Abstract
Expression of the brain–gut peptide cholecystokinin (CCK) in the developing olfactory–gonadotropin-releasing hormone-1 (GnRH-1) neuroendocrine systems was characterized, and the function of CCK in these systems was analyzed both in vivo and in vitro. We present novel data demonstrating that CCK transcript and protein are expressed in sensory cells in the developing olfactory epithelium and vomeronasal organ, with both ligand and receptors (CCK-1R and CCK-2R) found on olfactory axons throughout prenatal development. In addition, migrating GnRH-1 neurons in nasal regions express CCK-1R but not CCK-2R receptors. The role of CCK in olfactory–GnRH-1 system development was evaluated using nasal explants, after assessing that the in vivo expression of both CCK and CCK receptors was mimicked in this in vitro model. Exogenous application of CCK (10-7 m) reduced both olfactory axon outgrowth and migration of GnRH-1 cells. This inhibition was mediated by CCK-1R receptors. Moreover, CCK-1R but not CCK-2R antagonism caused a shift in the location of GnRH-1 neurons, increasing the distance that the cells migrated. GnRH-1 neuronal migration in mice carrying a genetic deletion of either CCK-1R or CCK-2R receptor genes was also analyzed. At embryonic day 14.5, the total number of GnRH-1 cells was identical in wild-type and mutant mice; however, the number of GnRH-1 neurons within forebrain was significantly greater in CCK-1R–/– embryos, consistent with an accelerated migratory process. These results indicate that CCK provides an inhibitory influence on GnRH-1 neuronal migration, contributing to the appropriate entrance of these neuroendocrine cells into the brain, and thus represent the first report of a developmental role for CCK.
Keywords: GnRH-1, CCK, CCK receptors, migration, olfactory system, development
Introduction
Hypothalamic hormone gonadotropin-releasing hormone-1 (GnRH-1) regulates anterior pituitary gonadotropes and as such is essential for reproduction. GnRH-1 neurons originate in the nasal placode (Wray, 2002) and migrate into brain along olfactory–vomeronasal axons (Wray et al., 1994). If olfactory axons do not penetrate brain, GnRH-1 neurons remain outside (Rugarli, 1999; MacColl et al., 2002), and if olfactory axons are redirected to non-CNS sites, GnRH-1 neurons are found at these aberrant locations (Gao et al., 2000; Murakami et al., 2000). Thus, development of the GnRH-1 neuroendocrine system is linked to development of the olfactory system.
Directed olfactory–vomeronasal axon outgrowth and GnRH-1 neuronal movement occur in nasal explants from embryonic day (E) 11.5 mice devoid of brain tissue (Fueshko and Wray, 1994; Kramer and Wray, 2000). This suggests that guidance cues in nasal regions are imprinted by other embryonic regions, at an age before E11.5 in mice, or are intrinsic to nasal regions, or both. Within nasal regions, several guidance molecules [neural cell adhesion molecule (NCAM) (Gong and Shipley, 1996), netrins (Shu et al., 2000), semaphorins (Gavazzi, 2001), and EphB2 (St. John and Key, 2001)] have been identified that influence olfactory–vomeronasal axon outgrowth. However, axons cross the nasal region and target the telencephalon before expression of, or during misexpression of, many of these molecules (Treloar et al., 1997; Yoshihara et al., 1997; Walz et al., 2002). Thus, nasal region signals influencing olfactory system development still remain to be identified.
Recently, attention has focused on intercellular signals influencing GnRH-1 migration and differentiation (Wray, 2002). To date, GABA (Fueshko et al., 1998; Bless et al., 2000; Heger et al., 2003), glutamate (Simonian and Herbison, 2001) and serotonin (Pronina et al., 2003a,b) have been shown to regulate GnRH-1 neuronal migration. Perturbation of these molecules, however, did not appear to alter initial GnRH-1 cell movement or olfactory axon outgrowth.
Other cell types have been identified within the developing nasal epithelium, including galanin (Matsuda et al., 1994; Key and Wray, 2000), somatostatin (Murakami and Arai, 1994), NPY (Hilal et al., 1996), tyrosine hydroxylase (Verney et al., 1996), PAM (peptidylglycine α-amidating monooxygenase), and PACAP (pituitary adenylyl cyclase-activating peptide) (Hansel et al., 2001), and calbindin d-28k (Toba et al., 2002). Several of these molecules modulate release of cholecystokinin (CCK) (Ghijsen et al., 2001), one of the most abundant neuropeptides in the CNS (Vanderhaeghen et al., 1975). Originally discovered in the gastrointestinal tract (Ivy and Oldberg, 1928), CCK is also expressed in several sensory systems (Kuljis et al., 1984; Atkinson and Shehab, 1986; Grothe and Unsicker, 1987; Herness et al., 2002). CCK modifies the migratory abilities of tumor astrocytes (De Hauwer et al., 1998; Lefranc et al., 2002) and lymphocytes (Medina et al., 1998), but a role for CCK in developing neuronal systems has not been examined.
This study evaluated the expression and role of CCK and its receptors (CCK-1R or CCK-2R) (Noble et al., 1999) in the developing olfactory–GnRH-1 systems in vivo and in vitro. We report CCK expression by olfactory receptor neurons and pheromone receptor neurons and on olfactory fibers along which GnRH-1 neurons migrate. We show that migrating GnRH-1 neurons express CCK-1R receptors, whereas both receptors are present on olfactory axons. In addition, perturbation of signals through the CCK–CCK-1R signal transduction pathway, both in knock-out (KO) mice and in nasal explants, alters GnRH-1 neuronal movement. Our results suggest a novel mechanism by which endogenous CCK modulates the GnRH-1 migratory process during prenatal development, ensuring appropriate GnRH-1 neuronal entrance into the CNS.
Materials and Methods
Animals. All mice were killed in accordance with National Institutes of Health–National Institute of Neurological Stroke and Disorders guidelines. NIH Swiss embryos were harvested at E11.5, E12.5, E14.5, and E17.5 (plug day, E0.5) and used for RNA isolation or immediately frozen and stored (–80°C) until laser capture microscopy, or they were postfixed (overnight; 4% paraformaldehyde in 0.1 m phosphate buffer, pH 7.4), cryoprotected, and then frozen and stored (–80°C) until processing for immunocytochemistry (ICC). CCK-1R- and CCK-2R-deficient mice (KO) and wild-type (WT) background control mice (C57BL/129) were provided by Dr. A. Kopin (Tufts University School of Medicine) (Langhans et al., 1997; Kopin et al., 1999). Embryos (E14.5), postnatal heads, and adult brains from these mice were harvested, frozen, and stored (–80°C) until processing.
Nasal explants. Nasal regions were cultured as described previously (Fueshko and Wray, 1994). Briefly, nasal pits of E11.5 NIH Swiss mice were isolated under aseptic conditions and adhered onto coverslips by a plasma (Cocalico Biologicals, Reamstown, PA)–thrombin (Sigma, St. Louis, MO) clot. Explants were maintained in defined serum-free medium (SFM) (Fueshko and Wray, 1994) at 37°C with 5% CO2. On culture day 3, fresh media containing fluorodeoxyuridine (8 × 10–5 m; Sigma) was given to inhibit proliferation of dividing olfactory neurons and non-neuronal explant tissue. For 7-d-old explants and older, the media was changed to fresh SFM on culture day 6 and every 2 d thereafter.
Transcript analyses
All primers were designed from published GenBank sequences and screened using BLAST to ensure specificity of binding. Primers were pretested on brain cDNA and thereafter used throughout the described protocols at a concentration of 250 nm. Amplified products were run on a 1.5% agarose gel.
RT-nested PCR of nasal region. Total RNA was isolated from noses obtained from E11.5–E17.5 mice using RNA STAT-60 (Tel-Test) following manufacturer's protocol. Briefly, tissue was homogenized (1 ml RNA STAT-60/50–100 mg tissue), chloroform was added (0.2 ml/ml homogenate), and the mixture was spun. Isopropanol was added (0.5 ml) to the aqueous layer to precipitate RNA. The RNA pellet was washed (75% ethanol), air-dried, and resuspended (DEPC-treated water). Total RNA from adult brain and whole E17.5 mice served as positive control tissues. One microgram of each sample was used for the RT-PCR reaction. The nested CCK primers used were as follows: outer forward primer (5′-GGCCCTGCTTGGAGGAGG-3′), outer reverse primer (5′-GGAAACACTGCC-TTCCGACC-3′), and CCK inner forward primer (5′-CCACACATACGACCCCTCG-3′). Additional CCK nested primers (Herness et al., 2002) were used to confirm results.
For the first RT-PCR reaction round, AccessQuick RT-PCR System (Promega, Madison, WI) was used with CCK outer forward and reverse primers. RT-PCR cycling was performed as follows: 48°C for 45 min; 94°C for 2 min; 40 cycles of 94°C for 30 sec, 65°C for 1 min, 68°C for 2 min, final extension 68°C for 7 min. A second round of PCR was conducted using 1 μl of undiluted RT-PCR product, which was amplified using CCK inner forward and reverse primers. For each reaction, 30.5 μl of nuclease-free H2O, 5 μl of 10× PCR GOLD buffer (Applied Biosystems, Foster City, CA), 4 μl of 25 mm MgCl2, 5 μl of dNTP mix (2.5 mm), and 0.5 μl of Amplitaq Gold (Applied Biosystems) were mixed. Primers (2 μl of each) and template cDNA (1 μl) were added to the mixture. The PCR program was as follows: 94°C pre-run for 10 min, 94°C for 30 sec, 65°C for 30 sec, 72°C for 2 min for 40 cycles, and 72°C post-run for 10 min. No products were amplified in water or brain RNA–RT (not reverse transcribed).
Laser capture microdissection and RT-nested PCR on tissue–specific regions. Laser capture microdissection (LCM) permits cells to be isolated (“captured”) from tissue sections for molecular analyses. In this study, olfactory epithelium (OE), vomeronasal organ (VNO) epithelium, and pituitary (Pit) were captured from E14.5 and E17.5 mouse frozen sections (see Fig. 1C,E) using a PALM LCM system (Zeiss, Oberkochen, Germany). The laser-microdissected tissues were popped into a sterile Microfuge cap containing 1 μl of 0.1% Triton X-100 and subsequently centrifuged for 1 min at 7500 × g to relocate material to the bottom of a sterile tube. Prime RNase inhibitor (Eppendorf; 7 μl diluted 1:100 in DEPC-treated water) was added. Captured tissue was used to synthesize first-strand cDNA using SuperScript III First-Strand Synthesis System for RT-PCR (Life Technologies, Gaithersburg, MD) following manufacturer's instructions. Controls without reverse transcriptase were performed to demonstrate the absence of contaminating genomic DNA. Brain and thymus total RNA were also reverse transcribed and used as positive and negative controls, respectively.
Figure 1.
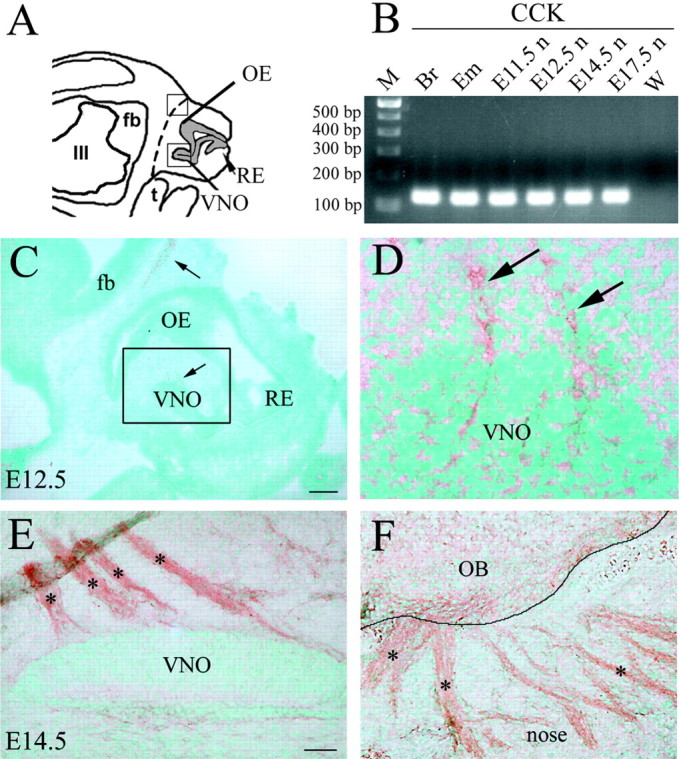
Olfactory system expresses CCK during embryonic development. A, Schematic of an E12.5 head; forebrain (fb), olfactory epithelium (OE), presumptive vomeronasal organ (VNO), respiratory epithelium (RE), tongue (t), and third ventricle (III) are depicted. Dashed line indicates boundary between nose and brain and represents region taken for nasal RNA isolation in B. B, Gel documentation of products produced by RT-PCR amplification using nested CCK primers. Total RNA was isolated from nose (E11.5n–E17.5n). Adult brain (Br) and E17.5 whole embryo (Em) were used as positive control tissues. A band of expected size (169 bp) was detected in positive control tissues after first-round PCR (data not shown). Second-round PCR, using 1 μl of first-round amplicons and second set of nested primers, yielded the predicted 135 bp product in all samples but water [w; a DNA ladder of 100 bp markers (m) was loaded in left lane]. C, Sagittal section of an E12.5 mouse embryo (16 μm) immunostained for CCK. CCK-immunopositive tracks extended from the presumptive VNO (D, see boxed area and high magnification), and a light signal was detected at the boundaries between the nasal region and the forebrain (nasal–forebrain junction; arrow). E, F, Immunohistochemistry for CCK on a sagittal section of an E14.5 embryo. At this stage, a notable increase in CCK immunoreactivity was evident throughout the axon bundles emerging from the VNO (E, asterisks) and the olfactory axons reaching the nasal–forebrain junction (F, asterisks). OB, Olfactory bulb. Scale bars: C, 150 μm; D, 15 μm; E, F, 50 μm.
PCR was performed for β-tubulin (general neuronal marker), early B-cell factor (EBF-2), an olfactory transcription factor and thus marker of olfactory/vomeronasal receptor neurons (Wang et al., 1997), and CCK following the protocol above, at 40 cycles on a thermocycler (30 sec denaturation at 94°C, 30 sec annealing at 55–65°C, 2 min elongation at 72°C). PCR primer pairs were as follows: β-tubulin forward primer (5′-GAGGACAGAGCCAAGTGGAC-3′) and reverse primer (5′-CAGGGCCAAGACAAGCAG-3′); EBF-2 forward primer (5′-TGCAGTAGTTGCTAACAGTGG-3′) and reverse primer (5′-TTTCCAATGCTAGAAGCCTAAC-3′). CCK primers were those described above. For β-tubulin and EBF-2, one round of PCR produced detectable products. A second round of PCR was necessary to visualize CCK products. The same profile as that used for the first PCR reaction was used for nested PCR. One microliter of cDNA amplified using CCK outer primers was added to the second standard 50 μl PCR mixture containing CCK inner forward and outer reverse primers and amplified with 40 cycles.
PCR on single GnRH-1 cell. At three time points [4.5, 7, and 28 d in vitro (DIV)], single GnRH-1 cells (n = 3, 5, and 5, respectively) were isolated from nasal explants, cDNA was produced, and PCR was amplified as described previously (Kramer et al., 2000; Sharifi et al., 2002). All cDNA pools were initially screened for GnRH-1 (correct cell phenotype), β-tubulin, and L19 (two house keeping genes: microtubule and ribosomal) using PCR. All cells used in this study were positive for all three transcripts. The primers sequences used were as follows: GnRH-1 (5′-GCTAGGCAGACAGAAACTTGC-3′), (5′-GCATCTACATCTTCTTCTGCC-3′), β-tubulin (described above), and L19 (5′-CCTGAAGGTCAAAGGGAATGTGTTC-3′), (5′-GGACAGAGTCTTGATGATCTCCTCC-3′). Each reaction mixture was generated as described above, and 2 μl of each primer and 1 μl template cDNA were added. The PCR program was as follows: 94°C pre-run for 10 min, 94°C for 30 sec, 55°Cor65°C (depending on primers) for 30 sec, 72°C for 2 min for 35 cycles, and 72°C post-run for 10 min. The same PCR profile was used for subsequent screening with the following primers: CCK-1R forward primer (5′-ATGCGAGGCCAGTAGCTAGA-3′), CCK-1R reverse primer (5′-CTTCTTACCCGGAGGCATTT-3′), CCK-2R forward primer (5′-TGAGCAGGTGTGGTTTATGG-3′), and CCK-2R reverse primer (5′-TTAACGATGGCACCAAATGA-3′). Specific bands were observed in total brain lanes, whereas no bands were seen in water or brain RNA–RT lanes.
Immunocytochemistry. The primary antisera used were against GnRH-1 (1:3000; SW-1) (Wray et al., 1988), peripherin (1:2000; Chemicon, Temecula, CA), CCK-8 (1:1000; ImmunoStar, Hudson, WI), anti-CCK-8 (Chemicon), CCK-1R, and CCK-2R receptors (1:4000; provided by Prof. P. Beart, University of Melbourne) (Mercer and Beart, 2004), and NCAM (1:60; Chemicon). Primary antibodies were rabbit polyclonals except for NCAM, which was a monoclonal antibody (mAb). Methods and Ab specificity were assessed by elimination of the primary Ab, inclusion of positive control tissue, and, in the case of CCK Ab, preabsorption of the primary Ab with a synthetic peptide [CCK-8S, 10–7 m (Research Plus), was incubated with primary Ab 1:1000, 4°C for 24 hr, before application to tissue sections or explants]. Duodenal tissue and cerebral cortex, known to contain CCK and CCK receptors (Beinfeld, 1983; Hokfelt et al., 1991; Wank, 1995; Liddle, 1997; Noble et al., 1999), were used as positive controls (data not shown). One should also note that colchicine is often required to detect CCK peptide in cell soma in vivo (Cho et al., 1983).
Mouse tissue sections or nasal explants were immunocytochemically stained as described previously (Fueshko and Wray, 1994). Fresh-frozen mouse embryos, postnatal heads, and adult brains were cryosectioned at 16 μm. These sections and explants were fixed with 4% formaldehyde for 1 hr before immunocytochemistry. Postfixed, cryoprotected, frozen embryos were cryosectioned at 12–16 μm. Briefly, sections or nasal explants were washed (PBS), incubated in 10% NGS/0.3% Triton X-100 (NGS/Tx-100) for 1 hr, washed several times in PBS, and placed in primary Ab (overnight at 4°C). The next day, tissues were washed (PBS), incubated in biotinylated secondary Ab [1 hr; 1:500 in PBS/0.3% Triton X-100; goat anti-rabbit biotinylated (GAR-Bt) (Vector Laboratories, Burlingame, CA); anti-mouse biotinylated (GAM-Bt) (Chemicon)], and processed using a standard avidin–biotin–horseradish peroxidase/3′, 3-diaminobenzidine (DAB) protocol. Double-label immunocytochemistry for GnRH-1–peripherin and GnRH-1–NCAM was performed using DAB (brown reaction) and SG substrate (Vector Laboratories; blue reaction) as described previously (Fueshko and Wray, 1994).
For double-immunofluorescence experiments, primary antisera were diluted as follows: anti-GnRH-1 (1:1000), anti-NCAM (1:60), and anti-CCK-1R, anti-CCK-2R, and anti-CCK (1:500). When two polyclonal primary antibodies were used (GnRH-1–CCK, GnRH-1–CCK-1R, GnRH-1–CCK-2R), staining of the first antigen–antibody complex was performed with goat anti-rabbit Alexa-Fluor 488 (1:500; Molecular Probes, Eugene, OR) secondary Ab. This step was followed by a blocking reaction with an anti-rabbit Fab fragment (80 ng/ml; Jackson ImmunoResearch, West Grove, PA) (Kramer et al., 2000) for 1 hr (room temperature) followed by PBS washes, fixation (4% formalin, 30 min), and washes (PBS) before application of the second primary Ab, which was visualized with conjugated fluorescent goat anti-rabbit CY-3 (1:800; Jackson ImmunoResearch). For CCK-1R–NCAM, CCK-2R–NCAM, and CCK–NCAM experiments, sections were incubated overnight (4°C) in a mixture of rabbit polyclonal antiserum and rat mAb to NCAM diluted in NGS/Tx-100 and visualized using an anti-rabbit CY-3 (1:800) and a biotin-conjugated anti-rat (Vector Laboratories) followed by avidin-FITC (1:400; Vector Laboratories).
Triple-immunostaining was performed for CCK-2R, NCAM, and GnRH-1 in explants. For this procedure, GnRH-1 and NCAM were incubated together and visualized using an anti-rabbit Alexa 488 (1:500; Molecular Probes) and a biotin-conjugated anti-rat (Vector Laboratories) followed by streptavidin–Alexa 350 (1:500; Molecular Probes). Before application of anti-CCK-2R, the blocking reaction (anti-rabbit Fab fragment; 80 ng/ml) (Kramer et al., 2000) was performed. A CY-3-conjugated anti-rabbit secondary Ab (1:800; Jackson ImmunoResearch) was used for visualization of the third antigen–antibody complex. Controls for single-, double-, or triple-staining protocols revealed no significant cross-epitope immunoreactivity among primary or secondary antibodies (data not shown).
Functional assays
Nasal explant. To determine the function of CCK in the developing olfactory–GnRH-1 systems, pharmacological perturbations were performed, and olfactory axon outgrowth and GnRH-1 cell migration were quantitated. Synthetic CCK-8 (Research Plus) was exogenously applied to nasal explants at four concentrations ranging from 10–5 to 10–12 m. CCK-1R antagonist (lorglumide; Sigma) and CCK-2R antagonist (L-365, 260; ML Laboratories, Liverpool, UK) were dissolved in DMSO (Sigma), aliquoted, and stored at –20°C. Each antagonist was tested from 10–5 to 10–9 m.
Nasal explants were treated at 3 DIV with pharmacological agents, fixed at 5 DIV, and processed by immunocytochemistry for GnRH-1–peripherin (see above). Quantitation of GnRH-1 cell migration and olfactory fiber outgrowth was performed on digitized images (Nikon Cool-Pix 5000 camera) that were overlaid on a calibration meter composed of concentric arcs 200 μm apart (see Fig. 7). In general, under all treatment conditions, the number of GnRH-1 cells decreases as a function of distance from the main tissue mass. GnRH-1 cell migration was calculated as the distance from the main tissue mass edge to the outer border of the cell migration front. The cell migration front does not represent the distance that the farthest GnRH-1 cell migrated but rather the most distal zone that contained multiple GnRH-1 cells. The average number of GnRH-1 cells detected in this outer 200 μm peripheral zone (front of migration) was consistent among treatment groups (control, CCK-treated, CCK plus CCK-1R antagonist treated, and CCK plus CCK-2R antagonist treated) and was ∼40 GnRH-1 cells. After the cell migration front, the GnRH-1 cell number in the next zone (usually 200 μm immediately farther than the front of migration) dropped to 10% in each group. These parameters were used to determine the front of GnRH-1 migration. Note that although the number of GnRH-1 cells in front of the migration zone was similar across groups (containing on average ∼40 GnRH cells), the absolute zone in which these cells were detected varied, and this was the value analyzed for cell migration. The total number of GnRH-1 cells was recorded as well, allowing GnRH-1 neuronal populations to be compared among treatment groups. For fiber outgrowth measurements, the distance from the border of the explant at which multiple peripherin-positive fibers ended was recorded; this method was chosen because the complex nature of the fiber network prevented quantitation of individual fiber lengths (Fueshko and Wray, 1994). A mean distance for cell migration and fiber outgrowth as well as a mean GnRH-1 cell total number were obtained for each treatment group; values are given ± SEM. All experiments used explants generated by different individuals on multiple culture dates.
Figure 7.

CCK alters olfactory axon outgrowth and GnRH-1 cell migration. A, Photomicrograph of nasal explant immunocytochemically labeled for GnRH-1 (brown) and peripherin (blue), at 5 DIV. Many GnRH-1 neurons were visualized within the main tissue mass and in the periphery. Images were digitized and overlaid to a calibration meter composed of concentric arcs. Olfactory axon outgrowth (left histograms) as well as front of GnRH-1 cell migration (right histograms) were calculated as a function of treatment. GnRH-1 neurons remained apposed to peripherin fibers (along which they migrate) under all conditions. B, C, CCK (100 nm) applied at 3 DIV significantly decreased, at 5 DIV, olfactory axon outgrowth (B) and the front of migration of GnRH-1 cells (C) compared with controls (significantly different from control; ap < 0.001). CCK was ineffective in the presence of the CCK-1R-selective antagonist (lorglumide 100 nm; significantly different from CCK; bp < 0.05). Such treatment returned both parameters to control values. Olfactory axon outgrowth and the front of migration were similar in explants treated with CCK versus CCK in the presence of the CCK-2R antagonist (L-365, 260, 100 nm). D, E, Endogenous CCK is functionally active in nasal explants and acts via CCK-1R. CCK-1R antagonist treatment did not alter axonal outgrowth (D) but significantly increased the front of GnRH-1 cell migration (E) (significantly different from control and CCK-2R antagonist treated groups; abp< 0.01). CCK-2R selective antagonist application did not alter either olfactory axon outgrowth (D) or the front of migration (E) compared with controls. Scale bars: A, 200 μm; inset, 15 μm.
Statistical comparisons of data were calculated with the statistical software Statview (SAS Institute, Cary, NC). ANOVA followed by Fisher's LSD post hoc analysis was used to compare groups (p < 0.05). In addition, χ2 analysis was performed; p < 0.01 was considered statistically significant.
Quantitative analysis of GnRH-1 neurons in CCK receptor mutant embryos. Serial sections (16 μm, four series) from CCK-1R–/–, CCK-2R–/–, and WT E14.5 mice (n = 3 for each group) were cut (Leica cryostat), and double-labeling for GnRH-1–peripherin was as described above (GnRH-1 immunoreactivity visualized using DAB and peripherin immunostaining using SG substrate). Quantitative analysis of GnRH-1 neuronal numbers was performed as a function of location (see Fig. 8), with GnRH-1 cells assigned to one of three regions (nasal region, nasal–forebrain junction, and CNS). The total number of cells counted in each region was calculated for each animal and combined to give group means ± SEM. Data for GnRH-1 cell distribution among the areas was compared by constructing contingency tables and applying the χ2 test for independence. Such an analysis determines whether the observed sample differences signify differences among populations or whether they are differences that one might expect among samples from the same population. If the χ2 analyses indicated significant differences, one-way ANOVA, followed by a Fisher's LSD post hoc test, was used to evaluate the number of GnRH-1 cells located within discrete compartments in the three animal groups. χ2 data were considered significantly different if p < 0.01. ANOVA data were considered significantly different if p < 0.05. All data are expressed as mean ± SEM.
Figure 8.
Migration of GnRH-1 neurons in CCK-1R–/– mutant embryos is accelerated. Quantitation of GnRH-1 cell number in three regions along the migratory pathway within sagittal and coronal sections from E14.5 WT, CCK-1R KO, and CCK-2R KO mice was performed. A, B, Photomicrographs showing GnRH-1 (brown)–peripherin (blue) immunoreactivity in a sagittal section of a CCK-1R KO embryo. The areas of analysis for GnRH-1 neuron location along the migratory pathway are shown in A. The boxed area = nasal–forebrain junction (N/FJ); cells located in areas outside this region were classified as in either the CNS (B) or nose. C, Analysis of GnRH-1 neuron location revealed an increase in the number of GnRH-1 neurons in the CNS of CCK-1R KO compared with WT and CCK-2R KO mice (**p < 0.01; by post hoc Fisher's LSD). Consistent with these data, fewer (although it did not reach significance; Fisher's LSD post hoc test; p=0.07) GnRH-1 cells were located in the N/FJ area of CCK-1R mutant embryos compared with WT and CCK-2R-deficient mice. Scale bars: A, 150 μm; B, 50 μm.
Results
CCK expression in the developing olfactory system
To identify whether CCK was expressed prenatally in nasal regions, where GnRH-1 neurons originate, nose tissue was removed (E11.5–E17.5) and RT-PCR experiments were performed (Fig. 1A,B). The first round of RT-PCR reaction failed to detect CCK product in nose tissue, whereas positive bands were found in control lanes (adult brain and E17.5 whole-embryo extracts), and β-tubulin primers produced strong bands in all lanes (data not shown). Because the OE and VNO comprise a small proportion of nasal region cells, a second round of PCR was performed. Using 1 μl of the first-round amplicons and the second set of nested CCK primers, a strong band of correct size was detected in all samples but water (Fig. 1B).
To characterize the cell types expressing CCK in the developing nasal region, immunocytochemistry was performed from E12.5 to E17.5. Throughout this period, CCK immunoreactivity was detected along olfactory axons exiting the VNO and OE and in axon bundles crossing the nasal mesenchyme. At E12.5, light CCK-immunopositive tracks extended from the OE and the developing VNO toward the presumptive olfactory bulb (Fig. 1C,D). By E14.5, the CCK-immunopositive signal in the nasal region was stronger, and numerous immunopositive CCK olfactory fibers were detected emerging from the VNO (Fig. 1E) and at the nasal–forebrain junction (Fig. 1F). Immunocytochemical results at E12.5 and E14.5 suggested that CCK-positive fibers spanning across the nasal region originated from both the OE and VNO structures; however, at these stages only scattered cell soma that were CCK immunoreactive were detected in the OE or VNO. In contrast, staining for CCK at E17.5 demonstrated that CCK was strongly expressed in the OE (Fig. 2A,B). CCK was detected in both cell soma and apical dendrites of olfactory receptor neurons (Fig. 2B, boxed picture). In addition, at E17.5, CCK protein was also detected in a subpopulation of VNO cells scattered in the central region of the neuroepithelium (Fig. 2C).
Figure 2.

Olfactory sensory neurons express CCK mRNA and protein during development. A, Schematic of E17.5 coronal section depicting location of VNO and OE. NMC, Nasal midline cartilage. B, C, Immunocytochemistry for CCK on E17.5 coronal mouse sections. At E17.5, olfactory receptor neurons were CCK positive (B). Perikarya (arrows; outlined enlarged picture), apical dendrites (arrowheads), and axons of these neurons (data not shown) were stained for CCK. In the VNO (C), CCK was expressed in a small population of sensory neurons, toward the central region of VNO neuroepithelium. This CCK labeling was in cell soma (inset) and occasionally in apical dendrites. D, Photographs of E14.5 and E17.5 sagittal sections after LCM. The representative pictures show two examples of microdissected OE (leftpanel) and pituitary (Pit; right panel). Total RNA isolated from dissected region of OE and VNO was subjected to RT-PCR followed by two PCR rounds using nested primers. A fragment of the expected size (135 bp) was detected for CCK in the OE but not in VNO or water at E14.5 (left panel). Expression of the olfactory marker, EBF-2 (165 bp), and β-tubulin (data not shown) confirmed the morphology of the dissected tissue. Total RNA isolated from dissected E17.5 OE, VNO, and Pit was processed as described for E14.5 tissue. A strong CCK band was evident in OE and Pit, whereas a weak but detectable product was present in VNO. No PCR product was observed in reactions that omitted either reverse transcriptase (data not shown) or starting material (water). PCR analysis using EBF-2 primers showed expected amplicons in OE and VNO but not in Pit (right panel). E, Sagittal section of an E14.5 mouse nose stained for GnRH-1 and peripherin (intermediate filament protein expressed in olfactory–vomeronasal axons). GnRH-1 neurons (blue staining) migrate along peripherin-positive axons (brown staining) through the nasal mesenchyme. Inset shows a high magnification of GnRH-1 neurons migrating out of the VNO (arrows indicate GnRH-1 cells associated with peripherin-immunoreactive fibers). F, Immunocytochemistry of an E14.5 mouse using GnRH-1 and CCK antisera. GnRH-1 neurons (green) emerge from the VNO and migrate along CCK-positive axons (red). Note that staining pattern resembles that in GnRH-1–peripher in double labeling (E). Scale bars: B, 10 μm; inset in B, 5 μm; C, 25 μm; inset in C, 6.25 μm; D, 50 μm.
To confirm these findings, in situ hybridization (ISH) was performed for CCK mRNA during embryonic development. Many regions known to express CCK mRNA and protein prenatally and postnatally (Cho et al., 1983; Itoh et al., 1998) were indeed positive at E14.5 and E17.5; hybridization signals were detected over cells in the hippocampus, forebrain, thalamus, ventromedial hypothalamic nucleus, and pituitary, and in the enteric nervous system (data not shown); however, no signal was detected in nasal regions. In taste receptors (Herness et al., 2002), robust CCK peptide expression has been documented, yet CCK mRNA expression levels were very low (detectable only after two rounds of RT-PCR). Because the expression pattern of CCK mRNA and protein in olfactory receptor neurons and pheromone receptor neurons resembled that reported for taste receptors, nasal tissues were analyzed using a similar RT-PCR strategy. Single punches from OE and VNO tissues were removed using LCM. Figure 2D shows an E14.5 sagittal section in which an area of the OE was laser captured. No other tissue was removed from the nasal section, and the remaining tissue was intact after the capture procedure. In the same manner, a region of the VNO was microdissected from an E14.5 section and processed for RT-PCR. After reverse transcription of the mRNA, PCR with specific primers for β-tubulin (positive control) and the olfactory marker Olf/early B-cell factor-2 was performed (Fig. 2D, left panel). As expected, a 158 bp band corresponding to β-tubulin (data not shown) and a 165 bp band corresponding to EBF-2 product were detected in both OE and VNO. Analysis of CCK mRNA expression in these olfactory structures was then performed. After the second-round PCR reaction, a CCK product was found in the OE but not in the VNO section (Fig. 2D, left panel). Because ICC detected CCK peptide in both nasal epithelia at E17.5, a similar analysis was performed on OE and VNO at this stage (Fig. 2D, right panel). Anterior pituitary (E17.5 Pit) laser-captured tissue was also included in this screen as a negative control for EBF-2 and as a positive control for CCK on the basis of our ISH results. EBF-2 PCR products were always detected in the OE and VNO, and in all cases no detectable bands were found in water or the Pit; however, a band of the expected size for CCK was detected in the Pit. In addition, at E17.5, a CCK band was again identified in the OE, and a light amplicon was now detected in the VNO as well (Fig. 2D, right panel).
Molecular analysis was performed to confirm CCK peptide localization in nasal epithelia. CCK-immunopositive axons were found exiting the OE and VNO from E12.5 to E17.5. Using RT-PCR, cells within the OE were CCK positive from E14.5 to E17.5, whereas cells in the VNO were positive at E17.5. Thus, although confirmation of the ICC data was obtained, low levels of CCK mRNA prevented additional information to be ascertained. Why CCK mRNA levels are so different from peptide levels during development is unclear, but they may reflect rapid changes that were outside the time points chosen or small cell populations dispersed throughout the region examined, or both. However, together these techniques allowed us to identify CCK expression in both olfactory receptor neurons and pheromone receptor neurons from E12.5 to E17.5, particularly those localized within olfactory fiber bundles.
Between E11.5 and E14.5, GnRH-1 cells emerge from olfactory pits in tracks that extend dorsocaudally through nasal regions toward the forebrain (Wray et al., 1989; Schwanzel-Fukuda and Pfaff, 1989). As the GnRH-1 cells move across this region and into the developing forebrain, they are associated with peripherin-positive olfactory fibers (Fueshko and Wray, 1994) (Fig. 2E). To determine whether the olfactory axons used for GnRH-1 migration expressed CCK, we performed double-label immunohistochemistry for GnRH-1 and CCK (Fig. 2F). At E12.5–E14.5, GnRH-1-positive–CCK-negative cells were detected migrating along CCK-immunoreactive fibers from the VNO to the nasal forebrain junction.
CCK-1R and CCK-2R receptor expression in the olfactory–GnRH-1 systems
To gain insight into the signaling pathway through which CCK may act prenatally in nasal regions, the expression pattern of CCK receptors (CCK-1R and CCK-2R) was examined from E12.5 to birth.
Olfactory–vomeronasal axons express NCAM (Calof and Chikaraishi, 1989; Miragall et al., 1989) and peripherin, whereas GnRH-1 cells in mouse do not (Fig. 3A–C) (Schwanzel-Fukuda et al., 1992; Fueshko and Wray, 1994). Immunocytochemistry for CCK-2R indicated that this receptor was expressed along olfactory tracks extending from the OE and the VNO to the presumptive olfactory bulb (Fig. 3D–F). To determine whether CCK-2R-immunopositive tracks were in fact olfactory axons, double-label immunofluorescence was performed for CCK-2R and NCAM. CCK-2R and NCAM expression overlapped on fibers emerging from the VNO at E12.5 (Fig. 3D) and were coexpressed in olfactory–vomeronasal axon bundles from the nasal tract to the medial surface of the forebrain throughout the analyzed stages (E12.5–E17.5). Because of low signal-to-noise levels in brain, we were unable to detect specific immunoreactivity for CCK-2R along the caudal nerve that GnRH-1 cells follow into the ventral forebrain (Yoshida et al., 1995). To determine whether migrating GnRH-1 neurons expressed CCK-2R, we performed double-label immunohistochemistry (Fig. 3E,F). At E12.5, GnRH-1-positive–CCK-2R-immunonegative neurons were detected migrating out of the VNO along CCK-2R-positive tracks (Fig. 3F). In all mice analyzed, migrating neurons located in the nasal compartment were single labeled for GnRH-1.
Figure 3.
CCK-receptors (CCK-1R, CCK-2R) are expressed on OE and VNO, whereas CCK-1R is expressed by GnRH-1 neurons in nasal regions. A–I, Sagittal sections of E12.5 mouse immunostained with indicated antibodies. GnRH-1 cells migrate in association with olfactory axons. A–C, GnRH-1 (blue) and NCAM (brown; A, B) or GnRH-1 and peripherin (brown; C). Note that these sections have been counterstained with methyl green. B, High magnification of boxed area in A shows GnRH-1-positive neurons migrating out of the developing VNO along NCAM-immunoreactive axons (arrows). C, Picture similar to that in B, showing GnRH-1 neurons associated with peripherin-positive vomeronasal axons (arrows). D, CCK-2R-immunoreactive fibers (red) colocalized with NCAM-immunopositive olfactory axons (green) crossing the nasalmesenchyme (arrows). The labeling pattern resembles those described in B and C. E, F, Double-label immunofluorescence for CCK-2R (red) and GnRH-1 (green). OE, VNO, and axons extending toward the telecephalon are CCK-2R positive (E; arrows indicate olfactory axons emerging from VNO). GnRH-1 neurons were CCK-2R negative (F, inset). G–I, Double-label immunofluorescence for CCK-1R (red) and GnRH-1 (green). CCK-1R immuno reactivity was detected in the OE, VNO, along olfactory fibers (arrows), and in CCK-1R-positive cells spanning the nasal mesenchyme. CCK-1R-immunoreactive cells coexpressed GnRH-1 (I, inset, arrowheads). Scale bars: A, 245 μm; B, C, 70 μm; D, E, G–I, 34 μm; inset in D, F, 20 μm; inset in F, inset in I, 10 μm.
Immunohistochemistry for CCK-1R revealed robust expression in the VNO and OE as well as on the olfactory axons emerging from these two structures (Fig. 3G). Double labeling for GnRH-1 and CCK-1R demonstrated that this receptor was expressed in GnRH-1 cells exiting the presumptive VNO and migrating through the cribriform plate (Fig. 3H,I). At all ages analyzed, GnRH-1 neurons located in nasal regions expressed CCK-1R. Once within the brain, it was difficult to determine whether GnRH-1 neurons maintained CCK-1R expression because of the high level of expression of this receptor in other CNS cells (data not shown). Thus, during embryonic development both CCK-1R and CCK-2R demarcate olfactory sensory fibers and the pathway on which the GnRH-1 neurons migrate; however, only CCK-1R is present on GnRH-1 neurons themselves.
Functional analyses
In vitro
It has been shown previously that the migration pattern of GnRH-1 neurons observed in vivo reproducibly occurs in nasal explants in vitro, with a shift in location of the GnRH-1 cell population from the olfactory pit epithelia (OPE) to the edge of the main tissue mass occurring from 1–3 DIV and continuing to more distant sites from 3–7 DIV (Fueshko and Wray, 1994). Thus, nasal explants represent a valuable tool to dissect apart spatial from temporal cues and focus on the properties of GnRH-1 neurons by controlling extracellular influences. To use nasal explants for functional studies, we first verified that this system retained expression of CCK and its receptors comparable with that observed in vivo. It should be noted that ICC on nasal explants is performed on the entire tissue; no sections are created. Thus, in nasal explants the detectable signal is often enhanced compared with tissue sections, and molecule localization is not limited to a single dimension. With the entire cell present and using conventional microscopy, membrane-localized molecules may not appear excluded from the nucleus, being detected throughout the cell surface. In addition, cells in different regions of the explant (periphery vs main tissue mass) undergo dimensional changes, with cells in the periphery often “flatter” than those on the explant; therefore, staining can appear to change with respect to cellular compartments depending on the explant age or location, or both.
Olfactory–vomeronasal axons express CCK. At 7 DIV, GnRH-1 neurons had migrated off the explant into the periphery along NCAM-positive olfactory fibers, resembling development in vivo (Fig. 4A,B). Examination by double immunofluorescence for NCAM and CCK revealed that NCAM-positive olfactory neurons in the OPE as well as NCAM-positive olfactory fibers coursing toward the nasal midline cartilage (Fig. 4C) were robustly labeled for CCK (Fig. 4D, merge image in E). Thus as in vivo, olfactory–vomeronasal axons expressed CCK in vitro.
Figure 4.
CCK expression in olfactory system in vitro mimics in vivo expression pattern. A, Schematic of a nasal explant removed from an E11.5 mouse and maintained in serum-free media for 7 DIV. Ovals represent olfactory pit epithelium (OPE); in center is nasal midline cartilage (NMC) and surrounding mesenchyme. GnRH-1 neurons (dots) migrate from OPE and follow olfactory axons to the midline and off the explant into the periphery. Boxed region 1 within schematic is area shown in B, whereas boxed region 2 is area shown in C–E. B, Double immunocytochemistry was performed using antibodies to GnRH-1 (brown) and NCAM (blue). Many GnRH-1 neurons are detected that migrated off the explant into the periphery along NCAM-immunoreactive olfactory fibers (see insert, arrow; asterisk indicates GnRH-1 neuron; NCAM-positive axon bundles). It should be noted that ICC on nasal explants is performed on the entire tissue; thus, cells in the periphery of the explant and in the inner tissue mass undergo dimensional changes. GnRH-1 cells located in the periphery migrate on a monolayer of fibroblast and appear flatter than those on the explant. C, D, Double immunofluorescence was performed on 7 DIV explants using antibodies to NCAM (red) and CCK (green). C, Immunostaining for NCAM revealed robust staining in OPE structures as well as along olfactory axons emerging from OPE and directed toward midline area. The OPE structures are contained in the inner tissue mass of the explant where the thickness is ∼300 μm. Cells located inside these areas appear more round, and cell density is very high; thus molecule localization is not limited to a single dimension. D, CCK immunoreactivity displayed similar labeling pattern as NCAM. E, Simultaneous visualization of both fluorescent wavelengths indicated that both OPE cell soma as well as processes were positive for CCK and NCAM. Scale bars: B, 27 μm; inset in B, E, 10 μm; C, D, 40 μm.
CCK-2R is expressed on olfactory fibers but not on GnRH-1 cells (Fig. 5). cDNA pools from single GnRH-1 cells removed from nasal explants at 4.5, 7, and 28 DIV (Kramer and Wray, 2000; Kramer et al., 2000) were examined for CCK-2R transcript. Although amplicons were detected in GnRH-1 cells from 4.5 DIV (one of three cells) and 7 DIV (five of five cells; data not shown), GnRH-1 cells were immunonegative for CCK-2R receptor protein at all ages examined. Immunocytochemistry for GnRH-1, CCK-2R, and NCAM was performed and revealed that GnRH-1-positive–CCK-2R-negative neurons (green) were tightly associated with olfactory fibers (pink) immunopositive for both CCK-2R (red) and NCAM (blue). Thus as in vivo, olfactory–vomeronasal axons expressed CCK-2R receptors.
Figure 5.
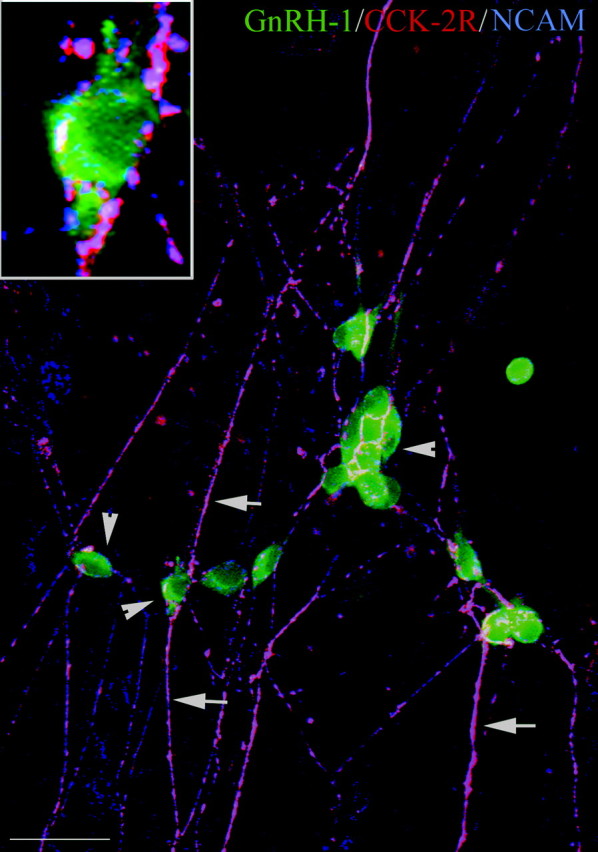
CCK-2R is expressed in olfactory axons but not GnRH-1 cells in nasal explants. Nasal explant at 7 DIV triple stained for GnRH-1 (green), CCK-2R (red), and olfactory axon antigen NCAM (blue) is shown. GnRH-1 neurons in the periphery of the explant are tightly associated with a network of olfactory fibers. Note that CCK-2R colocalized only with NCAM-positive fibers (arrows; pink) but not with GnRH-1 neurons (arrowheads; green). Scale bars: 20 μm; inset, 5 μm.
CCK-1R is expressed on olfactory fibers and GnRH-1 cells (Fig. 6). cDNA pools from GnRH-1 cells were examined, as described above, for the presence of CCK-1R transcript (Fig. 6F). At 4.5 and 7 DIV, two of three and five of five GnRH-1 cells expressed CCK-1R transcript, respectively. By 28 DIV, all GnRH-1 neurons (n = 5) were negative for this transcript. Double-immunofluorescence revealed colocalization of CCK-1R and NCAM in the OPE of nasal explants (Fig. 6A), and olfactory axons, along which GnRH-1 neurons migrated, exhibited CCK-1R staining as well (Fig. 6B,E). Hence, consistent with in vivo results, CCK-1R receptor demarcated the olfactory system. In addition, as in vivo, GnRH-1 cells expressed CCK-1R (Fig. 6B–E, arrows), and by 7 DIV, most GnRH-1 cells possessed characteristic CCK-1R plasma membrane immunoreactivity (Fig. 6C–E).
Figure 6.

GnRH-1 neurons express CCK-1R in vitro. Double immunofluorescence on explants using antibodies directed against CCK-1R and NCAM or GnRH-1 and CCK-1R is shown. A, OPE in the inner tissue mass of a 3 DIV nasal explant. Double labeling for CCK-1R–NCAM revealed colocalization in cells in the OPE as well as on olfactory fibers. B, At 3 DIV, a subpopulation of early migrating GnRH-1 neurons coexpressed the CCK-1R (arrows). CCK-1R robust staining was also evident along outgrowing axon bundles (arrowheads). At 7 DIV (C–E), in the periphery of the explant, CCK-1R-immunoreactivity was evident along the olfactory axon network (C). Bipolar neurons, associated with these fibers, appeared strongly labeled as well (C). At this in vitro stage, a large population of GnRH-1 neurons is located in the periphery of the explant (D). Double immunofluorescence showed that CCK-1R immunoreactivity was located on the majority of GnRH-1 cells and along olfactory fibers (E, merged image). Note in E that GnRH-1 is confined to the cytoplasmic compartment, whereas CCK-1R is distributed throughout the cell surface, resulting in colabeling in the cytoplasm (yellow) and red appearing over the nucleus (E). E, Inset, Immunocytochemistry for CCK-1R at 7 DIV using the DAB method and showing how CCK-1R immunoreactivity is localized throughout the cell surface of bipolar neurons associated with axons bundles in the periphery of the explant. F, Representative gel documentation of PCR products from single-cell RT-PCR performed on individual cells (4.5, 7, and 28 DIV) extracted from the periphery of nasal explants. Top row shows products produced by PCR amplification of the above cDNAs using primers specific for GnRH-1. A strong band of the expected size (320 bp) was evident in all samples, whereas no specific band was detected in water. The bottom row shows products produced by PCR amplification of the same cDNA samples using CCK-1R-specific primers. CCK-1R transcript (181 bp) was detected in primary GnRH-1 neurons from 4.5 to 7 DIV, whereas by 28 DIV no specific bands were detected in GnRH-1 neurons. Scale bars: A, C, D, 30 μm; B, E, inset in E, 10 μm.
In addition, immunocytochemical experiments confirmed single-cell PCR results showing a significant decrease of CCK-1R immunoreactivity in GnRH-1 cells by 14 DIV (data not shown). These data demonstrate that CCK-1R is expressed in GnRH-1 cells in a temporal window that covers the neuronal migratory process.
Perturbation of nasal explants. To determine whether CCK has an action on axon outgrowth and cell migration, four doses were tested. CCK application at 10–12 and 10–9 m had no effect on cell movement and olfactory axon outgrowth (mean front migration, ∼500 μm; olfactory axon outgrowth, ∼650 μm; n = 3 for each concentration tested) compared with controls (mean front migration, ∼515 μm; olfactory axon outgrowth, ∼634 μm). Higher concentrations of CCK (10–5 and 10–7 m) reduced both cell movement and olfactory axon outgrowth (mean front of migration, ∼325 and 340 μm; olfactory axon outgrowth, ∼400 and 420 μm, respectively; n = 4 for 10–5 m; n = 5 for 10–7 m). No significant differences were found between explants treated with 10–5 versus 10–7 m CCK. For explants treated with antagonists, similar dose results were obtained (CCK–1R and CCK-2R antagonist 10–9 m, mean front of migration, ∼550 μm; olfactory axon outgrowth, ∼650 μm; 10–7 m, front of migration, ∼650 and 500 μm; olfactory axon outgrowth, ∼700 and 680 μm, respectively; 10–5 m, front of migration, ∼600 and 520 μm; olfactory axon outgrowth, ∼680 and 600 μm, respectively; n = 3 for 10–9 and 10–5 m doses; n = 15 for CCK-1R antagonist 10–7 m; n = 17 for CCK-2R antagonist 10–7 m). Therefore, 10–7 m was chosen as the optimal concentration for both CCK and CCK receptor antagonists in our functional study. These same drug concentrations were used in previous electrophysiological studies (Herness et al., 2002).
To further characterize the mechanism by which CCK-8 regulates GnRH-1 cell migration and olfactory axon outgrowth, pharmacological manipulations were performed by exposing nasal explants (from 3–5 DIV) to CCK (100 nm), CCK plus CCK-1R antagonist (100 nm lorglumide) (Makovec et al., 1987) or CCK plus CCK-2R antagonist (100 nm L-365, 260) (Bock et al., 1989). At 5 DIV, explants were fixed and stained for GnRH-1–peripherin (marker of olfactory axons) (Fig. 7A), and total number of GnRH-1 cells, olfactory axon outgrowth, and front of migration of GnRH-1 neurons were analyzed (see Materials and Methods).
After exposure to CCK, total GnRH-1 cell number was un-changed (control = 173 ± 26; CCK treated = 153 ± 13), but both olfactory axon outgrowth and GnRH-1 cell migration were significantly reduced compared with controls (p < 0.001). To classify these effects as mediated by CCK-1R or CCK-2R receptors, selective pharmacological analysis with specific receptor antagonists was performed (Fig. 7B,C). When nasal explants were exposed to CCK + CCK-1R antagonist, both the olfactory axon outgrowth value and the front of migration value were restored to control conditions (p > 0.05) and differed from the CCK group (p < 0.05). With respect to these parameters, CCK-2R antagonist did not prevent changes induced by exogenous CCK (p > 0.2). These data indicated that CCK inhibited olfactory axonal outgrowth and GnRH-1 cell movement via CCK-1R and not CCK-2R.
To determine the role of endogenous CCK on GnRH-1–olfactory system development, we treated the explants with vehicle, CCK-1R, or CCK-2R antagonists. Again, no significant differences were found in total number of GnRH-1 cells (control = 173 ± 26; CCK-1R antagonist = 174 ± 33; CCK-2R antagonist = 226 ± 30). The average distance of peripherin-positive fibers was independent of either CCK-1R or CCK-2R blockage (Fig. 7D) (p > 0.05), whereas application of CCK-1R antagonist significantly increased the distance that GnRH-1 neurons moved from the border of the explant (25% farther; p < 0.003) compared with controls (Fig. 7E). In contrast to CCK-1R antagonist treatment, the values of GnRH-1 migration (Fig. 7E) (p > 0.5) did not change in CCK-2R antagonist-treated samples. These results indicate that endogenous CCK, acting via the CCK-1R receptor, can alter movement of GnRH-1 neurons.
CCK-1R and CCK-2R receptor-deficient mice
To determine whether the in vitro perturbation studies mimicked an in vivo scenario, CCK-1R and CCK-2R receptor knock-out postnatal and E14.5 mice were examined. The total number as well as the overall distribution of GnRH-1 neurons in postnatal and adult brains were similar among the three animal groups [postnatal day 3 (P3) WT, 756 ± 71; P3, CCK1RKO, 738 ± 24; adult CCK2RKO, 716 ± 37]. At E14.5, the general distribution of GnRH-1 neurons as well as olfactory axons in all embryos (WT, CCK-1R–/–, and CCK-2R–/–) was similar to that reported previously (Schwanzel-Fukuda et al., 1989; Wray et al., 1989, 1994); however, disruption of the CCK-1R receptor gene increased the number of GnRH-1 neurons in brain (Fig. 8). The total number of GnRH-1 neurons in these mice did not differ (n = 3 for each group; WT, 918.0 ± 37; CCK-1R–/–, 978.7 ± 55; CCK-2R–/–, 862.7 ± 99); however, χ2 analysis revealed significant differences among these three animal groups with respect to the distribution of the GnRH-1 population (χ2 = 268.0; p < 0.0001). Post hoc ANOVA revealed differences in GnRH-1 cell number within specific regions of the migratory pathway (nose, nasal–forebrain junction, and CNS) (Fig. 8) between WT and CCK1R-deficient mice. In CCK-1R–/– embryos there were significantly more GnRH-1 neurons in the CNS compartment, with 16–20% more cells than in WT and CCK-2R–/– animals (p < 0.01) (Fig. 8C). Concomitantly, although not significant, there were fewer (∼12%) GnRH-1 cells located in the nasal–forebrain junction of the CCK-1R–/– embryos (Fig. 8C) compared with their WT and CCK-2R–/– counterparts. These data are consistent with an acceleration of GnRH-1 neuronal migration.
Discussion
In this report, expression of CCK and its receptors in developing olfactory–GnRH-1 systems is characterized, and a functional role for CCK on GnRH-1 cell movement is identified: inhibition of cell migration via the CCK-1R receptor signal transduction pathway. These data indicate that CCK is an important developmental modulator of GnRH-1 neuronal entrance into brain.
Expression pattern of CCK and CCK receptors
CCK-expressing cells have been described throughout sensory systems in the CNS and PNS, including visual, gustatory, and olfactory systems (Kuljis et al., 1984; Miceli et al., 1987; Hokfelt et al., 1988; Savasta et al., 1988; Ingram et al., 1989; Schiffmann and Vanderhaeghen, 1991; Herness et al., 2002). In the olfactory system, CCK-containing neurons in olfactory bulb, anterior olfactory nucleus, and piriform cortex have been reported (Ingram et al., 1989; Schiffmann and Vanderhaeghen, 1991; Liu and Shipley, 1994). Here, we expand on CCK expression in the olfactory system to include OE and VNO. Using immunocytochemistry, we documented this peptide in developing olfactory and vomeronasal sensory cells. Although in situ hybridization signals were not above background in these regions, two rounds of RT-PCR reactions detected CCK product in noses (E11.5–E17.5) and micro-dissected tissues of OE (E14.5 and E17.5) and VNO (E17.5). In contrast to CCK expression in nasal regions, CCK transcript was detected in microdissected E17.5 anterior pituitary tissue after RT-PCR, and a signal for CCK mRNA was detected in anterior pituitary by ISH (as early as E14.5; data not shown). Immunocytochemistry revealed CCK protein in pituitary cells between E14.5 and E17.5 (data not shown). Although the function of CCK in this tissue prenatally is unknown, pituitary CCK protein has been documented postnatally in rodents (Korchak et al., 1984). Detection of CCK mRNA in pituitary prenatally indicates that technically our sensitivity and specificity was high but that CCK mRNA in nasal sensory receptor cells is extremely low. In a recent paper reporting CCK expression in rat taste receptor cells, Herness and coworkers (2002) described a similar apparent dichotomy between CCK peptide and mRNA expression as we observed in nasal sensory receptor cells. Consistent with Herness et al. (2002), we hypothesize that nasal sensory receptor cells rely on posttranslational regulation of CCK, keeping high levels of the peptide but low copies of mRNA.
Using two different antisera to CCK, immunopositive axons were detected exiting the presumptive OE and VNO as early as E12.5. The intensity of the signal increased by E14.5, and by E17.5 (the latest time point analyzed) robust staining was detected in the olfactory nerve entering the developing olfactory bulb. In mouse, most of the GnRH-1 neurons migrate from nasal regions into the telencephalon between E12.5 and E14.5 (Wray et al., 1989). From E14.5 to E17.5, axodendritic synapses start to form on the dendritic growth cones of mitral cells (Brunjes and Frazier, 1986). Thus, the spatiotemporal expression of CCK in nasal sensory cells correlates with the maturation of olfactory systems and migration of GnRH-1 neurons into the CNS.
To determine whether CCK in developing nasal regions could be functional, expression of CCK receptors was evaluated. CCK exerts various pharmacological effects via two G-protein-coupled receptors, CCK-1R and CCK-2R (for review, see Noble et al., 1999). CCK-1R is localized mainly in the gastrointestinal tract but also found in discrete brain regions (Moran et al., 1986; Mercer and Beart, 1997; Mercer et al., 1999). CCK-2R is widely distributed throughout the CNS (Moran et al., 1986; Mercer et al., 2000). In taste receptor cells, CCK-1R has been demonstrated to mediate the effect of CCK in the physiology of gustatory perception (Herness et al., 2002). In olfactory fibers, CCK-1R and CCK-2R receptors were both expressed from E12.5 to E17.5. Thus, olfactory axons coexpressed both ligand and receptors through prenatal development. In addition, we found that migrating GnRH-1 neuroendocrine cells located in the nasal compartment expressed CCK-1R but not CCK-2R. Thus, release of CCK in nasal regions during prenatal development could regulate olfactory sensory cells (olfactory receptor neurons as well as pheromone receptor neurons) and GnRH-1 neurons.
To address the role of CCK in olfactory–GnRH-1 system development, we turned to an in vitro model: nasal explants. Nasal explants maintain a large number of GnRH-1 neurons, migrating in a similar manner to GnRH-1 cells in vivo, as well as directed olfactory axon outgrowth (Fueshko and Wray, 1994). In addition, these explants are maintained in serum-free defined media, an important factor when conducting perturbation studies. Before using this model for functional studies, we characterized expression of CCK and its receptors. Expression of both CCK and CCK-receptors in olfactory–vomeronasal cells was mimicked in nasal explants. Primary GnRH-1 neurons expressed both CCK-1R transcript and protein. Interestingly, this expression pattern correlated with migration of these cells, being downregulated after 7 DIV, when GnRH-1 neurons stop migrating in vitro. Although CCK-2R mRNA expression in GnRH-1 cells was detected by RT-PCR on single cells, GnRH-1 neurons did not express CCK-2R protein, consistent with in vivo results.
Functional analysis
Exogenous application of CCK to nasal explants at 3 DIV reduced both olfactory axon outgrowth and the front of migration of GnRH-1 cells. Coapplication of CCK together with specific receptor antagonists identified CCK-1R receptor as the mediator of CCK actions, e.g., CCK effects were reversed when CCK was applied with the CCK-1R but not with the CCK-2R antagonist. Exposure to exogenous CCK-1R or CCK-2R antagonists alone did not alter olfactory axon outgrowth. Certainly we cannot rule out the possibility that the endogenous amount of CCK released in our explants is insufficient to alter olfactory axon outgrowth, but it may do so in vivo. CCK-1R antagonism did cause a shift in the location of GnRH-1 neurons, however, increasing the distance migrated by these cells. These data indicate that the main action of endogenous CCK is on cell movement and also suggests that an endogenous source of CCK, likely present in the nasal pits, is functional in the explants at the time of treatment (3 DIV).
Little is known about the signals that stimulate axon formation and outgrowth from olfactory sensory neurons (St. John et al., 2000). When reaching the rostral telencephalon, the olfactory sensory axons do not immediately enter the telencephalon; rather the axons defasciculate and branch at the boundaries between the nasal compartment and the developing olfactory bulb (Whitesides and LaMantia, 1996). This site is where GnRH-1 neurons also pause before entrance into the CNS (Fueshko et al., 1998; Mulrenin et al., 1999; Bless et al., 2000). The crossover from nasal region to CNS is a dramatic change in environment, and it has been suggested that such a pause may ensure appropriate maturation of GnRH-1 neurons, allowing them to respond to new molecular cues (for review, see Wray, 2002). To date, several molecules have been shown to alter the GnRH-1 migratory process, including GABAergic, serotoninergic, and glutamatergic signals (Fueshko et al., 1998; Bless et al., 2000; Simonian and Herbison, 2001; Heger et al., 2003; Pronina et al., 2003a).
The spatiotemporal expression of CCK and its receptors, together with our functional data, is consistent with CCK acting as an intercellular signal regulating the timing of entry into the brain of GnRH-1 cells and possibly olfactory axons. Previous studies have highlighted a role of CCK in cell migration and demonstrated that CCK has the ability to modify the migratory properties of other cell types, including human tumor astrocytes (De Hauwer et al., 1998; Lefranc et al., 2002) and lymphocytes (Medina et al., 1998), in the latter case, inhibiting the chemotaxis of those cells (Medina et al., 1998). To further investigate this hypothesis, analysis of GnRH-1 migration and olfactory axon outgrowth were examined in mice carrying a deletion of CCK-1R or CCK-2R. CCK-1R- and CCK-2R-deficient mice are both viable and fertile (Langhans et al., 1997; Kopin et al., 1999); however, we found that in CCK-1R–/– E14.5 mice, the number of GnRH-1 neurons that had entered the brain was significantly greater than that in WT and CCK-2R–/– mice of the same age. A concomitant reduction in GnRH-1 neurons in nasal regions was observed. These data are consistent with accelerated GnRH-1 neuronal migration into the forebrain and an earlier onset of the migratory process when the CCK signal transduction pathway via the CCK-1R receptor is abolished, supporting an inhibitory role for endogenous CCK in GnRH-1 cell migration. In postnatal and adult CCK-1R mutant mice, the number as well as the distribution of GnRH-1 neurons in brain appeared normal. These data suggest that although GnRH-1 cells entered the CNS earlier, compensatory cues established an appropriate distribution and maintained cell number in the mutant animals. The normal appearance of the GnRH-1 system in postnatal CCK-1R mutants is consistent with fertility in these mice. In accord with the in vitro perturbation studies, no major disruption of olfactory fibers was observed in the CCK-1R–/– mice. In this system, redundant mechanisms through the CCK-2R may occur developmentally.
To date, CCK in brain has been implicated in various functions postnatally (Crawley and Corwin, 1994; Beinfeld, 2001), but a developmental function for CCK has not been examined. In this report we documented a role for CCK in the developing GnRH-1 system and identified CCK-1R receptor as the mediator of the CCK effect. The in vivo and in vitro data presented here indicate that CCK can act prenatally to inhibit migration of GnRH-1 neurons from nose to brain, an essential event for reproductive competence.
Footnotes
This work was supported in part by Compagnia di San Paolo (P.G.) and National Institute of Diabetes and Digestive and Kidney Diseases Grant DK46767 (A.S.K.).P.M.B. and L.D.M. were supported by the National Health and Medical Research Council (Australia) and in part by a program grant. We thank Andree Reuss for technical assistance with these experiments and Kartik Pattabiraman for help in analyzing CCK-2R KO mice.
Correspondence should be addressed to Dr. Susan Wray, Cellular and Developmental Neurobiology Section, National Institute of Neurological Disorders and Stroke, National Institutes of Health, Building 36, Room 5A21, 36 Convent Drive–MSC 4156, Bethesda, MD 20892-4156. E-mail: wrays@ninds.nih.gov.
DOI:10.1523/JNEUROSCI.0649-04.2004
Copyright © 2004 Society for Neuroscience 0270-6474/04/244737-12$15.00/0
References
- Atkinson ME, Shehab SA (1986) Peripheral axotomy of the rat mandibular trigeminal nerve leads to an increase in VIP and decrease of other primary afferent neuropeptides in the spinal trigeminal nucleus. Regul Pept 16: 69–81. [DOI] [PubMed] [Google Scholar]
- Beinfeld MC (1983) Cholecystokinin in the central nervous system: a minireview. Neuropeptides 3: 411–427. [DOI] [PubMed] [Google Scholar]
- Beinfeld MC (2001) An introduction to neuronal cholecystokinin. Peptides 22: 1197–1200. [DOI] [PubMed] [Google Scholar]
- Bless EP, Westaway WA, Schwarting GA, Tobet SA (2000) Effects of gamma-aminobutyric acid(A) receptor manipulation on migrating gonadotropin-releasing hormone neurons through the entire migratory route in vivo and in vitro. Endocrinology 141: 1254–1262. [DOI] [PubMed] [Google Scholar]
- Bock MG, DiPardo RM, Evans BE, Rittle KE, Whitter WL, Veber DF, Anderson PF, Freidinger RM (1989) Benzodiazepin gastrin and brain cholecystokinin receptor ligands: L-365, 260. J Med Chem 32: 13–16. [DOI] [PubMed] [Google Scholar]
- Brunjes PC, Frazier LL (1986) Maturation and plasticity in the olfactory system of vertebrates. Brain Res 396: 1–45. [DOI] [PubMed] [Google Scholar]
- Calof AL, Chikaraishi DM (1989) Analysis of neurogenesis in a mammalian neuroepithelium: proliferation and differentiation of an olfactory neuron precursor in vitro. Neuron 3: 115–127. [DOI] [PubMed] [Google Scholar]
- Cho HJ, Shiotani Y, Shiosaka S, Inagaki S, Kubota Y, Kiyama H, Umegaki K, Tateishi K, Hashimura E, Hamaoka T, Tohyama M (1983) Ontogeny of cholecystokinin-8-containing neuron system of the rat: an immunohistochemical analysis. I. Forebrain and upper brainstem. J Comp Neurol 218: 25–41. [DOI] [PubMed] [Google Scholar]
- Crawley JN, Corwin RL (1994) Biological actions of cholecystokinin. Peptides 15: 731–755. [DOI] [PubMed] [Google Scholar]
- De Hauwer C, Camby I, Darro F, Migeotte I, Decaestecker C, Verbeek C, Danguy A, Pasteels JL, Brotchi J, Salmon I, Van Ham P, Kiss R (1998) Gastrin inhibits motility, decreases cell death levels and increases proliferation in human glioblastoma cell lines. J Neurobiol 37: 373–382. [DOI] [PubMed] [Google Scholar]
- Fueshko S, Wray S (1994) LHRH cells migrate on peripherin fibers in embryonic olfactory explant cultures: an in vitro model for neurophilic neuronal migration. Dev Biol 166: 331–348. [DOI] [PubMed] [Google Scholar]
- Fueshko SM, Key S, Wray S (1998) GABA inhibits migration of luteinizing hormone-releasing hormone neurons in embryonic olfactory explants. J Neurosci 18: 2560–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C, Noden DM, Norgren RB Jr (2000) LHRH neuronal migration: heterotypic transplantation analysis of guidance cues. J Neurobiol 42: 95–103. [PubMed] [Google Scholar]
- Gavazzi I (2001) Semaphorin-neuropilin-1 interactions in plasticity and regeneration of adult neurons. Cell Tissue Res 305: 275–284. [DOI] [PubMed] [Google Scholar]
- Ghijsen WE, Leenders AG, Wiegant VM (2001) Regulation of cholecystokinin release from central nerve terminals. Peptides 22: 1213–1221. [DOI] [PubMed] [Google Scholar]
- Gong Q, Shipley MT (1996) Expression of extracellular matrix molecules and cell surface molecules in the olfactory nerve pathway during early development. J Comp Neurol 366: 1–14. [DOI] [PubMed] [Google Scholar]
- Grothe C, Unsicker K (1987) Neuron-enriched cultures of adult rat dorsal root ganglia: establishment, characterization, survival, and neuropeptide expression in response to trophic factors. J Neurosci Res 18: 539–550. [DOI] [PubMed] [Google Scholar]
- Hansel DE, May V, Eipper BA, Ronnett GV (2001) Pituitary adenylyl cyclase-activating peptides and α-amidation in olfactory neurogenesis and neuronal survival in vitro J Neurosci 21: 4625–4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heger S, Seney M, Bless E, Schwarting GA, Bilger M, Mungenast A, Ojeda SR, Tobet SA (2003) Overexpression of glutamic acid decarboxylase-67 (GAD-67) in gonadotropin-releasing hormone neurons disrupts migratory fate and female reproductive function in mice. Endocrinology 144: 2566–2579. [DOI] [PubMed] [Google Scholar]
- Herness S, Zhao FL, Lu SG, Kaya N, Shen T (2002) Expression and physiological actions of cholecystokinin in rat taste receptor cells. J Neurosci 22: 10018–10029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilal EM, Chen JH, Silverman AJ (1996) Joint migration of gonadotropin-releasing hormone (GnRH) and neuropeptide Y (NPY) neurons from olfactory placode to central nervous system. J Neurobiol 31: 487–502. [DOI] [PubMed] [Google Scholar]
- Hokfelt T, Herrera-Marschitz M, Seroogy K, Ju G, Staines WA, Holets V, Schalling M, Ungerstedt U, Post C, Rehfeld JF (1988) Immunohistochemical studies on cholecystokinin (CCK)-immunoreactive neurons in the rat using sequence specific antisera and with special reference to the caudate nucleus and primary sensory neurons. J Chem Neuroanat 1: 11–51. [PubMed] [Google Scholar]
- Hokfelt T, Cortes R, Schalling M, Ceccatelli S, Pelto-Huikko M, Persson H, Villar MJ (1991) Distribution patterns of CCK and CCK mRNA in some neuronal and non-neuronal tissues. Neuropeptides 19[Suppl]: 31–43. [DOI] [PubMed] [Google Scholar]
- Ingram SM, Krause II RG Baldino Jr F, Skeen LC, Lewis ME (1989) Neuronal localization of cholecystokinin mRNA in the rat brain by using in situ hybridization histochemistry. J Comp Neurol 287: 260–272. [DOI] [PubMed] [Google Scholar]
- Itoh Y, Kozakai I, Toyomizu M, Ishibashi T, Kuwano R (1998) Mapping of cholecystokinin transcription in transgenic mouse brain using Escherichia coli beta-galactosidase reporter gene. Dev Growth Differ 40: 395–402. [DOI] [PubMed] [Google Scholar]
- Ivy AC, Oldberg E (1928) A hormone mechanism for gallbladder contraction and evacuation. Am J Physiol 86: 599–613. [Google Scholar]
- Key S, Wray S (2000) Two olfactory placode derived galanin subpopulations: leutinizing hormone releasing hormone (LHRH) neurons and vomeronasal cells. J Neuroendocrinol 12: 535–545. [DOI] [PubMed] [Google Scholar]
- Kopin AS, Mathes WF, McBride EW, Nguyen M, Al-Haider W, Schmitz F, Bonner-Weir S, Kanarek R, Beinborn M (1999) The cholecystokinin-A receptor mediates inhibition of food intake yet is not essential for the maintenance of body weight. J Clin Invest 103: 383–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korchak DM, Nilaver G, Beinfeld MC (1984) The development of motilin-like immunoreactivity in the rat cerebellum and pituitary as determined by radioimmunoassay. Neurosci Lett 48: 267–272. [DOI] [PubMed] [Google Scholar]
- Kramer PR, Wray S (2000) Novel gene expressed in nasal region influences outgrowth of olfactory axons and migration of luteinizing hormone-releasing hormone (LHRH) neurons. Genes Dev 14: 1824–1834. [PMC free article] [PubMed] [Google Scholar]
- Kramer PR, Krishnamurthy R, Mitchell PJ, Wray S (2000) Transcription factor activator protein-2 is required for continued luteinizing hormone-releasing hormone expression in the forebrain of developing mice. Endocrinology 141: 1823–1838. [DOI] [PubMed] [Google Scholar]
- Kuljis RO, Krause JE, Karten HJ (1984) Peptide-like immunoreactivity in anuran optic nerve fibers. J Comp Neurol 226: 222–237. [DOI] [PubMed] [Google Scholar]
- Langhans N, Rindi G, Chiu M, Rehfeld JF, Ardman B, Beinborn M, Kopin AS (1997) Abnormal gastric histology and decreased acid production in cholecystokinin-B/gastrin receptor-deficient mice. Gastroenterology 112: 280–286. [DOI] [PubMed] [Google Scholar]
- Lefranc F, Camby I, Belot N, Bruyneel E, Chaboteaux C, Brotchi J, Mareel M, Salmon I, Kiss R (2002) Gastrin significantly modifies the migratory abilities of experimental glioma cells. Lab Invest 82: 1241–1252. [DOI] [PubMed] [Google Scholar]
- Liddle RA (1997) Cholecystokinin cells. Annu Rev Physiol 59: 221–242. [DOI] [PubMed] [Google Scholar]
- Liu WL, Shipley MT (1994) Intrabulbar associational system in the rat olfactory bulb comprises cholecystokinin-containing tufted cells that synapse onto the dendrites of GABAergic granule cells. J Comp Neurol 346: 541–558. [DOI] [PubMed] [Google Scholar]
- MacColl G, Quinton R, Bouloux PM (2002) GnRH neuronal development: insights into hypogonadotrophic hypogonadism. Trends Endocrinol Metab 13: 112–118. [DOI] [PubMed] [Google Scholar]
- Makovec F, Bani M, Cereda R, Chiste R, Pacini MA, Revel L, Rovati LA, Rovati LC, Setnikar I (1987) Pharmacological properties of lorglumide as a member of a new class of cholecystokinin antagonists. Arzneimittelforschung 37: 1265–1268. [PubMed] [Google Scholar]
- Matsuda H, Tsukuda M, Kadota T, Kusunoki T, Kishida R (1994) Coexistence of galanin and substance P in the mouse nasal mucosa, including the vomeronasal organ. Neurosci Lett 173: 55–58. [DOI] [PubMed] [Google Scholar]
- Medina S, Del Rio M, Manuel Victor V, Hernanz A, De la Fuente M (1998) Changes with ageing in the modulation of murine lymphocyte chemotaxis by CCK-8S, GRP and NPY. Mech Ageing Dev 102: 249–261. [DOI] [PubMed] [Google Scholar]
- Mercer LD, Beart PM (1997) Histochemistry in rat brain and spinal cord with an antibody directed at the cholecystokininA receptor. Neurosci Lett 225: 97–100. [DOI] [PubMed] [Google Scholar]
- Mercer LD, Beart PM (2004) Immunolocalization of CCK1R in rat brain using a new anti-peptide antibody. Neurosci Lett 359: 109–113. [DOI] [PubMed] [Google Scholar]
- Mercer LD, Nunan J, Jones NM, Lawrence AJ, Cheung NS, Mercer JFB, Firth SD, Thompson P, Jarrott B, Finkelstein DI, Beart PM (1999) Immunohistochemistry with antibodies to the cholecystokinin-A receptor reveals a widespread distribution in rat and monkey central nervous system. In: Peptidergic G protein-coupled receptors from basic research to clinical applications (Geppetti P, ed), pp 175–180. Amsterdam: IOS.
- Mercer LD, Le VQ, Nunan J, Jones NM, Beart PM (2000) Direct visualization of cholecystokinin subtype2 receptors in rat central nervous system using anti-peptide antibodies. Neurosci Lett 293: 167–170. [DOI] [PubMed] [Google Scholar]
- Miceli MO, van der Kooy D, Post CA, Della-Fera MA, Baile CA (1987) Differential distributions of cholecystokinin in hamster and rat forebrain. Brain Res 402: 318–330. [DOI] [PubMed] [Google Scholar]
- Miragall F, Kadmon G, Schachner M (1989) Expression of L1 and N-CAM cell adhesion molecules during development of the mouse olfactory system. Dev Biol 135: 272–286. [DOI] [PubMed] [Google Scholar]
- Moran TH, Robinson PH, Goldrich MS, McHugh PR (1986) Two brain cholecystokinin receptors: implications for behavioral actions. Brain Res 362: 175–179. [DOI] [PubMed] [Google Scholar]
- Mulrenin EM, Witkin JW, Silverman AJ (1999) Embryonic development of the gonadotropin-releasing hormone (GnRH) system in the chick: a spatio-temporal analysis of GnRH neuronal generation, site of origin, and migration. Endocrinology 140: 422–433. [DOI] [PubMed] [Google Scholar]
- Murakami S, Arai Y (1994) Transient expression of somatostatin immunoreactivity in the olfactory-forebrain region in the chick embryo. Brain Res Dev Brain Res 82: 277–285. [DOI] [PubMed] [Google Scholar]
- Murakami S, Seki T, Rutishauser U, Arai Y (2000) Enzymatic removal of polysialic acid from neural cell adhesion molecule perturbs the migration route of luteinizing hormone-releasing hormone neurons in the developing chick forebrain. J Comp Neurol 420: 171–181. [DOI] [PubMed] [Google Scholar]
- Noble F, Wank SA, Crawley JN, Bradwejn J, Seroogy KB, Hamon M, Roques BP (1999) International Union of Pharmacology. XXI. Structure, distribution, and functions of cholecystokinin receptors. Pharmacol Rev 51: 745–781. [PubMed] [Google Scholar]
- Pronina T, Ugrumov M, Adamskaya E, Kuznetsova T, Shishkina I, Babichev V, Calas A, Tramu G, Mailly P, Makarenko I (2003a) Influence of serotonin on the development and migration of gonadotropin-releasing hormone neurones in rat foetuses. J Neuroendocrinol 15: 549–558. [DOI] [PubMed] [Google Scholar]
- Pronina T, Ugrumov M, Calas A, Seif I, Tramu G (2003b) Influence of monoamines on differentiating gonadotropin-releasing hormone neurones in foetal mice. J Neuroendocrinol 15: 925–932. [DOI] [PubMed] [Google Scholar]
- Rugarli EI (1999) Kallmann syndrome and the link between olfactory and reproductive development. Am J Hum Genet 65: 943–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savasta M, Palacios JM, Mengod G (1988) Regional localization of the mRNA coding for the neuropeptide cholecystokinin in the rat brain studied by in situ hybridization. Neurosci Lett 93: 132–138. [DOI] [PubMed] [Google Scholar]
- Schiffmann SN, Vanderhaeghen JJ (1991) Distribution of cells containing mRNA encoding cholecystokinin in the rat central nervous system. J Comp Neurol 304: 219–233. [DOI] [PubMed] [Google Scholar]
- Schwanzel-Fukuda M, Pfaff DW (1989) Origin of luteinizing hormone-releasing hormone neurons. Nature 338: 161–164. [DOI] [PubMed] [Google Scholar]
- Schwanzel-Fukuda M, Abraham S, Crossin KL, Edelman GM, Pfaff DW (1992) Immunocytochemical demonstration of neural cell adhesion molecule (NCAM) along the migration route of luteinizing hormone-releasing hormone (LHRH) neurons in mice. J Comp Neurol 321: 1–18. [DOI] [PubMed] [Google Scholar]
- Sharifi N, Reuss AE, Wray S (2002) Prenatal LHRH neurons in nasal explant cultures express estrogen receptor beta transcript. Endocrinology 143: 2503–2507. [DOI] [PubMed] [Google Scholar]
- Shu T, Valentino KM, Seaman C, Cooper HM, Richards LJ (2000) Expression of the netrin-1 receptor, deleted in colorectal cancer (DCC), is largely confined to projecting neurons in the developing forebrain. J Comp Neurol 416: 201–212. [DOI] [PubMed] [Google Scholar]
- Simonian SX, Herbison AE (2001) Differing, spatially restricted roles of ionotropic glutamate receptors in regulating the migration of GnRH neurons during embryogenesis. J Neurosci 21: 934–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St. John JA, Key B (2001) EphB2 and two of its ligands have dynamic protein expression patterns in the developing olfactory system. Brain Res Dev Brain Res 126: 43–56. [DOI] [PubMed] [Google Scholar]
- St. John JA, Tisay KT, Caras IW, Key B (2000) Expression of EphA5 during development of the olfactory nerve pathway in rat. J Comp Neurol 416: 540–550. [DOI] [PubMed] [Google Scholar]
- Toba Y, Horie M, Sango K, Tokashiki A, Matsui F, Oohira A, Kawano H (2002) Expression and immunohistochemical localization of heparan sulphate proteoglycan N-syndecan in the migratory pathway from the rat olfactory placode. Eur J Neurosci 15: 1461–1473. [DOI] [PubMed] [Google Scholar]
- Treloar H, Tomasiewicz H, Magnuson T, Key B (1997) The central pathway of primary olfactory axons is abnormal in mice lacking the N-CAM-180 isoform. J Neurobiol 32: 643–658. [DOI] [PubMed] [Google Scholar]
- Vanderhaeghen JJ, Signeau JC, Gepts W (1975) New peptide in vertebrate CNS reacting with antigastrin antibodies. Nature 257: 601–605. [DOI] [PubMed] [Google Scholar]
- Verney C, el Amraoui A, Zecevic N (1996) Comigration of tyrosine hydroxylase- and gonadotropin-releasing hormone-immunoreactive neurons in the nasal area of human embryos. Brain Res Dev Brain Res 97: 251–259. [DOI] [PubMed] [Google Scholar]
- Walz A, Rodriguez I, Mombaerts P (2002) Aberrant sensory innervation of the olfactory bulb in neuropilin-2 mutant mice. J Neurosci 22: 4025–4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SS, Tsai RY, Reed RR (1997) The characterization of the Olf-1/EBF-like HLH transcription factor family: implications in olfactory gene regulation and neuronal development. J Neurosci 17: 4149–4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wank SA (1995) Cholecystokinin receptors. Am J Physiol 269: G628–G646. [DOI] [PubMed] [Google Scholar]
- Whitesides III JG, LaMantia AS (1996) Differential adhesion and the initial assembly of the mammalian olfactory nerve. J Comp Neurol 373: 240–254. [DOI] [PubMed] [Google Scholar]
- Wray S (2002) Development of gonadotropin-releasing hormone-1 neurons. Front Neuroendocrinol 23: 292–316. [DOI] [PubMed] [Google Scholar]
- Wray S, Gahwiler BH, Gainer H (1988) Slice cultures of LHRH neurons in the presence and absence of brainstem and pituitary. Peptides 9: 1151–1175. [DOI] [PubMed] [Google Scholar]
- Wray S, Grant P, Gainer H (1989) Evidence that cells expressing luteinizing hormone-releasing hormone mRNA in the mouse are derived from progenitor cells in the olfactory placode. Proc Natl Acad Sci USA 86: 8132–8136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray S, Key S, Qualls R, Fueshko SM (1994) A subset of peripherin positive olfactory axons delineates the luteinizing hormone releasing hormone neuronal migratory pathway in developing mouse. Dev Biol 166: 349–354. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Tobet SA, Crandall JE, Jimenez TP, Schwarting GA (1995) The migration of luteinizing hormone-releasing hormone neurons in the developing rat is associated with a transient, caudal projection of the vomeronasal nerve. J Neurosci 15: 7769–7777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihara Y, Kawasaki M, Tamada A, Fujita H, Hayashi H, Kagamiyama H, Mori K (1997) OCAM: a new member of the neural cell adhesion molecule family related to zone-to-zone projection of olfactory and vomeronasal axons. J Neurosci 17: 5830–5842. [DOI] [PMC free article] [PubMed] [Google Scholar]





