Abstract
Gap junctions are intercellular channels that allow the passage of ions and small molecules between cells. In the nervous system, gap junctions mediate electrical coupling between neurons. Despite sharing a common topology and similar physiology, two unrelated gap junction protein families exist in the animal kingdom. Vertebrate gap junctions are formed by members of the connexin family, whereas invertebrate gap junctions are composed of innexin proteins. Here we report the cloning of two innexins from the leech Hirudo medicinalis. These innexins show a differential expression in the leech CNS: Hm-inx1 is expressed by every neuron in the CNS but not in glia, whereas Hm-inx2 is expressed in glia but not neurons. Heterologous expression in the paired Xenopus oocyte system demonstrated that both innexins are able to form functional homotypic gap junctions. Hm-inx1 forms channels that are not strongly gated. In contrast, Hm-inx2 forms channels that are highly voltage-dependent; these channels demonstrate properties resembling those of a double rectifier. In addition, Hm-inx1 and Hm-inx2 are able to cooperate to form heterotypic gap junctions in Xenopus oocytes. The behavior of these channels is primarily that predicted from the properties of the constituent hemichannels but also demonstrates evidence of an interaction between the two.
This work represents the first demonstration of a functional gap junction protein from a Lophotrochozoan animal and supports the hypothesis that connexin-based communication is restricted to the deuterostome clade.
Keywords: connexin, electrical synapse, gap junction, Hirudo medicinalis , innexin, leech, neuronal-glial communication, Xenopus oocytes
Introduction
Much of our knowledge about the basic properties of neuronal communication has been derived from the study of simple nervous systems such as the leech. Its large neurons may be studied using intracellular recording, and the synapses between many of its identified neurons have been characterized (Muller, 1979). Electrical coupling between leech neurons is common, allowing the fast propagation of impulses between cells (Baylor and Nicholls, 1969; Davis, 1989), and this coupling is important to maintain synchronicity in many neural circuits (Marder, 1984; Fernandez-de-Miguel et al., 2001; Rela and Szczupak, 2003). Electrical synapses are mediated by gap junctional coupling between neurons; similar structures are seen between non-neuronal cells.
As well as this role in neural networks, gap junctions are implicated in both homeostasis and embryonic development by their ability to mediate the passage of low molecular weight signaling molecules between cells. In the grasshopper embryo, path-finding neurons communicate with “guidepost” cells via gap junctions (Taghert et al., 1982), and inhibitory signaling mediated by transient gap junctional coupling is important during the development of connectivity within the leech CNS (Wolszon et al., 1994). Such coupling is also implicated in the ability of leech neurons to regenerate specific synapses after injury; temporary gap junctions connect the regenerating axon to its severed stump, leading to a fast recovery of function (Carbonetto and Muller, 1977).
Gap junctions appear to be common to all multicellular animals, but despite this functional conservation, a dichotomy exists at the molecular level. In deuterostomes, gap junctions are mediated by the connexin gene family (Kumar and Gilula, 1996; Harris, 2001), four-pass transmembrane proteins that aggregate to form hexameric hemichannels spanning each membrane. No connexin sequences have been found, however, in the now complete genomes of Drosophila and Caenorhabditis elegans. Gap junctional communication in these animals is instead mediated by members of the innexin family (Phelan et al., 1998b; Phelan and Starich, 2001). Innexins are also four-pass transmembrane proteins that appear to form multimeric channels but show no primary sequence homology to the connexins. Innexins have been cloned from molluscs (Alexopoulos et al., 2000; Kelmanson et al., 2002), flatworm (Panchin et al., 2000), and from a polychaete annelid (Potenza et al., 2002), suggesting that members of this gene family mediate gap-junction communication in all protostome animals. However, it has not been demonstrated experimentally that these Lophotrochozoan innexins form functional gap junction channels.
We wished to characterize the molecular basis of gap junctional communication in the leech CNS. Here we describe the cloning of two leech CNS innexins and demonstrate by in situ hybridization that Hm-inx1 is expressed in neurons, whereas Hm-inx2 is expressed in glia. To demonstrate that these proteins can form functional gap junctions we expressed them in paired Xenopus oocytes. This showed that each innexin is capable of forming homotypic channels and furthermore that the two innexins can interact to form a heterotypic channel. Each junction has distinct voltage-gating properties, and we have correlated these data with the in situ expression data to propose a function for these channels in vivo.
Materials and Methods
Molecular cloning. Putative leech innexin and connexin sequences were cloned by PCR amplification from a leech CNS cDNA library (a gift of Sergei Korneev, Sussex University) using degenerate and consensus-degenerate primers.
A degenerate connexin-specific primer was designed against the first extracellular loop region (consensus CNTxQP) of a multiple sequence alignment of eight connexins. This primer, GGYTGIWIIGTRTTRCA, was used together with a vector-specific primer to amplify putative connexin sequences from the leech CNS cDNA library.
A degenerate innexin-specific PCR primer was designed against the highly conserved second transmembrane domain of the innexin protein. A multiple sequence alignment of 12 C.elegans and 2 Drosophila innexins was generated using ClustalW (Thompson et al., 1994), and this was used to derive the consensus amino acid sequence of YYQWVP. This was reverse translated to give the degenerate primer GGIACCCAYTGRTARTA. This primer was used together with a vector-specific primer to amplify putative innexin sequences, resulting in the cloning of a partial Hm-inx1 sequence.
Hm-inx2 was cloned using consensus-degenerate primers designed against a multiple sequence alignment of five Lophotrochozoan innexins. In theory, these primers should amplify leech sequences more efficiently than primers designed against Ecdysozoan innexins. A forward primer (CAGTACGTCGGAGACCCAATCCACTGYTGGKKYCC) was designed against the conserved first transmembrane domain and a reverse primer (AGTACGTCGGAGACCCAATCCACTAYTAYCARTGG) against the fourth transmembrane domain. This primer combination was used to amplify a partial Hm-inx2 sequence.
Full-length sequences of both innexins were subsequently cloned using gene-specific primers.
PCR was performed in a total volume of 20 μl using between 2 and 4 μl of library. PCR conditions were optimized for each primer combination using control amplification of Dm-inx2, Dm-inx3, or Hm-inx1 as appropriate. PCR products were run on a 1.2% agarose gel. Bands of the expected size were extracted using a gel-extraction kit (Qiagen, Crawley, UK) and cloned into the TA-cloning vector pCRII (Invitrogen, Carlsbad, CA). DNA was prepared using Qiaprep spin columns and sequenced commercially by Genetix Ltd (New Milton, UK) and Sigma Genosystems (Pampisford, UK) using standard T7 and SP6 primers.
Sequence analysis. Sequence manipulations were performed using LaserGene software (DNAstar, Madison, WI). Multiple sequence alignments were generated using ClustalW (Thompson et al., 1994) and decorated using Boxshade 3.3.1 software. Phylogenetic analysis was performed with ClustalW using the neighbor-joining algorithm, and the resulting trees were plotted using TreeView 1.6.6 software (Page, 1996). Transmembrane domains were predicted using a dense alignment surface (DAS) algorithm (Cserzo et al., 1997).
In situ hybridization. Adult Hirudo medicinalis were purchased from Ricarimpex (Eysines, France). Leeches were anesthetized in 10% ethanol at 4°C for 20 min. The ventral nerve cord was removed under Ringer's solution (in mm: 115 NaCl, 1.8 CaCl2, 4 KCl, and 10 Tris maleate, pH 7.4). Midbody ganglia were taken and manually desheathed before being fixed in 4% paraformaldehyde, 50 mm PIPES, pH 7.0, 1 mm EDTA, 2 mm MgSO4, and 0.1% sodium deoxycholate for 1 hr at room temperature. Ganglia were washed five times for 5 min each in PBS plus 0.1% Tween 20 (PBT). They were then dehydrated through a methanol series and stored in 100% methanol at –20°C for up to 2 weeks.
After rehydration through a methanol series into PBT, ganglia were washed five times for 5 min in PBT. The tissue was then permeabilized by digestion with 20 μg/ml Proteinase K (Sigma Aldrich, Gillingham, UK) in PBT for 1 min at room temperature and then washed two times for 1 min each in PBT plus 2 mg/ml glycine followed by washing five times for 5 min each in PBT. Ganglia were post-fixed as above for 20 min at room temperature and then washed five times for 5 min each in PBT.
Digoxigenin (DIG)-labeled antisense cRNA probes were transcribed from Hm-inx1 and Hm-inx2 using a commercial kit (Roche Diagnostics, Lewes, UK). Ganglia were prehybridized in hybrix solution [50% (v/v) formamide, 5× SSC, 100 μg/ml tRNA, 50 μg/ml heparin, and 0.1% (v/v) Tween 20] and then hybridized overnight at 55°C with 500 ng/ml cRNA probe in hybrix solution. As a negative control some ganglia were hybridized with DIG-labeled sense cRNA probe. After hybridization, the ganglia were washed through the following series at 55°C: 3× 10 min 200 μl hybrix, 1× 10 min 500 μl hybrix: 2× SSC wash (1:1), 1× 30 min 500 μl 2× SSC wash [2× SSC, 50%(v/v) formamide, 0.2% (v/v) Tween 20]. Ganglia were then washed once at room temperature in 500 μl 2× SSC wash: PBT (1:1).
Ganglia were blocked by washing three times for 1 min each, three times for 20 min each, and one time for 1 hr in 1 ml of PBT with 0.2 mg/ml BSA on a rotating wheel. A polyclonal sheep anti-DIG alkaline-phosphatase conjugated antibody (Roche Diagnostics) was pre-absorbed against Drosophila embryos and then incubated with the ganglia for at least 1 hr at room temperature. After washing with five 15 min washes in 1 ml of PBT, they were stored overnight at 4°C in PBT. Antibody staining was detected using NBT and X-Phosphate following the manufacturer's instructions.
Transcription of mRNA. The coding sequences of Hm-inx1 and Hm-inx2 were subcloned into the transcription vector pSPJ2CL (a gift of Hugh Woodland, University of Warwick, UK) such that on transcription the resulting mRNA contained upstream and downstream UTRs of the Xenopus β-globin gene. Capped mRNA was transcribed using SP6 polymerase (Roche Diagnostics). The integrity of the mRNA was verified by in vitro translation of 35S-labeled protein with a rabbit reticulocyte lysate (Promega, Madison, WI).
Electrophysiology. Innexin mRNAs were translated in paired Xenopus oocytes using standard methods (Swenson et al., 1989). Stage V or VI oocytes were isolated and injected with 20 ng of Cx38 antisense DNA oligonucleotide (5′-CTGACTGCTCGTCTGTCCACACAG-3′) to eliminate coupling by endogenous connexin proteins. After incubation for 24 hr, oocytes were injected with 10 ng of innexin mRNA, the vitelline membrane was removed, and the oocytes were paired. Recordings were made 2–5 d after pairing using the dual voltage-clamp technique (Spray et al., 1981). Both cells were clamped at –80 mV before voltage steps were applied to the driver cell. The voltage-stepping protocol used to stimulate the driver cell consisted of steps of between 10 and 80 mV in 10 mV increments in both the depolarizing and hyperpolarizing directions. The current required to maintain the potential of the passive cell was recorded. Data was recorded using Axoclamp 500 amplifiers with Axodata 1.2.2 software and analyzed using Axodata 4 software (Axon Instruments, Union City, CA).
Results
Leeches have innexin but not connexin genes
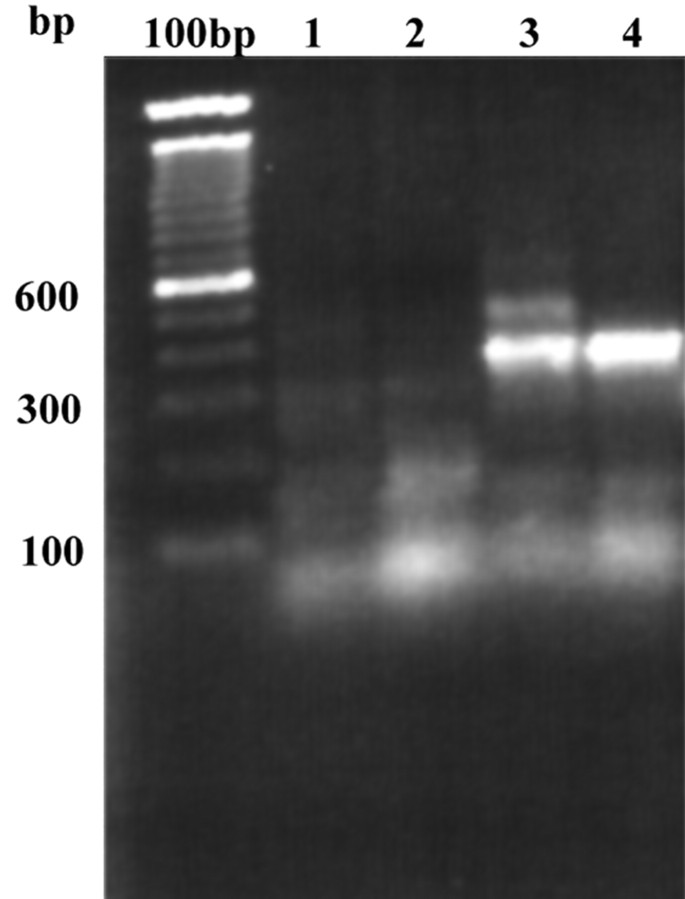
The connexin gene family form gap junctions in deuterostomes, whereas the innexin family form gap junctions in many invertebrates. It is not yet clear at what point in evolution this dichotomy arose and the leech, a Lophotrochozoan species, could therefore potentially use either innexins or connexins. To address this problem we designed degenerate primers against conserved regions of both families and used these primers to amplify putative-gap junction sequences from a leech CNS library by PCR. Figure 1 demonstrates the results of such an experiment. PCR with an innexin-specific primer resulted in the amplification of a clear 350 bp band (Fig 1, lanes 3, 4) which, on cloning and sequencing, was found to be highly homologous to the innexin gene family. Several PCR parameters such as primer annealing temperature and library concentration were varied in an attempt to amplify putative connexin sequences but, under our conditions, no clear bands were amplified (Fig 1, lanes 1, 2). We therefore concluded that connexins are likely to be absent from the leech genome.
Figure 1.
PCR products amplified from a leech CNS-specific cDNA library using degenerate primers designed to amplify putative connexins (lanes 1, 2) or putative innexins (lanes 3, 4). In lanes 1 and 3 the PCR reaction mix contained 10% library, whereas in lanes 2 and 4 this was increased to 20% without affecting the results. A bright band of ∼350 bp may be observed in lanes 3 and 4, whereas only primer dimers are seen in lanes 1 and 2. On cloning and sequencing, the 350 bp band was found to be an innexin.
A second set of degenerate primers was designed after the publication of Lophotrochozoan innexin sequences from mollusc and flatworm (Alexopoulos et al., 2000; Panchin et al., 2000), and a PCR reaction using these primers resulted in the amplification of a second partial innexin sequence from the leech CNS library (results not shown). The 5′ and 3′ sequence of these leech innexins were cloned by further rounds of PCR, and subsequently full-length sequences were amplified from the library using gene-specific primers.
Hm-inx1 encodes a protein of 414 amino acids with a predicted mass of 48.7 kDa, whereas Hm-inx2 encodes a protein of 398 amino acids with a predicted mass of 46.7 kDa. The two proteins are 41% identical and show 62% similarity. These proteins are clearly members of the innexin family and show most homology to innexins from other Lophotrochozoan species such as Clione. Figure 2 shows a multiple sequence alignment to illustrate this homology.
Figure 2.
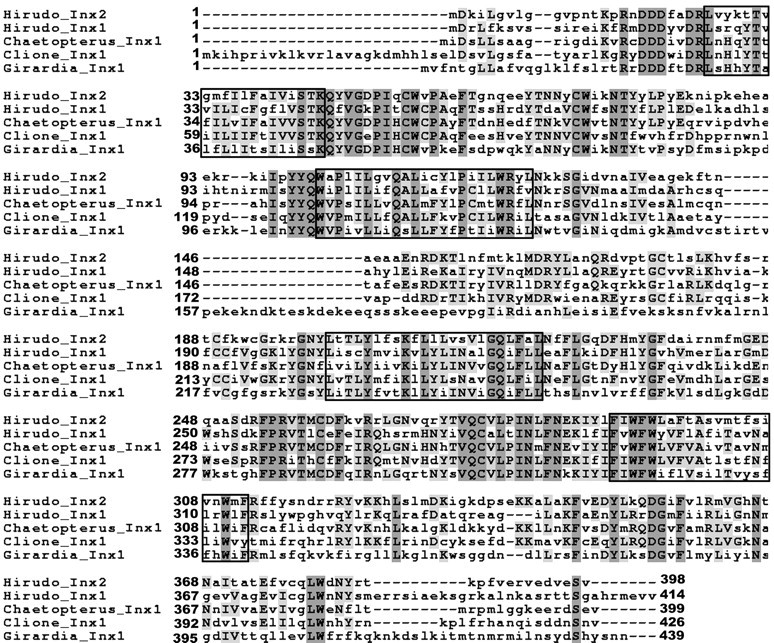
Sequence alignment of Hm-Inx1 and Hm-Inx2 to innexins cloned from other Lophotrochozoan species: the polychaete annelid Chaetopterus (Potenza et al., 2002), the platyhelminth Girardia (Panchin et al., 2000), and the mollusc Clione (Panchin et al., 2000). Shading indicates identical residues, whereas boxes indicate the predicted positions of the four transmembrane domains. GenBank accession numbers are as follows: Hm-Inx1: AJ512833; Hm-Inx2: AJ512834; Cv-Inx1: CAC69996; Gt-Inx1: AAF75840; and Cl-Inx1: AAF75839.
Evolution of the innexin family
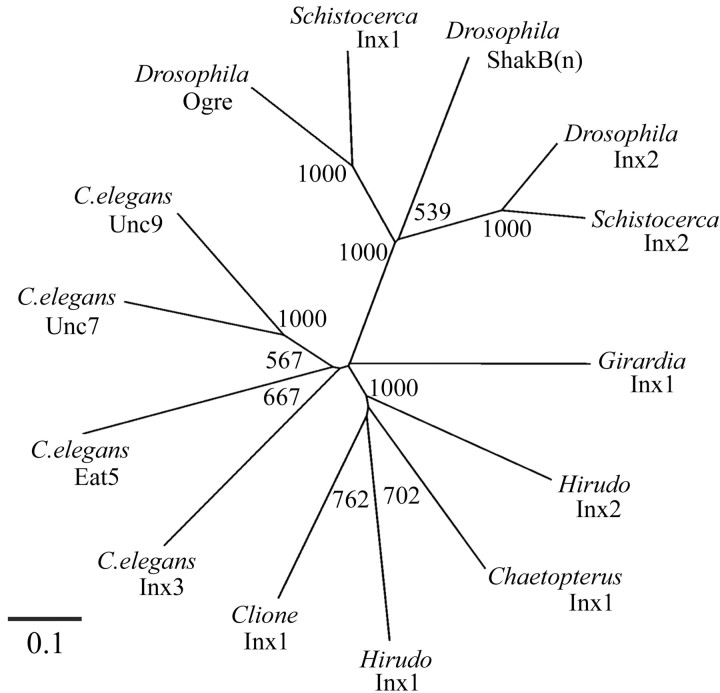
We performed a phylogenetic analysis to investigate the relationship of the leech innexins to previously characterized innexins. Figure 3 shows that the innexin family may be divided into three major clades representing the arthropod innexins (Drosophila and Schistocerca), the C. elegans innexins, and the Lophotrochozoan innexins (annelid and mollusc). It is interesting that the Ecdysozoan innexins are divided into these two unrelated clades, but it is not clear whether this reflects the faster rate of evolution noted in nematodes (Aguinaldo et al., 1997). Hm-inx1 and Hm-inx2 fall into the Lophotrochozoan clade along with the molluscan and polychaete sequences. The Girardia innexin lies outside of this clade, reflecting the high sequence divergence of this protein shown in Figure 2.
Figure 3.
Phylogenetic analysis of the relationship of Hm-Inx1 and Hm-Inx2 to Lophotrochozoan and representative Ecdysozoan innexins. The tree is unrooted. Numbers indicate bootstrap values per 1000 replicates. Note that the leech innexins form a robust clade along with the Clione and Chaetopterus innexins but that this clade does not include the Girardia innexin. Ecdysozoan innexins fall into two clades: a robust arthropod clade and a less robust C. elegans clade.
Several cross-species orthologs, such as Drosophila-inx2 and Schistocerca-inx2, may be identified within the arthropod clade by the short branch lengths and high bootstrap values. This suggests that gene duplication events have occurred within this phylum at a point before the divergence of these species. However, no such orthologs exist within the Lophotrochozoan clade: all four Lophotrochozoan innexins branch off the tree close to each other and have long branch lengths indicative of a long period of independent evolution. Therefore, we conclude that neither leech innexin is an ortholog of the innexin recently cloned from Chaetopterus, a polychaete annelid (Potenza et al., 2002) nor of any other known innexin.
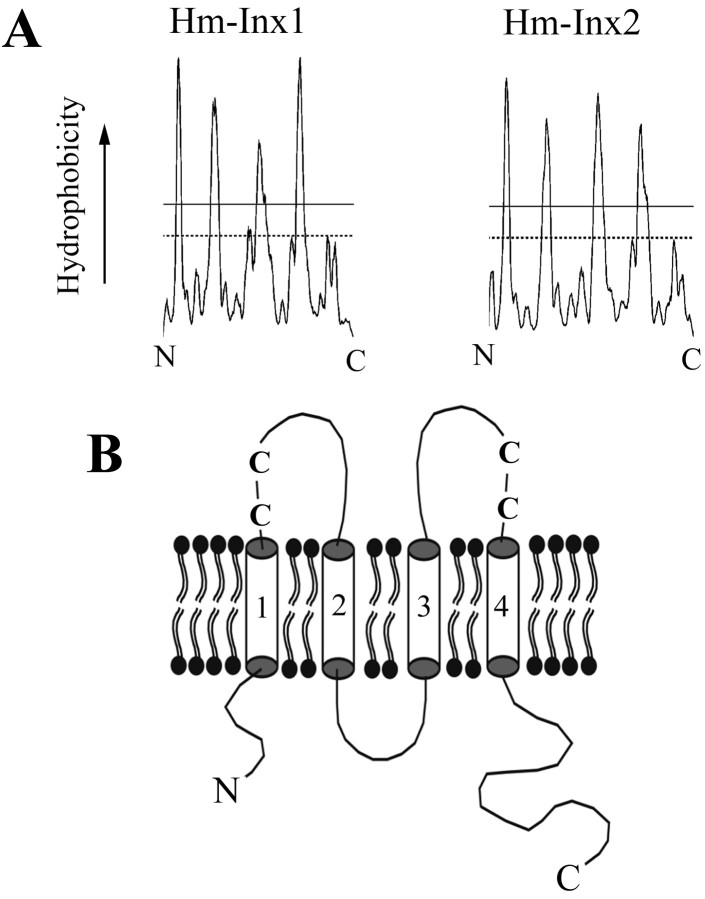
All innexins share a four transmembrane domain topology, and this is conserved in the leech sequences as shown in Figure 4, A and B. However, there is some variation in the length of intracellular and extracellular domains within the family. The Lophotrochozoan and C. elegans innexins have a relatively longer intracellular loop but slightly shorter extracellular loops than Drosophila innexins. Similarly, sequence motifs such as a DDD near the N-terminal are shared between Lophotrochozoans and C. elegans innexins but lacking in Drosophila. These data would seem to contradict the hypothesis that C. elegans innexins have evolved at a faster rate than others and suggests that the Ecdysozoan split may, in fact, be the result of a fast rate of arthropod evolution within the Ecdysozoa.
Figure 4.
A, Hydrophobicity plot of Hm-Inx1 and Hm-Inx2 derived using the DAS algorithm (Cserzo et al., 1997). Each spike on the plot that crosses the strict threshold indicates a predicted transmembrane domain. B, The predicted topology of an innexin protein. Innexins are thought to have four transmembrane domains (numbered cylinders) with intracellular N and C termini. The two extracellular loops each contain two completely conserved cysteine residues (C).
Expression in the CNS
Innexins are thought to oligomerize to form multimeric channels, most probably hexamers, which span the plasma membrane (Larsen, 1977; Shen et al., 2002), with two such hemichannels docking together to form a gap junction. A single gap junction channel, however, may be formed from more than one type of innexin protein. We therefore wished to compare the expression of the leech innexins to suggest the types of gap junction they may form.
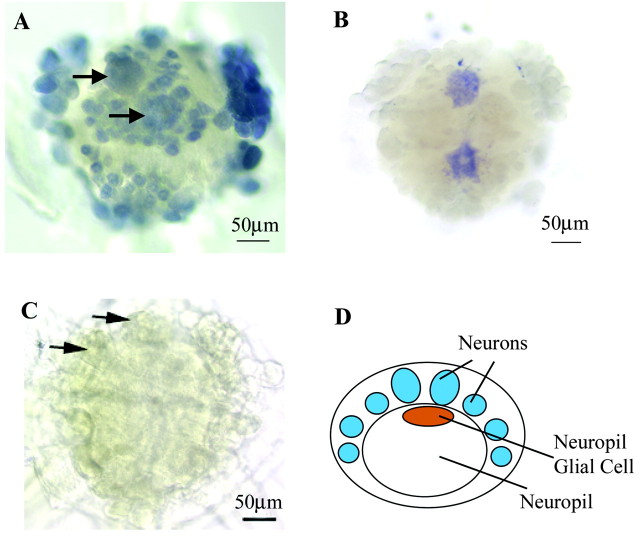
We studied the expression of the novel innexins in the leech CNS using in situ hybridization with DIG-labeled antisense RNA probes. Figure 5 shows that each innexin has a distinct expression pattern. Hm-inx1 is apparently expressed by all neurons (Fig. 5A). Many neurons in the leech CNS are connected by electrical synapses, and this expression therefore suggests that homotypic Hm-inx1 junctions may mediate this coupling, at least in part. Hm-inx2 is apparently only expressed by two cells in the CNS, the giant neuropil glial cells (Fig. 5B). The somata of these cells are located below the neuronal soma (Fig. 5D), and they have extensive projections within the neuropil. In addition to the neuropil glia, the leech also possesses six packet glia that serve to envelop the neuronal soma and two connective glia that envelop the interganglionic connectives. These glia are destroyed by the desheathing process, and it is therefore difficult to study their expression in whole-mount preparations. However, in situ hybridization may be performed to embryonic germinal plates without desheathing. Preliminary data show that Hm-inx2 is expressed by all three glial cell types in the embryonic CNS and confirms that the expression of Hm-inx1 is limited to neurons as in the adult (I. M. Dykes, unpublished observations). We therefore conclude that Hm-inx2 is highly likely also to be expressed in all glial cells in the adult CNS.
Figure 5.
Expression of Hm-inx1 and Hm-inx2 in the leech CNS. A–C, Whole-mount in situ hybridization to desheathed ganglia using DIG-labeled antisense RNA probes. Anterior is up. A, Hm-inx1 is expressed in all neurons, including the Retzius cells (arrows) but not in glia. B, Hm-inx2 is expressed in the two giant neuropil glial cells but not in neurons. C, Negative control showing staining with a sense Hm-Inx1 probe. Arrows show the position of the Retzius cells. D, Diagram of a transverse section through the ganglion showing the position of the neuropil glia relative to the neuronal cell bodies. Dorsal is up.
Heterologous expression of leech innexins
To demonstrate that Hm-inx1 and Hm-inx2 are sufficient to form functional gap junctions, we expressed innexin mRNA in paired Xenopus oocytes. Recordings were made using the dual voltage-clamp technique. This has become a standard assay for the function of both innexin and connexin-based gap junctions (Swenson et al., 1989; Phelan et al., 1998a; Landesman et al., 1999). Coupling caused by expression of endogenous Xenopus Cx38 was eliminated by the injection of an antisense oligonucleotide.
The expression pattern described above suggested that each innexin would be able to form homotypic junctions. To test this hypothesis 10 ng of either Hm-inx1 or Hm-inx2 mRNA was injected into each oocyte and similarly treated oocytes were paired. As shown in Table 1, only a subset of injected oocytes demonstrated coupling. Control oocytes injected with the Drosophila innexin shakB(lethal) coupled at a greater frequency.
Table 1.
Coupling efficiency and junctional conductance of innexin-expressing Xenopus oocyte pairs
|
Oocyte injection (cell 1/cell 2) |
Mass injected (ng/cell) |
Proportion of cells displaying coupling (%) |
n |
Maximum steady-state conductance of gap junction (μS) |
|---|---|---|---|---|
| Hm-inx1/Hm-inx1 | 10 | 25 | 36 | 4.06 ± 1.18 (9) |
| Hm-inx2/Hm-inx2 | 10 | 48 | 27 | 1.70 ± 0.49 (13) |
| Hm-inx1/Hm-inx2 | 10 | 44 | 18 | 1.54 ± 0.53 (8) |
| Dm-shakB(lethal)/Dm-shakB(lethal) | 2 | 77 | 13 | 13.19 ± 2.99 (10) |
| H2O/H2O |
N/A |
0 |
30 |
0.11 ± 0.04 (30) |
Oocytes were microinjected with an antisense Cx38 oligonucleotide and either innexin mRNA or water. The vitelline membrane was then removed, and oocytes were paired (see Materials and Methods). Recordings were made 2-6d after pairing using the dual voltage-clamp technique. Junctional conductance is shown as the maximum steady-state conductance observed, expressed as a mean ± SEM (number in parentheses indicates the number of cell pairs analyzed). Maximum conductance was observed on injection of a low depolarizing voltage step (homotypic Hm-inx1 and Dm-shakB(lethal)), a low hyperpolarizing voltage step (homotypic Hm-inx2), or a large hyperpolarizing step injected into the Hm-inx2-expressing cell (heterotypic junctions), as shown in the Gj/Vj plots (Figs. 5B, 6B, 7C).
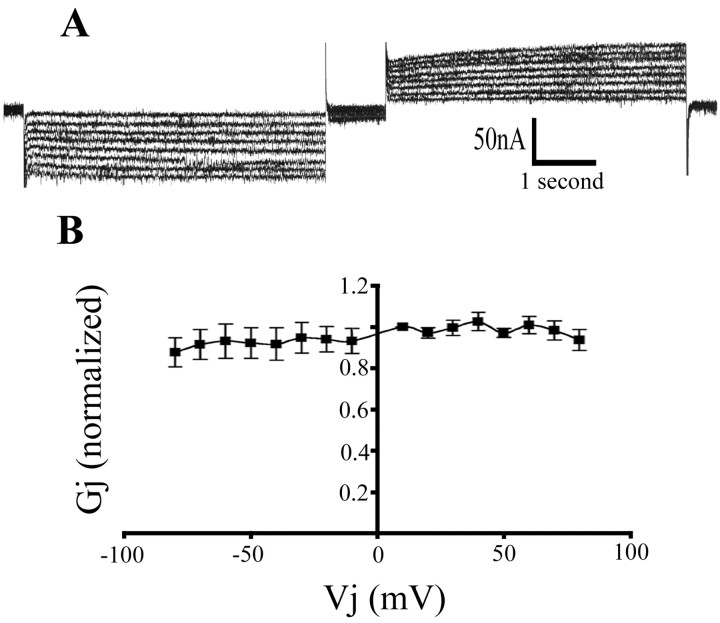
Homotypic Hm-inx1 channels
An example of a voltage-clamp recording from a pair of Hm-inx1-injected oocytes is shown in Figure 6A. The trace shows the current recorded in the passive cell in response to a series of voltage steps applied to the driver cell. Depolarizing (Fig. 6A, left) and hyperpolarizing (Fig. 6A, right) steps of between 10 and 80 mV in 10 mV increments were applied. In common with other characterized gap junction channels, the channel shows a large instantaneous conductance as soon as a voltage step is applied to the driver cell followed by a steady-state conductance. The channel is not greatly affected by transjunctional voltage (Vj) and remains open even at high voltages. Large Vj values have been observed to close Drosophila innexin channels (Phelan et al., 1998a; Stebbings et al., 2000), but this is not the case for Hm-Inx1 (Fig. 6B).
Figure 6.
Homotypic Hm-Inx1 gap junctions are not gated by junctional voltage. Voltage clamp recording from Hm-inx1-injected paired Xenopus oocytes. A, The trace shows the current in the passive cell required to maintain the holding potential of –80 mV when depolarizing and hyperpolarizing steps between ±10 and ±80 mV in 10 mV increments are applied to the driver cell. B, Graph of Gj versus Vj. Plot shows mean ± SEM; n = 5.
Homotypic Hm-inx2 channels
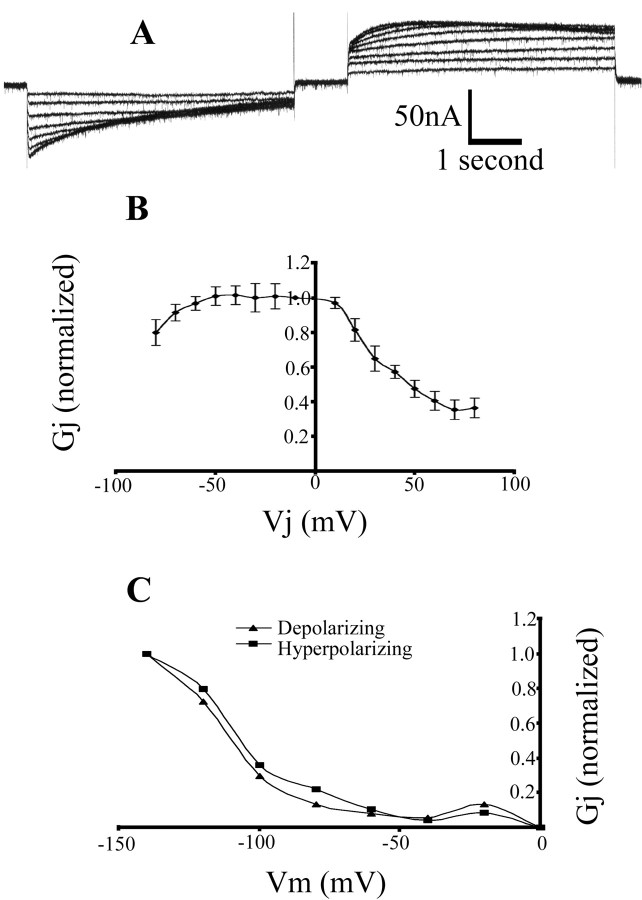
Hm-Inx2 also forms homotypic channels. An example of a voltage-clamp recording of this junction is shown in Figure 7A; the voltage step protocol was as described above. This channel demonstrates a highly asymmetrical response to depolarizing and hyperpolarizing voltage steps. At large depolarizing voltage steps (Fig. 7A, left), the channel shows a large instantaneous conductance but slowly closes with time to reach a low steady-state conductance. At large hyperpolarizing steps (Fig. 7A, right), the channel may be observed to initially open, reaching a maximum conductance at ∼1 sec after the start of the step, then closing slightly before reaching a high steady-state conductance. At low voltage steps the conductance of the channel is similar to voltage of both polarities and is not strongly gated. This response is illustrated graphically in Figure 7B. For steps of both polarities the steady-state conductance is maximal at values near 0 mV and decays when large voltage steps are applied.
Figure 7.
Homotypic Hm-Inx2 gap junctions are strongly gated by both Vj and Vm and behave as a double-rectifier. Voltage clamp recording from Hm-inx2-injected paired Xenopus oocytes. A, The trace shows the current in the passive cell required to maintain the holding potential of –80 mV when depolarizing and hyperpolarizing steps between ±10 and ±80 mV in 10 mV increments are applied to the driver cell. B, Graph of Gj versus Vj. Plot shows mean ± SEM; n = 4. C, The effect of membrane potential on Gj. A representative cell pair was clamped at a range of voltages between 0 and –140 mV. A 5 sec pulse of 40 mV was delivered to the driver cell, and the steady-state conductance across the junction was recorded. Both depolarizing (▴) and hyperpolarizing (▪) voltage steps were applied.
The observed gating properties of this channel may be the result of a transjunctional voltage-sensing mechanism, the product of a membrane potential sensor on the presynaptic (driver) side of the junction or a combination of the two. To distinguish between these possibilities we measured junctional conductance of a given voltage pulse over a range of holding potentials. Figure 7C demonstrates that the steady-state conductance of the junction is highly sensitive to membrane potential, and thus Vm sensitivity contributes to the characteristics of Hm-Inx2 channels. However, a comparison of Figure 7B with Figure 7C shows that Vm sensitivity is not sufficient to explain this behavior. Thus, the properties of Hm-Inx2 channels depend on both Vj and Vm.
Heterotypic channels
The processes of the two giant glial cells pervade the neuropil and come into contact with the processes of many neurons. Therefore, it is possible that Hm-Inx1 and Hm-Inx2 may interact to form a heterotypic gap junction, that is, a channel in which each hemichannel is composed of a different innexin. To test this, an oocyte injected with Hm-inx1 was paired with one injected with Hm-inx2. These oocytes displayed coupling with a similar frequency to the homotypic channels described above (Table 1). An example of a voltage-clamp recording of a heterotypic gap junction is shown in Figure 8. Figure 8A shows a recording from the Hm-inx1-injected oocyte while stimulating the Hm-inx2 oocyte, whereas Figure 8B shows a trace of the same cell pair recording from the Hm-inx2-injected oocyte while stimulating the Hm-inx1 oocyte. For comparison, the traces are shown to the same scale. The corresponding Gj versus Vj plots are shown in Figure 8, C and D, respectively.
Figure 8.
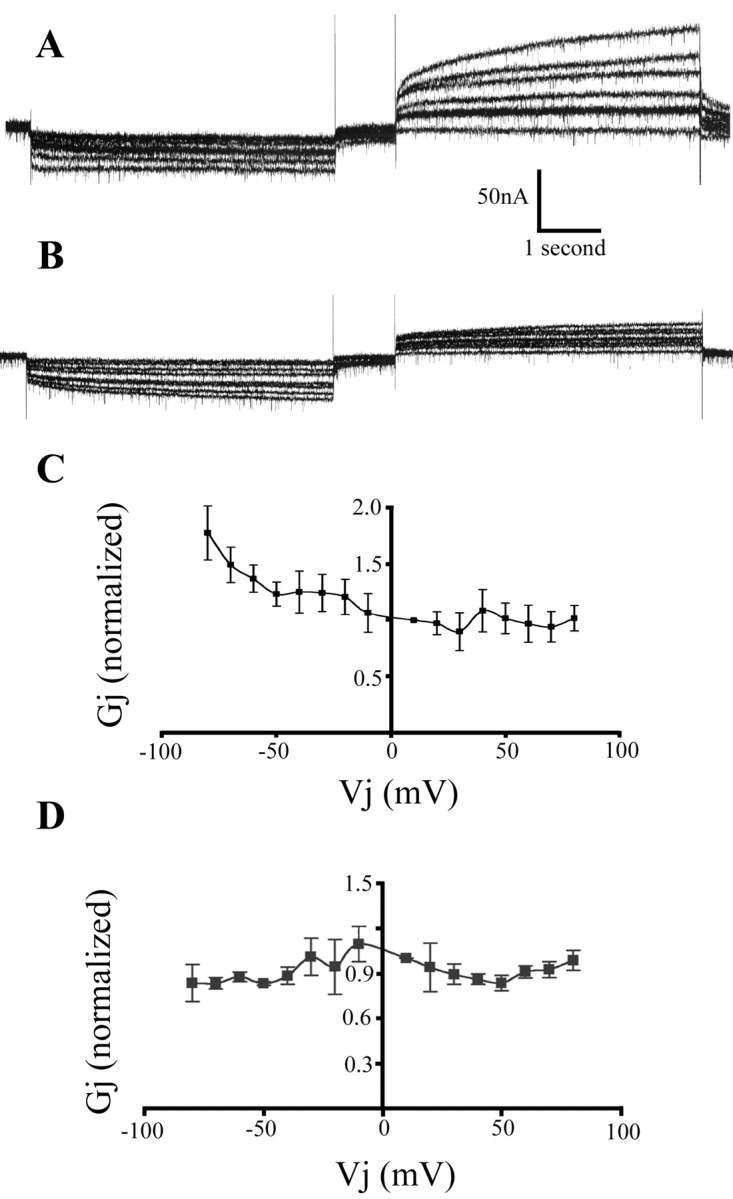
Heterotypic Hm-Inx1/Hm-Inx2 gap junctions display rectifying properties. Voltage-clamp recording from paired Hm-inx1 and Hm-inx2-injected Xenopus oocytes. A and B show traces demonstrating the current in the passive cell required to maintain the holding potential of –80 mV when depolarizing and hyperpolarizing steps between ±10 and ±80 mV in 10 mV increments are applied to the driver cell. C and D show graphs of Gj versus Vj. Plot shows mean ± SEM; n = 4. A, C, Recording from the Hm-inx1-injected oocyte while stimulating the Hm-inx2 oocyte. B, D, Recording from the Hm-inx2-injected oocyte while stimulating the Hm-Inx1 oocyte.
A comparison of Figure 8, A and B, shows that the behavior of this channel is determined primarily by the innexin on the presynaptic side of the junction. Hyperpolarizing steps delivered to the Hm-Inx2-expressing cell result in a dramatic increase in conductance in comparison to depolarizing steps, whereas conductance is always low when the Hm-Inx1-expressing cell is stimulated. Note that when the Hm-Inx2-expressing cell is stimulated, the behavior of the channel is almost entirely determined by the Vm sensitivity of the Hm-Inx2 hemichannel (Fig. 8C, compare Fig. 7C). These data are therefore consistent with the hypothesis that the conductance of the junction is determined primarily by a presynaptic Vm-sensitive gate rather than by a Vj-sensitive mechanism, and that Hm-Inx2 channels have a strong gate, whereas Hm-Inx1 channels do not.
However, the traces are not identical to the homotypic traces. For example, the conductance from the Hm-Inx1-expressing cell to the Hm-Inx2-expressing cell is lower than observed for homotypic Hm-Inx1 junctions. This indicates that Vm sensitivity is not sufficient to explain all of the data and therefore that there is likely to be some degree of interaction between the two hemichannels, leading to a change in the properties of the junction with respect to homotypic channels.
Discussion
We have cloned two innexins, Hm-inx1 and Hm-inx2, from the leech Hirudo medicinalis and shown that these have different expression patterns in the CNS. Hm-inx1 is neuronal-specific, whereas Hm-inx2 is glial-specific. We demonstrate that each of these innexins can form functional homotypic gap junctions in the paired Xenopus oocyte assay and that they can cooperate to form heterotypic junctions. Each of these junctions demonstrates novel properties not previously seen in this family. This work represents the first demonstration of gap junctional coupling mediated by a Lophotrochozoan innexin.
We found no evidence of connexins in the leech CNS. Together with the cloning of mollusc innexins in our laboratory (Alexopoulos et al., 2000) and by Panchin et al. (2000), this work suggests that the innexins are common to the protostome animals and that the connexins are restricted to the deuterostomes. It remains to be seen which of these families is the ancestral gap junction protein and why this ancestral protein was replaced by a second gap junction protein family in one lineage.
Although our data support the existence of a Lophotrochozoan clade of molluscs and annelids, the phylogenetic study presented here shows that the Lophotrochozoan innexins are more similar to the C. elegans clade of innexins than C. elegans innexins are to arthropod innexins. This suggests that the innexin family has undergone a rapid evolution in the arthropod lineage, and it therefore seems that the Drosophila innexins, the best characterized of the family (Stebbings et al., 2002), are also the most divergent from the typical innexin structure. This phylogeny suggests that the properties described for Drosophila innexins (Phelan et al., 1998a; Stebbings et al., 2000) may not be typical of the family as a whole and highlights the need for a functional analysis of the Lophotrochozoan innexins.
Electrical coupling is widespread in the leech CNS (Baylor and Nicholls, 1969; Fernandez-de-Miguel and Drapeau, 1995), and electrical synapses are generally described as rectifying (current passes preferentially in one direction) or non-rectifying (current is passed equally efficiently in both directions). The widespread expression of Hm-inx1 suggests that it may be a component of some or all of these synapses. Heterologous expression in oocytes demonstrated that homotypic Hm-Inx1 channels do not appear to have a gate sensitive to membrane potential. These junctions therefore display many of the properties one would expect to see in a nonrectifying synapse, and Hm-Inx1 channels must be considered as candidates to mediate this function. It has been proposed that rectifying electrical synapses contain voltage-dependent gates (Jaslove and Brink, 1986) and that the gap junctions forming such synapses are composed of two asymmetric hemichannels (Giaume et al., 1987). If true, then another innexin must be involved in these junctions. This cannot be Hm-Inx2 because it is not expressed in neurons. Drosophila has eight innexin genes, several of which are expressed in the nervous system (Stebbings et al., 2002), and C. elegans has 25 innexins, at least six of which have a neuronal expression pattern (Starich et al., 2001). It is therefore likely that other leech innexins await discovery and that different combinations of these innexins generates the diversity of electrical synapses seen in the leech CNS.
We see expression of Hm-Inx2 only in the two giant neuropil glial (NPG) cells. In addition, based on our unpublished observations in embryos, we propose that this innexin is also expressed in other glial cell types. These cells are known to be electrically coupled (Kuffler and Potter, 1964), and the presence of gap junctions has been demonstrated by electron microscopy (Coggeshall, 1974; Pumplin and Muller, 1983). These gap junctions pass dyes of up to 1.2 kDa (C. Lohr, personal communication). This is larger than any dye that has been shown to pass through electrical synapses between neurons and suggests that the molecular composition of neuronal and glial gap junctions may differ. Characterization of homotypic Hm-Inx2 channels in Xenopus oocytes showed that these junctions are highly sensitive to Vm, a property that correlates well with the physiology of the NPG cells. These cells are non-spiking, but oscillations in membrane potential are correlated with nervous activity (Deitmer et al., 1999). Therefore, we propose that homotypic Hm-Inx2 channels mediate gap junctional communication between the NPG cells and that these channels appear to be highly adapted to their function in the nervous system.
Although minor asymmetries in the response to voltage step polarity have been observed in Cx26 junctions (Barrio et al., 1991), Hm-Inx2 forms the first cloned gap junction channel to show such a large asymmetry in response. Homotypic Hm-Inx2 channels effectively behave as double-rectifiers, passing hyperpolarizing current preferentially in either direction. Double-rectifying electrical synapses have been described between touch sensory neurons in the leech (Acklin, 1988) and between pairs of SG neurons in the crayfish (Heitler et al., 1991). At these synapses, depolarization spreads in both directions, but hyperpolarization spreads poorly. In contrast, Hm-Inx2 homotypic junctions open on hyperpolarization but close when depolarizing current is applied.
In addition to forming homotypic channels, we demonstrate that Hm-Inx1 and Hm-Inx2 are compatible and can form heterotypic channels in Xenopus oocytes, which raises the possibility that such junctions exist in vivo. The expression patterns of the two innexins suggest that heterotypic Hm-Inx1/Hm-Inx2 channels may mediate neuronal–glial coupling. If heterotypic Hm-Inx1/Hm-Inx2 channels do indeed exist in vivo, then their physiology appears to be adapted for the transmission of hyperpolarizing signals from glia to neurons. Conductance of depolarizing signals would be low, as would be conductance of steps of either polarity from neurons (Hm-Inx1-expressing) to glia (Hm-Inx2-expressing). However, it should be noted that our recordings were made at a holding potential of –80 mV and that at a more physiological membrane potential, conductance of hyperpolarizing signals will be reduced because of the Vm sensitivity of Hm-Inx2 hemichannels.
Do these channels exist in vivo? Leech giant glial cells are among the best characterized of any glial cell yet despite almost 40 years of intense study, no neuronal-glial dye or electrical coupling has been described. Is it possible that such junctions will form between Xenopus oocytes in vitro but do not exist in vivo? Studies in leech cell culture have shown that novel electrical synapses not seen in CNS preparations will form in the dish (Fernandez-de-Miguel and Drapeau, 1995) presumably because of the lack of developmental constraints. Physiological differences exist between neurons and glia: glia have a more negative resting potential than neurons (Kuffler and Potter, 1964), and this is attributable to a different complement of ion channels. Perhaps it is advantageous to the leech to restrict the exchange of ions and metabolites between neurons and glia.
An alternative hypothesis is that such coupling might exist transiently in the embryo where it may be necessary to mediate pathfinding decisions but is lost in the adult nervous system. Transient gap junctions are known to be important in the development of the leech CNS (Wolszon et al., 1994). Neuronal–glial coupling has been demonstrated between astrocytes and neurons in neonatal rats (Alvarez-Maubecin et al., 2000). It will be interesting to test these hypotheses.
In conclusion, this work represents the first functional study of a Lophotrochozoan innexin. Our phylogenetic analysis has demonstrated the diversity within the innexin family and highlighted the need for comparative studies. The leech provides an excellent system in which to study the molecular basis of synaptic transmission between identified neurons. Neurons will form specific synapses when grown in vitro (Fernandez-de-Miguel and Drapeau, 1995) and are amenable to gene-knockdown using double-stranded RNA technology (Baker and Macagno, 2000). In addition the simple, metameric organization of the nervous system provides an ideal system in which to study the role of gap junctional communication in development and regeneration.
Footnotes
This work was supported by a grant from the Biotechnology and Biological Sciences Research Council (J.A.D.) and a research studentship from the same Research Council (I.D.). We thank Pauline Phelan and Cathy Holcroft for tutorials on the paired oocyte system and Rose Phillips for mRNA transcription.
Correspondence should be addressed to Jane A. Davies, Sussex Centre for Neuroscience, School of Biological Sciences, University of Sussex, Falmer, Brighton BN1 9QG, UK. E-mail: j.a.davies@sussex.ac.uk.
I. M. Dykes' present address: Division of Biology, University of California, San Diego, 9500 Gilman Drive, La Jolla, CA 92093-0368.
F. M. Freeman's present address: School of Science, Nottingham Trent University, Clifton, Nottingham NG11 8NS, UK.
DOI:10.1523/JNEUROSCI.3676-03.2004
Copyright © 2004 Society for Neuroscience 0270-6474/04/240886-09$15.00/0
References
- Acklin SE (1988) Electrical properties and anion permeability of doubly rectifying junctions in the leech central nervous system. J Exp Biol 137: 1–11. [Google Scholar]
- Aguinaldo AMA, Turbeville JM, Linford LS, Rivera MC, Garey JR, Raff RA, Lake JA (1997) Evidence for a clade of nematodes, arthropods and other moulting animals. Nature 387: 489–493. [DOI] [PubMed] [Google Scholar]
- Alexopoulos H, Dykes IM, Bacon JP, Davies JA (2000) Novel innexins in snails and leeches. Eur J Neurosci 12: S11:15. [Google Scholar]
- Alvarez-Maubecin V, Garcia-Hernandez F, Williams JT, Van Bockstaele EJ (2000) Functional coupling between neurons and glia. J Neurosci 20: 4091–4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker MW, Macagno ER (2000) RNAi of the receptor tyrosine phosphatase HmLAR2 in a single cell of an intact leech embryo leads to growth-cone collapse. Curr Biol 10: 1071–1074. [DOI] [PubMed] [Google Scholar]
- Barrio LC, Suchyna T, Bargiello T, Xu LX, Roginski RS, Bennett MV, Nicholson BJ (1991) Gap junctions formed by connexins 26 and 32 alone and in combination are differently affected by applied voltage. Proc Natl Acad Sci USA 88: 8410–8414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylor DA, Nicholls JG (1969) Chemical and electrical synaptic connexions between cutaneous mechanoreceptor neurones in the central nervous system of the leech. J Physiol (Lond) 203: 591–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonetto S, Muller KJ (1977) A regenerating neurone in the leech can form an electrical synapse on its severed axon segment. Nature 267: 450–452. [DOI] [PubMed] [Google Scholar]
- Coggeshall RE (1974) Gap junctions between identified glial cells in the leech. J Neurobiol 5: 463–467. [DOI] [PubMed] [Google Scholar]
- Cserzo M, Wallin E, Simon I, von Heijne G, Elofsson A (1997) Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng 10: 673–676. [DOI] [PubMed] [Google Scholar]
- Davis RL (1989) Voltage-dependent properties of electrical synapses formed between identified leech neurons in vitro. J Physiol (Lond) 417: 25–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deitmer JW, Rose CR, Munsch T, Schmidt J, Nett W, Schneider HP, Lohr C (1999) Leech giant glial cell: functional role in a simple nervous system. Glia 28: 175–182. [PubMed] [Google Scholar]
- Fernandez-de-Miguel F, Drapeau P (1995) Synapse formation and function: insights from identified leech neurons in culture. J Neurobiol 27: 367–379. [DOI] [PubMed] [Google Scholar]
- Fernandez-de-Miguel F, Vargas-Caballero M, Garcia-Perez E (2001) Spread of synaptic potentials through electrical synapses in Retzius neurones of the leech. J Exp Biol 204: 3241–3250. [DOI] [PubMed] [Google Scholar]
- Giaume C, Kado RT, Korn H (1987) Voltage-clamp analysis of a crayfish rectifying synapse. J Physiol (Lond) 386: 91–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris AL (2001) Emerging issues of connexin channels: biophysics fills the gap. Q Rev Biophys 34: 325–472. [DOI] [PubMed] [Google Scholar]
- Heitler WJ, Fraser K, Edwards DH (1991) Different types of rectification at electrical synapses made by a single crayfish neuron investigated experimentally and by computer simulation. J Comp Physiol [A] 169: 707–718. [DOI] [PubMed] [Google Scholar]
- Jaslove SW, Brink PR (1986) The mechanism of rectification at the electrotonic motor giant synapse of the crayfish. Nature 323: 63–65. [DOI] [PubMed] [Google Scholar]
- Kelmanson IV, Shagin DA, Usman N, Matz MV, Lukyanov SA, Panchin YV (2002) Altering electrical connections in the nervous system of the pteropod mollusc Clione limacina by neuronal injections of gap junction mRNA. Eur J Neurosci 16: 2475–2476. [DOI] [PubMed] [Google Scholar]
- Kuffler SW, Potter DP (1964) Glia in the leech central nervous system: physiological properties and neuron-glia relationship. J Neurophysiol 27: 290–310. [DOI] [PubMed] [Google Scholar]
- Kumar NM, Gilula NB (1996) The gap junction communication channel. Cell 84: 381–388. [DOI] [PubMed] [Google Scholar]
- Landesman Y, White TW, Starich TA, Shaw JE, Goodenough DA, Paul DL (1999) Innexin-3 forms connexin-like intercellular channels. J Cell Sci 112: 2391–2396. [DOI] [PubMed] [Google Scholar]
- Larsen WJ (1977) Structural diversity of gap junctions. A review. Tissue Cell 9: 373–394. [DOI] [PubMed] [Google Scholar]
- Marder E (1984) Roles for electrical coupling in neural circuits as revealed by selective neuronal deletions. J Exp Biol 112: 147–167. [DOI] [PubMed] [Google Scholar]
- Muller KJ (1979) Synapses between neurones in the central nervous system of the leech. Biol Rev Camb Philos Soc 54: 99–134. [DOI] [PubMed] [Google Scholar]
- Page RDM (1996) TREEVIEW: an application to display phylogenetic trees on personal computers. Comp Appl Biosci 12: 357–358. [DOI] [PubMed] [Google Scholar]
- Panchin Y, Kelmanson I, Matz M, Lukyanov K, Usman N, Lukyanov S (2000) A ubiquitous family of putative gap junction molecules. Curr Biol 10: R473–R474. [DOI] [PubMed] [Google Scholar]
- Phelan P, Starich TA (2001) Innexins get into the gap. Bioessays 23: 388–396. [DOI] [PubMed] [Google Scholar]
- Phelan P, Stebbings LA, Baines RA, Bacon JP, Davies JA, Ford C (1998a) Drosophila Shaking-B protein forms gap junctions in paired Xenopus oocytes. Nature 391: 181–184. [DOI] [PubMed] [Google Scholar]
- Phelan P, Bacon JP, Davies JA, Stebbings LA, Todman MG, Avery L, Baines RA, Barnes TM, Ford C, Hekimi S, Lee R, Shaw JE, Starich TA, Curtin KD, Sun YA, Wyman RJ (1998b) Innexins: a family of invertebrate gap-junction proteins. Trends Genet 14: 348–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potenza N, del Gaudio R, Rivieccio L, Russo GM, Geraci G (2002) Cloning and molecular characterization of the first innexin of the phylum annelida-expression of the gene during development. J Mol Evol 54: 312–321. [DOI] [PubMed] [Google Scholar]
- Pumplin DW, Muller KJ (1983) Distinctions between gap junctions and sites of intermediate filament attachment in the leech CNS. J Neurocytol 12: 805–815. [DOI] [PubMed] [Google Scholar]
- Rela L, Szczupak L (2003) Coactivation of motoneurons regulated by a network combining electrical and chemical synapses. J Neurosci 23: 682–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen DW, Lu QW, Wang L, Zhang RJ (2002) Ultrastructure of electrical synapses between nociceptive and anterior pagoda neurons in the CNS of the leech (Whitmania pigra). Invert Neurosci 4: 193–198. [DOI] [PubMed] [Google Scholar]
- Spray DC, Harris AL, Bennett MV (1981) Equilibrium properties of a voltage-dependent junctional conductance. J Gen Physiol 77: 77–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starich T, Sheehan M, Jadrich J, Shaw J (2001) Innexins in C. elegans. Cell Adhes Commun 8: 311–314. [DOI] [PubMed] [Google Scholar]
- Stebbings LA, Todman MG, Phelan P, Bacon JP, Davies JA (2000) Two Drosophila innexins are expressed in overlapping domains and cooperate to form gap-junction channels. Mol Biol Cell 11: 2459–2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbings LA, Todman MG, Phillips R, Greer CE, Tam J, Phelan P, Jacobs K, Bacon JP, Davies JA (2002) Gap junctions in Drosophila: developmental expression of the entire innexin gene family. Mech Dev 113: 197–205. [DOI] [PubMed] [Google Scholar]
- Swenson KI, Jordan JR, Beyer EC, Paul DL (1989) Formation of gap junctions by expression of connexins in Xenopus oocyte pairs. Cell 57: 145–155. [DOI] [PubMed] [Google Scholar]
- Taghert PH, Bastiani MJ, Ho RK, Goodman CS (1982) Guidance of pioneer growth cones: filopodial contacts and coupling revealed with an antibody to Lucifer Yellow. Dev Biol 94: 391–399. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolszon LR, Gao WQ, Passani MB, Macagno ER (1994) Growth cone “collapse” in vivo: are inhibitory interactions mediated by gap junctions? J Neurosci 14: 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]