Abstract
The mechanisms that regulate the development of the amygdaloid complex are as yet poorly understood. Here, we show that in the absence of the LIM-homeodomain (LIM-HD) gene Lhx2, a particular amygdaloid nucleus, the nucleus of the lateral olfactory tract (nLOT), is selectively disrupted. LIM family members are well suited for multiple roles in the development of complex structures because they participate in regulatory interactions that permit a diversity of function. To investigate the possible role for other LIM-HD genes as well as LIM-only (Lmo) genes in the developing amygdala, we examined their expression in the embryo. We show that amygdaloid nuclei upregulate distinct patterns of LIM gene expression from embryonic stages. This supports the hypothesis that LIM genes may participate in the mechanisms that control the development of the amygdala. The disruption of the nLOT in the Lhx2 mutant is the first evidence of a role for LIM-HD genes in the development of the amygdaloid complex. The combinatorial expression patterns of LIM genes suggest a comprehensive mechanism for patterning this structure.
Keywords: amygdala, LIM, nLOT, patterning, combinatorial, specification
Introduction
Several transcription factors have been identified that regulate the patterning of the dorsal and ventral telencephalon (for review, see Campbell, 2003). Little is known, however, about mechanisms that regulate the patterning of the amygdala, an interface structure consisting of both dorsal (pallial) as well as ventral (subpallial) telencephalic derivatives. There are different views about how the various amygdaloid nuclei may be categorized into distinct functional or cytoarchitectural groups (Puelles et al., 2000; Martinez-Garcia et al., 2002; McDonald, 2003). There is even controversy as to whether or not it is relevant to collectively refer to these nuclei as “amygdaloid” in the first place (Swanson and Petrovich, 1998). Resolving these issues is hampered by the complex morphology and intricate connectivities of the different nuclei, as well as a lack of evidence about the developmental mechanisms that underlie the formation of this structure.
The amygdala has cortical portions, whereas its deeper parts form proper nuclei (i.e., are non-cortical in morphology). Amygdaloid nuclei are located in a range of different positions: (1) juxtaposed to the olfactory cortex on the caudal aspect of the telencephalon; (2) more deeply, adjacent to the claustrum or the striatum; or (3) at extreme caudal positions, adjacent to the caudal end of the hippocampal formation. Because these sites are all fairly distant from the ventricular zone, precursors of amygdaloid nuclei must undergo extensive migration to form the mature structure. The complexity of the amydagloid nuclei suggests that a correspondingly complex set of regulatory mechanisms would be required for its proper development. This would involve the parcellation of migrating cells to the appropriate nucleus and the specification of these cells to distinct fates. To date, no comprehensive hypothesis has been proposed for mechanisms that may fulfill this role.
LIM-homeodomain (LIM-HD) genes play crucial roles in development and patterning in several systems. Twelve LIM-HD genes identified in the mouse are expressed in distinct, but partially overlapping, patterns throughout the CNS (Tsuchida et al., 1994; Nakagawa and O'Leary, 2001; Bulchand et al., 2003). In the spinal cord, a “LIM code” comprising specific combinations of LIM-HD genes specifies cell identity of discrete neuronal subtypes and acts at multiple stages of neuronal development (for review, see Shirasaki and Pfaff, 2002). A combinatorial expression pattern of LIM-HD genes has also been described in the developing thalamus, suggesting a role in the diversification of distinct thalamic nuclei during development (Nakagawa and O'Leary, 2001).
Here, we show that the absence of the LIM-HD gene Lhx2 disrupts the formation of one particular nucleus, whereas other nuclei appear unaffected. Furthermore, LIM-HD and LIM-only (Lmo) genes exhibit highly selective and partially overlapping domains of expression in the embryonic amygdala. We propose that this combinatorial expression may regulate the specification and differentiation of different amygdaloid nuclei. This is the first report of a role for an LIM-HD gene in the specification of the embryonic amygdala. The combinatorial expression of members of the LIM gene family suggests a broad mechanism for parcellating this structure.
Materials and Methods
Timed pregnant Swiss mice and Lhx2 +/- pregnant dams were obtained from the Tata Institute animal breeding facility. All procedures followed Institute Animal Ethics Committee guidelines and National Institutes of Health guidelines for the care and use of animals. In situ hybridization was performed as described previously (Bulchand et al., 2003). Briefly, the hybridization was performed overnight at 70°C in 4× SSC, 50% formamide, and 10% SDS. Posthybridization washes were at 70°C in 2× SSC and 50% formamide. We used the atlas Chemical Neuroanatomy of the Prenatal Rat Brain (Foster, 1998) as a guide to the morphology of the embryonic amygdaloid complex and for nomenclature: anterior amygdaloid area (AA), dorsal portion (AAD), ventral portion (AAV); the various “nuclei”: anterior cortical (ACo); basolateral (BL); basomedial (BM); central (Ce); lateral (La); medial (Me); posterodorsal portion (MeD); posteroventral portion (MeV); nucleus of the lateral olfactory tract, layers 1 and 2/3 (nLOT1 and nLOT2/3, respectively); posteromedial cortical (PMCo).
Results
As a starting point for analyzing whether the LIM gene family regulates the development of the amygdaloid complex, we focused on the Lhx2 mutant brain to examine whether the embryonic amygdala develops normally in the absence of this LIM-HD gene. We selected this gene because previous work had established Lhx2 as the only LIM-HD gene expressed in most of the ventricular zone of the dorsal and ventral telencephalon (Nakagawa et al., 1999; Bulchand et al., 2003). Because the origin of amygdaloid precursors is not completely understood, widespread expression of Lhx2 in the ventricular zone can be expected to include all sites from which amygdaloid precursors may arise, placing Lhx2 in a unique position to play a role in the development of much of the amygdaloid complex.
Furthermore, we examined the expression of other members of the LIM gene family in the embryonic amygdala. We selected embryonic day 15.5 (E15.5) to E17.5 for our studies because markers of amygdaloid nuclei are not detectable at earlier ages, whereas the expression patterns are strong at E15.5 and do not appear to change detectably for the next few days.
Lhx2 is expressed in a stream of cells emerging from the pallium-subpallium boundary
Neuroepithelial territories at the pallium-subpallium boundary (PSB) are thought to give rise to some amygdaloid nuclei (Puelles et al., 2000). Lhx2 labels a stream of cells that appears to arise from this region, which, by E12.5, has already reached the vicinity of the future site of the amygdaloid complex (Fig. 1a, arrowheads) (Yun et al., 2001). In addition, Lhx2 labels the ventricular zone of the medial and caudal ganglionic eminences (Fig. 1a and data not shown), which are also thought to be sources for amygdaloid precursors (Nery et al., 2002; Stenman et al., 2003).
Figure 1.
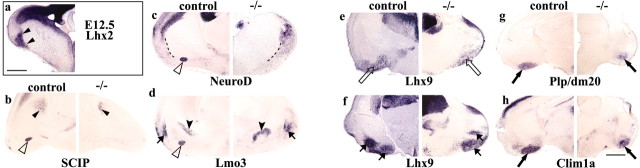
The embryonic amygdala is defective in the absence of Lhx2. At E12.5, control brains express Lhx2 in a stream of cells that arises from the PSB and continues into the region of the prospective amygdaloid complex (a, arrowheads). Lhx2 is also expressed in the entire telencephalic ventricular zone (a). At E15.5, the nLOT 2/3 is identified by expression of SCIP, NeuroD, and Lmo3 in control brains but is undetectable in the Lhx2 mutant (b-d, open arrowheads). In contrast, both control and mutant brains display expression of several amygdaloid and other ventral telencephalic markers: SCIP in a small patch in the striatum (b, arrowhead); NeuroD in a stretch of cells along the lateral telencephalon (c, dotted line); Lmo3 in the globus pallidus (d, arrowhead) and in the La (d, arrow); Lhx9 expression in the region of the AAV and the ACo (e, open arrows) and in the BM and MeV (f, arrows); Plp/dm20 and Clim1a expression in the PMCo (g, h, arrows). Scale bars, 200 μm.
An amygdaloid nucleus is disrupted in the absence of Lhx2
Although Lhx2 mutant embryos die at mid-gestation, we recovered 25 mutant brains at E15.5 and examined them, in parallel with littermate controls, for expression of a panel of markers that identify distinct amygdaloid nuclei from E15.5. The nLOT appears to be drastically perturbed in the Lhx2 mutant. Three independent markers of this nucleus are not detectable in Lhx2 mutant brains. SCIP, a POU-domain transcription factor, and NeuroD, a basic helix-loop-helix (bHLH) gene, mark layers 2/3 of the nLOT in control brains but do not reveal these in the mutant (Fig. 1b,c). This defect appears to be specific to the nLOT2/3, because SCIP expression is seen in other regions such as the striatum of both control and mutant brains. Likewise, NeuroD expression is seen in a stretch of scattered cells laterally that delineate the cortex from the striatum in both control and mutant brains, but the intense nLOT2/3 labeling is absent in the mutant. Finally, LIM-only gene Lmo3 is expressed normally in the mutant La and BL nuclei as well as in the globus pallidus but is strikingly absent from the site of the nLOT2/3, which would lie among these structures (Fig. 1d). Our panel of markers does not permit a conclusive analysis about the presence of nLOT1 in the mutant; however, based on three independent markers (SCIP, NeuroD, and Lmo3), we conclude that the nLOT is severely disrupted, at least in the specification of layers 2/3.
In contrast to the nLOT, several other amygdaloid nuclei appear to be specified in the absence of Lhx2. This is apparent despite the somewhat abnormal morphology of the mutant brains, which is attributable to the absence of much of the cortex (Bulchand et al., 2001). LIM-HD gene Lhx9 expression is detected in regions appropriate for the AAV and the ACo nucleus, the nuclei that flank the nLOT (Fig. 1e). Lhx9 expression also marks the BM and MeV nuclei in a characteristic pattern similar to that in control brains (Fig. 1f). Finally, the PMCo nucleus is marked in caudal sections of both control and mutant brains by cofactor of the LIM gene Clim1a as well as an unrelated gene encoding a proteolipid protein, Plp/dm20 (Fig. 1g,h).
The highly selective role of Lhx2 in the specification of the embryonic amygdala motivated an examination of the broader family of LIM-HD genes and LIM-only (Lmo) genes to provide an insight into possible interactions between these molecules in the development of this structure.
LIM gene expression parcellates the embryonic amygdala
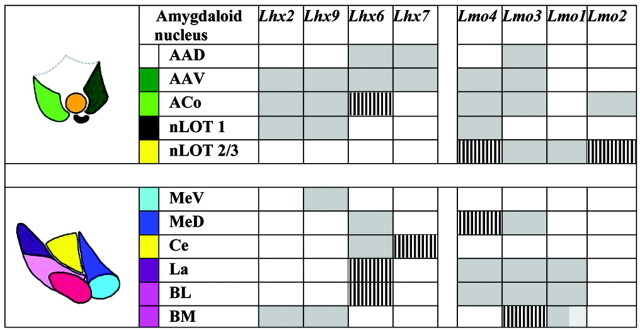
LIM genes exhibit combinatorial expression patterns in the amygdaloid complex (Table 1). Going from rostral to caudal in coronal sections, the nLOT is prominent at mid levels (Fig. 2, Level 1). The striking feature of nLOT labeling is that layers 2/3 of this structure express all four Lmo genes but are selectively excluded by the Lhx genes (Fig. 2). Lmo1 and Lmo3 appear to label the nLOT2/3 substantially, whereas Lmo2 and Lmo4 show a more limited labeling (Fig. 2, a,b,g,h). nLOT1, in contrast, expresses Lhx2 and Lhx9 as well as Lmo4 (Fig. 2c,f,h). Flanking the nLOT are the ACo and AA. Lmo2 is detected only in the ACo, whereas Lhx7 is present only in the AA (Fig. 2b,e). One interesting feature of AA labeling is that Lhx6-, Lhx7-, and Lmo3-labeled cells are present in both the dorsal (AAD) and ventral (AAV) portions of this nucleus (Fig. 2a,d,e), whereas Lhx2, Lhx9, and Lmo4 are restricted to the ventral region (Fig. 2c,f,h)
Table 1.
A LIM expression code in the embryonic amygdala
Shaded boxes represent expression; clear boxes, faint or undetectable expression; dual-shaded boxes, a gradient of expression; hatching represents expression in scattered cells within the structure.
Figure 2.
Combinatorial expression of LIM genes identifies distinct nuclei in the embryonic amygdaloid complex. a-h are sections of E17.5 brain, cut at Level 1 in the schematic. Lmo1-Lmo4 are expressed in the nLOT 2/3 (a, b, g, h, dotted circle). This structure is excluded by Lhx2, Lhx7, and Lhx9 expression, whereas Lhx6 is expressed in a few scattered cells (c-f, dotted circle). The nLOT1 expresses Lhx2, Lhx9, and Lmo4 (c, f, h, arrowhead). The ACo expresses all, except Lmo1 and Lhx7 (a-d, f, h, open arrowhead). The AAV expresses Lhx2, Lhx6, Lhx7, Lhx9, Lmo3, and Lmo4 (a, c-f, h, filled arrow), whereas the AAD expresses only Lhx6, Lhx7, and Lmo3 (a, d, e, open arrow). i-n are sections cut at Level 2 in the schematic. Lmo1, Lmo3, Lmo4, and Lhx6 are expressed in the La (i, j, k, n, filled arrowhead) and in the BL (i, j, k, n, open arrowhead). The BM expresses Lmo1 and Lmo3 in a gradient, Lhx9 throughout, and is excluded by Lmo4, Lhx6, and Lhx7 (i, j, k, m, n, dotted oval). The MeV expresses Lhx9 (m, open arrow), and the MeD expresses Lmo3 and Lhx6 (k, n, filled arrow). The Ce displays Lhx6 and Lhx7 expression but has greatly reduced expression of the other markers (i-l, n, asterisk, dashed lines). Scale bar, 200 μm.
At levels caudal to the nLOT, a cluster of six nuclei appears (Fig. 2, Level 2). On the lateral side are the La, BL, and BM. The Ce, MeD, and MeV are present medially. A striking pattern that emerges is that Lhx6 and Lhx9 expression marks mutually exclusive groups of nuclei: Lhx6 is expressed in all but the BM and the MeV, whereas Lhx9 exclusively labels these two nuclei (Fig. 2m,n). The individual nuclei can be identified by distinct combinations of LIM gene expression: the MeV is identified by Lhx9 expression; the MeD by expression of Lhx6 and also Lmo3; and the Ce by Lhx6 and Lhx7 (Fig. 2k-n). The La and BL both display the same combinations of gene expression: Lhx6, Lmo1, Lmo3, and Lmo4 all label these nuclei (Fig. 2I-k,n). The BM, in contrast, shows a gradient of Lmo1 expression, which overlaps with weak Lhx2 (data not shown) and strong Lhx9 (Fig. 2i,m) expression in this nucleus.
The PMCo appears at extreme caudal levels and is not labeled by any Lhx or Lmo gene expression (data not shown). However, Clim1a, a cofactor of LIM genes, selectively marks this nucleus (Fig. 1h).
Discussion
Patterning the vertebrate CNS requires that neuroepithelial cells be allocated into distinct groups of precursors, from which cells of specific structures will arise. For complex structures, comprising nuclei with distinct identities, there must be an additional parcellating mechanism that operates within this group of precursors, or alternatively, a specification mechanism that acts on groups of postmitotic cells, during or at the end of their migration. LIM-HD genes are well suited to play multiple roles in development because these molecules permit complex regulatory interactions that serve to modulate their functions. In this study, we report LIM gene expression early in the development of the amygdala. Furthermore, unique combinations of LIM gene expression identify distinct postmitotic amygdaloid nuclei. We propose that the actions of these proteins may underlie crucial aspects of the development of the amygdaloid complex. We also demonstrate the selective role of Lhx2 in the specification of one particular amygdaloid nucleus, the nLOT.
Of the 12 LIM-HD genes identified in the mouse, only Lhx2 is expressed in the ventricular zone of the telencephalon at locations thought to give rise to amygdaloid precursors. Although the precise sources for the different amygdaloid nuclei remain unclear, there is evidence to suggest that the amygdala is an interface structure with dorsal as well as ventral derivatives: a pallial component, consisting of the La and BL, and a subpallial component, consisting of the Ce and Me, has been proposed (Puelles et al., 2000; Stenman et al., 2003). Because LIM genes are expressed in the ventricular zone as well as in the final locations of the amydgaloid nuclei, these genes may play roles at multiple stages during the development of the embryonic amygdala.
The Lhx2 mutant reveals that much of the amygdaloid complex is specified but the nLOT is selectively disrupted
Although Lhx2 is in a position to regulate the development of much of the amygdaloid complex, several amygdaloid nuclei are not only specified in the Lhx2 mutant, but their relative positioning also appears remarkably well preserved. What might explain this surprisingly normal patterning? Regions of neuroepithelium located at the PSB are thought to give rise to some of these amygdaloid structures: the lateral pallium is thought to contribute to the BL nucleus and the ventral pallium to the La nucleus (Puelles et al., 2000; Stenman et al., 2003). Although Lhx2 is widely expressed in the telencephalic neuroepithelium, the expression level varies such that there is high medial-low lateral, high caudal-low rostral gradient (Nakagawa and O'Leary, 2001). This positions the PSB in a “low”-expressing region, which could explain why it is spared in the Lhx2 mutant (Vyas et al., 2003) and why the specification of the basolateral complex and the olfactory cortex appears unaffected. The origin of cells that form the nLOT has not yet been precisely identified, however. In fact, the nLOT is not considered to be part of the basolateral complex, but rather part of the olfactory cortex, because both the nLOT and the olfactory cortex receive input from the olfactory bulb (Swanson and Petrovich, 1998). However, the olfactory cortex appears specified normally in the Lhx2 mutant (Vyas et al., 2003). The selective loss of the nLOT therefore suggests that its precursors may arise from a different region of the neuroepithelium than the basolateral complex or the olfactory cortex.
A cell-autonomous role for Lhx2 would require that the protein act in the precursors of the nLOT2/3 before the cells have reached their final positions, because Lhx2 expression is not detected in the cells after they reach the nLOT2/3. It is not clear, however, why the defect in the Lhx2 mutant is specific to the nLOT. Analysis of mutant phenotypes of other genes involved in telencephalic patterning may provide more insight into why the nLOT is different from the other amygdaloid nuclei and the olfactory cortex.
Combinations: many ways to create differences
LIM-HD genes exhibit a “nested” expression pattern in the AAD, AAV, ACo, and nLOT. Other nuclei are marked by distinct single or paired expression of LIM-HD genes (Table 1). Together, these expression patterns provide a unique panel of markers that identify distinct amygdaloid nuclei in the embryo. Furthermore, they also raise the hypothesis that these molecules may act via combinatorial mechanisms, as have been identified in other systems (for review, see Shirasaki and Pfaff, 2002), to regulate the differentiation of these nuclei. Additionally, differential gene expression may provide a mechanism for subdividing nuclei (e.g., the dorsal and ventral portions of the Me and the AA express distinct combinations of LIM-HD genes). Also, the BM displays a gradient of expression of Lmo1 and Lmo3. Because these genes encode proteins that could compete with LIM-HD genes (Milan and Cohen, 1999), their graded expression could serve to differentially modulate the function of Lhx9, which is expressed throughout this nucleus. A point to be noted is that the combinatorial expression patterns we report do not particularly correlate with groupings of various amygdaloid nuclei, as have been suggested in the literature. For example, Lmo3 labels the olfactory cortex and the nLOT, which Swanson and Petrovich (1998) group together with the ACo and PCo as part of the olfactory cortex. However, the ACo and PCo display weak or no Lmo3 expression, whereas the La and BL, part of a different group, express this gene robustly. Interestingly, the ACo and PCo, both “cortical” nuclei, are not grouped by similar LIM gene expression, and neither are the medial and central nuclei, both thought to be derivatives of the striatum. This suggests that rather than specifying “group identity,” LIM genes may have a more complex role in regulating aspects of cell specification that may be independent of which group within the amygdaloid complex the cells belong to. The distinct neurochemical and connectional features of different nuclei within any group emphasizes the need for multiple control mechanisms. The data support the idea that LIM genes may constitute an important component of these mechanisms.
The potential roles of LIM-HD genes in the amygdala may include the regulation of neurotransmitter identity, as in the Drosophila nerve cord and in the vertebrate hindbrain (Benveniste et al., 1998; Ding et al., 2003). Indeed, loss of function of Lhx7 (also known as Lhx8) causes a drastic reduction in the cholinergic neuron population in the basal forebrain (Zhao et al., 2003), and Lhx6 has been implicated in regulating a subset of GABAergic interneurons arising from the medial ganglionic eminence (Marin et al., 2000).
LIM-HD genes also regulate axon pathfinding in the vertebrate spinal cord (Kania and Jessell, 2003) and in the Drosophila ventral nerve cord (Lundgren et al., 1995). This family may direct the development of the intricate connectivity (Pitkanen, 2003) of the amygdaloid complex. Indeed, a LIM-HD code has been proposed for an even wider role, based on comparative studies of thalamic and cortical expression across species: that of regulating not only the development, but also the evolution, of forebrain connectivity (Bachy et al., 2002).
Specificity of LIM domains and a role for Lmo genes
LIM-HD protein function is modulated by regulatory interactions via the LIM domains. Transcription factor activity of these proteins requires the LIM domains to be bound to a cofactors encoded by Clim genes (Jurata et al., 1998). Because Lmo proteins have only LIM domains, but no homeodomain, they can interfere with the function of LIM-HD proteins by competing for Clim molecules (Milan and Cohen, 1999). LIM-HD protein function can thus be modulated by the presence of the appropriate Lmo molecules.
Previous studies of the spinal cord and the thalamus did not include Lmo genes (Tsuchida et al., 1994; Nakagawa and O'Leary, 2001), which are expressed in distinct and selective patterns in the embryonic amygdala. The four mouse Lmo genes are all potential negative regulators of LIM-HD function, yet little is known about the functional differences among them. Each Lmo protein may have a selective ability to compete with particular LIM-HD proteins. Although no mechanisms have been identified by which LIM domains may confer selectivity of function, such a phenomenon has been demonstrated in the role of the Drosophila LIM-HD protein Apterous (Ap). A hybrid molecule made by interchanging the LIM domains of Ap with those of other LIM-HD proteins can rescue the wing phenotype of ap mutants but not the pathfinding defects, indicating that the LIM domains are functionally distinct (O'Keefe et al., 1998). This suggests a model in which, in different cells, context-specific interactions involving the LIM domains may be critical for distinct functions. Similarly, the LIM domains of Lmo genes may confer selective regulation of these interactions. Lmo gene expression within the amygdaloid complex supports distinct roles for these genes (Table 1). Finally, Lmo genes are known to interact or cooperate with other transcription factors, such as members of the bHLH family, to regulate transcription of downstream genes (Bao et al., 2000). In this context, it is interesting that the bHLH gene NeuroD is expressed in the nLOT2/3 (Fig. 1), where it may serve as a potential interacting partner for Lmo proteins in this structure. These mechanisms may add to the potential functions of Lmo genes in the embryonic amygdala.
In conclusion, the LIM-HD gene Lhx2 is required for the formation of one particular amygdaloid nucleus, the nLOT, providing the first evidence of LIM-HD gene function in the development of the amygdala. Furthermore, we report a LIM expression code in the embryonic amygdaloid complex that may underlie multiple aspects of its development. The distinct and overlapping expression patterns of LIM-HD and Lmo genes suggests complex roles for these molecules, either acting individually or in a combinatorial mechanism.
Footnotes
This work was supported by Wellcome Trust Senior Fellowship 056684/Z/99/Z (S.T.). We thank the following people for gifts of plasmid DNA: B. Anderson (Lmo4), I. Bach (Climax), R. Kageyama (NeuroD), G. Lemke (SCIP), R. Bansal (Plp/Dm20), Y. Nakagawa (Lhx9), F.D. Porter (Lhx2), T. Rabbitts (Lmo1,2,3), J. Rubenstein (Lhx6 and Lhx7), and Y. Zhao (Lhx5). We also thank F.D. Porter for giving us the Lhx2+/- breeding pairs, Y. Mane and S. Chavan for maintaining the Lhx2 mutant breeding colony, and V. Vaidya and members of the Tole laboratory for critical comments on this manuscript.
Correspondence should be addressed to Dr. Shubha Tole at the above address. E-mail: stole@tifr.res.in.
Copyright © 2004 Society for Neuroscience 0270-6474/04/246986-05$15.00/0
References
- Bachy I, Failli V, Retaux S (2002) A LIM-homeodomain code for development and evolution of forebrain connectivity. NeuroReport 13: A23-A27. [DOI] [PubMed] [Google Scholar]
- Bao J, Talmage DA, Role LW, Gautier J (2000) Regulation of neurogenesis by interactions between HEN1 and neuronal LMO proteins. Development 127: 425-435. [DOI] [PubMed] [Google Scholar]
- Benveniste RJ, Thor S, Thomas JB, Taghert PH (1998) Cell type-specific regulation of the Drosophila FMRF-NH2 neuropeptide gene by Apterous, a LIM homeodomain transcription factor. Development 125: 4757-4765. [DOI] [PubMed] [Google Scholar]
- Bulchand S, Grove EA, Porter FD, Tole S (2001) LIM-homeodomain gene Lhx2 regulates the formation of the cortical hem. Mech Dev 100: 165-175. [DOI] [PubMed] [Google Scholar]
- Bulchand S, Subramanian L, Tole S (2003) Dynamic spatiotemporal expression of LIM genes and cofactors in the embryonic and postnatal cerebral cortex. Dev Dyn 226: 460-469. [DOI] [PubMed] [Google Scholar]
- Campbell K (2003) Dorsal-ventral patterning in the mammalian telencephalon. Curr Opin Neurobiol 13: 50-56. [DOI] [PubMed] [Google Scholar]
- Ding YQ, Marklund U, Yuan W, Yin J, Wegman L, Ericson J, Deneris E, Johnson RL, Chen ZF (2003) Lmx1b is essential for the development of serotonergic neurons. Nat Neurosci 6: 933-938. [DOI] [PubMed] [Google Scholar]
- Foster GA (1998) Chemical neuroanatomy of the prenatal rat brain. Oxford: Oxford UP.
- Jurata LW, Pfaff SL, Gill GN (1998) The nuclear LIM domain interactor NLI mediates homo- and heterodimerization of LIM domain transcription factors. J Biol Chem 273: 3152-3157. [DOI] [PubMed] [Google Scholar]
- Kania A, Jessell TM (2003) Topographic motor projections in the limb imposed by LIM homeodomain protein regulation of Ephrin-A:EphA interactions. Neuron 38: 581-596. [DOI] [PubMed] [Google Scholar]
- Lundgren SE, Callahan CA, Thor S, Thomas JB (1995) Control of neuronal pathway selection by the Drosophila LIM homeodomain gene apterous. Development 121: 1769-1773. [DOI] [PubMed] [Google Scholar]
- Marin O, Anderson SA, Rubenstein JL (2000) Origin and molecular specification of striatal interneurons. J Neurosci 20: 6063-6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Garcia F, Martinez-Marcos A, Lanuza E (2002) The pallial amygdala of amniote vertebrates: evolution of the concept, evolution of the structure. Brain Res Bull 57: 463-469. [DOI] [PubMed] [Google Scholar]
- McDonald AJ (2003) Is there an amygdala and how far does it extend? An anatomical perspective. Ann NY Acad Sci 985: 1-21. [DOI] [PubMed] [Google Scholar]
- Milan M, Cohen SM (1999) Regulation of LIM homeodomain activity in vivo: a tetramer of dLDB and apterous confers activity and capacity for regulation by dLMO. Mol Cell 4: 267-273. [DOI] [PubMed] [Google Scholar]
- Nakagawa Y, O'Leary DD (2001) Combinatorial expression patterns of LIM-homeodomain and other regulatory genes parcellate developing thalamus. J Neurosci 21: 2711-2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nery S, Fishell G, Corbin JG (2002) The caudal ganglionic eminence is a source of distinct cortical and subcortical cell populations. Nat Neurosci 5: 1279-1287. [DOI] [PubMed] [Google Scholar]
- O'Keefe DD, Thor S, Thomas JB (1998) Function and specificity of LIM domains in Drosophila nervous system and wing development. Development 125: 3915-3923. [DOI] [PubMed] [Google Scholar]
- Pitkanen A (2003) Connectivity of the rat amygdaloid complex. In: The amygdala, A functional analysis (Aggleton J, ed), pp 31-117. Oxford: Oxford UP.
- Puelles L, Kuwana E, Puelles E, Bulfone A, Shimamura K, Keleher J, Smiga S, Rubenstein JL (2000) Pallial and subpallial derivatives in the embryonic chick and mouse telencephalon, traced by the expression of the genes Dlx-2, Emx-1, Nkx-2.1, Pax-6, and Tbr-1. J Comp Neurol 424: 409-438. [DOI] [PubMed] [Google Scholar]
- Shirasaki R, Pfaff SL (2002) Transcriptional codes and the control of neuronal identity. Annu Rev Neurosci 25: 251-281. [DOI] [PubMed] [Google Scholar]
- Stenman J, Yu RT, Evans RM, Campbell K (2003) Tlx and Pax6 co-operate genetically to establish the pallio-subpallial boundary in the embryonic mouse telencephalon. Development 130: 1113-1122. [DOI] [PubMed] [Google Scholar]
- Swanson LW, Petrovich GD (1998) What is the amygdala? Trends Neurosci 21: 323-331. [DOI] [PubMed] [Google Scholar]
- Tsuchida T, Ensini M, Morton SB, Baldassare M, Edlund T, Jessell TM, Pfaff LS (1994) Topographic organization of embryonic motor neurons defined by expression of LIM homeobox genes. Cell 79: 957-970. [DOI] [PubMed] [Google Scholar]
- Vyas A, Saha B, Lai E, Tole S (2003) Paleocortex is specified in mice in which dorsal telencephalic patterning is severely disrupted. J Comp Neurol 466: 545-553. [DOI] [PubMed] [Google Scholar]
- Yun K, Potter S, Rubenstein JL (2001) Gsh2 and Pax6 play complementary roles in dorsoventral patterning of the mammalian telencephalon 128: 193-205. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Marin O, Hermesz E, Powell A, Flames N, Palkovits M, Rubenstein JL, Westphal H (2003) The LIM-homeobox gene Lhx8 is required for the development of many cholinergic neurons in the mouse forebrain. Proc Natl Acad Sci USA 100: 9005-9010. [DOI] [PMC free article] [PubMed] [Google Scholar]