Abstract
The LIM homeodomain family of transcription factors is involved in many processes in the developing CNS, ranging from cell fate specification to connectivity. A member of this family of transcription factors, lhx6, is expressed in the medial ganglionic eminence (MGE) of the ventral telencephalon, where the vast majority of cortical interneurons are generated. Its expression in the GABA-containing MGE cells that migrate to the cortex suggests that this gene uniquely or in combination with other transcription factors may play a role in the neurochemical identity and migration of these neurons. We performed loss of function studies for lhx6 in mouse embryonic day 13.5 brain slices and dissociated MGE neuronal cultures using Lhx6-targeted small interfering RNA produced by a U6 promoter-driven vector. We found that silencing lhx6 impeded the tangential migration of interneurons into the cortex, although it did not obstruct their dispersion within the ganglionic eminence. Blocking lhx6 expression in dissociated MGE cultured neurons did not interfere with the production of GABA or its synthesizing enzyme. These results indicate that lhx6, unlike the closely related member lhx7, does not regulate neurotransmitter choice in interneurons but plays an important role in their migration from the ventral telencephalon to the neocortex.
Keywords: cerebral cortex, interneurons, migration, GABA, RNA interference, RNAi, rodent
Introduction
It is now established that the subpallium of the embryonic forebrain gives rise to a variety of neuronal types destined for different regions of the brain. In particular, the lateral ganglionic eminence (LGE) generates the striatal projection neurons (Deacon et al., 1994) as well as cells of the olfactory bulb and some of the interneurons of the pallium (cortex) (Anderson et al., 2001, 2002). The vast majority of cortical and striatal interneurons arise in the medial ganglionic eminence (MGE) and caudal ganglionic eminence (CGE) (Lavdas et al., 1999; Nery et al., 2002; Valcanis and Tan, 2003; Xu et al., 2004). Genetic perturbation studies in recent years have helped to establish boundaries between progenitor domains in the subpallium (Marin and Rubenstein, 2001). However, it is not yet understood how the fates of cells that arise in particular progenitor pools are specified. For example, how are interneurons destined for the cerebral cortex and hippocampus sorted from striatal interneurons in the MGE?
A feature common to MGE cells that migrate to the cortex is the expression of the LIM homeodomain (LIM-hd) gene lhx6 (Lavdas et al., 1999). Overlapping domains of lhx6 expression and of the structurally related lhx7 have been detected in the MGE of mouse and rat embryos (Grigoriou et al., 1998; Marin et al., 2000; Asbreuk et al., 2002). Tracer labeling experiments (Lavdas et al., 1999) indicate that the expression of lhx6 in the cortex defines a population of cells that originate in the MGE and migrate dorsally crossing the corticostriatal boundary. Although the function of lhx6 and lhx7 during mammalian embryogenesis is currently unknown, their expression patterns in the ganglionic eminence (GE), together with the previously established role of other LIM-hd proteins in cell fate determination and differentiation, suggests that they have a role in the generation of the neuronal diversity in the basal forebrain. Results from a number of studies (Marin et al., 2000; Asbreuk et al., 2002; Zhao et al., 2003) now point to an important role of lhx7 in the development of cholinergic neurons in the forebrain. As for lhx6, its expression in migrating interneurons suggests that this gene uniquely or in combination with other transcription factors might play a role in the decision of MGE cells to differentiate in situ or to migrate dorsally to the cortex. Recent studies have also speculated that lhx6 may regulate the development of the GABAergic neurons in the striatum (Marin et al., 2000). Here, we used the novel method of RNA interference (RNAi) to test the hypothesis that lhx6 plays a role in the neurochemical identity of cortical interneurons and in their migration from the ventral telencephalon. We show that lhx6 does not regulate the expression of GABA in MGE neurons, but it does control their migration from the subpallium.
Materials and Methods
Dissociated cell cultures. The brains of embryonic day (E) 13.5 mouse embryos were removed (E0.5, day vaginal plug was found), and the desired forebrain regions were microdissected and placed in a solution containing 0.05% trypsin and 100 μg/ml DNaseI in Neurobasal medium (Invitrogen, Paisley, UK). After 15 min incubation (at 5% CO2, 37°C), they were washed in Neurobasal medium with 10% fetal bovine serum and dissociated with a fire-polished Pasteur pipette. The resulting suspension was centrifuged at 3000 rpm for 3 min. The cell pellet was resuspended in medium containing Neurobasal medium, 2 mm l-glutamine, penicillin/streptomycin, and 1:50 dilution of B-27 (Invitrogen). A total of 2 × 104 cells were plated onto each 13 mm diameter glass coverslip, according to the instructions of the manufacturer (Invitrogen).
Construction of vectors. We used a vector-based RNAi approach to intracellularly produce a short hairpin double stranded RNA (dsRNA) from a DNA template under the RNA polymerase III promoter. Annealed oligonucleotides were cloned into the HincII-NcoI sites of the pTZ-U6 + 1 vector (a kind gift from Dr. John Rossi, City of Hope, Duarte, CA). Our construct, pTZ-U6-Lhx6, was designed to target the lhx6 mRNA at position 173-192 from the sequences published by Kimura et al. (1999). This sequence is devoid of secondary structures, is outside of the conserved LIM or homeobox domains, and gives a specific hit only for lhx6 in the GenBank database. The oligonucleotides (Sigma Genosys, London, UK) were annealed at a concentration of 10 μm in 10 mm Tris HCl, pH 7.5, 50 mm NaCl, 1 mm EDTA at 95°C for 1 min. The solution was then cooled to 70°C followed by slow cooling to 4°C for 2.5 hr. The sequences of the oligonucleotides were as follows (underlined letters denote hairpin loop):
Sense AGACGCAGAGGCCTTGGTTCAAGAGACCAAGGCCTCTGCGTCTGACTTTTTC and antisense CATGGAAAAAGTCAGACGCAGAGGCCTTGGTCTCTTGAACCAAGGCCTCT GCGTCT.
As a control, we used the empty pTZ-U6 + 1 vector or a small interfering RNA (siRNA)-producing vector on the basis of the same lhx6 sequence but containing a few base mutations (shown below in underlined bold lowercase letters; deletions are denoted by empty parentheses). The identity of both constructs was verified by sequencing.
Sense AGACGCcAGAGGCCTTGGTTCAAGAGACCAAGGa(_)CTg-TGCGTCTGACTTTTTC and antisense CATGGAAAAAGTCAGACGCAcAG(_)tCCTTGGTCTCTTGAACCAAGGCCTCT gGCGTCT.
Two groups of investigators (Grigoriou et al., 1998; Kimura et al., 1999) have cloned lhx6 independently, and although the reported sequences were similar, there were several differences at the 5′ and 3′ ends of the coding region. To choose the correct target sequence for our dsRNA construct, we cloned lhx6 from an E13.5 brain cDNA library. Our sequencing data agreed with the sequence published by Kimura et al. (1999) (95% identity at the nucleotide level).
Transfections. Dissociated MGE cells were transfected at 2 d postplating, because this time was found to render the best transfection efficiency (10-20% on average). The transfection was performed using Lipofectamine 2000 according to the guidelines of the manufacturer (Invitrogen). For each coverslip, 3 μl of Lipofectamine 2000 and 1 μg of DNA were used. To visualize the transfected cells, the experimental constructs were cotransfected with pEGFP-N1 vector (BD Biosciences, Palo Alto, CA; vector referred to as pEGFP) in a ratio 5:1. The cultures were processed for immunohistochemistry on day 4 postplating.
Immunohistochemistry. The immunohistochemistry procedure was performed as described previously (Lavdas et al., 1999). The primary antibodies used were anti-Lhx6 (rabbit polyclonal, 1:2000; a kind gift from Dr. V. Pachnis, National Institute for Medical Research, London, UK), anti-GAD 65/67 (rabbit polyclonal, 1:250; Affiniti Research Products, Exeter, UK), anti-calbindin (rabbit polyclonal, 1:10,000; Swant, Bellinzona, Switzerland), anti-GABA (rabbit polyclonal, 1:1000; Sigma), and anti-MAP2 (mouse monoclonal, 1:500; Sigma). The anti-rabbit or anti-mouse FITC or rhodamine-conjugated secondary antibodies (Molecular Probes, Eugene, OR) were used at a dilution of 1:500. The confocal microscope images were reconstructed using MetaMorph imaging software (Universal Imaging Corporation, West Chester, PA).
Electroporation of brain slices. Slices were prepared from E13.5 mouse embryos as described previously (Lavdas et al., 1999). Medial coronal slices were mounted onto nitrocellulose filters (0.45 μm; Millipore, London, UK) and were subsequently incubated with 5 μg of DNA for 1 min. To focally electroporate the GE, a vertically oriented platinum wire electrode (negative pole) was positioned over the GE while a horizontal electrode was positioned underneath the nitrocellulose filter (positive pole). The electrodes were connected to a dual-pulse isolated stimulator (Intracel, Herts, UK). The following electroporation conditions were used: 100 Hz, 50 V, 5 msec width of pulse, three to five pulses. Slices were then transferred in defined culture medium containing DMEM:F12 (Sigma), 5% heat-inactivated fetal bovine serum, 1× N-2, 100 μm l-glutamine, 2.4 gm/l d-glucose (Sigma), and 5 U/ml penicillin and 5 mg/ml streptomycin in a humidified 5% CO2 incubator at 37°C. All materials were purchased from Invitrogen, unless otherwise stated. Labeled slices were fixed with 4% paraformaldehyde in 0.1 m phosphate buffer, pH 7.4, at room temperature for 2 hr, mounted onto glass slides, and examined with a confocal microscope (Leica, Nussloch, Germany). The images were reconstructed using MetaMorph imaging software.
Results
Lhx6 plays a role in the tangential migration of cortical interneurons
Previous studies reported lhx6 expression in the subventricular and submantle zones of the MGE and in tangentially arranged corridors of cells in the marginal, intermediate, and subventricular zones of the cortex (Grigoriou et al., 1998; Lavdas et al., 1999). These lhx6-expressing cells were shown to be tangentially migrating cortical interneurons that had arisen in the MGE (Lavdas et al., 1999). Based on these observations, we sought to investigate the role of lhx6 in interneuron migration by studying the loss of lhx6 function in MGE neurons using RNA interference. We chose to silence lhx6 in MGE neurons by focally electroporating brain slices with a U6 promoter-driven vector producing double-stranded siRNA, because this method provides the advantage of genetic manipulation in a controlled spatiotemporal manner.
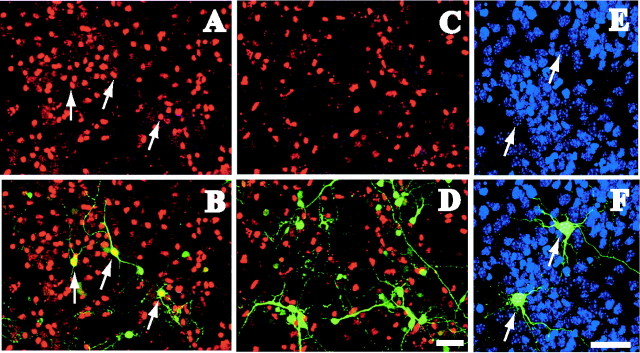
To visualize transfection and to demonstrate that our hairpin siRNA could efficiently hinder lhx6 expression, we cotransfected the siRNA construct or control vectors (empty or mutated siRNA vector) with a pEGFP vector (5:1 molar ratio to ensure that all green fluorescent protein (GFP)-expressing cells had been cotransfected with the siRNA plasmid) in cells dissociated from the MGE of E13.5 mouse brains. The cells were fixed 32-72 hr post-transfection and immunostained for Lhx6. Dissociated MGE cells cotransfected with the control vector showed a number of GFP-expressing cells also positive for Lhx6 (Fig. 1A,B). This finding confirmed that the transfection procedure and the backbone vectors did not interfere with the expression of Lhx6. Dissociated MGE cells cotransfected with the siRNA and the pEGFP vectors showed no GFP-expressing cells positive for Lhx6 (Fig. 1C,D), indicating that the siRNA efficiently disrupted the expression of lhx6. Transfection with siRNA did not affect cell viability as indicated by 4′, 6′-diamidino-2-phenylindole (DAPI) staining (Fig. 1E,F). We confirmed that the siRNA vector conferred loss of lhx6 function in preparations analyzed 32-72 hr post-transfection. Although recent reports (Omi et al., 2004) have indicated that siRNA can have a long-lasting effect in mammalian cell cultures, we performed our analysis 2-3 d post-transfection for best visualization of transiently transfected cells by means of GFP expression.
Figure 1.
RNAi effectively blocks the expression of lhx6 in GE neurons. A-D, Dissociated MGE cells taken from E13 mice were transfected with pEGFP and control vector (A, B) or pEGFP and the siRNA vector (C, D) and immunostained for Lhx6 (red). A high proportion of MGE cells transfected with pEGFP/control vector were also positive for Lhx6 (A, B, arrows). Dissociated MGE cells transfected with pEGFP/siRNA vector did not exhibit Lhx6 immunoreactivity, indicating that the production of RNAi in these cells inhibited lhx6 expression (n = 5 experiments). E, F, siRNA transfection did not affect cell viability as shown with DAPI nuclear staining (arrows). Scale bars, 50 μm.
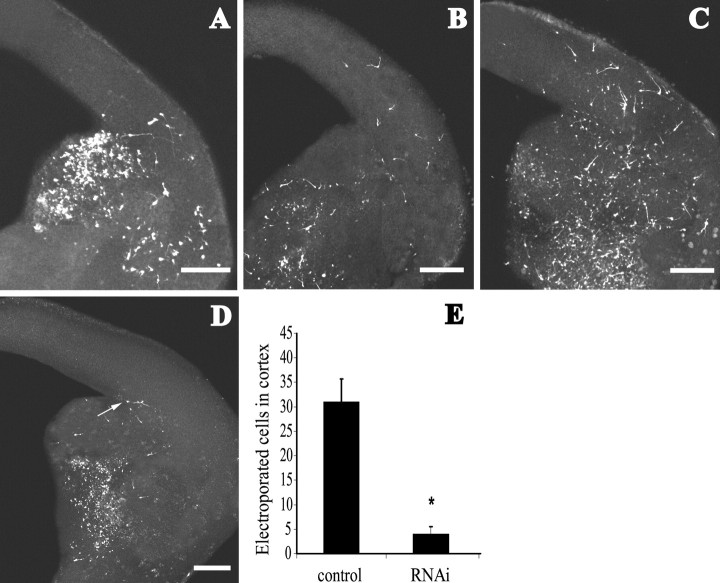
Before delivering the siRNA construct in brain slices by electroporation, we first examined the kinetics of electroporated neurons. We used a set-up similar to the one described by Marin et al. (2001) to electroporate pEGFP alone or together with a control vector to the GE of E13.5 brain slices and analyzed the migratory behavior of neurons after 24, 48, and 72 hr (Fig. 2A-C). The electroporated cells displayed the typical bipolar morphology of tangentially migrating neurons. We observed that 24 hr post-transfection the first GFP-positive neurons had already reached the lateral cortex. At 48 hr, electroporated cells had migrated further, and by 72 hr, many cells were found in the dorsomedial cortex, indicating that electroporated cells advanced into the cortex through time. Confocal image analysis showed that electroporation was not superficial, because many cells had been transfected deeply into the tissue (200-250 μm in a 300 μm slice). Furthermore, although transfection by electroporation delivered a construct only to a group of cells at any one time and not to successive cohorts of cells (as is the case with fluorescent dye labeling), the efficiency of transfection achieved was successful enough to provide a sufficient number of cells to be monitored (Fig. 2E, control).
Figure 2.
Electroporation of lhx6 silencing constructs in E13.5 mouse brain slices. A-C, The GE was electroporated with control vector/pEGFP, and tangential migration was visualized after 24 hr (A), 48 hr (B), and 72 hr (C). More GFP cells were found in the cortex with time. Electroporation of siRNA vector/pEGFP resulted in labeled neurons dispersing within the GE but not entering the cortex (D). Sometimes, neurons (arrow) would accumulate at the corticostriatal boundary. Scale bars, 200 μm. E, Graph depicts quantification of cell migration from the GE electroporated with control vector (31 ± 4.7 cells within the cortex) and siRNA vector (4 ± 1.5 cells at corticostriatal boundary; Student's t test; p < 0.0001).
Having established that our siRNA vector hindered lhx6 expression efficiently and that electroporation did not inhibit tangential migration of transfected cells, we combined the two methods to examine whether lhx6 is involved in the migration of neurons from the MGE to the cortex. Focal electroporation of the siRNA and the pEGFP vectors (surplus 5:1 molar ratio) in brain slices resulted in neurons dispersing from the site of electroporation comparable with control experiment (empty or mutated siRNA vector), suggesting that the motility of neurons electroporated with the siRNA vector was not affected (Fig. 2D). However, no GFP-labeled neurons were detected in the cortex 48-72 hr post-transfection (n = 30 brain slices). On a few occasions, a few GFP-labeled cells had accumulated at the corticostriatal boundary (4 ± 1.5 cells) (Fig. 2D, arrow, E) but, contrary to the tangentially migrating neurons, they did not display the typical bipolar morphology of actively migrating cells. The failure of tangentially migrating neurons to enter the cortex suggests that lhx6 is necessary for those neurons to migrate past the corticostriatal notch.
Lhx6 does not regulate the GABAergic phenotype
Early in forebrain development, the MGE is nearly the sole contributor of GABA-containing interneurons to the rodent cortex (Anderson et al., 2001, 2002). The majority of these migrating interneurons express Lhx6 (93.3 ± 6.7% of dye-labeled MGE cells in the cortex express Lhx6 at E13.5), suggesting that this gene might regulate their GABAergic phenotype. Double immunostaining experiments for Lhx6 and GABA in dissociated E13.5 cortical cultures showed that only a proportion of GABAergic neurons contain Lhx6 and vice versa (65.2 ± 2.2% of Lhx6 cells express GABA and 72 ± 2.4% of GABA cells express Lhx6) (Fig. 3A-C), suggesting that lhx6 is not associated with the expression of the neurotransmitter in these neurons. To exclude the possibility that Lhx6 cells in the cortex are glia precursors, we performed double immunolabeling analysis of Lhx6-expressing cells with MAP2, a marker of differentiated neurons, in dissociated cortical cultures. We found that 95.3 ± 4.7% of Lhx6 cells express MAP2, confirming their neuronal identity (Fig. 3A,D,E). Similar experiments performed to assess colocalization of Lhx6 with GABA in subpallial neurons also showed that this gene is not closely associated with GABA expression (49 ± 3.6% of GABA cells express Lhx6 and 25.7 ± 3.5% of Lhx6 cells express GABA in dissociated cultures from mouse E13.5 MGE) (Fig. 3A,F,G).
Figure 3.
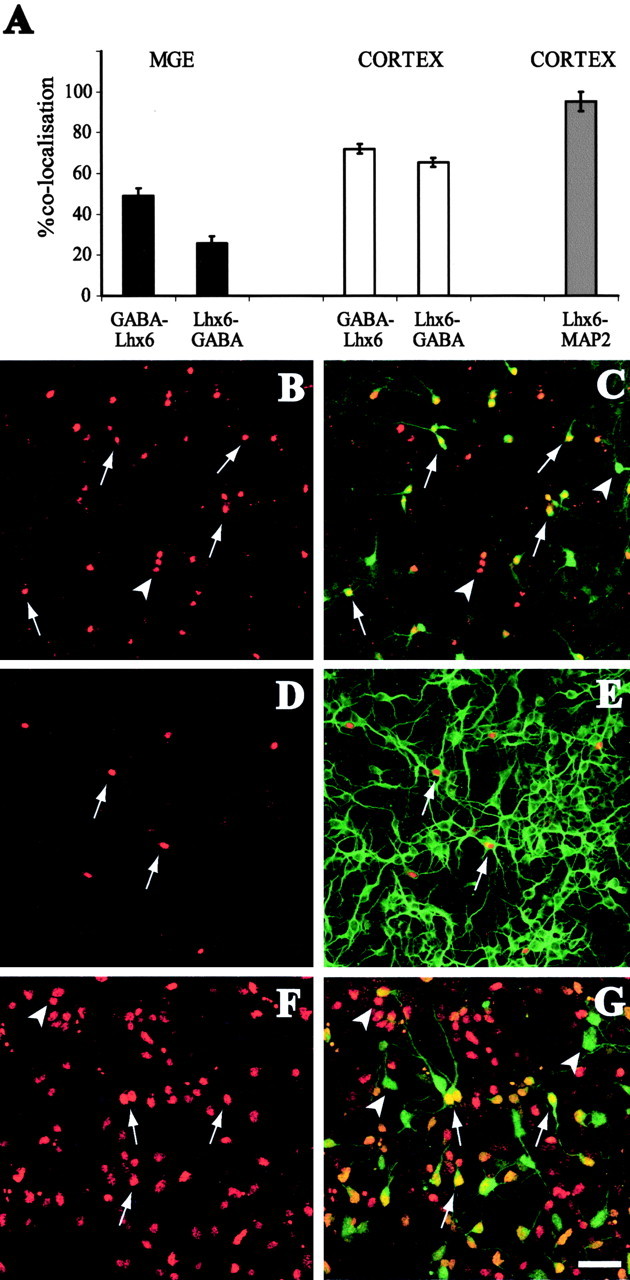
Expression of Lhx6 in dissociated cultures from mouse E13.5 brain. A, The graph depicts percentage colocalization of Lhx6 with various markers in dissociated cultures (n = 3 litters). B, C, Lhx6 (B, red) colocalizes with GABA (C, green) in some cells in dissociated cortical cultures (arrows). GABA cells that do not colocalize with Lhx6 and many Lhx6 cells that do not express GABA are also observed (arrowheads). D, E, Lhx6 (D, red) in cortical dissociated cultures colocalizes (arrows) with MAP2 (E, green). F, G, Lhx6 (red) colocalizes with GABA in MGE cell cultures (arrows), but there are also GABA cells that do not express Lhx6 and vice versa (arrowheads). Scale bar, 50 μm.
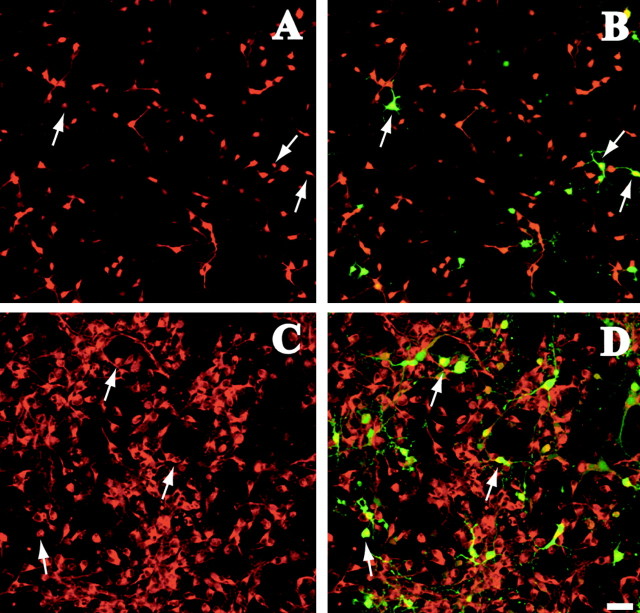
To investigate whether Lhx6 can regulate the GABAergic phenotype, we transfected our siRNA vector and examined the expression of GABA and GAD65/67 in dissociated cell cultures prepared from E13.5 mouse MGE (32-72 hr post-transfection). The results were normalized against the control in every experiment to eliminate variability in transfection efficiency. We found that loss of lhx6 function did not affect the expression of the neurotransmitter or its synthesizing enzyme in MGE neurons (Fig. 4A-D) (GABA, -3.8 ± 11.4%; GAD65/67, 1.3 ± 9.3% different from control). Similarly, silencing lhx6 in MGE cultures did not affect calbindin expression (the colocalization of siRNA transfected neurons with calbindin was 2.1 ± 8% different from control). These observations confirmed the results of the double-immunostaining studies described above, which showed that the GABAergic phenotype of MGE and cortical neurons is independent of lhx6. In addition, our results indicate that the siRNA vector interfered specifically with the expression of Lhx6, leaving other developmental pathways such as the expression of GAD and GABA intact.
Figure 4.
lhx6 silencing in dissociated cultures from mouse E13.5 brain. Dissociated MGE cultures were transfected with lhx6-directed siRNA-producing vector and pEGFP. A-D, Silencing of lhx6 did not affect expression of GABA (A, B, red) or GAD65/67 (C, D, red), because several siRNA-transfected cells (B, D, green) were positive for these markers (arrows). Scale bars, 50 μm. n = 3 litters.
Discussion
The role of lhx6 in tangential migration of cortical interneurons
Experimental evidence points to an important role of LIM-hd proteins in many developmental processes in the nervous system ranging from cell lineage and phenotype determination to axonal guidance and patterning (for review, see Hobert and Westphal, 2000). The present study focused on the role of one such transcription factor, lhx6, in the migration and phenotype specification of cortical interneurons by performing loss of gene function experiments with the aid of double-stranded siRNA.
The method of dsRNA as a tool for silencing target genes was initially developed and used in C. elegans and later in D. melanogaster (Hannon, 2002). Its use in mammalian systems was initially limited, because the presence of long dsRNA triggers general transcription shutdown and subsequent cell death (for review, see Kumar and Carmichael, 1998). This problem has been overcome with the use of siRNAs that are either chemically synthesized and introduced into cells exogenously (Elbashir et al., 2001) or produced intracellularly by DNA templates (Lee et al., 2002). The main limitation in using the former siRNAs has been their short half-life within the cell. The development of DNA vectors that produce siRNA has made it possible to silence a gene for an extended period of time (Brummelkamp et al., 2002). Here, we used such a vector to produce a short hairpin dsRNA in focally electroporated brain slices after confirming that the construct hampers Lhx6 expression effectively in dissociated MGE neurons. Silencing lhx6 in brain slices resulted in cells dispersing normally within the GE but not migrating into the cortex, indicating that lhx6 is necessary for the migration of cortical interneurons from the MGE.
The restricted expression of lhx6 in the MGE suggests that this transcription factor does not account for the migration of all subpallial neurons destined for the cortex. In the later stages of development, the LGE, which is primarily devoid of lhx6, provides a significant number of tangentially migrating neurons to the cortex (Anderson et al., 2001). Moreover, in Nkx2.1 mutants, where the MGE is respecified to LGE and lhx6 expression is lost (Sussel et al., 1999; Marin et al., 2000; Nery et al., 2002), cells from the expanded LGE still migrate to the cortex. Together, these findings suggest that lhx6 does not account for the migration of all subpallial neurons destined for the cortex, and that other transcription factors may be required to provide neurons of the GE with the necessary instructions to migrate toward the cortex. Furthermore, lhx6 might not solely regulate the migration of MGE-derived cortical interneurons. It is possible that a rich combinatorial code of LIM-hd proteins, similar to the one described in the spinal cord (for review, see Shirasaki and Pfaff, 2002), also exists during forebrain development (i.e., lhx6 might form functionally diverse protein complexes with other proteins, including transcription factors of the same family). Likely binding partners include lhx1, 2, 5, and 7, which occupy regions of the dorsal and ventral telencephalon where lhx6 is also found (Shawlot and Behringer, 1995; Porter et al., 1997; Zhao et al., 1999; Monuki et al., 2001; Asbreuk et al., 2002). It is possible that lhx6 functions in synergy with these or other factors as part of a protein complex that, if disrupted, tangential migration is arrested.
Lhx6 does not specify the GABAergic phenotype of interneurons
It has been suggested that the expression of lhx6 and lhx7 in the ventral telencephalon defines distinct cell types in this region of the developing brain (Marin et al., 2000). Recent genetic evidence supports an important role for lhx7 in the development of cholinergic neurons in the basal forebrain (Asbreuk et al., 2002; Zhao et al., 2003). The expression of lhx6 in complementary subpallial regions and in migrating cortical interneurons has prompted speculation that it may be involved in the specification of their GABAergic phenotype. Other LIM-hd transcription factors have been implicated in neurotransmitter choice (e.g., the C. elegans lim-6 has been found to regulate GAD expression and to control the morphology of GABAergic neurons) (Hobert et al., 1999).
The observation that there is no extensive colocalization of GABA with Lhx6 in dissociated cortical and MGE cultures suggests that lhx6 does not regulate the expression of GABA in these neurons, although a role for this gene in the specification of distinct subsets of GABAergic neurons cannot be excluded. Our lhx6-silencing experiments confirmed that the expression of GABA and its synthesizing enzyme GAD65/67 as well as calbindin are not controlled by this transcription factor. Furthermore, in DLX1/2 mutants, where there is almost complete loss of GAD65 cells (Anderson et al., 1997a,b), a number of lhx6-positive cells survive (Marin et al., 2000). These observations, in conjunction with the fact that GABAergic projection neurons and interneurons arise not only from the MGE but also other subpallial regions, clearly show that lhx6 is not directly associated with the GABAergic phenotype.
In summary, the results presented here show that lhx6 does not specify the GABAergic phenotype of cortical interneurons, but it regulates their migration from the subpallium.
Footnotes
The work was supported by the Wellcome Trust.
Correspondence should be addressed to John G. Parnavelas, Department of Anatomy and Developmental Biology, University College London, Gower Street, London WC1E 6BT, UK. E-mail: j.parnavelas@ucl.ac.uk.
Copyright © 2004 Society for Neuroscience 0270-6474/04/245643-06$15.00/0
P.A. and A.L. contributed equally to this work.
References
- Anderson SA, Eisenstat DD, Shi L, Rubenstein JL (1997a) Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes. Science 278: 474-476. [DOI] [PubMed] [Google Scholar]
- Anderson SA, Qiu M, Bulfone A, Eisenstat DD, Meneses J, Pedersen R, Rubenstein JL (1997b) Mutations of the homeobox genes Dlx-1 and Dlx-2 disrupt the striatal subventricular zone and differentiation of late born striatal neurons. Neuron 19: 27-37. [DOI] [PubMed] [Google Scholar]
- Anderson SA, Marin O, Horn C, Jennings K, Rubenstein JL (2001) Distinct cortical migrations from the medial and lateral ganglionic eminences. Development 128: 353-363. [DOI] [PubMed] [Google Scholar]
- Anderson SA, Kaznowski CE, Horn C, Rubenstein JL, McConnell SK (2002) Distinct origins of neocortical projection neurons and interneurons in vivo Cereb Cortex 12: 702-709. [DOI] [PubMed] [Google Scholar]
- Asbreuk CH, van Schaick HS, Cox JJ, Kromkamp M, Smidt MP, Burbach JP (2002) The homeobox genes Lhx7 and Gbx1 are expressed in the basal forebrain cholinergic system. Neuroscience 109: 287-298. [DOI] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R (2002) A system for stable expression of short interfering RNAs in mammalian cells. Science 296: 550-553. [DOI] [PubMed] [Google Scholar]
- Deacon TW, Pakzaban P, Isacson O (1994) The lateral ganglionic eminence is the origin of cells committed to striatal phenotypes: neural transplantation and developmental evidence. Brain Res 668: 211-219. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411: 494-498. [DOI] [PubMed] [Google Scholar]
- Grigoriou M, Tucker AS, Sharpe PT, Pachnis V (1998) Expression and regulation of Lhx6 and Lhx7, a novel subfamily of LIM homeodomain encoding genes, suggests a role in mammalian head development. Development 125: 2063-2074. [DOI] [PubMed] [Google Scholar]
- Hannon GJ (2002) RNA interference. Nature 418: 244-251. [DOI] [PubMed] [Google Scholar]
- Hobert O, Westphal H (2000) Functions of LIM-homeobox genes. Trends Genet 16: 75-83. [DOI] [PubMed] [Google Scholar]
- Hobert O, Tessmar K, Ruvkun G (1999) The Caenorhabditis elegans lim-6 LIM homeobox gene regulates neurite outgrowth and function of particular GABAergic neurons. Development 126: 1547-1562. [DOI] [PubMed] [Google Scholar]
- Kimura N, Ueno M, Nakashima K, Taga T (1999) A brain region-specific gene product Lhx6.1 interacts with Ldb1 through tandem LIM-domains. J Biochem (Tokyo) 126: 180-187. [DOI] [PubMed] [Google Scholar]
- Kumar M, Carmichael GG (1998) Antisense RNA: function and fate of duplex RNA in cells of higher eukaryotes. Microbiol Mol Biol Rev 62: 1415-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavdas AA, Grigoriou M, Pachnis V, Parnavelas JG (1999) The medial ganglionic eminence gives rise to a population of early neurons in the developing cerebral cortex. J Neurosci 19: 7881-7888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee NS, Dohjima T, Bauer G, Li H, Li MJ, Ehsani A, Salvaterra P, Rossi J (2002) Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat Biotechnol 20: 500-505. [DOI] [PubMed] [Google Scholar]
- Marin O, Rubenstein JL (2001) A long, remarkable journey: tangential migration in the telencephalon. Nat Rev Neurosci 2: 780-790. [DOI] [PubMed] [Google Scholar]
- Marin O, Anderson SA, Rubenstein JL (2000) Origin and molecular specification of striatal interneurons. J Neurosci 20: 6063-6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin O, Yaron A, Bagri A, Tessier-Lavigne M, Rubenstein JL (2001) Sorting of striatal and cortical interneurons regulated by semaphorin-neuropilin interactions. Science 293: 872-875. [DOI] [PubMed] [Google Scholar]
- Monuki ES, Porter FD, Walsh CA (2001) Patterning of the dorsal telencephalon and cerebral cortex by a roof plate-Lhx2 pathway. Neuron 32: 591-604. [DOI] [PubMed] [Google Scholar]
- Nery S, Fishell G, Corbin JG (2002) The caudal ganglionic eminence is a source of distinct cortical and subcortical cell populations. Nat Neurosci 5: 1279-1287. [DOI] [PubMed] [Google Scholar]
- Omi K, Tokunaga K, Hohjoh H (2004) Long-lasting RNAi activity in mammalian neurons. FEBS Lett 558: 89-95. [DOI] [PubMed] [Google Scholar]
- Porter FD, Drago J, Xu Y, Cheema SS, Wassif C, Huang SP, Lee E, Grinberg A, Massalas JS, Bodine D, Alt F, Westphal H (1997) Lhx2, a LIM homeobox gene, is required for eye, forebrain, and definitive erythrocyte development. Development 124: 2935-2944. [DOI] [PubMed] [Google Scholar]
- Shawlot W, Behringer RR (1995) Requirement for Lim1 in head-organizer function. Nature 374: 425-430. [DOI] [PubMed] [Google Scholar]
- Shirasaki R, Pfaff SL (2002) Transcriptional codes and the control of neuronal identity. Annu Rev Neurosci 25: 251-281. [DOI] [PubMed] [Google Scholar]
- Sussel L, Marin O, Kimura S, Rubenstein JL (1999) Loss of Nkx2.1 homeobox gene function results in a ventral to dorsal molecular respecification within the basal telencephalon: evidence for a transformation of the pallidum into the striatum. Development 126: 3359-3370. [DOI] [PubMed] [Google Scholar]
- Valcanis H, Tan SS (2003) Layer specification of transplanted interneurons in developing mouse neocortex. J Neurosci 23: 5113-5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Cobos I, De La Cruz E, Rubenstein JL, Anderson SA (2004) Origins of cortical interneuron subtypes. J Neurosci 24: 2612-2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Sheng HZ, Amini R, Grinberg A, Lee E, Huang S, Taira M, Westphal H (1999) Control of hippocampal morphogenesis and neuronal differentiation by the LIM homeobox gene Lhx5. Science 284: 1155-1158. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Marin O, Hermesz E, Powell A, Nuria F, Palkovits M, Rubenstein JLR, Westphal H (2003) The LIM-homeobox gene Lhx8 is required for the development of many cholinergic neurons in the mouse forebrain. Proc Natl Acad Sci USA 100: 9005-9010. [DOI] [PMC free article] [PubMed] [Google Scholar]