Abstract
Two inbred rat lines have been developed that show either high (HAB) or low (LAB) anxiety-related behavior. The behavioral phenotype correlates with arginine vasopressin (AVP) expression at the level of the hypothalamic paraventricular nucleus (PVN), but not supraoptic nucleus, with HAB animals overexpressing the neuropeptide in both magnocellular and parvocellular subdivisions of the PVN. We detected a number of single nucleotide polymorphisms (SNPs) in the AVP locus that differ between the HAB and LAB animals, two of which were embedded in cis-regulatory elements. The HAB-specific allele of the AVP gene promoter occurs in 1.5% of outbred Wistar rats and is more transcriptionally active in vivo, as revealed by allele-specific transcription studies in cross-mated HAB/LAB F1 animals. Interestingly, one specific SNP [A(-1276)G] conferred reduced binding of the transcriptional repressor CArG binding factor A (CBF-A) in the HAB allele, the consequent differential regulation of the AVP promoter resulting in an overexpression of AVP in vitro and in vivo. Furthermore, CBF-A is highly coexpressed in AVP-containing neurons of the PVN supporting an important role for regulation of AVP gene expression in vivo. Taken together, our results demonstrate a role for an AVP gene polymorphism and CBF-A in elevated AVP expression in the PVN of HAB rats likely to contribute to their behavioral and neuroendocrine phenotype.
Keywords: anxiety, depression, common variant/common disease hypothesis, arginine vasopressin, hypothalamic-pituitary-adrenal axis, CBF-A
Introduction
Anxiety disorders and major depression represent two of the most common and debilitating psychiatric disorders, and despite decades of intensive research, the cause of these remain elusive (Nestler et al., 2002). Functional studies have yet to identify a focal brain region or particular neurotransmitter system as the primary site of the abnormality. Furthermore, genetic studies have failed to generate replicable evidence for specific culpability genes (Segurado et al., 2003) and allelic variations that show robust associations in most populations, nor have they explained a major fraction of the risk (Lohmueller et al., 2003).
To gain more insight into the molecular mechanisms underlying anxiety and depression, we have established an animal model by selectively and bidirectionally breeding Wistar rats for either high (HAB) or low (LAB) anxiety-related behavior (Wigger et al., 2004). Trait selection leads to homozygosity at loci conferring trait anxiety and represents a powerful strategy to identify genes underlying complex behaviors. Based on previous experimental evidence, the neuropeptide arginine vasopressin (AVP) plays a major role in behavioral and neuroendocrine features typical of trait anxiety and comorbid depression in the HAB line. First, AVP mRNA expression, as well as release of the neuropeptide, are higher in the paraventricular nucleus (PVN) of HAB rats compared with LAB rats. Second, intra-PVN treatment with an AVP V1 receptor antagonist resulted in a decrease in anxiety/depression-related behavior (Wigger et al., 2004). Third, both the hyper-reactive hypothalamic-pituitary-adrenal axis and the pathological outcome of the dexamethasone-corticotropin-releasing hormone challenge test in HAB animals are driven by AVP overexpression and release (Keck et al., 2002). Moreover, treatment with the antidepressant paroxetine normalized AVP mRNA expression, the hypothalamic-pituitary-adrenal axis, and depression-like behavior but was without any effect in LAB rats (Keck et al., 2003). Therefore, to pursue further this issue and to gain insight into the molecular mechanism underlying the differential expression and release of AVP in the HAB and LAB lines, we decided to investigate the AVP locus as a candidate gene for AVP overexpression.
We demonstrate here that the HAB-specific allele of the AVP gene promoter contains a number of single nucleotide polymorphisms (SNPs); one in particular [A(-1276)G] disrupts cognate binding of the transcriptional repressor CArG binding factor A (CBF-A), consequently leading to AVP overexpression.
Materials and Methods
Animals. All HAB and LAB rats tested were bred in the animal facilities of the Max-Planck-Institute of Psychiatry in Munich, Germany (Wigger et al., 2004). Briefly, inbred HABs and LABs were selected based on the results obtained in the elevated plus-maze test at the age of 10 weeks. This test is based on creating a conflict between the rat's exploratory drive and its innate fear of open and exposed areas. The elevated plus-maze consists of a plus-shaped platform with two closed (with 40 cm walls) and two open arms, connected at the center by a neutral zone and elevated 73 cm above the floor. During the 5 min exposure, the following main parameters were recorded by a video/computer system: (1) percentage of time spent on the open arms; (2) number of entries into open and closed arms; and (3) latency to the first entry into an open arm (Liebsch et al., 1998).
DNA analysis. DNA was purified from the tails of rats using the NucleoSpin Tissue kit (Macherey-Nagel). Plasmids and PCR products were sequenced using the ThermoSequenase Cycle sequencing kit (Pharmacia).
Reporter and cDNA constructs. Fragments of the rat AVP gene were generated by PCR from genomic DNA purified from either a HAB or LAB animal. AVP promoter fragments were inserted into the polylinker site of pGL2, a promoterless luciferase expression vector (Promega, Madison, WI). A proximal promoter fragment of 1146 bp containing sequence -860 bp (-775 bp after digestion) to +282 bp relative to the start codon was amplified (forward primer, ACATCCACTTCCCTCCTGC; reverse primer, CGTTCTGGGAAACCCATCTA). To include additional upstream sequences, genomic DNA was amplified (forward primer, GTACCCCATCCACACAGACC; reverse primer, ATATAAGATAACTGCTTCCTTC) to give a product of 3119 bp harboring 2797 bp upstream from the AVP start codon. Pfu polymerase (NEB) was used in an amplification reaction of 15 cycles at 94°C for 40 sec, 58°C for 40 sec, and 72°C at 40 sec, followed by 20 cycles at 94°C for 40 sec, 54°C for 40 sec, and 68°C for 2 min, ending with 68°C for 20 min. Both PCR products were purified using Qiagen (Hilden, Germany) spin columns; the proximal promoter fragment was digested with KpnI and ScaI to give 931 bp, and the distal one by KpnI and HincII (Fermentas) giving a 2950 bp product. After separation by agarose gel electrophoresis, the proximal and distal promoter fragments, from the HAB and LAB animals, were cloned into the pBSKII vector digested with KpnI and EcoRV (Fermentas). Recombinant plasmids were sequence verified, and inserts were released using KpnI and SmaI and then inserted into the pGL2 vector digested with KpnI and SmaI.
Vectors containing various copies of the CArG elements from either the HAB or LAB alleles were generated by serially cloning oligonucleotides containing the CArG box (LAB sense, ctctagagagctcccactgtCCTTATTAGGaagaggcagct; HAB sense, ctctagagagctcccactgtCCTTATTGGGaagaggcagct) into the KpnI and SacI site of the pGL2.TATA vector upstream of the TATA element derived from E1B (Hoffmann et al., 2003). The pRK7-FLAG.CBF-A expression vector was made by cloning the mouse cDNA for CBF-A (a kind gift from Drs. Akira Inoue and Shinji Kamada, Department of Oncogene Research, Osaka University, Osaka, Japan) into the pRK7 vector downstream the cytomegalovirus promoter. CBF-A was first released from pSR-CBF-A-N (Kamada and Miwa, 1992) using Tth111I and SacI, after which the ends were blunted by T4 DNA polymerase and cloned into the SmaI site of pBSKII. The CBF-A fragment was further released by a partial BamHI digest and EcoRI digest and ligated into the BamHI and EcoRI sites of pRK7-FLAG. pGEX-2TKCBF-A was made by releasing CBF-A from pBSKII, by a partial BamHI and EcoRI digest and inserting into pGEX-2TK (Pharmacia) digested with BamHI and EcoRI. The series of CBF-A deletion constructs were a kind gift from Drs. Thomas Leanderson and Alaitz Aranburu (Section of Immunology, Department of Cell and Molecular Biology, Lund University, Lund, Sweden). These constructs contain deletions of either, or both, of the RNA-binding domains (RBDs) (Bemark et al., 1998).
Allele-specific transcription in vivo. RNA was extracted from the PVN of the hypothalamus of F1 animals, heterozygous for the two AVP promoter alleles, using the acidic phenol method. Oxytocin heterogeneous RNAs (hnRNAs) and AVP hnRNAs were reverse transcribed and amplified using gene-specific primers (OneStep RT-PCR kit; Qiagen). Primer pairs were designed to anneal to the RNA from oxytocin and the pre-spliced hnRNA from AVP. The forward primer for oxytocin locates to the 5′ untranslated region (ATCTCGGACTGAACACCAACGC) and the gene-specific primer to the first intron (CTGTGCACAATCCATATCGGGAC). The forward primer and the gene-specific primer for AVP derived from sequences in exon 1 and the first intron, respectively (forward, CCGACATGGAGCTGAGAC; reverse, TGAGCCTAGTGACTGGATTCCC). PCR was performed under the following conditions: cDNA synthesis at 50°C for 30 min and 94°C for 15 min; amplification for 20 cycles at 94°C for 30 sec, 55°C for 30 sec, and 68°C for 30 sec. An additional 20 cycles of PCR were performed on 1 μl of the previous reaction using a nested primer and Pfu polymerase (NEB). Products were purified and cloned into the EcoRV site of the vector pBSKII (Stratagene, La Jolla, CA). After transformation into DH5α bacteria, DNA was isolated from single colonies (Nucleospin miniprep kit; Macherey-Nagel) and sequenced to track the allele from which the RNA was transcribed.
Cell culture and transfection experiments. Human osteosarcoma-like cells, Saos-2 (ATTC HTB-85), were grown in DMEM supplemented with 10% fetal calf serum. Human small cell lung cancer cells, Lu-165 (a kind gift from Dr. Takeo Terasaki, Pathology Division, National Cancer Center Hospital, Tokyo, Japan), were grown in RPMI 1640 with l-glutamine supplemented with 10% fetal bovine serum. Saos-2 cells were seeded 12 hr before electroporation. They were harvested and resuspended in 1× electroporation buffer (EP) (Hoffmann et al., 2003); 50 μl of this mixture, containing 1 × 10 6cells, was added to a 100 μl mixture containing 1 μg of AVP-luciferase vector, 0.2 μg of pRK7.β-galactosidase, 4 μl of MgSO4 (1 m), and 3 μg of pGEM4 filling DNA in 1× EP buffer, which was then subjected to electroporation. Lu-165 cells were transfected as described previously (Coulson et al., 1999). Luciferase activity was measured and standardized on β-galactosidase values as described previously (Hoffmann et al., 2003).
In vitro DNA double-strand assay. Between 1 and 10 ng of glutathione S-transferase (GST)-CBF-A or GST protein were incubated with 20,000 cpm of end-labeled LAB probe for 30 min at room temperature. Fifty units of S1 nuclease (Amersham) were added to the sample, which was then incubated for 90 sec at room temperature. The reaction was stopped by incubating for 1 min at room temperature with 1 μl of proteinase K (100 μg/ml; Sigma, St. Louis, MO). An equal volume of formamide loading buffer was added, and the whole sample was boiled for 5 min to denature the DNA. The samples were then placed immediately on ice for 1 min, heated at 72°C for 2 min, then separated on a denaturing 8% polyacrylamide sequencing gel and exposed to x-ray film (Fujifilm).
In situ hybridization. To generate a CBF-A riboprobe, the plasmid pBSK-N-CBF-A, containing the 326 bp N-terminal sequence of the CBF-A cDNA, was linearized with EcoRV and XbaI to produce antisense and sense probes. In situ hybridization reactions were performed as described previously (Cota et al., 2003).
Immunohistochemistry. Immunohistochemistry was performed as described previously (Salchner and Singewald, 2002). Double immunofluorescence staining was used to visualize CBF-A and AVP immunoreactivity simultaneously. Vibratome sections (50 μm) were preincubated for 30 min in immunobuffer containing 5% normal donkey serum (Jackson ImmunoResearch, West Grove, PA) to block nonspecific binding sites. After washing in immunobuffer, sections were incubated for 48 hr at room temperature in a polyclonal rabbit antibody to CBF-A (dilution, 1:1000) and a polyclonal guinea pig antibody to AVP (GHC 8103; dilution, 1:10,000; Peninsula, Belmont, CA) in immunobuffer containing 5% normal donkey serum. The anti-N-CBF-A antibody, raised in rabbit toward the first low conserved 75 amino acids of the N-terminal part of CBF-A, was a generous gift from Drs. T. Leanderson and A. Aranburu. After rinsing in immunobuffer, sections were exposed for 2.5 hr at room temperature to the species-specific fluorophore-conjugated secondary antibodies [Cy2-conjugated donkey anti-rabbit IgG (dilution, 1:100), Cy3-conjugated donkey anti-guinea pig IgG (dilution, 1:400), and 7-amino-4-methylcoumarin-3-acetic acid (AMCA)-conjugated donkey anti-guinea pig IgG (dilution, 1:100); Jackson ImmunoResearch] diluted in immunobuffer containing 1% normal donkey serum. After rinsing in 50 mm Tris-HCl, pH 7.4, sections were mounted onto slides and dried overnight. Mounted sections were cleared in 100% ethanol and Histo-clear (National Diagnostics, Atlanta, GA) and coverslipped. Negative controls were performed by omission of the primary antibodies. No staining was observed in any of these control sections. Images of AVP (see below) and double-labeling experiments were recorded on a fluorescence microscope (Olympus BX-51) equipped with adequate filter systems using a digital camera (Olympus DP50) and analySIS image processing software (Soft Imaging Systems, Münster, Germany). Original fluorescence signals from Cy2 and Cy3 in double-labeling experiments were maintained, whereas a pseudocolor (green) was used for AMCA to facilitate evaluation of double labeling.
For correlation analyses, five HAB, five NAB (Wistar controls with normal anxiety-related behavior) and six LAB animals were phenotyped on the elevated plus-maze and killed 2 d later for AVP immunohistochemistry. Semiquantification analysis was performed by means of image analysis using optical software (Optimas 5.2; Optimas Corp., Silver Spring, MD).
Results
Identification of SNPs in the AVP gene promoter of HAB rats
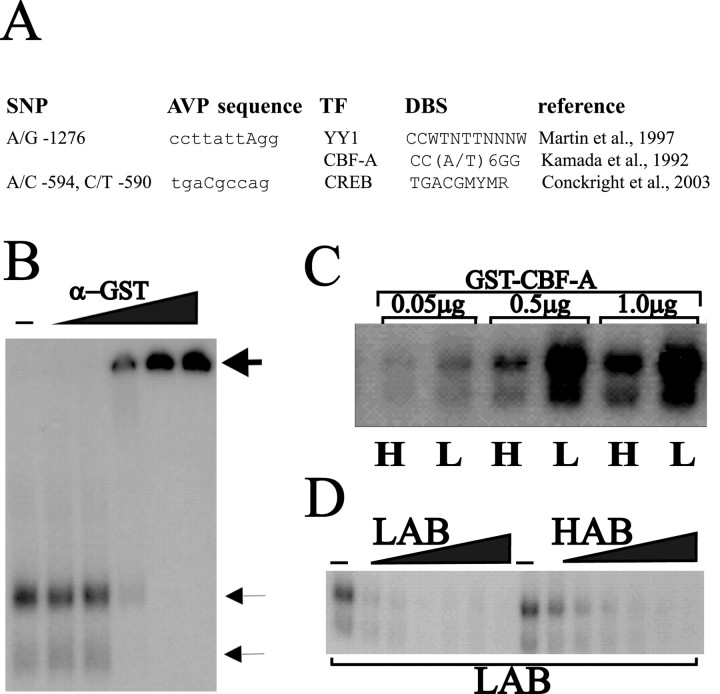
Sequencing of the AVP locus revealed 10 SNPs within a region of 2.7 kb of the AVP promoter sequence differing between the HAB and LAB animals (Fig. 1A). In the AVP gene itself, a single base pair substitution was found in the first intron [T(+549)C], and one more SNP was identified in the sequence 245 bp downstream of the AVP gene. HAB animals were homozygous for all these SNPs, whereas LAB animals were homozygous for the published nucleotide sequence (AF112362). This led us to ask whether LAB animals represent the common wild-type allele and HAB animals a less frequent allele. To clarify this issue, we tested 100 outbred rats (Charles River, Sulzfeld, Germany) for the presence of these polymorphisms. Three were found to be heterozygous, indicating a gene frequency of 1.5% for the HAB allele in the outbred Wistar rat population. Selective inbreeding enriches for genetic information associated with a particular trait, which shifts the animals' phenotype from the population mean (Falconer and Mackay, 1996). Therefore, our data suggest that inbreeding selected for this HAB allele, which in turn might contribute to increased anxiety- and depression-like behavior in this line. Indeed, all three heterozygous rats showed a twofold higher anxiety-related behavior on the elevated plus-maze than the population mean (7.2% vs 13.6 ± 1.5% time spent on the open arms of the elevated plus-maze; data not shown).
Figure 1.
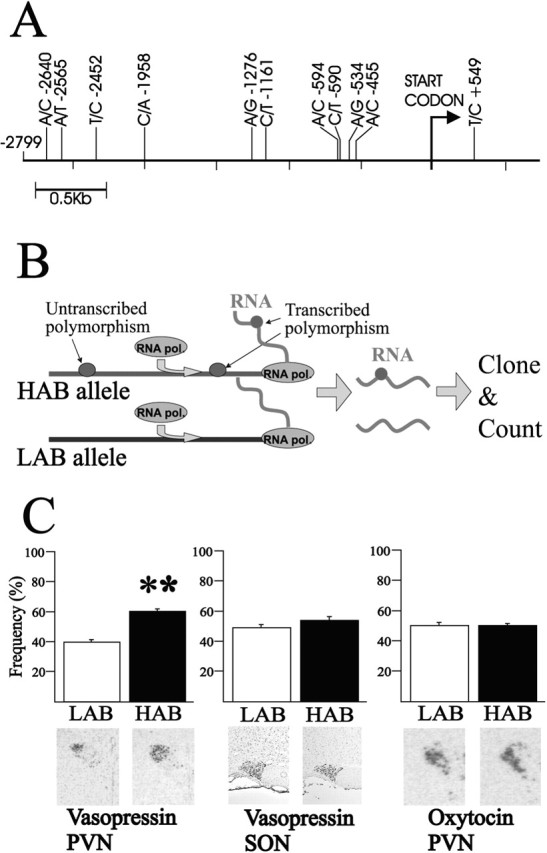
SNPs in the HAB/LAB AVP promoter confer differential allele-specific transcription in vivo. A, Scheme of the rat AVP promoter region. This region, containing 2799 bp, harbors 10 SNPs. The T(+549)C polymorphism, which was used to determine allelic-specific transcription in vivo, locates in intron 1. B, Scheme of the allele-specific transcription method. A heterozygote animal contains a promoter allele from both the HAB and LAB lines expressed in the same PVN neurons. A transcribed polymorphism localized to the same allele as a putative regulatory polymorphism in the promoter can be used to track the promoter allele from which the mRNA was transcribed. RNA extracted from the PVN is used in reverse transcription-PCR and subcloning, and sequencing of the resulting products yields a precise measure of the efficiency by which either allele was transcribed in vivo. C, Allelic transcription rate for AVP in the PVN evidenced a 60:40 ratio in favor of the HAB allele. In situ hybridization data from the HAB and LAB PVN also demonstrate higher levels of AVP mRNA in the HAB animal, which correlates well with the higher activity of the AVP promoter. **p < 0.005. D, Allelic transcription rate for oxytocin in the PVN demonstrated a 50:50 ratio for the oxytocin transcription rates, in agreement with the in situ hybridization data demonstrating no difference between the HAB and LAB lines.
HAB allele confers increased AVP transcription in vivo
The above findings raised the question of whether AVP overexpression in the PVN may be attributable to regulation in cis, caused by the polymorphisms in the regulatory region of the AVP gene, or, alternatively, attributable to regulation in trans resulting from different cellular environments in the PVN of the HAB and LAB animals, respectively. To discern between these possibilities, we studied both promoter alleles within the same cellular environment. This is the case for a heterozygote animal derived from crossing homozygous HAB and LAB lines. As schematically shown in Figure 1B, the representation of polymorphisms transcribed by the coding part of the gene is a result of the transcriptional activity of the respective promoter allele, which in turn is critically dependent on the presence of regulatory polymorphisms in the promoter. Therefore, by extracting RNA from the PVN of a heterozygote animal and cloning the reverse transcribed-specific cDNA, it is possible to determine the frequency of plasmid clones containing either transcribed polymorphism and to track precisely which RNA species is more prevalent in vivo. The PVN of two male and two female F1 heterozygote rats was used to test for the expression of AVP and oxytocin as a control. We sequenced 100 randomly picked plasmid clones harboring the respective cDNA from each animal. The AVP data show a distinct difference between the two alleles (p < 0.005), with the HAB AVP promoter showing a 50% increase in activity (Fig. 1C). Interestingly, when the experiment was performed in the supraoptic nucleus (SON) of F1 animals, the differential expression disappeared, supporting previous data for a strict nucleus-specific regulation of the AVP gene in the HAB animals. As a control for the in vivo transcription experiment, we chose the oxytocin gene, which localizes 11 kb away from the AVP locus. We measured an equal contribution of the HAB and LAB oxytocin alleles, corresponding well with previous data showing no significant line-specific differences in expression in the PVN (Wigger et al., 2004). Evidently, this difference in AVP in the heterozygous rat will enlarge under the homozygous condition present in the HAB animal, leading us to conclude that the differences in the expression of AVP in vivo are, predominantly, attributable to differences in cis regulation of the AVP promoter allele. This further prompted us to study exactly which SNP(s) could be key to this regulation.
The SNP A(-1276)G in the CArG element of the HAB allele reduces CBF-A binding
We addressed this issue by first analyzing each SNP to identify those that were embedded in putative DNA-binding sites using the transcription element search system program (http://www.cbil.upenn.edu/tess). We then correlated these findings with a detailed survey of the literature regarding the expression of regulatory factors in the PVN of the rat hypothalamus. Based on these criteria, the SNPs listed in Figure 2A appeared to be of potential interest. With respect to the putative cAMP-responsive element (CRE) site, present solely in the LAB animal C(-590)T allele, our in vitro studies (see below) failed to evidence any difference in basal or in cAMP-dependent AVP promoter activity between the LAB and the HAB promoters. This finding was not unexpected because the AVP promoter contains at least two additional CREs (Iwasaki et al., 1997), which suggests that this putative distal CRE site does contribute to CRE-dependent expression of the AVP promoter. In further support of this view, a recent genome-wide survey suggested that functional CREs are preferentially located more proximal (Conkright et al., 2003). More interestingly, the A to G substitution 1276 bp upstream from the AVP start codon is predicted to disrupt a CArG element (Treisman, 1987) in the HAB promoter. This cis-regulatory element, also known as a serum response element (SRE), is bound by serum response factor (SRF) (Treisman, 1987), YY1 (Martin et al., 1997), and CBF-A (Kamada and Miwa, 1992). SRF, although expressed in the brain, is noticeably absent from the PVN (Blume et al., 1998). YY1 bound to the AVP CArG element 100-fold less efficient when compared with a known consensus site (Hyde-DeRuyscher et al., 1995), and, more importantly, the SNP failed to effect this binding in an electrophoretic mobility shift assay (EMSA) (data not shown). In agreement, YY1 weakly repressed the AVP promoter activity in a dose-dependent manner but did not distinguish between the HAB or LAB alleles (data not shown). Because CBF-A is highly expressed in the PVN (see below), we performed additional experiments to determine in detail whether CBF-A binds to the CArG box and whether CBF-A differentiates between the CArG boxes from HAB and LAB alleles. After incubation of a full-length CBF-A-GST fusion protein with the AVP CArG element, two distinct protein-DNA complexes were observed in an EMSA (Fig. 2B). The addition of GST antibody caused a supershift of both the major low mobility and the minor higher mobility complexes. In contrast, preincubation with control IgGs or BSA failed to elicit a supershift attesting to the specificity of the observed DNA-protein complexes (data not shown). In addition, we performed an EMSA with nuclear extracts from Saos-2 cells transfected with a CBF-A expression vector, and again two complexes were seen that were completely shifted by the CBF-A antibody (data not shown). These results show that CBF-A purified from either eukaryote or prokaryote cells binds similarly to the AVP CArG element with high affinity. We next investigated whether CBF-A discriminates between binding to the HAB and LAB CArG element. In fact, we observed, for different concentrations of GST-CBF-A, a strikingly stronger binding to the CArG box derived from the LAB allele when compared with the HAB allele in three independent experiments (Fig. 2C). To substantiate our findings, we performed a competition assay, and in this experiment, the LAB oligonucleotide was clearly more efficient than the HAB oligonucleotide in competing for CBF-A binding, as evidenced by the rapidly decreasing radioactive DNA-protein complex (Fig. 2D).
Figure 2.
CBF-A binds differentially to CArG boxes from LAB and HAB lines. A, SNPs embedded in putative consensus DNA-binding sites (DBS) as predicted by the TESS program. Only those DBS are listed in which the respective transcription factor (TF) is known to be expressed in the PVN. B, Recombinant CBF-A protein binds specifically to the CArG element from the AVP promoter of the LAB line forming two complexes in an EMSA. The major low mobility and minor higher mobility complexes (thin arrows) are both supershifted (thick arrow) with increasing concentrations of GST antibody, as indicated at the top. C, Increasing concentrations of CBF-A reveal a higher affinity for the LAB (L) double-stranded CArG box than for the HAB (H) sequence in EMSA. D, The unlabeled LAB oligonucleotide competes more efficiently than the unlabeled HAB oligonucleotide for CBF-A binding to the LAB sequence, in EMSA, demonstrating that the CArG element from the LAB animal binds CBF-A more efficiently. The results in B-D are representative of three individual experiments.
CBF-A binds to single-stranded DNA element
In the EMSA experiments so far, we have used double-stranded DNA (dsDNA) as a probe, however CBF-A has also been reported to bind to single-stranded DNA (ssDNA) with an even higher affinity (Kamada and Miwa, 1992), which might be important to its function in RNA metabolism and additionally to its function in transcriptional regulation (see below). To address this topic, and to obtain additional insight into the consequences of CBF-A binding to the CArG-box element of the AVP promoter, we competed the LAB probe with increasing concentrations of double-stranded self-oligonucleotide or, alternatively, with increasing concentrations of single-stranded sense or antisense self-oligonucleotide. As shown in Figure 3A, the ssDNA probes were clearly more efficient in this assay than the double stranded. Of additional interest was the finding that CBF-A bound to the antisense probe with higher affinity than the sense probe, showing an additional level of specificity in the recognition of the CArG element. In view of the high affinity of CBF-A for ssDNA, we examined further the possibility that CBF-A could induce or stabilize single-stranded regions within the AVP promoter CArG element. To test this hypothesis, we used the specificity of S1 nuclease to preferentially digest ssDNA (Fig. 3B). CBF-A protein was incubated with the double-stranded CArG element, after which S1 nuclease was added, cleaving any ssDNA regions rapidly into mononucleotides. Therefore, in case CBF-A supports the formation of any ssDNA regions in the dsDNA CArG probe, S1 nuclease cleavage will be enhanced, as evidenced by the appearance of mononucleotides at the expense of dsDNA. Different concentrations of CBF-A were supplemented with the GST protein alone to keep total protein concentrations in each reaction constant. The results in Figure 3C show that the increase of S1 nuclease-mediated degradation correlates with the amount of CBF-A and not with GST alone. This suggests that CBF-A enhances melting of the dsDNA CArG box. We further tested to which extent this DNA melting is dependent on DNA binding of CBF-A by the RBDs by using a series of CBF-A deletion constructs lacking either or both of the RBDs (Fig. 3D). Previous work has shown that the N terminus and C-terminal regions of the CBF-A protein have little or no effect on DNA binding (Bemark et al., 1998), which in contrast critically depends on the central RBDs. The EMSA experiment in Figure 3E supports these findings in demonstrating that the presence of the RBD domains of CBF-A are crucial for the DNA binding. Specifically, however, our results support the notion that RBD1 of CBF-A appears to be of particular importance for DNA binding. The results of the S1 nuclease assay revealed that DNA binding strictly correlated with S1 nuclease sensitivity because the CBF-A protein containing a deletion of both RBDs is unable to enhance S1-mediated DNA digestion (Fig. 3F). The recombinant CBF-A proteins lacking either one of the RBDs, however, both increased S1 nuclease sensitivity, suggesting that despite reduced DNA binding in the absence of one of the RBDs, the proteins were still able to efficiently promote S1 nuclease sensitivity.
Figure 3.
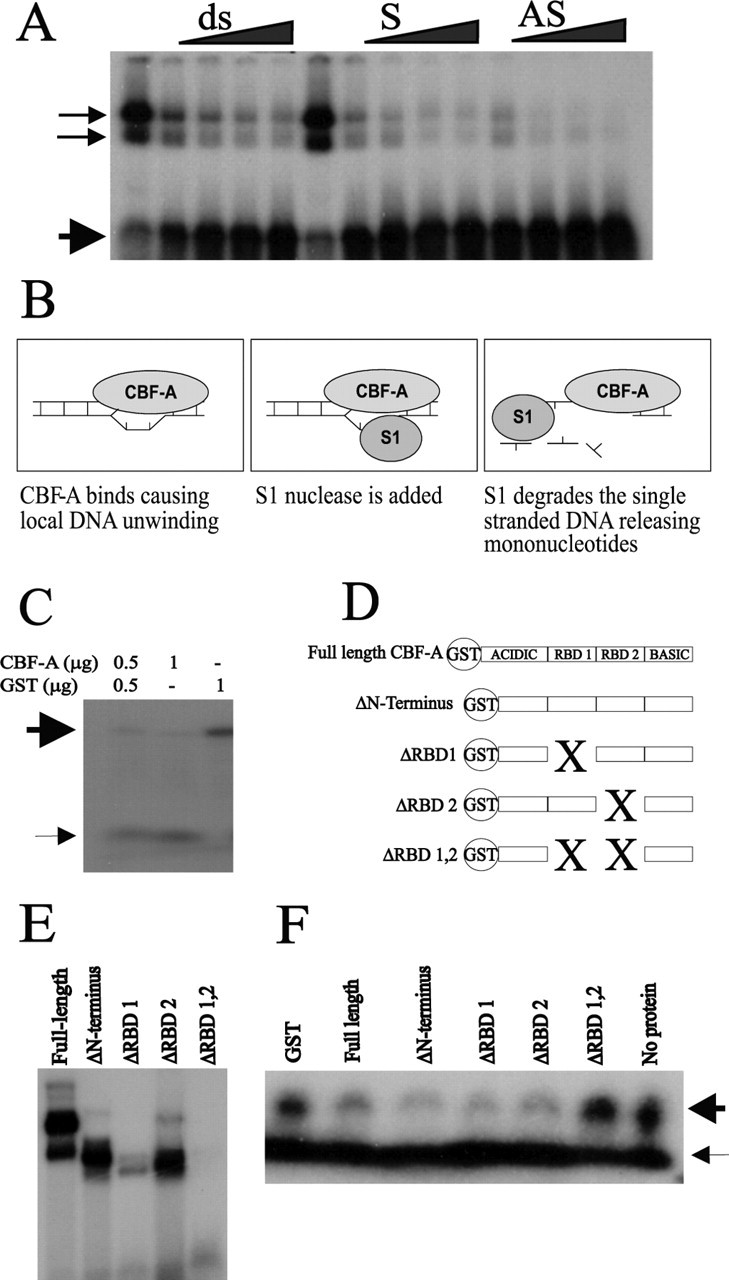
High-affinity binding of CBF-A to single-stranded CArG box causes local DNA strand separation. A, Sense (S) and antisense (AS) DNA probes of the LAB CArG compete more efficiently for CBF-A DNA binding than the dsDNA element (ds). Additionally, CBF-A DNA binding to the dsDNA CArG box is preferentially competed with the antisense DNA of the CArG element. The thin arrows denote probe bound by protein, and thick arrows denote the free probe. B, Scheme of the S1 nuclease assay for the detection of dsDNA melting. CBF-A binding to dsDNA is predicted to cause local strand separation and to confer increased sensitivity to S1 nuclease, which cleaves the DNA into mononucleotides. C, An S1 nuclease assay reveals that increasing amounts of CBF-A cause dsDNA cleavage into mononucleotides (thin arrow), whereas GST alone causes no cleavage of the dsDNA probe (thick arrow). D, Scheme of RBD deletion mutants. The two RBDs, labeled RBD 1 and RBD 2, localize to the central part of CBF-A and are flanked by an acidic N-terminal and a basic C-terminal region. E, The RBD motifs are crucial for DNA binding as evidenced in EMSA by full-length CBF-A DNA binding compared with the ΔRBD 1,2 protein. The presence of RBD 1 is more important for DNA binding than RBD 2. F, The presence of either of the RBD motifs leads to S1 nuclease sensitivity. In contrast, the ΔRBD 1,2 protein lacking DNA binding prevented S1 nuclease-mediated digestion. The results in A, C, E, and F are representative of at least three individual experiments.
HAB CArG element impairs CBF-A-induced repression leading to overexpression of AVP in vitro
Reporter gene assays were used to delineate which regions of the AVP promoter were important for the differential expression seen in the allele-specific transcription in vivo (Fig. 1C) and whether above differences in CBF-A binding translate into different transcriptional activities. Promoter plasmids containing 0.8 kb (proximal) and 2.7 kb (distal) upstream sequences of the HAB and LAB AVP gene revealed no differences in expression between either alleles (Fig. 4A) when tested in the osteosarcoma cell line Saos-2. Similar results were obtained when these constructs were transfected into the mouse neuronal cell line Neuro2a and the AVP expressing small cell lung cancer cell line Lu-165 (Coulson et al., 1999) (data not shown). This was also the case when promoter activity was stimulated by coexpression of the catalytic subunit of protein kinase A (data not shown). Our transfection experiments confirmed, however, that the construct containing the distal promoter shows a third of the activity of the proximal promoter (Fig. 4A), as has been reported previously (Iwasaki et al., 1997), supporting the idea that the region 2.7-0.9 kb upstream from the start codon contains regulatory elements important for repression of the AVP gene. Therefore, the absence of allele-specific differences suggests that either the cell lines do not express the specific transcription factor important for enhanced AVP gene activity in HAB animals, which might be expressed only in the PVN, or that other factors in the cell lines, such as SRF (see below), might even mask the effect of the regulatory SNPs on AVP promoter expression in these experiments. Similarly, both the distal and proximal AVP promoter constructs contain multiple regulatory elements that could override the effects of any single polymorphism in the context of transformed cell lines. CBF-A has been reported to confer both repression (Kamada and Miwa, 1992) and activation (Mikheev et al., 2000) at the CArG box. Therefore, we determined the role of CBF-A in relation to AVP promoter activity by overexpressing CBF-A in the AVP-containing cell line Lu-165 and the Saos-2 cell line. In fact, increasing concentrations of CBF-A repressed promoter activity in both the Lu-165 (Fig. 4B) and Saos-2 (data not shown) cell lines. Interestingly, we consistently measured dose-dependent differences in CBF-A-mediated repression between the HAB and LAB constructs, with the HAB AVP sequence being less prone to repression than the LAB construct at similar CBF-A concentrations. To clarify whether this differential repression of the HAB and LAB constructs might be mediated by CBF-A through the polymorphic CArG element at position -1276, we created promoter constructs carrying only the CArG element, flanked by 8 bp upstream and downstream of the neighboring AVP sequence, in front of a TATA element. First, we wanted to determine whether the CArG element behaves as a regulatory element in such a less complex system; second, whether the polymorphism might influence this function; and third, whether CBF-A confers transcriptional regulation through this element. The CArG element has been well documented as a potent activating sequence (Scott et al., 2002), with SRF being one of the major transcription factors to bind (Treisman, 1987). Expressing the LAB CArG element in osteosarcoma cells, which contain SRF, indeed showed the element to function as a potent enhancer. Moreover, insertion of additional copies of the CArG element in front of the TATA element increased the activity by double and triple the expression (Fig. 4C), in accordance with previous studies (Scott et al., 2002). When comparing the HAB and the LAB CArG elements, however, we observed that the LAB sequence conferred twice the activity of the HAB, demonstrating that the polymorphism affects the function of the CArG element (Fig. 4C).
Figure 4.
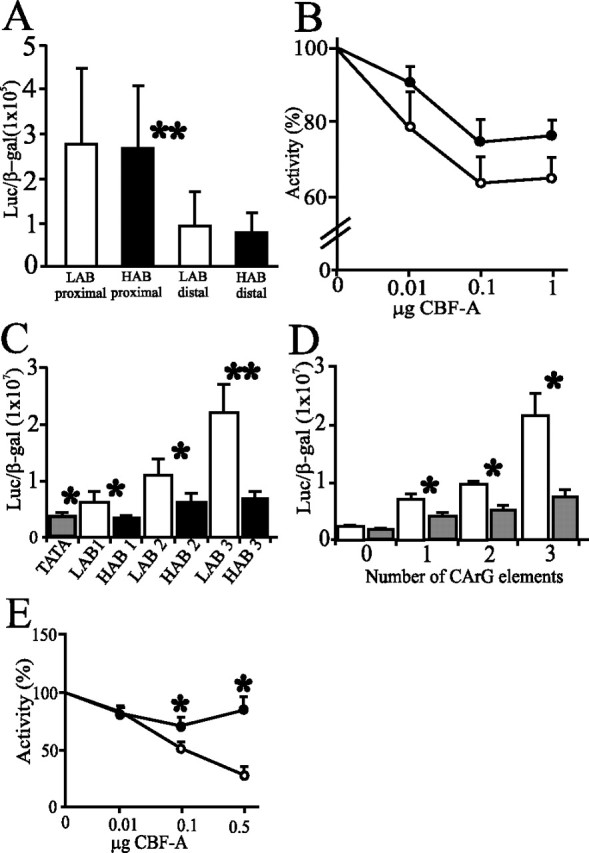
AVP promoter activity in vitro is differentially regulated by CBF-A. A, Cotransfected AVP promoter constructs lack significant differences between HAB and LAB alleles in Saos-2 cells. Note, however, that the activity of the distal promoter is one-third of the activity of the proximal. B, HAB and LAB distal promoters constructs are repressed by increasing concentrations of a cotransfected expression vector for CBF-A (pRK7.CBF-A) in Lu-165 cells. The LAB promoter is more efficiently repressed to a higher degree. C, One, two, or three copies of the CArG element from the LAB allele, inserted in front of a TATA element, conferred potent transactivation when expressed in Saos-2 cells, which is impaired in the presence of the A(-1276)G polymorphism derived from the HAB allele. D, CBF-A represses minimal reporter constructs dependent on the number of CArG elements. Minimal reporter constructs with one, two, or three copies of the LAB CArG box were cotransfected with (filled boxes) or without (open boxes) pRK7.CBF-A (0.1 μg) in Saos-2 cells. E, A minimal reporter construct containing three copies of the LAB CArG element (filled circles) is significantly more repressed than in the case of the HAB element (filled squares) when cotransfected with increasing amounts of pRK7.CBF-A in Lu-165 cells. Data are ±SD. *p < 0.05; **p < 0.005. All results were taken from three individual experiments.
When Saos-2 cells were cotransfected with a constant amount of an expression vector for CBF-A and the LAB-specific minimal CArG promoter plasmids, it was evident that CBF-A repressed promoter activity and that this repression was closely related to the number of CArG elements present (Fig. 4D). Next, we cotransfected increasing amounts of CBF-A with the minimal promoter construct encoding three copies of either the LAB or HAB allele-derived CArG boxes. We observed at the highest doses of CBF-A, a striking fourfold repression of the LAB line-derived CArG box when compared with the HAB line (Fig. 4E). Together, these findings demonstrate that impaired CBF-A binding to the HAB sequence results in a weakened repression of the HAB allele by CBF-A, thus leading to overexpression of AVP.
CBF-A colocalizes with AVP in the PVN
Collectively, our results so far support a role for CBF-A in repression of the AVP gene promoter in vitro and suggest a similar function for CBF-A in vivo. Therefore, we next investigated CBF-A expression in relation to AVP-containing neurons in the PVN. A previous study reported that CBF-A mRNA is intensely expressed in the PVN of the hypothalamus (Rushlow et al., 1999), although it remained unanswered to which extent CBF-A colocalizes with AVP in the different populations of neurons that contribute to the PVN. Double fluorescence immunohistochemistry experiments for CBF-A and AVP expression in the PVN of the hypothalamus evidenced broadly overlapping AVP and CBF-A expression in both the magnocellular and parvocellular subdivisions of the PVN (Fig. 5A). Moreover, AVP and CBF-A intensely colocalized at the cellular level in PVN neurons (Fig. 5B). In addition, data from in situ hybridization and immunohistochemistry revealed no differences in CBF-A expression and content in the PVN between the HAB and LAB animals (data not shown). This finding rules out the possibility that lower levels of CBF-A could underlie the increased AVP expression present in the HAB AVP neurons and further strengthens the importance of the cis-regulatory CArG polymorphism.
Figure 5.
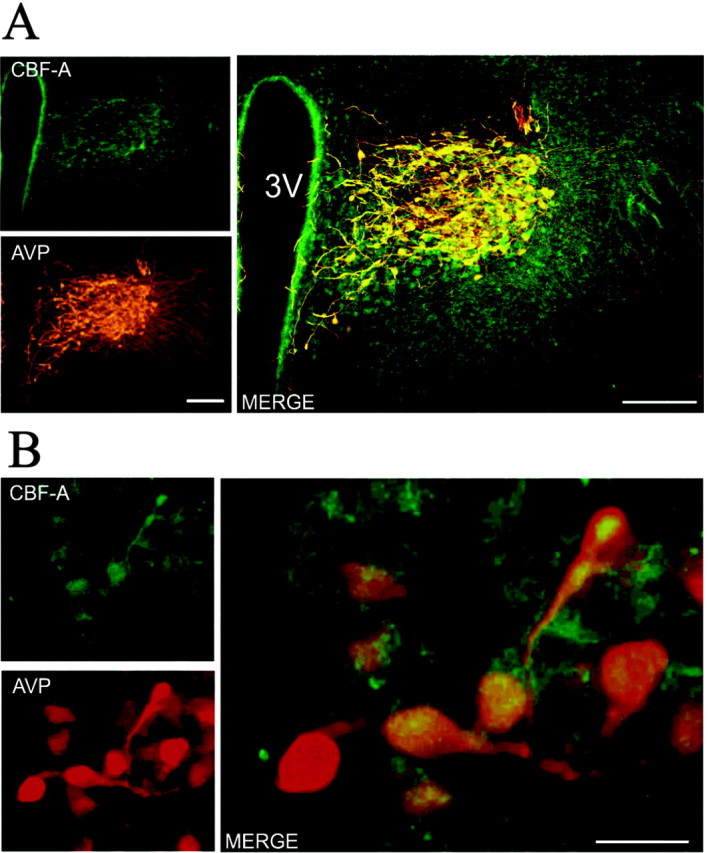
CBF-A and AVP expression broadly overlaps at the hypothalamic level. A, Fluorescence immunohistochemistry for CBF-A (top left) and AVP (bottom left) expression in the rat PVN. A merge of CBF-A and AVP immunofluorescence (right) is shown. Scale bar, 200 μm. B, Cellular colocalization of CBF-A and AVP. Fluorescence immunohistochemistry of CBF-A (top left) and AVP (bottom left) in magnocellular neurons of the rat PVN. A merge of CBF-A and AVP immunofluorescence (right) is shown. Scale bar, 25 μm.
AVP levels in the PVN correlate with anxiety-related behavior
HAB, LAB, and NAB (normal Wistar controls) Wistar rats were phenotyped for their behavior on the elevated plus-maze and further subjected to immunohistochemistry to determine relative amounts of AVP in the parvocellular and magnocellular compartments of the PVN (Fig. 6). In both subdivisions, AVP peptide levels were significantly correlated with anxiety-related behavior (i.e., negatively with percentage of time spent on the open arms of the elevated plus-maze as indicated in Fig. 6A,B).
Figure 6.
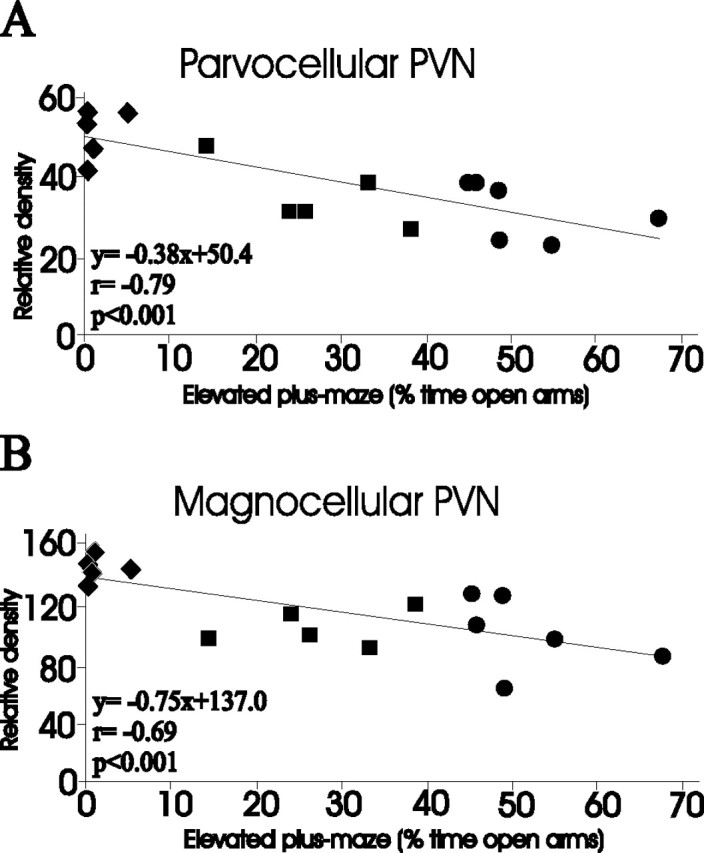
Correlation between AVP immunohistochemistry and anxiety-related behavior. Immunohistochemistry (relative gray density), in parvocellular (A) and magnocellular (B) subdivisions of the PVN, was correlated with anxiety-related behavior (percentage of time spent on the open arms of the elevated plus-maze) in HAB (n = 5; filled diamonds), NAB (Wistar controls; n = 5; filled squares), and LAB (n = 6; filled circles) rats.
Discussion
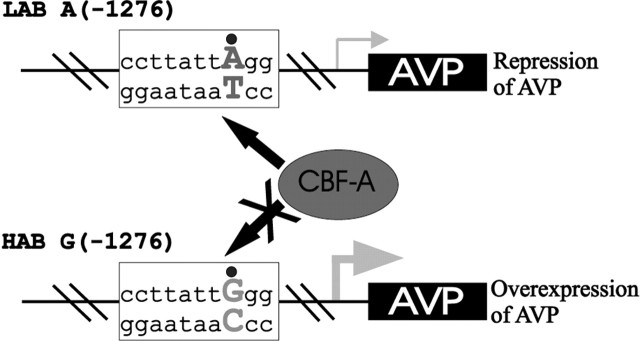
HAB animals have been well documented to show increased AVP mRNA expression, as well as elevated dendritic release of the neuropeptide in the PVN, which correlates with a hyper-reactive hypothalamic-pituitary-adrenal axis, pronounced anxiety, and comorbid depression-like behavior (Keck et al., 2003; Wigger et al., 2004). We now obtained direct evidence for line-specific differences in the sequence of the AVP promoter playing a causal role in this differential AVP expression. The A(-1276)G polymorphism results in decreased binding of CBF-A, leading to impaired transcriptional repression of the AVP gene in HAB rats (Fig. 7). This would result in the observed overexpression of AVP in magnocellular and parvocellular neurons of the PVN under nonstressed basal conditions. In the heterozygous rat PVN, the HAB AVP promoter shows a 50% increased activity compared with the LAB allele, which lacks all of the HAB-specific SNPs. The fact that the allele-specific transcription rate was measured in vivo circumvents the limitations associated with the analysis of cell-specific genes in cultures and adds additional relevance to these findings. This HAB-specific allele occurs with a frequency of 1.5% in outbred Wistar rats, compatible with the common disease-common variant hypothesis (Lohmueller et al., 2003), and supports the idea that trait selection leads to homozygosity at loci conferring trait anxiety/depression. Moreover, we observed that the LAB-specific AVP allele was identical to published sequences from outbred rats in accordance with normal levels of hypothalamic AVP gene expression. Hence, the LAB and HAB lines represent distinct disease entities, with AVP behaving as a vulnerability gene solely in the latter. The line, neuropeptide, and site specificity of AVP overexpression in the PVN of HAB animals is underscored by the findings that (1) the expression of oxytocin, the structurally and functionally related neuropeptide, did not differ between the lines, and (2) AVP expression in the SON, in contrast to the PVN, did not differ between the lines. Additional studies are necessary to identify the mechanism(s) responsible for this nucleus specificity (see below).
Figure 7.
Model for impaired repression at the AVP promoter by the A(-1276)G polymorphism. The A(-1276)G polymorphism impairs binding of the transcriptional repressor CBF-A, resulting in overexpression of AVP in magnocellular and parvocellular neurons of the PVN under nonstressed basal conditions.
Of the 10 SNPs found in the distal promoter of the AVP gene, only two mapped to known consensus DNA-binding sites, of which only the CArG element appears an attractive candidate, regarding differential regulation of the AVP gene. CArG boxes form the core of the SRE, which is bound by the transcription factor SRF (Treisman, 1987), and SREs have been characterized as the major controlling element in a large number of immediate-early genes (IEGs) (for review, see Treisman, 1990). IEG expression in the brain responds to a variety of stimuli including stress, growth factors, mechanical injury, seizures, and ischemia as well as numerous chemical agents and receptor agonists and antagonists. Importantly, CBF-A binds equally well to the CArG element, hereby possibly attenuating gene expression in the refractory period of c-fos induction after repeated stimulation (Rushlow et al., 1999). In support of such a mechanism, transactivation by SRF at the AVP-derived CArG boxes contained in the minimal reporter plasmid was efficiently repressed when CBF-A protein levels were increased in Saos-2 cells. Interestingly, SRF, although moderately expressed in the hypothalamus (Herdegen et al., 1997; Stringer et al., 2002), is distinctly absent in the PVN (Blume et al., 1998). Conversely, CBF-A shows an intense expression in the PVN in this study and in a previous study (Rushlow et al., 1999), in addition to being widely expressed in many other areas of the brain including the olfactory bulb, hippocampus, cerebellum, and hindbrain and peripheral tissues such as the spleen, lung, and thymus. In the PVN, therefore, CBF-A seems to act independently of SRF possibly as a constitutive repressor or alternatively competing with an, as yet unknown, PVN-specific activating transcription factor operating at the CArG box. It should also be further noted that our immunohistochemistry experiments revealed CBF-A expression in the SON (data not shown). This raises the issue why AVP expression does not differ in this nucleus despite the presence of CBF-A and leads us to propose that an additional level of control, perhaps involving posttranslational modification of CBF-A, is at work in the SON of the hypothalamus. Indeed, different isoforms of CBF-A exist, representing either differential phosphorylation or glycosylation (Bemark et al., 1998), and these isoforms appear in cells in response to certain cellular stimuli (Umar et al., 2003). This might explain the differential activity of CBF-A irrelevant of its similar expression/concentration in the PVN and SON, as measured by in situ hybridization and immunohistochemistry (data not shown).
The high affinity binding of CBF-A to the specific ssDNA appears to be the driving force for local DNA strand separation at this element. These alterations in DNA conformation and topology may affect the DNA-binding activities of transactivating factors operating through cis-regulatory elements located within or next to these structures. Such a mechanism has been described for a number of ssDNA-binding proteins (Kelm et al., 2003) and could then be used by CBF-A to modulate the binding of dsDNA-dependent regulators to the CArG element or adjacent sequences.
Although the A(-1276)G SNP clearly underlies AVP overexpression in the PVN of HAB animals, we can only hypothesize that this overexpression is critically involved in the hyperanxious HAB phenotype. First, AVP levels in the magnocellular and parvocellular subdivisions of the PVN correlate significantly with trait anxiety (Fig. 6). Importantly, these immunohistochemistry-based correlations support recent AVP in situ hybridization, AVP microdialysis (Wigger et al., 2004), and Fos expression data (Salome et al., 2004), the latter reflecting neuronal activation in the magnocellular and parvocellular PVN after weak stimulation, being stronger in HAB than in LAB animals. Second, administration of an AVP V1a/b receptor antagonist directly into the PVN of freely behaving HAB rats, via inverse microdialysis, normalized their behavioral peculiarities (Wigger et al., 2004), thus supporting the anxiogenic and depression-inducing potency of the central AVP system (Landgraf et al., 1995; Pitkow et al., 2001; Griebel et al., 2002); similarly, peripheral administration of the antagonist normalized neuroendocrine aberrations in HAB animals (Keck et al., 2002). Third, the mitigating effects of the antidepressant paroxetine on both behavioral and neuroendocrine aberrations of HAB rats were associated with a normalization of AVP expression in both the magnocellular and parvocellular subdivisions of the PVN (Keck et al., 2003). All these findings support the hypothesis that the SNP A(-1276)G underlying AVP overexpression, as described here, contributes to the behavioral and neuroendocrine phenotype of HAB rats. In this context, it is of interest to note the growing literature linking AVP expression to affective disorders (Scott and Dinan, 2002; Millan, 2003).
In conclusion, the HAB-specific AVP promoter represents a natural model for AVP overexpression and highlights, in turn, cognate molecular pathways that potentially fuel the resulting neuroendocrine and behavioral aberrations. Specifically, our finding that the SNP in position 1276 of the AVP gene promoter underlies AVP overexpression in the PVN of HAB rats, makes this SNP a potential target for additional studies aimed at improving therapeutic tools. Our present work exemplifies that selective inbreeding for behavioral traits and combined phenotypic and molecular analyses of candidate genes is an important tool to address these issues and to elucidate molecular mechanisms underlying neuropeptide expression in distinct brain areas, anxiety-related behavior, and comorbid depression.
Footnotes
This work was supported by the Deutsche Forschungsgemeinschaft (SP 386/4-1). We are very thankful to A. Hoffmann and M. Zimbelmann for technical support. The generous gifts of CBF-A cDNA, CBF-A antibody, and Lu165 cells were from A. Inoue, S. Kamada, A. Aranburu, T. Leanderson, T. Terasaki, and J. Coulson. Advice from R. Treisman was also appreciated.
Correspondence should be addressed to Dr. Dietmar Spengler, Department of Molecular Neuroendocrinology, Max-Planck-Institute of Psychiatry, Kraepelinstrasse 2-10, 80804 Munich, Germany. E-mail: spengler@mpipsykl.mpg.de.
Copyright © 2004 Society for Neuroscience 0270-6474/04/247762-09$15.00/0
References
- Bemark M, Olsson H, Heinegard D, Leanderson T (1998) Purification and characterization of a protein binding to the SP6 kappa promoter. A potential role for CArG-box binding factor-A in kappa transcription. J Biol Chem 273: 18881-18890. [DOI] [PubMed] [Google Scholar]
- Blume A, Seifert K, Lebrun CJ, Mollenhoff E, Gass P, Unger T, Herdegen T (1998) Differential time course of angiotensin-induced AP-1 and Krox proteins in the rat lamina terminalis and hypothalamus. Neurosci Lett 241: 87-90. [DOI] [PubMed] [Google Scholar]
- Conkright MD, Canettieri G, Screaton R, Guzman E, Miraglia L, Hogenesch JB, Montminy M (2003) TORCs: transducers of regulated CREB activity. Mol Cell 12: 413-423. [DOI] [PubMed] [Google Scholar]
- Cota D, Marsicano G, Tschop M, Grubler Y, Flachskamm C, Schubert M, Auer D, Yassouridis A, Thone-Reineke C, Ortmann S, Tomassoni F, Cervino C, Nisoli E, Linthorst AC, Pasquali R, Lutz B, Stalla GK, Pagotto U (2003) The endogenous cannabinoid system affects energy balance via central orexigenic drive and peripheral lipogenesis. J Clin Invest 112: 423-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulson JM, Fiskerstrand CE, Woll PJ, Quinn JP (1999) E-box motifs within the human vasopressin gene promoter contribute to a major enhancer in small-cell lung cancer. Biochem J 344: 961-970. [PMC free article] [PubMed] [Google Scholar]
- Falconer DS, Mackay TFC (1996) Introduction to quantitative genetics. Essex, UK: Longman.
- Griebel G, Simiand J, Serradeil-Le Gal C, Wagnon J, Pascal M, Scatton B, Maffrand JP, Soubrie P (2002) Anxiolytic- and antidepressant-like effects of the non-peptide vasopressin V1b receptor antagonist, SSR149415, suggest an innovative approach for the treatment of stress-related disorders. Proc Natl Acad Sci USA 99: 6370-6375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herdegen T, Blume A, Buschmann T, Georgakopoulos E, Winter C, Schmid W, Hsieh TF, Zimmermann M, Gass P (1997) Expression of activating transcription factor-2, serum response factor and cAMP/Ca response element binding protein in the adult rat brain following generalized seizures, nerve fibre lesion and ultraviolet irradiation. Neuroscience 81: 199-212. [DOI] [PubMed] [Google Scholar]
- Hoffmann A, Ciani E, Boeckardt J, Holsboer F, Journot L, Spengler D (2003) Transcriptional activities of the zinc finger protein Zac are differentially controlled by DNA binding. Mol Cell Biol 23: 988-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde-DeRuyscher RP, Jennings E, Shenk T (1995) DNA binding sites for the transcriptional activator/repressor YY1. Nucleic Acids Res 23: 4457-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki Y, Oiso Y, Saito H, Majzoub JA (1997) Positive and negative regulation of the rat vasopressin gene promoter. Endocrinology 138: 5266-5274. [DOI] [PubMed] [Google Scholar]
- Kamada S, Miwa T (1992) A protein binding to CArG box motifs and to single-stranded DNA functions as a transcriptional repressor. Gene 119: 229-236. [DOI] [PubMed] [Google Scholar]
- Keck ME, Wigger A, Welt T, Muller MB, Gesing A, Reul JM, Holsboer F, Landgraf R, Neumann ID (2002) Vasopressin mediates the response of the combined dexamethasone/CRH test in hyper-anxious rats: implications for pathogenesis of affective disorders. Neuropsychopharmacology 26: 94-105. [DOI] [PubMed] [Google Scholar]
- Keck ME, Welt T, Muller MB, Uhr M, Ohl F, Wigger A, Toschi N, Holsboer F, Landgraf R (2003) Reduction in hypothalamic vasopressinergic hyper-drive contributes to clinically relevant behavioural and neuroendocrine effects of chronic paroxetine treatment in a psychpathological rat model. Neuropsychopharmacology 28: 235-243. [DOI] [PubMed] [Google Scholar]
- Kelm Jr RJ, Wang SX, Polikandriotis JA, Strauch AR (2003) Structure/function analysis of mouse Purbeta, a single-stranded DNA-binding repressor of vascular smooth muscle alpha-actin gene transcription. J Biol Chem 278: 38749-38757. [DOI] [PubMed] [Google Scholar]
- Landgraf R, Gerstberger R, Montkowski A, Probst JC, Wotjak CT, Holsboer F, Engelmann M (1995) V1 vasopressin receptor antisense oligodeoxynucleotide into septum reduces vasopressin binding, social discrimination abilities, and anxiety-related behavior in rats. J Neurosci 15: 4250-4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebsch G, Montkowski A, Holsboer F, Landgraf R (1998) Behavioural profiles of two Wistar rat lines selectively bred for high or low anxiety-related behaviour. Behav Brain Res 94: 301-310. [DOI] [PubMed] [Google Scholar]
- Lohmueller KE, Pearce CL, Pike M, Lander ES, Hirschhorn JN (2003) Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nat Genet 33: 177-182. [DOI] [PubMed] [Google Scholar]
- Martin KA, Gualberto A, Kolman MF, Lowry J, Walsh K (1997) A competitive mechanism of CArG element regulation by YY1 and SRF: implications for assessment of Phox1/MHox transcription factor interactions at CArG elements. DNA Cell Biol 16: 653-661. [DOI] [PubMed] [Google Scholar]
- Mikheev AM, Mikheev SA, Zhang Y, Aebersold R, Zarbl H (2000) CArG binding factor A (CBF-A) is involved in transcriptional regulation of the rat Ha-ras promoter. Nucleic Acids Res 28: 3762-3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millan MJ (2003) The neurobiology and control of anxious states. Prog Neurobiol 70: 83-244. [DOI] [PubMed] [Google Scholar]
- Nestler EJ, Barrot M, DiLeone RJ, Eisch AJ, Gold SJ, Monteggia LM (2002) Neurobiology of depression. Neuron 34: 13-25. [DOI] [PubMed] [Google Scholar]
- Pitkow LJ, Sharer CA, Ren X, Insel TR, Terwilliger EF, Young LJ (2001) Facilitation of affiliation and pair-bond formation by vasopressin receptor gene transfer into the ventral forebrain of a monogamous vole. J Neurosci 21: 7392-7396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushlow WJ, Rajakumar N, Flumerfelt BA, Naus CC (1999) Characterization of CArG-binding protein A initially identified by differential display. Neuroscience 94: 637-649. [DOI] [PubMed] [Google Scholar]
- Salchner P, Singewald N (2002) Neuroanatomical substrates involved in the anxiogenic-like effect of acute fluoxetine treatment. Neuropharmacology 43: 1238-1248. [DOI] [PubMed] [Google Scholar]
- Salome N, Salchner P, Viltart O, Sequeira H, Wigger A, Landgraf R, Singewald N (2004) Neurobiological correlates of high (HAB) versus low anxiety-related behavior (LAB): differential Fos expression in HAB and LAB rats. Biol Psychiatry 55: 715-723. [DOI] [PubMed] [Google Scholar]
- Scott LV, Dinan TG (2002) Vasopressin as a target for antidepressant development: an assessment of the available evidence. J Affect Disord 72: 113-124. [DOI] [PubMed] [Google Scholar]
- Scott SD, Joiner MC, Marples B (2002) Optimizing radiation-responsive gene promoters for radiogenetic cancer therapy. Gene Ther 9: 1396-1402. [DOI] [PubMed] [Google Scholar]
- Segurado R, Detera-Wadleigh SD, Levinson DF, Lewis CM, Gill M, Nurnberger Jr JI, Craddock N, DePaulo JR, Baron M, Gershon ES, Ekholm J, Cichon S, Turecki G, Claes S, Kelsoe JR, Schofield PR, Badenhop RF, Morissette J, Coon H, Blackwood D, et al. (2003) Genome scan meta-analysis of schizophrenia and bipolar disorder, part III: bipolar disorder. Am J Hum Genet 73: 49-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringer JL, Belaguli NS, Iyer D, Schwartz RJ, Balasubramanyam A (2002) Developmental expression of serum response factor in the rat central nervous system. Brain Res Dev Brain Res 138: 81-86. [DOI] [PubMed] [Google Scholar]
- Treisman R (1987) Identification and purification of a polypeptide that binds to the c-fos serum response element. EMBO J 6: 2711-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman R (1990) The SRE: a growth factor responsive transcriptional regulator. Semin Cancer Biol 1: 47-58. [PubMed] [Google Scholar]
- Umar A, Luider TM, Berrevoets CA, Grootegoed JA, Brinkmann AO (2003) Proteomic analysis of androgen-regulated protein expression in a mouse fetal vas deferens cell line. Endocrinology 144: 1147-1154. [DOI] [PubMed] [Google Scholar]
- Wigger A, Sanchez MM, Mathys KC, Ebner K, Frank E, Liu D, Kresse A, Neumann ID, Holsboer F, Plotsky PM, Landgraf R (2004) Alterations in central neuropeptide expression, release, and receptor binding in rats bred for high anxiety: critical role of vasopressin. Neuropsychopharmacology 29: 1-14. [DOI] [PubMed] [Google Scholar]