Abstract
SynGAP is a brain-specific ras GTPase-activating protein that is an abundant component of the signaling complex associated with the NMDA-type glutamate receptor. We generated mutant mice lacking synGAP to study its physiological role. Homozygous mutant mice die in the first few days after birth; however, neurons from mutant embryos can be maintained in culture. Here, we report that spine and synapse formation are accelerated in cultured mutant neurons, and the spines of mature mutant neurons are significantly larger than those of wild type. Clusters of PSD-95 and subunits of AMPA-type and NMDA-type glutamate receptors accumulate in spines of mutant neurons by day 10 in vitro, whereas in wild-type neurons they are still mostly located in dendritic shafts. The frequency and amplitude of miniature EPSCs are larger in mutant neurons at day 10 in vitro, confirming that they have more functional synapses. At day 21 in vitro, the spines of mutant neurons remain significantly larger than those of wild type. The mutant phenotype at day 10 in vitro can be rescued by introduction of recombinant wild-type synGAP on day 9. In contrast, introduction of mutant synGAP with a mutated GAP domain or lacking the terminal domain that binds to PSD-95 does not rescue the mutant phenotype, indicating that both domains play a role in control of spine formation. Thus, the GAP activity of synGAP and its association with PSD-95 are important for normal regulation of spine and synapse formation in hippocampal neurons.
Keywords: synaptogenesis, postsynaptic, NMDA, AMPA, PSD-95, synapsin, filopodia, Ras
Introduction
The NMDA-type glutamate receptor (NMDA receptor) regulates synapse formation during development (Constantine-Paton et al., 1990) and synaptic plasticity in the adult brain (Malenka and Nicoll, 1993). Influx of Ca2+ through the activated receptor into the postsynaptic spine can trigger a variety of biochemical responses, depending on the timing and magnitude of the Ca2+ flux (Sjostrom and Nelson, 2002). Our goal is to understand the organization of signaling pathways that respond to Ca2+ influx into the spine. A core complex of signaling proteins associated with the NMDA receptor at excitatory synapses is found in the postsynaptic density (PSD) fraction (Kennedy, 1997, 2000; Sheng and Sala, 2001). We and others have identified many of the proteins in this complex (Kennedy, 1998; Sheng and Kim, 2000; Xiao et al., 2000). Among the most prominent for which functions are still unknown is the Ras GTPase-activating protein synGAP (Chen et al., 1998; Kim et al., 1998). SynGAP is a major component of the PSD at glutamatergic postsynaptic terminals, where it associates with the PDZ domains of PSD-95 via its C-terminal t-T/SXV domain (Chen et al., 1998; Kim et al., 1998; Zhang et al., 1999), competing with a variety of other potential binding partners for PSD-95 (Kennedy, 2000; Sheng and Sala, 2001). Together, the four splice variants of synGAP are approximately as abundant in the PSD fraction as PSD-95 itself (Chen et al., 1998), suggesting that it is an effective competitor in vivo. Ca2+/calmodulin-dependent protein kinase II (CaMKII), another abundant protein in the PSD complex, phosphorylates synGAP (Chen et al., 1998) and thereby increases its GAP activity (Oh et al., 2002). Because CaMKII is activated by Ca2+ ions flowing through the NMDA receptor, synGAP may act as a point of integration between NMDA receptor activation and Ras signaling pathways.
We generated mice with a deletion in the synGAP gene to enable identification of downstream pathways that are deregulated in its absence. As shown by other groups (Komiyama et al., 2002; Kim et al., 2003), we found that mice with the homozygous synGAP deletion [knock-out (ko)] die shortly after birth; however, neurons from ko embryos can be maintained in culture. Synaptic plasticity is altered in adult mice heterozygous for a synGAP deletion (Komiyama et al., 2002; Kim et al., 2003), and the ratio of AMPA to NMDA receptors is increased in neurons cultured from synGAP ko embryos (Kim et al., 2003). Here, we further investigate the cellular phenotype of neurons with a synGAP deletion. We report that spines and synapses form earlier, and spines ultimately become larger in ko than in wt neurons. Clusters of PSD proteins appear in spines earlier in the ko neurons. These alterations in spine development are rescued by expression of wild-type synGAP (wt SynGAP) in ko neurons. However, mutation of the GAP domain or removal of the t-T/SXV domain renders synGAP unable to rescue the ko cellular phenotype. Our results indicate that both GAP activity and interaction of synGAP with PSD-95 help to determine the timing of spine formation and the size of mature spines.
Materials and Methods
Gene targeting construct. Twelve synGAP genomic DNA fragments were isolated by screening a λFIX II mouse genomic library (129 SvJ strain) with a cDNA probe encoding the plekstrin homology (PH) and C2 domains of the synGAP protein. Genomic sequences used to generate the final targeting construct were derived from a single ∼17 kb genomic segment. The starting targeting vector pPNTloxPv2 (kindly provided by Dr. Jeong Kyo Yoon, California Institute of Technology, Pasadena, CA) was a modified version of pPNT (Tybulewicz et al., 1991) in which the Neo cassette is flanked by two loxP sites. The short arm of the synGAP targeting construct (Fig. 1 A) was a 2.4 kb fragment of intron 3 of the synGAP genomic DNA. It was inserted at the 5′-end of the Neo cassette upstream of the first loxP site in pPNTloxPv2. The long arm included a 5.6 kb genomic fragment containing exons 4-9 with intervening introns inserted at the 3′-end of the Neo cassette downstream of the second loxP site. This sequence was followed by a 136 bp fragment from the vector pRAY2 (gift from P. Seeburg, Max Planck Institute, Heidelberg, Germany) containing a third loxP site and 1.5 kb of the synGAP gene, including introns 9-11 and exons 10 and 11.
Figure 1.
Construction of a synGAP knock-out mouse. A, The targeting construct included a Neo cassette flanked by LoxP sites inserted into intron 3 of the synGAP gene and an additional downstream LoxP site within intron 9. Expression of Cre recombinase in recombinant ES cells resulted in deletion of exons 4-9 of the synGAP gene (ko) in a subset of ES cells, two of which were used to construct synGAP knock-out lines. B, Example of PCR products from DNA of wt, het, and ko embryos. C, D, Immunoblots comparing levels of synGAP (C) and other synaptic proteins (D) in hippocampal homogenates of wt, het, and ko mice at P0, and in hippocampal neurons at 10 DIV cultured from wt, het, and ko embryos, as described in Materials and Methods. Expression of all four isoforms of synGAP protein is absent in ko neurons and reduced in het neurons compared with wt.
Generation of mouse embryonic stem cells for injection into blastocysts. Twenty micrograms of linearized targeting construct were electroporated into 1 × 107 cells/ml of mouse embryonic stem (ES) (CJ7) cells. The transfected ES cells were grown in the presence of the antibiotics G418 (180 μg/ml) and fialuridine (0.2 μm) for 9 d to select for homologous recombinants. Twelve recombinant clones were identified and confirmed by genomic Southern blots with probes flanking the targeted genomic sequences. Two of these clones were expanded for generation of synGAP knock-out and floxed mutant ES cells. They were transfected with the Cre expressing vector pOG231 (kindly provided by Dr. Henry Lester, California Institute of Technology, Pasadena, CA) by electroporation. Twenty-eight G418-sensitive clones were identified. The genotypes of these clones were determined by PCR analyses. Two of the clones had complete deletion of sequences between exons 4 and 9 of synGAP (ko). Two clones with the ko genotype, both of which had normal karyotypes, were used for injection into blastocysts to generate chimeras. Injections and breeding of chimeras were performed by the Transgenic Mouse Core Facility at Caltech (Pasadena, CA). The synGAP ko mutation described here is maintained in a heterozygote line and has been back-crossed onto a C57BL/6 background. Crossing synGAP heterozygous (het) mice results in progeny with a Mendelian distribution (1 wt:2.4 het:1.3 ko; n = 138).
Genotyping. Genomic DNA was isolated from mouse tails and used for PCR with a set of three primers: one recognizing a 3′ synGAP sequence (MGIN-11; 5′-GAGAGAGATGGAGGGTCACTTGAG-3′) and two recognizing 5′ sequences, either within synGAP (MGEX9-1; 5′-CGGATGCTATGTGCAGTGCTGGA-3′) or within the LoxP site (Lox-DS; 5′-GAAGAGGAGTTTACGTCCAGCCAAGCT-3′), in wt or knockout (ko), respectively. PCR cycles started with denaturation of DNA at 94°C for 2 min followed by 35 cycles consisting of denaturation (95°C; 30 sec), annealing (58°C; 30 sec), and extension (72°C; 2 min) followed by a final extension at 72°C for 10 min. The products were a 1.8 kb fragment from wt and a 1.7 kb fragment from ko (Fig. 1 B).
Primary neuronal culture and immunocytochemistry. Hippocampi from individual mice at embryonic day 16 (E16) or E17 were dissected, dissociated by trypsin treatment and mechanical trituration, and plated onto polylysine/laminin-coated (Sigma, St. Louis, MO) glass coverslips (12 mm in diameter) at a density of ∼200 neurons/mm2. Cultures were maintained in neurobasal medium (Invitrogen, Grand Island, NY) complemented with B27, glutamate, and glutamax-I (Invitrogen) as described previously (Brewer et al., 1993). To confirm that cell density was similar between wt and ko cultures, cell counts of Hoechst-stained (Molecular Probes, Eugene, OR) nuclei were done on a set of 21-d-in vitro (DIV) cultures (data not shown).
After 10 or 21 d in vitro, coverslips were rinsed briefly in ice-cold PBS and fixed with ice-cold 4% paraformaldehyde and 4% sucrose in PBS for 5-10 min. Coverslips were rinsed in ice-cold PBS followed by ice-cold methanol, incubated in methanol at -20°C for 20 min, washed once with PBS for 15 min, and blocked with 5% NGS, 0.05% Triton X-100 in H-PBS (450 mm NaCl and 20 mm phosphate buffer, pH 7.4) for at least 1 hr at 4°C. Primary antibodies were added in blocking buffer, and cultures on coverslips were incubated overnight at 4°C. The following antibodies and dilutions were used for immunofluorescent staining: rabbit anti-synGAP, mouse anti-PSD-95, mouse anti-NMDA receptor subunit 1 (NR1) (1:500; Affinity BioReagents, Golden, CO), rabbit anti-GluR1 (1: 100; Chemicon, Temecula, CA), and rabbit anti-synapsin I (Cho et al., 1992) (1:1000). After three washes in blocking buffer, coverslips were incubated in goat anti-rabbit or goat anti-mouse secondary antibodies conjugated to Alexa 568 or Alexa 680 (1:200 each; Molecular Probes) at room temperature for 1 hr. Coverslips were washed once in blocking buffer for 15 min, twice in PBS for 15 min, postfixed for 5 min with 2% paraformaldehyde in PBS for 5-10 min, washed twice in PBS for 10 min, and mounted on slides with a drop of ProLong antifade reagent (Molecular Probes).
Immunoblotting. The hippocampus was dissected from postnatal day 1 (P1) brains and homogenized at 900 rpm in Teflon-glass homogenizers in 200 μl of RIPA (radioimmunoprecipitation assay) lysis buffer (50 mm Tris, pH 8, 2 mm EDTA, pH 8, 150 mm NaCl, 1% Nonidet P-40, 0.5% deoxycholate, 0.1% SDS, 0.5 mm DTT) containing a mixture of protease inhibitors (Complete; Roche, Indianapolis, IN). Cultured hippocampal neurons maintained for 10 d in vitro were lysed in the same buffer. Protein concentration was determined by the bicinchoninic acid method (Pierce, Rockford, IL) using bovine serum albumin as a standard. Protein samples (8-10 μg) were fractionated by SDS-PAGE and electrophoretically transferred to nitrocellulose filters (Schleicher & Schuell, Dassel, Germany). Filters were blocked for 1 hr in TBS, 0.1% Tween 20, and 5% milk at room temperature and then incubated with primary antibodies in blocking buffer overnight at 4°C. The antibodies were used at the following dilutions: rabbit anti-synGAP, 1:2000; mouse anti-PSD-95, 1:2500; mouse anti-α-CaMKII, 1:2000 (all from Affinity BioReagents, Golden, CO), rabbit anti-ERK (extracellular signal-regulated kinase) (Cell Signaling Technology, Beverly, MA), rabbit anti-NR1, 1:2000; mouse anti-MAP2, 1:500; mouse anti-synapsin I, 1:2000 (all from Chemicon); rabbit anti-GluR1, 1:1000; rabbit anti-GluR2/3, 1:1000 (all from Upstate Biotechnology, Lake Placid, NY); polyclonal anti-NR2A, 1:1000; and rabbit anti-NR2B antisera, 1:1000. Blots were then washed three times in blocking buffer and incubated in goat anti-rabbit IgG conjugated to either horseradish peroxidase (1:2000; Chemicon) or Alexa 700 or Alexa 800 (Molecular Probes). Protein bands were visualized by chemiluminescence using SuperSignal (Pierce) or with a fluorescence scanner (LICOR Biosciences, Lincoln, NE), according to the instructions of the manufacturer.
Viral constructs and infections. Sindbis-enhanced green fluorescent protein (eGFP) virus was provided by E. Schuman (California Institute of Technology, Pasadena, CA), and nsp2S-eGFP (nsp2S is a variant of Sindbis virus) was provided by P. Seeburg. Sindbis viruses expressing eGFP and FLAG epitope-tagged synGAPs were constructed as follows. Sequences encoding eGFP, internal ribosome entry site 2 (IRES2), and synGAP were inserted into viruses in tandem to permit independent expression of the two proteins with stronger expression of eGFP than of synGAP. DNA sequence encoding eGFP preceded by a Kozak sequence was obtained from pLP-IRES2-eGFP (BD Biosciences, San Jose, CA) by PCR with high fidelity Taq polymerase (Roche), a 5′-oligo containing a ClaI and XbaI linker, and a 3′-oligo containing a PstI linker. Similarly, DNA sequence encoding an IRES2 was obtained from pLP-IRES2-eGFP by PCR with a 5′-oligo upstream of a BamHI site in the vector and a 3′-oligo containing a NotI site. This procedure resulted in inclusion of a Kozak sequence at the 3′-end of the IRES. For construction of the virus expressing wild-type synGAP, a cDNA encoding flag-synGAP (gift from Dr. Jeong Oh, California Institute of Technology, Pasadena, CA) was amplified by PCR with a 5′-oligo containing a NotI site and a 3′-oligo containing a SacII site. The eGFP, IRES2, and synGAP segments were subcloned in tandem into the multiple cloning site of pBSK II (-) (eGFP-IRES2-flag-synGAP). DNA sequencing of this construct revealed no mutations. It was digested with XbaI and SacII, and the SacII site was blunted with ExoT (New England Biolabs, Beverly, MA) and ligated into the XbaI and StuI sites of pSinRep5 (Invitrogen) or nsp2S. Viral particles encoding eGFP-IRES2-wtSynGAP were produced according to the procedures provided by Invitrogen. Sindbis and nsp2S viral particles gave similar results and were used interchangeably.
The ΔSXV mutant was created by amplifying the flag-synGAP cDNA with a 5′-oligo containing a NotI site and a 3′-oligo designed to substitute a stop codon followed by a SacII site for the terminal 5 amino acids (QQTRV) and incorporated into nsp2S as described above.
To construct a virus expressing synGAP with an inactive GAP domain, three point mutations were created in the flag-synGAP sequence by site-directed mutagenesis: R470A, L580I, R581E. Mutations homologous to these three mutations reduce the catalytic activity of the GAP domain of p120 RasGAP severalfold (Skinner et al., 1991; Ahmadian et al., 1997). Mutagenic oligonucleotides (18-25 bases) that contained codons for the indicated mutations were synthesized at the Caltech Oligonucleotide Synthesis Laboratory. The oligonucleotides were phosphorylated at the 5′-end by T4 kinase and then annealed to the denatured synGAP plasmid (pSinRep5) at room temperature for 30 min. The oligonucleotides were extended with T4 DNA polymerase and T4 DNA ligase in vitro to generate a hemimethylated, double-stranded DNA molecule. A restriction digestion was performed with Dpn-1 to eliminate nonmutant plasmid DNA (those with two methylated strands). The DNA molecules were then transformed into the Escherichia coli mutS strain (deficient in the methylation-specific repair system), and colonies were screened by DNA sequencing for plasmids containing the desired mutations (Kramer et al., 1984). Viral particles were produced as described above.
Neurons were infected by adding the viruses (0.3-0.5 μl) to the culture medium on the ninth or 20th day in vitro. The neurons were incubated in normal growing conditions for 16-20 hr after infection and then fixed as described above.
Light, epifluorescence, and confocal microscopy. A Zeiss (Thornwood, NY) Axiovert 200 microscope was used for epifluorescence and light microscopy. Images were acquired with a Plan-Apochromat 63×/1.4 oil objective and a high-resolution CCD camera (Axiocam MRm) under the control of a computer equipped with AxioVision 3.1 (Zeiss). Exposure times were set so that pixel brightness was not saturated and were held constant during acquisition of all images (1300 × 1030 pixels) for each experiment. For some experiments, a Zeiss 510 laser-scanning microscope was used for confocal fluorescence microscopy. The pinhole aperture of the 510 microscope was set to 0.8 μm, and the zoom was set to 2.9. Images were 512 × 100-200 pixels; three to six optical sections, 1 μm each, were acquired for each view. The parameters for each channel (laser intensity, brightness, contrast, and gain) were set so that pixel brightness was not saturated and were held constant during acquisition of all images for each experiment. We found no difference between results acquired with the 510 confocal microscope and the Axiovert 200, so results with the two microscopes were pooled. In all experiments, neurons were first viewed with a green-fluorescence filter, and pyramidal-shaped neurons expressing GFP were imaged. Images of dendrites were acquired in two to four channels, as appropriate.
Image analysis. For images acquired with the Zeiss 510, Z-sections were first summed and projected in the z dimension (NIH ImageJ software). For analysis of area and brightness of fluorescent puncta of double- or triple-stained neurons, KS 300 software was used to create a mask of the dendrite (dendrite mask) from the images acquired in the GFP channel. The dendrite mask was applied to the corresponding images of PSD-95, NR1, GluR1, or synapsin immunofluorescence to exclude fluorescence from noninfected neurons in the field. The masked images were then automatically thresholded with the use of KS300 software to include only immunolabeled puncta in the analysis. The resulting binary images were saved for additional analysis. (In an independent analysis, the threshold was kept constant for all images in an experiment, and similar results were obtained.) Protrusions along the masked dendrites were outlined individually (Adobe Photoshop, Adobe Systems, San Jose, CA) by an investigator blinded to the genotype, and a second mask (protrusion mask) was created containing only the outlined protrusions.
The number of puncta and protrusions per 50 μm of dendrite and the area and mean pixel brightness of each punctum were obtained with the use of KS300 software. To obtain the width of protrusions, the protrusion mask was applied to each GFP image, and then the software drew a rectangle bounding the mask around each protrusion, and the length and width of each rectangle was recorded. Note that this method of automatic analysis tends to underestimate the magnitude of differences in width of spine heads between wt and ko, because bends in filopodia increase their recorded width. To obtain the percentage of puncta in protrusions or the percentage of protrusions containing a punctum (cluster), the intersection of the protrusion mask and the binary images of immunostained puncta was obtained. Then, the number of protrusions in the intersection was divided by the total number of protrusions, or the number of puncta in the intersection was divided by the total number of puncta. Values for width, area, and brightness were normalized to wt, and all values were imported into Prism software (GraphPad, San Diego, CA) for statistical analysis and graphing. All statistical analyses were performed using unpaired Student's t test or one-way ANOVA, as indicated. Significance was accepted as p < 0.05.
Although wtSynGAP was constructed with a FLAG-tag at the N terminus, we were unable to detect the tag by immunocytochemistry. Therefore, to estimate the expression of synGAP in neurons infected with Sindbis virus expressing recombinant synGAP, we applied the dendrite mask (described above) to the images of synGAP fluorescence in infected neurons. We measured the mean brightness of synGAP fluorescence within the mask (total brightness divided by area). We corrected these values by subtracting the mean brightness of ko neurons. Corrected average values for neurons expressing recombinant synGAP were normalized to the corrected average value for wt.
Electrophysiological recordings. Hippocampal neurons were maintained in culture for 9-10 d. Miniature EPSCs (mEPSCs) were recorded with a patch electrode (4-5 MΩ) in the whole-cell voltage-clamp mode (Axopatch 200; Axon Instruments, Foster City, CA). The bathing solution contained the following (in mm): 145 NaCl, 5 KCl, 0.5 MgCl, 2 CaCl2, 5 HEPES, 10 glucose, pH 7.4. Glutamatergic mEPSCs were pharmacologically isolated by adding tetrodotoxin (0.5 μm) and picrotoxin (30 μm) to the bath solution. The pipette solution contained the following (in mm): 145 potassium gluconate, 10 KCl, 5 NaCl, 2 MgCl2, 0.1 CaCl2, 5 EGTA, 5 HEPES, 2 ATP, pH 7.2. Recordings of mEPSCs were obtained for 3 min at a potential of -70 mV. Access resistance (15-18 MΩ) of the electrode was measured throughout the recordings and did not change >10%. Records were filtered at 2 kHz and digitized at 10 kHz. Parameters of mEPSCs were analyzed with custom-made software. Asymmetric events with a rise time shorter than the decay time and amplitude >4 pA (the threshold for event detection) were chosen for the analysis. Data were acquired from 10 wt and 9 ko neurons arising from three and four different embryos, respectively. Data from each group were averaged, and statistical significance of differences of means was determined by unpaired Student's t test.
Results
Construction and phenotype of synGAP ko mice
ES cells were transfected with a targeting construct that contained three loxP sites enclosing a neomycin selection cassette and genomic DNA, including exons 4-9 of synGAP (Fig. 1A). After selection for homologous recombination, mutant ES cells were transfected with a vector that transiently expresses Crerecombinase (see Materials and Methods). We screened these ES cells for mutants with a complete deletion of exons 4-9(ko). Two distinct ES cell colonies bearing the ko mutation were used to develop two independent lines of synGAP ko mice. Both of the lines express the same phenotypes, and we use them interchangeably. The genotype of individual mice is determined by PCR of genomic DNA as described in Materials and Methods (Fig. 1B).
The synGAP ko mutation results in a recessive lethal phenotype. On P0, homozygous ko mice are initially similar in size to wt, and a milk spot is often observed in their abdomens, suggesting that mortality is not caused by immediate malnutrition. Blood sugar levels are similar among littermates (data not shown). However, the ko mice rapidly develop a “failure-to-thrive” phenotype. They do not grow well and, by P1, become noticeably smaller than their littermates. They begin to weaken, display impaired motor skills and trembling, and generally die on P1 or by early P2. In contrast, cultured neurons derived from ko embryos appear healthy in vitro and are indistinguishable by phase microscopy from cultured neurons derived from wt littermates (data not shown). The neurons display levels of synGAP expected from their genotype (Fig. 1C). Heterozygous neurons have approximately half as much synGAP, and synGAP is not detectable in homozygotes. We did not detect differences in the amount of several other synaptic proteins in brain homogenates at P0 or in extracts of hippocampal cultures at 10 DIV (Fig. 1D), suggesting that expression of synaptic proteins is not altered by the synGAP deletion. Kim et al. (2003) also found no difference at P4 in the expression of synaptic proteins in their synGAP knock-out pups, which express a low level of short synGAP-like proteins from cryptic start sites and live until P5.
Development of spines in neuronal cultures
To investigate whether loss of synGAP affects the maturation of synapses in culture, we examined spine formation in dissociated hippocampal neurons cultured from individual E16-E17 embryos. We found that mature-looking spines appear earlier on dendrites of ko neurons than on those of wt neurons. We examined the morphology of dendritic protrusions on pyramidal shaped neurons (Fig. 2A) at 10 DIV by imaging neurons filled with heterologously expressed GFP (see Materials and Methods). There is a small increase in the number of protrusions on dendrites of ko neurons compared with wt. However, the more dramatic change is a shift in the distribution of morphologies of the protrusions in ko neurons toward that of mature spines with wide heads (Fig. 2A, arrowhead) and away from that of thin filopodia (Fig. 2A, arrow).
Figure 2.
SynGAP ko neurons have larger dendritic protrusions. A, Top, Representative confocal images of cultured hippocampal neurons at 10 DIV after expression of eGFP on 9 DIV (see Materials and Methods). Bottom, Projected z-sections of dendritic segments (∼40 μm) imaged by confocal microscopy. The population of protrusions on dendrites of ko neurons is shifted toward those with large heads resembling mature spines (right arrow), compared with the distribution of protrusions on dendrites of wt neurons. In wt neurons, thinner, elongated filopodia (left arrow) predominate. Scale bar, 5 μm. B, Quantification of number, width, and area of dendritic protrusions at 10 DIV. The number of protrusions in projected z-sections of 50 μm dendritic segments increased from 8.27 ± 0.52 in wt (n = 79) to 10.0 ± 0.48 in ko (n = 87; *p = 0.016). Widths and areas of protrusions normalized to wt are represented as cumulative histograms with bins of 0.2 normalized units. Both width and area are shifted toward higher values in ko neurons [width of protrusions (mean ± SEM); wt, 1.0 ± 0.01 vs ko, 1.12 ± 0.01, p < 0.0005, area of protrusions (mean ± SEM); wt, 1.0 ± 0.02 vs ko, 1.22 ± 0.02, p < 0.0005]. Note that the automated measurement of width underestimates the differences in head width between wt and ko (see Materials and Methods). Measurements were made of 653 protrusions on 79 dendrites from 29 wt neurons and 870 protrusions on 87 dendrites from 27 ko neurons.
Synaptic proteins
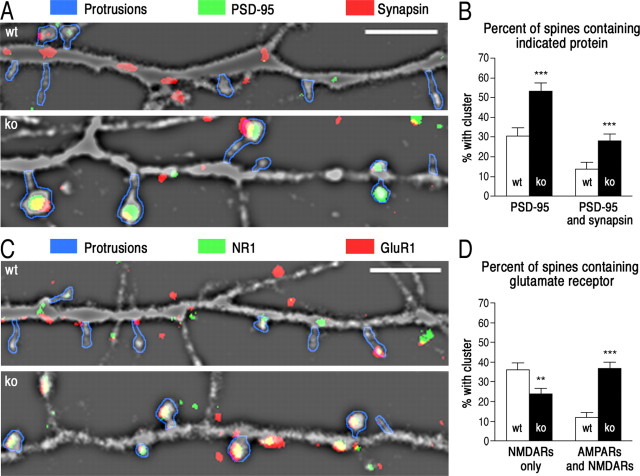
The hypothesis that spine maturation occurs earlier in ko than in wt neurons is supported by the sizes of clusters and subcellular distributions of postsynaptic proteins. Clusters of PSD-95 are present in dendrites of both wt and ko neurons (Fig. 3A, top); however, the clusters are significantly larger in ko neurons (Fig. 3B). The same is true for clusters of subunits of the NMDA-type (NR1) and AMPA-type (GluR1) glutamate receptors (Fig. 3A, bottom, B). The clusters of these proteins are also brighter in ko neurons (NR1, 1.10 ± 0.02-fold; GluR1, 1.49 ± 0.02-fold; PSD-95, 1.35 ± 0.02-fold), supporting the interpretation that, on average, each cluster in ko neurons contains more protein. In contrast, the areas of clusters of the presynaptic protein synapsin are only slightly altered in ko neurons (Fig. 3A,B).
Figure 3.

Clusters of postsynaptic proteins are larger and moved into protrusions in ko neurons. A, Representative images of dendritic segments (∼40 μm) from eGFP-expressing wt and ko neurons at 10 DIV prepared as described in Figure 2 and immunostained for PSD-95 and synapsin (top) or for NR1 and GluR1 (bottom), as described in Materials and Methods. PSD-95, NR1, and GluR1 immunostained puncta are larger in ko neurons. Scale bar, 5 μm. B, Areas of immunostained puncta in ko and wt neurons. Images were masked and thresholded as described in Materials and Methods. The mean areas of puncta in ko neurons were normalized to those of wt puncta. Area of NR1 puncta: wt, 1.0 ± 0.05; ko, 1.26 ± 0.05. Area of GluR1 puncta: wt, 1.0 ± 0.06; ko, 1.53 ± 0.06. Measurements were made of 506 NR1 puncta and 651 GluR1 puncta on 43 dendrites from 13 wt neurons and of 605 NR1 puncta and 666 GluR1 puncta on 52 dendrites from 12 ko neurons. Area of PSD-95 puncta: wt, 1.00 ± 0.09; ko, 1.92 ± 0.10. Area of synapsin puncta: wt, 1.0 ± 0.03; ko, 1.06 ± 0.04 (p = 0.228). Measurements were made of 264 PSD-95 puncta and 992 synapsin puncta on 36 dendrites from 16 wt neurons, and 246 PSD-95 puncta and 712 synapsin puncta on 35 dendrites from 12 ko neurons. C, Movement of postsynaptic proteins from the dendritic shaft into protrusions in ko neurons and wt neurons. We measured the percentage of PSD-95, NR1, and GluR1 puncta that overlap with dendritic protrusions (defined by the protrusion mask described in Materials and Methods). Percentage of PSD-95 clusters in protrusions (mean ± SEM): wt, 31.3 ± 4.6; ko, 67.8 ± 4.6; NR1 in protrusions: wt, 26.0 ± 3.1; ko, 53.5 ± 3.4; GluR1: wt, 30.0 ± 3.3; ko, 53.6 ± 3.3 (***p < 0.0005; two-tailed Student's t test).
Although the numbers of clusters of PSD-95, NR1, and GluR1 in dendrites are approximately the same in wt and ko neurons at 10 DIV, many more of the clusters are located at the tips of protrusions in ko neurons than in wt (Fig. 3A,C). Conversely, a larger proportion of protrusions contain a cluster of PSD-95 in ko neurons than in wt neurons at this age, and more of these protrusions are associated with a presynaptic synapsin puncta, indicating the presence of a synaptic junction (Fig. 4A,B). Finally, more protrusions in ko neurons contain both AMPA-type and NMDA-type glutamate receptors as opposed to NMDA-type receptors alone (Fig. 4C,D). Thus, by this criterion, the proportion of “silent” synapses is reduced in ko neurons compared with wt neurons at 10 DIV. Together, these data show that at 10 DIV, the population of protrusions in ko neurons has shifted substantially toward a mature spine phenotype when compared with wt neurons.
Figure 4.
At 10 DIV, more protrusions on ko neurons have characteristics of mature spines than on wt neurons. A, In ko neurons, more spines have a cluster of PSD-95 associated with a presynaptic terminal indicated by a synapsin puncta. Representative images of dendrites stained for PSD-95 (green) and synapsin (red). Immunostained puncta were identified as in Figure 3. The GFP image was made translucent with a high-pass filter for a better view of the distribution of PSD-95 (green) and synapsin (red) in dendrites. Protrusions were outlined by hand (blue for clarity). In ko neurons, more spines have a cluster of PSD-95 associated with a presynaptic terminal indicated by a synapsin puncta. B, The percentage of protrusions overlapping with a PSD-95 punctum or with both a PSD-95 and a synapsin puncta was determined as described in Materials and Methods. The percentage of protrusions containing PSD-95 increased from 30.5 ± 4.1% in wt neurons to 53.2 ± 4.4% in ko neurons. The percentage containing both PSD-95 and synapsin increased from 14 ± 3.4% in wt neurons to 28.0 ± 3.7% in ko neurons. Measurements were made on the same set of data analyzed in Figure 3. C, In ko neurons, more spines contain both AMPA and NMDA-type glutamate receptors than contain NMDA-type receptors alone. Representative images of dendrites stained for NR1 (green) and GluR1 (red) are presented as in Figure 4. Immunostained puncta were identified as in Figure 3. D, The percentage of protrusions containing only NMDA receptors decreased from 36 ± 3.5% in wt neurons to 24 ± 2.8% in ko neurons. Conversely, the percentage of spines containing both AMPA and NMDA receptors was greater in ko neurons (37 ± 3.0%) than in wt neurons (12 ± 2.5%). Measurements were made on the same set of data analyzed in Figure 3 (*p < 0.05, **p < 0.005, ***p < 0.0005; Student's t test).
Structure of mature spines in neuronal cultures at 21 d in vitro
To investigate whether the absence of synGAP has an effect on the structure of spines that have matured in culture, we examined the size and number of spines in hippocampal neuronal cultures at 21 DIV after infecting them with virus expressing GFP on day 20 (see Materials and Methods). Spines are significantly larger in synGAP ko neurons even at this late stage in culture (Fig. 5A,B). ko neurons have spines with a mean head width 1.2 times larger and a mean area 1.34 times larger than those in wt neurons (Fig. 5B). We did not find a significant difference in the density of spines per dendrite (15.5 ± 0.6 per 50 μm in wt vs 16.2 ± 0.8 per 50 μm in ko). We also stained neuronal cultures at 21 DIV with fluorescent antibody against PSD-95. As at 10 DIV, immunostained puncta were significantly larger (Fig. 5A,C) and brighter (data not shown) in ko neurons than in wt. These data indicate that synGAP plays a role in limiting the size of spines and of the postsynaptic density in mature synapses as well as in developing synapses.
Figure 5.
Spines and PSD-95 puncta remain larger in ko neurons than in wt neurons at 21 DIV. A, Representative confocal images of dendritic segments (∼50 μm) from cultured hippocampal neurons stained for PSD-95 on 21 DIV after expressing GFP on 20 DIV (see Materials and Methods). B, Quantification of the number, widths, and areas of spines. Widths and areas of spines are normalized to wt and represented as a cumulative histogram. Both are shifted toward larger values in ko neurons compared with wt neurons. Width of spines: (mean ± SEM) wt, 1.0 ± 0.01 versus ko, 1.2 ± 0.01. Area of spines: (mean ± SEM) wt, 1.0 ± 0.02 versus ko, 1.34 ± 0.03; p < 0.0005. Measurements were made of 876 protrusions on 57 dendrites from 13 wt neurons and 881 protrusions on 54 dendrites from 15 ko neurons. C, Quantification of area of PSD-95 puncta. PSD-95 images were masked and thresholded as described in Materials and Methods. Areas were measured with KS300 software and normalized to wt values. Area (mean ± SEM): wt, 1.0 ± 0.02 versus ko, 1.5 ± 0.04; ***p < 0.0005 (two-tailed Student's t test). Measurements were made of 3090 puncta on 50 dendrites from 13 wt neurons and 2533 puncta on 43 dendrites from 15 ko neurons.
mEPSCs in neuronal cultures
The morphological characteristics of ko neurons at 10 DIV suggests that they have more active synapses than wt neurons at the same age. To test this hypothesis, we compared the frequency and amplitude of mEPSCs in ko neurons to those in wt neurons at 10 DIV (Fig. 6). We found a higher frequency of mEPSCs in the ko neurons (Fig. 6C), confirming that more synaptic terminals release glutamate onto receptor clusters in ko neurons than in wt neurons at this age. In addition, the amplitude of the mEPSCs in ko neurons was considerably larger than in wt neurons, indicating that physiologically responsive glutamate receptor clusters are larger at synapses in ko neurons (Fig. 6A,B).
Figure 6.
Frequency and amplitude of mEPSCs are increased in cultured synGAP ko neurons at 10 DIV. A, Representative electrophysiological traces of spontaneous synaptic activity in wt and ko neurons. Three minute recordings of mEPSCs were obtained at a potential of -70 mV. B, Averaged mEPSCs illustrating the increase in peak amplitude in ko neurons compared with wt controls. C, Frequencies and peak amplitudes of mEPSCs. The mean frequency was higher in ko neurons (wt, 0.2 ± 0.03 Hz vs ko, 0.5 ± 0.1 Hz; **p < 0.005). The entire distribution of mEPSC amplitudes shown in the cumulative histogram is shifted to higher values in ko neurons (wt, 11.7 ± 0.5 pA vs ko, 19.8 ± 0.5 pA; p < 0.0001). Measurements were made of 181 mEPSCs from 10 wt neurons and 435 mEPSCs from 9 ko neurons isolated from 3 wt embryos and 4 ko embryos.
Rescue of the mutant phenotype by recombinant wild-type synGAP
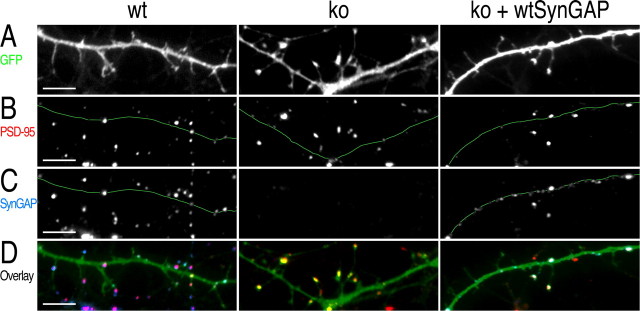
To test whether reintroduction of synGAP protein, or mutant versions of synGAP protein, into ko neurons can rescue the mutant cellular phenotypes we have described, we infected ko neurons with Sindbis viruses engineered to express both GFP and synGAP. To accomplish this, we introduced an IRES2 promotor driving expression of recombinant synGAP into the Sindbis virus following the GFP open-reading frame (see Materials and Methods). Eighteen hours after infection of neurons on day 9 in vitro with this virus, we observed expression levels of synGAP similar to those of endogenous synGAP in wt neurons (Figs. 7C, 8E), although the levels of expression were highly variable (Fig. 8E,SD bars). The location in spines of recombinant synGAP expressed in ko neurons was indistinguishable from that of endogenous synGAP in wt neurons (Figs. 7C, 8C).
Figure 7.
Transient expression of synGAP partially rescues precocious spine formation in hippocampal neurons at 10 DIV. Cultured hippocampal neurons were infected at 9 DIV with Sindbis viruses engineered to express the indicated proteins, as described in Materials and Methods. Neurons were fixed at 10 DIV and stained with antibodies against PSD-95 and synGAP. Quantitative analysis of the data from which the images are taken is shown in Figure 8. A, Representative images of dendritic segments from wt neurons expressing GFP, ko neurons expressing GFP, and ko neurons expressing both GFP and wtSynGAP. Transient expression of synGAP in ko neurons shifts the appearance of dendritic protrusions back toward that of wt. B, Immunostaining for PSD-95 in the dendritic segments shown in A. The location of the dendritic shaft is marked by a thin green line. In the ko neurons expressing synGAP, the percentage of protrusions containing PSD-95 puncta is reduced toward that of wt neurons. In contrast, the size and brightness of PSD-95 puncta still resembles that of ko neurons. C, Immunostaining for synGAP in the dendritic segments shown in A. The dendritic shaft is marked as in B. Antibodies against synGAP recognize recombinant wtSynGAP. The expression of wtSynGAP in dendrites of infected ko neurons was similar to that of endogenous synGAP in wt neurons. D, Overlay of GFP (green), PSD-95 (red), and synGAP (blue) images. Scale bars, 5 μm.
Figure 8.
Both the GAP domain and the PDZ domain-binding motif of synGAP are necessary to rescue precocious spine formation in hippocampal neurons at 10 DIV. A, Representative images of dendritic segments from ko neurons expressing GFP and either wtSynGAP, synGAP with a mutated GAP domain (GAP*), or synGAP with a deletion of the t-T/SXV domain (ΔSXV). Expression of wtSynGAP, but not GAP* or ΔSXV in ko neurons shifts the appearance of dendritic protrusions back toward that of wt. B, Immunostaining for PSD-95 in the dendritic segments shown in A. The location of the dendritic shaft is marked by a thin green line. The percentage of protrusions containing PSD-95 was shifted back toward that of wt after expression of wtSynGAP but not after expression of ΔSXV. In contrast, expression of GAP* caused a slight shift of the percentage of protrusions containing PSD-95 back toward wt that was not statistically significant (G). Finally, the brightness of PSD-95 puncta was not shifted at all toward wt by expression of wtSynGAP, GAP*, or ΔSXV. C, Immunostaining for synGAP in the dendritic segments shown in A. The dendritic shaft is marked as in B. The synGAP antibody recognizes recombinant wtSynGAP, GAP*, and ΔSXV. The expression of recombinant synGAP in dendrites of infected ko neurons was similar to that of endogenous synGAP in wt neurons. Also note that the t-T/SXV motif of synGAP is not necessary to target synGAP to dendritic protrusions. D, Overlay of GFP (green), PSD-95 (red), and synGAP (blue) images. Scale bars, 5 μm. E, Quantification of fluorescent staining for synGAP in dendrites measured as described in Materials and Methods. SynGAP brightness (percentage of wt; mean ± SD): wt, 100.0 ± 45.8; ko, 0.0 ± 12.3; ko plus wtSynGAP, 93.0 ± 53.1; ko plus GAP*, 90.0 ± 75.2; ko plus ΔSXV, 126.0 ± 55.2. Measurements were made of dendrites from 31 wt neurons prepared from five wt embryos and of dendrites of 29 ko neurons, 32 ko plus wtSynGAP neurons, 22 ko plus GAP* neurons, and 17 ko plus ΔSXV neurons from four ko embryos. Note that expression levels of synGAP in individual neurons are highly variable. F, Quantification of width of protrusions. Measurements were made as described in Materials and Methods. Protrusion widths (normalized to wt; mean ± SEM): wt, 1.0 ± 0.0; ko, 1.144 ± 0.085; ko plus wtSynGAP, 1.027 ± 0.023; ko plus GAP*, 1.110 ± 0.036; ko plus ΔSXV, 1.182 ± 0.017. Measurements were made of 3608 protrusions from 31 wt neurons prepared from five wt embryos and 3434 protrusions from 29 ko neurons, 3486 protrusions from 32 ko plus wtSynGAP neurons, 2068 protrusions from 22 ko plus GAP* neurons, and 1421 protrusions from 17 ko plus ΔSXV neurons prepared from four ko embryos. The experiment was repeated at least three times for each viral construct (wtSynGAP, GAP*, and ΔSXV), and similar results were obtained in each experiment. Therefore, the mean widths from all experiments were averaged. A p value of 0.0002 was obtained in a one-way ANOVA test, indicating a low probability that all of the means for each condition are the same. A Newman-Keuls post-test was performed to estimate the p values between each group. For wt versus ko, p < 0.01; for wt versus ko plus GAP*, p < 0.05; for wt versus ko plus ΔSXV, p < 0.01; for ko versus ko plus wtSynGAP, p < 0.05; and for ko plus wtSynGAP versus ko plus ΔSXV, p < 0.01. All other comparisons had p > 0.05 and thus are considered not significantly different. The same p value estimates were obtained using the Tukey test, except those for wt versus ko plus GAP*, which was p > 0.05. G, Quantification of percentage of protrusions containing PSD-95. Measurements were made as described in Materials and Methods. Percentage of protrusions containing PSD-95 (mean ± SEM): wt, 29.97 ± 2.38; ko, 46.51 ± 2.82; ko plus wtSynGAP, 28.47 ± 2.50; ko plus GAP*, 38.37 ± 3.55; ko plus ΔSXV, 48.97 ± 3.75. Measurements were made of 2530 PSD-95 puncta in dendrites of 31 wt neurons prepared from five wt embryos and 2023 PSD-95 puncta in dendrites of 29 ko neurons, 1974 PSD-95 puncta in dendrites of 32 ko plus wtSynGAP neurons, 2973 PSD-95 puncta in dendrites of 22 ko plus GAP* neurons, and 2509 PSD-95 puncta in dendrites of 17 ko plus ΔSXV neurons prepared from four ko embryos. A p value of <0.0001 was obtained in a one-way ANOVA test. A Newman-Keuls post-test was performed to estimate the p values between each group. For wt versus ko, p < 0.001; for ko versus ko plus wtSynGAP, p < 0.001; for wt versus ko plus ΔSXV, p < 0.01; for ko plus wtSynGAP versus ko plus ΔSXV, p < 0.01. All other comparisons had p > 0.05. The same p value estimates were obtained using the Tukey test. H, Quantification of brightness of PSD-95 puncta. Brightness of fluorescent PSD-95 puncta (normalized to wt; mean ± SEM): wt, 1.0 ± 0.046; ko, 1.461 ± 0.088; ko plus wtSynGAP, 1.386 ± 0.064; ko plus GAP*, 1.447 ± 0.077; ko plus ΔSXV, 1.844 ± 0.097. Measurements were made on the same data set used in G. A p value of <0.0001 was obtained in the one-way ANOVA test. A Newman-Keuls post-test was performed to estimate the p values between each group. For wt versus ko, ko plus wtSynGAP, ko plus GAP*, or ko plus ΔSXV, p < 0.001; for ko plus ΔSXV versus ko plus wtSynGAP, p < 0.001; for ko plus ΔSXV versus ko, or ko plus GAP*, p < 0.01. All other comparisons had p > 0.05. The same p value estimates were obtained using the Tukey test.
To test whether altered spine formation in the ko neurons could be reversed by transient expression of wt synGAP, we compared neurons infected at day 9 with Sindbis viruses engineered to express both wtSynGAP and GFP or GFP alone (Fig. 7). The reintroduction of synGAP into ko neurons shifted the appearance of protrusions toward that of wt neurons. The mean width of protrusions decreased from 1.140 ± 0.085-fold to 1.027 ± 0.023-fold of wt (Figs. 7, 8F). In addition, the percentage of protrusions that contained a cluster of PSD-95 was reduced from 46.5 ± 2.8% in ko neurons back to 28.5 ± 2.5%, the same level as in wt neurons (Figs. 7, 8G). Similarly, the proportion of total PSD-95 clusters that had moved into protrusions was reduced from 2.30 ± 0.15-fold of wt (p < 0.001) (Fig. 3C) back to 1.50 ± 0.20-fold of wt (p > 0.05; not statistically different from wt). These results indicate either that the process of spine maturation is highly dynamic and thus mostly reversible in <18 hr, or that the process began at approximately the same time as the expression of recombinant synGAP reached significant levels.
Interestingly, the transient expression of wtSynGAP did not reverse the effect of the ko mutation on the size (data not shown) and brightness of immunostained PSD-95 puncta (Figs. 7, 8H). This result suggests that formation of PSD-95 clusters occurred before significant expression of wtSynGAP began on day 9. Functional protein clusters exist in dynamic equilibrium with monomers; therefore, this result also suggests that the rate of exchange between large clusters of PSD-95 and monomers or small clusters must be quite slow. Thus, 18 hr is not long enough for newly introduced synGAP to shift the equilibrium back toward the smaller clusters observed in wt neurons on day 10.
Role of the GAP domain and the t-T/SXV motif in spine formation
To determine whether the Ras GTPase-activating activity of synGAP or its ability to bind to the PDZ domains of PSD-95 are important for spine formation, we introduced two mutations into the synGAP sequence of the Sindbis virus. The GAP domain was mutated at three residues important for its enzymatic function as described in Materials and Methods (GAP⋆). Mutation of homologous residues in p120 RasGAP dramatically reduces its GAP catalytic rate and its affinity for Ras (Skinner et al., 1991; Ahmadian et al., 1997). We also deleted the t-T/SXV domain of synGAP, which binds PSD-95, by removing the five C-terminal residues as described in Materials and Methods (ΔSXV). Hippocampal neurons were infected at 9 DIV with the viruses expressing each of the mutant synGAPs, and their effects were compared with those of wtSynGAP expressed in ko neurons (ko plus wtSynGAP) (Fig. 8).
The ΔSXV mutation did not alter the targeting of recombinant synGAP to spines (Fig. 8C). This result means that binding to PSD-95 is not necessary to target synGAP to dendritic spines. Nevertheless, the ΔSXV mutation renders synGAP unable to rescue most of the spine phenotypes observed in ko neurons at 10 DIV. Expression of ΔSXV in ko neurons does not decrease the width of protrusions at 10 DIV (Fig. 8A,F) and does not decrease the proportion of protrusions that contain PSD-95 (Fig. 8B,G). It also does not reduce the proportion of total PSD-95 clusters that have moved into protrusions (2.44 ± 0.27-fold of wt; p < 0.001). These data mean that an intact t-T/SXV domain is necessary for synGAP to retard widening of spine heads and movement of PSD-95 clusters into spines.
The GAP⋆ mutation also renders synGAP unable to rescue the cellular phenotype of the ko neurons, although its effects differ from those of the ΔSXV mutant. Expression of the GAP⋆ mutant of synGAP in ko neurons only slightly reduces the width of protrusions, in contrast to wtSynGAP, which reduces the width of protrusions to wt levels (Fig. 8A,F). Thus, a fully active GAP domain is necessary for the full reversal of spine width by synGAP. Expression of the GAP⋆ mutant in ko neurons reduces the mean percentage of spines containing PSD-95 approximately half as far as does expression of wtSynGAP (Fig. 8B,G). Thus, loss of GAP activity partially interferes with the ability of wtSynGAP to retard movement of PSD-95 into spines.
Expression of the GAP⋆ mutant in ko neurons has the same effect as expression of wtSynGAP on the proportion of total PSD-95 clusters found in protrusions at 10 DIV (1.49 ± 0.18-fold of that of wt), indicating that the GAP domain is not required for this effect of synGAP. Thus, the GAP domain appears less crucial than the t-T/SXV domain for the ability of synGAP to retard spine formation. It is important to keep in mind, however, that GAP activity is reduced but not abolished completely by the mutations that we introduced into the GAP domain (Skinner et al., 1991; Ahmadian et al., 1997). Therefore, residual GAP activity could account for the relatively weak effects of these mutations on spine formation. Nonetheless, the results indicate that both the t-T/SXV and the GAP domains are necessary for maximum rescue of precocious spine formation; however, the mechanisms by which they act are likely to be at least partially distinct.
Like expression of wtSynGAP, expression of the GAP⋆ mutant does not reverse the larger size of PSD-95 clusters (Fig. 8B,H; note that brightness correlates with area in these images). Notably, expression of the ΔSXV mutant actually increases their average size further. This result supports the conclusion that the GAP domain and the t-T/SXV domains affect different mechanisms during spine formation.
Discussion
Effects of synGAP deletion on spine formation and structure
Overexpression of certain PSD proteins can increase the size of spine heads (El-Husseini et al., 2000; Pak et al., 2001; Sala et al., 2001). In contrast, we found that elimination of expression of synGAP leads to precocious synapse formation in developing neurons in culture and an increase in the size of spine heads in mature neurons. This result suggests that synGAP normally retards synapse formation and decreases the size of spine heads, perhaps reflecting its regulatory function.
We have shown that neurons with a deletion of synGAP grow well in culture, and that spines and glutamatergic synapses form earlier in ko than in wt neurons cultured under the same conditions. This effect is most apparent at 10 DIV when most of the dendritic protrusions in wt neurons still have the appearance of fine filopodia; however, in ko neurons, a larger proportion have the appearance of mature spines with wide heads. Immunolabeled clusters of PSD-95, AMPA, and NMDA receptors are larger in ko neurons than in wt neurons, and many more of the clusters are located in protrusions that resemble spines, rather than in the dendritic shaft. In ko neurons, more spines are associated with presynaptic terminals containing synapsin, and more of them contain both AMPA and NMDA receptors, rather than NMDA receptors alone. Finally, the frequency and amplitude of mEPSCs are higher in ko neurons compared with wt neurons at 10 DIV. All of these observations demonstrate that spine and synapse formation is accelerated in cultured neurons in the absence of synGAP. The effect of loss of synGAP on the appearance of spines is not confined to developmental stages. In mature neurons at 21 DIV, spines and their associated clusters of PSD-95 are considerably larger in ko neurons than in wt.
Some of the effects on spine structure at 10 DIV can be reversed by transient expression of wt synGAP in ko neurons at 9 DIV. Eighteen hours after infection with a virus expressing synGAP, the level of synGAP in dendrites has returned to that of wt, and the population of protrusions has shifted back toward those resembling thin filopodia rather than mature spines. Thus, the timing of spine formation is directly affected by the absence or presence of synGAP on days 9 and 10, rather than indirectly affected by a change in the neurons caused by the absence of synGAP before day 9. This result means that the processes governing spine formation in young neurons are dynamic and can be shifted rapidly toward more or fewer mature spines.
Mechanisms of spine formation that are altered by synGAP deletion
Maximum reversal of precocious spine formation by synGAP requires both a fully active GAP domain and a t-T/SXV domain. Thus, both regulation of Ras inactivation by synGAP and its precise localization in the NMDA receptor signaling complex are important for controlling spine formation. However, the effects of mutations in these two domains on the reversal by synGAP are different, suggesting that they influence spine formation by at least partially different mechanisms.
SynGAP affects at least three distinct processes involved in spine formation; clustering of PSD proteins, widening of the spine head, and movement of PSD proteins into the spine head (Fig. 9). The absence of synGAP leads to formation of larger clusters of PSD-95, AMPA receptors, and NMDA receptors. This effect at day 10 DIV is not reversible by transient expression of synGAP at 9 DIV. Thus, formation of the clusters likely begins earlier than day 9 in vitro, and the clusters are stable enough that they do not decrease in size in response to expression of synGAP for 24 hr. Transient expression of the ΔSXV mutant dramatically increases the size of PSD-95 clusters. As yet, we have no mechanistic explanation for this apparent dominant negative effect.
Figure 9.
Cartoon of effects of synGAP on spine formation. Top, Left, At 10 DIV, most protrusions in wt neurons resemble filopodia. PSD-95 clusters are small and localized in the dendritic shaft. Right, Absence of synGAP (ko) enhances at least three distinct processes involved in spine formation, widening of the spine head, clustering of PSD proteins (represented by PSD-95), and movement of PSD proteins into the spine head. Bottom, Left, Reintroduction of wtSynGAP into ko neurons at 9 DIV was sufficient to reverse two of these processes but not clustering of PSD-95. Middle, Mutating the GAP domain (GAP*) prevents reversal of the effects on spine head width. Right, Deletion of the t-T/SXV domain (ΔSXV) prevents reversal of effects on both spine head width and on movement of PSD-95 into spines.
The absence of synGAP also leads to wider spine heads beginning at 10 DIV and continuing until at least 21 DIV. Transient expression of wtSynGAP at 9 DIV reverses this effect at 10 DIV; however, mutation of either the t-T/SXV or the GAP domain blocks the reversal. Thus, both domains participate in the role of synGAP in retarding widening of spine heads and prolonging the period of filopodial extension.
The GAP domain increases the rate of inactivation of GTP-bound Ras. In the absence of synGAP regulation of Ras in the postsynaptic compartment is likely disrupted. In many cells, Ras activation can lead to activation of the Rho family of GTP-binding proteins and to activation of phosphatidylinositol 3 kinase, both of which regulate the actin cytoskeleton (Kodaki et al., 1994; Rodriguez-Viciana et al., 1994, 1997; Hall, 1998). Thus, the importance of the GAP domain for broadening of the spine head might be explained by its putative effects on the cytoskeleton.
The absence of synGAP in ko neurons accelerates movement of PSD-95 into spine heads; reintroduction of synGAP or of the GAP⋆ mutant into ko neurons at 9 DIV reverses this effect. In contrast, introduction of the ΔSXV mutant produces no reversal of the effect and increases the size and brightness of clusters of PSD-95. One potential explanation is that synGAP competes for binding to the PDZ domains of PSD-95 with another protein that influences the size of PSD-95 clusters and/or their movement into spines. In the absence of the t-T/SXV domain of synGAP, more of this hypothetical protein might bind to PSD-95 and accelerate its clustering and/or movement into spines. This hypothesis is related to the more general one that altering the ratio among PSD proteins in dendrites can shift the equilibrium of spine size and/or size of protein clusters. This general hypothesis is supported by studies showing that overexpression of PSD-95 (El-Husseini et al., 2000), shank (Sala et al., 2001), and SPAR (Pak et al., 2001) in cultured neurons increases the size of spine heads and of clusters of PSD proteins. When it is present, SynGAP may compete for binding to PSD-95 with other proteins, and this competition may ultimately limit the size of clusters of PSD-95 and its associated proteins and their movement into spines.
Another possibility is that if synGAP cannot bind directly to PSD-95, its ability to effectively downregulate Ras is reduced. An increase in active Ras may lead to increased accumulation of PSD-95 clusters. Zhu et al. (2002) found that when Ras activity was transiently increased in hippocampal slices, AMPA but not NMDA receptor currents increased. According to their model, reduction of the GAP activity of synGAP would produce a higher level of active Ras at synapses and thus an increase in AMPA receptor currents. We do observe an increase in AMPA receptor current in developing cultured ko neurons. However, we also observe an increase in the size and number of clusters of NMDA receptors and PSD-95 at spines in these same neurons. Thus, the role of Ras in control of receptor numbers at synapses is complex.
SynGAP has other domains that may contribute to its functions, including a PH domain followed by a C2 domain located near the GAP domain. The PH domain of the related family member, p120 RasGAP, participates in regulation of its GAP activity (Drugan et al., 2000); thus, the PH domain of synGAP may also be an important functional determinant. We have not yet explored the role of these additional domains in control of spine formation.
Other synGAP knock-out mutations
Two other laboratories have made deletion mutations of synGAP. One mutation deletes parts of the gene encoding the C2 and GAP domain (Komiyama et al., 2002). Adult mice heterozygous for this mutation have abnormal long-term potentiation. The other mutation removes exons 7 and 8, which encode portions of the PH and C2 domains (Kim et al., 2003). A low level (∼2% of wt) of synGAP splice variants containing a GAP domain is still expressed in these mice. They begin to weaken at P3 and die at P5 (Kim et al., 2003), whereas mice homozygous for our deletion of exons 4-9 die by P2. The small amount of synGAP expression in the exon 7 and 8 deletion mutant may account for this difference. Kim et al. (2003) reported that cultured cortical neurons homozygous for the exon 7 and 8 deletion had 32% more AMPA receptor clusters at 18-20 DIV than wt neurons and ∼20% fewer silent synapses that contain only NMDA receptors and no AMPA receptors. This observation may reflect the same alterations in regulation of spine formation that we have described.
The findings presented here set the stage for a more detailed mechanistic analysis of the roles of Ras, PSD-95, and of various domains of synGAP in regulating spine formation and the size of the PSD.
Footnotes
This work was supported by National Institutes of Health Grants NS17660 and NS28710 to M.B.K., MH49176 to M.B.K. and H. A. Lester, and 1T32MH20069 to L.E.V. and by Swiss National Foundation Fellowship 823A-064694 to I.K. We thank Dr. Jeong Oh for the gift of recombinant synGAP, Dr. Henry Lester for help with electrophysiology, Drs. Peter Seeburg and Erin Schuman for gifts of plasmids, and Adeline Seah, Margaret Hainline, Dr. Gilberto Hernandez, Shannon O'Dell, and Alan Rosenstein for technical assistance. We also thank Edoardo Marcora for revisions of this manuscript and members of the Kennedy laboratory for many helpful discussions.
Correspondence should be addressed to Mary B. Kennedy, Division of Biology 216-76, California Institute of Technology, Pasadena, CA 91125. E-mail: kennedym@its.caltech.edu.
H.-J. Chen's present address: AmCyte Inc., 2825 Santa Monica Boulevard, Suite 200, Santa Monica, CA 90404-2429.
I. Knuesel's present address: Swiss Federal Institute of Technology, Laboratory of Behavioral Neurobiology, Schorenstrasse 16, 8603 Schwerzenbach, Switzerland.
Copyright © 2004 Society for Neuroscience 0270-6474/04/248862-11$15.00/0
References
- Ahmadian MR, Stege P, Scheffzek K, Wittinghofer A (1997) Confirmation of the arginine-finger hypothesis for the GAP-stimulated GTP-hydrolysis reaction of Ras. Nat Struct Biol 4: 686-689. [DOI] [PubMed] [Google Scholar]
- Brewer GJ, Torricelli JR, Evege EK, Price PJ (1993) Optimized survival of hippocampal neurons in B27-supplemented Neurobasal, a new serum-free medium combination. J Neurosci Res 35: 567-576. [DOI] [PubMed] [Google Scholar]
- Chen H-J, Rojas-Soto M, Oguni A, Kennedy MB (1998) A synaptic Ras-GTPase activating protein (p135 SynGAP) inhibited by CaM kinase II. Neuron 20: 895-904. [DOI] [PubMed] [Google Scholar]
- Cho KO, Hunt CA, Kennedy MB (1992) The rat brain postsynaptic density fraction contains a homolog of the Drosophila discs large tumor supressor protein. Neuron 9: 929-942. [DOI] [PubMed] [Google Scholar]
- Constantine-Paton M, Cline HT, Debski E (1990) Patterned activity, synaptic convergence, and the NMDA receptor in developing visual pathways. Annu Rev Neurosci 13: 129-154. [DOI] [PubMed] [Google Scholar]
- Drugan JK, Rogers-Graham K, Gilmer T, Campbell S, Clark GJ (2000) The Ras/p120 GTPase-activating protein (GAP) interaction is regulated by the p120 GAP pleckstrin homology domain. J Biol Chem 275: 35021-35027. [DOI] [PubMed] [Google Scholar]
- El-Husseini AE, Schnell E, Chetkovich DM, Nicoll RA, Bredt DS (2000) PSD-95 involvement in maturation of excitatory synapses. Science 290: 1364-1368. [PubMed] [Google Scholar]
- Hall A (1998) Rho GTPases and the actin cytoskeleton. Science 279: 509-514. [DOI] [PubMed] [Google Scholar]
- Kennedy MB (1997) The postsynaptic density at glutamatergic synapses. Trends Neurosci 20: 264-268. [DOI] [PubMed] [Google Scholar]
- Kennedy MB (1998) Signal transduction molecules at the glutamatergic postsynaptic membrane. Brain Res Brain Res Rev 26: 243-257. [DOI] [PubMed] [Google Scholar]
- Kennedy MB (2000) Signal-processing machines at the postsynaptic density. Science 290: 750-754. [DOI] [PubMed] [Google Scholar]
- Kim JH, Liao D, Lau L-F, Huganir RL (1998) SynGAP: a synaptic RasGAP that associates with the PSD-95/SAP90 protein family. Neuron 20: 683-691. [DOI] [PubMed] [Google Scholar]
- Kim JH, Lee HK, Takamiya K, Huganir RL (2003) The role of synaptic GTPase-activating protein in neuronal development and synaptic plasticity. J Neurosci 23: 1119-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodaki T, Woscholski R, Hallberg B, Rodriguez-Viciana P, Downward J, Parker PJ (1994) The activation of phosphatidylinositol 3-kinase by Ras. Curr Biol 4: 798-806. [DOI] [PubMed] [Google Scholar]
- Komiyama NH, Watabe AM, Carlisle HJ, Porter K, Charlesworth P, Monti J, Strathdee DJ, O'Carroll CM, Martin SJ, Morris RG, O'Dell TJ, Grant SG (2002) SynGAP regulates ERK/MAPK signaling, synaptic plasticity, and learning in the complex with postsynaptic density 95 and NMDA receptor. J Neurosci 22: 9721-9732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer B, Kramer W, Fritz HJ (1984) Different base/base mismatches are corrected with different efficiencies by the methyl-directed DNA mismatch-repair system of E. coli Cell 38: 879-887. [DOI] [PubMed] [Google Scholar]
- Malenka RC, Nicoll RA (1993) NMDA-receptor-dependent synaptic plasticity-multiple forms and mechanisms. Trends Neurosci 16: 521-527. [DOI] [PubMed] [Google Scholar]
- Oh JS, Chen H-J, Rojas-Soto M, Oguni A, Kennedy MB (2002) Erratum. Neuron 33: 151. [DOI] [PubMed] [Google Scholar]
- Pak DT, Yang S, Rudolph-Correia S, Kim E, Sheng M (2001) Regulation of dendritic spine morphology by SPAR, a PSD-95-associated RapGAP. Neuron 31: 289-303. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Viciana P, Warne PH, Dhand R, Vanhaesebroeck B, Gout I, Fry MJ, Waterfield MD, Downward J (1994) Phosphatidylinositol-3-OH kinase as a direct target of Ras. Nature 370: 527-532. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Viciana P, Warne PH, Khwaja A, Marte BM, Pappin D, Das P, Waterfield MD, Ridley A, Downward J (1997) Role of phosphoinositide3-OH kinase in cell transformation and control of the actin cytoskeleton by Ras. Cell 89: 457-467. [DOI] [PubMed] [Google Scholar]
- Sala C, Piech V, Wilson NR, Passafaro M, Liu G, Sheng M (2001) Regulation of dendritic spine morphology and synaptic function by Shank and Homer. Neuron 31: 115-130. [DOI] [PubMed] [Google Scholar]
- Sheng M, Kim E (2000) The Shank family of scaffold proteins. J Cell Sci 113: 1851-1856. [DOI] [PubMed] [Google Scholar]
- Sheng M, Sala C (2001) PDZ domains and the organization of supramolecular complexes. Annu Rev Neurosci 24: 1-29. [DOI] [PubMed] [Google Scholar]
- Sjostrom PJ, Nelson SB (2002) Spike timing, calcium signals and synaptic plasticity. Curr Opin Neurobiol 12: 305-314. [DOI] [PubMed] [Google Scholar]
- Skinner RH, Bradley S, Brown AL, Johnson NJE, Rhodes S, Stammer DK, Lowe PN (1991) Use of the Glu-Glu-Phe C-terminal epitope for rapid purification of the catalytic domain of normal and mutant ras GTPase-activating proteins. J Biol Chem 266: 14163-14166. [PubMed] [Google Scholar]
- Tybulewicz VL, Crawford CE, Jackson PK, Bronson RT, Mulligan RC (1991) Neonatal lethality and lymphopenia in mice with a homozygous disruption of the c-abl proto-oncogene. Cell 65: 1153-1163. [DOI] [PubMed] [Google Scholar]
- Xiao B, Tu JC, Worley PF (2000) Homer: a link between neural activity and glutamate receptor function. Curr Opin Neurobiol 10: 370-374. [DOI] [PubMed] [Google Scholar]
- Zhang W, Vazquez L, Apperson M, Kennedy MB (1999) Citron binds to PSD-95 at glutamatergic synapses on inhibitory neurons in the hippocampus. J Neurosci 19: 96-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JJ, Qin Y, Zhao M, Van Aelst L, Malinow R (2002) Ras and Rap control AMPA receptor trafficking during synaptic plasticity. Cell 110: 443-455. [DOI] [PubMed] [Google Scholar]