Abstract
Regulation of AMPA receptors (AMPARs) at synapses plays a critical role in alterations of synaptic strength in the brain. Stargazin, an AMPAR-interacting protein, is critical for clustering and regulation of synaptic AMPARs. Stargazin interacts with AMPARs via its extracellular domain and with PDZ [postsynaptic density-95 (PSD-95)/Discs large (Dlg)/zona occludens-1 (ZO-1)] proteins via its C-terminal PDZ-binding motif, and these interactions are necessary for stargazin and AMPAR synaptic targeting. By studying the expression of stargazin mutant constructs in cultured hippocampal neurons, we identified a novel domain corresponding to residues 243-283 within the cytoplasmic C terminus of stargazin that is also required for stargazin and AMPAR synaptic clustering. To identify proteins that interact with this stargazin synaptic clustering domain, we performed a yeast two-hybrid assay and found that this stargazin domain binds to nPIST (neuronal isoform of protein-interacting specifically with TC10), a Golgi-enriched protein implicated in trafficking of transmembrane proteins. Using in situ hybridization, immunohistochemistry, coimmunoprecipitation studies, and biochemical fractionation, we found that stargazin and nPIST colocalize and interact in the brain. Finally, by studying AMPAR clustering in transfected hippocampal neurons, we found that overexpression of nPIST enhances AMPAR synaptic clustering, whereas transfection of a dominant-negative nPIST construct attenuates AMPAR synaptic clustering. These studies identify a novel stargazin domain necessary for synaptic clustering of AMPARs and suggest that nPIST and stargazin interactions play a critical role in AMPAR trafficking to the synapse.
Keywords: postsynaptic density, acidic cluster, synapse, trafficking, PSD-95, GluRδ2
Introduction
Glutamate receptors mediate the majority of excitatory synaptic transmission in the brain. The two major classes of ionotropic receptors at glutamatergic synapses, NMDA receptors (NMDARs) and AMPA receptors (AMPARs), are highly concentrated at the postsynaptic density (PSD) of excitatory synapses, but they are differentially anchored. Whereas NMDARs are relatively stable components of the PSD, AMPARs recycle rapidly, and this dynamic nature of AMPAR expression at the synapse underlies its critical role in manifesting changes in synaptic strength (Nishimune et al., 1998; Song et al., 1998; Luthi et al., 1999; Ehlers, 2000; Liu and Cull-Candy, 2000; Luscher et al., 2000; Malinow et al., 2000; Man et al., 2000).
The divergent behavior of AMPAR versus NMDAR proteins at the synapse is, in part, mediated by differential tethering to the PSD. The cytosolic C-terminal tails of AMPARs and NMDARs associate with distinct PDZ [postsynaptic density-95 (PSD-95)/Discs large (Dlg)/zona occludens-1 (ZO-1)] domain-containing synaptic scaffolding proteins (Kornau et al., 1997; Ziff, 1997; Garner et al., 2000). With respect to AMPAR subunits, the C terminus of glutamate receptor (GluR) 2 binds to PDZ domains from several synaptic proteins including PICK1 (protein interacting with C kinase 1), GRIP (glutamate receptor-interacting protein), and ABP (AMPAR-binding protein) (Srivastava et al., 1998; Dev et al., 1999; Kim and Huganir, 1999), and the C terminus of GluR1 binds to synaptic proteins such as SAP97 (synapse-associated protein with a molecular weight of 97 kDa) (Sans et al., 2001). Despite the capacity of AMPAR subunits to bind constituents of the PSD, AMPARs are not trafficked to synapses in cerebellar granule cell neurons lacking stargazin (Chen et al., 2000), the four-pass membrane protein mutated in epileptic stargazer mice (Letts et al., 1998). The stargazin cytosolic C terminus contains a PDZ ligand that interacts with PSD-95. The PDZ ligand is critical for stargazin and AMPAR synaptic clustering and function, because synaptic AMPARs can be rescued by transfection of stargazer mutant granule cells with constructs coding for stargazin but not when transfected with stargazin lacking the C-terminal PDZ-binding domain.
Many intracellular proteins contain C-terminal PDZ-binding motifs that can interact with synaptic scaffolding molecules such as PSD-95, yet are not enriched at the synapse. Thus, despite the necessity of the stargazin PDZ-binding motif for stargazin and AMPAR synaptic targeting, we reasoned that the stargazin PDZ-binding motif is unlikely to be sufficient for this targeting. By analyzing synaptic clustering of stargazin deletion mutants in transfected hippocampal neurons, we found that a region of the intracellular C terminus of stargazin corresponding to aa 243-283 is necessary for stargazin and AMPAR synaptic targeting. By yeast two-hybrid screening, we identified a Golgi-resident PDZ protein, nPIST (neuronal isoform of protein-interacting specifically with TC10), that interacts with stargazin amino acid residues 243-283, implicating nPIST in the mechanism of stargazin-AMPAR synaptic targeting. To further elucidate the role of nPIST in AMPAR trafficking, we engineered a deletion construct of nPIST that binds stargazin but lacks a functional PDZ ligand. When overexpressed in cultured hippocampal neurons, this construct reduced AMPAR synaptic clustering. Interestingly, by coimmunoprecipitating nPIST from brain homogenates, we found that nPIST coimmunoprecipitates with both stargazin and PSD-95. These data support the hypothesis that nPIST interaction with stargazin mediates AMPAR synaptic targeting and suggests a model for AMPAR trafficking in which nPIST, by binding to both stargazin and synaptic proteins such as PSD-95, serves as a chaperone in the pathway between AMPAR synthesis and ultimate synaptic localization.
Materials and Methods
Antibodies. The following primary antibodies were used: rabbit polyclonal antibodies to PSD-95 (Brenman et al., 1996); stargazin [either serum provided by Verity Letts (The Jackson Laboratory, Bar Harbor, ME) or Chetkovich et al. (2002)]; pan-TARP (tetramembrane-spanning AMPA receptor-associated protein), which recognizes stargazin as well as the closely related family members calcium channel (cc) γ-3, ccγ-4, and ccγ8 (Tomita et al. 2003); GluR1 (Chemicon, Temecula, CA); guinea pig antibodies to green fluorescent protein (GFP) (El-Husseini et al. 2000); monoclonal antibodies to PSD-95 (number 046; Affinity Bioreagents, Golden, CO), GFP (Becton Dickinson Biosciences, Palo Alto, CA), and synaptophysin (Sigma, St. Louis, MO); NMDAR subunit 1 (NR1; BD Biosciences PharMingen, San Diego, CA). For Western blotting, protein extracts were resolved by SDS-PAGE and transferred to polyvinylidene difluoride membranes. Primary antibodies were diluted in block solution containing 3% BSA and 0.1% Tween 20 in TBS and incubated with membranes overnight at 4°C. Labeled bands were visualized using ECL (Amersham Biosciences, Piscataway, NJ).
cDNA cloning and mutagenesis. Constructs coding for stargazin, stargazin(Δ319-323), and PSD-95 as well as their GFP and hemagglutinin (HA) fusions have been described previously (Topinka and Bredt, 1998; Craven et al., 1999; Chen et al., 2000). Stargazin deletion constructs were created by first amplifying base pairs 1-609 by PCR and cloning into the expression vector GW1 at HindIII and KpnI sites to make GW1-stargazin(1-203). To prepare stargazin(Δ204-311)-GFP, GFP with a nested, in-frame XmaI site and base pairs coding for the last 12 amino acids of stargazin in the 3′ primer was amplified and inserted into GW1-stargazin(1-203) at KpnI and EcoRI. Stargazin(Δ244-311)-GFP and stargazin(Δ284-311) were produced by PCR amplification of the appropriate fragments and inserted into GW1-stargazin(Δ204-311) at HindIII and KpnI. Stargazin(Δ204-243)-GFP and stargazin(Δ204-283) were produced by insertion of the appropriate amplification fragments into the nested XmaI site and EcoRI. The C-terminal stargazin yeast two-hybrid bait constructs and nPIST yeast two-hybrid prey constructs were produced by PCR amplification of the appropriate base pairs of stargazin or nPIST and subcloned into pGBKT7 or pGADT7 (BD Biosciences Clontech, Palo Alto, CA), respectively, at EcoRI/BamHI sites. Full-length nPIST was generated by reverse transcription (RT)-PCR from mouse brain cDNA using the following primers: 5′-TATGGTACCGCCGCCATGTCGGCG and 3′-TATGGATCCGTAGGCCTTCTTCTG. The resultant 1.4 kb fragment was inserted into the KpnI and BamHI site of the mammalian expression vector pEGFP-N3 (BD Biosciences Clontech). nPIST(1-312)-GFP was produced by PCR and inserted into the same vector at the same sites. The proper introduction of all mutations and deletions was verified by DNA sequencing.
Yeast two-hybrid assays. Yeast two-hybrid assays were performed using the Matchmaker kit (Becton Dickinson Biosciences), according to the manufacturer's protocols. Briefly, stargazin(204-323) was subcloned into the bait vector, pGBKT7. Yeast (AH109) was transformed with the bait vector under appropriate selection. A rat cDNA library in the prey vector, pGADT7 (Becton Dickinson Biosciences), was then screened by transformation into AH109 cells carrying bait plasmid and plated onto selective media. Initial screening yielded mostly PDZ proteins, so a second construct was prepared [stargazin(204-323)T321E] that contained a mutated residue to prevent PDZ-ligand interactions. Two identical clones of nPIST were obtained as positive interactors with this bait. Directed yeast two-hybrid assays were performed by cotransforming engineered plasmids into competent yeast, followed by plating onto selective media.
In situ hybridization. RNA was isolated using the guanidine isothiocyanate/CsCl method, and mRNA was selected using oligo dT Sepharose. For Northern blotting, mRNA was separated on a formaldehyde agarose gel and transferred to a nylon membrane. The filter was sequentially hybridized with random primed [32P] probes, which were generated using a 120 bp probe corresponding to the C terminus of PIST. In situ hybridization using [35S]-labeled RNA probes was performed as described (Sassoon and Rosenthal, 1993). An antisense probe to the unique C terminus of PIST (372-463) was synthesized from the pBluescript vector. Tissue sections were exposed to x-ray film for 4 d.
Cell transfection and immunoprecipitation. For COS-7 cell experiments, cells were grown in DMEM containing 10% fetal bovine serum, penicillin, and streptomycin. Cells were transfected using Lipofectamine reagent according to the manufacturer's protocol (Invitrogen, Carlsbad, CA). Cells were washed with ice-cold PBS and resuspended in 0.4 ml of lysis buffer containing TEE (50 mm Tris-HCl, pH 7.4, 1 mm EDTA, and 1 mm EGTA) and 150 mm NaCl. To this lysate we added 0.1 ml of SDS-PAGE sample buffer lacking β-mercaptoethanol, and 20 μl was loaded on 10% SDS-PAGE gels. For immunoprecipitation experiments, the samples were lysed in TEE containing 1% Triton X-100, 10 μg/ml aprotinin (Sigma), 10 μg/ml leupeptin (Sigma), and 1 mm PMSF (Sigma), then incubated with 2 μg of the appropriate antibodies for 1 hr at 4°C. After the addition of 20 μl of protein A-Sepharose beads (Sigma), samples were incubated for 1 hr at 4°C. Immunoprecipitates were washed four times with wash buffer containing TEE, 150 mm NaCl, and 1% Triton X-100, boiled in SDS-PAGE sample buffer with 1 mm DTT for 2 min, and resolved by SDS-PAGE.
To obtain tissue homogenates, adult rat tissues were homogenized in 10 volumes (w/v) of 50 mm HEPES, pH 7.5, containing 10 μg/ml aprotinin (Sigma), 10 μg/ml leupeptin (Sigma), and 1 mm PMSF (Sigma). Nuclei were removed by centrifugation at 1000 × g. Protein concentrations were determined by the Bradford method, and the volume was adjusted to normalize protein concentration. Sample buffer was added, and 20 μl (∼10 mg total protein) was resolved by SDS-PAGE and immunoblotted for PIST. For immunoprecipitation experiments, adult rat brains were homogenized as above. Nuclei were removed by centrifugation at 1000 × g. Crude membranes were obtained by centrifugation of the postnuclear homogenate at 100,000 × g and were resuspended in 1% SDS in 50 mm Tris-HCl, pH 7.5. This step was followed by a 10-fold dilution in buffer containing 1% Triton X-100. Samples were then incubated with 2 μg of primary antibodies for 1 hr at 4°C. After the addition of 20 μl of protein A-Sepharose beads (Sigma), samples were incubated for 1 hr at 4°C. Immunoprecipitates were washed four times with wash buffer containing TEE, 150 mm NaCl, and 0.1% Triton X-100, boiled in SDS-PAGE sample buffer, and resolved by SDS-PAGE.
Primary neuronal culture and transfection. Hippocampal cultures were transfected as described previously (Craven et al., 1999). Briefly, acutely dissociated hippocampal neurons from embryonic day 18 rats were transfected in suspension by lipid-mediated gene transfer based on the protocol described by Kaech et al. (1996). Cells were then plated at a density of 600/mm2 on glass coverslips (Fisher Scientific, Pittsburgh, PA) and maintained in Neurobasal media supplemented with B27 (Invitrogen). For some experiments, neurons were transfected after 10-17 d in vitro using Effectene (Qiagen, Valencia, CA) according to manufacturer's protocols.
Immunofluorescence labeling and quantitation of AMPAR clustering. Coverslips were removed from culture wells and fixed in 2% paraformaldehyde for 15 min or in 100% ice-cold methanol for 10 min. After washing with PBS containing 0.1% Triton X-100 (PBST) three times for 5 min, cells were incubated in PBST containing 3% normal goat serum for 1 hr at room temperature. Primary antibodies were added in block solution for 1 hr at room temperature, followed by donkey anti-mouse or goat anti-rabbit secondary antibodies conjugated to Alexa-488 (Molecular Probes, Eugene, OR) or Cy3 (Jackson ImmunoResearch, West Grove, PA), diluted 1:200 in block solution, for 1 hr at room temperature. Coverslips were then mounted on slides (Superfrost/Plus slides; Fisher Scientific) with Fluoromount-G (Southern Biotechnology, Birmingham, AL). Images were taken under fluorescence microscopy with a 63× oil-immersion objective (numerical aperture, 1.4) affixed either to an Axiovert S100 TV inverted microscope (Zeiss, Thornwood, NY) equipped with a Hammamatsu 12 bit ORCA, an interline CCD camera (Technical Instruments, San Francisco, CA), an excitation and emission filter-wheel (Sutter, Novato, CA), and a MetaMorph Imaging system (Universal Imaging, Downingtown, PA); or with Zeiss Axiovert 200M inverted microscope with Axiovision 3.0 software-driven controls, equipped with Axio Cam HRm camera.
For analysis, images of both control and transfected neurons were taken within the same visual field. Images for each construct were obtained from a minimum of three separate preparations. For each neuron studied, the largest caliber proximal dendrite was analyzed. Data were analyzed with NIH Image software. To determine the synaptic localization of transfected stargazin-GFP constructs, we assessed colocalization with PSD-95. Cultured neurons were costained for both the desired protein, such as stargazin-GFP, and PSD-95. Cluster colocalization was determined using the “colocalization” plugin for NIH Image J, which takes the two different channel source image clusters to generate a novel image that corresponds to overlapping pixels that are at least 50% above source channel threshold, and this image corresponds to colocalized (synaptic) puncta. Puncta at least 4 pixels in diameter were quantitated with the “Analyze Particle” feature of Image J, which identified at least 95% of dendritic PSD-95 containing puncta and eliminated identification of much of the particulate background material. By dividing the number of stargazin-GFP puncta by the number of colocalized puncta, we generated a synaptic cluster percentage, which is a measure of the percentage of clusters that are synaptic (colocalized with PSD-95).
For determination of endogenous stargazin/TARP, GluR1, and NR1 clustering, the average cluster intensity from 30 to 50 dendritic puncta recorded sequentially along the primary dendrite was expressed as a cluster intensity ratio (CIR), which is the average cluster intensity divided by the average fluorescence intensity of the dendritic shaft. To ensure that clusters were attributed to the proper parent dendrite (transfected versus nontransfected), puncta were counted in regions of the coverslip in which there were clearly no dendrites from untransfected cells, confirmed by examining background fluorescence of the parent dendritic shaft. Average CIR values for five to nine transfected neurons were compared with that of paired untransfected control neurons for each of the constructs using a paired t test with Microsoft Excel software. Data were normalized for tabular presentation by dividing the average CIR of the transfected cells by the CIR of their respective paired controls. For GluR1 clustering, the majority of GluR1 puncta detected under permeabilized conditions are synaptic (Carroll et al., 1999).
Results
A novel region in the stargazin C terminus is necessary for synaptic targeting
The C-terminal four amino acids of stargazin specify a consensus sequence for PDZ binding (-TTPV) that is necessary for stargazin synaptic targeting. To determine whether other domains in the stargazin C terminus are also necessary for synaptic targeting, we transfected hippocampal neurons with GFP-tagged stargazin constructs and examined synaptic clustering by immunostaining for GFP and synaptic markers. Whereas GFP-tagged stargazin (stargazin-GFP) is clustered at synapses (Fig. 1A-C, Table 1), a stargazin construct lacking the C-terminal PDZ-binding domain [stargazin(Δ319-323)-GFP] is diffusely localized (Fig. 1D-F, Table 1). We next prepared a stargazin construct in which all but the last 12 amino acids of the intracellular C-terminal tail of stargazin are replaced by GFP [stargazin(Δ204-311)-GFP]. When transfected into hippocampal neurons, stargazin(Δ204-311)-GFP is diffusely localized, similar to the expression pattern of stargazin(Δ319-323)-GFP (Fig. 1G-I, Table 1). This stargazin construct, which includes the PDZ-binding domain, retains the ability to bind to and be clustered by PSD-95 when expressed in heterologous cells (Figure 1J-M). These findings suggest that the stargazin PDZ-binding domain is necessary but not sufficient to mediate synaptic targeting of stargazin and implicate other structural elements within the C terminus of stargazin as being important for stargazin synaptic targeting. To identify which novel region of stargazin is critical for synaptic targeting, we transfected hippocampal neurons with stargazin constructs containing the PDZ-binding domain as well as different regions of the stargazin C terminus. Although retaining the ability to bind PSD-95, stargazin(Δ204-283)-GFP and stargazin(Δ244-311)-GFP showed diffuse staining in neurons, whereas stargazin(Δ204-243)-GFP and stargazin(Δ284-311)-GFP were clearly clustered at synapses (Table 1). These findings indicate that in addition to the PDZ-binding domain, stargazin synaptic targeting is dependent on the presence of amino acid residues between 244 and 283 of the stargazin C terminus.
Figure 1.
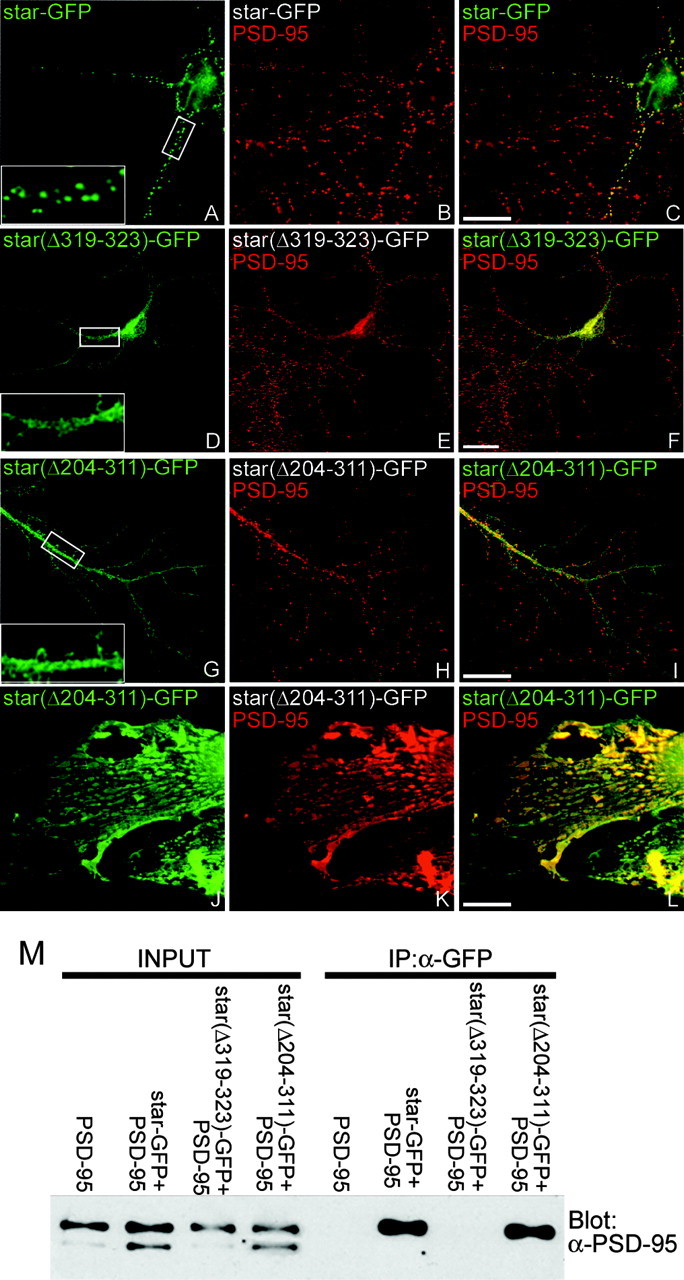
The stargazin PDZ ligand and C terminus are both necessary for stargazin synaptic targeting. Constructs encoding stargazin-GFP (star-GFP), stargazin(Δ319-323)-GFP [star(Δ319-323)-GFP], or stargazin(Δ204-311)-GFP [star(Δ204-311)-GFP] were transfected into neurons (A-I), which were then fixed (at 11-17 d in vitro) and stained for GFP (green) and PSD-95 (red). A-C, Stargazin-GFP clusters at synaptic sites. Stargazin lacking the PDZ-binding domain, stargazin(Δ319-323)-GFP, does not target to synapses (D-F). Similar to the construct lacking the PDZ-binding domain, a construct lacking all of the intracellular C-terminal tail, except the last 11 amino acids, stargazin(Δ204-311)-GFP was also diffusely localized in neurons (G-I). In contrast, when cotransfected with PSD-95 in COS-7 cells, stargazin(Δ204-311)-GFP was clustered by PSD-95 (J-L). Merged images are shown in C, F, I, and L. The insets in A, D, and G are digitally magnified 3.5×. Scale bar, 10 μm. M, COS-7 cells were transfected alone or with the indicated combinations of PSD-95, stargazin-GFP (star-GFP), stargazin(Δ319-323)-GFP [star(Δ319-323)-GFP], or stargazin(Δ204-311)-GFP [star(Δ204-311)-GFP]; cell lysates were prepared, and GFP was immunoprecipitated (IP). Although PSD-95 interacted strongly with stargazin-GFP and stargazin(Δ204-311)-GFP, stargazin(Δ319-323)-GFP failed to coimmunoprecipitate. Input corresponds to 5% of total lysate.
Table 1.
Stargazin and AMPAR synaptic clustering is dependent on the stargazin PDZ-binding site as well as stargazin amino acid residues 244-283
|
|
PSD-95 binding |
Synaptic clustering |
||||
|---|---|---|---|---|---|---|
| Construct |
GFP |
GluR1 |
NR1 |
|||
| Stargazin-GFP | + | 65.4 ± 5.8 | 93.0 ± 5.1 | 89.4 ± 7.7 | ||
| Stargazin(Δ319-323)-GFP | − | 9.6 ± 1.8* | 57.6 ± 4.7* | 100.1 ± 5.6 | ||
| Stargazin(Δ204-311)-GFP | + | 7.5 ± 0.9* | 71.8 ± 5.1* | 87.4 ± 6.2 | ||
| Stargazin(Δ204-243)-GFP | + | 56.8 ± 3.2 | 99.1 ± 4.8 | ND | ||
| Stargazin(Δ204-283)-GFP | + | 7.6 ± 0.9* | 67.2 ± 4.7* | 96.9 ± 6.1 | ||
| Stargazin(Δ244-311)-GFP | + | 8.1 ± 0.8* | 64.5 ± 4.2* | ND | ||
| Stargazin(Δ284-311)-GFP |
+ |
46.2 ± 8.3 |
91.6 ± 4.0 |
ND |
||
PSD-95 binding was determined by coimmunoprecipitation using anti-GFP antisera from COS-7 cells transfected with PSD-95 and the indicated stargazin construct. To determine clustering of stargazin, GluR1, and NR1, constructs encoding stargazin-GFP or the indicated stargazin deletion mutations were transfected into neurons, which were then fixed (at 11-17 d in vitro) and stained for GFP, GluR1, or NR1. Clustering of stargazin-GFP constructs was determined by calculating a synaptic clustering percentage (SC) (see Materials and Methods), whereas GluR1 and NR1 clustering was determined by calculating the CIR of stained puncta in neurons as described in Materials and Methods. For stargazin-GFP constructs, the SCP of 30-50 puncta from four to five cells were quantitated (SCP ± SEM) and synaptic clustering of different constructs was compared with stargazin-GFP by ANOVA, followed by Dunnett's multiple comparison test. For GluR1 clustering, t tests were performed using CIR values of 30-50 puncta from five to nine pairs of transfected and control neurons for each construct and are expressed as percentage of control GluR1 or NR1 CIR of paired untransfected neurons within the same image ± SEM. *p < 0.01. ND, Not determined experimentally.
Stargazin synaptic targeting controls AMPAR synaptic targeting
We previously demonstrated that stargazin is necessary for proper AMPAR synaptic function. Transfection of hippocampal neurons with wild-type stargazin does not affect synaptic AMPAR clustering, whereas transfection with a diffusely targeted stargazin(Δ319-323)-GFP construct dramatically downregulates synaptic AMPAR currents (Chen et al., 2000). This effect is likely attributable to the retained ability of diffusely localized stargazin(Δ319-323)-GFP to bind AMPAR subunits, preventing their synaptic localization. To determine whether stargazin affects AMPAR function by regulating synaptic AMPAR clustering, we performed immunostaining for the AMPAR GluR1 subunit in hippocampal neurons transfected with stargazin constructs and quantitated the GluR1 synaptic CIR, which is the ratio of average GluR1 puncta fluorescence intensity and the average dendritic fluorescence intensity (Craven et al., 1999). Whereas stargazin-GFP had no effect on GluR1 clustering (Fig. 2A-C,J; Table 1), stargazin(Δ319-323)-GFP and stargazin(Δ204-311)-GFP both display diminished GluR1 clustering compared with nontransfected cells (Fig. 2D-J, Table 1). Furthermore, other stargazin constructs that were able to target to synapses, stargazin(Δ204-243)-GFP and stargazin(Δ283-311)-GFP, did not effect GluR1 synaptic clustering, whereas the other diffusely targeted stargazin constructs, stargazin(Δ204-283)-GFP and stargazin(Δ243-311)-GFP, diminished GluR1 synaptic clustering (Table 1; also see supplemental Fig. 1, available at www.jneurosci.org/cgi/content/full/24/34/9491/DC1). Despite effects on AMPAR clustering, several of the diffusely targeted stargazin constructs showed no alteration of NR1 clustering, suggesting that the stargazin deletions specifically impair AMPAR synaptic clustering (Table 1). These findings confirm that targeting of stargazin controls the clustering of AMPAR at the synapse and furthermore demonstrate that (1) the stargazin C-terminal PDZ domain is necessary but not sufficient to mediate AMPAR synaptic clustering, and (2) structural elements within amino acid residues 243-283 of the stargazin C terminus are required for proper AMPAR synaptic clustering.
Figure 2.
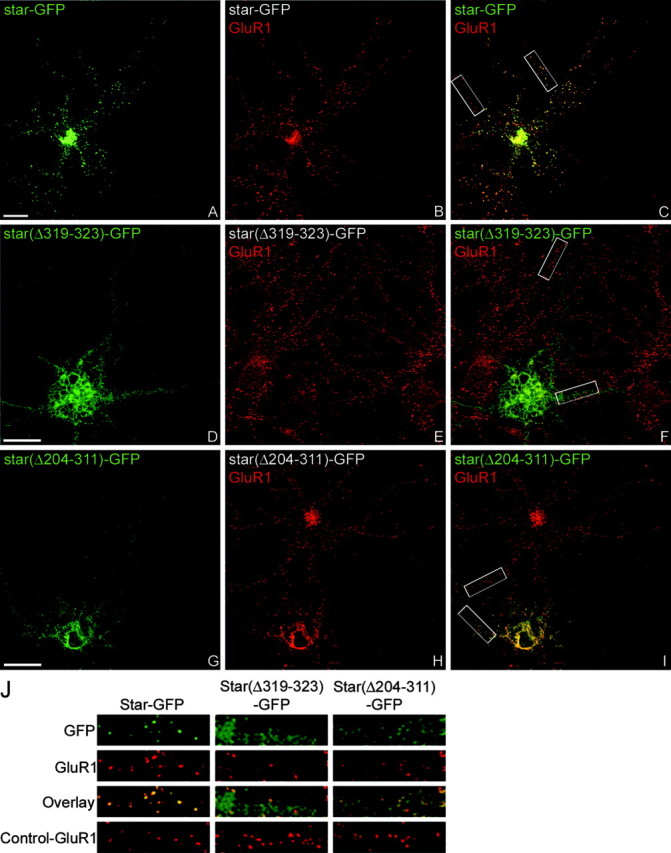
The C terminus of stargazin exclusive of the last 12 amino acids is necessary for AMPAR synaptic targeting. Constructs encoding stargazin-GFP (star-GFP), stargazin(Δ319-323)-GFP [star(Δ319-323)-GFP], or stargazin(Δ204-311)-GFP [star(Δ204-311)-GFP] were transfected into neurons, which were then fixed (at 11-17 d in vitro) and stained for GFP (green) and GluR1 (red). GluR1 remains clustered at synaptic sites in neurons transfected with stargazin-GFP (A-C, J). In neurons transfected with stargazin(Δ319-323)-GFP, GluR1 shows reduced clustering compared with adjacent untransfected cells (D-F, J). Similar to the reduction of GluR1 clustering seen with the construct lacking the PDZ-binding domain, clustering of GluR1 is also reduced in neurons transfected with stargazin(Δ204-311)-GFP (G-J). Merged images are shown in C, F, and I. The white boxes in C, F, and I are enlarged and presented in J. Scale bar, 10 μm.
Stargazin interacts with the Golgi-preferring PDZ protein nPIST
Because stargazin and AMPAR synaptic clustering are dependent on the non-PDZ-binding residues of the stargazin C-terminal tail, we reasoned that molecules other than PDZ proteins such as PSD-95 interact with stargazin and are necessary for proper synaptic targeting. To address this possibility, we screened a rat brain yeast two-hybrid library with the C terminus of stargazin. A total of 5 × 106 clones were screened yielding ∼1500 interacting clones. Forty-six of the interacting clones were sequenced. Of these, 24 were unlikely positives based on their cellular localization (secreted or extracellular versus the intracellular tail of stargazin), and 22 clones were known PDZ proteins. To screen more specifically for non-PDZ interactions, we used the stargazin C terminus that contains a point mutation (T321E) that blocks PDZ binding (Chetkovich et al., 2002). This screen of 2 × 106 clones yielded 150 interacting clones, of which 16 were isolated and sequenced. Fourteen clones were unlikely positives, including multiple different clones of carboxypeptidase E, a secreted protein. We also obtained two identical clones corresponding to amino acids (183-463) of the neuronal isoform of PIST. PIST is a widely expressed Golgi-preferring protein that has been implicated in receptor protein trafficking and contains two coiled-coil domains, a PDZ domain, and an acidic cluster (AC) region (Fig. 3A) (Neudauer et al., 2001). The neuronal isoform of PIST, nPIST, differs from PIST by an eight amino acid insertion within the second coiled-coil domain and is the unique isoform expressed in neuronal tissues (Yao et al., 2001; Cheng et al., 2002; Yue et al., 2002). The yeast two-hybrid clone obtained included half of the second coiled-coil domain, a linker region, the PDZ domain, and C-terminal tail containing the AC domain (Fig. 3A).
Figure 3.

nPIST interacts with the stargazin C terminus independent of its PDZ-binding domain. A, Yeast two-hybrid screening with screening with the stargazin C terminus that contains a point mutation in the PDZ-binding domain [stargazin(203-323)T321E] yielded a clone of nPIST containing half of a predicted coiled-coil (CC), linker region between the last CC and PDZ domains, and PDZ and AC (AC) domains. B, Directed yeast two-hybrid interactions with the nPIST prey [nPIST(183-463)] or the PDZ domains of PSD-95 [PSD-95 (PDZ I-III)] and the cytoplasmic tails of NR2A [NR2A(838-1465)], Kv1.4 [Kv1.4(571-656)], and deletions of stargazin revealed that nPIST binds NR2A and that stargazin residues between amino acids 262 and 283 are necessary for nPIST binding, whereas stargazin binding to PSD-95 requires an intact stargazin PDZ-binding domain. C, ClustalW formatted alignments of stargazin(243-283), ccγ3(239-277), ccγ4(289-389), and ccγ8(280-321), showing a highly conserved region within the stargazin nPIST-binding domain (residues 249-277). D, Deletions within the nPIST prey showed binding to the stargazin C terminus (203-323) and the stargazin C terminus with a missing (203-319) or mutated (T321E) PDZ-binding domain occurred between nPIST amino acid residues 211 and 312 and required the first part of the nPIST PDZ domain independent of a traditional PDZ ligand-binding groove, whereas the complete PDZ domain was required for interaction with the cytoplasmic tail of NR2A (residues 838-1465, NR2A).
PIST interacts with several proteins via its coiled-coil domains (Charest et al., 2001; Neudauer et al., 2001). Additionally, the PIST PDZ domain is known to bind to and modulate the trafficking of several transmembrane proteins (Yao et al. 2001; Cheng et al., 2002; Yue et al., 2002). Our nPIST construct contained a functional PDZ domain, and in directed yeast two-hybrid interactions, nPIST bound to the C terminus of the NMDAR subunit NR2A but not the C terminus of the shaker-type potassium channel Kv1.4 (Fig. 3B). Although our screening stargazin bait contained a mutation known to disrupt PDZ-ligand interactions, PDZ proteins such as PICK1 can interact with phosphorylated substrates (Chung et al., 2000), suggesting the PDZ domain of nPIST might bind to the phosphorylation-mimic stargazinT321E C terminus. To test this possibility, we deleted the last four amino acids of stargazin from the bait construct [stargazin(204-319)]. Although this construct failed to interact with a construct containing the three PDZ domains from PSD-95 (PDZ I-III), it interacted robustly with nPIST, confirming non-PDZ binding of nPIST to stargazin (Fig. 3B). Using a combination of overlapping deletion constructs from both the N and C terminus of the stargazin bait, we narrowed the nPIST interaction domain of stargazin to residues 262-282 (Fig. 3B), which is included in the region required for synaptic targeting. This stargazin region contained no known structural domains, however stargazin residues 249-277 are highly conserved between stargazin homologs known to bind AMPARs ccγ-3, ccγ-4, and ccγ8 (Tomita et al., 2003). Indeed, although the entire stargazin C terminus shows 27% identity and 75% homology with AMPAR-binding stargazin homologs, within residues 249-277 there is 41% identity and 93% homology, suggesting conservation of function for this domain (Fig. 3C). Using a similar approach for nPIST, we found that binding to stargazin required a region of nPIST corresponding to amino acid residues 211-312, which is highly conserved between species (100% identity between mouse, rat, and human and 70% identity to Caenorhabditis elegans PIST; data not shown). This included the linker between the second coiled-coil and the PDZ domain and the first 15 amino acids of the PDZ domain. A functional nPIST PDZ domain was neither necessary nor sufficient to mediate interaction with stargazin by yeast two-hybrid assay (Fig. 3D). In contrast, the PDZ domain of nPIST was required for interaction with the C terminus of NR2A. These results demonstrate that nPIST-stargazin interactions are independent of nPIST PDZ-ligand interactions and suggest a model in which nPIST can simultaneously bind to stargazin and PDZ domain ligands.
Although the stargazin C terminus bound to nPIST in yeast, we wondered whether this interaction occurs in mammalian cells. To examine the interaction of stargazin and nPIST in cells, we performed RT-PCR from mouse brain RNA and obtained a full-length nPIST cDNA. This cDNA was subcloned as a C-terminal GFP fusion (nPIST-GFP). When transfected alone in COS-7 cells, stargazin shows diffuse membrane staining as well a perinuclear accumulation (Fig. 4A). Similar to other transmembrane proteins expressed in heterologous cells, the perinuclear accumulation of stargazin colocalizes with the transferrin receptor endosomal marker (data not shown). In contrast, nPIST-GFP expresses both diffusely and in a perinuclear distribution that colocalizes with the Golgi marker TGN38 (Fig. 4B-E); this is consistent with the localization described for endogenous PIST in non-neuronal tissues (Charest et al., 2001; Yao et al., 2001). When transfected together with nPIST-GFP, stargazin colocalizes with nPIST-GFP within the same perinuclear compartment (Fig. 4F-H). As predicted by the yeast two-hybrid experiments, an nPIST construct lacking a functional PDZ domain and C terminus, nPIST(1-312)-GFP, still colocalizes with stargazin (Fig. 4I,J). In contrast, an nPIST construct lacking a stargazin binding domain [nPIST(289-463)-GFP] does not influence coexpressed stargazin localization (Fig. 4L; and supplemental Fig. 2, available at www.jneurosci.org/cgi/content/full/24/34/9491/DC1), and the localization of a stargazin construct lacking the nPIST-binding domain [stargazin(Δ244-323)-GFP] is not affected by coexpression with nPIST (Fig. 4M; and supplemental Fig. 2, available at www.jneurosci.org/cgi/content/full/24/34/9491/DC1).
Figure 4.
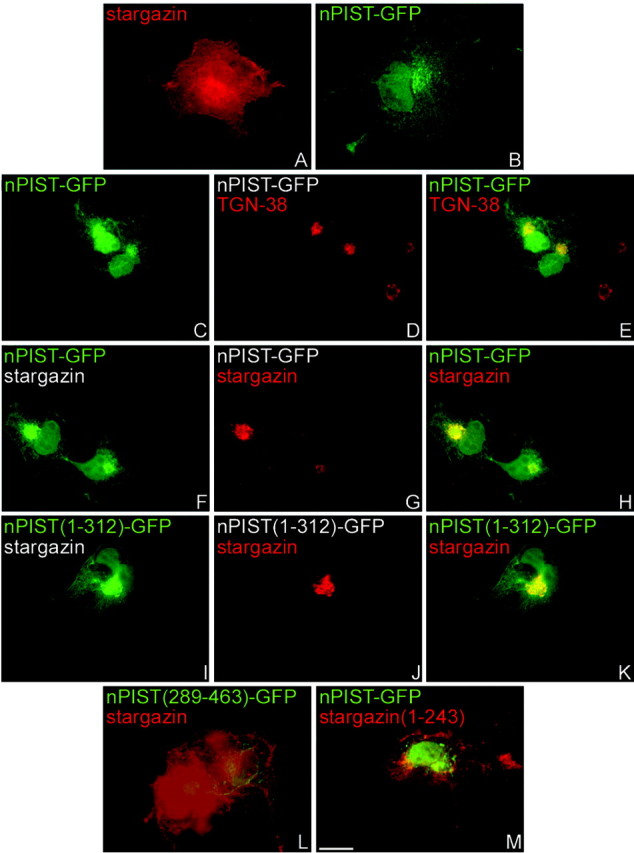
nPIST localizes to the TGN and colocalizes with stargazin when cotransfected in COS-7 cells. COS-7 cells were transfected with the indicated constructs and immunostained as indicated. A, Stargazin (red) transfected alone shows diffuse membrane and perinuclear staining. B-E, nPIST-GFP (green) transfected alone shows diffuse staining as well as perinuclear accumulations that colocalize with the trans-Golgi network marker, TGN38 (red). F-H, When stargazin (red) is expressed with nPIST-GFP (green), stargazin accumulates with nPIST-GFP in a perinuclear compartment. I-K, An nPIST construct that lacks a complete PDZ and AC domain [nPIST(1-312)-GFP, green] also accumulates stargazin in a perinuclear compartment. In contrast, cotransfection of stargazin (red) with an nPIST construct lacking the predicted stargazin binding site [nPIST(289-463)-GFP, green] (L) or transfection of nPIST-GFP (green) with a stargazin construct lacking the nPIST binding site [stargazin(1-243)] (M) results in no perinuclear colocalization. Scale bar, 10 μm.
nPIST interacts with stargazin in brain
We next asked whether nPIST and stargazin are found in similar brain regions by in situ hybridization. Examining embryonic rat tissues, we found that nPIST is enriched in the developing brain and peripheral neurons (Fig. 5A-C). Furthermore, in postnatal day 30 rat brain, nPIST is enriched in multiple brain regions, including neurons in the cerebellum, hippocampus, and forebrain and shows a similar distribution as stargazin (Fig. 5D,E). To characterize the localization of nPIST protein in the brain, we subcloned the C terminus of nPIST into the PET32 protein expression vector and generated a PIST-specific antibody. Although this antibody recognizes both neuronal and nonneuronal PIST isoforms, the unique presence of nPIST in brain tissue (Yao et al., 2001; Cheng et al., 2002; Yue et al., 2002) permits detection of the nPIST isoform when performing immunostaining or Western blotting on brain tissues or slices. Western blotting of rat tissue homogenates shows that PIST is expressed widely and that nPIST is enriched in brain tissues (Fig. 5F). Furthermore, immunohistochemical staining reveals that the nPIST protein localizes to neurons in the forebrain, hippocampus, and cerebellum in a pattern similar to that observed with in situ hybridization (Fig. 5G-I). In non-neuronal cells, PIST localizes predominantly to the Golgi apparatus (Charest et al., 2001; Cheng et al., 2002). To characterize the subcellular localization of nPIST in neurons, we performed immunohistochemical staining in cultured hippocampal neurons. This staining demonstrates marked enrichment of nPIST in the Golgi, confirmed by colocalization with the Golgi-specific antigens GM130 and TGN38 (Fig. 6A-F), and nPIST is also present diffusely as well as in tubulovesicular structures in dendrites (Fig. 6J,K). Although the cis-Golgi marker GM130 is predominantly perinuclear with very little dendritic staining (Fig. 6E), the pan-Golgi marker TGN-38 stains dendritic vesicular structures in addition to perinuclear Golgi (Fig. 6B,J). Interestingly, dendritic staining by TGN-38 only partially colocalizes with the dendritic staining of nPIST (Fig. 6J, arrows), suggesting that some of the puncta containing nPIST in the dendrites represent a post-Golgi vesicular fraction. Indeed, the punctate structures stained by nPIST juxtapose or overlap with the synaptic marker PSD-95 (Fig. 6L, arrows). These findings are consistent with the observation that nPIST is enriched in the PSD by biochemical fractionation (Yue et al., 2002).
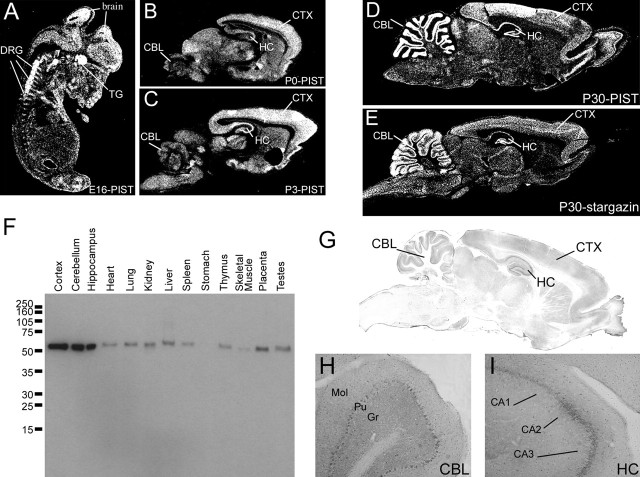
Figure 5.
nPIST is enriched in developing and adult rat brain. A, In situ hybridization of sagittal sections of embryonic day 16 rat embryos shows that nPIST is expressed in diverse populations of neurons and is enriched in brain, dorsal root ganglia (DRG), and trigeminal ganglia (TG). Both nPIST and stargazin occur at high levels in the cerebellum (CBL), hippocampus (HC), and cerebral cortex (CTX) in postnatal day 0 (P0) (B), P3 (C), and P30 (D, E) rat brain. F, Western blotting using anti-PIST antibodies shows widespread PIST expression with enrichment in brain tissues, corresponding to the unique presence of the neuronal isoform nPIST in the brain. G, Immunohistochemistry shows high levels of nPIST expression in the hippocampus (HC), cortex (CTX), and cerebellum (CBL), consistent with in situ hybridization data. H, Higher-power magnification demonstrates higher nPIST expression in the Purkinje cell layer (Pu) than the molecular (Mol) or granule cell (Gr) layer of the cerebellum. I, In the hippocampus, nPIST staining is enriched in the pyramidal cell layers CA1, CA2, and CA3.
Figure 6.
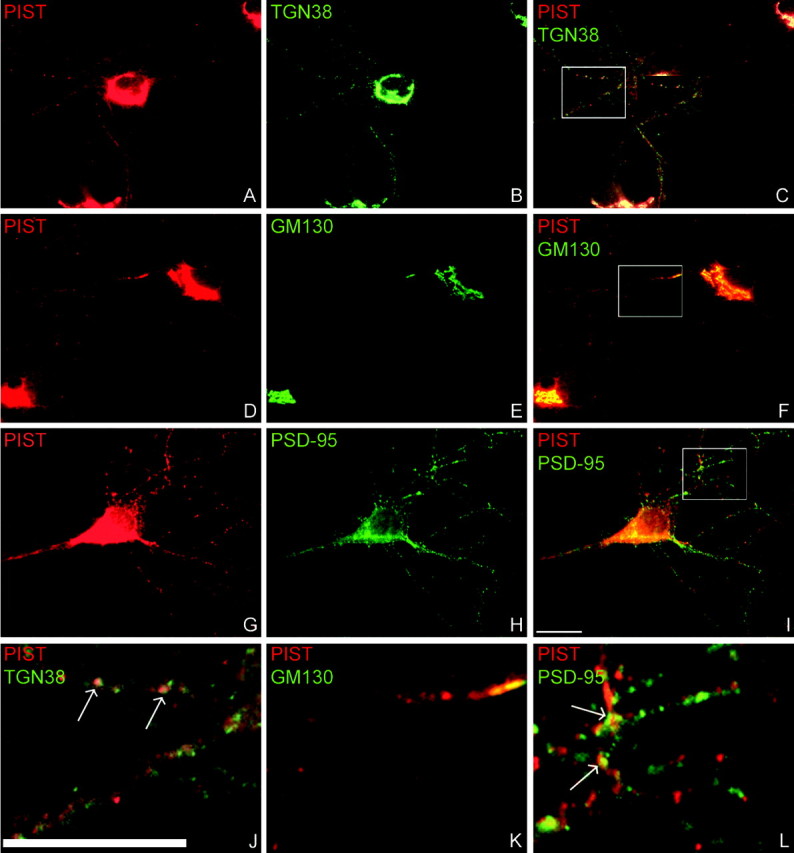
Endogenous nPIST is localized in the Golgi and tubulovesicular structures of dendrites in hippocampal neurons. PIST-specific antibodies, the Golgi markers TGN38 and GM130, and the synaptic marker PSD-95 were used to stain endogenous proteins in cultured hippocampal neurons. A-C, J, nPIST staining (red) colocalizes with TGN38 (green) in the Golgi and in a subset of vesicles in the distal dendrites (arrows). D-F, K, The cis-Golgi marker GM 130 (green) does not stain dendritic puncta but partially colocalizes with nPIST (red) in the perinuclear region and proximal dendrites. G-I, L, nPIST and PSD-95 staining juxtapose or overlap (arrows) in puncta in the distal dendrites. Scale bar, 10 μm.
That nPIST is enriched in the PSD and partially colocalizes with PSD-95 suggests that nPIST might chaperone stargazin from Golgi to synaptic structures. To address this possibility, we performed immunoprecipitation studies with PIST-specific antibodies using brain homogenates. We found that antibodies to nPIST coimmunoprecipitated stargazin and a stargazin-specific antibody coimmunoprecipitated nPIST, consistent with the predicted interaction of nPIST with stargazin (Fig. 7A). Additionally, stargazin antibodies coimmunoprecipitated both GluR1 and PSD-95 but not synaptophysin (Fig. 7A), consistent with the known interaction between stargazin and both the PSD-95 and GluR subunits. Interestingly, nPIST antibodies also coimmunoprecipitated both PSD-95 and GluR1 (Fig. 7A). This coimmunoprecipitation of PSD-95 and GluR1 by nPIST antibodies could result from immunoprecipitation of stargazin bound to PSD-95 and GluR1. However, the efficiency of coimmunoprecipitation of PSD-95 by anti-PIST antisera appeared to be better than for stargazin (see supplemental Table 1, available at www.jneurosci.org/cgi/content/full/24/34/9491/DC1), suggesting that nPIST may interact directly with PSD-95.
Figure 7.
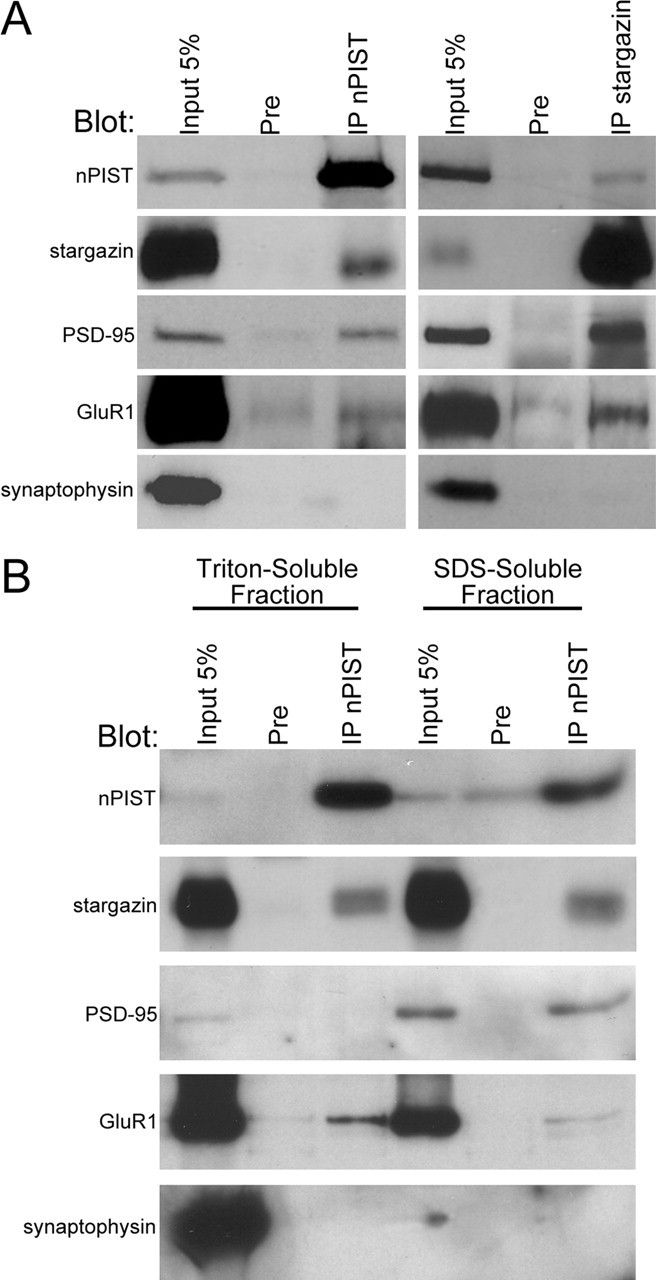
nPIST and stargazin cofractionate and coimmunoprecipitate from brain. A, Stargazin and nPIST coimmunoprecipitate from the brain. Rat brain membranes were solubilized with SDS. After the addition of Triton X-100, extracts were immunoprecipitated with antibodies to either PIST (IP nPIST), preimmune sera (Pre), or stargazin-specific antibodies (IP stargazin). After SDS-PAGE, blots were probed with antibodies to PIST (nPIST), stargazin, PSD-95, GluR1, or synaptophysin. PIST antisera avidly precipitated nPIST and also coimmunoprecipitated stargazin, PSD-95, and GluR1 but not synaptophysin. Stargazin antiserum immunoprecipitated stargazin and also precipitated nPIST, PSD-95, and GluR1 but not synaptophysin. B, nPIST cofractionates with stargazin in brain membranes and coimmunoprecipitates GluR1 and PSD-95 from different fractions. Crude membranes were solubilized with Triton X-100 or SDS as described in Materials and Methods. nPIST and stargazin were present in both Triton X-100-soluble and SDS-soluble fractions, whereas PSD-95 was enriched in SDS-soluble and GluR1 was enriched in Triton X-100-soluble membranes. PIST antisera (IP nPIST) effectively coimmunoprecipitated stargazin from both fractions, PSD-95 from SDS soluble membranes, and GluR1 from Triton-soluble membranes. Synaptophysin was present in the Triton-soluble fraction but was not coimmunoprecipitated with PIST antiserum. For quantitation, see supplementary Table 1 (available at www.jneurosci.org/cgi/content/full/24/34/9491/DC1).
Synaptic proteins such as stargazin, AMPARs, and PSD-95 fractionate differentially. Whereas PSD-95 is a relatively insoluble component of the PSD and requires SDS for solubilization, stargazin and AMPAR are mostly Triton extractable, perhaps reflecting their dynamic regulation at the PSD (Park et al., 1999). To further explore the interaction between nPIST and stargazin, we fractionated brain membranes and evaluated the distribution in and immunoprecipitation from Triton-soluble and -insoluble membranes. nPIST and stargazin appeared evenly distributed between the two fractions, whereas GluR1 was enriched in the Triton-soluble membranes and PSD-95 was present at higher levels in the Triton-insoluble membranes (Fig. 7B, Input lanes). Coimmunoprecipitation using anti-PIST antisera revealed coimmunoprecipitation of stargazin with nPIST in both Triton-soluble and -insoluble fractions. In contrast, GluR1 coimmunoprecipitated with nPIST primarily in Triton-soluble membranes, and PSD-95 coimmunoprecipitated with nPIST from the Triton-insoluble fraction (Fig. 7B; supplemental Table 1, available at www.jneurosci.org/cgi/content/full/24/34/9491/DC1). These findings suggest that nPIST may exist in distinct biochemical pools at the PSD, a relatively soluble fraction that binds to stargazin and AMPARs, and a less soluble fraction that interacts with stargazin and PSD-95 at the synapse.
Full-length nPIST is necessary for stargazin-AMPAR synaptic targeting
To further explore the possibility that nPIST-stargazin interactions may be critical for AMPAR function, we cotransfected nPIST-GFP and stargazin-HA into cultured hippocampal neurons. Stargazin-HA expressed alone is targeted to synapses (Chen et al., 2000). Similar to endogenous nPIST, nPIST-GFP expressed alone primarily localizes to the Golgi but also shows both diffuse and punctate dendritic staining (Fig. 8A-C). When stargazin-HA is cotransfected with nPIST-GFP in neurons, both proteins colocalize in the perinuclear area as well as in proximal and distal dendrites (Fig. 8D-F), confirming that nPIST and stargazin can interact in cultured neurons. Because we hypothesize that nPIST may serve a chaperone role for stargazin-AMPARs in neurons, we reasoned that nPIST overexpression might affect stargazin-AMPAR synaptic clustering. We therefore evaluated GluR1 clustering in neurons transfected with nPIST. We found that GluR1 staining in hippocampal neurons transfected with nPIST-GFP showed a small, but significant, increase in cluster intensity compared with control neurons (120.1 ± 5.5% control CIR; n = 7; p < 0.05) (Fig. 8G-I, M; also see supplementary Fig. 3, available at www.jneurosci.org/cgi/content/full/24/34/9491/DC1), consistent with a chaperone role for nPIST in stargazin-AMPAR synaptic targeting. Because PDZ domains are critical for synaptic targeting of PSD-95, and protein AC motifs can regulate membrane trafficking, we reasoned that the C terminus of nPIST, which contains both a PDZ and AC domain, could function in the delivery of stargazin-AMPARs to the synapse. We therefore transfected neurons with nPIST(1-312)-GFP, a construct that retains stargazin binding and Golgi localization in COS-7 cells but lacks the C-terminal PDZ and AC domains that could be important for synaptic targeting. We reasoned that this construct could function as a “dominant negative” by binding to endogenous stargazin, thereby reducing interaction with endogenous nPIST and preventing synaptic targeting. Transfection of neurons with nPIST(1-312)-GFP resulted in Golgi staining similar to nPIST-GFP; however, GluR1 synaptic clustering was dramatically decreased (64.3 ± 6.9% control CIR; n = 5; p < 0.05) (Fig. 8J-M; also see supplementary Fig. 3, available at www.jneurosci.org/cgi/content/full/24/34/9491/DC1). Although stargazin-AMPAR interactions can be dissociated by adding glutamate or AMPA to cultured neurons (Tomita et al., 2004), under basal conditions such as those used in these experiments, endogenous stargazin/TARPs are almost 100% colocalized with GluR1 clusters (Tomita et al., 2003). Indeed, by using an antibody that recognizes stargazin and other TARPs, we found that nPIST(1-312)-GFP diminished and nPIST-GFP increased endogenous stargazin/TARP clustering, consistent with nPIST-mediated effects on stargazin-AMPARs synaptic targeting [121.3 ± 2.6% control CIR for nPIST-GFP (n = 3; p < 0.01) and 64.5 ± 4.1% control CIR for nPIST(1-312)-GFP (n = 5; p < 0.01)] (supplementary Fig. 3, available at www.jneurosci.org/cgi/content/full/24/34/9491/DC1). Neither nPIST-GFP nor nPIST(1-312)-GFP had any effect on NR1 clustering [89.4 ± 9.7% control CIR for nPIST-GFP (n = 5; p > 0.05) and 87.4 ± 7.8% control CIR for nPIST(1-312)-GFP (n = 5; p > 0.05)] (supplementary Fig. 3, available at www.jneurosci.org/cgi/content/full/24/34/9491/DC1), nor did they appear to have nonspecific effects on synaptogenesis because PSD-95 and synaptophysin staining were unchanged (data not shown). These findings suggest that nPIST serves as a selective chaperone for stargazin-AMPARs in the delivery of AMPAR to established PSD-95-containing synapses and that the nPIST PDZ or AC domains are required for this function.
Figure 8.
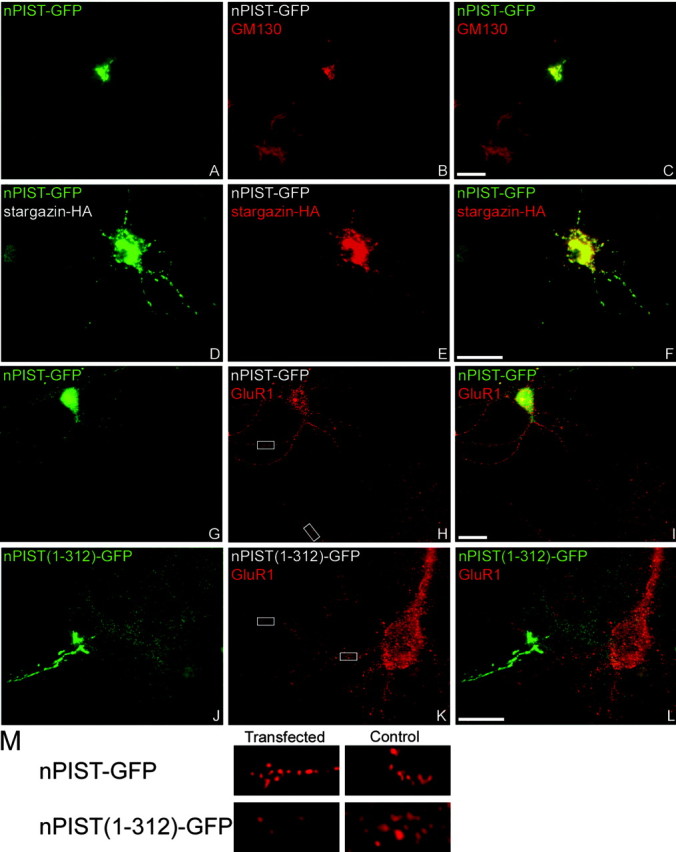
nPIST regulates AMPAR clustering. Constructs encoding nPIST-GFP, nPIST(1-312)-GFP, and stargazin-HA were transfected into neurons, which were then fixed (at 11-17 d in vitro) and stained for GFP (green) or GM130, HA, or GluR1 (red). When transfected alone, nPIST-GFP (green) colocalizes with GM130 (red) in the Golgi as well as in puncta and diffusely within dendrites (A-C). When cotransfected with stargazin-HA (red), both proteins colocalize perinuclearly and in dendrites (D-F). nPIST-GFP, when transfected alone, results in a small but significant increase in AMPAR clustering (red) (G-I). In contrast, nPIST(1-312)-GFP (green) dramatically reduces the intensity of GluR1 synaptic clusters (red) (J-L). Merged images are shown in C, F, I, and L. Scale bar, 10 μm. M, Insets from H and K showing 6× digital magnification of GluR1 staining in transfected or control cells for nPIST-GFP and nPIST(1-312)-GFP.
Discussion
This study demonstrates the presence of a novel domain within the C terminus of stargazin necessary for the synaptic targeting of stargazin and AMPARs and implicates nPIST as a chaperone for stargazin-AMPAR synaptic targeting. Interactions between AMPARs and scaffolding molecules at the synapse have been studied in detail previously. This present work illustrates a distinct component of the AMPAR trafficking pathway that could play a role in the development and activity-dependent modulation of AMPAR-mediated currents at synapses.
AMPARs are tetramers assembled from four subunits, GluR1-GluR4, with most AMPARs containing a GluR1 and/or a GluR2 subunit. The C terminus of GluR1 contains a PDZ-binding motif that is important for calcium/calmodulin-dependent protein kinase II-dependent insertion of AMPARs at the synapse, but the synaptic binding partner of GluR1 necessary for this insertion is not known (Leonard et al., 1998; Hayashi et al., 2000). The C terminus of GluR2 binds to GRIP/ABP (Dong et al., 1997; Srivastava et al., 1998), as well as PICK1 (Matsuda et al., 1999; Xia et al., 1999; Chung et al., 2000), and the phosphorylation of the GluR2 PDZ-binding site by protein kinase C inhibits binding of GluR2 to GRIP/ABP but does not affect PICK1 binding (Matsuda et al., 1999; Chung et al., 2000). Phosphorylation of GluR2 subunits might underlie certain forms of synaptic plasticity because recruitment of AMPARs to silent synapses requires an AMPAR interaction with GRIP (Li et al., 1999), and induction of long-term depression (LTD) in the cerebellum and hippocampus is associated with phosphorylation of the PDZ-binding site of GluR2, favoring an interaction with PICK1 (Xia et al., 2000; Kim et al., 2001; Seidenman et al., 2003). In addition to PDZ binding, the C terminus of GluR2 interacts with the membrane fusion protein NSF (N-ethylmaleimide-sensitive fusion protein) (Nishimune et al., 1998; Osten et al., 1998; Song et al., 1998; Noel et al., 1999), an interaction that may also regulate expression of AMPARs at the synapse (Luthi et al., 1999; Luscher et al., 2000).
Although regulation of interactions between AMPAR subunits and their binding partners is clearly implicated in the stabilization of AMPARs at the synapse and synaptic plasticity, much less is known about the mechanisms of AMPAR trafficking from its site of synthesis in the endoplasmic reticulum to the synapse. AMPAR targeting to the synapse requires interaction with the transmembrane protein stargazin (Chen et al., 2000), likely through interaction of GluR subunits with the transmembrane domain of stargazin (Tomita et al., 2004). The cytosolic COOH-terminal tail of stargazin contains a PDZ-binding site that binds to type 1 PDZ domains from PSD-95 and related proteins. This PDZ-binding site is critical for synaptic function of AMPARs, because transfecting mutant granule cells with a stargazin construct lacking the extreme C terminus, stargazin(Δ319-323), does not rescue synaptic responses. Interestingly, stargazin(Δ319-323), which also binds to AMPARs, rescues extrasynaptic AMPAR responses (Chen et al., 2000), suggesting that AMPAR targeting to the synapse progresses by two distinct stargazin-dependent mechanisms. First, the stargazin transmembrane domains interact with AMPARs and regulate the delivery of AMPARs to the cell surface. Second, interaction of the C-terminal PDZ-binding site of stargazin with PSD-95 or a related PDZ protein targets AMPARs to synapses. The focus of our current study sheds additional light on the mechanism of stargazin-dependent AMPAR targeting to synapses. Although the C-terminal PDZ-binding domain is necessary for synaptic targeting, it is not sufficient.
Interestingly, the stargazin residues exclusive of the PDZ-binding domain necessary for synaptic targeting are highly conserved between AMPAR-targeting stargazin homologs and interact with the neuronal isoform of the cytoplasmic protein PIST, a cytosolic protein that contains two coiled-coil domains, a PDZ domain and an AC region (Neudauer et al., 2001). The nonneuronal isoform of PIST is widely expressed and predominantly localized to the Golgi apparatus. Additionally, the coiled-coil domain of PIST binds to the Q-SNARE (Q-soluble N-ethylmaleimide-sensitive factor attachment protein receptor) syntaxin-6, implicating PIST in membrane vesicle trafficking (Charest et al., 2001). Furthermore, PIST overexpression decreases the membrane trafficking of cystic fibrosis transmembrane conductance regulator (Cheng et al., 2002; Swiatecka-Urban et al., 2002) and is implicated in membrane expression of frizzled (Yao et al., 2001), effects mediated by interaction of the respective proteins with the PDZ domain of PIST. Given the likely role of PIST in modulating vesicular protein trafficking, our findings that nPIST binds stargazin at the same region necessary for stargazin synaptic targeting and that the dominant-negative nPIST construct attenuates AMPAR clustering strongly suggests that nPIST interaction with stargazin is critical for AMPAR trafficking and synaptic delivery.
The nPIST C terminus is critical for stargazin-AMPAR synaptic targeting. Whereas nPIST PDZ domain interactions may underlie this function, the AC domain is also a potential site for regulation of nPIST intracellular trafficking between the Golgi and synapse. In neurons, the Golgi apparatus is present in the cell body region as well as in the dendritic shaft, and this distributed network of secretory organelles at distant sites in neurons may allow for localized control over protein sorting and delivery of synaptic components (Horton and Ehlers, 2003). Interestingly, AC domains regulate trafficking between the Golgi apparatus and the plasma membrane in several proteins (Molloy et al., 1999). The trafficking of one such protein, furin, is regulated by phosphorylation of the AC domain by casein kinase 2 (CK2) and dephosphorylation by protein phosphatase 2A (PP2A) (Jones et al., 1995; Molloy et al., 1998). CK2, which promotes surface expression of furin, has been shown to be upregulated in hippocampal long-term potentiation (Charriaut-Marlangue et al., 1991), whereas PP2A, which downregulates furin membrane expression, has been implicated in hippocampal LTD (Mulkey et al., 1993). Future studies will address the role of the nPIST AC domain in synaptic targeting and explore whether nPIST AC domain phosphorylation during synaptic plasticity controls the trafficking of the stargazin-AMPAR complex between the Golgi and synapses or surface membrane.
In addition to regulating vesicular trafficking of transmembrane proteins, nPIST has been implicated in the mechanism of autophagy in the lurcher mouse via interactions with GluRδ2 and Beclin-1 (Yue et al., 2002). Activation of autophagy results in budding of vesicles from the Golgi and plasma membrane, a phenomenon consistent with the described role of PIST in vesicular trafficking of transmembrane proteins in extra-neuronal tissues. That the PDZ domain of nPIST interacts with GluRδ2 is particularly interesting in the context of the role of stargazin in synaptic targeting. Additionally, we find that the nPIST PDZ domain interacts with the C terminus of NR2A, another molecule enriched at synapses. Our data support a model in which nPIST interacts with the stargazin-AMPAR complex and serves as a chaperone during trafficking between departure from the Golgi apparatus and ultimate delivery to the synapse (Fig. 9A). GluRδ2 or NR2A may serve as a docking site for nPIST in the delivery of the stargazin-AMPAR to scaffolding molecules such as PSD-95 at the synapse. In contrast, our finding that nPIST coimmunoprecipitates efficiently with PSD-95 in the brain and suggests direct binding of nPIST to PSD-95 at the PSD. Additional studies are needed to elucidate whether nPIST binding to GluRδ2, NR2A, or PSD-95 might influence stargazin-AMPAR synaptic targeting.
Figure 9.
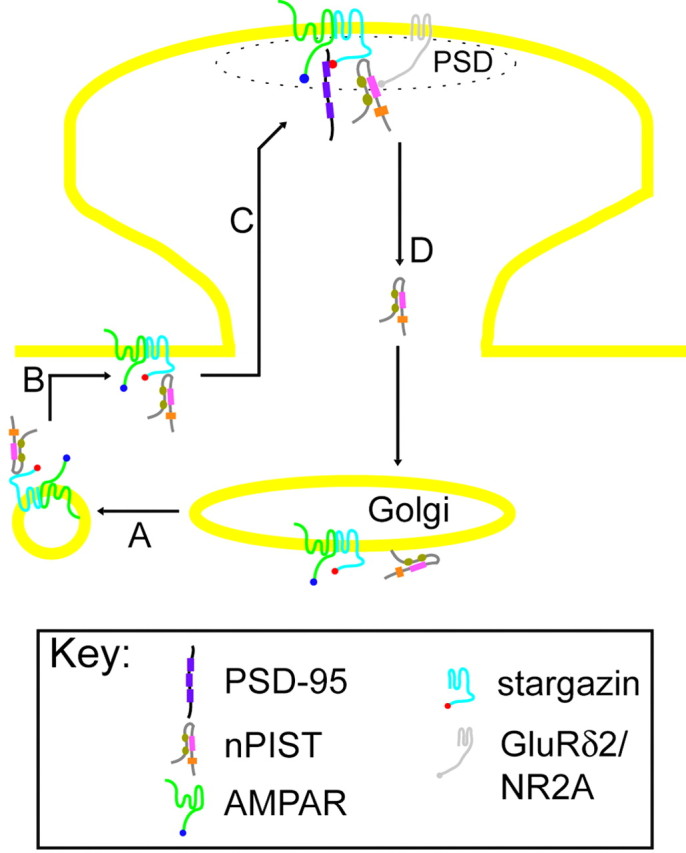
Model demonstrating regulation of AMPAR at synapses. Stargazin-AMPAR complexes interact with nPIST in the Golgi and exit the Golgi in vesicles (A). The vesicles fuse at extrasynaptic sites (B), and nPIST chaperones the stargazin-AMPAR complex to the PSD, where it interacts with synaptic proteins such as GluRδ2, NR2A, or directly with PSD-95, allowing the stargazin C terminus to bind PSD-95 and stabilize the AMPAR at the synapse (C). nPIST is recycled to the Golgi to repeat the cycle (D).
We show evidence that nPIST is necessary for proper AMPAR targeting to the synapse and suggest a model in which nPIST releases the stargazin-AMPAR complex to synaptic proteins present at the PSD. The binding of stargazin to synaptic PDZ proteins such as PSD-95 could serve as an anchoring point for functional stargazin-AMPARs at the synapse. Indeed, PSD-95-mediated enhancement of AMPAR currents at the synapse is dependent on an intact stargazin PDZ-binding domain (Schnell et al., 2002). Alternatively, stargazin binding to PSD-95 at the synapse could serve as a requisite and limiting “point of entry” for stargazin-AMPAR complexes at the synapse. After introduction of the stargazin-AMPAR complex to the synapse through stargazin binding to PSD-95, constitutive or activity-dependent modifications such as phosphorylation of AMPAR subunits or stargazin might regulate the transfer of the stargazin-AMPAR complex between physiologically active sites, or perhaps between active and silent sites by favoring interactions with different stargazin or AMPAR subunit binding proteins. Along these lines, stargazin binding to PSD-95 is blocked by phosphorylation by protein kinase A (PKA) (Chetkovich et al., 2002), a protein kinase implicated in synaptic plasticity. Phosphorylation of stargazin by PKA at the synapse could release the stargazin-AMPAR complex from its binding to PSD-95, which would allow introduction of more nonphosphorylated stargazin-AMPAR to PSD-95 at the synapse. Additionally, release of stargazin-AMPARs from PSD-95 to synaptic GRIP/ABP would allow regulation of AMPAR function to be determined by GluR1 or GluR2 interactions with their respective binding partners such as GRIP/ABP or PICK1. Direct phosphorylation of GluR subunits could thereby modulate AMPAR function at the synapse under conditions of synaptic plasticity. Whether nPIST-stargazin interactions function in the shuttling of AMPAR between PSD-95 and GRIP/ABP or PICK1 at the synapse remains to be investigated. It is our goal that these and future studies aimed at understanding the processes of AMPAR targeting and trafficking will elucidate mechanisms underlying development and synaptic plasticity and may shed light on the disruption of synaptic function thought to underlie neurological diseases such as epilepsy and neurodegenerative conditions such as Alzheimer's disease.
Footnotes
This work was supported by grants from the Human Frontiers Science Program and the Christopher Reeves Paralysis Foundation to D.S.B., from the National Institutes of Health to D.S.B. and D.M.C., and from the National Association for Research on Schizophrenia and Depression to D.M.C. D.S.B. is an established investigator of the American Heart Association, and D.M.C. is a fellow of the Howard Hughes Medical Institute. We thank Drs. Jack Kessler and Jaime Garcia-Anoveros for comments on this manuscript and Tim Stocker and Adrienne Wang for technical support with preliminary experiments in the preparation of this manuscript.
Correspondence should be addressed to Dr. Dane M. Chetkovich, Davee Department of Neurology and Clinical Neuroscience, Northwestern University Medical School, 303 East Chicago Avenue, Ward 10-201, Chicago, IL 60611-3008. E-mail: d-chetkovich@northwestern.edu.
Copyright © 2004 Society for Neuroscience 0270-6474/04/249491-12$15.00/0
A.E.C. and S.-H.K contributed equally to this work.
References
- Brenman JE, Chao DS, Gee SH, McGee AW, Craven SE, Santillano DR, Wu Z, Huang F, Xia H, Peters MF, Froehner SC, Bredt DS (1996) Interaction of nitric oxide synthase with the postsynaptic density protein PSD-95 and alpha1-syntrophin mediated by PDZ domains. Cell 84: 757-767. [DOI] [PubMed] [Google Scholar]
- Carroll RC, Lissin DV, von Zastrow M, Nicoll RA, Malenka RC (1999) Rapid redistribution of glutamate receptors contributes to long-term depression in hippocampal cultures. Nat Neurosci 2: 454-460. [DOI] [PubMed] [Google Scholar]
- Charest A, Lane K, McMahon K, Housman DE (2001) Association of a novel PDZ domain-containing peripheral Golgi protein with the Q-SNARE (Q-soluble N-ethylmaleimide-sensitive fusion protein (NSF) attachment protein receptor) protein syntaxin 6. J Biol Chem 276: 29456-29465. [DOI] [PubMed] [Google Scholar]
- Charriaut-Marlangue C, Otani S, Creuzet C, Ben-Ari Y, Loeb J (1991) Rapid activation of hippocampal casein kinase II during long-term potentiation. Proc Natl Acad Sci USA 88: 10232-10236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Chetkovich DM, Petrailia R, Sweeney N, Kawaski Y, Wenthold R, Bredt DS, Nicoll RA (2000) Stargazin mediates synaptic targeting of AMPA receptors by two distinct mechanisms. Nature 408: 936-943. [DOI] [PubMed] [Google Scholar]
- Cheng J, Moyer BD, Milewski M, Loffing J, Ikeda M, Mickle JE, Cutting GR, Li M, Stanton BA, Guggino WB (2002) A Golgi-associated PDZ domain protein modulates cystic fibrosis transmembrane regulator plasma membrane expression. J Biol Chem 277: 3520-3529. [DOI] [PubMed] [Google Scholar]
- Chetkovich DM, Chen L, Stocker TJ, Nicoll RA, Bredt DS (2002) Phosphorylation of the postsynaptic density-95 (PSD-95)/Discs large/zona occludens-1 binding site of stargazin regulates binding to PSD-95 and synaptic targeting of AMPA receptors. J Neurosci 22: 5791-5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung HJ, Xia J, Scannevin RH, Zhang X, Huganir RL (2000) Phosphorylation of the AMPA receptor subunit GluR2 differentially regulates its interaction with PDZ domain-containing proteins. J Neurosci 20: 7258-7267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craven SE, El-Husseini AE, Bredt DS (1999) Synaptic targeting of the postsynaptic density protein PSD-95 mediated by lipid and protein motifs. Neuron 22: 497-509. [DOI] [PubMed] [Google Scholar]
- Dev KK, Nishimune A, Henley JM, Nakanishi S (1999) The protein kinase C alpha binding protein PICK1 interacts with short but not long form alternative splice variants of AMPA receptor subunits. Neuropharmacology 38: 635-644. [DOI] [PubMed] [Google Scholar]
- Dong H, O'Brien RJ, Fung ET, Lanahan AA, Worley PF, Huganir RL (1997) GRIP: a synaptic PDZ domain-containing protein that interacts with AMPA receptors. Nature 386: 279-284. [DOI] [PubMed] [Google Scholar]
- Ehlers MD (2000) Reinsertion or degradation of AMPA receptors determined by activity-dependent endocytic sorting. Neuron 28: 511-525. [DOI] [PubMed] [Google Scholar]
- El-Husseini AE, Schnell E, Chetkovich DM, Nicoll RA, Bredt DS (2000) PSD-95 involvement in maturation of excitatory synapses. Science 290: 1364-1368. [PubMed] [Google Scholar]
- Garner CC, Nash J, Huganir RL (2000) PDZ domains in synapse assembly and signalling. Trends Cell Biol 10: 274-280. [DOI] [PubMed] [Google Scholar]
- Hayashi Y, Shi SH, Esteban JA, Piccini A, Poncer JC, Malinow R (2000) Driving AMPA receptors into synapses by LTP and CaMKII: requirement for GluR1 and PDZ domain interaction. Science 287: 2262-2267. [DOI] [PubMed] [Google Scholar]
- Horton AC, Ehlers MD (2003) Dual modes of endoplasmic reticulum-to-Golgi transport in dendrites revealed by live-cell imaging. J Neurosci 23: 6188-6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones BG, Thomas L, Molloy SS, Thulin CD, Fry MD, Walsh KA, Thomas G (1995) Intracellular trafficking of furin is modulated by the phosphorylation state of a casein kinase II site in its cytoplasmic tail. EMBO J 14: 5869-5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech S, Kim JB, Cariola M, Ralston E (1996) Improved lipid-mediated gene transfer into primary cultures of hippocampal neurons. Brain Res Mol Brain Res 35: 344-348. [DOI] [PubMed] [Google Scholar]
- Kim CH, Chung HJ, Lee HK, Huganir RL (2001) Interaction of the AMPA receptor subunit GluR2/3 with PDZ domains regulates hippocampal long-term depression. Proc Natl Acad Sci USA 98: 11725-11730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Huganir RL (1999) Organization and regulation of proteins at synapses. Curr Opin Cell Biol 11: 248-254. [DOI] [PubMed] [Google Scholar]
- Kornau HC, Seeburg PH, Kennedy MB (1997) Interaction of ion channels and receptors with PDZ domain proteins. Curr Opin Neurobiol 7: 368-373. [DOI] [PubMed] [Google Scholar]
- Leonard AS, Davare MA, Horne MC, Garner CC, Hell JW (1998) SAP97 is associated with the alpha-amino-3-hydroxy-5-methylisoxazole-4-propionic acid receptor GluR1 subunit. J Biol Chem 273: 19518-19524. [DOI] [PubMed] [Google Scholar]
- Letts VA, Felix R, Biddlecome GH, Arikkath J, Mahaffey CL, Valenzuela A, Bartlett 2nd FS, Mori Y, Campbell KP, Frankel WN (1998) The mouse stargazer gene encodes a neuronal Ca2+-channel gamma subunit. Nat Genet 19: 340-347. [DOI] [PubMed] [Google Scholar]
- Li P, Kerchner GA, Sala C, Wei F, Huettner JE, Sheng M, Zhuo M (1999) AMPA receptor-PDZ interactions in facilitation of spinal sensory synapses. Nat Neurosci 2: 972-977. [DOI] [PubMed] [Google Scholar]
- Liu SQ, Cull-Candy SG (2000) Synaptic activity at calcium-permeable AMPA receptors induces a switch in receptor subtype. Nature 405: 454-458. [DOI] [PubMed] [Google Scholar]
- Luscher C, Nicoll RA, Malenka RC, Muller D (2000) Synaptic plasticity and dynamic modulation of the postsynaptic membrane. Nat Neurosci 3: 545-550. [DOI] [PubMed] [Google Scholar]
- Luthi A, Chittajallu R, Duprat F, Palmer MJ, Benke TA, Kidd FL, Henley JM, Isaac JT, Collingridge GL (1999) Hippocampal LTD expression involves a pool of AMPARs regulated by the NSF-GluR2 interaction. Neuron 24: 389-399. [DOI] [PubMed] [Google Scholar]
- Malinow R, Mainen ZF, Hayashi Y (2000) LTP mechanisms: from silence to four-lane traffic. Curr Opin Neurobiol 10: 352-357. [DOI] [PubMed] [Google Scholar]
- Man YH, Lin JW, Ju WH, Ahmadian G, Liu L, Becker LE, Sheng M, Wang YT (2000) Regulation of AMPA receptor-mediated synaptic transmission by clathrin-dependent receptor internalization. Neuron 25: 649-662. [DOI] [PubMed] [Google Scholar]
- Matsuda S, Mikawa S, Hirai H (1999) Phosphorylation of serine-880 in GluR2 by protein kinase C prevents its C terminus from binding with glutamate receptor-interacting protein. J Neurochem 73: 1765-1768. [DOI] [PubMed] [Google Scholar]
- Molloy SS, Thomas L, Kamibayashi C, Mumby MC, Thomas G (1998) Regulation of endosome sorting by a specific PP2A isoform. J Cell Biol 142: 1399-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molloy SS, Anderson ED, Jean F, Thomas G (1999) Bi-cycling the furin pathway: from TGN localization to pathogen activation and embryogenesis. Trends Cell Biol 9: 28-35. [DOI] [PubMed] [Google Scholar]
- Mulkey RM, Herron CE, Malenka RC (1993) An essential role for protein phosphatases in hippocampal long-term depression. Science 261: 1051-1055. [DOI] [PubMed] [Google Scholar]
- Neudauer CL, Joberty G, Macara IG (2001) PIST: a novel PDZ/coiled-coil domain binding partner for the rho-family GTPase TC10. Biochem Biophys Res Commun 280: 541-547. [DOI] [PubMed] [Google Scholar]
- Nishimune A, Isaac JT, Molnar E, Noel J, Nash SR, Tagaya M, Collingridge GL, Nakanishi S, Henley JM (1998) NSF binding to GluR2 regulates synaptic transmission. Neuron 21: 87-97. [DOI] [PubMed] [Google Scholar]
- Noel J, Ralph GS, Pickard L, Williams J, Molnar E, Uney JB, Collingridge GL, Henley JM (1999) Surface expression of AMPA receptors in hippocampal neurons is regulated by an NSF-dependent mechanism. Neuron 23: 365-376. [DOI] [PubMed] [Google Scholar]
- Osten P, Srivastava S, Inman GJ, Vilim FS, Khatri L, Lee LM, States BA, Einheber S, Milner TA, Hanson PI, Ziff EB (1998) The AMPA receptor GluR2 C terminus can mediate a reversible, ATP-dependent interaction with NSF and alpha- and beta-SNAPs. Neuron 21: 99-110. [DOI] [PubMed] [Google Scholar]
- Park HT, Jeon ES, Bae KW (1999) Postnatal development of detergent-insoluble properties of NMDA and AMPA receptor subunits in the rat brain synaptic membrane. Brain Res Dev Brain Res 115: 83-87. [DOI] [PubMed] [Google Scholar]
- Sans N, Racca C, Petralia RS, Wang YX, McCallum J, Wenthold RJ (2001) Synapse-associated protein 97 selectively associates with a subset of AMPA receptors early in their biosynthetic pathway. J Neurosci 21: 7506-7516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassoon D, Rosenthal N (1993) Detection of messenger RNA by in situ hybridization. Methods Enzymol 225: 384-404. [DOI] [PubMed] [Google Scholar]
- Schnell E, Sizemore M, Karimzadegan S, Chen L, Bredt DS, Nicoll RA (2002) Direct interactions between PSD-95 and stargazin control synaptic AMPA receptor number. Proc Natl Acad Sci USA 99: 13902-13907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidenman KJ, Steinberg JP, Huganir R, Malinow R (2003) Glutamate receptor subunit 2 Serine 880 phosphorylation modulates synaptic transmission and mediates plasticity in CA1 pyramidal cells. J Neurosci 23: 9220-9228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song I, Kamboj S, Xia J, Dong H, Liao D, Huganir RL (1998) Interaction of the N-ethylmaleimide-sensitive factor with AMPA receptors. Neuron 21: 393-400. [DOI] [PubMed] [Google Scholar]
- Srivastava S, Osten P, Vilim FS, Khatri L, Inman G, States B, Daly C, DeSouza S, Abagyan R, Valtschanoff JG, Weinberg RJ, Ziff EB (1998) Novel anchorage of GluR2/3 to the postsynaptic density by the AMPA receptor-binding protein ABP. Neuron 21: 581-591. [DOI] [PubMed] [Google Scholar]
- Swiatecka-Urban A, Duhaime M, Coutermarsh B, Karlson KH, Collawn J, Milewski M, Cutting GR, Guggino WB, Langford G, Stanton BA (2002) PDZ domain interaction controls the endocytic recycling of the cystic fibrosis transmembrane conductance regulator. J Biol Chem 277: 40099-40105. [DOI] [PubMed] [Google Scholar]
- Tomita S, Chen L, Kawasaki Y, Petralia RS, Wenthold RJ, Nicoll RA, Bredt DS (2003) Functional studies and distribution define a family of transmembrane AMPA receptor regulatory proteins. J Cell Biol 161: 805-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita S, Fukata M, Nicoll RA, Bredt DS (2004) Dynamic interaction of stargazin-like TARPs with cycling AMPA receptors at synapses. Science 303: 1508-1511. [DOI] [PubMed] [Google Scholar]
- Topinka JR, Bredt DS (1998) N-terminal palmitoylation of PSD-95 regulates association with cell membranes and interaction with K+ channel, Kv1.4. Neuron 20: 125-134. [DOI] [PubMed] [Google Scholar]
- Xia J, Zhang X, Staudinger J, Huganir RL (1999) Clustering of AMPA receptors by the synaptic PDZ domain-containing protein PICK1. Neuron 22: 179-187. [DOI] [PubMed] [Google Scholar]
- Xia J, Chung HJ, Wihler C, Huganir RL, Linden DJ (2000) Cerebellar long-term depression requires PKC-regulated interactions between GluR2/3 and PDZ domain-containing proteins. Neuron 28: 499-510. [DOI] [PubMed] [Google Scholar]
- Yao R, Maeda T, Takada S, Noda T (2001) Identification of a PDZ domain containing Golgi protein, GOPC, as an interaction partner of frizzled. Biochem Biophys Res Commun 286: 771-778. [DOI] [PubMed] [Google Scholar]
- Yue Z, Horton A, Bravin M, DeJager P, Selimi F, Heintz N (2002) A novel protein complex linking the delta2 glutamate receptor and autophagy. Implications for neurodegeneration in lurcher mice. Neuron 35: 921-933. [DOI] [PubMed] [Google Scholar]
- Ziff EB (1997) Enlightening the postsynaptic density. Neuron 19: 1163-1174. [DOI] [PubMed] [Google Scholar]