Abstract
TRPV1 (transient receptor potential vanilloid receptor subtype 1) is a member of the TRP channel family gated by vanilloids, protons, and heat. Structurally, TRPV1 appears to be a tetramer formed by the assembly of four identical subunits around a central aqueous pore. The molecular determinants that govern its subunit oligomerization remain elusive. Here, we report the identification of a segment comprising 684Glu-721Arg (referred to as the TRP-like domain) in the C terminus of TRPV1 as an association domain (AD) of the protein. Purified recombinant C terminus of TRPV1 (TRPV1-C) formed discrete and stable multimers in vitro. Yeast two-hybrid and pull-down assays showed that self-association of the TRPV1-C is blocked when segment 684Glu-721Arg is deleted. Biochemical and immunological analysis indicate that removal of the AD from full-length TRPV1 monomers blocks the formation of stable heteromeric assemblies with wild-type TRPV1 subunits. Deletion of the AD in a poreless TRPV1 subunit suppressed its robust dominant-negative phenotype. Together, these findings are consistent with the tenet that the TRP-like domain in TRPV1 is a molecular determinant of the tetramerization of receptor subunits into functional channels. Our observations suggest that the homologous TRP domain in the TRP protein family may function as a general, evolutionary conserved AD involved in subunit multimerization.
Keywords: ion channel, oligomerization, TRP domain, nociceptors, sensory transduction, synaptic transmission
Introduction
TRPV1 (transient receptor potential vanilloid receptor subtype 1) is a polymodal detector of noxious chemical and thermal stimuli (Caterina and Julius, 2001). An expression-cloning strategy, using capsaicin as an specific agonist, unveiled its molecular identity (Caterina et al., 1999). Heterologous expression of TRPV1 cDNA results in capsaicin-activated inward currents that recapitulate most of the functional properties described for the capsaicin and heat-activated receptors of dorsal root ganglion neurons (Caterina et al., 2000). Receptor functional analysis demonstrated that TRPV1 is a nonselective cation channel activated by vanilloids and temperatures higher than 42°C, which exhibits a time- and Ca2+-dependent outward rectification, followed by a long-lasting refractory state (during which the cell does not respond to capsaicin or other stimuli). In addition, mild extracellular acidic pH (∼6.5) potentiates TRPV1 channel activation by noxious heat and vanilloids, whereas acidic conditions (pH < 6) directly activate the channel. Several studies have demonstrated a critical role of the receptor in acute thermal nociception and hyperalgesia and neurogenic inflammation (Caterina et al., 2000; Davis et al., 2000, García-Martínez et al., 2002; Ji et al., 2002).
The TRPV1 protein is a member of the large TRP ion channel family (Clapham et al., 2001; Minke and Cook, 2002; Montell et al., 2002; Birnbaumer et al., 2003; Clapham, 2003). TRPV1 subunits are membrane proteins with a predicted relative molecular mass of 95 kDa (Caterina and Julius, 2001; Kedei et al., 2001). Structurally, TRPV1 monomers display a hydrophilic intracellular N-terminus domain containing three-conserved ankyrin repeats and several kinase consensus sequences (see Fig. 1A). Phosphorylation of this domain by PKA notably affects the rate of capsaicin-induced receptor desensitization (Bhave et al., 2002). Hydrophobicity analysis of the protein reveals the presence of six putative membrane-spanning segments (S1-S6) and a stretch linking the fifth and sixth segments that contains an amphipathic fragment denoted as the P-loop, which contributes to its permeation properties (García-Martínez et al., 2000). The protein also exhibits a cytosolic C terminus that harbors phosphoinositide and calmodulin binding domains, and PKC consensus sites (Numazaki et al., 2002, 2003; Bhave et al., 2003; Prescott and Julius, 2003;). In addition, this region shows a TRP-like domain containing a TRP box with similarity to the highly conserved TRP domain present in the TRP protein family (Clapham et al., 2001; Minke and Cook, 2002; Montell et al., 2002; Birnbaumer et al., 2003; Clapham, 2003). Functional studies have attributed to the C terminus an important role on inflammation-induced TRPV1 potentiation, as well as the receptor thermal sensitivity (Premkumar and Ahern, 2000; Numazaki et al., 2002, 2003; Bhave et al., 2003; Prescott and Julius, 2003).
Figure 1.
Identification of a potential association domain in the C terminus of TRPV1. A, Molecular model of TRPV1 subunits showing the N-terminus and C-terminus cytosolic domains, the six membrane-spanning segments, and the amphipathic stretch connecting the fifth and sixth transmembrane segments. The amino acid sequence of the C terminus, along with the secondary structure prediction, is shown at the bottom. Cylinders and arrows denote predicted α-helices and β-strands, respectively. Secondary structure prediction was performed by the consensus secondary structure prediction method (www.expasy.org). B, Coiled-coil prediction of the C terminus of TRPV1. Analysis was performed with the program Coils (www.ch.embnet.org) (Lupas et al., 1991; Lupas, 1996). C, Sequence alignment of TRPV1 C terminus of the TRPV family members. Shaded areas denote amino acid conservation. The TRP box and the calmodulin and PIP2 binding domains are depicted (Clapham, 2003; Numazaki et al., 2003; Prescott and Julius, 2003). Amino acid alignment was performed with the program Antheprot version 4.7.
TRPV1 monomers appear to assemble with fourfold symmetry around a central aqueous pore, giving rise to a tetrameric stoichiometry for the functional TRPV1 homomer (Kedei et al., 2001; Kuzhikandathil et al., 2001). The molecular determinants that define TRPV1 subunit multimerization are still elusive. Recent studies have identified protein motifs in the C terminus of K+-selective, voltage-gated channels that act as association domains (ADs) of channel monomers (Zerangue et al., 2000; Quirk and Reinhart, 2001; Jenke et al., 2003). These protein domains, also referred to as “tetramerization coiled-coils” (or TCCs), are precisely located immediately carboxyl of the pore region of each subunit and encompass an stretch of ≤45 amino acids (Jenke et al., 2003). A hallmark of ADs is their high propensity to adopt an α-helical secondary structure and to form coiled-coil multimers (Jenke et al., 2003). Here, we report that the TRP-like domain comprising 684Glu-721Arg between the core channel domain and the C-terminal modulatory region fulfills these properties (see Fig. 1A). Our results indicate that this segment is an AD implicated in the tetramerization of TRPV1 subunits into functional channels.
Materials and Methods
Plasmids and deletion constructs. The C terminus of rat TRPV1 (amino acids 682-830) was cloned into the pET-22b plasmid (Novagen, Madison, WI) with NdeI/HindIII to obtain a His-tagged fusion protein with the 6xHis incorporated at the C end, into the pGEX-4T1 plasmid (Amersham Biosciences, Arlington Heights, IL) with NotI/SalI to obtain a glutathione S-transferase (GST)-fusion protein, into the pGBKT7 plasmid (Clontech, Cambridge, UK) with NdeI/EcoRI, and into the pACT2 plasmid (Clontech) with NdeI/SmaI and Klenow for yeast two-hybrid assays.
Deletions were obtained by one-step inverse PCR with the proofreading Pfu turbo DNA polymerase and two restriction digestions (Cabedo et al., 2002). PCR products were digested (in the amplification buffer) with 20 U of DpnI overnight at 37°C to remove the methylated plasmid templates, subsequently purified, digested, and ligated. Designed deletions were verified by restriction digestion and by automatic sequencing.
His-tagged fusion protein expression and purification. His-tagged C terminus of TRPV1 (TRPV1-C) fusion protein in pET-22b was transformed into BL21-CodonPlus (RIL) Escherichia coli (Stratagene, La Jolla, CA). Bacterial cultures were incubated at 37°C with agitation until reaching an OD600 of 0.6. Protein expression was induced with 0.1 mm isopropyl-β-d-thiogalactopyranoside at 37°C for 4 hr. Bacteria were collected by centrifugation, resuspended in 100 mm Tris, pH 8.8, buffer, and lysed by sonication in a Branson (Danbury, CT) Sonifier Cell Disruptor 250 at 4°C. Inclusion bodies containing the His-fusion protein were purified by centrifugation from the bacterial lysates, solubilized with 6.0 m guanidinum chloride in 100 mm Tris, pH 8.8, and incubated with Ni-NTA agarose (Qiagen, Hilden, Germany). To facilitate the refolding of the protein, the guanidinium chloride was slowly removed with a linear gradient of urea (Suenaga et al., 1998). His-tagged TRPV1-C protein was eluted from the column with 250 mm Imidazol, 500 mm NaCl, 100 mm NaH2PO4, 10mm β-mercaptoethanol, 20% glycerol, 0.2 m arginine, and 2% 3-[(3-cholamidopropyl) dimethylammonio]-1-propanesulfonate, pH 8.0. Purity of the protein was evaluated by SDS-PAGE.
Yeast two-hybrid assay. The TRPV1-C and different deletion mutants were fused to the Gal4 DNA binding domain (pGBKT7) and/or the Gal4 DNA activation domain (pACT2) and transformed into the PJ69-2A strain of Saccaromyces cerevisiae (Bai and Elledge, 1997). Positive interactions were identified by the ability of yeast to grow on selective medium (-Trp/-Leu) and for α-galactosidase expression.
In vitro pull-down assay. GST-fusion proteins were affinity purified on gluthation-Sepharose 4D columns (Amersham Biosciences). Immobilized fusion proteins (∼10 μg) were incubated with in vitro translated [35S]TRPV1-C or [35S]Δ(684-707) or [35S]Δ(684-721) (TNT T7 Quick Coupled Transcription/Translation System; Promega, Madison, WI) in binding buffer (10 mm Tris-HCl, 300 mm NaCl, 16 mm 2-mercaptoethanol, 1% Triton X-100, and 10 mm gluthation, pH 8.0) for 2 hr at 22°C. After three washes with binding buffer, GST-bound complexes were eluted from the resin and denaturated with SDS-PAGE sample buffer at 60°C for 30 min. Proteins complexes were resolved by SDS-PAGE gels and visualized by autofluorography.
Immunocytochemistry. The TRPV1 wild-type subunit was tagged with an hemagglutinin (HA) epitope in the extracellular loop that connects transmembrane domains S1 and S2 by recombinant PCR and verified by DNA sequencing. Insertion of the HA epitope did not alter the TRPV1 channel properties (data not shown).
HEK293 cells were cultured in DMEM supplemented with 10% fetal calf serum and 2 mm l-glutamine and 1% penicillin-streptomycin solution. Before transfection, the cells were replated at a density of ∼1 × 105 cells/cm2 onto poly-d-lysine-coated coverslips for immunofluorescence. Cells were transfected using the Lipofectamine 2000 (Invitrogen, San Diego, CA) using the recommended protocol of the manufacturer and was used for experiments 48 hr after transfection. Transfected cells were incubated with 1:250 dilution of anti-HA antibody (clone 12CA5; Roche Products, Hertfordshire, UK) in DMEM supplemented with serum for 1 hr at 4°C. After extensive washing, cells were fixed in 4% paraformaldehyde and 4% sucrose for 20 min. Cells were then washed and incubated with Alexa488 anti-mouse IgG (Molecular Probes, Eugene, OR) secondary antibody, washed, embedded, and analyzed by confocal microscopy (model TCS; Leica, Nussloch, Germany).
Biotin labeling of surface proteins. Transfected HEK293 cells were harvested 48 hr after transfection, incubated with sulfo-NHS-biotin (Sigma, St. Louis, MO) for 30 min at 4°C, and lysed with lysis buffer (50 mm HEPES, pH 7.4, 150 mm NaCl, 10% glycerol, 1% Triton X-100, 2 mm phenylmethylsulfonyl fluoride, and 5 mm iodoacetamide) for 20 min at 22°C. Biotin-conjugated cell surface proteins were purified with streptavidin-agarose (Sigma). Affinity-purified protein complexes were denaturated with SDS-PAGE sample buffer containing 4 m urea (75°C, 20 min), separated by SDS-PAGE, and probed by Western immunoblotting using anti-TRPV1 serum (1:2000) overnight at 4°C. Immunoreactive bands were visualized using the ECL method.
Oocyte electrophysiology. Amphibian oocytes were harvested and microinjected with cRNA encoding the rat TRPV1 channel and deletion species as described previously (García-Martínez et al., 2000, 2002). Whole-cell currents from oocytes were recorded with a two-microelectrode voltage-clamp amplifier. Oocytes were continuously perfused (2 ml/min) with Mg2+-Ringer's solution (in mm: 10 HEPES, pH 7.4, 115 NaCl, 2.8 KCl, 0.1 BaCl2, and 2.0 MgCl2) at 20°C. TRPV1 currents were activated with acidic solution (Mg2+-Ringer's solution with 10 mm MES, pH 6.0) or capsaicin at the indicated concentrations. The holding potential was kept at -80 mV. Recordings were performed 3-5d after injection. Data acquisition and processing was performed with the Pulse/PulseFit version 8.5 software package (HEKA Elektronik, Lambrecht/Pfalz, Germany).
Circular dichroism measurements. Circular dichroism (CD) spectra were collected on a Jasco (Easton, MD) J810 spectropolarimeter fitted with a thermostated cell holder. Far-UV measurements (200-260 nm) were recorded with a 1 mm path length cell containing 0.25 mg/ml synthetic peptide encoding part of the predicted AD (NETVNKIAQESKNIWKLQRAITILDTEKSFLKG) in 20 mm sodium phosphate and 150 mm NaCl, pH 7.5. All spectra were recorded at 25°C and at 50 nm/min (response time of 1 sec), averaged (five scans), and corrected for the buffer contribution. CD signals [in millidegrees (mdeg)] were converted to mean ellipticity (θ, deg/cm2/dmol) using the relationship θv = 0.1 × CD signal/(C × N × I), were C denotes the peptide concentration, N is the number of residues, and I is the path length. Secondary structure elements were inferred by fitting the CD spectra as described previously (Blanes-Mira et al., 2001).
Molecular modeling. The model was constructed using the structure of C terminus of the hyperpolarized, cyclic nucleotide-gated channel HCN2 in the presence of cGMP [Protein Data Bank (PDB) accession number 1Q3E, at 1.9 Å resolution] as the template. The multiple sequence alignment of several members of the transport proteins family [HCN1, HCN2, SPIH (poliketide synthase), and CNGA1 (cyclic nucleotide gated channel α1)] and TPRV1 C terminal (see Fig. 6 A) was made with ClustalW at European Bioinformatics Institute web page (http://www.ebi.ac.uk/) (Higgins et al., 1994). The homology modeling was performed in the Swiss-Model Protein Modeling Server at ExPASy Molecular Biology web server (http://kr.expasy.org/) (Schwede et al., 2003). Structure visualization was made using WHATIF and Swiss PDB viewer version 3.7 (Vriend, 1990; Guex and Peitsch, 1997). The resulting PDB files containing the model were energy minimized using GROMOS 43B1 implemented in Swiss-PDB Viewer version 3.7 and evaluated in terms of energy by FOLD-X web page (http://fold-x.embl.de) (Guerois et al., 2002).
Figure 6.
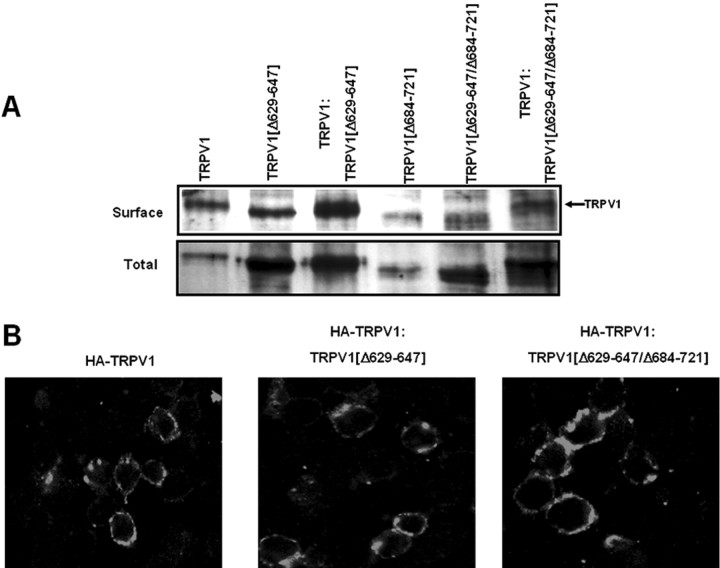
Surface expression of TRPV1 is not affected by deletion mutants TRPV1(Δ629-647) and TRPV1(Δ629-647/Δ684-721). A, Cell surface and total expression of TRPV1 and deletion mutants expressed alone or coexpressed in HEK293 cells. Biotinylated surface proteins were affinity purified with agarose-streptavidin. Total, Total protein present in whole-cell extracts before affinity purification. The presence of TRPV1 was probed by Western immunoblots using an anti-TRPV1 antibody. B, Confocal immunofluorescence images of HA-tagged TRPV1 expressed alone (left) or coexpressed with TRPV1(Δ629-647) (middle) or with TRPV1(Δ629-647/Δ684-721) (right). Coexpression was performed using a cDNA ratio of 1:1 (w/w). Conditions are as described in Figure 4.
Results
Sequence analysis of the C terminus of TRPV1
To identify a protein motif in TRPV1-C involved in subunit oligomerization, we evaluated its propensity to adopt an α-helical secondary conformation and to form coiled-coil structures, two hallmarks of ADs of channel proteins (Jenke et al., 2003). As displayed in Figure 1, A and B, the stretch-encompassing residues 684Glu-721Arg, immediately after the S6 transmembrane segment, are predicted to fold into an α-helix and to form coiled-coil structures. The C-terminal region of TRPV1-C exhibits the presence of mixture of short α-helical and β-strand secondary elements and the complete absence of coiled-coil peaks. Notably, amino acid sequence alignments of TRPV family members reveals an evolutionary conservation of segment 684Glu-721Arg (Fig. 1C). In marked contrast, the modulatory protein domain containing the phosphoinositide and calmodulin (769Val-818Thr) interacting sites in TRPV1 is poorly preserved in other TRPV channels. These observations suggest that the region 684Glu-721Arg may function as ADs involved in TRPV1 subunit multimerization. Because of the homologous protein location of segment 684Glu-721Arg and the so-called TRP domain, a highly conserved 25 amino acid motif in the TRPC (TRP, canomical), TRPN (TRP, ankyrin-repeat), TRPP (TRP, polycystic), TRPM (TRP, melastatin) protein subfamilies (Clapham et al., 2001; Minke and Cook, 2002; Montell et al., 2002; Birnbaumer et al. 2003; Clapham, 2003), we denote it in TRPV1 as the TRP-like domain. Note that the TRP-like domain also contains a conserved TRP box (IWKLQR).
The TRP-like domain is a molecular determinant of protein oligomerization
We first evaluated whether recombinant TRPV1-C is able to form stable oligomers in vitro. Recombinant TRPV1-C was purified from inclusion bodies and in vitro refolded. As depicted in Figure 2A, bacterially expressed, monomeric His-tagged TRPV1-C displays an electrophoretic mobility of ∼16 kDa, consistent with the protein estimated molecular weight of 18.6 kDa. However, we noticed that in vitro refolded TRPV1-C monomers were unstable in solution and tended rapidly (≤1 hr) to aggregate at 23°C. Intriguingly, SDS-PAGE analysis of the aggregated solution revealed the presence of individual, SDS-resistant multimers of 30, 55, and 80 kDa apparent molecular weights, which might correspond to dimers, trimers, and tetramers of TRPV1-C monomers. However, because protein aggregates may have anomalous electrophoretic mobility, these assemblies could represent higher-order oligomers. Nonetheless, independently of the stoichiometry of the in vitro aggregates, their formation implies that TRPV1-C self-associates in solution.
Figure 2.
TRPV1-C self-associates in vitro. A, Multimerization of affinity-purified His-tagged TRPV1-C protein in solution. Protein concentration was 1 mg/ml. Arrows indicate the monomeric, dimeric, trimeric, and tetrameric assemblies of the fusion protein. B, Schematic representation of the TRPV1-C deletion species designed the role as AD of the 684Glu-721Arg region. Truncation mutant Δ(684-707) deletes the first predicted α-helix, whereas deletion species Δ(684-721) removes both α-helices (Fig. 1 A). C, Yeast two-hybrid analysis of the interaction between the full-length TRPV1-C and deletion species in selective medium. Blue indicates X-α-Gal degradation by the reporter gene α-galactosidase. D, Pull-down assays of in vitro translated [35S]TRPV1-C, [35S]Δ(684-707), and [35S]Δ(684-721) with GST-TRPV1-C and GST-Δ(684-707).
To investigate the role of the TRP-like domain in the protein self-association, we analyzed the protein-protein interactions by yeast two-hybrid assays using the entire TRPV1-C as bait. The preys were the full-length TRPV1-C and the deletion forms Δ(684-707) and Δ(684-721) (Fig. 2B), which suppress the long α-helix and the predicted coiled-coil domain, respectively (Fig. 1A). Figure 2C shows yeast growth and α-galactosidase activity when the TRPV1-C was used as bait and prey, corroborating that the C terminus of TRPV1 self-associates. The interaction with TRPV1-C appears preserved for deletion mutant Δ(684-707), which also exhibits self-association. In contrast, yeast growth and α-galactosidase activity were impaired when the truncation species Δ(684-721) was used as prey, indicating that its interaction with the TRPV1-C was abolished. Therefore, segment 684Glu-721Arg appears to be a molecular determinant of TRPV1-C self-association.
The strength of the monomer-monomer interaction was evaluated by in vitro pull-down assays. GST-tagged fusion proteins of wild-type and deletion mutant Δ(684-707) were immobilized onto gluthation beads and incubated with in vitro translated [35S]TRPV1-C, or [35S] Δ(684-707) or [35S] Δ(684-721) (Fig. 2D). As illustrated, the [35S]TRPV1-C was retained by GSTTRPV1-C and GST-Δ(684-707) but not by immobilized GST alone. Deletion mutant [35S]Δ(684-707) was only pulled down by GST-TRPV1-C, whereas [35S]Δ(684-721) was not retained by either fusion protein. Collectively, all of these findings indicate that the TRP-like domain is an AD of TRPV1-C implicated in the protein multimerization.
A peptide patterned after the TRP-like domain displays propensity to fold into an α-helix
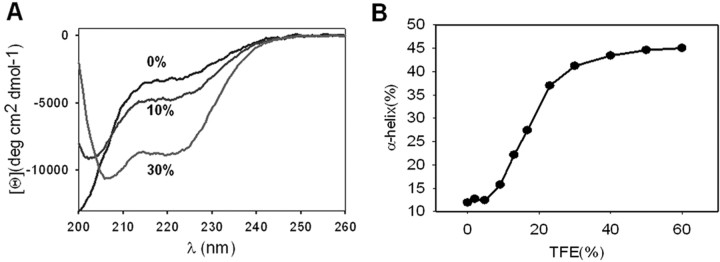
Secondary structure prediction algorithms forecast a high probability of stretch 684Glu-721Arg to adopt anα-helical structure (Fig. 1A). To validate this property, we inferred the secondary structure of a 32-mer peptide (Ac-GETVNKIAQESKNIWKLQRAITILDTEKSFLKG-NH2) mimicking most of the amino acid sequence of the oligomerization domain by CD. Analysis of CD spectra showed that the sequence exhibited a low content in α-helix secondary structure in aqueous buffer that was increased by addition of trifluoroethanol (TFE), as evidenced by the appearance of CD minima at 207 and 220 nm (Fig. 3A). As illustrated in Figure 3B, TFE augmented the α-helical content of the 32-mer peptide in a dose-dependent manner. Therefore, these results suggest that segment 684Glu-721Arg in the TRPV1-C exhibits a propensity to adopt an α-helical arrangement, in agreement with the predicted secondary structure (Fig. 1A, bottom). We could not observe, however, the formation of stable coiled-coil structures with the 32-mer peptide in solution by size-exclusion chromatography (data not shown), presumably because additional sequences in the TRVP1-C are necessary to thermodynamically and/or kinetically stabilize higher-order oligomers. In support of this tenet, TCCs that form stable multimers were longer than 40 amino acids (Jenke et al., 2003).
Figure 3.
A peptide patterned after the TRP-like segment exhibits propensity to adopt an α-helical conformation. A, Far-UV CD spectra of the peptide at increasing percentages of TFE (0, 10, and 30%). Peptide concentration was 100 μm in 20 mm Tris, pH 8.0.CD spectra represent the average of five scans and were corrected for the buffer contribution. B, Quantification of the α-helical content as a function of the percentage of TFE. α-Helical values were inferred as described previously (Blanes-Mira et al., 2001).
Deletion of the TRP-like domain abrogates TRPV1 channel function
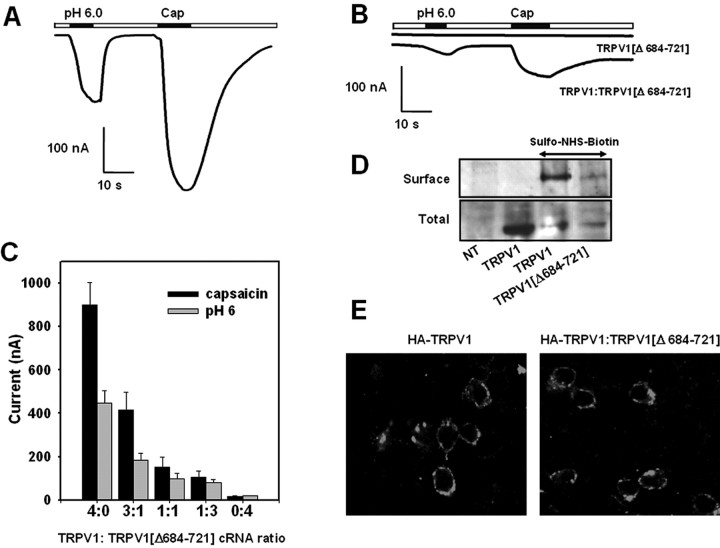
The functional relevance of the TRP-like domain on TRPV1 channel activity was next addressed in Xenopus oocytes. For this purpose, the amino acid segment 684Glu-721Arg was deleted from TRPV1 subunits [TRPV1(Δ684-721)]. Figure 4B, displays that, at variance with wild-type channels (Fig. 4A), the deletion mutant TRPV1(Δ684-721) failed to respond to both capsaicin (up to 100 μm) and extracellular acidic solution (pH 6.0). Biochemical analysis of transfected cells reveals that the total and surface expression of the deleted form TRPV1(Δ684-721) is reduced compared with the wild-type channels (Fig. 4D).
Figure 4.
Deletion of the TRP-like domain in TRPV1 abolishes channel function and affects channel expression. A, B, Acidic extracellular solution (pH 6.0) and 10 μm capsaicin (Cap) evoked responses from wild-type TRPV1 and deletion species TRPV1(Δ684-721), respectively, expressed in Xenopus oocytes. Ionic currents evoked from oocytes coinjected with cRNA encoding both receptor species at a ratio 1:1 (w/w) is also shown in B. C, Magnitude of the average ionic currents evoked by pH 6.0 and 10 μm capsaicin as a function of the cRNA ratio (w/w) encoding TRPV1 and TRPV1(Δ684-721) coinjected into oocytes. Ionic currents were obtained in Mg2+-Ringer's solution at a holding potential of -80 mV. Data are given as mean ± SEM, with n ≥ 6 (number of oocytes). D, Cell surface and total expression of TRPV1 and TRPV1(Δ684-721) expressed in HEK293 cells. Surfaced expressed proteins were biotinylated with sulfo-NHS-biotin and affinity purified with agarose-streptavidin. Total expression denotes the expressed protein in the whole-cell extracts before affinity purification. The presence of TRPV1 species was probed with an anti-TRPV1 antibody by Western immunoblot. NT, Nontransfected cells. E, TRPV1(Δ684-721) does not prevent the surface expression of HA-tagged TRPV1 channels. Confocal immunofluorescence images of HEK293 cells transfected with HA-tagged TRPV1 (left) and cotransfected with HA-tagged TRPV1 and TRPV1(Δ684-721) at a cDNA ratio of 1:1 (w/w) (right). Nonpermeabilized cells were incubated with an anti-HA monoclonal antibody, extensively washed, and probed with Alexa488 anti-mouse IgG.
In coexpression assays of TRPV1(Δ684-721) with TRPV1, the magnitude of the capsaicin- and pH-elicited ionic currents was decreased as the cRNA ratio TRPV1(Δ684-721)/TRPV1 was augmented (Fig. 4B,C), although TRPV1 responses were not completely abolished by the deletion species. Inhibition of TRPV1 activity by TRPV1(Δ684-721) was not attributable to downregulation of the surface expression of wild-type monomers, as evidenced by the immunocytochemical detection of HA-tagged TRPV1 subunits in the plasma membrane of cells cotransfected with both monomers (Fig. 4E). The capsacin- and pH-evoked ionic currents recorded from putative heteromeric TRPV1(Δ684-721)/TRPV1 channels were virtually identical to those elicited from wild-type channels. As depicted, the relationship Icap/IpH was not significantly changed, nor was it the shape of the I-V relationships or the reversal potential of the ionic currents [2 ± 3 mV (n = 3) for TRPV1 and -1 ± 2 mV for TRPV1/TRPV1(Δ684-721) (1:1) (n = 3)]. Thus, the ionic currents recorded from oocytes coinjected with TRPV1(Δ684-721) and TRPV1 cRNAs appear to be primarily performed by wild-type channels. Together, these data suggest that deletion species TRPV1(Δ684-721) may not coassemble with TRPV1. In addition, our findings imply that the TRP-like domain is important for efficient expression of functional channels. Suppression of this domain seems to create truncated monomers incapable of self-associating, as well as to interact with wild-type subunits.
Deletion of the TRP-like domain from a poreless TRPV1 subunit suppresses its robust dominant-negative phenotype
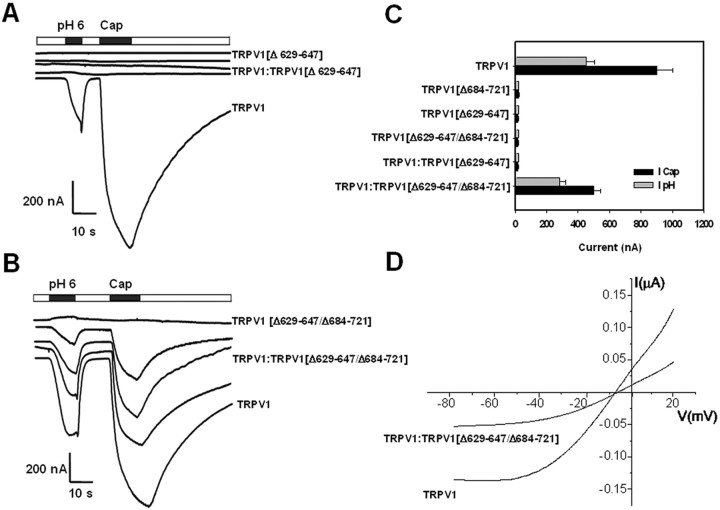
To demonstrate that the TRP-like domain is critical for subunit oligomerization, we designed a rescue experiment of TRPV1 channel activity from the robust negative dominance of a poreless TRPV1 mutant monomer. Similar to Quirk and Reinhart (2001), we deleted the P-loop in the pore domain of TRPV1 (amino acids 629-647), giving rise to the nonfunctional deletion mutant TRPV1(Δ629-647) (also referred here to as the poreless TRPV1) (Fig. 5A,C). It is worth noting that removal of the P-loop did not affect the total and surface expression of the truncated protein, indicating that this protein domain is not involved in subunit multimerization (Fig. 6A). Coexpression of the poreless subunit with full-length TRPV1 monomers fully abolished the functional expression of wild-type channels at all cRNA ratios explored, indicating a strong dominant-negative phenotype of the poreless subunit (Fig. 5A,C). The poreless TRPV1 monomer and the wild-type subunit were expressed in the plasma membrane, as deduced from biotinylation of both protein subunits (Fig. 6A). The delivery of HA-tagged TRPV1 to the plasma membrane was not disturbed by the presence of poreless subunits (Fig. 6B). Thus, together, these results imply that TRPV1 and poreless TRPV1 monomers coassemble, giving rise to nonfunctional heteromeric channels, and suggest that a single poreless monomer in the final oligomer suffices to abrogate channel function.
Figure 5.
Removal of TRP-like domain in a poreless TRPV1 channel revokes its robust negative dominance over wild-type channels. A, Coinjection of TRPV1 channels with the poreless mutant TRPV1(Δ629-647) produces nonfunctional channels. B, Coexpression of TRPV1 with the double-deletion mutant TRPV1(Δ629-647/Δ684-721) gives rise to functional channels. Ionic currents were elicited by extracellular acidic conditions (pH 6.0) and 10 μm capsaicin (Cap) at a holding potential of -80 mV. Deletion species and wild-type monomers were coinjected at the following cRNA ratios: 3:1, 2:2, and 1:3 (w/w) (middle traces in A, B). C, Magnitude of the pH 6.0 and 10 μm capsaicin evoked responses for the different TRPV1 species assayed. Coexpression refers to the cRNA ratio 1:1 (w/w). Data are given as mean ± SEM, with n ≥ 6 (number of oocytes). D, Representative current-to-voltage characteristics evoked by pH 6.0 from oocytes injected with TRPV1 alone and coinjected with TRPV1(Δ629-647/Δ684-721)(1:1) cRNAs. Oocytes were held at -80 mV and depolarized to 20 mV in 5 sec (20 mV/sec) using a ramp protocol. Leak currents were obtained at extracellular pH 7.4 and subtracted from the agonist-evoked ionic currents (García-Martínez et al., 2000, 2002). Each trace is representative of at least three oocytes.
We reasoned that, if the TRP-like domain is an AD, its removal from the poreless mutant subunit should block its interaction with TRPV1 monomer, thus suppressing its negative dominance and allowing the expression of functional TRPV1 channels in a coinjection experiment. We subsequently deleted the TRP-like domain in the poreless TRPV1 subunit, therefore creating the deletion form TRPV1(Δ629-647/Δ684-721). As expected, deletion mutant TRPV1(Δ629-647/Δ684-721) was not functional (Fig. 5B). Noteworthy, its coexpression with TRPV1 subunits at different cRNA ratios did not prevent the expression of functional TRPV1 channels (Fig. 5B). The capsacin- and pH-evoked ionic currents in these coexpression assays were virtually identical to those characteristic of wild-type TRPV1 channels. The sensitivity to capsaicin, the ionic permeability, and the ratio Icap/IpH were undistinguishable from TRPV1 (Fig. 5C,D), suggesting that the channel activity recorded from oocytes co-injected with TRPV1 and TRPV1(Δ629-647/Δ684-721) correspond to homomeric wild-type channels.
To demonstrate that the TRPV1(Δ629-647/Δ684-721) does not multimerize with TRPV1 subunits, we analyzed the surface expression of wild-type subunits with two complementary strategies. First, we investigated biochemically the surface expression of two different receptor subunits when expressed alone or coexpressed with wild-type channels (Fig. 6A). As seen, removal of the TRP-like domain from the poreless subunit decreased the total and surface expression of the protein (Fig. 6A), in accord with results obtained with deletion mutant TRPV1(Δ684-721) (Fig. 4D). Noteworthy, coexpression of TRPV1(Δ629-647/Δ684-721) with TRPV1 did not alter the expression of the wild-type channels in the cell surface (Fig. 6A). However, the mutant protein TRPV1(Δ629-647/Δ684-721) does not seem to be present in the cell surface when coexpressed with wild-type monomers (Fig. 6A), implying that it does not associate with TRPV1 subunits. Therefore, these observations are consistent with the notion that the ionic current recorded in the coexpression experiments of TRPV1(Δ629-647/Δ684-721) and TRPV1 is mediated by homomeric wild-type channels.
This conclusion was further supported by the observation that mutant TRPV1(Δ629-647/Δ684-721) did not preclude the delivery of the HA-tagged TRPV1 channels to the plasma membrane (Fig. 6B). Because a single subunit of the poreless mutant in the oligomer abrogates channel function, these data strongly suggest that suppression of the TRP-like domain in the poreless TRPV1 prevents its interaction with TRPV1 subunits. Collectively, our findings demonstrate that the TRP-like domain is an AD involved in the multimerization of TRPV1 subunits.
Discussion
TRPV1 channels are oligomeric membrane proteins composed of four identical subunits assembled around a central aqueous pore (Kedei et al., 2001; Kuzhikandathil et al., 2001). Multimerization of channel monomers is mediated by oligomerization-association domains that determine the final subunit stoichiometry of the functional channel, as well as the subunit composition (Zerangue et al., 2000; Quirk and Reinhart, 2001; Jenke et al., 2003). Thus far, the molecular identity of this important domain in TRPV1 monomers has remained elusive.
The salient contribution of this study is the discovery that the TRP-like domain (comprising 684Glu-721Arg) adjacent to the inner pore helix is an AD involved in the tetramerization of TRPV1 channel subunits into functional channels. We found that the C terminus of TRPV1 self-associates in vitro, giving rise to stable multimers. Yeast two-hybrid and in vitro pull-down assays show that the TRP-like domain is necessary for the self-association of TRPV1-C monomers. Deletion of the AD in full-length TRPV1 subunits prevented their assembly into active homomeric channels. Functional analysis of coexpression experiments suggests that AD-less subunits are unable to stably associate with TRPV1 monomers, as implied by the virtually identical channel properties of homomeric and putative heteromeric assemblies. Furthermore, truncation of the AD in a poreless TRPV1 subunit fully suppressed its robust negative dominance over TRPV1 monomers when both subunit forms were coexpressed in Xenopus oocytes. Biochemical and immunological studies demonstrate that removal of the AD from TRPV1 notably affected the surface expression of the protein and blocked the formation of stable heteromeric assemblies with wild-type TRPV1 subunits. The functional role of AD appears specific because deletion of the extracellular P-loop that conforms the ionic pore abrogated channel function without affecting subunit oligomerization or membrane delivery. Together, these data provide compelling evidence that the TRP-like domain is a tetramerization motif of TRPV1 subunits. Because of the evolutionary conservation of the TRP protein family (Harteneck et al., 2000; Clapham, 2003), our results suggest a functional role in the elusive TRP domain as an AD that may delimit the selectivity of subunit multimerization of TRP channel subunits. Noteworthy, ADs are also located adjacent to the inner pore helix of voltage-gated K+ channels and hyperpolarized and cyclic-nucleotide-gated channels (HCN and GCN) (Paoletti et al., 1999; Wainger et al., 2001; Zagotta et al., 2003). Hence, the presence of ADs C terminal of the channel pore appears a preserved feature of ion channels, at least for those displaying the canonical Shaker-like channel topology.
Recently, Vlachová et al. (2003) have proposed a molecular model for the C terminus of TRPV1 on the basis of its presumed primary sequence similarity to the FHIT (fragile histidine triad) protein. According to this model, the TRP-like domain of TRPV1 adopts an anti-parallel β-strand conformation. Clearly, this structural arrangement is incompatible with the high propensity of the AD to fold into an α-helix secondary structure. Furthermore, a comparison of the secondary structure prediction of the FHIT and the C terminus of TRPV1 reveals notable secondary structural divergences, specially in the N end of TRPV1-C (Fig. 7A). In light of this information, a revision of the currently proposed molecular model seems warranted. For this task, we took advantage of the notion that TRP channels appear to be cast in the same evolutionary mold as voltage-gated K+ and HCN channels (Zagotta et al., 2003). Besides the similar topology, three additional features support this tenet. (1) The C terminus of TRPV1 and HCN appears to exhibit a modular structural arrangement consisting of binding domain (cyclic nucleotide for HCN and phosphoinositides for TRPV1) and a C-linker region that connects the binding domain to the channel pore. (2) It also consists of a fairly conserved pattern of secondary structural elements of the entire C-terminus domain, including the α-helical conformation of the region contiguous to the inner pore helix. Indeed, comparison of the secondary structure prediction of TRPV1-C and the C terminus of HCN channels reveals significant similarities (Fig. 7B). It is worth noting that only two secondary structure elements are missing in the TRPV1-C, namely the second α-helix and β-strand in the C-linker and binding domain, respectively. (3) The C-linker region that connects the binding domain and the pore region in both channel types functions as a tetramerization domain. Accordingly, the x-ray crystallographic structure of the C-terminal fragment of HCN2 may be considered as a realistic structural scaffold for modeling the TRPV1-C (Zagotta et al., 2003).
Figure 7.
TRPV1-C shows structural similarities to C terminus of HCN channels. A, Primary and secondary structures of the human FHIT protein (Swissprot P49789). B, Amino acid alignment of the C terminus of cyclic nucleotide-gated (HCN2, GenBank accession number NP_001185; HCN1, GenBank accession number NP_066550; SPIH, GenBank accession number CAD_43449; CNGA1, Swissprot accession number P29973) ion channels and TRPV1. Bars and arrows denote α-helices and β-strands secondary structure elements, respectively. Predicted elements for TRPV1 were compared with those present in the structure of HCN2 (Zagotta et al., 2003).
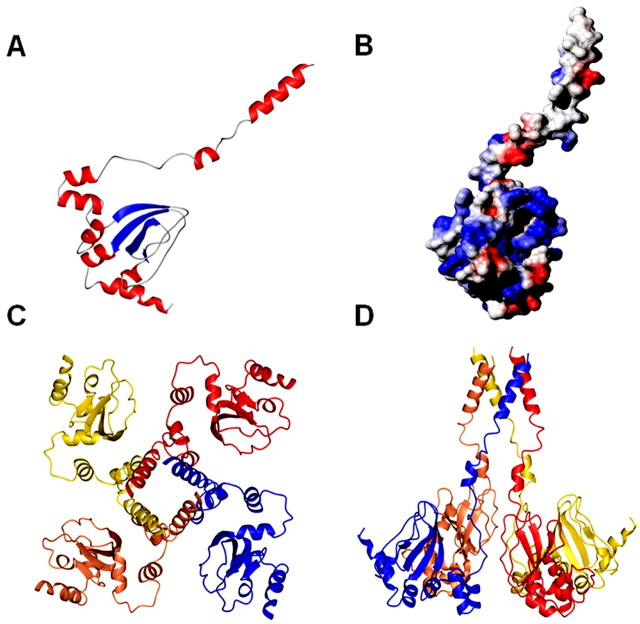
The molecular model of TRPV1-C is composed of two distinct domains, the C-linker corresponding to the TRP-like domain and the regulatory region harboring the phosphatidylinositol-4, 5-biphosphate (PIP2) and calmodulin binding sites (Fig. 8A). Whereas the binding domain shows a compact structure created by a mixture of four α-helices and four anti-parallel β-strands, the TRP-like domain consists of an amphipatic α-helix connected to the modulatory domain by a flexible loop and short α-helix. The surface electrostatic potential shows the presence of an apolar side for the α-helix that structures the AD, consistent with a multimerization role (Fig. 8B). The length and amphipaticity of helix A is compatible with the formation of a four-helix bundle structure that holds together the subunits to produce the homotetramer (Fig. 8C,D). The four-helix bundle arrangement structures a wide pore that may be relevant for ion permeation, similar to the HCN and shaker-like K+ channels (Cortes et al., 2001; Zagotta et al., 2003). Note that the TRP domain mediates most of the subunit-subunit interactions in the tetramer. In contrast, the regulatory domain is not involved in the formation of the subunit-subunit contacts. Several data are consistent with this model: (1) deletion of the modulatory domain does not affect the expression of functional channels (Vlachová et al., 2003), whereas removal of the AD abrogates subunit interaction and channel activity; (2) PKC phosphorylation sites 709Thr and 800Ser are solvent accessible; and (3) the region 769Val-818Thr structures a small cavity with an overall positive surface potential in which PIP2 could bind (Numazaki et al., 2003; Prescott and Julius, 2003) (Fig. 8B). Residues 781Lys and 785Arg, implicated in PIP2 binding (Prescott and Julius, 2003), are major providers of the positive electrostatic potential. Thus, although caution must be exercised when inferring protein structure from functional assays, our data suggest a protein fold for the C terminus of TRPV1 similar to that of the distantly related HCN protein family. Additional experimental work will demonstrate the accuracy of the proposed molecular model for TRPV1-C.
Figure 8.
Molecular model for the C terminus of TRPV1. A, Molecular model of a TRPV1-C monomer. The structure of the mouse HCN2 C-linker and cyclic nucleotide binding domain construct bound to cGMP was used as a scaffold (PDB 1Q3E) (Zagotta et al., 2003). B, Electrostatic surface potential of monomeric TRPV1-C. C, D, TRPV1-C putative tetramer representation parallel to the fourfold axis of symmetry from the intracellular side and normal to the plane of the membrane, respectively.
In conclusion, we reported that the TRP-like domain in TRPV1 acts as an AD that contributes to subunit oligomerization, thus suggesting a role for the highly conserved TRP domain in TRP channels. These results imply that the limited mixing of TRP subunits to give rise to heteromeric assemblies may be defined by the molecular compatibility of this protein domain. Noteworthy, the location of the AD as a linker of the channel pore and C-terminal regulatory regions suggests an additional role of this region as a transducer of conformational changes in the C terminus resulting from protein phosphorylation and/or protein-protein interactions to the channel gate. As HCN proteins (Paoletti et al., 1999; Wainger et al., 2001; Zagotta et al., 2003), TRP channels appear to use a general, evolutionary conserved mechanism to couple modulatory domains to the core gating structure.
Footnotes
This work was supported by Spanish Ministry of Science and Technology Grant SAF2003-0509, Instituto de la Salud Carlos III Grant FIS-01/1162, and Fundación La Caixa Grant 01/085-00 (A.F.-M.). We thank Marco Caprini for technical assistance and Reme Torres for cDNA and cRNA preparation, oocyte injection, and manipulation. We are indebted to Dr. Davis Julius for the cDNA encoding rat TRPV1.
Correspondence should be addressed to Antonio Ferrer-Montiel, Instituto de Biología Molecular y Celular, Universidad Miguel Hernández, Avenida Ferrocarril s/n, 03202 Elche (Alicante), Spain. E-mail aferrer@umh.es.
Copyright © 2004 Society for Neuroscience 0270-6474/04/245307-08$15.00/0
References
- Bai C, Elledge SJ (1997) Searching for interacting proteins with the two-hybrid system I. In: The yeast two-hybrid system (Bartel PL, Fields S, eds), pp 11-28. Oxford: Oxford UP.
- Bhave G, Zhu W, Wang H, Brasier DJ, Oxford GS, Gereau IV RW (2002) cAMP-dependent protein kinase regulates desensitization of the capsaicin receptor (VR1) by direct phosphorylation. Neuron 35: 721-731. [DOI] [PubMed] [Google Scholar]
- Bhave G, Hu H-J, Glauner KS, Zhu W, Wang H, Brasier DJ, Oxford GS, Gereau IV R (2003) Protein kinase C phosphorylation sensitizes but does not activate the capsaicin receptor transient receptor potential vanilloid 1 (TRPV1). Proc Natl Acad Sci USA 100: 12480-12485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaumer L, Yidirim E, Abramowitz J (2003) A comparison of the genes encoding for canonical TRP channels an their M, V and P relatives. Cell Calcium 33: 419-432. [DOI] [PubMed] [Google Scholar]
- Blanes-Mira C, Ibañez C, Fernandez-Ballester G, Planells-Cases R, Perez-Paya E, Ferrer-Montiel A (2001) Thermal stabilization of the catalytic domain of botulinum neurotoxin E by phosphorylation of a single tyrosine residue. Biochemistry 40: 2234-2242. [DOI] [PubMed] [Google Scholar]
- Cabedo H, Luna C, Fernandez AM, Gallar J, Ferrer-Montiel A (2002) Molecular determinants of the sensory and motor neuron-derived factor insertion into plasma membrane. J Biol Chem 277: 19905-19912. [DOI] [PubMed] [Google Scholar]
- Caterina MJ, Julius D (2001) The vanilloid receptor: a molecular gateway to the pain pathway. Annu Rev Neurosci 24: 487-517. [DOI] [PubMed] [Google Scholar]
- Caterina MJ, Rosen TA, Tominaga M, Brake AJ, Julius D (1999) A capsaicin receptor: a heat-activated ion channel in the pain pathway. Nature 389: 816-824. [DOI] [PubMed] [Google Scholar]
- Caterina MJ, Leffler A, Malmberg AB, Martin WJ, Trafton J, Petersen-Zeitz KR, Koltzenburg M, Basbaum AI, Julius D (2000) Impaired nociception and pain sensation in mice lacking the capsaicin receptor. Science 288: 306-313. [DOI] [PubMed] [Google Scholar]
- Clapham DE (2003) TRP channels as cellular sensors. Nature 426: 517-524. [DOI] [PubMed] [Google Scholar]
- Clapham DE, Runnels LW, Strübing C (2001) The TRP ion channel family. Nat Rev Neurosci 2: 387-396. [DOI] [PubMed] [Google Scholar]
- Cortes DM, Cuello LG, Perozo E (2001) Molecular architecture of full-length KcsA: role of cytoplasmic domains in ion permeation and activation gating. J Gen Physiol 117: 165-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JB, Gray J, Gunthorpe MJ, Hatcher JP, Davey PT, Overend P, Harries MH, Latcham J, Clapham C, Atkinson K, Hughes SA, Rance K, Grau E, Harper AJ, Pugh PL, Rogers DC, Bingham S, Randall A, Sheardown SA (2000) Vanilloid-receptor-1 is essential for inflammatory thermal hyperalgesia. Nature 405: 183-187. [DOI] [PubMed] [Google Scholar]
- García-Martínez C, Morenilla-Palao C, Planells-Cases R, Merino JM, Ferrer-Montiel A (2000) Identification of an aspartic residue in the P-loop of the Vanilloid receptor that modulates pore properties. J Biol Chem 275: 32552-32558. [DOI] [PubMed] [Google Scholar]
- García-Martínez C, Humet M, Planells-Cases R, Gomis A, Caprini M, Viana F, De La Pena E, Sanchez-Baeza F, Carbonell T, De Felipe C, Perez-Paya E, Belmonte C, Messeguer A, Ferrer-Montiel A (2002) Attenuation of thermal nociception and hyperalgesia by VR1 blockers. Proc Natl Acad Sci USA 99: 2374-2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerois R, Nielsen JE, Serrano L (2002) Predicting changes in the stability of proteins and protein complexes: a study of more than 1000 mutations. J Mol Biol 320: 369-387. [DOI] [PubMed] [Google Scholar]
- Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modelling. Electrophoresis 18: 2714-2723. [DOI] [PubMed] [Google Scholar]
- Harteneck C, Plant TD, Schultz G (2000) From worm to man: three sub-families of TRP families. Trends Neurosci 23: 159-166. [DOI] [PubMed] [Google Scholar]
- Higgins D, Thompson J, Gibson T, Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenke M, Sánchez A, Monje F, Sthümer W, Weseloh RM, Pardo LM (2003) C-terminal domains implicated in the functional surface expression of potassium channels. EMBO J 22: 395-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji RR, Samad TA, Jin SX, Schmoll R, Woolf CJ (2002) p38 MAPK activation by NGF in primary sensory neurons after inflammation increases TRPV1 levels and maintains heat hyperalgesia. Neuron 36: 57-68. [DOI] [PubMed] [Google Scholar]
- Kedei N, Szabo T, Lile JD, Treanor JJ, Olah Z, Iadarola MJ, Blumberg PM (2001) Analysis of the native quaternary structure of vanilloid receptor 1. J Biol Chem 276: 28613-28619. [DOI] [PubMed] [Google Scholar]
- Kuzhikandathil EV, Wang H, Szabo T, Morozova N, Blumberg PM, Oxford GS (2001) Functional analysis of capsaicin receptor (vanilloid receptor subtype 1) multimerization and agonist responsiveness using a dominant negative mutation. J Neurosci 21: 8697-8706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A (1996) Coiled-coils: new structures and new functions. Trends Biochem Sci 21: 375-382. [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J (1991) Predicting coiled coils form protein sequences. Science 252: 1162-1164. [DOI] [PubMed] [Google Scholar]
- Minke B, Cook B (2002) TRP channel proteins and signal transduction. Physiol Rev 82: 429-472. [DOI] [PubMed] [Google Scholar]
- Montell C, Birnbaumer L, Flockerzi V (2002) The TRP channels, a remarkably functional family. Cell 108: 595-598. [DOI] [PubMed] [Google Scholar]
- Numazaki M, Tominaga T, Toyooka H, Tominaga M (2002) Direct phosphorylation of capsaicin receptor VR1 by protein kinase Cϵ and identification of two target serine residues. J Biol Chem 277: 13375-13378. [DOI] [PubMed] [Google Scholar]
- Numazaki M, Tominaga T, Takeuchi K, Murayama N, Toyooka H, Tominaga M (2003) Structural determinants of TRPV1 desensitization interacts with calmodulin. Proc Natl Acad Sci USA 100: 8002-8006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoletti P, Young EC, Siegelbaum SA (1999) C-linker of cyclic nucleotide-gated channels controls coupling of ligand binding to channel gating. J Gen Physiol 113: 17-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premkumar LS, Ahern GP (2000) Induction of vanilloid receptor channel activity by protein kinase C. Nature 408: 985-990. [DOI] [PubMed] [Google Scholar]
- Prescott ED, Julius D (2003) A modular PIP2 binding site as a determinant of capsaicin receptor sensitivity. Science 300: 1284-1288. [DOI] [PubMed] [Google Scholar]
- Quirk JC, Reinhart PH (2001) Identification of a novel tetramerization domain in large conductance KCa channels. Neuron 32: 13-23. [DOI] [PubMed] [Google Scholar]
- Schwede T, Kopp J, Guex N, Peitsch MC (2003) SWISS-MODEL: an automated protein homology-modelling server. Nucleic Acids Res 31: 3381-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suenaga M, Ohmae H, Tsuji S, Itoh T, Nishimura O (1998) Renaturation of recombinant human neurotrophin-3 from inclusion bodies using an aggregation suppressor. Biotechnol Appl Biochem 28: 119-124. [PubMed] [Google Scholar]
- Vlachová V, Tisinger J, Suŝánková K, Lyfenko A, Ettrich R, Vyklický L (2003) Functional role of the C-terminal cytoplasmic tail of rat vanilloid receptor 1. J Neurosci 23: 1340-1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vriend G (1990) WHAT IF: a molecular modeling and drug design program. J Mol Graph 8: 51-55. [DOI] [PubMed] [Google Scholar]
- Wainger BJ, Degennaro M, Santoro B, Siegelbaum SA, Tibbs GR (2001) Molecular mechanism of cAMP modulation of HCN pacemaker channels. Nature 411: 805-810. [DOI] [PubMed] [Google Scholar]
- Zagotta WN, Olivier NB, Black KD, Young EC, Olson R, Gouaux E (2003) Structural basis for modulation and agonist specificity of HCN pacemaker channels. Nature 425: 200-205. [DOI] [PubMed] [Google Scholar]
- Zerangue N, Jan YN, Jan LY (2000) An artificial tetramerization domain restores efficient assembly of functional Shaker channels lacking T1. Proc Natl Acad Sci USA 97: 3591-3595. [DOI] [PMC free article] [PubMed] [Google Scholar]