Abstract
Fragile X syndrome is caused by the absence of the mRNA-binding protein Fragile X mental retardation protein (FMRP), which may play a role in activity-regulated localization and translation of mRNA in dendrites and at synapses. We investigated whether neuronal activity and glutamatergic signals regulate trafficking of FMRP and its encoding Fmr1 mRNA into dendrites or at synapses. Using high-resolution fluorescence and digital imaging microscopy in cultured hippocampal neurons, FMRP and Fmr1 mRNA were localized in granules throughout dendrites and within spines. KCl depolarization rapidly increased FMRP and Fmr1 mRNA levels in dendrites. Metabotropic glutamate receptor (mGluR) activation, in particular mGluR5 activation, was necessary for localization of FMRP into dendrites. Blockade of either PKC or internal calcium prevented mGluR-dependent localization of both FMRP and Fmr1 mRNA in dendrites. The activity-dependent localization of FMRP was not dependent on protein synthesis. Fluorescence recovery after photobleaching analysis of live neurons transfected with enhanced green fluorescent protein–FMRP revealed increased granule trafficking in response to KCl depolarization. In contrast to its dendritic localization, mGluR activation diminished FMRP, but not Fmr1 mRNA, localization at synapses. These results demonstrate regulation of FMRP and Fmr1 mRNA trafficking in dendrites and synapses in response to specific glutamatergic signals.
Keywords: FMRP, Fmr1 mRNA, mRNA localization, synaptic plasticity, mGluR, dendritic spine, synapse
Introduction
Fragile X syndrome (FXS), the most common inherited form of mental retardation, is caused by the blocked expression of Fragile X mental retardation protein (FMRP), an mRNA-binding protein believed to play a role in the regulation of local protein synthesis and possibly mRNA localization in dendrites (for review, see Antar and Bassell, 2003). FMRP target mRNAs identified by microarray analysis encode proteins important for neuronal development and plasticity (Brown et al., 2001; Miyashiro et al., 2003). FMRP-associated mRNAs display altered translational profiles in human FX cells and brain fractions from Fmr1 knockout (KO) mice, which do not express FMRP (Brown et al., 2001; Zalfa et al., 2003). These studies suggest diverse functions for FMRP in dendrites, including mRNA localization (Miyashiro et al., 2003) and translation (Zalfa et al., 2003).
The FX phenotype includes defects in synaptic structure and plasticity. FX-affected individuals and FMR1 KO mice display defects in dendritic spine morphology characterized by long, thin, and overabundant spines (Irwin et al., 2000a), suggesting that impaired regulation of mRNA localization and translation in FXS may affect spine maturation or synaptic pruning. Spine structure is linked to long-term synaptic plasticity (Bonhoeffer and Yuste, 2002), and increased FMRP expression in response to activity has been observed in synaptosomes after exposure to glutamate receptor (GluR) agonists (Weiler et al., 1997) or whisker-pad stimulation in vivo (Todd and Mack, 2000). Hippocampal levels of FMRP increase in animals reared in a stimulating environment (Irwin et al., 2000b). Also, the FMR1 KO exhibits enhanced metabotropic GluR (mGluR)-dependent hippocampal long-term depression (LTD) (Huber et al., 2002; Snyder et al., 2001). Because this specific form of LTD is known to involve postsynaptic protein synthesis (Huber et al., 2000), these studies suggested that FMRP might be regulated by an mGluR signaling pathway. Collectively, these studies demonstrate activity-dependent regulation of FMRP expression and point to impaired long-term synaptic plasticity resulting from the absence of FMRP. Such defects could result from impaired delivery of FMRP ribonucleoprotein granules (mRNPs) into dendrites or deficits in their synaptic translation.
Although the cellular expression of FMRP is known to be regulated by activity (Weiler et al., 1997; Todd and Mack, 2000), it is unknown whether activity modulates trafficking of FMRP and its cargo mRNAs in dendrites. The regulated trafficking of FMRP mRNPs may provide an important means for influencing local protein synthesis and synaptic plasticity. The regulation of FMRP localization in neurons has not been studied previously. We used high-resolution fluorescence and digital imaging microscopy in cultured hippocampal neurons to analyze the dendritic and synaptic localization of FMRP and its encoding Fmr1 mRNA in response to specific glutamatergic pathways. These studies identify the role of an mGluR signaling pathway in the localization of FMRP and Fmr1 mRNA in dendrites. In contrast to its dendritic localization, mGluR activation diminished FMRP but not Fmr1 mRNA localization at synapses. Such observations suggest that defects in synaptic plasticity in FXS may be caused partly by impaired trafficking of FMRP–mRNA complexes in response to distinct glutamatergic signaling pathways.
Materials and Methods
Hippocampal culture and drug treatments. Embryonic day 18 (E18) rat hippocampi were cultured as described (Goslin and Banker, 1998). Cells were plated (90,000–120,000 cells per square centimeter) on poly-l-lysine-coated coverslips (1.0 mg/ml) in minimal essential medium (MEM) with FBS (10%) for 2 hr, inverted onto dishes containing astroglia, and grown in defined N2-conditioned medium (Goslin and Banker, 1998). Cells were fixed with 4% paraformaldehyde in 1× PBS at room temperature for 18 min or treated at various time points before fixation with 30 mm KCl (10 min, except where indicated), 50 μm APV (5 or 15 min; Tocris, Ellisville, MO), 100 μm CNQX (15 min; Tocris), or 1 mm (S)-α-methyl-4 carboxyphenylglycine (MCPG) (30 min; Tocris) added to KCl-containing media. Additionally, 10 μm 2-methyl-6-(phenylethynyl) pyridine hydrochloride (MPED) (Tocris) and 5 μm (S)-(+)-α-amino-4-carboxy-2-methylbenzene acetic acid (LY)-367385 (Tocris) were added to KCl along with APV and CNQX. Other experimental sets were conducted with 50–100 μm (S)-3,5-dihydroxyphenylglycine (DHPG) (5 min; Tocris; with 50 μm APV, 100 μm CNQX) or 50 μm AMPA (15 min, with APV). Matching experiments included the PKC inhibitor RO-32-0432 (1 μm; 30 min; Calbiochem, San Diego, CA) or BAPTA-AM (25 μm; 15 min; Sigma, St. Louis, MO), both in the presence of APV and CNQX. Finally, 1 mg/μl cycloheximide (30 min; Sigma) was used in the presence of APV and CNQX.
Immunofluorescence. Immunofluorescence (IF) was performed as described (Tiruchinapalli et al., 2003). For primary antibody incubations (1 hr, room temperature), FMRP was detected with mouse antibody (1C3, 1:1000; Chemicon, Temecula, CA) and synapsin with rabbit antibody (1:500; Sigma). For secondary antibody incubations (1 hr, room temperature), Cy3 and Cy5 fluorochrome-conjugated anti-mouse and rabbit antibodies were used (1:1500 and 1:750, respectively; Jackson ImmunoResearch, West Grove, PA). Alexa 488-conjugated phalloidin detected F-actin (1:500; Molecular Probes, Eugene, OR).
Fluorescence in situ hybridization with digoxigenin-labeled probes. Probes were designed to the coding region of mouse Fmr1 mRNA using DNAsis and Oligo 4 and checked for homology to other mRNAs by BLAST (basic local alignment search tool) search. Amino-modified oligonucleotides were synthesized on a DNA synthesizer and labeled with a digoxigenin succinimide ester (Roche Molecular Biochemicals, Indianapolis, IN) as described previously (Bassell et al., 1998).
Fixed cells were subjected to fluorescence in situ hybridization (FISH) with the digoxigenin-labeled Fmr1 probes [method described as for β-actin probes (Bassell et al., 1998)]. Hybridized probes were detected by IF using a Cy3-conjugated mouse anti-digoxigenin antibody (1:1500) and a Cy3-conjugated anti-mouse IgG antibody (Jackson ImmunoResearch) blocked in bovine serum albumin. Alternatively, digoxigenin-labeled probes were detected using sheep anti-digoxigenen antibody (1: 10; Roche Molecular Biochemicals) and a Cy2-conjugated anti-sheep antibody (1:500; Roche Molecular Biochemicals) blocked in normal donkey serum.
Data analysis, digital imaging, and image reconstruction. IF images were visualized using a 60× or 40× Plan-Neofluar objective, 100 W mercury arc lamp, and HiQ bandpass filters (Chroma Tech) on a Nikon Eclipse inverted microscope. Images were captured on a cooled CCD camera (Quantix, Photometrics) using IP Lab software (Scanalytics). For indicated experiments, Z-stacks were acquired (11 sections at 0.2 μm each) and deconvolved (Power Microtome). Volume rendering and three-dimensional (3-D) reconstruction were performed using Imaris software (Bitplane, Inc.).
Mean IF intensity for FMRP or Fmr1 mRNA was analyzed within a defined region of interest (ROI) traced along a dendrite, selected 10 μm from the cell body for proximal dendrites, or from secondary dendrites (where noted). Total IF intensity of the ROI was then divided by area of the ROI to normalize for differences in dendritic area. Each experiment was repeated a minimum of three times, imaging at least 15 cells, three dendrites per cell per experiment, using a paired Student's t test in all cases except with analysis of the mGluR blockers MPEP and LY (see above), which were analyzed with an unpaired Student's t test.
The percentage of synapsin puncta that contained FMRP, Fmr1 mRNA, or FMRP and Fmr1 mRNA-positive synapses were analyzed. Spines were defined as dendritic protrusions associated with synapsin puncta; filopodia were defined as protrusions lacking synapsin puncta. Quantification was performed on images of equal exposure threshold values. At least 45 dendrites, 60–120 synapses per 20 μm per dendrite, were analyzed for each variable.
Enhanced green fluorescent protein–FMRP transfection. Enhanced green fluorescent protein (pEGFP)–FMRP was generated by inserting PCR products of the open reading frame of murine FMRP into the SacI and EcoRI sites of the pEGFP-C1 vector containing a cytomegalovirus promoter (BD Biosciences, San Diego, CA). Cultured neurons were transfected using CaPO4 as described (Kohrman et al., 1999), and EGFP–FMRP was expressed overnight and then processed as above.
Fluorescence recovery after photobleaching. Rat hippocampal neurons (E18) were cultured on 40 mm coverslips (Bioptechs, Butler, PA) and transfected with EGFP–FMRP. Transfected cells were imaged within 12 hr of transfection in L15 medium (Invitrogen, Grand Island, NY) supplemented with 20 mm HEPES, pH 7.2, FBS (0.5%), and N2 supplements, with or without 20 mm KCl. Images were captured on an Olympus Fluoview 500 confocal microscope using an argon blue (488 nm) laser. Capture and analysis have been described previously (Zhang et al., 2001) with these modifications: each fluorescence recovery after photobleaching (FRAP) time point (1–5 min) reflects the timed average fluorescence intensity of three sequential frames (10 sec apart) to minimize noise. For the average graph of all cells measured (see Fig. 3E), the recovery values at each indicated time point (1–5 min) were averaged for all FRAP analyses in a treatment, pooling all cells treated for <1 hr. These values represent experiments from 13 unstimulated cells (25 different neurites) and 10 stimulated cells (19 different neurites).
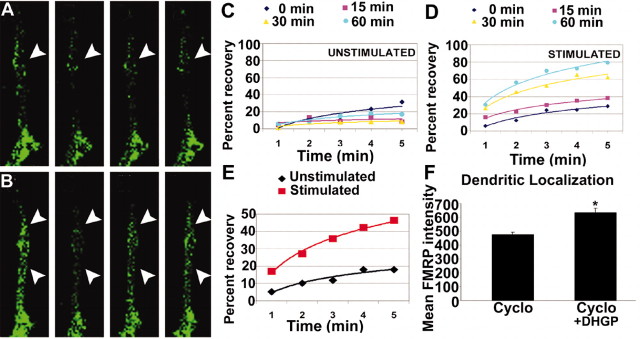
Figure 3.
A–E, FRAP analysis of increased EGFP–FMRP granule trafficking in response to KCl stimulation. Hippocampal neurons were transfected with EGFP–FMRP and imaged with either no added stimulus(A, C) or 20 mm KCl (B, D). The recovery of each photobleached region was followed for 5 min at 1 min intervals. A, The first panel (from left) shows the EGFP–FMRP signal in an unstimulated neurite before photobleach, and the second panel shows immediately after photobleach. Subsequent panels show signal at 3 min (3rd panel) and 5 min (4th panel) after photobleach. Arrowheads (A, B) point to regions that exemplify the reduced recovery of EGFP–FMRP granules in the absence of stimulation (A) compared with those in the same cell treated with 20 mm KCl (B). Quantitative analysis of recovery of the EGFP–FMRP signal in this cell is shown at three time points (15, 30, 60 min) after perfusion with normal media as a control (C) or media with KCl (D) treatment. A summary of all FRAP analyses (E) compares the percentage recovery of EGFP–FMRP in neurites of several cells measured for each treatment within 1 hr of stimulation or mock treatment. F, Dendritic localization of FMRP was not protein synthesis dependent. Bath application of cycloheximide for 30 min to cultured neurons before stimulation (5 min DHPG) did not affect mGluR-dependent FMRP localization.
Results
Visualization of FMRP and Fmr1 mRNA granules in dendrites and spines using high-resolution fluorescence microscopy
The prevalence of FMRP at synapses has not been quantified, although electron microscopy has confirmed the presence of postsynaptic FMRP in vivo (Feng et al., 1997). The synaptic localization of FMRP was quantified on cultured hippocampal neurons using IF and digital imaging analysis. In neurons cultured for 3 weeks (mature), to allow elaboration of the dendritic arbor, spines, and synapses (Goslin et al., 1998), FMRP was found in the cell body, and it extended into neuronal processes in a granular pattern (Fig. 1B,C), which has also been described in Castren et al. (2001). Double-labeling of FMRP (red) and synapsin, as a marker for presynaptic boutons (blue), revealed significant colocalization of FMRP with synapsin puncta (54%) (Fig. 1B,C). In immature [8 d in vitro (DIV)] neurons, 75% of synapsin puncta were colocalized with FMRP signal (Fig. 1A,C). There is a seven-fold increase in synapse density during this culture period as estimated by synapsin staining (Fig. 1D). We observed no change in total dendritic FMRP by IF and digital imaging analysis during this developmental period (Fig. 1E). These data show that FMRP is frequently localized at synapses, and its presence is particularly prominent during early development when synapses are being formed.
Figure 1.
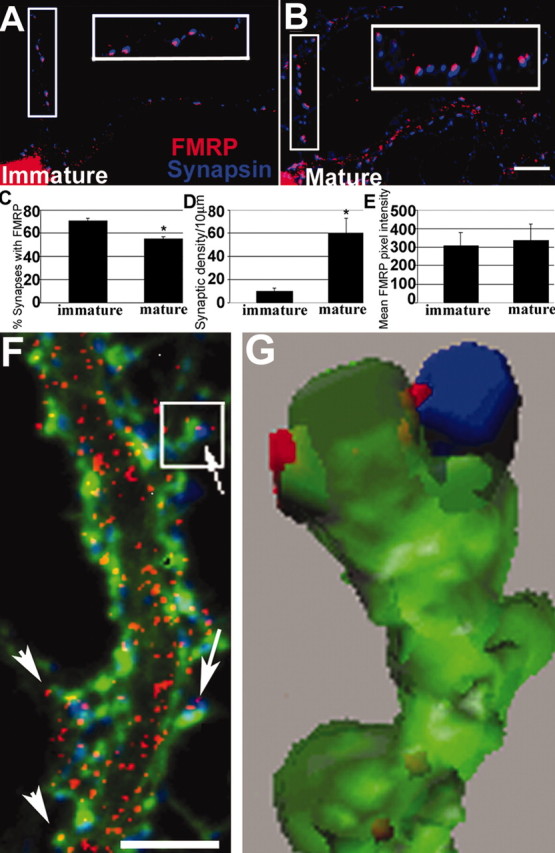
Developmental prevalence of FMRP at synapses. A, IF of an 8 DIV neuron shows frequent FMRP (red) colocalization with synapsin (blue; see enlarged inset). B, Same as A but a mature 19 DIV neuron. C, Histogram showing that 75% of synapsin puncta colocalized with FMRP at 8 DIV; only 54% colocalized at 19 DIV (*p < 0.05; Student's t test). D, Density of synapses increased as neurons matured (p < 0.05). E, There was no difference in FMRP levels in dendritic shafts of 8 and 19 DIV cultures. Scale bar, 10 μm. F, Triple-label IF detects FMRP (red), synapsin (blue), and F-actin (green). FMRP granules are distributed throughout dendrites, spine synapses (arrows), and filopodia (arrowheads). Scale bar, 5μm. G, 3-D reconstruction of boxed spine from F shows FMRP granules in spine neck and head, apposed to synapsin.
FMRP granules extended throughout the dendrite and into spines, which were identified as actin-rich protrusions that colocalized with synapsin (Fig. 1F, arrows). Triple-labeling (FMRP, red; synapsin, blue; F-actin, green) distinguished shaft synapses from dendritic spines (Fig. 1F). 3-D reconstruction (Fig. 1G) after deconvolution of a z-series (acquired from Fig. 1F, box) revealed FMRP throughout the spine compartment: in neck and head and apposed to the presynaptic interface (Fig. 1G). FMRP granules were in 71% of dendritic spines of mature cultures. FMRP was also in dendritic filopodia, which are actin-rich protrusions not associated with synapsin puncta (Fig. 1F, arrowheads).
In vitro and in vivo binding assays demonstrate that one target of FMRP is its own encoding Fmr1 mRNA (Ashley et al., 1993; Ceman et al., 1999; Schaeffer et al., 2001). It is unknown whether FMRP is associated with Fmr1 mRNA in dendrites. Past efforts did not reveal Fmr1 mRNA localization in dendrites (for review, see Steward and Schuman, 2003), but these analyses were done 2 hr after high-frequency stimulation, and although able to induce Arc mRNA localization, this time scale or paradigm may not have captured or induced Fmr1 mRNA localization (Valentine et al., 2000).
Combining FISH and IF, we examined the localization of Fmr1 mRNA with FMRP. We used digoxigenin-labeled oligonucleotide probes and FISH on cultured hippocampal neurons to demonstrate that Fmr1 mRNA was localized to dendrites of cultured hippocampal neurons (Fig. 2A, green) and was often colocalized (yellow granules, arrows) with FMRP (red) in both primary and secondary dendrites. In mature cultures, Fmr1 mRNA was found in ∼56% of total synapses and ∼76% of dendritic spines (n = 95 dendrites; data not shown). Although the most of the synapses in these cultured neurons are shaft synapses, both FMRP and Fmr1 mRNA were more common (on a percentage basis) in spine synapses. We also transfected neurons with an EGFP–FMRP construct to visualize the colocalization of FMRP and Fmr1 mRNA. FISH analysis of transfected neurons showed that EGFP–FMRP also colocalized with Fmr1 mRNA (Fig. 2B). Quantitative analysis of endogenous FMRP with Fmr1 mRNA demonstrated that ∼26% of the FMRP-positive puncta colocalized with Fmr1 mRNA.
Figure 2.
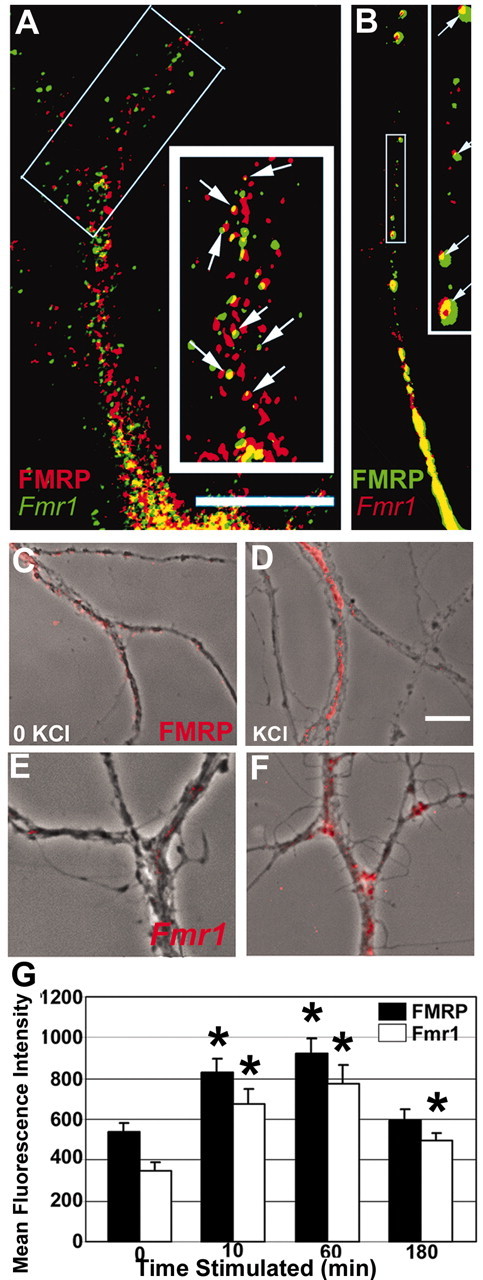
Localization of FMRP and Fmr1 mRNA granules in dendritic compartments and their KCl-induced localization. A, FISH followed by IF shows that FMRP (red) and Fmr1 mRNA (green) localize to dendrites and colocalize (yellow; see arrows in enlarged inset) with each other: 26% FMRP-positive pixels also contain Fmr1 mRNA. Scale bar, 10μm. B, Transfected rat hippocampal neurons show EGFP–FMRP granules colocalized with Fmr1 mRNA (red); see also enlarged inset (arrows). C–G, Time course of KCl-induced localization of FMRP and Fmr1 mRNA in dendrites. C, IF detection of FMRP (red) granules in secondary dendrites from unstimulated neurons (phase). Stimulation with KCl (D, G) caused FMRP IF intensity in secondary dendrites to increase. Increases were sustained for 1 hr and then subsided. E, FISH detection of Fmr1 mRNA granules (red) was apparent in secondary dendrites of unstimulated neurons (phase). After KCl treatment (F, G), Fmr1 mRNA IF intensity in dendrites increased dramatically. Note: fluorescence intensity was thresholded to just above background in unstimulated neurons (C, E) to illustrate the marked increase in intensity on stimulation (C–F). Scale bar, 10 μm.
Activity-dependent trafficking of FMRP requires metabotropic glutamate receptors
Depolarization with KCl was used to determine whether neuronal activity can influence FMRP localization. After 10 min of KCl stimulation, FMRP IF intensity in dendrites increased by 54% (Fig. 2G) and was maintained for 1 hr (Fig. 2C,D,G), returning to baseline in 3 hr (Fig. 2G). A 5 min KCl pulse also caused an increase (69%) in FMRP in 10 min, which dropped to baseline 30 min later (data not shown), indicating a sharper rise and a faster fall in FMRP dendritic localization.
FMRP redistribution may relocalize associated mRNAs, such as Fmr1 mRNA, in response to activity. We examined Fmr1 mRNA localization in dendrites 10 min, 1 hr, and 3 hr after KCl stimulation and showed that, consistent with FMRP distribution, KCl stimulation increased Fmr1 mRNA localization in dendrites of hippocampal neurons cultured for 19 DIV (Fig. 2E–G) by 94% after the first 10 min, peaking at 1 hr and then declining sharply (Fig. 2G), with kinetics similar to those of FMRP, by 3 hr. Because Fmr1 mRNA localization is rapid and declines between the first and third hours, its localizing response to activity may have been previously undetected at a 2 hr time point (Valentine et al., 2000).
The rapid kinetics of KCl-mediated increases in dendritic FMRP levels suggests that the effects were caused by trafficking as opposed to de novo local translation. Although movement of EGFP–FMRP granules in PC12 cells has been demonstrated (De Diego et al., 2002), we have examined the regulated trafficking of EGFP–FMRP. To test this, EGFP–FMRP was transfected into hippocampal neurons and subjected to quantitative FRAP analysis. FMRP recovery was measured over 5 min intervals at several time points before (Fig. 3A) and after (Fig. 3B) stimulation. Because newly synthesized EGFP takes at least 30 min to fold and acquire fluorescence (Aakalu et al., 2001), the rapid increase in fluorescence recovery (during the 5 min analysis) was most likely caused by increased motility of FMRP into the bleached zone. FRAP analysis revealed that KCl-treated neurons exhibited persistently higher recovery rates than untreated cells (Fig. 3C,D). Analysis of FRAP experiments indicated a ∼35-fold increase in recovery rate in the presence of KCl compared with unstimulated neurons (Fig. 3E), indicating that KCl increases the abundance and dynamic movement of FMRP in dendrites. Furthermore, the protein synthesis inhibitor cycloheximide did not prevent the regulated localization of FMRP in dendrites (Fig. 3F). These data suggest that the increased levels of FMRP in dendrites, in response to stimulation, reflect trafficking into dendrites from a preexisting somatic pool.
Because mGluR stimulation elevates FMRP expression in synaptosomal preparations (Weiler et al., 1997) and mGluR-dependent long-term plasticity is altered in FMR1 KO mice (Huber et al., 2002), we investigated whether FMRP trafficking is specifically regulated by mGluRs. KCl-induced change in FMRP dendritic localization was completely blocked by the group 1/2 mGluR antagonist MCPG, whereas the AMPAR and NMDAR antagonists CNQX (Fig. 4C) and APV (data not shown), respectively, had no effect. The mGluR5 antagonist MPEP significantly blocked the KCl-dependent localization of FMRP, whereas the mGluR antagonist LY-367385 impaired but did not significantly block this response (Fig. 4D).
Figure 4.
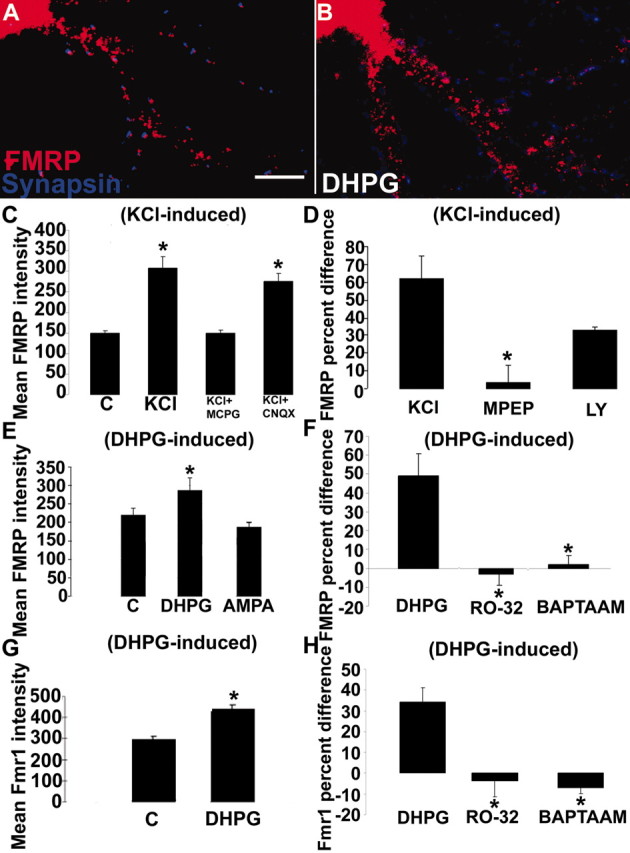
mGluRs regulate FMRP and Fmr1 mRNA localization in dendrites. A, B, E, DHPG stimulation (5 min) resulted in increased FMRP levels in dendrites. C, KCl stimulation (10 min) also increased FMRP levels (p < 0.05) in dendrites. This response was entirely inhibited by an mGluR antagonist, MCPG, but not by the AMPAR antagonist, CNQX. D, Bath application of mGluR5 antagonist MPEP blocked (p < 0.05) the KCl-regulated increase of FMRP in dendrites whereas mGluR1 antagonist LY-367385 did not. Bars indicate percentage difference of control compared with KCl application (KCl), MPEP compared with MPEP plus KCl (MPEP), and LY compared with LY plus KCl (LY). E, Treatment of neurons with the group I mGluR agonist DHPG resulted in increased dendritic FMRP levels (p < 0.05), a response not observed with AMPA. F, Blockade of the mGluR pathway through either PKC inactivation (RO-32) or chelating internal calcium (BAPTA-AM) abolished the dendritic localization of FMRP in response to mGluR activation. G, Fmr1 mRNA also demonstrated mGluR-dependent dendritic trafficking. DHPG activation of mGluR receptors localized the mRNA in dendrites (p < 0.05). H, Blockade of the mGluR pathway, as in F, also prevented Fmr1 mRNA localization. Histograms depict mean FMRP or Fmr1 mRNA IF intensity in response to various stimuli or the percentage difference in immunofluorescence intensity of an antagonist before and after stimulus. Unstimulated cultures in each histogram are labeled “C.” All culture conditions contain antagonists APV and CNQX, except where testing AMPA or NMDA. Scale bar, 10 μm.
We used specific glutamatergic agonists to confirm the role of mGluRs in FMRP trafficking (Fig. 4E). The group I mGluR receptor agonist DHPG and the ionotropic AMPAR agonist AMPA were bath applied to cultures. DHPG was applied in the presence of the NMDA antagonist APV and the AMPA antagonist CNQX, and AMPA was applied in the presence of APV alone. DHPG activation of group I mGluRs (5 min) caused a significant increase in FMRP localization into proximal dendrites (48%) (Fig. 4A,B,E), indicating a role for an mGluR-driven pathway signaling FMRP granule localization. In contrast, AMPA did not alter the localization of FMRP in dendrites. A similar result was obtained in DHPG-stimulated rat hippocampal neurons transiently transfected with human EGFP–FMRP (data not shown).
Group I mGluRs are defined by their coupling to phospholipase C (PLC). Signal transduction occurs through the Gq protein, thereby stimulating PKC and releasing intracellular stores of calcium (Valenti et al., 2002). To determine which components of the mGluR pathway were important for FMRP trafficking, we bath-applied PKC inhibitor RO-32-0432 (1 μm; 30 min before stimulation) or chelator of intracellular calcium BAPTA-AM, (25 μm; 15 min before stimulation) (Fig. 4F) to our cultures. Blockade of either pathway of the mGluR cascade resulted in a loss of mGluR-dependent dendritic localization of FMRP.
Fmr1 mRNA localization in dendrites is also regulated by mGluR activation
Because Fmr1 mRNA demonstrated a KCl activity dependence with kinetics similar to FMRP, we tested whether Fmr1 mRNA also demonstrated mGluR-dependent localization after 5 min DHPG stimulation. Fmr1 mRNA localization in dendrites increased 37% after DHPG stimulation (Fig. 4G). As with FMRP dendritic localization in response to mGluR stimulation, Fmr1 mRNA localization was also blocked by RO-32-0432 and BAPTA-AM (Fig. 4F,H), suggesting that similar mechanisms regulate FMRP and Fmr1 mRNA dendritic localization.
FMRP, but not Fmr1 mRNA, localization at the synapse is reduced after mGluR activation
Although KCl and DHPG increased FMRP localization in dendrites (see above), application of either stimulus, as well as AMPA, resulted in a surprising decrease in its localization to synapses (Fig. 5A,C) (decreases: 42% DHPG; 51% AMPA). This measured loss of FMRP from synapses was not attributed to reduced levels of FMRP in dendrites (Fig. 4E) or accompanied by increases in synapse density (Fig. 5D) that may have been caused by DHPG stimulation. Single-label in situ hybridization coupled with double-label immunofluorescence allowed simultaneous visualization and quantification of the FMRP protein along with Fmr1 mRNA at synapses (Fig. 5B,E). Subsequent to KCl depolarization, FMRP localization at synapses was diminished (from 79 to 53%) (Fig. 5E); however, Fmr1 mRNA localization at synapses was not significantly affected by KCl depolarization (from 64 to 59%) (Fig. 5E). Accordingly, the percentage of synapses containing both FMRP and Fmr1 mRNA was significantly diminished (from 42 to 19%) (Fig. 5E). Because there was no loss of Fmr1 mRNA from synapses after either KCl depolarization (Fig. 5E) or mGluR activation (Fig. 5F), we demonstrate that glutamatergic activity differentially regulates the ratio of FMRP to Fmr1 mRNA at synapses. These data suggest a local uncoupling or divergence between FMRP and Fmr1 mRNA at synapses in response to stimulation.
Figure 5.
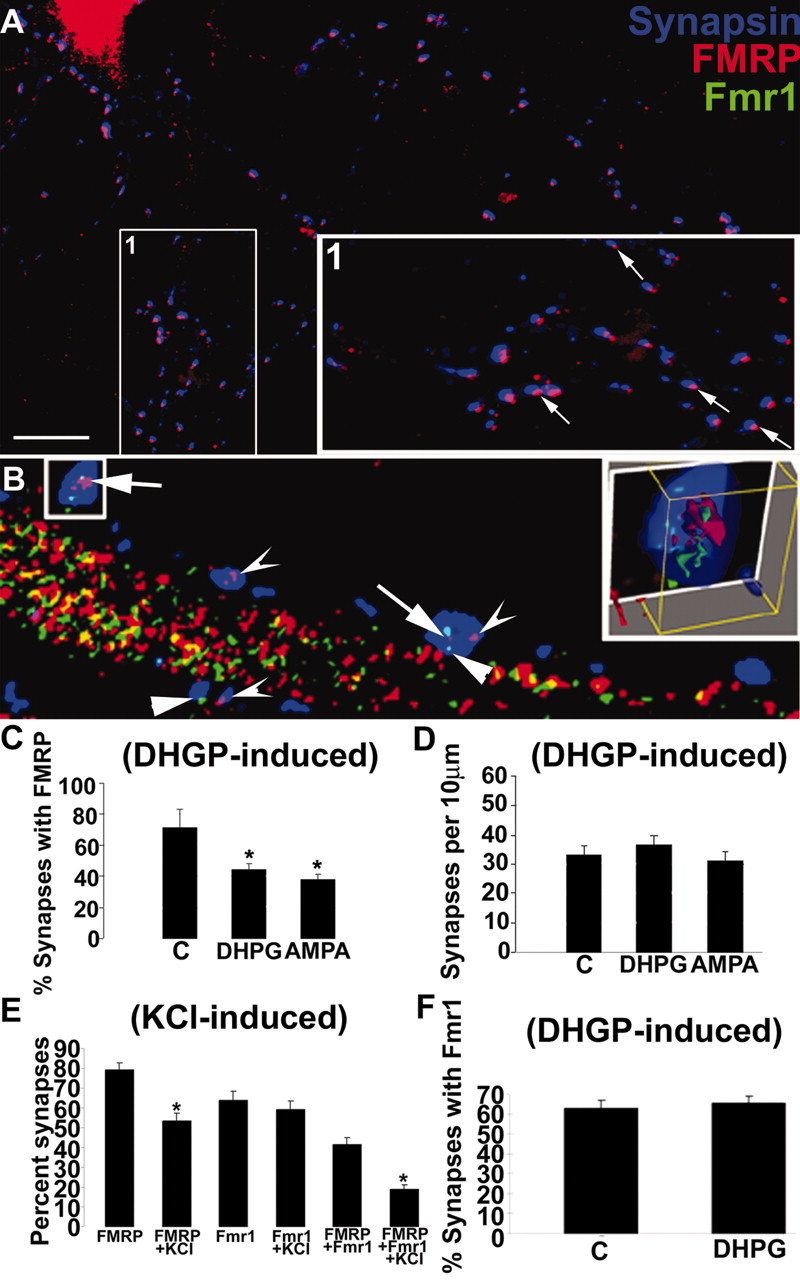
mGluRs regulate FMRP and Fmr1 mRNA localization in synapses. A, Rat (8 DIV) rat hippocampal cultures with double-label IF showing extensive FMRP (red) colocalization with synapsin puncta (blue) along neuronal processes. Boxed inset (1) is rotated and enlarged (at side). Arrows in enlarged inset depict examples of FMRP–synapsin colocalization. B, A combination FISH and double-label IF shows that FMRP and Fmr1 mRNA localize and colocalize in synapses. Concave arrowheads show FMRP localized to synapses; straight arrowheads show Fmr1 mRNA localized to synapses. Full arrows show FMRP and Fmr1 mRNA colocalization within a synapse (see E for quantification). Histogram in C quantified the percentage of synapses with an FMRP punctum associated with them before and after DHPG (5 min) and AMPA (15 min) stimulation (p < 0.05). D, Histogram shows that these short glutamatergic treatments do not affect synaptic density (p < 0.05). E, Histogram depicts changes in percentage synaptic localization of FMRP and Fmr1 mRNA after KCl depolarization in same cells using single-label FISH combined with double-label IF. Bars 1 and 2 show decreased FMRP localization at synapses after KCl stimulation (∼80 to ∼50%). Bars 3 and 4 show that Fmr1 mRNA is not diminished at synapses after KCl stimulation (∼60 to ∼60%). Bars 5 and 6 show percentage synapses with both FMRP and Fmr1 mRNA in them before and after KCl depolarization (∼45 to ∼20%). The loss of FMRP–Fmr1 mRNA colocalization at synapses after stimulation appears to be attributable to the activity-dependent loss of FMRP from synapses. F, mGluR stimulation via DHPG does not affect Fmr1 mRNA localization at dendrites despite the effect of DHPG to diminish FMRP at synapses (C). Scale bar, 10 μm.
Discussion
High-resolution imaging provides a new view of FMRP and Fmr1 mRNA regulation
We have used high-resolution fluorescence microscopy and digital imaging analysis to investigate the spatial localization and trafficking of FMRP and Fmr1 mRNA in dendrites and spines of hippocampal neurons. One advantage of this approach is that it affords both the sensitivity and resolution to permit quantitative analysis of dynamic changes of both FMRP and Fmr1 mRNA levels and localization in dendrites and at synapses in response to pharmacological manipulations. Although FMRP is known to be localized to postsynaptic sites and spines (Feng et al., 1997), it is unclear how frequent the synaptic localization really is and whether it can be modulated during neuronal development or in response to activity. Here we find that FMRP is expressed abundantly in dendrites throughout development, but is particularly highly localized first at developing synapses and later in mature spines. These data are consistent with morphologic observations suggesting failure of spine maturation in the absence of FMRP expression (Irwin et al., 2000), but they also suggest an ongoing role of FMRP in the regulation or maintenance of fully developed synapses.
In addition, our quantitative fluorescence imaging studies have shown that glutamatergic activity can directly and differentially modulate the dendritic and synaptic localization of FMRP and the Fmr1 mRNA. After stimulation, FMRP levels rose transiently in the dendritic processes. Our observations of increased FMRP mobility after stimulation, analyzed by FRAP (Fig. 3), suggest that activity-dependent pathways may affect the transport kinetics of FMRP, within dendrites, possibly providing an increased fraction of FMRP associated with motors, or an increased processivity of the motor associated with the mRNPs. This could facilitate the movement of FMRP and its target mRNAs in dendrites. Importantly, Fmr1 mRNA levels in dendrites increased with similar kinetics after stimulation, providing evidence that activity-driven changes in the localization of FMRP serve to redistribute its target mRNAs as well. These findings suggest that neuronal activity can, through the modulation of the subcellular localization of FMRP, impact local translation of important dendritic and synaptic proteins.
Regulated FMRP and Fmr1 mRNA localization shares characteristics with synaptic plasticity
mGluR activation was shown to be both necessary and sufficient for the increased dendritic localization of FMRP granules (Fig. 4C,E). The group I/II mGluR antagonist MCPG blocked KCl-dependent trafficking of FMRP in dendrites (Fig. 4C). Furthermore, DHPG-dependent mGluR activation was sufficient to localize FMRP (Fig. 4E). Interestingly, this same treatment has been found to induce a form of LTD in the hippocampus, which is altered in FMRP-deficient mice (Huber et al., 2002). This suggests that regulation of dendritic and synaptic FMRP localization may influence synaptic plasticity.
Group I mGluRs, mGluR5 and to a lesser extent mGluR1, are abundant in dendrites and dendritic spines of hippocampal neurons (Lujan et al., 1996; Patel et al., 2003). Group I mGluRs couple to PLC, causing downstream activation of PKC and a release of calcium from intracellular stores (Oliet et al., 1997). We found that blockage of either arm of this pathway disrupted the dendritic localization of both FMRP and Fmr1 mRNA (Fig. 4F,H), suggesting a coordinated role of these signals in this process. mGluR-dependent LTD similarly requires both PKC activation and elevations in postsynaptic calcium (Oliet et al., 1997), further supporting a link between plasticity and the regulation of FMRP–Fmr1 mRNA localization. Because the mGluR5 antagonist MPEP significantly blocked FMRP localization in dendrites, this suggests a predominant role for mGluR5 in FMRP and Fmr1 mRNA localization in hippocampal dendrites (Fig. 4D). This is in accord with a predominant role for mGluR5s in postsynaptic hippocampal LTD (Oliet et al., 1997; Huber et al., 2002).
mGluR and NMDAR signals may regulate the localization and translation of distinct mRNAs
Our findings suggest that the regulation of FMRP and Fmr1 mRNA localization by mGluRs may provide a pathway for regulation of mRNA localization distinct from those reported previously. A number of studies have pointed to a prominent role for NMDA receptors in the regulation of both mRNA localization and translation. High-frequency stimulation in vivo was shown to stimulate the dendritic localization of Arc mRNA, and this response was dependent on NMDA receptors (Steward and Worley, 2001). In cultured hippocampal neurons, NMDA receptors were required for the localization of a β-actin mRNP complex with zip code binding protein to dendrites of cultured hippocampal neurons (Tiruchinapalli et al., 2003). NMDA receptors also stimulated the translation of EGFP-reporter constructs that contain the 3′UTR cytoplasmic polyadenylation element (CPE) of CaMKIIα mRNA (Wells et al., 2001).
Despite the importance of NMDA receptors in the regulation of these mRNAs and binding proteins, our findings show that mGluRs, and not NMDARs, play a critical role in the regulation of the dynamic trafficking of FMRP and Fmr1 mRNA. Our data are consistent with findings that mGluRs signal the synthesis of FMRP in synaptosomes (Weiler et al., 1997). Although our studies do not rule out the possibility that NMDARs may play a role in other aspects of FMRP expression, they suggest that mGluR and NMDAR signals may regulate distinct mRNA targets or different steps of the mRNA localization and translation pathway. Such concepts may be important for understanding how NMDAR-dependent long-term plasticity differs from mGluR-dependent long-term plasticity. Although both are known to require protein synthesis in the postsynaptic neuron (dendrite), it remains unclear whether NMDA and mGluR signals regulate a common or different set of mRNAs.
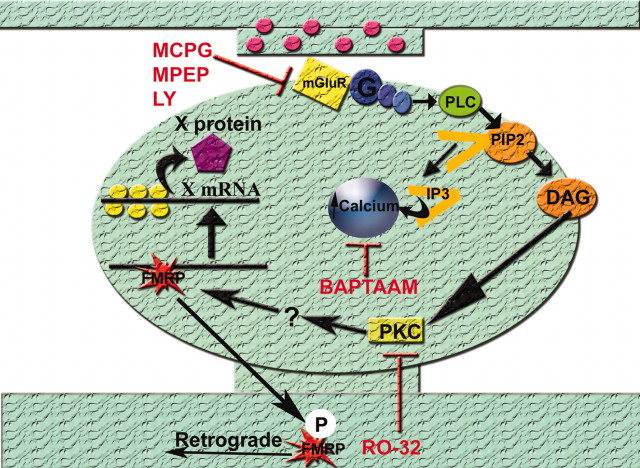
mRNA-binding proteins are potentially direct targets of glutamatergic regulation. The molecular mechanism of translational regulation of CaMKIIα mRNA in synaptosomes was shown to involve NMDA receptor-dependent phosphorylation of the mRNA-binding protein cytoplasmic polyadenylation element binding protein (CPEB) by Aurora kinase (Huang et al., 2002). Our studies indicate the presence of an mGluR signaling pathway that involves activation of PKC and release of internal calcium (Fig. 6). Although it is unknown whether PKC phosphorylation affects FMRP directly or indirectly, we note that the different phosphorylation states of FMRP have been shown previously to regulate its association with polyribosomes (Ceman et al., 2003). Future work is needed to identify how mGluR signals may affect the binding, transport, or translation of mRNAs by FMRP.
Figure 6.
Model for FMRP and Fmr1 mRNA localization in dendrites and at synapses. Glutamatergic stimulation activates the mGluR, triggering release of internal calcium and activation of PKC, a cascade of events that localizes FMRP and Fmr1 mRNA in dendrites. FMRP and its target mRNA colocalize at the synapse, in a complex where FMRP represses translation. Synaptic activation causes the dissociation of FMRP from the target mRNA, relieving the repression, and the loss of FMRP from the synapse, perhaps because of PKC phosphorylation. Although there is no direct evidence that PKC phosphorylates FMRP, it is known that there is a phosphorylation site on the FMRP protein (Ceman et al., 2003). Phosphorylated FMRP could become a retrograde messenger, returning to the cell body. Meanwhile, the mRNA is now derepressed and able to translate proteins that are important for synaptic structure and plasticity.
Glutamatergic regulation stimulates loss of FMRP from synapses
In contrast to what was observed in dendritic processes, at synapses there was a loss of FMRP after stimulation of either mGluRs or AMPA receptors. This was quantified as a decrease in the percentage of synapses (synapsin puncta) that had detectable FMRP signal in them (Fig. 5C). This decrease in FMRP-positive synapses was not attributed to an increase in the density of synapses (Fig. 5D). The loss of FMRP from synapses may be stimulated by any rises in local calcium levels, triggered by activation of either mGluRs (via internal stores) or AMPARs (via voltage-gated calcium channels). FMRP either may be leaving the synapse in response to synaptic signaling or may be locally degraded at the synapse through a ubiqitination pathway, as was shown recently to regulate PSD-95 (postsynaptic density-95) in response to glutamatergic stimulation (Colledge et al., 2003). In either case, the fact that synaptic Fmr1 mRNA levels were not reduced after stimulation suggests that the protein is uncoupled physically and functionally from its cargo under these conditions. We speculate that the regulated loss of FMRP from synapses observed here may have the effect of releasing repressed synaptic mRNAs so they can now be translated (Fig. 6).
Our findings of a loss of FMRP at synapses in cultured neurons can be reconciled with data that mGluR activation increased synthesis of FMRP in synaptosomal preparations (Weiler et al., 1997). Because these synaptosomes are biochemically isolated and sealed, it is not possible to track the fate of newly synthesized FMRP or a change in its localization after stimulation. Our studies, by direct visual examination of FMRP after mGluR stimulation, suggest that synaptically activated FMRP, localized or newly synthesized, may become a retrograde signal and localize back to the cell body only to return to the synapse carrying specific mRNAs important for synaptic structure and long-term plasticity (Fig. 6).
FMRP is known to be a repressor of translation in vitro (Laggerbauer et al., 2001). In Fragile X syndrome, the absence of FMRP-mediated repression is believed to result in runaway or excessive protein synthesis at synapses (Zalfa et al., 2003); however, an alternative model emerges if one considers that FMRP may act like CPEB, which is known to play a dual role in both mRNA localization in dendrites and translational derepression at synapses (Huang et al., 2002, 2003). In the absence of FMRP in FXS, one might predict impaired mRNA localization and, consequently, reduced translation at synapses. Future work is clearly needed to understand the full scope of FMRP interactions with its numerous mRNA targets and the molecular deficits in mRNA regulation in dendrites and at synapses in FXS.
Summary
The high-resolution microscopic approach used here has provided new information on the regulated localization of FMRP and Fmr1 mRNA in dendrites and at synapses. We show that synaptic activation through the mGluR5 receptor regulates the trafficking of FMRP and Fmr1 mRNA to dendrites via a signaling pathway that involves PKC and rise in internal calcium. Such findings will motivate further studies to identify in particular whether FMRP and its associated proteins are substrates for this regulation and in general whether mRNA binding proteins regulate localization and localized translation by divergent cues. Because mGluR-dependent LTD and postsynaptic protein synthesis are known to be dependent on mGluR5s (Oliet et al., 1997; Huber et al., 2002), and because it is impaired in FX (Huber et al., 2002), our results suggest a role for trafficking of FMRP and its cargoes in the regulation of mGluR-dependent LTD.
This work motivates investigation into defects in activity-dependent trafficking of FMRP-associated mRNAs in dendrites and at synapses that may underlie the synaptic deficits of FXS.
Toward the long-term goals of pharmacologic intervention in FX, our findings of regulation of FMRP through mGluR5 have important implications in the design of agents that may modulate specific glutamatergic signaling pathways that are imbalanced in Fragile X with respect to other forms of synaptic modification.
Footnotes
This work was largely supported by FRAXA Research Foundation (fellowships to L.N.A. and J.B.D.). We thank Concqer Fragile X Foundation, the Kirby Foundation, and the National Institutes of Health (Grant EB002088) for support. We acknowledge training support to L.N.A. (NIH T32 DK07513-14) and J.B.D. (Albert Einstein College of Medicine Scholar Award). We thank Adam Hartly and Alexadra Fayer for assistance with 3-D reconstructions, Steve Warren for sending FMRP cDNA, and Taesun Eom for valuable construct preparation. We also thank Honglai Zhang for expertise and helpful discussions.
Correspondence should be addressed to Gary Bassell, Department of Neuroscience, Rose Kennedy Center for Mental Retardation, Albert Einstein College of Medicine, 1400 Pelham Parkway, Bronx, NY 10461. E-mail: bassell@aecom.yu.edu.
DOI:10.1523/JNEUROSCI.0099-04.2004
Copyright © 2004 Society for Neuroscience 0270-6474/04/242648-08$15.00/0
R.A. and J.B.D. contributed equally to this work.
References
- Aakalu G, Smith WB, Jiang C, Nguyen N, Schuman EM (2001) Dynamic visualization of local protein synthesis in hippocampal neurons. Neuron 30: 489–502. [DOI] [PubMed] [Google Scholar]
- Antar LN, Bassell GJ (2003) Sunrise at the synapse: the FMRP mRNP shaping the synaptic interface. Neuron 555–558. [DOI] [PubMed]
- Ashley CT, Wilkinson KD, Reines D, Warren ST (1993) FMR1 protein: conserved RNP family domains and selective RNA binding. Science 262: 563–566. [DOI] [PubMed] [Google Scholar]
- Bassell GJ, Zhang HL, Byrd AL, Femino AM, Singer RH, Taneja KL, Lifshitz LM, Herman IM, Koisk KS (1998) Sorting of β actin mRNA and protein to neurites and growth cones in culture. J Neurosci 18: 251–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonhoeffer T, Yuste R (2002) Spine motility. Phenomenology, mechanisms, and function. Neuron 35: 1019–1027. [DOI] [PubMed] [Google Scholar]
- Brown V, Jin P, Ceman S, Darnell JC, O'Donnell WT, Tenenbaum SA, Jin X, Feng Y, Wilkinson KD, Keene JD (2001) Microarray identification of FMRP associated brain mRNAs and altered mRNA translational profiles in Fragile X syndrome. Cell 107: 477–487. [DOI] [PubMed] [Google Scholar]
- Castren M, Haapasalo A, Oostra BA, Castren E (2001) Subcellular localization of Fragile X mental retardation protein with the I304N mutation in the RNA-binding domain in cultured hippocampal neurons. Cell Mol Neurobiol 1: 29–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceman S, Brown V, Warren ST (1999) Isolation of an FMRP-associated messenger ribonucleoprotein particle and identification of nucleolin and the Fragile X-related proteins as components of the complex. Mol Cell Biol 19: 7925–7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceman S, O'Donnell WT, Reed M, Patton S, Pohl J, Warren ST (2003) Phosphorylation influences the translation state of FMRP-associated polyribosomes. Hum Mol Genet 12: 3295–3305. [DOI] [PubMed] [Google Scholar]
- Colledge M, Snyder EM, Crozier RA, Soderling JA, Jin Y, Langeberg LK, Lu H, Bear MF, Scott JD (2003) Ubiquitination regulates PSD-95 degradation and AMPA receptor surface expression. Neuron 40: 595–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Diego Otero Y, Severijnen LA, van Cappellen G, Schrier M, Oostra B, Willemsen R (2002) Transport of fragile X mental retardation protein via granules in neuritis of PC12 cells. Mol Cell Biol 23: 8332–8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Gutekunst CA, Eberhart DE, Yi H, Warren ST, Hersch SM (1997) FMRP: nucleocytoplasmic shuttling and association with somatodendritic polyribosomes. J Neurosci 17: 1539–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goslin K, Banker G (1998) Rat hippocampal neurons in low density culture. In: Culturing nerve cells, Ed 2 (Banker G, Goslin K, eds), pp 339–371. Cambridge, MA: MIT.
- Huang YS, Jung MY, Sarkissian M, Richter JD (2002) N-methyl-d-aspartate receptor signaling results in Aurora kinase-catalyzed CPEB phosphorylation and alpha CaMKII mRNA polyadenylation at synapses. EMBO J 21: 2139–2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YS, Carson JH, Barbarese E, Richter JD (2003) Facilitation of dendritic mRNA transport by CPEB. Genes Dev 17: 638–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber KM, Kayser MS, Bear MF (2000) Role for rapid dendritic protein synthesis in hippocampal mGluR-dependent long-term depression. Science 288: 1254–1257. [DOI] [PubMed] [Google Scholar]
- Huber KM, Gallagher SM, Warren ST, Bear MF (2002) Altered synaptic plasticity in a mouse model of Fragile x mental retardation. Proc Natl Acad Sci USA 99: 7746–7750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin SA, Galvez R, Greenough WT (2000a) Dendritic spine structural anomalies in Fragile X mental retardation syndrome. Cereb Cortex 10: 1038–1044. [DOI] [PubMed] [Google Scholar]
- Irwin SA, Swain RA, Christmon CA, Chakravarti A, Weiler IJ, Greenough WT (2000b) Evidence for altered Fragile-X mental retardation protein expression in response to behavioral stimulation. Neurobiol Learn Mem 74: 87–93. [PubMed] [Google Scholar]
- Kohrman M, Lui M, Kaether C, DesGroseillers L, Dotti CG, Kiebler MA (1999) Microtubule dependent recruitment of Staufen-GFP into large RNA containing granules and dendritic transport in living hippocampal neurons. Mol Biol Cell 10: 2945–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laggerbauer B, Ostareck D, Keidel EM, Ostaeereck-Lederer A, Fischer U (2001) Evidence that Fragile X mental retardation protein is a negative regulator of translation. Hum Mol Genet 10: 329–338. [DOI] [PubMed] [Google Scholar]
- Lujan R, Nusser Z, Roberts JD, Shigemoto R, Somogyi P (1996) Perisynaptic localization of metabotropic glutamate receptors mGluR1 and mGluR5 on dendrites and dendritic spines in the rat hippocampus. Eur J Neurosci 8: 1488–1500. [DOI] [PubMed] [Google Scholar]
- Miyashiro KY, Beckel-Mitchener A, Purk TP, Becker KG, Barret T, Liu L, Carbonetto S, Weiler IJ, Greenough WT, Eberwine J (2003) RNA cargoes associating with FMRP reveal deficits in cellular functioning in Fmr1 null mice. Neuron 37: 417–431. [DOI] [PubMed] [Google Scholar]
- Oliet SHR, Malenka RC, Nicoll RA (1997) Two distinct forms of long-term depression coexist in CA1 hippocampal pyramidal cells. Neuron 18: 969–982. [DOI] [PubMed] [Google Scholar]
- Patel S, Krause SM, Hamill T, Chaudhary A, Burns DH, Gibson RA (2003) In vitro characterization of [3H]methoxyPyEP, an mGluR5 selective radioligand. Life Sci 73: 371–379. [DOI] [PubMed] [Google Scholar]
- Schaeffer C, Bardoni B, Mandel JL, Ehresmann B, Ehresmann C, Moine H (2001) The Fragile X mental retardation protein binds specifically to its mRNA via a purine quartet motif. EMBO J 20: 4803–4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder EM, Philpot BD, Huber KM, Dong X, Fallon JR, Bear MF (2001) Internalization of ionotropic glutamate receptors in response to mGluR activation. Nat Neurosci 4: 1079–1085. [DOI] [PubMed] [Google Scholar]
- Steward O, Schuman EM (2003) Compartmentalized synthesis and degradation of proteins in neurons. Neuron 40: 347–359. [DOI] [PubMed] [Google Scholar]
- Steward O, Worley PF (2001) Selective targeting of newly synthesized Arc mRNA to active synapses requires NMDA receptor activation. Neuron 30: 227–240. [DOI] [PubMed] [Google Scholar]
- Tiruchinapalli DM, Oleynikov Y, Kelic S, Shenoy SM, Hartley A, Stanton PK, Singer RH, Bassell GJ (2003) Activity-dependent trafficking and dynamic localization of zip code binding protein 1 and β-actin mRNA in dendrites and spines of hippocampal neurons. J Neurosci 23: 3251–3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd PK, Mack KJ (2000) Sensory stimulation increases cortical expression of FMRP in vivo. Mol Brain Res 14: 17–25. [DOI] [PubMed] [Google Scholar]
- Valenti O, Conn PJ, Marino MJ (2002) Distinct physiological roles of the Gq-coupled metabotropic glutamate receptors co-expressed in the same neuronal populations. J Cell Physiol 191: 125–137. [DOI] [PubMed] [Google Scholar]
- Valentine G, Chakravarty S, Sarvev J, Bramham C, Herkenham M (2002) Fragile X (fmr1) mRNA expression is differentially regulated in two adult models and activity-dependent gene expression. Brain Res Mol Brain Res 75: 337–341. [DOI] [PubMed] [Google Scholar]
- Weiler IJ, Irwin SA, Klintsova AY, Spencer CM, Eberwine J, Greenough WT (1997) Fragile X mental retardation protein is translated near synapses. Proc Natl Acad Sci USA 94: 5395–5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells DG, Dong X, Quinlan EM, Huang YS, Bear MF, Richter JD, Fallon JR (2001) A role for the cytoplasmic polyadenylation element in NMDA receptor-regulated mRNA translation in neurons. J Neurosci 21: 9541–9548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalfa F, Giorgi M, Primerano B, Moro A, Di Penta A, Reis S, Oostra B, Bagni C (2003) The Fragile X syndrome protein FMRP associates with BC1 RNA and regulates the translation of specific mRNAs at synapses. Cell 112: 317–327. [DOI] [PubMed] [Google Scholar]
- Zhang HL, Eom T, Oleynikov Y, Shenoy SM, Lieblet DA, Dictenberg JB, Singer RH, Bassell GJ (2001) Neurotrophin induced transport of b-actin mRNP complex increases β-actin levels and stimulates growth cone motility. Neuron 31: 261–275. [DOI] [PubMed] [Google Scholar]