing neurons in vitro. Brain Res Dev Brain Res 90: 92-101. [DOI] [PubMed] [Google Scholar]
- Nimchinsky EA, Sabatini BL, Svoboda K (2002) Structure and function of dendritic spines. Annu Rev Physiol 64: 313-353. [DOI] [PubMed] [Google Scholar]
- Nisenbaum LK, Webster SM, Chang SL, McQueeney KD, LoTurco JJ (1998) Early patterning of prelimbic cortical axons to the striatal patch compartment in the neonatal mouse. Dev Neurosci 20: 113-124. [DOI] [PubMed] [Google Scholar]
- Perez-Navarro E, Canudas AM, Akerund P, Alberch J, Arenas E (2000) Brain-derived neurotrophic factor, neurotrophin-3, and neurotrophin-4/5 prevent the death of striatal projection neurons in a rodent model of Huntington's disease. J Neurochem 75: 2190-2199. [DOI] [PubMed] [Google Scholar]
- Regeur L, Pakkenberg B (1989) Optimizing sampling designs for volume measurements of components of human brain using a stereological method. J Microsc 155: 113-121. [DOI] [PubMed] [Google Scholar]
- Reiner A, Albin RL, Anderson KD, D'Amato CJ, Penney JB, Young AB (1988) Differential loss of striatal projection neurons in Huntington disease. Proc Natl Acad Sci USA 85: 5733-5737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidtke K, Manner H, Kaufmann R, Schmolck H (2002) Cognitive procedural learning in patients with fronto-striatal lesions. Learn Mem 9: 419-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal M, Andersen P (2000) Dendritic spines shaped by synaptic activity. Curr Opin Neurobiol 10: 582-586. [DOI] [PubMed] [Google Scholar]
- Seroogy KB, Lundgren KH, Tran TM, Guthrie KM, Isackson PJ, Gall CM (1994) Dopaminergic neurons in rat ventral midbrain express brain-derived neurotrophic factor and neurotrophin-3 mRNAs. J Comp Neurol 342: 321-334. [DOI] [PubMed] [Google Scholar]
- Sheth AN, McKee ML, Bhide PG (1998) The sequence of formation and development of corticostriate connections in mice. Dev Neurosci 20: 98-112. [DOI] [PubMed] [Google Scholar]
- Tyler WJ, Pozzo-Miller L (2003) Miniature synaptic transmission and BDNF modulate dendritic spine growth and form in rat CA1 neurones. J Physiol (Lond) 553: 497-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dellen A, Deacon R, York D, Blakemore C, Hannan AJ (2001) Anterior cingulate cortical transplantation in transgenic Huntington's disease mice. Brain Res Bull 56: 313-318. [DOI] [PubMed] [Google Scholar]
- van den Akker E, Reijnen M, Korving J, Brouwer A, Meijlink F, Deschamps J (1999) Targeted inactivation of Hoxb8 affects survival of a spinal ganglion and causes aberrant limb reflexes. Mech Dev 89: 103-114. [DOI] [PubMed] [Google Scholar]
- Ventimiglia R, Mather PE, Jones BE, Lindsay RM (1995) The neurotrophins BDNF, NT-3 and NT-4/5 promote survival and morphological and biochemical differentiation of striatal neurons in vitro. Eur J Neurosci 7: 213-222. [DOI] [PubMed] [Google Scholar]
- Vigers AJ, Baquet ZC, Jones KR (2000) Expression of neurotrophin-3 in the mouse forebrain: insights from a targeted LacZ reporter. J Comp Neurol 416: 398-415. [DOI] [PubMed] [Google Scholar]
- von Bartheld CS, Byers MR, Williams R, Bothwell M (1996) Anterograde transport of neurotrophins and axodendritic transfer in the developing visual system. Nature 379: 830-833. [DOI] [PubMed] [Google Scholar]
- West MJ, Slomianka L, Gundersen HJ (1991) Unbiased stereological estimation of the total number of neurons in the subdivisions of the rat hippocampus using the optical fractionator. Anat Rec 231: 482-497. [DOI] [PubMed] [Google Scholar]
- Widmer HR, Hefti F (1994) Neurotrophin-4/5 promotes survival and differentiation of rat striatal neurons developing in culture. Eur J Neurosci 6: 1669-1679. [DOI] [PubMed] [Google Scholar]
- Wu K, Xu JL, Suen PC, Levine E, Huang YY, Mount HT, Lin SY, Black IB (1996) Functional trkB neurotrophin receptors are intrinsic components of the adult brain postsynaptic density. Brain Res Mol Brain Res 43: 286-290. [DOI] [PubMed] [Google Scholar]
- Zuccato C, Ciammola A, Rigamonti D, Leavitt BR, Goffredo D, Conti L, MacDonald ME, Friedlander RM, Silani V, Hayden MR, Timmusk T, Sipione S, Cattaneo E (2001) Loss of huntingtin-mediated BDNF gene transcription in Huntington's disease. Science 293: 493-498. [DOI] [PubMed] [Google Scholar]