Abstract
The neurons generated at the germinal rhombic lip undergo long distance migration along divergent pathways to settle in widely dispersed locations within the hindbrain, giving rise to cerebellar granule cells and precerebellar nuclei. Neurotrophin-3 (NT-3) signaling has been shown to be required for proper migration and survival of cerebellar granule cells. The molecular bases that govern NT-3 expression within the cerebellum, however, remain unknown at present. Here we report that, during early mouse neurogenesis, the Barhl1 homeobox gene is highly expressed by the rhombic lip and rhombic lip-derived migratory neurons. Its expression is later restricted to cerebellar granule cells and precerebellar neurons extending mossy fibers, two groups of neurons that synaptically connect in the adult cerebellar system. Loss of Barhl1 function causes cerebellar phenotypes with a striking similarity to those of NT-3 conditional null mice, which include attenuated cerebellar foliation as well as defective radial migration and increased apoptotic death of granule cells. Correlating with these defects, we find that NT-3 expression is dramatically downregulated in granule cells of the posterior lobe of Barhl1–/– cerebella. Moreover, in the precerebellar system of Barhl1–/– mice, all five nuclei that project mossy fibers fail to form correctly because of aberrant neuronal migration and elevated apoptosis. These results suggest that Barhl1 plays an essential role in the migration and survival of cerebellar granule cells and precerebellar neurons and functionally link Barhl1 to the NT-3 signaling pathway during cerebellar development.
Keywords: Barhl1, homeobox gene, neurotrophin-3, cerebellum, neuronal migration, apoptosis, rhombic lip, pontine gray nucleus
Introduction
The cerebellum must integrate cortical commands with sensory input information to coordinate motor activities. These commands and information are relayed to the cerebellum via the precerebellar system, which consists of six pairs of bilaterally symmetrical, but topographically separate nuclei: the pontine gray (PGN) and reticulotegmental (RTN) nuclei in the pons, and the vestibular (VN), external cuneate (ECN), lateral reticular (LRN), and inferior olivary (ION) nuclei within the medulla. The neurons in all precerebellar nuclei extend excitatory afferent fibers. However, they give rise to two distinct fiber systems that innervate different target cells in the cerebellum and have different functional roles. The inferior olivary neurons extend climbing fibers that directly innervate Purkinje cells, whereas the neurons in all other precerebellar nuclei form mossy fibers that influence Purkinje cells indirectly through synapses with the granule cells.
During neurogenesis, cerebellar granule cells and precerebellar neurons have been shown to originate from the germinal rhombic lip, a region of incomplete closure of the dorsal neural tube at the fourth ventricle. It is generally thought that the anterior rhombic lip gives rise to cerebellar granule cell progenitors and the posterior rhombic lip to precerebellar neuron precursors (Altman and Bayer, 1987d; Hatten et al., 1997; Wingate and Hatten, 1999; Rodriguez and Dymecki, 2000; Wingate, 2001). In the chick, however, the anterior rhombic lip has been shown to contribute a small number of progenitors to the PGN in addition to all cerebellar granule cells (Wingate and Hatten, 1999). Granule cells migrate over the surface of the cerebellar cortex to form the external granule layer (EGL). After exit from the cell cycle, they then migrate radially into the cerebellar cortex to form the internal granule layer (IGL). Once generated by the neuroepithelium of the posterior rhombic lip, precerebellar neurons also undergo long distance migration along divergent pathways to settle in discrete precerebellar nuclei in the pons and medulla (Altman and Bayer, 1987a,b,c; Hatten, 2002).
Although recent molecular genetic studies have identified a number of genes involved in the development of migratory cerebellar and precerebellar neurons (Hatten et al., 1997; Wingate, 2001), the molecular mechanisms underlying their migration, differentiation, and maintenance remain largely unknown. Barhl1 is a mammalian homolog of the Drosophila BarH genes, which encode homeodomain transcription factors that are required for normal development of the compound eye and external sensory organs (Kojima et al., 1991; Higashijima et al., 1992a,b; Bulfone et al., 2000; Li et al., 2002). During mouse embryogenesis, Barhl1 is expressed in the CNS and inner ear hair cells (Bulfone et al., 2000; Li et al., 2002). Our recent genetargeting study has demonstrated an essential role for Barhl1 in the long-term maintenance of cochlear hair cells (Li et al., 2002). To understand if Barhl1 also has a role during CNS development, in this work, we analyzed the expression pattern of Barhl1 during CNS development and investigated CNS defects in Barhl1 null mice. We found that Barhl1 displayed a distinct expression pattern in the cerebellar and precerebellar systems. The absence of Barhl1 caused a dramatic downregulation of NT-3 expression in cerebellar granule cells, resulting in attenuated foliation and hypotrophy of the cerebellum, as well as aberrant radial migration and increased death of granule cells. Similarly, it caused anomalous migration and loss of mossy fiber-extending precerebellar neurons. Therefore, our data have uncovered a crucial role for Barhl1 in the control of migration and survival of cerebellar and precerebellar neurons and identified NT-3 as a major Barhl1 downstream gene during cerebellar development.
Materials and Methods
Animals. The Barhl1 knock-out mice were generated previously (Li et al., 2002) and maintained in our laboratory. The stage of mouse embryos was determined by taking the morning when the copulation plug was shown as embryonic day 0.5 (E0.5). All genotypes described were confirmed by PCR (Li et al., 2002).
Real-time quantitative RT-PCR and Northern blot analysis. Cerebella from five each of P6 Barhl1+/+, Barhl1+/–, Barhl1–/– animals were dissected in RNAlater solution (Ambion, Austin, TX), and total RNA was extracted using TRIzol (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. RNA was resuspended in RNase-free ddH2O and further purified and treated with DNase I using the RNeasy total RNA isolation kit following the manufacturer's instructions (Qiagen, Valencia, CA). QRT-PCR was performed in duplicate for each RNA sample (100 ng) using the QuantiTect SYBR green one-step RT-PCR kit (Qiagen). The following sequence-specific primers were designed using the MacVector software (Accelrys, San Diego, CA): Barhl1,5′-CAAAGTGAAGGAGGAGGGCG-3′ and 5′-GTGTCGGTGAGGTTGAGCGA-3′; NT-3, 5′-GATTGATGACAAACACTGGAAC-3′ and 5′-CACAGGAAGTGTCTATTCGTATC-3′; BDNF, 5′-CGGGACGGTCACAGTCCTA-3′ and 5′-GGGATTACACTTGGTCTCGTAGAAATAC-3′; and GAPDH, 5′-TCACCACCATGGAGAAGGC-3′ and 5′-GCTAAGCAGTTGGTGGTGCA-3′. PCR products were monitored in real time (Mx4000 multiplex quantitative PCR system; Stratagene, La Jolla, CA), and the threshold cycles (Ct) were determined using the Mx4000 software. For each set of primers, a no template control and a no reverse amplification control were included. Postamplification dissociation curves were performed to verify the presence of single amplification product in the absence of DNA contamination. Relative quantities of copy numbers were calculated from known quantities of input copy numbers of cloned Barhl1, NT-3, BDNF, and GAPDH cDNA plasmids using the comparative threshold cycle number of each sample fitted to a seven-point standard curve (r2 = 0.99) (Overbergh et al., 1999). All data were tested for significance using two sample Student's t test with unequal variances. Northern blot analysis was performed according to standard methods.
RNA in situ hybridization. RNA in situ hybridization was performed as previously described (Sciavolino et al., 1997) using digoxigenin-labeled riboprobes prepared following the manufacturer's protocol (Roche Diagnostics, IN). Probes: Barhl1 was a previously isolated mouse cDNA clone (Li et al., 2002); NT-3 was a coding segment amplified by RT-PCR from mouse cerebellar RNA; Math1 and NeuroD coding sequences were amplified by PCR from mouse genomic DNA; the human PAX6 plasmid was described by Singh et al. (1998); the mouse Netrin-1 by Serafini et al. (1996), the rat DCC and Neogenin by Keino-Masu et al. (1996), the rat Unc5h1, Unc5h2 and Unc5h3 by Leonardo et al. (1997), the mouse EphB2 by Lu et al. (2001), and the human Ephrin-B2 by Yue et al. (1999).
β-Galactosidase staining. β-Galactosidase staining was conducted essentially as described (Ben-Arie et al., 2000; Eng et al., 2001). Briefly, for staining of whole-mount embryos and brains, animals were fixed in 4% paraformaldehyde–PBS at 4°C for 2–12 hr depending on the stages and then rinsed for 20 min in PBS containing 0.02% Nonidet P-40 and 0.01% sodium deoxycholate. Staining was performed overnight either at 30°C or 37°C in PBS buffer containing 0.02% Nonidet P-40, 0.01% sodium deoxycholate, 5 mm potassium ferricyanide, 5 mm potassium ferrocyanide, and 0.5 mg/ml X-gal. Some whole-mount-stained embryos were dehydrated in graded ethanol and cleared in 1:2 benzyl alcohol–benzyl benzoate. Section staining was performed following the same procedure as whole-mount staining except that all sections were counterstained with Fast Red (Vector Laboratories, Burlingame, CA).
Generation of polyclonal anti-Barhl1 antibody and immunohistochemistry. DNA fragment corresponding to amino acids 3–92 of the mouse Barhl1 protein was amplified by PCR and inserted into the pGEMEX (Promega, Madison, WI) and pMAL-cR1 (New England Biolabs, Beverly, MA) vectors to express fusion proteins with the bacteriophage T7 gene 10 protein and bacterial maltose-binding protein, respectively. Antibody production and affinity purification were performed as described previously by Xiang et al. (1993, 1995).
For immunohistochemistry, cryosections were treated in methanol with 3% of hydrogen peroxide for 3 min to quench endogenous peroxidase activity. After three washes in PBS, they were blocked in 5% of normal goat serum for 1 hr before overnight incubation at 4°C with primary antibodies [anti-Barhl1, 1:10; anti-Brn3a (Xiang et al., 1995), 1:5; anti-active caspase-3 (BD Pharmingen, San Diego, CA), 1:100]. The sections were then washed in PBS for three times, 7 min each, incubated with biotinylated goat anti-rabbit IgG (1:200; Vector Laboratories) for 1 hr, and subsequently processed using the ABC kit (Vector Laboratories). Color reaction was performed using the NovaRed substrate kit (Vector Laboratories). For double staining, postnatal day 5 (P5) brain sections were first immunostained with the anti-Brn3a antibody, rinsed in PBS for three times, and then stained for β-galactosidase activity for 2 hr at 37°C as described above. The labeled sections were dehydrated and mounted with Permount (Fisher Scientific, Springfield, NJ).
Quantitation of neuron number and cell death assay. To quantify the number of neurons in the PGN and RTN, serial coronal sections (16 μm) of P100 mouse brains were stained with cresyl violet. Images of the PGN and RTN were then captured using a SPOT digital camera (Diagnostic Instruments Inc., Sterling Heights, MI), and the number of neurons was scored from captured images. Only neurons with a clear nucleus and nucleoli were counted. Every other section was scored, and three or four nuclei were counted for each genotype. All data were tested for significance using two sample Student's t test with unequal variances.
Cell death assay was performed by measuring the expression of the proteolytically activated form of caspase-3 as an indicator of apoptosis (Panchision et al., 2001). To determine the number of apoptotic cells in the PGN, coronal cryosections from E16.5, P0, P2, and P4 brains were immunostained with an antibody against the active caspase-3 as described above and then counterstained with methyl green. The caspase-3-positive cells were counted in all serial sections of the PGN and a Student's t test was used to determine the significance. Four samples were collected for each genotype. To measure the number of apoptotic cells in the cerebellum, sagittal sections of P8 cerebella were similarly analyzed.
BrdU and DiI labeling. BrdU labeling was performed as described (Borghesani et al., 2002). Retrograde DiI (1.1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine) labeling was performed essentially as described (Bloch-Gallego et al., 1999). In brief, P1 brains were dissected out of the skull following fixation of neonates with 4% paraformaldehyde–PBS. Several DiI crystals were then inserted into one of both hemicerebella using a glass pipette tip under a dissection microscope. The DiI-inserted brains were incubated in 2% paraformaldehyde–PBS at 37°C for 4 weeks in the dark. They were then rinsed in PBS, embedded in 3% agarose, and cut at a thickness of 100 μm with a vibratome. The sections were mounted in Aqua polymount (Polysciences, Warrington, PA) and sealed with nail polish.
Results
Expression pattern of Barhl1 in the cerebellar and precerebellar systems
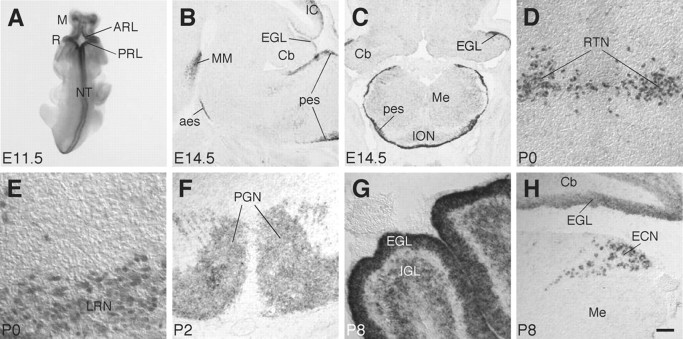
To understand the role of the Barhl1 gene during CNS development, we systemically analyzed its spatial and temporal expression patterns in the mouse. As revealed by RNA in situ hybridization, Barhl1 is prominently expressed in the developing cerebellar and precerebellar systems. From E11.5 to early postnatal stages, strong Barhl1 expression is observed in the rhombic lip as well as in most rhombic lip-derived migratory neurons and hindbrain structures (Fig. 1). During cerebellar development, Barhl1 expression is first found in the anterior rhombic lip; then in granule cells derived from it, initially located in the EGL and later in the IGL (Fig. 1A–C,G,H). In the developing precerebellar system, there is a strong expression of Barhl1 in the posterior rhombic lip at E11.5 (Fig. 1A). As development progresses, Barhl1 expression is seen in the anterior extramural migratory stream and its derivative precerebellar nuclei, the PGN and RTN (Fig. 1B,F). Similarly, Barhl1 is abundantly expressed in the posterior extramural migratory stream and its derivative precerebellar nuclei, the VN, LRN, and ECN (Fig. 1B,C,H). However, Barhl1 does not appear to be expressed in the ION (Fig. 1C). To examine tissue distribution of the Barhl1 protein, we developed a specific polyclonal anti-Barhl1 antibody (see Materials and Methods) and could show by immunohistochemistry that the Barhl1 protein was also specifically localized to the cerebellar granule cells and precerebellar neurons that extend mossy fibers (Fig. 1D,E) (data not shown). Consistent with it being a transcription factor, Barhl1 is nuclear (Fig. 1D,E). Elsewhere in the developing CNS, we found that Barhl1 is expressed in the diencephalon, mesencephalon and neural tube, as described in a previous report (Fig. 1A,B) (Bulfone et al., 2000).
Figure 1.
Expression of Barhl1 in developing cerebellar and precerebellar systems. A, The Barhl1 transcript was detected by in situ hybridization in a whole-mount embryo at E11.5. B, C, Localization of Barhl1 transcripts in sagittal (B) and coronal (C) sections through brains of E14.5 embryos. D,E, Neurons within the reticulotegmental and lateral reticular nuclei in coronal sections through brains of P0 mice were immunolabeled with an anti-Barhl1 antibody. F–H, Localization of Barhl1 transcripts in coronal sections through brains of P2 (F) and P8 (G, H) animals. Barhl1 expression is seen in restricted areas within the diencephalon, mesencephalon, cerebellum, brainstem, and neural tube. Note the strong expression of Barhl1 in the rhombic lip, external granule layer, anterior and posterior extramural migratory streams, and precerebellar nuclei extending mossy fibers (A–H), but not in the inferior olivary neurons (C). aes, Anterior extramural migratory stream; ARL, anterior rhombic lip; Cb, cerebellum; ECN, external cuneate nucleus; EGL, external granule layer; IC, inferior colliculus; IGL, internal granule layer; ION, inferior olivary nucleus; LRN, lateral reticular nucleus; M, mesencephalon; Me; medulla; MM, mammillary region; NT, neural tube; pes, posterior extramural migratory stream; PGN, pontine gray nucleus; PRL, posterior rhombic lip; R, rhombencephalon; RTN, reticulotegmental nucleus. Scale bar: E, 25 μm; D, G, 50 μm; F, H, 100 μm; B, C, 250 μm; A, 385 μm.
To further investigate the expression patterns of Barhl1 during cerebellar and precerebellar development, we made use of a lacZ reporter knocked in the Barhl1 locus (Li et al., 2002). Similar to the inner ear (Li et al., 2002), our analysis of β-galactosidase activity in Barhl1+/- mice showed that the knock-in lacZ reporter could recapitulate the expression pattern of the endogenous Barhl1 gene in all CNS structures. For instance, β-galactosidase staining of whole-mount embryos revealed the same spatial and temporal expression pattern of Barhl1 within the diencephalon, mesencephalon, rhombencephalon, and spinal cord as detected by RNA in situ hybridization (Figs. 1A, 2A–C). In the developing cerebellar and precerebellar systems, strong β-galactosidase activity was observed in the anterior and posterior rhombic lips, granule cells of the EGL and IGL, anterior and posterior extramural migratory streams, and all neurons within the PGN, RTN, VN, LRN, and ECN (Fig. 2B,D–I). Thus, the knock-in lacZ reporter confirms a strong Barhl1 expression in cerebellar granule cells and precerebellar neurons, suggesting that Barhl1 may play an important developmental role in these neurons.
Figure 2.
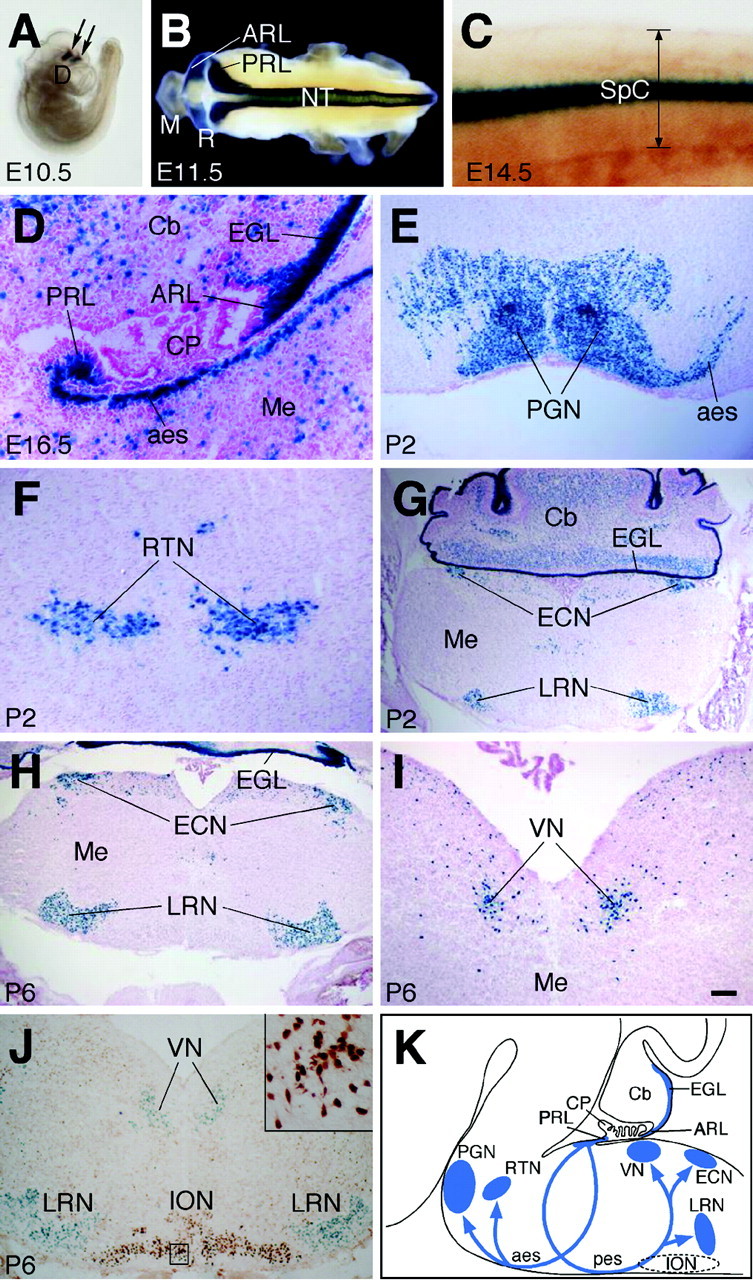
Expression of the lacZ reporter during CNS development in Barhl1+/– mice. A–I, β-galactosidase activity was visualized by X-gal staining in whole-mount embryos (A–C) and in brain sections from various stages counterstained with Fast Red (D–I). The embryo in C was cleared in benzyl alcohol–benzyl benzoate after X-gal staining. lacZ was expressed in two small stripes (indicated by arrows in A) within the diencephalon at E10.5. Its expression in the neural tube was seen in the dorsal tip at E11.5 (B), but localized in two symmetric lateral columns in the middle of the spinal cord (its thickness is indicated in C) at E14.5. Within the cerebellar and precerebellar systems, strong β-galactosidase activity was found in the rhombic lip, the external granule cells of the cerebellum, anterior and posterior extramural migratory streams, and all precerebellar neurons extending mossy fibers (D–I). J, A medullary section from a P6 Barhl1+/– pup was double-stained with X-gal and an anti-Brn3a antibody. lacZ-expressing cells (blue) were restricted in lateral reticular and vestibular nuclei, which did not overlap with those that expressed Brn3a (brown) in the inferior olivary nucleus. The inset indicates that all Brn3a+ cells are free from blue stain. K, Schematic diagram illustrating the expression pattern of Barhl1 in the cerebellar and precerebellar systems. The inferior olivary nucleus (dashed oval) lacks any Barhl1 expression. CP, choroid plexus; D, diencephalon; SpC, spinal cord; VN, vestibular nucleus. Scale bar: D, F, 50 μm; E, I, 100 μm; C, 125 μm; J, 192 μm; G, H, 250 μm; A, 400 μm; B, 500 μm.
Although all neurons that project climbing fibers within the ION also originate from the posterior rhombic lip, no Barhl1 expression was observed in this nucleus and the intramural migratory stream throughout CNS development by in situ hybridization, immunohistochemistry or the lacZ reporter, indicating a specificity of Barhl1 to mossy fiber-extending neurons. To further demonstrate this specificity, we double-stained P6 Barhl1+/- medullary sections for β-galactosidase activity and Brn3a immunoreactivity, a marker for ION neurons (McEvilly et al., 1996; Xiang et al., 1996). We found that none of the Barhl1-expressing cells (β-gal+) overlapped with any inferior olivary neurons (Brn3a+) (Fig. 2J). Therefore, within the precerebellar system, Barhl1 transcript and protein are exclusively expressed in nuclei whose neurons make mossy fiber projections to cerebellar granule cells, where Barhl1 is also prevalently expressed (Fig. 2K).
Defects in migration and survival of Barhl1–/– cerebellar granule cells
Given the strong expression of Barhl1 in granule cells of the developing cerebellum, we asked whether the absence of Barhl1 would cause any cerebellar abnormalities. We first examined the gross morphology of cerebella in Barhl1 wild-type and null mutant mice at P23 and P63, when all external granule cells had migrated into the IGL (Hatten et al., 1997). At P23, although all Barhl1+/+ cerebella (10 of 10) displayed a normal foliation pattern (Fig. 3A,C), the vermis lobule VII and the intercrural fissure that separates it from lobule VI were absent from most of Barhl1–/– cerebella (12 of 16) (Fig. 3B,D). A much smaller portion of Barhl1+/- cerebella (6 of 20) exhibited a similar foliation anomaly. In addition to the foliation abnormality, we found that all Barhl1-/- cerebella exhibited hypotrophy with a visible reduction in their overall size compared to those of Barhl1+/+ cerebella (Fig. 3A,B). To determine the cellular basis of size reduction of the mutant cerebellum, we measured apoptotic cell death by assaying for the active caspase-3 immunoreactivity in P8 control and null mice. In lobule VI, a 42% increase in the density of apoptotic cells was observed within the IGL of Barhl1–/– cerebella (wild-type mean ± SD, 100.8 ± 11.9 cells/mm2, n = 3; mutant, 143.2 ± 33.2 cells/mm2, n = 3). Interestingly, the cerebellar phenotypes observed in Barhl1 null mice, including the lack of vermis lobule VII, hypotrophy and increased granule cell death, most closely resemble those present in the cerebella of NT-3 (neurotrophin-3) and BDNF (brain-derived neurotrophic factor) null mice (Schwartz et al., 1997; Bates et al., 1999; Borghesani et al., 2002; Carter et al., 2003).
Figure 3.
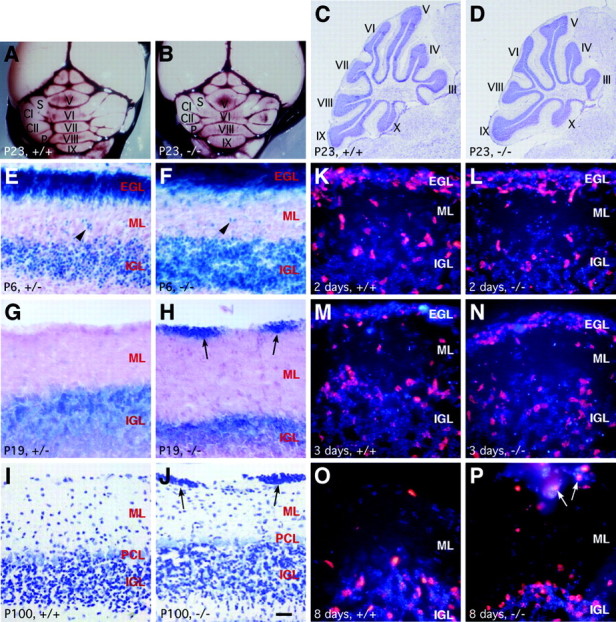
Abnormalities in the cerebella of Barhl1–/– mice. A–D, Attenuated foliation and size reduction in the cerebella of P23 Barhl1–/– mice. Macroscopic views of ink-stained wild-type (A) and mutant (B) cerebella showed a diminution in foliation and size of the Barhl1–/– cerebellum. Cresyl violet labeling of sagittal cerebellar sections further revealed the loss of vermis lobule VII in the Barhl1–/– cerebellum (C, D). The vermis lobules are indicated by numerals. E–P, Defective migration of granule cells in postnatal Barhl1–/– cerebella. E–J, Cerebellar sections from the indicated stages and genotypes were stained with X-gal (E–H) or cresyl violet (I, J). Compared with the wild-type and heterozygote, many clusters of granule cells (indicated by black arrows) get stuck on the surface of the mutant cerebellum at P19 and P100 (G–J). The arrowheads point to migrating granule cells in the molecular layer (E, F). K–P, Dividing granule cells in wild-type and Barhl1–/– littermates were pulse-labeled at P9 by BrdU and then visualized by BrdU immunohistochemistry (red) along with a weak DAPI counterstain at the indicated times after injection. All images were taken from lobules VI and VII. In the mutant, there are fewer BrdU+ cells within the IGL at 2 d after BrdU labeling and more BrdU+ cells within the EGL at 3 d post-BrdU labeling (K–N). Some BrdU+ cells (indicated by white arrows) persist in granule cell ectopias even 8 d after BrdU labeling in the mutant (O, P). CI, crus I; CII, crus II; EGL, external granule layer; IGL, internal granule layer; ML, molecular layer; P, paramedian lobule; PCL, Purkinje cell layer; S, simplex. Scale bar: E-J, 25 μm; K-P, 16.7 μm; C, D, 400 μm; A, B, 927 μm.
We used the knock-in lacZ as a marker to follow the generation, differentiation, and migration of cerebellar granule cells in developing Barhl1-/- mice. During embryogenesis and early postnatal development, lacZ-positive cells were abundantly present in the EGL of both Barhl1+/– and Barhl1–/– animals (Fig. 3E,F). By P6 in Barhl1–/– cerebella, similar to those of heterozygotes, numerous lacZ-positive granule cells already migrated into the IGL, whereas many lacZ-positive cells, apparently radially migrating granule cells, were also seen within the molecular layer (Fig. 3E,F). By P19 in the Barhl1+/– cerebellum, all lacZ-positive cells were located in the IGL, and the molecular layer and its surface were free of any lacZ-positive cells (Fig. 3G). By contrast, in the mutant cerebellum, many lacZ-positive cells were still present on the surface although the molecular layer was free of lacZ-positive cells (Fig. 3H). This phenomenon was observed most prevalently in the posterior lobe, indicating a regional specificity. By cresyl violet staining of cerebellar sections, we found that P100 Barhl1-/- mice formed a rather normal laminar cerebellar structure containing granule, Purkinje and molecular layers; however, many clusters of granule cells in the posterior lobe stalled and formed ectopias on the surface of the cerebellum (Fig. 3I,J). Therefore, the majority of granule cells appear to be generated in the Barhl1-/- cerebellum, being able to differentiate and migrate; however, a small fraction of granule cells may fail to initiate radial migration.
To examine more directly whether the ectopic granule cells resulted from migration defects in Barhl1 null cerebella, we pulse-labeled a cohort of granule cells in P9 wild-type and mutant animals with BrdU and monitored their migration for an 8 d period (Fig. 3K–P). In mutant cerebella, fewer cells appeared to have migrated into the IGL by 2 d after BrdU labeling, and more BrdU+ cells were seen within the EGL at 3 d after BrdU labeling (Fig. 3K–N). By 8 d after BrdU labeling, however, the great majority of BrdU+ cells had migrated into the IGL in both wild-type and mutant cerebella (Fig. 3O,P), indicating that some mutant granule cells were delayed for migration from the EGL. Interestingly, a minor number of BrdU+ cells in Barhl1–/– cerebella were still retained within some superficial ectopias even 8 d after BrdU labeling (Fig. 3P), suggesting that the dislocated granule cells in adult mutants resulted from a failure for some granule cells to initiate radial migration. Consistent with this notion, the ectopic cells in Barhl1–/– cerebella were not caused by persistent proliferation of some progenitor cells because none of them could be labeled by a pulse of BrdU at P30 (data not shown).
Downregulation of NT-3 expression in cerebella of Barhl1 mutants
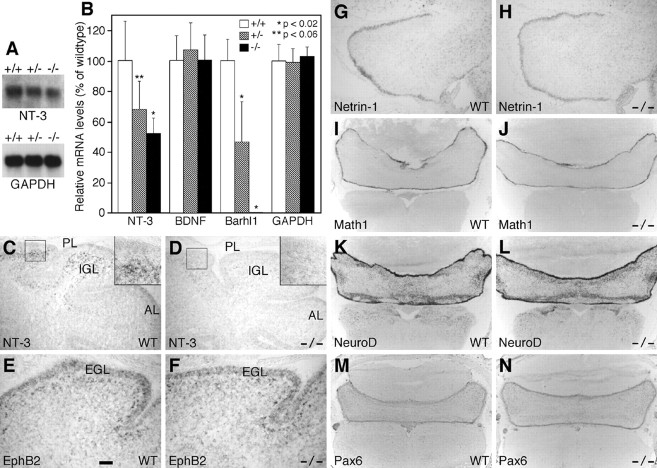
Given the similar cerebellar phenotypes present between Barhl1 and NT-3 or BDNF mutant mice (Schwartz et al., 1997; Bates et al., 1999; Borghesani et al., 2002; Carter et al., 2003), we investigated whether loss of Barhl1 function would affect NT-3 and BDNF expression in the cerebellum. Compared with the wild-type, Northern blot analysis showed a substantial reduction of NT-3 mRNA levels in cerebella of P6 Barhl1–/– and Barhl1+/- animals (Fig. 4A). To confirm this observation, we performed real-time quantitative RT-PCR, using total RNA isolated from P6 Barhl1+/+, Barhl1+/–, and Barhl1–/– cerebella. In Barhl1+/– and Barhl1–/– cerebella, we found that NT-3 mRNA levels were reduced to ∼70 and 50% of wild-type levels, respectively (Fig. 4B). As a control, however, GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNA levels were not altered in cerebella of any of the three different genotypes (Fig. 4B). Consistent with a null mutation, no Barhl1 mRNA was detected in the Barhl1-/- cerebellum and ∼50% of wild-type levels of Barhl1 mRNA was present in the Barhl1+/- cerebellum (Fig. 4B). Contrary to the significant downregulation of NT-3 expression, however, real-time RT-PCR did not detect any change in BDNF expression levels in Barhl1 mutant cerebella (Fig. 4B), indicating a lack of regulation of BDNF by Barhl1.
Figure 4.
Downregulation of NT-3 expression within the cerebella of Barhl1–/– mice. A, Northern blot comparing NT-3 mRNA levels in P6 cerebella of Barhl1+/+, Barhl1+/–, and Barhl1–/– animals. The blot was stripped and rehybridized with a GAPDH (glyceraldehyde-3-phosphate dehydrogenase) probe as shown below. B, Real-time RT-PCR analysis of NT-3, BDBF, Barhl1, and GAPDH mRNA levels in P6 cerebella of three genotypes. Each histogram represents the mean ± SD for five cerebella. C–N, P6 (C, D), P3 (E,F), and P1 (G–N) cerebellar sections from wild-type and mutant mice were hybridized with the indicated riboprobes. The inset in C or D is a higher magnification of the boxed area in each panel. Note the strong NT-3 signal within the IGL of the posterior lobe (PL) in the wild-type cerebellum (C) but the near absence of NT-3 signal in both anterior (AL) and posterior lobes of the mutant (D). In the EGL, comparable expression levels of EphB2 (E, F), netrin-1 (G, H), Math1 (I, J), NeuroD (K, L), and Pax6 (M, N) were seen between the wild-type and mutant cerebella. Scale bar: E, F, 50 μm; C, D, G, H, 100 μm; I-N, 250 μm.
In the postnatal cerebellum, NT-3 expression has been demonstrated to display a regional specificity; high levels of expression are found only in granule cells of the posterior lobe (Tojo et al., 1995). In agreement, as observed in P6 cerebellar sections hybridized with a specific NT-3 riboprobe, there was prominent expression of NT-3 mRNA in granule cells of the IGL within the posterior lobe of wild-type cerebella (Fig. 4C). By contrast, this high level of NT-3 expression was nearly abolished in the posterior lobe of Barhl1–/– cerebella (Fig. 4D), indicating that in Barhl1–/– cerebella the twofold overall reduction in NT-3 mRNA levels results primarily from a drastic downregulation of NT-3 expression in the posterior lobe.
It has been shown that Math1, NeuroD, Pax6, netrin-1 signaling, and ephrin-B signaling all play a role in the determination, migration, or maintenance of cerebellar granule cells (Serafini et al., 1996; Ben-Arie et al., 1997; Fazeli et al., 1997; Przyborski et al., 1998; Bloch-Gallego et al., 1999; Engelkamp et al., 1999; Miyata et al., 1999; Alcantara et al., 2000; Goldowitz et al., 2000; Lu et al., 2001; Yamasaki et al., 2001). We thus investigated by RNA in situ hybridization whether loss of Barhl1 function would affect the expression of these molecules. We found that Math1, NeuroD, Pax6, Netrin-1, DCC, neogenin, Unc5h2, Unc5h3, EphB2, and Ephrin-B2 were all essentially normally expressed in cerebellar granule cells of Barhl1 null animals (Fig. 4E–N) (data not shown), suggesting that Barhl1 may not regulate multiple signaling pathways in the control of granule cell migration and survival.
Defects in migration and survival of Barhl1–/– precerebellar neurons
To analyze defects in the precerebellar system of Barhl1–/– mice, we followed destinations of lacZ-positive cells in all precerebellar nuclei of embryonic and postnatal animals. At P5 and P19, β-galactosidase staining of whole-mount brains revealed that the mutant PGN became aberrantly narrow while the mutant LRN was dramatically reduced in size (Fig. 5A,B). Consistent with this observation, as analyzed in coronal sections, the ECN, LRN, and VN, which were derived from the posterior extramural migratory stream, were all substantially diminished in size in Barhl1–/– mice (Fig. 5C,D,G,H). Moreover, their neurons tended to be more scattered than in the control (Fig. 5A–D,G,H). In the Barhl1+/– brain, neurons from the anterior extramural migratory stream gave rise to two symmetrical and separate RTNs (Fig. 5E); however, they appeared to intermingle to form a single nucleus with a greatly reduced size in the Barhl1–/– pons (Fig. 5F). Thus, the absence of Barhl1 appeared to cause improper migration and significant loss of neurons within all precerebellar nuclei that project mossy fibers. Consistent with the absence of Barhl1 expression in the ION, however, no abnormality in ION laminar structure and size was observed in Barhl1–/– mice (data not shown).
Figure 5.
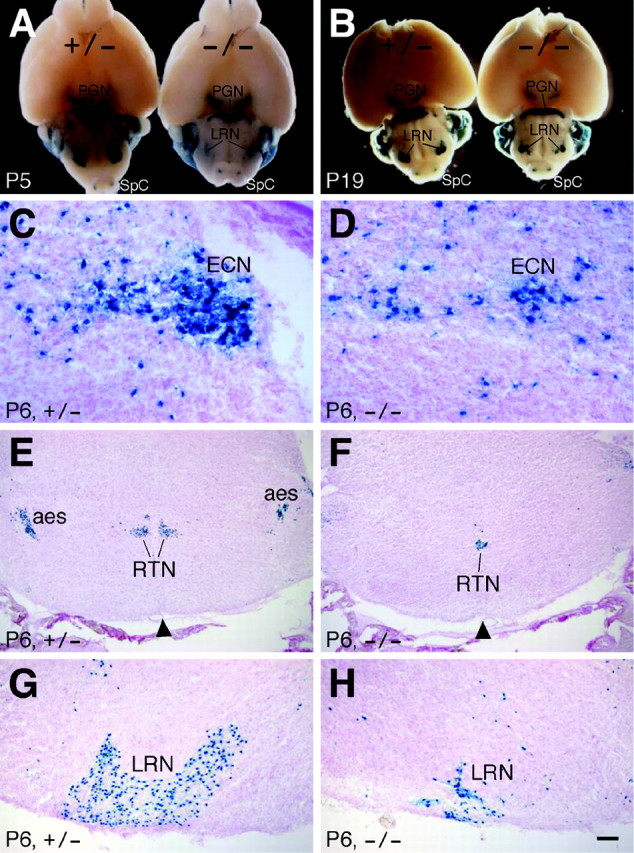
Abnormalities in all precerebellar nuclei that extend mossy fibers in Barhl1–/– mice. A, B, X-gal staining of P5 and P19 Barhl1+/– and Barhl1–/– whole-mount brains. C–H, X-gal staining of P6 coronal brain sections from Barhl1+/– (C, E, G) and Barhl1–/– (D, F, H) mice. The pontine gray (PGN), reticulotegmental (RTN), external cuneate (ECN), and lateral reticular (LRN) nuclei are all reduced in size in Barhl1–/– mice (A–H). Instead of two in the control (E), a single fused RTN is present in the mutant (F). The arrowheads point to midlines of brain sections (E, F). Scale bar: C, D, 50 μm; G, H, 100 μm; E, F, 250 μm; A, 1000 μm; B, 1429 μm.
A more detailed analysis of the mutant PGN was performed to better understand precerebellar abnormalities of Barhl1 null mice. At P19, β-galactosidase staining of Barhl1+/– whole-mount brains revealed two large peach-shaped clusters of lacZ-expressing cells that were symmetrically located medioventrally in the pons. Whereas in Barhl1–/– mice, these clusters essentially became two narrow strips, significantly elongated laterally (Fig. 6A,B). Moreover, although there was a clear midline that was free of lacZ-expressing cells between the two PGNs in the Barhl1+/– brain, the midline was barely seen in the Barhl1–/– brain (Fig. 6A,B). These defects were also evident in null mutants at early postnatal stages (Fig. 6C–F), indicating that many PGN neurons fail to migrate into their proper positions in the absence of Barhl1. Similar to other mossy fiber-extending nuclei, we also noted a great size reduction of the Barhl1–/– PGN, by comparing brain sections from control and mutant mice labeled by β-galactosidase activity or cresyl violet (Fig. 6E–H). At P100, a quantitation of neurons showed a 70% decrease in the total number of neurons present in the Barhl1–/– PGN compared with the control Barhl1+/– nucleus (Fig. 6I). Similarly, a 70% reduction was also observed in the total number of neurons within the Barhl1–/– RTN (Fig. 6I), demonstrating a dramatic loss of neurons in the PGN and RTN of Barhl1–/– mice.
Figure 6.
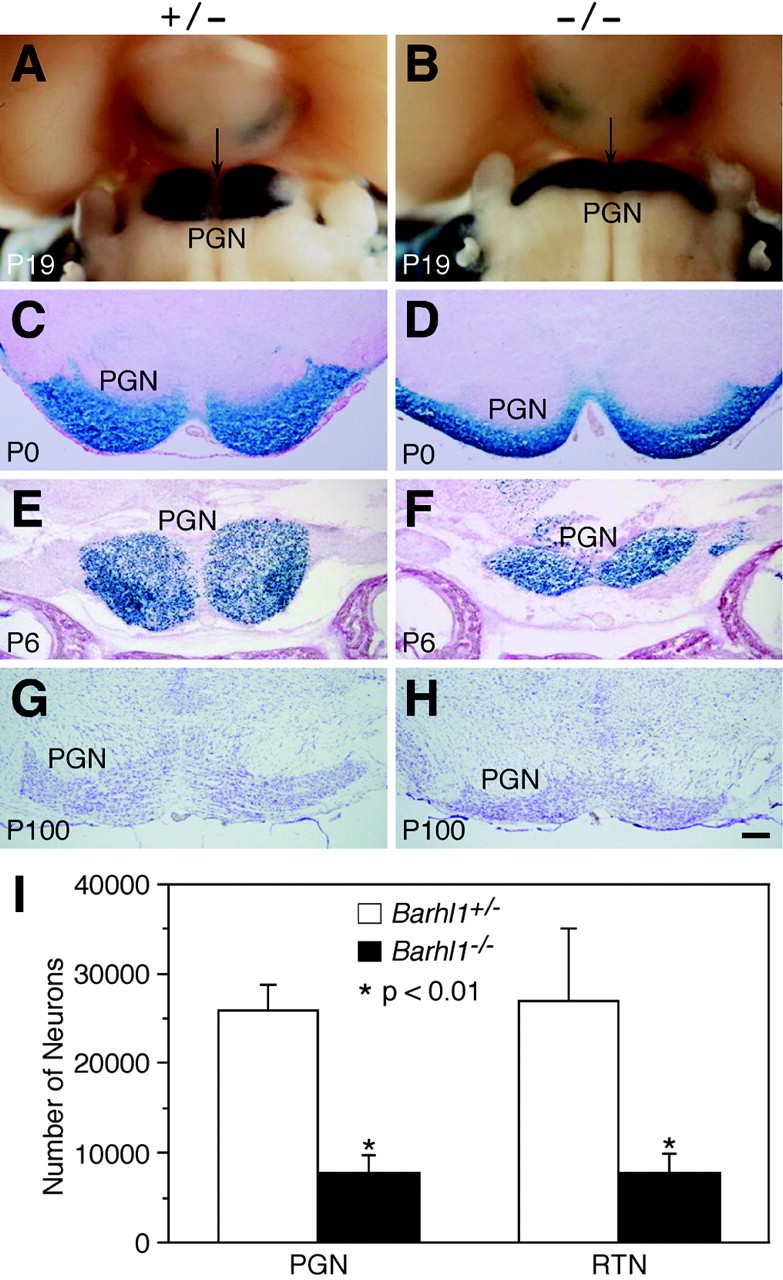
Abnormalities in the pontine gray (PGN) and reticulotegmental (RTN) nuclei of Barhl1–/– mice. A, B, P19 whole-mount brains were stained with X-gal. Compared with Barhl1+/– mice (A), the pontine gray nuclei in Barhl1–/– mice (B) were smaller, elongated, and incompletely separated at the midline (indicated by arrows). C–H, Coronal brain sections from postnatal animals at the indicated developmental stages were stained with X-gal (C–F) or cresyl violet (G, H). In the pontine gray nuclei of Barhl1–/– mice, lacZ-expressing cells failed to form tight clusters at P0 (C, D), and many of them became lost by P6 (E, F). At P100, the mutant PGN became much smaller than the control (G, H). I, Quantitation of neuron numbers in pontine gray and reticulotegmental nuclei of P100 Barhl1+/– and Barhl1–/– mice. Each histogram represents the mean ± SD for four nuclei. Scale bar: C, D, 156 μm; G, H, 200 μm; E, F, 250 μm; A, B, 400 μm.
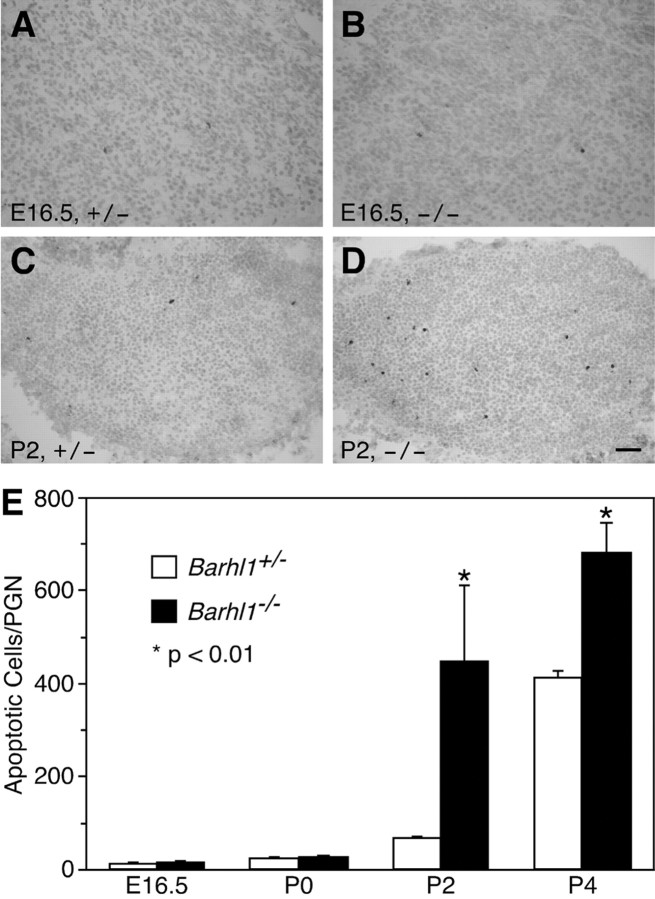
To determine the mechanism that led to neuron reduction in the mutant PGN, we measured apoptotic cell death in control and null mice during embryonic and postnatal stages (Fig. 7). As visualized by immunoreactivity for the active caspase-3, there was a very low level of apoptotic cell death in the Barhl1+/– PGN at E16.5 and P2 (Fig. 7A,C). In the Barhl1–/– PGN, there was a similar low level of apoptotic cell death at E16.5 but a greatly elevated cell death at P2 (Fig. 7B,D). By quantitation, we found that although there was no difference in the number of neurons undergoing apoptosis in the PGN between control and null mutants at E16.5 and P0, the number of neurons undergoing apoptosis in the mutant increased ∼700% at P2 and ∼70% at P4 (Fig. 7E). Therefore, most of the neuron loss in the Barhl1–/– PGN occurred via apoptosis during early postnatal stages. Consistent with this observation, the mutant PGN is already greatly reduced in size as early as P5–P6 (Figs. 5A, 6E,F).
Figure 7.
Loss of neurons by apoptotic cell death in Barhl1–/– pontine gray nuclei. A–D, Cells undergoing apoptosis were immunostained with an anti-active caspase-3 antibody in E16.5 (A, B) and P2 (C, D) Barhl1+/– (A, C) and Barhl1–/– (B, D) pontine gray nuclei. A significant increase of apoptotic neurons was observed in the Barhl1–/– pontine gray nucleus at P2 (C, D). E, Quantitation of apoptotic cell death in Barhl1–/– and controlBarhl1+/– pontine gray nuclei during development. Each histogram represents the mean ± SD for four nuclei. Scale bar: A, B, 25 μm; C, D, 50 μm.
Formation and pathfinding of mossy fibers are not altered in Barhl1–/– cerebellar and precerebellar systems
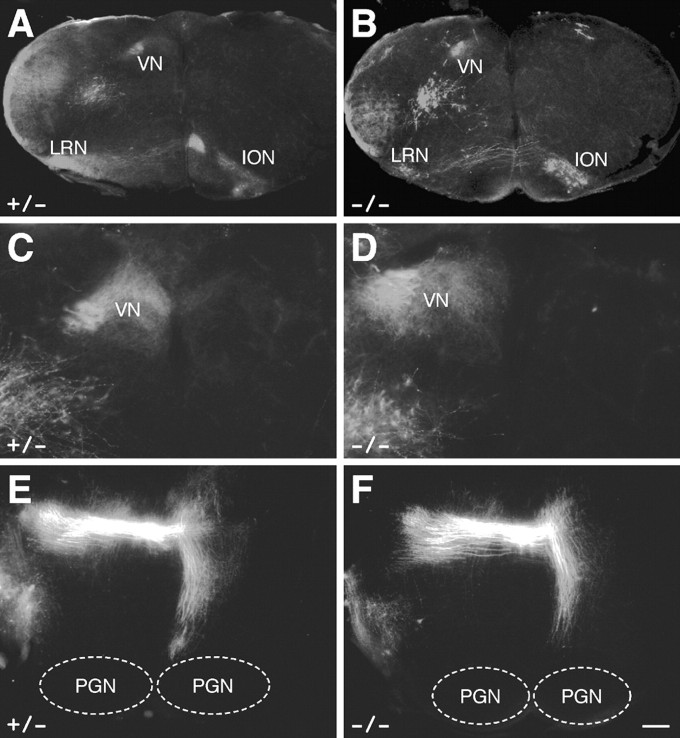
Given the strong expression of Barhl1 in granule cells and mossy fiber-extending neurons in the developing cerebellar and precerebellar systems, we investigated whether loss of Barhl1 function had any effect on the formation and navigation of mossy fibers. To visualize and trace mossy fiber projections to the cerebella of Barhl1–/– mice, we unilaterally inserted DiI crystals in hemispheres of P1 control and mutant cerebella. In the medulla of control and null mice, we found that neurons within the VN and LRN were ipsilaterally labeled, whereas those within the ION were contralaterally labeled without significant difference between the two genotypes (Fig. 8 A–D). Similarly, unilateral retrograde tracing primarily labeled contralateral pontocerebellar fiber bundles in both control and null mice (Fig. 8 E,F). Thus, the absence of Barhl1 does not appear to affect the formation and guidance of cerebellar mossy fibers.
Figure 8.
Retrograde labeling of precerebellar nuclei in control and mutant newborn mice after unilateral DiI injections in the cerebellum. In both control and null mice, the vestibular (VN) and lateral reticular nuclei (LRN) were ipsilaterally labeled, whereas the inferior olivary nuclei (ION) were contralaterally labeled (A–D). Unilateral retrograde tracing also similarly labeled pontocerebellar fibers derived from the contralateral pontine gray nuclei (PGN) in control and null mice (E, F). Scale bar: C, D, 100 μm; A, B, E, F, 250 μm.
Discussion
The experiments described in this report aimed to investigate the expression pattern and biological function of Barhl1 during CNS development. We provide evidence to show that Barhl1 is strongly expressed within the rhombic lip and rhombic lipderived migratory neurons in developing cerebellar and precerebellar systems. Its expression is later confined to cerebellar granule cells and precerebellar neurons extending mossy fibers, two groups of neurons that synaptically connect in the adult. Targeted disruption of Barhl1 in mice results in attenuated foliation and hypotrophy of the cerebellum caused by deficiencies in radial migration and survival of granule cells. Moreover, it causes inappropriate migration of mossy fiber-extending precerebellar neurons and a dramatic loss of these neurons by apoptotic cell death. Notably, a search of candidate Barhl1 downstream genes in the cerebellum has identified NT-3, whose expression is greatly downregulated by the absence of Barhl1 in the posterior cerebellar lobe and which has been shown to be required for radial migration and survival of cerebellar granule cells. Thus, our data together reveal a key role for Barhl1 in the control of migration and survival of cerebellar and precerebellar neurons and identify NT-3 as a major downstream gene that mediates the crucial function of Barhl1 during cerebellar development.
Barhl1 is expressed in migratory cells fated to become cerebellar granule cells and precerebellar neurons extending mossy fibers
During murine embryogenesis, cerebellar granule cells and precerebellar neurons are all derived from the germinal rhombic lip. The anterior rhombic lip gives rise to cerebellar granule cells while the posterior rhombic lip to precerebellar neurons. RNA in situ hybridization and β-galactosidase staining of Barhl1+/– embryos and sections indicate that Barhl1 is expressed within both anterior and posterior rhombic lips in early embryos, in migratory cells generated by rhombic lips, as well as in their descending cerebellar granule cells and precerebellar neurons. Therefore, Barhl1 may be involved in the determination, differentiation, and/or maintenance of cerebellar and precerebellar neurons because it is expressed by progenitor cells as well as by differentiating and differentiated neurons in the developing cerebellar and precerebellar systems. Indeed, our data have demonstrated that Barhl1 is required for the migration and survival of cerebellar and precerebellar neurons. However, it appears to be dispensable for their fate determination as cerebellar granule cells and precerebellar neurons are both produced and largely differentiated in Barhl1 null mice, as indicated by β-galactosidase staining, histochemical labeling, retrograde DiI tracing, as well as marker gene expression (Figs. 3, 4, 5, 6, 7, 8). In the inner ear, we have shown previously that Barhl1 is similarly not required for fate commitment of sensory hair cells (Li et al., 2002).
In the precerebellar system, neurons extending mossy fibers or climbing fibers not only have different innervation targets but they also follow distinct migration pathways to reach discrete precerebellar nuclei during development. Cells of extramural migratory streams take either an anteroventral subpial route to settle in the PGN and RTN (Altman and Bayer, 1987a), or a posteroventral subpial path to settle in the VN, LRN, and ECN (Altman and Bayer, 1987b; Rodriguez and Dymecki, 2000). All neurons in the ION that project climbing fibers, in contrast, are derived from migratory cells following an intramural circumferential migration path (Altman and Bayer, 1987c). Interestingly, Barhl1 is found only in precerebellar neurons extending mossy fibers where it regulates their migration and survival. Because the onset of Barhl1 expression is seen in rhombic lips and migratory streams before cell migration even takes place (Figs. 1, 2), we identify Barhl1 as an early marker for mossy fiber-extending precerebellar neurons. Similarly, a fate-mapping study has shown that Wnt-1 expression demarcates a progenitor pool that gives rise only to precerebellar neurons that extend mossy fibers (Rodriguez and Dymecki, 2000). In contrast, the two closely related POU domain transcription factors Brn3a and Brn3b are expressed only in cells of the intramural migratory stream and ION (McEvilly et al., 1996; Xiang et al., 1996). The absence of Brn3a in mice has been shown to result in disorganization and loss of several component units of the ION, suggesting a role for Brn3a in the migration and/or survival of neurons extending climbing fibers (McEvilly et al., 1996; Xiang et al., 1996).
Barhl1 controls radial migration and survival of cerebellar granule cells by regulation of NT-3 expression
In the cerebellum, we have uncovered a critical role of Barhl1 in the control of granule cell migration. Our BrdU labeling and tracing experiments have revealed that there is a delay in migration of some granule cells out of the EGL in Barhl1 null cerebella. This defect is similar to but milder than that in mice deficient for BDNF or the peroxisome assembly gene PEX2 (Schwartz et al., 1997; Borghesani et al., 2002; Faust, 2003). Given ectopias of granule cells are found only on the surface but not within the molecular layer of adult Barhl1–/– cerebella, we propose that Barhl1 is additionally involved in the initial step of radial migration of a small fraction of granule cells. In agreement with this speculation, we are able to show that ectopic cells in adult mutants are nonproliferative and that some granule cells fail to exit EGL even 8 d after BrdU labeling (Fig. 3P). Because the large majority of granule cells are still normally located within the IGL in the mutant cerebellum, in addition to Barhl1, there must be other regulatory factors involved in the initial step of radial migration. A most likely candidate is the other Barhl family member, Barhl2/MBH1 (Saito et al., 1998; Bulfone et al., 2000), which we have shown to be expressed in cerebellar granule cells and thus may play a redundant role (Mo et al., 2004).
Barhl1 is a homeoprotein that most likely exerts its function by transcriptionally regulating the expression of its target genes. Our work has identified NT-3 as a major effector gene directly or indirectly regulated by Barhl1 during cerebellar development. Analysis of the 5′-flanking sequence of NT-3 reveals at least two potential Barhl1 binding sites containing CTAATTG. In the developing cerebellum, NT-3 is prominently expressed in the IGL and within the premigratory zone of the EGL in the posterior lobe at early postnatal stages when Barhl1 is also highly expressed (Rocamora et al., 1993; Tojo et al., 1995). Administration of exogenous NT-3 in vivo has been shown to promote granule cell migration and survival, whereas depletion of the endogenous NT-3 has the opposite effect (Neveu and Arenas, 1996; Doughty et al., 1998; Katoh-Semba et al., 2000). By conditional gene targeting, it has been demonstrated that NT-3 acts as a survival factor for a subset of cerebellar granule cells (Bates et al., 1999). As well, analysis of mice deficient for neurotrophin receptors including the NT-3 receptor TrkC has uncovered a critical role for NT-3 signaling in radial migration and survival of granule cells (Minichiello and Klein, 1996). Thus, the delay and failure for some granule cells to initiate radial migration in Barhl1 null cerebella can be attributed to the significant downregulation of NT-3 expression. Similarly, the substantial size reduction of Barhl1 null cerebella by apoptosis appears to result from NT-3 downregulation because apoptotic death of granule cells is also increased in mice deficient for NT-3 or TrkC (Minichiello and Klein, 1996; Bates et al., 1999). Notably, loss of Barhl1 function almost abrogates the high-level NT-3 expression in the posterior lobe of the cerebellum including lobules VI and VII (Fig. 4C,D), where foliation pattern is specifically disrupted in the mutant. Thus, although the overall level of NT-3 expression is downregulated only by 50% in Barhl1 null cerebella, the dramatic region-specific downregulation of NT-3 expression is expected and has been observed to cause more severe phenotypes in Barhl1 null mice than in NT-3 heterozygotes.
In Barhl1 null cerebella, we observed a loss of folium VII and the intercrural fissure, defective radial migration of granule cells as well as a reduction of the cerebellar size caused by elevated apoptotic death of granule cells. These phenotypes closely mimic the cerebellar phenotypes present in NT-3 and BDNF null mice as well as in a rat model of hypothyroidism that causes great down-regulation of cerebellar NT-3 and BDNF expression (Neveu and Arenas, 1996; Schwartz et al., 1997; Bates et al., 1999; Borghesani et al., 2002; Carter et al., 2003). The cerebellar phenotypes of Barhl1 null mice also display some similarities to those present in mice deficient for PEX2, the orphan receptor gene rev-erbAα, or the putative glycosyltransferase gene Large (Chomez et al., 2000; Holzfeind et al., 2002; Faust, 2003). However, we could not detect by real-time RT-PCR any alteration in expression levels of BDNF, PEX2, rev-erbAα, or Large in Barhl1 null cerebella (Fig. 4B) (data not shown), suggesting that the major functions of Barhl1 during cerebellar development may be mediated by NT-3. In addition, consistent with this notion, the absence of Barhl1 does not appear to affect the expression of netrin-1 or ephrin-B signaling molecules which have been implicated in the control of directional migration of cerebellar granule cells (Alcantara et al., 2000; Lu et al., 2001). Despite our demonstration of the regulation of NT-3 expression by Barhl1, the expression of NT-3, like many other genes, appear to be controlled by multiple signaling pathways. For instance, its expression has been shown to be regulated by the thyroid hormone, BDNF and MEF2 (Neveu and Arenas, 1996; Shalizi et al., 2003).
At present, the transcriptional cascade that controls cerebellar granule cell development remains largely unknown. Our study implicates Barhl1 as a key regulator in the cascade that controls proper migration and survival of granule cells. Math1 is expressed in the rhombic lip as early as E9.5 before the onset of Barhl1 expression (Akazawa et al., 1995; Ben-Arie et al., 1996, 2000; Helms and Johnson, 1998; Helms al., 2000), and has been shown to be required for the specification of cerebellar granule cells (Ben-Arie et al., 1997; Helms et al., 2001). In this work, we show that the expression of Math1 is not altered in Barhl1–/– cerebella. Hence, during cerebellar development, Barhl1 must act genetically downstream of Math1 and may be one of its direct target genes as suggested in the inner ear hair cells and spinal cord (Bermingham et al., 1999; Bermingham et al., 2001; Li et al., 2002). Barhl1 may act also downstream of Pax6 given that: (1) the absence of Barhl1 does not affect Pax6 expression, (2) loss of either Barhl1 or Pax6 function causes defects in granule cell migration (Engelkamp et al., 1999; Yamasaki et al., 2001), and (3) Barhl1 and Pax6 share very similar spatial and temporal expression patterns in the developing and mature granule cells (Engelkamp et al., 1999; Yamasaki et al., 2001). The epistatic relationship between Barhl1 and NeuroD, however, is less clear because NeuroD is expressed late in development only in postmitotic granule cells and yet its expression is not altered in Barhl1–/– cerebella (Miyata et al., 1999).
Barhl1 controls migration and survival of mossy fiber-extending precerebellar neurons
The differentiation of precerebellar neurons is characterized by their lengthy migration to settle into their remote target sites in the pons and medulla. Our current work suggests that Barhl1 plays a major role in the programmed migration of mossy fiber-extending neurons derived from the extramural migratory streams. In particular, Barhl1 appears to be involved in the final stage of the migration program for fine positioning neurons migrating into target precerebellar nuclei, because most, or perhaps all of mossy fiber-extending neurons in Barhl1 null mutants can reach the proximity of their target sites but many fail to migrate within the target nuclei. Thus, in the absence of Barhl1, many mossy fiber-extending neurons may fail to respond to local attractive and/or repulsive guidance cues necessary for migration into a proper position. Such a failure can readily explain the elongated PGN or fused RTN in the mutant (Figs. 5, 6). Similar to Barhl1, Pax6 has been shown to play a key role in the control of programmed migration of the precerebellar neurons, however, unlike Barhl1, Pax6 appears to be required for the initiation of their migration (Engelkamp et al., 1999). In Pax6 null mice, migration defects of precerebellar neurons lead to an enlarged lower rhombic lip, dramatically reduced PGN, and severely disorganized ECN and LRN (Engelkamp et al., 1999).
Aside from its role in migration, Barhl1 plays an essential role in the maintenance of precerebellar neurons that project mossy fibers. The loss of 70% of all neurons in the mutant PGN primarily occurs at early postnatal stages via apoptosis, resulting in dramatically reduced size of PGN as early as P5. However, neuron loss does not appear to be a direct result of improper migration as many of the remaining dislocated neurons survive in the adult mutant PGN (Fig. 6). In the rodent, the majority of mossy fiber-extending neurons have been shown to project into the cerebellum to make synapses with granule cells during the perinatal and early postnatal period (Ashwell and Zhang, 1992). Like many other axon-projecting neurons, they appear to require target-derived neurotrophic factors for survival because we can show that many PGN neurons die naturally in control mice during early postnatal stages when they would compete for granule cell-derived survival factors (Fig. 7E). Thus, there are at least two possible explanations for the increased cell death in the mutant PGN. These two possibilities are not necessarily mutually exclusive. First, there may be a reduced supply of neurotrophic factors from mutant cerebellar granule cells. This is likely because we have revealed a significant reduction of NT-3 expression in the mutant cerebellum. Second, loss of Barhl1 function may render PGN neurons nonresponsive to target-derived survival factors.
Although to date, little is known about the molecular mechanisms underlying the migration of precerebellar neurons, the existence of stereotyped migratory pathways followed by precerebellar neurons indicates that their migration may be under the regulation of specific guidance cues. Recent evidence implicates that some axon guidance molecules such as netrin-1 and its receptor DCC play a role in the control of directional migration of precerebellar neurons (Alcantara et al., 2000). However, the absence of Barhl1 does not appear to alter the expression of netrin-1 and its receptors in the brainstem or cause misrouting of mossy fibers (Fig. 8) (data not shown). Thus, Barhl1 may regulate the expression of other unidentified signaling molecules to control proper migration of mossy fiber-extending neurons.
Footnotes
This work was supported by National Institutes of Health Grants DC04594 and EY12020 (to M.X.). We are grateful to Dr. Marc Tessier-Lavigne for providing cDNA probes for netrin-1 and its receptors, Dr. John Flanagan for the EphB2 cDNA, Dr. Renping Zhou for the Ephrin-B2 plasmid, and Dr. Grady Saunders for the PAX6 cDNA. We thank Drs. Michael Shen and James Millonig for thoughtful comments on this manuscript.
Correspondence should be addressed to Dr. Mengqing Xiang, Center for Advanced Biotechnology and Medicine, 679 Hoes Lane, Piscataway, NJ 08854. E-mail: xiang@cabm.rutgers.edu.
DOI:10.1523/JNEUROSCI.4444-03.2004
Copyright © 2004 Society for Neuroscience 0270-6474/04/243104-11$15.00/0
References
- Akazawa C, Ishibashi M, Shimizu C, Nakanishi S, Kageyama R (1995) A mammalian helix-loop-helix factor structurally related to the product of Drosophila proneural gene atonal is a positive transcriptional regulator expressed in the developing nervous system. J Biol Chem 270: 8730–8738. [DOI] [PubMed] [Google Scholar]
- Alcantara S, Ruiz M, De Castro F, Soriano E, Sotelo C (2000) Netrin-1 acts as an attractive or as a repulsive cue for distinct migrating neurons during the development of the cerebellar system. Development 127: 1359–1372. [DOI] [PubMed] [Google Scholar]
- Altman J, Bayer SA (1987a) Development of the precerebellar nuclei in the rat: IV. The anterior precerebellar extramural migratory stream and the nucleus reticularis tegmenti pontis and the basal pontine gray. J Comp Neurol 257: 529–552. [DOI] [PubMed] [Google Scholar]
- Altman J, Bayer SA (1987b) Development of the precerebellar nuclei in the rat: III. The posterior precerebellar extramural migratory stream and the lateral reticular and external cuneate nuclei. J Comp Neurol 257: 513–528. [DOI] [PubMed] [Google Scholar]
- Altman J, Bayer SA (1987c) Development of the precerebellar nuclei in the rat: II. The intramural olivary migratory stream and the neurogenetic organization of the inferior olive. J Comp Neurol 257: 490–512. [DOI] [PubMed] [Google Scholar]
- Altman J, Bayer SA (1987d) Development of the precerebellar nuclei in the rat: I. The precerebellar neuroepithelium of the rhombencephalon. J Comp Neurol 257: 477–489. [DOI] [PubMed] [Google Scholar]
- Ashwell KW, Zhang LL (1992) Ontogeny of afferents to the fetal rat cerebellum. Acta Anat 145: 17–23. [DOI] [PubMed] [Google Scholar]
- Bates B, Rios M, Trumpp A, Chen C, Fan G, Bishop JM, Jaenisch R (1999) Neurotrophin-3 is required for proper cerebellar development. Nat Neurosci 2: 115–117. [DOI] [PubMed] [Google Scholar]
- Ben-Arie N, McCall AE, Berkman S, Eichele G, Bellen HJ, Zoghbi HY (1996) Evolutionary conservation of sequence and expression of the bHLH protein Atonal suggests a conserved role in neurogenesis. Hum Mol Genet 5: 1207–1216. [DOI] [PubMed] [Google Scholar]
- Ben-Arie N, Bellen HJ, Armstrong DL, McCall AE, Gordadze PR, Guo Q, Matzuk MM, Zoghbi HY (1997) Math1 is essential for genesis of cerebellar granule neurons. Nature 390: 169–172. [DOI] [PubMed] [Google Scholar]
- Ben-Arie N, Hassan BA, Bermingham NA, Malicki DM, Armstrong D, Matzuk M, Bellen HJ, Zoghbi HY (2000) Functional conservation of atonal and Math1 in the CNS and PNS. Development 127: 1039–1048. [DOI] [PubMed] [Google Scholar]
- Bermingham NA, Hassan BA, Price SD, Vollrath MA, Ben-Arie N, Eatock RA, Bellen HJ, Lysakowski A, Zoghbi HY (1999) Math1: an essential gene for the generation of inner ear hair cells. Science 284: 1837–1841. [DOI] [PubMed] [Google Scholar]
- Bermingham NA, Hassan BA, Wang VY, Fernandez M, Banfi S, Bellen HJ, Fritzsch B, Zoghbi HY (2001) Proprioceptor pathway development is dependent on Math1. Neuron 30: 411–422. [DOI] [PubMed] [Google Scholar]
- Bloch-Gallego E, Ezan F, Tessier-Lavigne M, Sotelo C (1999) Floor plate and netrin-1 are involved in the migration and survival of inferior olivary neurons. J Neurosci 19: 4407–4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghesani PR, Peyrin JM, Klein R, Rubin J, Carter AR, Schwartz PM, Luster A, Corfas G, Segal RA (2002) BDNF stimulates migration of cerebellar granule cells. Development 129: 1435–1442. [DOI] [PubMed] [Google Scholar]
- Bulfone A, Menguzzato E, Broccoli V, Marchitiello A, Gattuso C, Mariani M, Consalez GG, Martinez S, Ballabio A, Banfi S (2000) Barhl1, a gene belonging to a new subfamily of mammalian homeobox genes, is expressed in migrating neurons of the CNS. Hum Mol Genet 9: 1443–1452. [DOI] [PubMed] [Google Scholar]
- Carter AR, Berry EM, Segal RA (2003) Regional expression of p75NTR contributes to neurotrophin regulation of cerebellar patterning. Mol Cell Neurosci 22: 1–13. [DOI] [PubMed] [Google Scholar]
- Chomez P, Neveu I, Mansen A, Kiesler E, Larsson L, Vennstrom B, Arenas E (2000) Increased cell death and delayed development in the cerebellum of mice lacking the rev-erbA(α) orphan receptor. Development 127: 1489–1498. [DOI] [PubMed] [Google Scholar]
- Doughty ML, Lohof A, Campana A, Delhaye-Bouchaud N, Mariani J (1998) Neurotrophin-3 promotes cerebellar granule cell exit from the EGL. Eur J Neurosci 10: 3007–3011. [DOI] [PubMed] [Google Scholar]
- Eng SR, Gratwick K, Rhee JM, Fedtsova N, Gan L, Turner EE (2001) Defects in sensory axon growth precede neuronal death in Brn3a-deficient mice. J Neurosci 21: 541–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelkamp D, Rashbass P, Seawright A, van Heyningen V (1999) Role of Pax6 in development of the cerebellar system. Development 126: 3585–3596. [DOI] [PubMed] [Google Scholar]
- Faust PL (2003) Abnormal cerebellar histogenesis in PEX2 Zellweger mice reflects multiple neuronal defects induced by peroxisome deficiency. J Comp Neurol 461: 394–413. [DOI] [PubMed] [Google Scholar]
- Fazeli A, Dickinson SL, Hermiston ML, Tighe RV, Steen RG, Small CG, Stoeckli ET, Keino-Masu K, Masu M, Rayburn H, Simons J, Bronson RT, Gordon JI, Tessier-Lavigne M, Weinberg RA (1997) Phenotype of mice lacking functional Deleted in colorectal cancer (Dcc) gene. Nature 386: 796–804. [DOI] [PubMed] [Google Scholar]
- Goldowitz D, Hamre KM, Przyborski SA, Ackerman SL (2000) Granule cells and cerebellar boundaries: analysis of Unc5h3 mutant chimeras. J Neurosci 20: 4129–4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatten ME (2002) New directions in neuronal migration. Science 297: 1660–1663. [DOI] [PubMed] [Google Scholar]
- Hatten ME, Alder J, Zimmerman K, Heintz N (1997) Genes involved in cerebellar cell specification and differentiation. Curr Opin Neurobiol 7: 40–47. [DOI] [PubMed] [Google Scholar]
- Helms AW, Johnson JE (1998) Progenitors of dorsal commissural interneurons are defined by MATH1 expression. Development 125: 919–928. [DOI] [PubMed] [Google Scholar]
- Helms AW, Abney AL, Ben-Arie N, Zoghbi HY, Johnson JE (2000) Autoregulation and multiple enhancers control Math1 expression in the developing nervous system. Development 127: 1185–1196. [DOI] [PubMed] [Google Scholar]
- Helms AW, Gowan K, Abney A, Savage T, Johnson JE (2001) Overexpression of MATH1 disrupts the coordination of neural differentiation in cerebellum development. Mol Cell Neurosci 17: 671–682. [DOI] [PubMed] [Google Scholar]
- Higashijima S, Michiue T, Emori Y, Saigo K (1992a) Subtype determination of Drosophila embryonic external sensory organs by redundant homeo box genes BarH1 and. BarH2. Genes Dev 6: 1005–1018. [DOI] [PubMed] [Google Scholar]
- Higashijima S, Kojima T, Michiue T, Ishimaru S, Emori Y, Saigo K (1992b) Dual Bar homeo box genes of Drosophila required in two photoreceptor cells, R1 and R6, and primary pigment cells for normal eye development. Genes Dev 6: 50–60. [DOI] [PubMed] [Google Scholar]
- Holzfeind PJ, Grewal PK, Reitsamer HA, Kechvar J, Lassmann H, Hoeger H, Hewitt JE, Bittner RE (2002) Skeletal, cardiac and tongue muscle pathology, defective retinal transmission, and neuronal migration defects in the Large(myd) mouse defines a natural model for glycosylation-deficient muscle-eye-brain disorders. Hum Mol Genet 11: 2673–2687. [DOI] [PubMed] [Google Scholar]
- Katoh-Semba R, Takeuchi IK, Semba R, Kato K (2000) Neurotrophin-3 controls proliferation of granular precursors as well as survival of mature granule neurons in the developing rat cerebellum. J Neurochem 74: 1923–1930. [DOI] [PubMed] [Google Scholar]
- Keino-Masu K, Masu M, Hinck L, Leonardo ED, Chan SS, Culotti JG, Tessier-Lavigne M (1996) Deleted in Colorectal Cancer (DCC) encodes a netrin receptor. Cell 87: 175–185. [DOI] [PubMed] [Google Scholar]
- Kojima T, Ishimaru S, Higashijima S, Takayama E, Akimaru H, Sone M, Emori Y, Saigo K (1991) Identification of a different-type homeobox gene, BarH1, possibly causing Bar(B) and Om(1D) mutations in Drosophila. Proc Natl Acad Sci USA 88: 4343–4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardo ED, Hinck L, Masu M, Keino-Masu K, Ackerman SL, Tessier-Lavigne M (1997) Vertebrate homologues of C. elegans UNC-5 are candidate netrin receptors. Nature 386: 833–838. [DOI] [PubMed] [Google Scholar]
- Li S, Price SM, Cahill H, Ryugo DK, Shen MM, Xiang M (2002) Hearing loss caused by progressive degeneration of cochlear hair cells in mice deficient for the Barhl1 homeobox gene. Development 129: 3523–3532. [DOI] [PubMed] [Google Scholar]
- Lu Q, Sun EE, Klein RS, Flanagan JG (2001) Ephrin-B reverse signaling is mediated by a novel PDZ-RGS protein and selectively inhibits G protein-coupled chemoattraction. Cell 105: 69–79. [DOI] [PubMed] [Google Scholar]
- McEvilly RJ, Erkman L, Luo L, Sawchenko PE, Ryan AF, Rosenfeld MG (1996) Requirement for Brn-3.0 in differentiation and survival of sensory and motor neurons. Nature 384: 574–577. [DOI] [PubMed] [Google Scholar]
- Minichiello L, Klein R (1996) TrkB and TrkC neurotrophin receptors cooperate in promoting survival of hippocampal and cerebellar granule neurons. Genes Dev 10: 2849–2858. [DOI] [PubMed] [Google Scholar]
- Miyata T, Maeda T, Lee JE (1999) NeuroD is required for differentiation of the granule cells in the cerebellum and hippocampus. Genes Dev 13: 1647–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo Z, Li S, Yang X, Xiang M (2004) Role of the Barhl2 homeobox gene in the specification of glycinergic amacrine cells. Development 131: 1607–1618. [DOI] [PubMed] [Google Scholar]
- Neveu I, Arenas E (1996) Neurotrophins promote the survival and development of neurons in the cerebellum of hypothyroid rats in vivo. J Cell Biol 133: 631–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbergh L, Valckx D, Waer M, Mathieu C (1999) Quantification of murine cytokine mRNAs using real time quantitative reverse transcriptase PCR. Cytokine 11: 305–312. [DOI] [PubMed] [Google Scholar]
- Panchision DM, Pickel JM, Studer L, Lee SH, Turner PA, Hazel TG, McKay RD (2001) Sequential actions of BMP receptors control neural precursor cell production and fate. Genes Dev 15: 2094–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przyborski SA, Knowles BB, Ackerman SL (1998) Embryonic phenotype of Unc5h3 mutant mice suggests chemorepulsion during the formation of the rostral cerebellar boundary. Development 125: 41–50. [DOI] [PubMed] [Google Scholar]
- Rocamora N, Garcia-Ladona FJ, Palacios JM, Mengod G (1993) Differential expression of brain-derived neurotrophic factor, neurotrophin-3, and low-affinity nerve growth factor receptor during the postnatal development of the rat cerebellar system. Brain Res Mol Brain Res 17: 1–8. [DOI] [PubMed] [Google Scholar]
- Rodriguez CI, Dymecki SM (2000) Origin of the precerebellar system. Neuron 27: 475–486. [DOI] [PubMed] [Google Scholar]
- Saito T, Sawamoto K, Okano H, Anderson DJ, Mikoshiba K (1998) Mammalian BarH homologue is a potential regulator of neural bHLH genes. Dev Biol 199: 216–225. [DOI] [PubMed] [Google Scholar]
- Schwartz PM, Borghesani PR, Levy RL, Pomeroy SL, Segal RA (1997) Abnormal cerebellar development and foliation in BDNF-/- mice reveals a role for neurotrophins in CNS patterning. Neuron 19: 269–281. [DOI] [PubMed] [Google Scholar]
- Sciavolino PJ, Abrams EW, Yang L, Austenberg LP, Shen MM, Abate-Shen C (1997) Tissue-specific expression of murine Nkx3.1 in the male urogenital system. Dev Dyn 209: 127–138. [DOI] [PubMed] [Google Scholar]
- Serafini T, Colamarino SA, Leonardo ED, Wang H, Beddington R, Skarnes WC, Tessier-Lavigne M (1996) Netrin-1 is required for commissural axon guidance in the developing vertebrate nervous system. Cell 87: 1001–1014. [DOI] [PubMed] [Google Scholar]
- Shalizi A, Lehtinen M, Gaudilliere B, Donovan N, Han J, Konishi Y, Bonni A (2003) Characterization of a neurotrophin signaling mechanism that mediates neuron survival in a temporally specific pattern. J Neurosci 23: 7326–7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S, Tang HK, Lee JY, Saunders GF (1998) Truncation mutations in the transactivation region of PAX6 result in dominant-negative mutants. J Biol Chem 273: 21531–21541. [DOI] [PubMed] [Google Scholar]
- Tojo H, Takami K, Kaisho Y, Nakata M, Abe T, Shiho O, Igarashi K (1995) Neurotrophin-3 is expressed in the posterior lobe of mouse cerebellum, but does not affect the cerebellar development. Neurosci Lett 192: 169–172. [DOI] [PubMed] [Google Scholar]
- Wingate RJ (2001) The rhombic lip and early cerebellar development. Curr Opin Neurobiol 11: 82–88. [DOI] [PubMed] [Google Scholar]
- Wingate RJ, Hatten ME (1999) The role of the rhombic lip in avian cerebellum development. Development 126: 4395–4404. [DOI] [PubMed] [Google Scholar]
- Xiang M, Zhou L, Peng YW, Eddy RL, Shows TB, Nathans J (1993) Brn-3b: a POU domain gene expressed in a subset of retinal ganglion cells. Neuron 11: 689–701. [DOI] [PubMed] [Google Scholar]
- Xiang M, Zhou L, Macke JP, Yoshioka T, Hendry SH, Eddy RL, Shows TB, Nathans J (1995) The Brn-3 family of POU-domain factors: primary structure, binding specificity, and expression in subsets of retinal ganglion cells and somatosensory neurons. J Neurosci 15: 4762–4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang M, Gan L, Zhou L, Klein WH, Nathans J (1996) Targeted deletion of the mouse POU domain gene Brn-3a causes selective loss of neurons in the brainstem and trigeminal ganglion, uncoordinated limb movement, and impaired suckling. Proc Natl Acad Sci USA 93: 11950–11955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki T, Kawaji K, Ono K, Bito H, Hirano T, Osumi N, Kengaku M (2001) Pax6 regulates granule cell polarization during parallel fiber formation in the developing cerebellum. Development 128: 3133–3144. [DOI] [PubMed] [Google Scholar]
- Yue Y, Widmer DA, Halladay AK, Cerretti DP, Wagner GC, Dreyer JL, Zhou R (1999) Specification of distinct dopaminergic neural pathways: roles of the Eph family receptor EphB1 and ligand ephrin-B2. J Neurosci 19: 2090–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]