Abstract
In Drosophila, the neuropeptide pigment-dispersing factor (PDF) is required to maintain behavioral rhythms under constant conditions. To understand how PDF exerts its influence, we performed time-series immunostainings for the PERIOD protein in normal and pdf mutant flies over 9 d of constant conditions. Without pdf, pacemaker neurons that normally express PDF maintained two markers of rhythms: that of PERIOD nuclear translocation and its protein staining intensity. As a group, however, they displayed a gradual dispersion in their phasing of nuclear translocation. A separate group of non-PDF circadian pacemakers also maintained PERIOD nuclear translocation rhythms without pdf but exhibited altered phase and amplitude of PERIOD staining intensity. Therefore, pdf is not required to maintain circadian protein oscillations under constant conditions; however, it is required to coordinate the phase and amplitude of such rhythms among the diverse pacemakers. These observations begin to outline the hierarchy of circadian pacemaker circuitry in the Drosophila brain.
Keywords: pigment-dispersing factor, circadian rhythm, Drosophila, lateral neurons, nuclear accumulation, period
Introduction
The organizing principles for the neuronal networks underlying circadian oscillations are essentially unknown. Which cells are the critical oscillators for particular output functions, what is their hierarchical organization, and how are synchronizing signals coordinated among pacemaker groups to provide coherent circadian output? In the Drosophila brain, ∼100 pacemaker neurons are defined by expression of period (per) and other genes essential for circadian rhythmicity (Kaneko and Hall, 2000). These clock cells are divided into the lateral (LN) and dorsal (DN) neural groups (Helfrich-Förster, 2003). Mosaic analysis suggests that LNs, but not DNs, are necessary to establish locomotor rhythms (Frisch et al., 1994; Vosshall and Young, 1995). LNs are segregated into distinct dorsal (LNd) and ventral (LNv) groups; the latter is divided into small and large subgroups. The LNv clock neurons express the neuropeptide pigment-dispersing factor (pdf) (Helfrich-Förster, 1998). Genetic ablation of the entire LNv group produces a syndrome similar to that observed in pdf01 mutant flies (Renn et al., 1999). Such flies anticipate light-to-dark transition events but are phase advanced; in constant darkness, ∼70% lose their locomotor rhythms, and the remainder display only weak rhythms.
The rhythmic nature of single pacemaker cells has traditionally been ascribed to individual cell properties (Michel et al., 1993; Welsh et al., 1995; Liu et al., 1997; Herzog et al., 1998). Recent evidence, however, suggests that interneuronal communication may be required to sustain basic molecular rhythms. Manipulations that alter pacemaker membrane excitability or disrupt transmitter signaling between pacemakers result in severe dampening of molecular rhythms (Harmar et al., 2002; Nitabach et al., 2002; Colwell et al., 2003; Lee et al., 2003). Likewise, Peng et al. (2003) recently found that transcript rhythms of the clock genes timeless (tim) and cryptochrome (cry) were mostly damped in pdf01 mutants of Drosophila. They suggested that neural network interactions are required to maintain basic molecular rhythms in the absence of environmental cues; however, transcript and protein rhythms do not function equally in establishing the circadian clock: PER and TIM proteins display strong rhythms of protein accumulation in the absence of rhythmic per and tim transcription (Cheng and Hardin, 1998). Likewise, flies lacking rhythmic per and tim transcription can also maintain rhythmic locomotor activity (Yang and Sehgal, 2001). Thus, we have asked whether the PER protein displays different patterns of daily accumulation in control versus pdf01 mutant flies. In contrast to the conclusions of Peng et al. (2003), we find that in pdf01 mutants, PER molecular rhythms persist within individual LN pacemaking neurons under conditions in which behavioral rhythms are absent. PDF is required, however, for the normal synchronization of oscillations among different pacemaker neurons. The precise details in the molecular phenotype of pdf01 mutants suggest that PDF-dependent hierarchical communication occurs from the LNv to the LNd pacemakers and that a phase-delaying action of PDF serves as the primary coordinating force. These observations thus begin to define the precise organizational properties binding the heterogeneous circadian pacemakers in Drosophila.
Materials and Methods
Fly stocks. y w;; pdf01 female mutant flies were used from the stock designated W15 (Renn et al., 1999); these flies are PDF protein nulls. Control female flies were used from the W33 stock (y w), which possesses a functional pdf gene within a genetic background similar to that of W15 flies.
Behavioral analysis. Young adult females (1-5 d old) were monitored at 25°C in Trikinetics monitors (Waltham, MA) (Hamblen et al., 1998). Flies were entrained in 12 hr light/dark (LD) conditions for 6 d and then released into constant darkness (DD) for 9 d. Activity counts were accumulated in half-hour bins. To evaluate rhythmicity of locomotor behavior, χ2 periodogram analysis was used. Rhythm periods with power <10 and width <2 were considered arrhythmic (Ewer et al., 1992). Females were used because they display a slightly more severe phenotype (W. Li and P. H. Taghert, unpublished observations). In several experiments conducted under constant conditions during the past 9 months, 64% of y w;; pdf01 males (n = 598) and 72% of y w;; pdf01 females (n = 504) were arrhythmic. Activity figures were generated with Matlab modules (Levine et al., 2002).
Immunofluorescence staining. Immunostaining was performed as described previously (Kaneko et al., 1997; Shafer et al., 2002). Flies were entrained in LD for 5 d and released into constant darkness. On DD days 3, 6, and 9, flies were collected at circadian times 5, 11, 17, and 23 in darkness, anesthetized on ice, and then dissected under normal light conditions. Such conditions have been shown previously not to affect quantification of PER levels in the fly brain (Kaneko et al., 1997). Dissections were performed on dry ice in fixative [3.7% formaldehyde (Sigma, St. Louis, MO), 100 mm PIPES, 1 mm MgCl2, 1 mm EGTA, pH 6.9]. Fixation was continued for 1 hr at room temperature. Rabbit anti-PER antibody was used at a dilution of 1:1000 (Kaneko et al., 1997). Alex-aFluor 488-conjugated goat anti-rabbit secondary antibody (Molecular Probes, Eugene, OR) was used at 1:500 dilution. Brains were mounted in Vectashield anti-fade mounting medium between two coverslips. All brains were mounted in the same orientation, such that when imaging, the ventral lateral neurons were closer to the objective lens than the dorsal neural groups.
Confocal imaging and identification of neural groups. Samples were imaged on the Olympus FVX confocal microscope using a 40× water immersion objective (numerical aperture, 1.15). The following procedures allowed quantification and comparison of staining intensities. As described above, all brains were mounted in the same orientation so that the light path that traveled through tissue to reach each neural group was relatively constant between brains. Confocal settings (photomultiplier sensitivity, laser strength, aperture widths) were kept constant for all images captured. One brain was randomly chosen to serve as a control and imaged at each imaging session. Subsequently, the stained PER intensities were compared between the sessions to confirm that intersession system variability was <10%.
Confocal image intensity quantification. Images were captured in 12-bit grayscale space, and all subsequent analysis preserved this dynamic range. To quantify the intensity of PER staining for one neural group of one brain (e.g., small LNvs), the following procedure was used. The slices of the confocal stack that contained images of the neural group were identified. A maximum projection image was created from this subset of slices. The boundaries of the neural group in the maximum projection image were manually delimited using the “select contiguous regions” tool of the GIMP image processing program (http://www.gimp.org). The 12-bit pixel intensities within the marked boundaries were averaged and tallied as the PER-stained intensity of the neural group. The pixels within five pixels outside of the marked boundaries were averaged and tallied as background and subtracted from the PER-stained intensity. Code for intensity quantification from images was written in Perl and C.
Scoring PER subcellular distributions. Individual slices from confocal stacks were arrayed such that the entire depth of a neuronal group could be viewed simultaneously. Cytoplasmic or nuclear calls were made blindly by two observers for all neuronal cell bodies that were positive for PER immunostaining. Cytoplasmic calls were made for ring-shaped staining patterns; nuclear calls encompassed exclusively nuclear as well as nuclear with cytoplasmic staining.
Results
The loss of locomotor behavior in pdf01 mutants was observed in ∼70% of flies and manifested gradually (Renn et al., 1999); in all flies, residual rhythms were observed for 2-3 d after release into constant conditions (Fig. 1). Therefore, to assess molecular oscillations during periods of behavioral arrhythmicity, we performed anti-PER immunostaining in control and pdf01 adult fly brains on days 3, 6, and 9 of constant darkness (DD days 3, 6, and 9). Despite the presumed persistence of ∼30% rhythmic flies in the pdf01 population, we detected changes in molecular oscillations of the pdf01 ventral and dorsal LNs as compared with control flies (below).
Figure 1.
After light entrainment, dissipation of locomotor rhythms in pdf01 mutants is gradual. Young female flies were entrained for 6 d in 12 hr LD cycles and then released into DD for 9 d. The representative data are double plotted, with the last 24 hr repeated on the next row. Rhythmic (R) and arrhythmic (AR) designations refer to behavioral analysis results from periodogram analysis for DD days 1-9. Most control flies (y w;;) remain rhythmic in DD. pdf01 flies (y w;; pdf01) often retain residual rhythms for 2-3 d of DD before becoming arrhythmic. Approximately 30% of pdf01 flies retain weak, fast-running rhythms in DD (example in the rightmost plot).
For this study, we focused on the ventral and dorsal LNs because they are the only clock neurons essential for establishing locomotor circadian rhythms under constant conditions (Frisch et al., 1994; Vosshall and Young, 1995; Helfrich-Förster, 1997; Blanchardon et al., 2001; Veleri et al., 2003). Expression of the period gene in these neurons is sufficient to drive locomotor rhythms in period mutant flies (Frisch et al., 1994). Furthermore, among the LNv neurons, we focused only on the “small” subset. In agreement with previous reports (Herzog et al., 1998; Helfrich-Förster, 2001; Yang and Sehgal 2001; Shafer et al., 2002; Peng et al., 2003; Veleri et al., 2003), we did not observe consistent PER intensity rhythms in the large LNvs over the DD sampling period (data not shown). Importantly, PER subcellular distribution in the large LNvs of both control and pdf01 flies remained nuclear at all time points (Shafer et al., 2002).
Here, we report on two measures of PER protein oscillations: a rhythm of subcellular distribution and a rhythm of staining intensity. Because PER appears in the cytoplasm of clock neurons for only a brief period of time in early subjective night (Shafer et al., 2002), this marker serves to indicate the rhythmic phase of individual neurons and individual flies (Curtin et al., 1995; Kloss et al., 2001; Martinek et al., 2001; Lin et al., 2002; Akten et al., 2003). The daily rhythm in the staining intensity of PER is the more traditional measure of its molecular oscillation (Veleri et al., 2003) and provides an independent confirmation of overall rhythmic expression.
Phase dispersal in the small LNvs
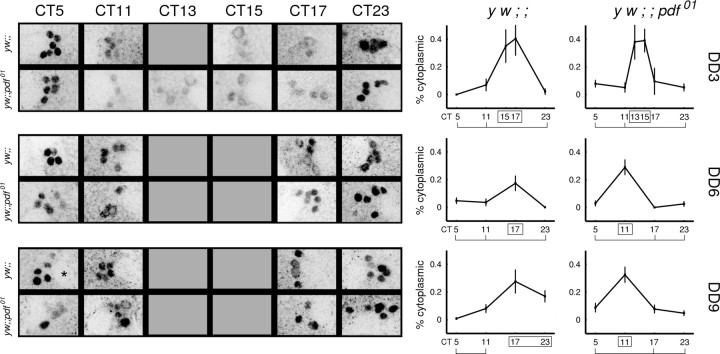
Consistent with previous observations (Curtin et al., 1995; Martinek et al., 2001; Shafer et al., 2002), we found that the distribution of PER in the small LNvs in control (y w) flies was predominantly nuclear for most of the circadian cycle at CT5, CT11, and CT23 (Fig. 2). At CT17 on all observed days, however, we observed individual small LNv neurons with distinct cytoplasmic PER and little or no nuclear PER. A daily pattern of cytoplasmic PER accumulation was also clearly observed as late as DD day 9 in the small LNv of pdf01 flies; however, although the subcellular localization rhythm of PER persisted in prolonged darkness, its phasing among this group of neurons was disrupted in pdf01. At CT17, PER was predominantly cytoplasmic in the small LNvs of control flies, but in pdf01, PER displayed a mixture of cytoplasmic and nuclear distributions at CT17. Such a mixture of subcellular distribution was not merely a population effect, because it was observed within individual pdf01 brains (Fig. 2).
Figure 2.
The phasing of the small LNv neurons disperses over time. On the right, each plot shows the percentage of cells expressing PER in their cytoplasm or nucleus across a circadian cycle from control (y w;;) and pdf01 mutants (y w;; pdf01). The y-axis represents the average percentage of cytoplasmic PER cells in the small LNv cluster of one brain hemisphere. An average of 13 hemispheres are represented in each time point. Error bars indicate SEM. Cells were visible and scored at all time points, except at CT13 of DD6 in both control and pdf01 flies, when low PER immunoreactivity precluded identification of the small LNvs (indicated as breaks in the graph). All plots showed statistically significant rhythms by ANOVA testing (p < 0.05). x-axis represents CT. Markings denote statistically equivalent time points by the Tukey multiple comparison test: boxed time points have peak values, and bracketed time points have trough values. Example images of small LNvs are shown on the left. For both control and mutant genotypes, the neurons displayed nuclear PER at CT5 and CT23 across all days. Control flies display predominantly cytoplasmic PER at CT17, whereas individual brains of pdf01 mutants have mixed cytoplasmic (“doughnuts”) and nuclear distributions of PER at CT17. The overall intensity of these images was adjusted to facilitate viewing of the PER signal.
To assess whether the window of predominantly cytoplasmic PER accumulation occurred at a novel phase in pdf01, we immunostained brains at additional time points (CT13, CT15, CT17, and CT19) on DD day 6. At CT13, the small LNvs could not be reliably identified in either control or pdf01 flies because of low levels of PER staining intensity; however, the observation of staining in other neurons at CT13 indicated that the PER immunostaining procedure was successful (data not shown). In individual brains of control flies at CT15 and CT17, PER was predominantly cytoplasmic (Fig. 2); however, at CT15 and later time points (CT17 and CT19), the small LNvs within individual brains in pdf01 presented a mixed population of cells with cytoplasmic or nuclear PER. On day 6, the differences between control and pdf01 genotypes at CT15 and CT17 were statistically signficant (t test; p < 0.05).
The mixed phasing among the small LNvs of pdf01 became more severe with time in DD (Fig. 2). By DD day 9, cytoplasmic PER was detected as early as CT11 in pdf01 flies, whereas control flies maintained a predominant peak of cytoplasmic PER at CT17. Despite these changes, the persistence of PER within the nucleus of sLNvs of pdf01 at CT5 and CT23 throughout all 9 d in DD indicates that individual neurons maintained molecular rhythms in the absence of the PDF neuropeptide. Taken together, these results show that PER molecular rhythms persist in the small LNvs of pdf01 mutants, days after the loss of behavioral rhythms. In the absence of PDF, however, the rhythmic phase of small LNvsin pdf01 mutants dispersed relative to each other over the several days in constant darkness. Strikingly, the dispersal was unidirectional: bidirectional dispersion would have resulted in the concomitant observation of cytoplasmic PER at CT23 in pdf01 by DD day 9.
Phase advancement in LNds of pdf01 mutants
The LNds also maintained a rhythm of PER subcellular distribution during the DD days under observation. Similar to the small LNvs, PER assumed a nuclear distribution throughout most of the circadian cycle in the LNds of control flies (Fig. 3). Because we observed this peak of cytoplasmic PER at CT17 throughout all 9 d of DD in control flies, it served as a phase marker of LNd neuronal rhythms. There was one difference in this rhythm between the two LN pacemaker groups: small LNv neurons uniformly displayed cytoplasmic distribution of PER early in subjective night, whereas fewer than half of the LNds displayed cytoplasmic PER at this time (compare Figs. 2, 3).
Figure 3.
The LNds of pdf01 mutants phase advance in DD. Please refer to Figure 2 for details on figure format. In the seneurons, cytoplasmic PER is seen at CT17 of control flies but earlier in pdf01 mutants. Across all DD days, the LNds have predominantly nuclear PER at CT5 and CT23. Low PER staining in control brains stained at CT13 of DD3 precluded identification of LNds. An average of 13 hemispheres are represented in each time point. All plots showed statistically significant rhythms by ANOVA testing (p < 0.05). An asterisk on the representative image of y w on CT5 of DD9 marks a sinus-like feature, which has not been described previously, around which the LNds were consistently observed.
In pdf01, LNd neurons also maintained PER oscillations, but these oscillations displayed significant differences from those of control flies. As early as DD day 3, the LNd cytoplasmic peak was phase advanced compared with control flies (Fig. 3), appearing at CT13 and CT15. By DD days 6 and 9, this cytoplasmic peak in pdf01 brains advanced to CT11. Parenthetically, we noted that LNd cell bodies were in association with or near a structure resembling a sinus (Fig. 3). To our knowledge, this structure has not been described previously. Its functions and potential relation to LNd neuronal function are not known.
Rhythms in PER staining intensity
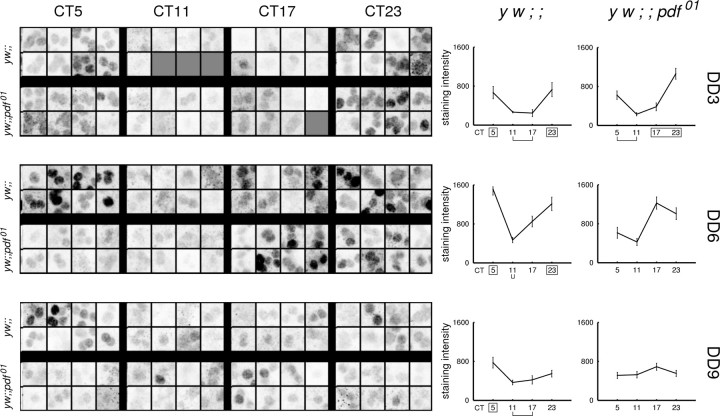
We also measured rhythms of PER staining intensity in the small LNv and LNd neurons to provide independent measures of their molecular rhythms. In the small LNvs of both control and pdf01 flies, rhythms in staining intensity declined in amplitude over the course of 9 d in DD. Statistical analysis indicated that the rhythm in both genotypes persisted throughout this time (Fig. 4). In both genotypes, peak intensity values were observed in the early subjective morning time points (CT5 and CT23). We noted one difference: although control flies maintained low staining at CT17 across the DD days, the staining intensity at CT17 in pdf01 mutants was not statistically distinguishable from the peak values at CT5 and CT23 on DD days 6 and day 9 (Fig. 4). These observations are consistent with the results seen in the subcellular distribution rhythms in the small LNvs (Fig. 2). The gradual phase advancement of a portion of small LNvs in pdf01 mutants is reflected as an increase in staining intensity within that group at CT17.
Figure 4.
PER intensity rhythms persist in the small LNvs of pdf01 mutants. Confocal images from a subset of brains are shown; quantified intensity values are shown in the graphs to the right. At some time points, PER staining was too low to allow for reliable identification of small LNvs. In these panels, such as CT11 of DD day 3 in control flies, blank squares exist to indicate that fewer than eight examples could be found. On average, 15 brain hemispheres per time point were imaged. All plots showed statistically significant rhythms by ANOVA testing (p < 0.05). In the graphs, the y-axis represents absolute intensity values of a maximum of 4095. x-axis represents CT. Markings denote statistically equivalent time points by the Tukey multiple comparison test: boxed time points have peak values, and bracketed time points have trough values.
In LNd neurons of control flies, a rhythm of staining intensity was present in all 9 d of DD, but it declined in amplitude (Fig. 5). Peak values were also observed in the early morning time points CT23 and CT5. This pattern of staining was present on DD day 3 in pdf01 mutants. By DD day 6, however, a phase advancement was observed such that the normally high-staining time point of CT5 became a trough, whereas CT17 became a peak point. This advancement of phase was consistent with that seen in LNd PER subcellular distribution (Fig. 3). By DD day 9, the rhythms in PER staining intensity no longer displayed statistically significant variations with time in the LNds of pdf01 mutants (Fig. 5), although the rhythm in subcellular distribution persisted in these neurons (Fig. 3). Surprisingly, these observations suggest that although pdf is not required to maintain coordinated molecular rhythms among the LNds, it is required for the normal phasing and robust amplitude of such rhythms.
Figure 5.
PER intensity rhythms phase advance and then diminish in the LNds of pdf01 mutants. Please refer to Figure 2 for details on figure format. All plots showed statistically significant rhythms by ANOVA testing (p < 0.05), except for the data from y w;; pdf01, DD day 9.
Discussion
The mechanism by which multi-oscillator coordination occurs in the fly circadian system is essentially unknown. Previously, PDF was hypothesized to couple bilateral pacemakers because PDF neurons project to multiple clock centers within the fly brain (Helfrich-Förster, 1998; Renn et al., 1999) and because injections of a related peptide, pigment-dispersing hormone, produced a phase-delaying signal in the cockroach circadian system (Petri and Stengl, 1997). To investigate the role of PDF in Drosophila, clock activity in pdf01 mutants was assessed by monitoring the expression of PER, the protein level of which is an essential state variable of the fly clock (Yang and Sehgal, 2001). Because of the graduated, continuous nature of fluctuations in PER protein levels, phase assessment is difficult using the standard two or four time-point sampling of molecular rhythms. Therefore, we used the discrete marker of subcellular localization to evaluate the phase of individual neurons within neural subgroups (Shafer et al., 2002). Alterations in the phasing of this cytoplasmic window have been correlated previously with numerous clock gene mutant phenotypes with similar temporal resolution (Curtin et al., 1995; Kloss et al., 2001; Martinek et al., 2001; Lin et al., 2002; Akten et al., 2003).
Using this indicator of PER subcellular distribution, we observe that in the absence of light and pdf function, the phases of the small LNv clocks gradually disperse (Fig. 2). In normal flies, these neurons display a persistent peak of cytoplasmic PER in the middle of subjective night, indicating that they are phase locked. Early in free run, the magnitude of the cytoplasmic peak in pdf01 mutants is comparable with that of normal flies, but it progressively blunts with continued days in DD. Among possible explanations, we favor the hypothesis that these neurons are proceeding with normal rhythms but with a progressive desynchrony. That interpretation is supported by two observations. First, PER remains predominantly nuclear at the flanking time points of CT5 and CT23. This, with cytoplasmic PER seen only at discrete time points, indicates that normal rhythms in subcellular distribution are progressing within individual neurons. The preservation of PER staining intensity rhythms further supports the interpretation of normal rhythmic progression (Fig. 4). Second, the entrance of these neurons into the cytoplasmic phase is observed at progressively earlier times, by CT11 of DD day 9; however, only some of the neurons become phase advanced, because neurons with cytoplasmic PER are observed at the normal time point of CT17 across all 9 d of DD. This heterogeneous phase advancement might be a stochastic process or might indicate further differentiation of circadian function among the small LNvs. Together these results suggest that PDF (released by LNv neurons) provides feedback that normally helps to synchronize the eight autonomous small LNvs within individual brains. We postulate that this synchronization supports the circadian fluctuations of PDF at the terminals of the small LNvs (Park et al., 2000).
Compared with the small LNvs, the LNds responded in a different manner to the absence of pdf. They exhibited a coherent phase advancement of the PER cytoplasmic window and reverted to short-period, low-amplitude cycling (Figs. 3, 5). That phase advance was paralleled by an early peak in the rhythm of LNd PER staining intensity at CT17 of DD day 6. Interestingly, the aggregate PER intensity rhythm was severely damped by DD day 9, despite the maintained rhythm in the nuclear translocation within individual LNd neurons. In control flies, the PER intensity rhythm also damped during DD, but not to as great an extent. PDF is therefore required to maintain high-amplitude PER intensity rhythms in the LNds beyond DD day 6. One possibility is that PDF signaling entrains LNd neurons by delaying the entrance of PER into the nucleus. The delay would allow greater levels of PER to accumulate before its translocation to the nucleus and would generate a more forceful negative feedback on the clock system to result in a greater amplitude of molecular rhythms (Leloup and Goldbeter, 2003).
We propose that these complex roles of pdf help to explain its contributions in maintaining behavioral rhythms (Fig. 6). PDF release represents the essential signaling function of the small LNv neurons, because the pdf null syndrome is phenocopied by the ablation of PDF-expressing cells (Renn et al., 1999). This PDF output may proceed directly to influence premotor centers that regulate behavioral rhythms. A nonexclusive alternative is that the premotor centers are controlled by an auxiliary clock center (e.g., the LNds) that is itself subject to small LNv (i.e., PDF) influence. The pdf01 mutant population includes an ∼30% minority of flies that display residual, short-period rhythmicity past DD day 3 (Renn et al., 1999). Our results provide evidence for the continued activity of a phase-advanced, low-amplitude clock within LNd neurons. We speculate that LNd neurons therefore may represent the persistent, rhythmic center responsible for that durable, rhythmic aspect of pdf01 behavior. Furthermore, we propose that the gradual winding down of free-running behavioral rhythms in pdf01 flies may result from the gradual loss of normal PER intensity rhythms that we observe in the LNds after DD day 3. By this day, the amplitude of these rhythms may have approached a threshold level required for overt behavioral rhythmicity. The requirement of a molecular amplitude threshold for locomotor rhythms is supported by the phenotype of the Clkar mutant, which is behaviorally arrhythmic despite the presence of low-amplitude molecular cycling (Allada et al., 2003). We therefore hypothesize that the LNd pacemaker group is a significant participant in the neuronal circuitry underlying Drosophila behavioral rhythms under constant conditions (Helfrich-Förster, 1998; Kaneko and Hall, 2000; Peng et al., 2003) and that its molecular rhythms are influenced by the small LNv group via its PDF output.
Figure 6.
A model for interactions between the two lateral neuron pacemaking centers in the Drosophila brain. In wild-type flies (top), the small LNv center (s-LNv) maintains the normal phase and amplitude of molecular rhythms in the LNd center as well as its own synchronicity through PDF communication (possibly autocrine). Projections from the s-LNvs and/or the LNds regulate premotor centers (PMC) and organize behavioral rhythms. In the absence of PDF (in pdf01 mutants, bottom), a syndrome of effects is observed: the s-LNvs become desynchronized, but because they are effectively silenced by the pdf01 mutation, the effects of this decoupling are unlikely to influence the PMC. With the lack of PDF entrainment, the LNds revert to a low-amplitude, fast-running clock; however, PDF is not required for their synchronization. The loss of a strong circadian signal from the LNd and/or the s-LNv into the PMC leads to behavioral arrhythmicity. Shades of gray represent differences in phase and/or period. The graphs to the left represent the decreased amplitude of PER staining rhythms in LNds and the desynchrony of staining rhythms in the LNvs of pdf01 mutants.
The importance of neuronal interactions and network properties for molecular oscillations within pacemakers is a central problem in circadian physiology. The application of tetrodotoxin (TTX) to cultured mice suprachiasmatic nucleus (SCN) slices results in the diminution of mPER rhythms (Lee et al., 2003). In contrast, the behavioral rhythms disrupted with in vivo TTX application to the rat SCN reemerge with the preservation of the entrance phase after TTX washout, indicating the continuation of an underlying clock (Schwartz et al., 1987). Thus, similar to our findings in Drosophila, these results suggest that the high-amplitude molecular fluctuations that are required for behavioral rhythms are an emergent property of a multi-oscillator SCN network. Both in mammals and in the fly, however, the intrinsic molecular clockwork appears to persist in the absence of cellular interactions. The persistence of clock function in fly pdf01 mutants was reflected in the maintained rhythm of PER nuclear translocation. The rhythmic, gated nuclear translocation of the mCRY proteins has been reported to occur in mammalian liver (Lee et al., 2001). An evaluation of such translocation rhythms may apply to the study of SCN clockworks as well, under conditions during which the intensity rhythms may otherwise be lost.
In mammals, these same neuronal interactions appear equally important to coordinate the phases of heterogeneous cell types of the regionally diverse SCN. The intact SCN displays coherent, topographically stratified waves of mPer1 transcription; however, when the ventral SCN subregion is separated from the dorsal, the dorsal pacemakers exhibit altered phases and/or periods, whereas the ventral pacemakers maintain their phase properties (Yamaguchi et al., 2003). These observations suggest that the SCN is composed of several pacemaking centers, which must interact via reproducible network connections to coordinate each others' rhythmic output. The neuroanatomy of the Drosophila circadian system likewise suggests that it is composed of multiple pacemaking centers, and our results outline rules by which they may interact: the small LNv center maintains the LNd center and delays it and also feeds back to delay itself by entraining component neurons (Fig. 6). A phase-delaying function of a PDF-related neuropeptide has been described previously (Petri and Stengl, 1997). These interactions between LNs require PDF and may occur directly or indirectly through interneurons.
At the level of single pacemakers, the functions of PDF may be explained by a common regulation of the rate at which PER enters the nucleus from the cytoplasmic phase. Such regulation may be achieved, for example, through kinases such as doubletime (Kloss et al., 1998, 2001; Price et al., 1998), shaggy (Martinek et al., 2001), or the casein kinase 2 holoenzyme (Lin et al., 2002; Akten et al., 2003). This hypothesis presents a framework within which detailed cellular analysis of interactions within a hierarchy of circadian pacemakers may be pursued.
Footnotes
Y.L. and G.D.S. are supported by National Institutes of Health Grant HG00249. P.H.T. is supported by National Institute of Mental Health Grant MH067122 and the Human Frontiers Scientific Program Organization. We thank Susan Renn and Mei Han for contributions to preliminary experiments in this effort; Jeff Hall for PER antibodies; Erik Herzog, Russell Van Gelder, Orie Shafer, and Joel Levine for their comments on this manuscript; and Shin Lin for help with statistical analysis.
Correspondence should be addressed to Paul H. Taghert, Department of Anatomy and Neurobiology, Box 8108, Washington University Medical School, 660 South Euclid Avenue, St. Louis, MO 63110. E-mail: taghertp@pcg.wustl.edu.
Copyright © 2004 Society for Neuroscience 0270-6474/04/247951-07$15.00/0
References
- Akten B, Jauch E, Genova GK, Kim EY, Edery I, Raabe T, Jackson FR (2003) A role for CK2 in the Drosophila circadian oscillator. Nat Neurosci 6: 251-257. [DOI] [PubMed] [Google Scholar]
- Allada R, Kadener S, Nandakumar N, Rosbash M (2003) A recessive mutant of Drosophila clock reveals a role in circadian rhythm amplitude. EMBO J 22: 3367-3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchardon E, Grima B, Klarsfeld A, Chelot E, Hardin PE, Preat T, Rouyer F (2001) Defining the role of Drosophila lateral neurons in the control of circadian rhythms in motor activity and eclosion by targeted genetic ablation and PERIOD protein overexpression. Eur J Neurosci 13: 871-888. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Hardin PE (1998) Drosophila photoreceptors contain an autonomous circadian oscillator that can function without period mRNA cycling. J Neurosci 18: 741-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colwell CS, Michel S, Itri J, Rodriguez W, Tam J, Lelievre V, Hu Z, Liu X, Waschek JA (2003) Disrupted circadian rhythms in VIP- and PHI-deficient mice. Am J Physiol Regul Integr Comp Physiol 285: 939-949. [DOI] [PubMed] [Google Scholar]
- Curtin KD, Huang ZJ, Rosbash M (1995) Temporally regulated nuclear entry of the Drosophila period protein contributes to the circadian clock. Neuron 14: 365-372. [DOI] [PubMed] [Google Scholar]
- Ewer J, Frisch B, Hamblen-Coyle MJ, Rosbash M, Hall JC (1992) Expression of the period clock gene within different cell types in the brain of Drosophila adults and mosaic analysis of these cells' influence on circadian behavioral rhythms. J Neurosci 12: 3321-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch B, Hardin PE, Hamblen-Coyle MJ, Rosbash M, Hall JC (1994) A promoterless period gene mediates behavioral rhythmicity and cyclical per expression in a restricted subset of the Drosophila nervous system. Neuron 12: 555-570. [DOI] [PubMed] [Google Scholar]
- Hamblen MJ, White NE, Emery PT, Kaiser K, Hall JC (1998) Molecular and behavioral analysis of four period mutants in Drosophila melanogaster encompassing extreme short, novel long, and unorthodox arrhythmic types. Genetics 149: 165-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmar AJ, Marston HM, Shen S, Spratt C, West KM, Sheward WJ, Morrison CF, Dorin JR, Piggins HD, Reubi JC, Kelly JS, Maywood ES, Hastings MH (2002) The VPAC(2) receptor is essential for circadian function in the mouse suprachiasmatic nuclei. Cell 109: 497-508. [DOI] [PubMed] [Google Scholar]
- Helfrich-Förster C (1997) Development of pigment-dispersing hormone-immunoreactive neurons in the nervous system of Drosophila melanogaster. J Comp Neurol 380: 335-354. [DOI] [PubMed] [Google Scholar]
- Helfrich-Förster C (1998) Robust circadian rhythmicity of Drosophila melanogaster requires the presence of lateral neurons: a brain-behavioral study of disconnected mutants. J Comp Physiol [A] 182: 435-453. [DOI] [PubMed] [Google Scholar]
- Helfrich-Förster C (2003) The neuroarchitecture of the circadian clock in the brain of Drosophila melanogaster. Microsc Res Tech 62: 94-102. [DOI] [PubMed] [Google Scholar]
- Helfrich-Förster C, Winter C, Hofbauer A, Hall JC, Stanewsky R (2001) The circadian clock of fruit flies is blind after elimination of all known photoreceptors. Neuron 30: 249-261. [DOI] [PubMed] [Google Scholar]
- Herzog ED, Takahashi JS, Block GD (1998) Clock controls circadian period in isolated suprachiasmatic nucleus neurons. Nat Neurosci 1: 708-713. [DOI] [PubMed] [Google Scholar]
- Kaneko M, Hall JC (2000) Neuroanatomy of cells expressing clock genes in Drosophila: transgenic manipulation of the period and timeless genes to mark the perikarya of circadian pacemaker neurons and their projections. J Comp Neurol 422: 66-94. [DOI] [PubMed] [Google Scholar]
- Kaneko M, Helfrich-Förster C, Hall JC (1997) Spatial and temporal expression of the period and timeless genes in the developing nervous system of Drosophila: newly identified pacemaker candidates and novel features of clock gene product cycling. J Neurosci 17: 6745-6760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloss B, Price JL, Saez L, Blau J, Rothenfluh A, Wesley CS, Young MW (1998) The Drosophila clock gene double-time encodes a protein closely related to human casein kinase Iepsilon. Cell 94: 97-107. [DOI] [PubMed] [Google Scholar]
- Kloss B, Rothenfluh A, Young MW, Saez L (2001) Phosphorylation of period is influenced by cycling physical associations of double-time, period, and timeless in the Drosophila clock. Neuron 30: 699-706. [DOI] [PubMed] [Google Scholar]
- Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM (2001) Post-translational mechanisms regulate the mammalian circadian clock. Cell 107: 855-867. [DOI] [PubMed] [Google Scholar]
- Lee HS, Billings HJ, Lehman MN (2003) The suprachiasmatic nucleus: a clock of multiple components. J Biol Rhythms 18: 435-449. [DOI] [PubMed] [Google Scholar]
- Leloup JC, Goldbeter A (2003) Toward a detailed computational model for the mammalian circadian clock. Proc Natl Acad Sci USA 100: 7051-7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine JD, Funes P, Dowse HB, Hall JC (2002) Advanced analysis of a cryptochrome mutation's effects on the robustness and phase of molecular cycles in isolated peripheral tissues of Drosophila. BMC Neurosci 3: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JM, Kilman VL, Keegan K, Paddock B, Emery-Le M, Rosbash M, Allada R (2002) A role for casein kinase 2alpha in the Drosophila circadian clock. Nature 420: 816-820. [DOI] [PubMed] [Google Scholar]
- Liu C, Weaver DR, Strogatz SH, Reppert SM (1997) Cellular construction of a circadian clock: period determination in the suprachiasmatic nuclei. Cell 91: 855-860. [DOI] [PubMed] [Google Scholar]
- Martinek S, Inonog S, Manoukian AS, Young MW (2001) A role for the segment polarity gene shaggy/GSK-3 in the Drosophila circadian clock. Cell 105: 769-779. [DOI] [PubMed] [Google Scholar]
- Michel S, Geusz ME, Zaritsky JJ, Block GD (1993) Circadian rhythm in membrane conductance expressed in isolated neurons. Science 259: 239-241. [DOI] [PubMed] [Google Scholar]
- Nitabach MN, Blau J, Holmes TC (2002) Electrical silencing of Drosophila pacemaker neurons stops the free-running circadian clock. Cell 109: 485-495. [DOI] [PubMed] [Google Scholar]
- Park JH, Helfrich-Förster C, Lee G, Liu L, Rosbash M, Hall JC (2000) Differential regulation of circadian pacemaker output by separate clock genes in Drosophila. Proc Natl Acad Sci USA 97: 3608-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Stoleru D, Levine JD, Hall JC, Rosbash M (2003) Drosophila free-running rhythms require intercellular communication. PLoS Biol 1: E13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petri B, Stengl M (1997) Pigment-dispersing hormone shifts the phase of the circadian pacemaker of the cockroach Leucophaea maderae J Neurosci 17: 4087-4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price JL, Blau J, Rothenfluh A, Abodeely M, Kloss B, Young MW (1998) double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell 94: 83-95. [DOI] [PubMed] [Google Scholar]
- Renn SC, Park JH, Rosbash M, Hall JC, Taghert PH (1999) A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell 99: 791-802. [DOI] [PubMed] [Google Scholar]
- Schwartz WJ, Gross RA, Morton MT (1987) The suprachiasmatic nuclei contain a tetrodotoxin-resistant circadian pacemaker. Proc Natl Acad Sci USA 84: 1694-1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafer OT, Rosbash M, Truman JW (2002) Sequential nuclear accumulation of the clock proteins period and timeless in the pacemaker neurons of Drosophila melanogaster J Neurosci 22: 5946-5954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veleri S, Brandes C, Helfrich-Förster C, Hall JC, Stanewsky R (2003) A self-sustaining, light-entrainable circadian oscillator in the Drosophila brain. Curr Biol 13: 1758-1767. [DOI] [PubMed] [Google Scholar]
- Vosshall LB, Young MW (1995) Circadian rhythms in Drosophila can be driven by period expression in a restricted group of central brain cells. Neuron 15: 345-360. [DOI] [PubMed] [Google Scholar]
- Welsh DK, Logothetis DE, Meister M, Reppert SM (1995) Individual neurons dissociated from rat suprachiasmatic nucleus express independently phased circadian firing rhythms. Neuron 14: 697-706. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Isejima H, Matsuo T, Okura R, Yagita K, Kobayashi M, Okamura H (2003) Synchronization of cellular clocks in the suprachiasmatic nucleus. Science 302: 1408-1412. [DOI] [PubMed] [Google Scholar]
- Yang Z, Sehgal A (2001) Role of molecular oscillations in generating behavioral rhythms in Drosophila. Neuron 29: 453-467. [DOI] [PubMed] [Google Scholar]