Abstract
Understanding how the hippocampus processes information critical for establishing spatial and declarative memories will benefit greatly from determining not only what kind of information the hippocampus registers, but also how this information is processed across the different hippocampal subfields. We addressed this question using a novel immediate-early gene-based brain-imaging method (Arc/H1a catFISH) that allows comparisons of neuronal ensembles activated by two experiences separated by ∼30 min. Rats exposed to the same environment twice activated CA3 and CA1 ensembles with a similarly high degree of overlap. Changing the identity or configuration of local cues, or changing distal cues, activated CA3 and CA1 ensembles with reduced overlap. Yet, the overlap was greater in CA3 than in CA1. In contrast, rats exposed to two completely different environments activated CA3 and CA1 ensembles with low overlap, and this overlap was even lower in CA3 compared with CA1. Thus, CA3 has a discontinuous, whereas CA1 has a graded, population response to alterations of an environment. Additionally, as indicated by the percentage of active neurons, the context representation was more sparse in CA3 (∼18%) than in CA1 (∼35%). Finally, CA3 and CA1 activity levels were not correlated within a session, arguing against a simple coactivation of these regions. Instead, the within-rat ratio of CA3/CA1 cell activity was correlated across sessions, suggesting that the balance of CA3/CA1 activity is individual specific. Taken together, these findings suggest that CA3 and CA1 neuronal ensembles perform distinct, yet complementary, functions in the processing of spatial and contextual information.
Keywords: Arc, Homer 1a, hippocampus, context discrimination, place cell, memory
Introduction
The hippocampus is critical for acquiring, consolidating, and retrieving declarative memories (for review, see Squire and Knowlton, 1994; Eichenbaum, 2001; Nadel and Moscovitch, 2001). However, the relative contributions of the hippocampal subregions (including CA3 and CA1) to these memory processes are not well understood. Theoretical considerations first developed by Marr (1971) have placed emphasis on the sparsity of coding necessary to store the large number of representations that the hippocampus encodes into memory (for review, see Skaggs and McNaughton, 1992). Sparsely encoded patterns are proposed to originate in CA3 after highly correlated patterns of activity arriving from the entorhinal cortex have been filtered extensively by the dentate gyrus (McNaughton and Nadel, 1990; Treves and Rolls, 1994; Vinogradova, 2001). Additionally, because of its extensive network of recurrent collaterals, CA3 is proposed to be an autoassociator (McNaughton and Nadel, 1990; Treves and Rolls, 1994) and comparator (Vinogradova, 2001) capable of retrieving entire patterns from partial or degraded input and comparing them with processed sensory input arriving from the entorhinal cortex. Thus, CA3 can act as a filter controlling the information transmitted to CA1. The role of CA1 in pattern generation/modification, however, is less well understood.
The current study investigates how CA3 and CA1 process modified contextual information by comparing, within an animal, the CA3 and CA1 neuronal ensembles activated by two sequential behavioral experiences. We used a novel cellular imaging method, Arc/Homer 1a catFISH [cellular analysis of temporal activity by fluorescence in situ hybridization (Vazdarjanova et al., 2002)], that provides a histological readout of the activity history of hippocampal and neocortical networks. This method capitalizes on the different structure of two immediate-early genes (IEGs), Arc and Homer 1a (H1a), and the precise onset and shutoff of synaptic activity-driven IEG transcription in neurons. Whereas Arc mRNA is derived from a short primary transcript (∼3.5 kb), H1a mRNA is generated from a longer and more complex primary transcript (∼45 kb) (Bottai et al., 2002). Thus, when brain sections are double labeled with a H1a 3′-untranslated region (UTR)-specific riboprobe and a full-length Arc riboprobe, one can separate neurons activated ∼2-15 min before sacrifice (cells containing Arc transcription foci) and neurons activated ∼25-40 min earlier (cells containing H1a transcription foci) (Vazdarjanova et al., 2002). By quantifying Arc/H1a cell staining, we determined the responses of CA3 and CA1 neuronal ensembles to several modifications of an environment. We report that the CA3 population response is distinct from that of CA1 in two important ways: (1) CA3 produces a discontinuous response to increasing modifications of an environment, whereas CA1 generates a graded response; and (2) the CA3 ensemble representation of an environment is sparser than that of CA1. Furthermore, we find that the ratio of CA3 to CA1 activity is highly correlated across sessions within an animal regardless of behavioral condition, suggesting that the balance of CA3-CA1 network activity is a property of the individual animal. These data provide evidence that CA3 and CA1 neuronal ensembles perform distinct, yet complementary, functions in the processing of contextual and spatial information.
Materials and Methods
Subjects and behavioral procedures. Twenty-four 3-month-old male Sprague Dawley rats (275-300 gm on arrival; Harlan Sprague Dawley, Indianapolis, IN) were used. The rats were handled daily for 1 week before the experiment to habituate them to the general handling procedure. To ensure active exploration of a novel environment, 1 d before the experiment, the rats were allowed to explore for 6 min a white plastic box (35 × 45 cm; 13 cm high) situated in a room different from the two rooms used during the experiment. On the day of the experiment, each rat was exposed to novel environment A for 6 min (epoch 1), returned to its home cage in the colony room for 20 min, and then exposed for 6 more min (epoch 2) to either environment A (n = 4), environment A modified in one of three ways (Aobj, Aconf, and Ab; n = 4 for each group), or a completely different environment (B; n = 4) (Fig. 1a). The environments were cleaned thoroughly with 20% ethyl alcohol between subjects (for environments A, Aobj, Aconf, and Ab), or dilute acetic acid solution (for environment B). Environment A was an open box (61 × 61 cm) with 24 cm high walls, one of which contained a white cue card. The box had a Plexiglas floor divided into nine squares with four identical balls (3 cm in diameter) fixed in place (Fig. 1b). Environment Aobj was environment A in which the local cues (balls) were substituted with other local cues (cubes with 4 cm sides). In environment Aconf, the local cues (balls) were moved to a different configuration. Environment Ab was environment A moved to a different room with different distal cues. Environment B was a cylinder (75 cm in diameter) located in the same room as environment Ab (Fig. 1b). Exploration sessions were videotaped and later analyzed for the number of zone transitions, object approaches, and rearings of each rat. A “zone transition” was scored when the rear legs of a rat crossed the line separating any two-grid squares (Fig. 1b). An “object approach” was scored when a rat approached an object such that its nose either touched or was within ∼2 cm of the object. A “rearing” was scored when a rat stood on its hind legs. All behavioral testing was conducted during the dark half of the light/dark daily cycle, which is the active period for the rats. Thus, rats from the caged control group were not in a deep sleep but were quietly awake before they were killed.
Figure 1.
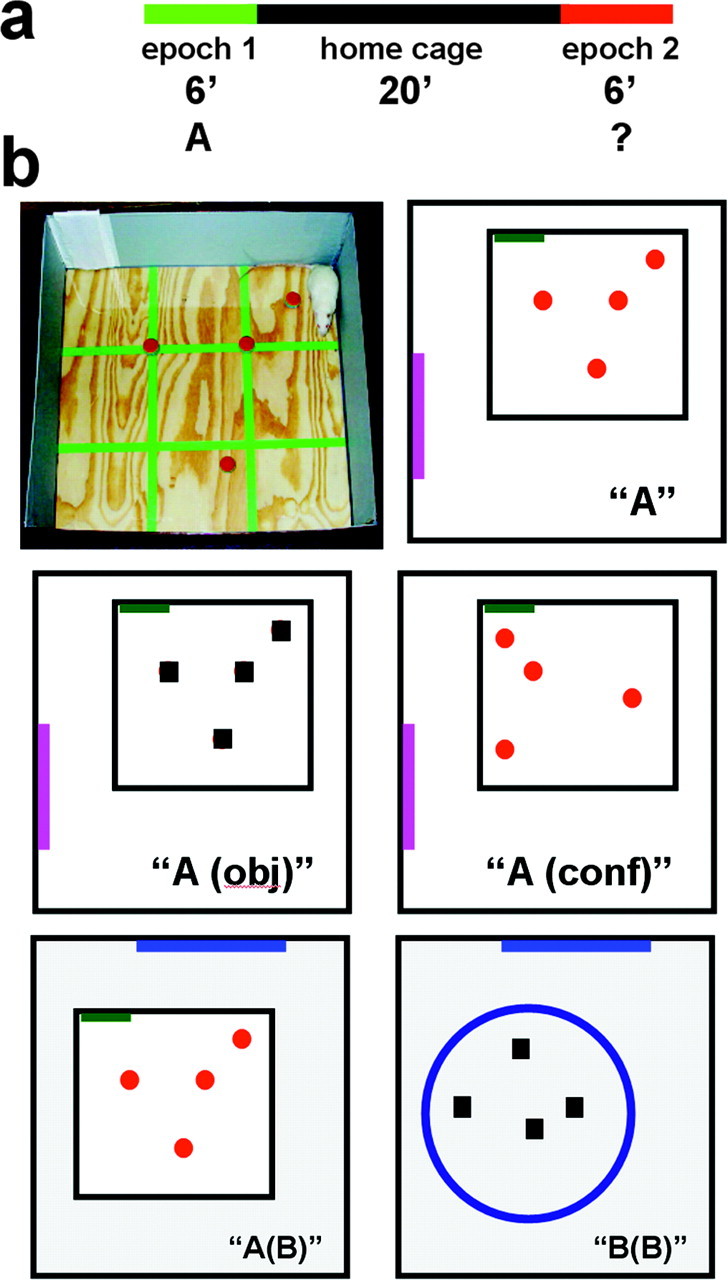
Schematic representation of the time course (a) and types of novel environments (b) used in the experiment. Animals were allowed to freely explore the novel environment A (b, top row) for 6 min (epoch 1). After a 20 min rest period in the home cage, the rats were exposed to either environment A again or to one of the other four environments schematically represented in b (epoch 2). Three of these environments were modified A environments: in Aobj, the local cues were changed from balls to cubes; in Aconf, the same local cues (balls) were rearranged in a different cue configuration; and in Ab, the entire local environment was moved into a different room with different distal cues. Environment B was in the second room and was as different as possible from environment A, yet contained the same number of local cues to ensure a similar level of complexity.
At the end of epoch 2, the rats were killed by decapitation, and the brains were removed rapidly and flash frozen in isopentane (at ∼-50°C). Additionally, four rats were killed directly from the home cage, without any behavioral handling, at various times during the experiment to establish baseline levels of IEG expression. The frozen brains were then grouped, using optimal cutting temperature medium (VWR Scientific, West Chester, PA) in a block such that one brain from each behavioral group was present in a block. The resulting four blocks were cryosectioned to obtain 20-μm-thick coronal sections, which were collected on Superfrost Plus slides (VWR Scientific).
Fluorescence in situ hybridization. Double-label fluorescence in situ hybridization (FISH) for the IEGs Arc and Homer 1a (Arc/H1a catFISH) was performed as described in detail previously (Guzowski et al., 1999; Vazdarjanova et al., 2002). Briefly, Arc and H1a hapten-labeled antisense riboprobes were hybridized together with the tissue overnight. Then, the digoxigenin-labeled Arc riboprobe was detected with anti-digoxigenin-HRP conjugate (Roche Applied Science, Indianapolis, IN) and revealed with a cyanine-3 substrate kit (CY3 Direct FISH; PerkinElmer Life Sciences, Emeryville, CA). Subsequently, the slides were treated with 2% H2O2 to quench any residual HRP activity, and the fluorescein-labeled probe targeting the 3′-UTR of H1a was detected with anti-fluorescein-HRP conjugate (Roche Applied Science) and a cyanine-5 substrate kit (CY5 Direct FISH; PerkinElmer Life Sciences). Nuclei were counterstained with either YOYO-1 or Sytox-Green (Molecular Probes, Eugene, OR).
Image acquisition and analysis. Images were acquired on a Leica (Nussloch, Germany) TCS-4D confocal microscope equipped with a krypton-argon laser. The pinhole, photomultiplier tube (PMT) gain and contrast settings were kept constant for all image stacks acquired from a slide. The settings were optimized for detecting only strong intranuclear signals and for minimizing weaker cytoplasmic signals by adjusting the laser power settings and PMT gain accordingly (Vazdarjanova et al., 2002) (see Fig. 3a). Confocal z-stacks composed of 1-μm-thick optical sections were collected in regions CA3 and CA1 of the dorsal hippocampus (anteroposterior, ∼-3.6 mm from bregma) from three slides/animal and stored for off-line analysis. Stacks for CA3 (one stack per slide) were collected with a 20× objective, whereas stacks for CA1 (two stacks/slide) were collected with a 40× oil objective. Thus, comparable numbers of cells per region were collected for each rat.
Figure 3.
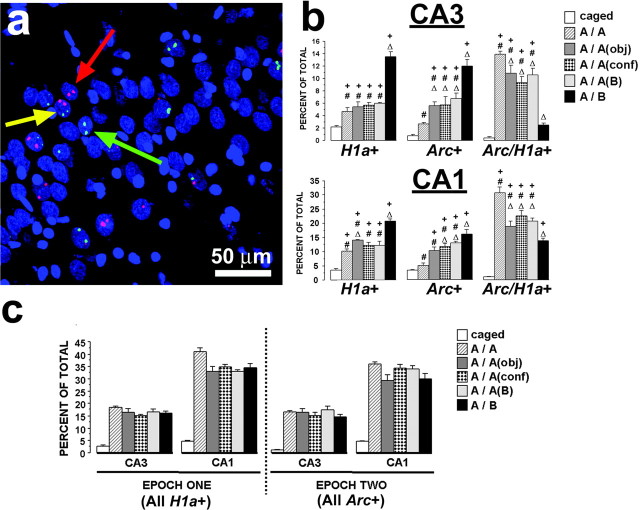
Summary of the Arc/H1a catFISH data. a, A confocal projection image (of 20 1 μm optical slices compressed into a single plane) from CA3 showing the four possible cell staining profiles (nuclei are shown in blue): the green arrow points to a H1a+ neuron, which has only H1a transcription foci (green) and thus was activated only during epoch 1; the red arrow points to an Arc+ neuron, which has only Arc transcription foci (red color) and thus was activated only during epoch 2; and the yellow arrow points to a Arc/H1a+ neuron that contains both Arc and H1a transcription foci and thus was activated during both epoch 1 and epoch 2. Neurons that had no foci were classified as negative. Scale bar, 50 μm. b, Staining profiles of CA3 and CA1 neuronal ensembles of the different behavioral groups expressed as percentages of total counted neurons. In both CA3 and CA1, the A/A group had the lowest percentage of H1a+ and Arc+ neurons and the highest percentage of Arc/H1a+, indicating the highest level of overlap of neuronal ensembles activated by epoch 1 and epoch 2. Conversely, the A/B group had the lowest level of overlap of activated neuronal ensembles. All other behavioral groups had an intermediate, but mutually equal, degree of overlap. +p < 0.05 relative to caged controls; #p < 0.05 relative to group A/B; Δ, p < 0.05 relative to group A/A. c, Size of the CA3 and CA1 neuronal ensembles active during epochs 1 and 2, expressed as the percentage of total counted neurons. The activated CA1 neuronal ensembles in all behavioral groups were significantly greater than those of CA3, and both CA3 and CA1 ensembles of the exploration groups were dramatically larger than those of caged controls.
Non-neuronal cells, identified as small cells (∼5 μm in diameter) with intensely bright and uniformly stained nuclei, were excluded from the analysis. We use the term neuronal nuclei to describe the large, diffusely stained nuclei present in the sections, and only these nuclei were included in the analysis. Our recent work has shown that cells containing these large nuclei in the hippocampus stain either with α-CaMKII (α calcium/calmodulin-dependent protein kinase II) or GAD65/67 in double-label FISH experiments, indicating pyramidal neurons or interneurons, respectively (Vazdarjanova et al., 2003). In addition, we included only those neuronal nuclei that were present in the median 20% of the stack in our analyses. This optical dissector technique minimizes sampling errors attributable to partial cells and stereological concerns because variations in cell volumes do not influence sampling frequencies (West, 1993). Neuronal nuclei from the pyramidal cell body layer of CA3 and CA1 were classified as negative (containing no transcription foci), H1a+ (containing only H1a transcription foci), Arc+ (containing only Arc transcription foci), or Arc/H1a+ (containing transcription foci for both Arc and H1a) by an experimenter blind to the relationship between the image stacks and the behavioral conditions they represented. The neurons in the latter three classes were most likely pyramidal neurons and not interneurons, because we have shown that exploration of a novel environment, similar to environment A in this study, induces Arc expression exclusively in α-CaMKII-positive cells and not GAD65/67-positive cells in the CA3 and CA1 (Vazdarjanova et al., 2003).
Calculation of similarity scores. In Arc/H1a catFISH imaging experiments involving two behavioral experiences (epochs), it is helpful to reduce the complex cell staining data to a value that can be used to compare cell activity patterns across multiple brain regions. The similarity score takes the four measured cell-staining values (negative, Arc+, H1a+, and Arc/H1a+; see above) and reduces them to a single value. With this method, a value of 1 represents a single neuronal population faithfully activated in both epochs. A value of 0 indicates that two statistically independent cell populations were activated during the two epochs.
The similarity score is derived as follows: (1) Epoch 1 active cells = fraction of total H1a-positive cells [(H1a+ plus Arc/H1a+)/total cells]. (2) Epoch 2 active cells = fraction of total Arc positive cells [(Arc+ plus Arc/H1a+)/total cells]. (3) p(E1E2) = epoch 1 active cell fraction × epoch 2 active cell fraction. This is the probability of cells being active in both epochs (Arc/H1a+ cells), assuming the two epochs activated statistically independent neuronal ensembles. (4) diff(E1E2) = (Arc/H1a+) - p(E1E2). This is a measure of deviation from the independence hypothesis. (5) Least epoch = the smaller of the ensembles activated by epoch 1 or epoch 2. (6) Similarity score = diff(E1E2)/(least epoch - p(E1E2). This normalizes the diff(E1E2) fraction to a perfect A/A condition. A perfect A/A is 1, and a perfect A/B is 0.
Statistical analyses. Group differences for the behavioral measures, as well as similarity scores, were analyzed with ANOVA, followed by Scheffe's post hoc tests. Differences within the same group across the two behavioral epochs were compared with paired t tests. For testing whether independent cell ensembles were activated during epochs 1 and 2, a two-tailed one-group t test (with a hypothesized mean of 0) was applied to the similarity score data (see Fig. 4). For all tests, the null hypotheses were rejected at the 0.05 level of significance.
Figure 4.
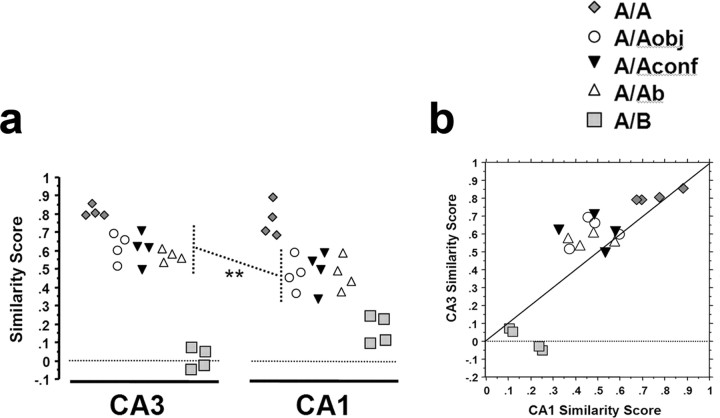
Analysis of similarity scores of rats from the different behavioral groups. A similarity score of 1 indicates a complete overlap of the neuronal ensembles activated by epoch 1 and epoch 2 (no remapping), whereas a similarity score of 0 indicates no overlap beyond that predicted by chance (complete remapping; see Materials and Methods). a, In both CA3 and CA1, the similarity scores from rats of the A/Aobj, A/Aconf, and A/Ab groups were significantly different from the A/A and A/B groups (p < 0.02) but were not different from each other (p > 0.82). Thus, in both CA3 and CA1, all alterations to environment A (here termed A′ and comprising groups A/Aobj, A/Aconf, and A/Ab) produced a similar effect on the neural ensemble response in that cell population. Notably, the similarity score of the A/A′ rats was greater in CA3 compared with CA1 (**p = 0.001). b, Correlation between the CA3 and CA1 similarity scores for each rat. Note that the CA3 similarity scores for the A/A′ rats tended to be higher than those for CA1, as indicated by the majority of points being above the 45 ° line. Furthermore, the CA3 similarity scores of the A/B group were lower than those of CA1 in the same rats (all points of the A/B group are below the 45 ° line). These findings, as visualized in a and b, indicate that CA3 separates well dissimilar representations (A vs B), but when the threshold for remapping is not met (as in the A/A and A/A′ groups), then the initiated pattern completion will de-emphasize mild perturbations of the environment. In CA1, in contrast, differences in the environment are represented more continuously (see Results and Discussion for details).
Results
To examine how discrete changes to an environmental context influence the ensemble activation patterns of CA3 and CA1 neuronal networks, 3-month-old male rats were exposed sequentially to two similar, but modified, environments, separated by a rest interval of 20 min (Fig. 1) (see Materials and Methods for additional details). As controls, separate groups of rats (n = 4 per group) were exposed sequentially to the same environment twice, or to two entirely different environments. The environments were defined by both local (“intramaze”) and distal (“extramaze”) cues. In addition, four rats were killed directly from the home cage, at different times throughout the course of the experiment, without any new behavioral experience, to determine the basal levels of Arc and Homer 1a (H1a) expression. The cell populations active during each distinct experience were detected using Arc/H1a catFISH (Vazdarjanova et al., 2002). As an operational definition, we use the term “activity” to refer to the proportion of neurons containing either Arc or H1a for a given behavioral epoch. Although the relationship between neural activity to IEG transcriptional activity is not precisely defined at present, there is a strong correspondence between “place cell” activity and Arc and H1a transcriptional activity under the behavioral conditions used here (Wilson and McNaughton, 1993; Guzowski et al., 2001; Vazdarjanova et al., 2002), thus making this a reasonable first-order approximation.
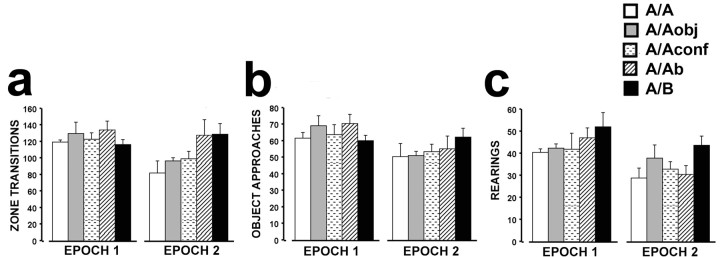
Understanding how features of an environment influence ensemble responses of CA3 and CA1 neurons requires that the experimental subjects explored all aspects of the environment. Figure 2 shows the mean number of zone transitions, object approaches, and rearings for rats from each group for each behavioral experience (epoch). The overall exploration of the intramaze cues (Fig. 2a,b), as well as extramaze cues (Fig. 2c), was high during both epochs for all groups. However, there was a slight decrease in exploratory activity during epoch 2 when all groups, which were not statistically different from each other within an epoch, were collapsed into one group (paired t tests; zone transitions: mean difference (diff.) = 17.60, p = 0.01; object approaches: mean diff. = 10.55, p < 0.01; rearings: mean diff. = 10.05, p < 0.001). Such differences within individual groups were less obvious, most likely because of the lack of statistical power resulting from the small number of rats in each group (n = 4 per group). In contrast to the trend of decreased exploratory activity during epoch 2 for rats of the A/A, A/Aobj, A/Aconf, or A/Ab groups, rats in the A/B group did not exhibit decreased exploration during epoch 2.
Figure 2.
Summary of the behavioral data per epoch. For all epochs, there were no significant differences in the measures of exploratory behavior between groups as revealed by overall ANOVAs. Consequently, behavioral measures within an epoch were collapsed across groups for subsequent analyses. Although all rats explored actively the novel environments during both epoch 1 and epoch 2, as evidenced by the high number of zone transitions, object approaches and number of rearings, during epoch 2 there was a small, but statistically significant, decrease in exploratory behavior evident in all three behavioral measures when all groups were collapsed (see Results).
Figure 3a shows the different cell staining profiles observed using Arc/Homer 1a catFISH. As characterized previously in Arc/H1a catFISH (Vazdarjanova et al., 2002), nuclei containing H1a transcription foci (Fig. 3a, green) indicate neurons active ∼30 min before the animal's death. Nuclei with Arc transcription foci (Fig. 3a, red) indicate neurons active within ∼10 min of the animal's death. Nuclei containing both H1a and Arc foci were active both ∼30 and ∼10 min before the animal's death. Thus, neurons active only during epoch 1 contain only H1a foci (Fig. 3b, H1a+), neurons active only during epoch 2 contain only Arc foci (Fig. 3b, Arc+), and neurons active during both epochs 1 and 2 have both H1a and Arc foci (Fig. 3b, Arc/H1a+). The mean number of neurons analyzed per rat was 242.3 ± 6.7 SEM (range, 154-300) for CA3 and 259.5 ± 4.1 SEM (range, 225-301) for CA1.
As illustrated in Figure 3c, the percentage of neurons with H1a foci was very low in rats from the caged control group (∼5%), whereas exploring the novel environment A during epoch 1 resulted in a dramatic increase of the proportion of both CA3 and CA1 neurons with H1a foci (ANOVA for CA3: F(5,18) = 33.94, p < 0.0001; for CA1: F(5,18) = 83.06, p < 0.0001; all post hoc comparisons of caged to any of the exploration groups had p < 0.0001). Similarly, exploring a novel environment during epoch 2 resulted in a substantial increase in the proportion of CA3 and CA1 neurons with Arc foci compared with caged controls (ANOVA for CA3: F(5,18) = 25.35, p < 0.0001; for CA1: F(5,18) = 50.49, p < 0.0001; all post hoc comparisons of caged to any of the exploration groups had p < 0.0001). Furthermore, the percentage of neurons active during epoch 2 was comparable with that active during epoch 1 for both CA3 and CA1 for all exploration groups (ANOVA for CA3: F(4,15) = 0.76, p = 0.57; for CA1: F(4,15) = 2.52, p = 0.09). Thus, levels of neural activity within a given subfield did not change across epochs in either CA3 or CA1. This finding contrasts the slightly decreased behavioral activity from epoch 1 to epoch 2 and suggests that Arc/H1a expression does not simply reflect behavioral activity.
As noted above, exploring an environment encountered once before (A/A group), an environment not encountered but similar to that encountered during epoch 1 (A/Aobj, A/Aconf, and A/Ab groups), or a completely novel environment (A/B group) activated similar proportions of neurons compared with epoch 1, for both CA3 and CA1 (Fig. 3c). However, differences in the ensemble representations were evident between the different behavioral groups (Fig. 3b). These ensemble differences for epochs 1 and 2 were characterized by varying degrees of overlap. Among the exploration groups, the rats from the A/A group had the highest degree of overlap of ensembles active during epoch 1 and epoch 2, as evidenced by the highest percentage Arc/H1a+ neurons and lowest percentage of H1a+ and Arc+ neurons. The rats from the A/B group had the lowest overlap, as evidenced by the lowest percentage of Arc/H1a+ neurons and highest percentage of H1a+ and Arc+ neurons. The rats from the remaining groups had intermediate degrees of overlap. These results are expressed in summary in Figure 4 as similarity scores (see Materials and Methods). Note that catFISH can only detect whether cells were active in an epoch or not, unlike electrophysiological recording methods that can differentiate cells that were active in two epochs, but which changed their spatial pattern of activity. Thus, catFISH similarity scores may slightly overestimate the actual ensemble similarity, if alterations of firing fields occur across sessions.
In both CA3 and CA1, exploring the same environment during both epoch 1 and epoch 2 activated primarily the same neuronal ensemble, as evidenced by a similarity score close to 1 for the A/A group (mean ± SEM: CA3, 0.81 ± 0.03; CA1, 0.76 ± 0.10). Exploring two very different environments during epoch 1 and epoch 2 (A/B group), in contrast, activated two statistically independent CA3 neuronal ensembles, as evidenced by a similarity score close to 0 (mean ± SEM, 0.02 ± 0.06; p = 0.61). Like CA3, the similarity score for CA1 of the A/B group was dramatically lower than that of the A/A group (mean ± SEM, 0.18 ± 0.08), although this value was significantly >0 (p = 0.02). Interestingly, although the similarity scores of the A/Aobj, A/Aconf, and A/Ab groups were different from both the A/A and A/B groups (ANOVA for CA3: F(4,15) = 91.95, p < 0.0001; for CA1: F(4,15) = 19.75, p < 0.0001; all appropriate post hoc comparisons had p < 0.02), they were not different from each other (p > 0.82 for all three comparisons). Thus, changing the type or configuration of intramaze cues, as well as changing the extramaze cues, of environment A activated CA3 and CA1 neuronal ensembles that only partially overlapped with the respective neuronal ensembles activated by the original environment A. Furthermore, any of these perturbations of environment A resulted in a similar degree of overlap with the neuronal ensembles initially activated by environment A.
Despite the similarities in staining profiles and overlap ratios (similarity scores) observed for CA3 and CA1, there were three notable differences. First, the proportion of active cells comprising the CA3 ensemble was significantly less than that of CA1 for any of the exploration groups both for epoch 1 (ANOVA range: F(1,6) = 38.35-240.94; p < 0.001 in all cases) and epoch 2 (ANOVA range: F(1,6) = 19.76-238.85; p < 0.005 in all cases) (Fig. 3b,c). Second, the similarity scores of the A/Aobj, A/Aconf, and A/Ab groups combined (collectively termed A/A′) were significantly greater for CA3 than those for CA1 (ANOVA: F(1,22) = 14.16; p = 0.001) (Fig. 4a). Thus, the partially modified environments were treated as more similar to the original environment in CA3 than in CA1, where differences between the environments were emphasized. This point is visualized on Figure 4b, where for the majority of the rats in the A/A and A/A′ groups, the CA3 similarity scores were higher than the CA1 similarity scores, as evidenced by the points above the 45° line. Third, the similarity score of the A/B group was significantly lower in CA3 compared with CA1 (F(1,6) = 11.00; p = 0.016), suggesting that the representation of environment B was more orthogonal in CA3 as compared with CA1.
We investigated the possible interaction between CA3 and CA1 by examining whether overall cell activity levels in these regions were correlated within a behavioral epoch. Such a finding would be consistent with a unitary “hippocampal activation” involving both CA3 and CA1. The cells active during epoch 1 were defined as the total percentage of H1a+ neurons, regardless of Arc staining. Conversely, the cells active during epoch 2 were defined as the total percentage of Arc+ cells, regardless of H1a staining. For both epoch 1 (Fig. 5a) and epoch 2 (Fig. 5b), CA3 and CA1 activity levels were not correlated (epoch 1: r2 = 0.04, p = 0.43; epoch 2: r2 = 0.01, p = 0.66), arguing against a simple coactivation of these regions during behavior. In contrast, activity levels within a rat for both CA3 (Fig. 5c) and CA1 (Fig. 5d) were modestly correlated across behavioral epochs (CA3: r2 = 0.46, p < 0.001; CA1: r2 = 0.41, p < 0.002), indicating that for each rat, a similar proportion of CA3 or CA1 cells were active during two closely spaced exploration sessions.
Figure 5.
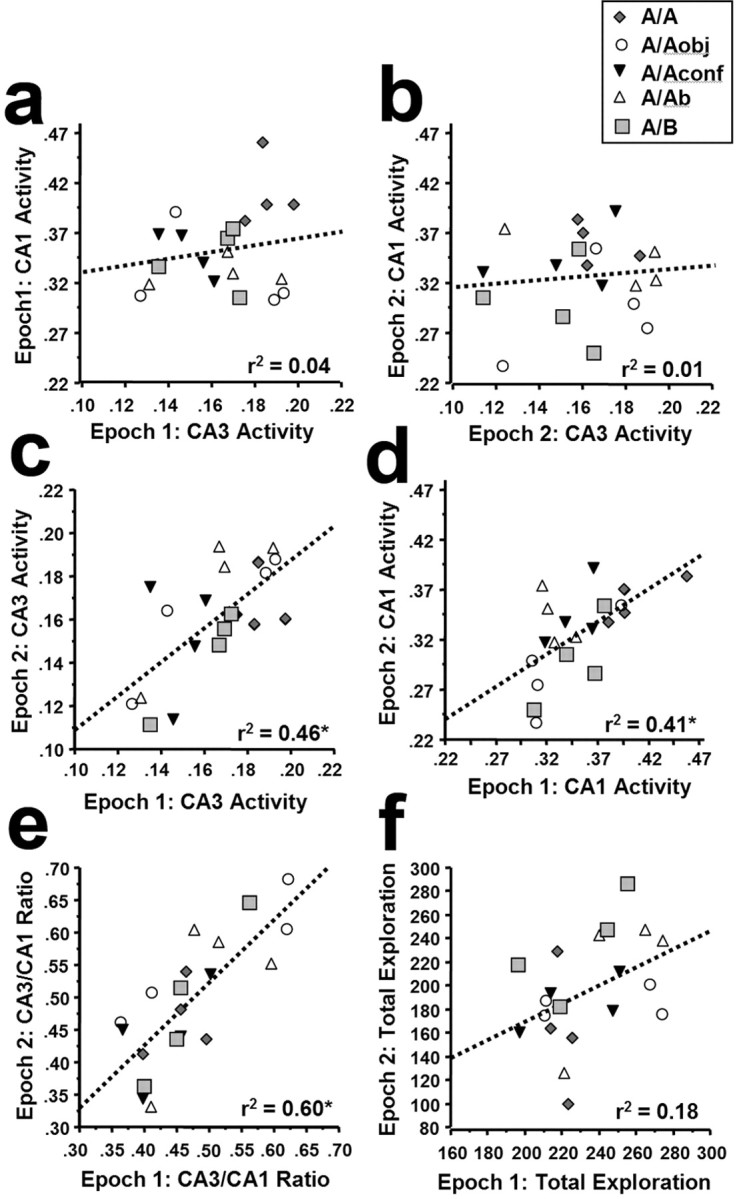
Within-animal correlations of neuronal ensemble activity between regions (CA3 and CA1) and across behavioral epochs. Epoch 1 ensemble activity is defined as the fraction of H1a+ cells (H1a+ plus Arc/H1a+) of the total counted cells, and epoch 2 ensemble activity is defined as the fraction of Arc+ cells (Arc+ plus Arc/H1a+) of the total counted cells (see Materials and Methods and Results for additional details). Ensemble activity levels between CA3 and CA1 were not correlated for either epoch 1 (a) or epoch 2 (b). This finding indicates that there is not a simple coactivation of these cell populations during exploratory behavior. In contrast, levels of activity within both CA3 (c) and CA1 (d) were modestly correlated across behavioral epochs. Furthermore, the within-rat ratio of CA3 to CA1 activity was strongly correlated across behavioral epochs (e). The fact that the CA3 and CA1 activity per epoch was not correlated across rats (a, b), but that the CA3/CA1 within-rat ratio was correlated across sessions, suggests that the balance of CA3 to CA1 activity is highly specific to the individual rat. The CA3/CA1 ratio was not related to the nature of the experience, because all groups were evenly distributed along the regression line (e), nor was it related to levels of general exploratory behavior, because the total exploration measures across epochs 1 and 2 were not significantly correlated (f). See Results for additional details.
We next examined whether the ratio of CA3/CA1 activity was constant across behavioral epochs within a rat. This value was generated by dividing the proportion of CA3 cells active per epoch by the proportion of CA1 cells active within that same epoch. Thus, a higher number indicates a greater bias toward CA3 activity, and a lower value indicates a bias toward CA1 activity. The CA3/CA1 ratio within a rat was highly correlated across epochs (Fig. 5e) (r2 = 0.60; p < 0.0001; slope = 0.97). This correlation was not simply related to exploratory activity because: (1) exploratory activity was not correlated within an animal across epochs (Fig. 5f) (r2 = 0.18; p > 0.05); (2) the mean, SD, and range of total exploratory activity varied considerably from epoch 1 to epoch 2 (Fig. 5f) (epoch 1: range, 196-275 counts; mean ± SD, 233.8 ± 25.0; epoch 2: range, 98-287 counts; mean ± SD, 195.6 ± 45.6), whereas the CA3/CA1 ratios did not (Fig. 5e) (epoch 1: range, 0.36-0.62; mean ± SD, 0.47 ± 0.08; epoch 2: range, 0.33-0.68; mean ± SD, 0.49 ± 0.10); and (3) the distribution of CA3/CA1 ratios for the different groups did not cluster in any way for epoch 2 (Fig. 5e), despite the fact that the input was widely different and detected in the ensemble responses of both CA3 and CA1. Furthermore, additional analyses (data not shown) indicate that none of the gene activity or behavioral measures was correlated with time of day. Thus, the relative balance of CA3/CA1 activity was rat specific, although the factors that govern this balance are not clear at present.
Discussion
This study was performed to characterize how defined changes to an environment modify neural ensemble representations within CA3 and CA1 regions of the hippocampus. An ensemble was defined as those neurons expressing either Arc or H1a transcription foci, determined using Arc/H1a catFISH. Thus defined, ensembles were specific for one of the two context exposures and revealed differences both within and between CA3 and CA1 population responses within an animal. As detailed below, the current findings provide compelling evidence that CA3 and CA1 ensembles perform distinct, yet complementary, functions in the processing of spatial and contextual information.
Before proceeding, it is important to emphasize that both Arc and H1a are components of the postsynaptic density and can play prominent roles in enabling lasting synaptic modification. Arc (Lyford et al., 1995), also identified as Arg3.1 (Link et al., 1995), is a component of NMDA receptor complexes (Husi et al., 2000) and can be targeted to active dendrites (Steward et al., 1998). Inhibiting Arc protein expression in the hippocampus disrupts long-term potentiation and long-term spatial memory (Guzowski et al., 2000). H1a is a synaptic activity-regulated isoform of the Homer 1 gene, which can act as a dominant-negative regulator of constitutively expressed Homer 1 isoforms (Homer 1b/c) in coupling membrane events with intracellular calcium release (Xiao et al., 2000; Yuan et al., 2003). Thus, these genes, induced in hippocampal networks after learning about an environmental context, may play a critical role in stabilizing these networks during memory consolidation for retrieval of such information at a later time.
CA3 and CA1 ensemble activity is controlled by local cues, local cue configurations, and distal cues
When rats explored the same environment twice (group A/A), the activated CA3 and CA1 ensembles were mostly overlapping (Figs. 3b, 4). Such stability of the population response in a static environment is consistent with a previous report using an earlier catFISH method (Guzowski et al., 1999) and in electrophysiological studies of CA3 (Tanila, 1999) and CA1 (Barnes et al., 1997; Skaggs and McNaughton, 1998) place cells. In contrast, rats from the A/B group showed little overlap of CA3 and CA1 neuronal ensembles active in different environments (Figs. 3b, 4). Changing the type (group A/Aobj) or the configuration (group A/Aconf) of the local cues, as well as changing the distal cues (group A/Ab), resulted in an intermediate degree of overlap in both CA3 and CA1. In both regions, this intermediate overlap was indistinguishable between the three groups, suggesting that in both CA3 and CA1, the configuration and type of local cues, as well as the distal cues, are equally capable of inducing “partial remapping” of the ensemble representation. This partial remapping was not induced by differences in exploratory activity because the latter was similar between all groups (Fig. 2).
These catFISH findings are consistent with electrophysiological studies demonstrating changes in the hippocampal representation of environments (place cell firing) in which the local and/or distal cues were either rotated (Shapiro et al., 1997; Tanila et al., 1997a; Knierim, 2002) or changed (Tanila et al., 1997b). Similarly, the geometric arrangement of local cues strongly influences place field positions provided they are not positioned near the center of the environment (Cressant et al., 1999; Muller et al., 1999). In other studies, hippocampal neurons have been shown to fire in response not just to a specific location in an environment, but also to auditory, visual, and olfactory cues when these cues are discriminative stimuli in discrimination tasks (Wible et al., 1986; Eichenbaum et al., 1987; Sakurai, 1990; Young et al., 1994; Wood et al., 1999). Furthermore, a subpopulation of CA1 neurons fires in response to landmarks/goal, regardless of the latter's location in the environment (Gothard et al., 1996a,b). Such recording data, and the catFISH data presented here, are consistent with the memory indexing theory of hippocampal function, which states, “the role of the hippocampus is to form and retain an index of neocortical areas activated by experimental events” (Teyler and DiScenna, 1986). Thus, whenever new cues or cue configurations activate new neocortical modules, the index code in CA3 and CA1 is modified to incorporate these changes in neocortical activation.
Altering the environmental context produces discontinuous CA3, and graded CA1, ensemble responses
Despite the similarities in CA3 and CA1 responses noted above, the current results demonstrate, for the first time, differences in CA3 and CA1 ensemble responses to modifications of an environment. First, the proportion of CA3 neurons activated during a given epoch was less than that of CA1. This relative sparsity of encoding in CA3 is consistent with the hypothesis that CA3, with its recurrent collaterals, is part of an associative memory network capable of retrieving entire memory patterns from partial or degraded inputs via pattern completion (Marr, 1971; McNaughton and Morris, 1987). By this hypothesis, which is corroborated by findings from electrophysiological (McNaughton et al., 1989; Barnes et al., 1990), lesion (Brun et al., 2002), and genetic (Nakazawa et al., 2003) studies, the sparse encoding in CA3 is necessary for storing/retrieving a large number of representations (Marr, 1971; McNaughton and Morris, 1987).
The Marr hypothesis can also explain the second difference observed in the CA3 and CA1 ensemble responses: CA3 had a higher degree of overlap, compared with CA1, of activated neuronal ensembles in the A/A′ groups. Thus, when rats explore environment Aobj, Aconf, or A/Ab, they encounter familiar features of the recently experienced environment A and the CA3 system may be attempting to retrieve the representation of environment A (pattern completion). This system, however, also processes information about features of the environment that arrives through direct cortical inputs to CA3 (Yeckel and Berger, 1990; Witter et al., 2000; Cooper and Mizumori, 2001). Integrating the two kinds of input (retrieval of the closest matching representation via the recurrent collaterals and information on various features of the environment arriving through the perforant path) results in a representation that is very similar, yet only partially overlapping with that of environment A. This modest partial remapping is conveyed to CA1, where it is further integrated with information from the perforant path arriving directly to CA1 (Yeckel and Berger, 1990; Witter et al., 2000; Muir and Bilkey, 2001). This additional processing may emphasize the differences with the representation of the original environment A, resulting in a decreased overlap of activated CA1 neuronal ensembles compared with CA3 (pattern separation). A recent electrophysiological study reported similar findings of CA3 pattern completion and CA1 pattern separation to environmental modifications (Lee et al., 2004).
Another notable difference between CA3 and CA1 ensemble responses was seen in the A/B group: the ensemble representations of two different environments were more orthogonal in CA3 as compared with CA1. This finding is consistent with a recent study showing that the proportion of CA1 neurons with related place fields in two distinct environments was above chance, whereas that of CA3 neurons was not (Leutgeb et al., 2003). A possible explanation for these findings is that pattern completion in CA3 increases the threshold for remapping compared with CA1. This explanation can also account for the modest remapping in CA3 of partial changes in an environment (A/A′ conditions) and the more dramatic remapping in the A/B condition. If the partial environment modifications were not sufficiently strong, the threshold for remapping in CA3 was not reached and the resulting representations were very similar to those of the A/A condition. When the environment encountered during epoch 2 was sufficiently different from that encountered during epoch 1 (A/B), the threshold was met, and the CA3 representations were completely orthogonal. Thus, CA3 has a discontinuous, whereas CA1 has a graded, response to changes in the environment. An alternative explanation is that the geometry differences of environments A and B influence CA3 neurons more strongly than they do CA1 neurons. Previous studies have shown geometry differences of an environment have a major influence on place cell responses (Muller and Kubie, 1987; Sharp, 1997; Wilson et al., 2003).
Conclusion
Here, we used Arc and H1a as sensitive “genomic timers” of neural activity history to address how contextual and spatial information is processed by CA3 and CA1 neuronal ensembles. Our findings provide insight into the cellular basis of two competing functions of the hippocampus, pattern completion and pattern separation (O'Reilly and Rudy, 2000). When confronted with two different environments (A/B), CA3 produced orthogonal representations for each environment (pattern separation). However, when input was sufficiently similar (A/A and A/A′ groups), pattern completion in CA3 de-emphasized the mild perturbations of the environment. These two functions produced the discontinuous distribution of similarity scores (Fig. 4). In contrast, the varying environmental modifications were represented continuously by CA1 ensembles. Moreover, the relative weighting of CA3/CA1 activity was variable, but stable, within an individual across epochs (Fig. 5e). Thus, integration of the distinct CA3 and CA1 ensemble representations may provide a highly refined representation of context. Future studies combining catFISH with systems and behavioral neuroscience approaches will help elucidate the network interactions involved in learning and memory.
Footnotes
This work was supported by National Institutes of Health Grants 1 R01 MH60123 (J.F.G.) and 1 F32 MH064357 (A.V.). We thank Drs. Thane Plummer, Teiko Miyashita, Kate Jeffery, and Carol Barnes and the two reviewers for helpful comments on previous versions of this manuscript. We also thank Dr. Gail Lewandowski for database design, implementation, and management, and Beth Takacs and Shannon Salamone for technical assistance.
Correspondence should be addressed to Dr. John F. Guzowski, Department of Neurosciences, Basic Medical Sciences Building, Room 145, University of New Mexico Health Sciences Center, Albuquerque, NM 87131-5223. E-mail: jguzowski@salud.unm.edu
Copyright © 2004 Society for Neuroscience 0270-6474/04/246489-08$15.00/0
References
- Barnes CA, McNaughton BL, Mizumori SJ, Leonard BW, Lin LH (1990) Comparison of spatial and temporal characteristics of neuronal activity in sequential stages of hippocampal processing. Prog Brain Res 83: 287-300. [DOI] [PubMed] [Google Scholar]
- Barnes CA, Suster MS, Shen J, McNaughton BL (1997) Multistability of cognitive maps in the hippocampus of old rats. Nature 388: 272-275. [DOI] [PubMed] [Google Scholar]
- Bottai D, Guzowski JF, Schwarz MK, Kang SH, Xiao B, Lanahan A, Worley PF, Seeburg PH (2002) Synaptic activity-induced conversion of intronic to exonic sequence in Homer 1 immediate-early gene expression. J Neurosci 22: 167-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brun VH, Otnass MK, Molden S, Steffenach HA, Witter MP, Moser MB, Moser EI (2002) Place cells and place recognition maintained by direct entorhinal-hippocampal circuitry. Science 296: 2243-2246. [DOI] [PubMed] [Google Scholar]
- Cooper BG, Mizumori SJ (2001) Temporary inactivation of the retrosplenial cortex causes a transient reorganization of spatial coding in the hippocampus. J Neurosci 21: 3986-4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cressant A, Muller RU, Poucet B (1999) Further study of the control of place cell firing by intra-apparatus objects. Hippocampus 9: 423-431. [DOI] [PubMed] [Google Scholar]
- Eichenbaum H (2001) The hippocampus and declarative memory: cognitive mechanisms and neural codes. Behav Brain Res 127: 199-207. [DOI] [PubMed] [Google Scholar]
- Eichenbaum H, Kuperstein M, Fagan A, Nagode J (1987) Cue-sampling and goal-approach correlates of hippocampal unit activity in rats performing an odor-discrimination task. J Neurosci 7: 716-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gothard KM, Skaggs WE, Moore KM, McNaughton BL (1996a) Binding of hippocampal CA1 neural activity to multiple reference frames in a landmark-based navigation task. J Neurosci 16: 823-835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gothard KM, Skaggs WE, McNaughton BL (1996b) Dynamics of mismatch correction in the hippocampal ensemble code for space: interaction between path integration and environmental cues. J Neurosci 16: 8027-8040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzowski JF, McNaughton BL, Barnes CA, Worley PF (1999) Environment-specific expression of the immediate-early gene Arc in hippocampal neuronal ensembles. Nat Neurosci 2: 1120-1124. [DOI] [PubMed] [Google Scholar]
- Guzowski JF, Lyford GL, Stevenson GD, Houston FP, McGaugh JL, Worley PF, Barnes CA (2000) Inhibition of activity-dependent arc protein expression in the rat hippocampus impairs the maintenance of long-term potentiation and the consolidation of long-term memory. J Neurosci 20: 3993-4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzowski JF, McNaughton BL, Barnes CA, Worley PF (2001) Imaging neural activity with temporal and cellular resolution using FISH. Curr Opin Neurobiol 11: 579-584. [DOI] [PubMed] [Google Scholar]
- Husi H, Ward MA, Choudhary JS, Blackstock WP, Grant SG (2000) Proteomic analysis of NMDA receptor-adhesion protein signaling complexes. Nat Neurosci 3: 661-669. [DOI] [PubMed] [Google Scholar]
- Knierim JJ (2002) Dynamic interactions between local surface cues, distal landmarks, and intrinsic circuitry in hippocampal place cells. J Neurosci 22: 6254-6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Yoganarasimha D, Rao G, Knierim JJ (2004) Comparison of population coherence of place cells in hippocampal subfields CA1 and CA3. Nature, in press. [DOI] [PubMed]
- Leutgeb S, Kjelstrup KG, Treves A, Moser M, Moser EI (2003) Differential representation of context in hippocampal areas CA3 and CA1. Soc Neurosci Abstr 29: 91.5. [Google Scholar]
- Link W, Konietsko U, Kauselmann G, Krug M, Schwanke B, Frey U, Kuhl D (1995) Somatodendritic expression of an immediate-early gene is regulated by synaptic activity. Proc Natl Acad Sci USA 92: 5734-5738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyford GL, Yamagata K, Kaufmann WE, Barnes CA, Sanders LK, Copeland NG, Gilbert DJ, Jenkins NA, Lanahan AA, Worley PF (1995) Arc, a growth factor and activity-regulated gene, encodes a novel cytoskeleton-associated protein that is enriched in neuronal dendrites. Neuron 14: 433-445. [DOI] [PubMed] [Google Scholar]
- Marr D (1971) Simple memory: a theory for archicortex. Philos Trans R Soc Lond B Biol Sci 262: 23-81. [DOI] [PubMed] [Google Scholar]
- McNaughton BL, Morris RGM (1987) Hippocampal synaptic enhancement and information storage within a distributed memory system. Trends Neurosci 10: 408-415. [Google Scholar]
- McNaughton BL, Nadel L (1990) Hebb-Marr networks and the neurobiological representation of action in space. In: Neuroscience and connectionist theory (Gluck MA, Rumelhart DE, eds), pp 1-64. Hillsdale, NJ: Erlbaum.
- McNaughton BL, Barnes CA, Meltzer J, Sutherland RJ (1989) Hippocampal granule cells are necessary for normal spatial learning but not for spatially-selective pyramidal cell discharge. Exp Brain Res 76: 485-496. [DOI] [PubMed] [Google Scholar]
- Muir GM, Bilkey DK (2001) Instability in the place field location of hippocampal place cells after lesions centered on the perirhinal cortex. J Neurosci 21: 4016-4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller RU, Kubie JL (1987) The effects of changes in the environment on the spatial firing of hippocampal complex-spike cells. J Neurosci 7: 1951-1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller RU, Poucet B, Fenton AA, Cressant A (1999) Is the hippocampus of the rat part of a specialized navigational system? Hippocampus 9: 413-422. [DOI] [PubMed] [Google Scholar]
- Nadel L, Moscovitch M (2001) The hippocampal complex and long-term memory revisited. Trends Cogn Sci 5: 228-230. [DOI] [PubMed] [Google Scholar]
- Nakazawa K, Sun LD, Quirk MC, Rondi-Reig L, Wilson MA, Tonegawa S (2003) Hippocampal CA3 NMDA receptors are crucial for memory acquisition of one-time experience. Neuron 38: 305-315. [DOI] [PubMed] [Google Scholar]
- O'Reilly RC, Rudy JW (2000) Computational principles of learning in the neocortex and hippocampus. Hippocampus 10: 389-397. [DOI] [PubMed] [Google Scholar]
- Sakurai Y (1990) Hippocampal cells have behavioral correlates during the performance of an auditory working memory task in the rat. Behav Neurosci 104: 253-263. [DOI] [PubMed] [Google Scholar]
- Shapiro ML, Tanila H, Eichenbaum H (1997) Cues that hippocampal place cells encode: dynamic and hierarchical representation of local and distal stimuli. Hippocampus 7: 624-642. [DOI] [PubMed] [Google Scholar]
- Sharp PE (1997) Subicular cells generate similar spatial firing patterns in two geometrically and visually distinctive environments: comparison with hippocampal place cells. Behav Brain Res 85: 71-92. [DOI] [PubMed] [Google Scholar]
- Skaggs WE, McNaughton BL (1992) Computational approaches to hippocampal function. Curr Opin Neurobiol 2: 209-211. [DOI] [PubMed] [Google Scholar]
- Skaggs WE, McNaughton BL (1998) Spatial firing properties of hippocampal CA1 populations in an environment containing two visually identical regions. J Neurosci 18: 8455-8466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squire LR, Knowlton BJ (1994) Memory, hippocampus, and brain systems. In: The cognitive neurosciences (Gazzaniga M, ed), pp 825-837. Cambridge, MA: MIT.
- Steward O, Wallace CS, Lyford GL, Worley PF (1998) Synaptic activation causes the mRNA for the IEG Arc to localize selectively near activated postsynaptic sites on dendrites. Neuron 21: 741-751. [DOI] [PubMed] [Google Scholar]
- Tanila H (1999) Hippocampal place cells can develop distinct representations of two visually identical environments. Hippocampus 9: 235-246. [DOI] [PubMed] [Google Scholar]
- Tanila H, Shapiro ML, Eichenbaum H (1997a) Discordance of spatial representation in ensembles of hippocampal place cells. Hippocampus 7: 613-623. [DOI] [PubMed] [Google Scholar]
- Tanila H, Sipila P, Shapiro M, Eichenbaum H (1997b) Brain aging: impaired coding of novel environmental cues. J Neurosci 17: 5167-5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teyler TJ, DiScenna P (1986) The hippocampal memory indexing theory. Behav Neurosci 100: 147-154. [DOI] [PubMed] [Google Scholar]
- Treves A, Rolls ET (1994) Computational analysis of the role of the hippocampus in memory. Hippocampus 4: 374-391. [DOI] [PubMed] [Google Scholar]
- Vazdarjanova A, McNaughton BL, Barnes CA, Worley PF, Guzowski JF (2002) Experience-dependent coincident expression of the effector immediate-early genes Arc and Homer 1a in hippocampal and neocortical neuronal networks. J Neurosci 22: 10067-10071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazdarjanova A, Ramirez-Amaya V, Sutherland VL, Chaula MK, Worley PF, Barnes CA, Guzowski JF (2003) Behavior induces expression of the plasticity-related immediate-early gene Arc in excitatory hippocampal and cortical neurons, but not in astrocytes or inhibitory neurons. Soc Neurosci Abstr 29: 519.7. [Google Scholar]
- Vinogradova OS (2001) Hippocampus as comparator: role of the two input and two output systems of the hippocampus in selection and registration of information. Hippocampus 11: 578-598. [DOI] [PubMed] [Google Scholar]
- West MJ (1993) New stereological methods for counting neurons. Neurobiol Aging 14: 275-285. [DOI] [PubMed] [Google Scholar]
- Wible CG, Findling RL, Shapiro M, Lang EJ, Crane S, Olton DS (1986) Mnemonic correlates of unit activity in the hippocampus. Brain Res 399: 97-110. [DOI] [PubMed] [Google Scholar]
- Wilson IA, Ikonen S, McMahan RW, Gallagher M, Eichenbaum H, Tanila H (2003) Place cell rigidity correlates with impaired spatial learning in aged rats. Neurobiol Aging 24: 297-305. [DOI] [PubMed] [Google Scholar]
- Wilson MA, McNaughton BL (1993) Dynamics of the hippocampal ensemble code for space. Science 261: 1055-1058. [DOI] [PubMed] [Google Scholar]
- Witter MP, Naber PA, van Haeften T, Machielsen WC, Rombouts SA, Barkhof F, Scheltens P, Lopes da Silva FH (2000) Cortico-hippocampal communication by way of parallel parahippocampal-subicular pathways. Hippocampus 10: 398-410. [DOI] [PubMed] [Google Scholar]
- Wood ER, Dudchenko PA, Eichenbaum H (1999) The global record of memory in hippocampal neuronal activity. Nature 397: 613-616. [DOI] [PubMed] [Google Scholar]
- Xiao B, Tu JC, Worley PF (2000) Homer: a link between neural activity and glutamate receptor function. Curr Opin Neurobiol 10: 370-374. [DOI] [PubMed] [Google Scholar]
- Yeckel MF, Berger TW (1990) Feedforward excitation of the hippocampus by afferents from the entorhinal cortex: redefinition of the role of the trisynaptic pathway. Proc Natl Acad Sci USA 87: 5832-5836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young BJ, Fox GD, Eichenbaum H (1994) Correlates of hippocampal complex-spike cell activity in rats performing a nonspatial radial maze task. J Neurosci 14: 6553-6563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan JP, Kiselyov K, Shin DM, Chen J, Shcheynikov N, Kang SH, Dehoff MH, Schwarz MK, Seeburg PH, Muallem S, Worley PF (2003) Homer binds TRPC family channels and is required for gating of TRPC1 by IP3 receptors. Cell 114: 777-789. [DOI] [PubMed] [Google Scholar]