Abstract
In Drosophila, the transcription factor cAMP response element-binding protein 2 (dCREB2) has been reported to modulate the formation of long-term olfactory memory (LTM). Overexpression of a repressor isoform of CREB (dCREB2-b) under the control of a heat-shock promoter was reported to block LTM, whereas overexpression of an activator isoform (dCREB2-a) was reported to enhance LTM. A ratiometric model based on these results predicts that the balance of functional dCREB2-a and dCREB2-b provides a switch for memories to remain labile or to become enduring. We show here that the dCREB2-a transgene originally reported to enhance LTM carries a mutation that produces a translational reading-frame shift with the consequent formation of a stop codon at predicted amino acid position 79. Overexpression of this mutant dCREB2-a transgene or a corrected dCREB2-a transgene failed to show any enhancement of LTM. Overexpression of the dCREB2-b repressor transgene, in contrast, produced the anticipated block in LTM formation. We discuss the implications of these findings and propose an alternative model for the role of dCREB in Drosophila LTM.
Keywords: Drosophila, CREB, learning, memory, olfactory conditioning, behaviors
Introduction
A surprising observation was reported in 1995 (Yin et al., 1995a) that overexpression in Drosophila of the transcription factor cAMP response element-binding protein 2-a (dCREB2-a) markedly enhanced the formation of long-term olfactory memory (LTM). This form of dCREB is thought to activate changes in gene expression in response to intracellular elevations of cAMP and phosphorylation by protein kinase A (PKA). The critical behavioral experiments used transgenic animals in which a dCREB2-a coding cassette was expressed from a heat-shock promoter. Heat-shocked transgenic flies that were conditioned with a single training trial exhibited an enhancement of LTM that persisted for up to 7 d, whereas the memory of nonheat-shocked flies lasted only 1 d. In parallel studies (Yin et al., 1994), overexpression of a dCREB repressor (dCREB2-b) was reported to have the opposite effect, blocking the formation of LTM induced by a spaced training protocol. These data supported a model in which dCREB acts as a molecular switch for the formation of LTM in fruit flies, dependent on the ratio of functional dCREB activator and repressor in the relevant neurons (Yin and Tully, 1996; Belvin and Yin, 1997; Tully, 1997). The abundance of dCREB2-a was interpreted to be limiting for the formation of LTM such that its overexpression would lead to dCREB2-a-mediated gene transcription and long-lasting behavioral plasticity. The idea that dCREB functions as a switch for gating the formation of LTM has formed the rationale for attempts at developing new pharmacological therapies to treat different forms of memory dysfunction (Friedrich, 2000; Scott et al., 2002; Tully et al., 2003).
The dCREB2-a and dCREB2-b isoforms are two of at least seven different forms predicted from the alternative splicing of the primary transcript of the dCREB2 gene (Yin et al., 1995b), which is also known as dCREB-B (Usui et al., 1993). Three of these forms, dCREB2-b, -c, and -d, were isolated as cDNA clones. Four other splice variants (dCREB2-a, -q, -r, and -s) were identified by reverse transcription-mediated PCR (RT-PCR) after analysis of the genomic sequence revealed additional putative coding exons. The PKA-dependent transcriptional activity of the dCREB2-a and dCREB2-b isoforms was tested in F9 cells. The data suggested that the dCREB2-a isoform functioned as a PKA-dependent activator, whereas dCREB2-b functioned as a repressor of PKA-dependent activation by the dCREB2-a. These data were consistent with the view that the two isoforms exhibited opposing actions on gene expression.
One major unanswered question concerns the neurons that use dCREB2 activity for LTM. Many studies have documented the role of mushroom body (MB) neurons for short-term olfactory memory (STM) (Davis, 1996; Roman and Davis, 2001), and a recent study implicated one class of MB neurons for LTM (Pascual and Preat, 2001). In addition, a newly isolated set of enhancer detector lines that are putative new LTM mutants exhibit reporter expression within MB neurons (Dubnau et al., 2003). These observations are consistent with both STM and LTM being mediated by the same set of neurons (Isabel et al., 2004). We therefore began experiments using the GAL4-upstream activation sequence (UAS) system to direct expression of dCREB2-a and dCREB2-b to different regions of the Drosophila brain to define the neurons that require these molecules for their roles in LTM. In the process of creating reagents for our studies, we surprisingly uncovered a mutation in the heat-shock promoter-driven dCREB2-a transgene (hs-dCREB2-a) used in the original study. This mutation produces a frame shift and early termination in the open reading frame of the hs-dCREB2-a transgene. We report here the steps that led to the detection of this mutation and our attempts to replicate the critical behavioral experiments using original hs-dCREB2-a-containing lines. In addition, we report the behavioral characterization of two newly constructed UAS-dCREB2-a transgenic lines corrected for the original mutation.
Materials and Methods
Transgenic Drosophila and heat shock induction. Transgenic lines 17-2, C28, and C30 were maintained in the R. L. Davis and T. Preat Laboratory stock collections. After discovery of the deletion mutation in the Davis Laboratory copy of C28 and C30, additional copies of the stocks were requested and received from the following laboratories: C28 and C30 from Dr. J. Yin (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY) and C28 from Dr. J. Hirsh (University of Virginia, Charlottesville, VA). Forty to sixty flies per food bottle were cultured at 18°C. Flies were collected 2 or 3 d after eclosion, transferred to food vials, and kept overnight at 18°C before undergoing the heat-shock treatment as described previously (Yin et al., 1995a).
Molecular cloning. Genomic DNA from C28 and C30 fly lines was prepared following a quick protocol, as described in the Purogene DNA isolation Kit (Gentra Systems, Minneapolis, MN). To isolate the dCREB2-a cassette, the 23 base primer 5′-TAACCAGCAACCAAGTAATCAA-3′, which is specific to the hsp70 region of the P-element, was used as a forward primer, and the primer 5′-TCTAGATCAATCGTTCTTGGTCTGAC-3′ was used as the reverse primer. PCR products were subcloned into the topoisomerase (TOPO) vector pCR 2.1 and sequenced. The UAS-dCREB2-a mutant transgene was constructed by isolating the dCREB2-a coding cassette with a base deletion at amino acid 38 from the C30 line by PCR using the followingforward and reverse primers: forward, 5′-GCGGCCGCAACATGGACAACAGCATC-3′; reverse, 5′-TCTAGATCAATCGTTCTTGGTCTGAC-3′.
The restriction sites NotI and XbaI were engineered at the 5′ ends of the primers to facilitate cloning into the expression vector pPBretU (Roman et al., 1999). The corrected dCREB2-a transgene was constructed by replacing the first 139 bp (from the start codon to BstEII site) of the mutated dCREB2-a cassette with the identical sequence, but without the deletion, isolated from the dCREB2-b sequence. Standard procedures were used for germline transformation. The expression of UAS-dCREB2-a transgene was driven from the heat-shock GAL4 line 82-1-2.
A λZAPII cDNA library (5.3 × 106 independent clones) made from Drosophila heads was used in PCR reactions using the same primers listed above. PCR products were electrophoresed in 0.8% agarose gels, and a doublet band between 0.8 and 1.1 Kb was gel purified and cloned into pTOPO (Invitrogen, San Diego, CA). Colonies were screened by PCR for the presence of the different CREB isoforms. Three different classes of products were observed. Sequence analyses of the corresponding plasmids demonstrated these to represent the dCREB2-b, -c, and -d isoforms.
Total RNA was isolated from 50-100 Drosophila heads by using 1 ml of triazol (Invitrogen) per 100 mg of tissue. After homogenization, the samples were incubated at room temperature for 5 min. Two hundred microliters of chloroform were added per milliliter of triazol. Samples were centrifuged for 15 min at 12,000 × g. The aqueous phase was then precipitated with 0.5 ml of isopropanol at room temperature for 10 min and centrifuged at 12,000 × g for 10 min at 4°C. After washing with 75% ethanol, the pellet was air dried. RT-PCR was performed according to standard procedures. The following pair of 24-mers representing sequences in exons 1 and 7, respectively, was used for RT-PCR: forward, 5′-CAACTCGTCGGCGGCATCGGGCTC-3′; reverse, 5′-CCAGGCACTTGATGTACTCCTTCT-3′.
Western analysis. For protein analysis, flies were frozen in liquid nitrogen. Ten heads, separated by sieving, were homogenized directly in 100 μl of 1× SDS loading buffer. Samples were boiled for 5 min, and the debris was eliminated by centrifugation before loading. The equivalent of two heads was loaded in each well of a 15% SDS-polyacrylamide gel. Standard protocols were used for blotting. The membranes were probed with a 1:20 dilution of an anti-dCREB2-b monoclonal antibody, generously provided by Dr. J. Yin. We used a horseradish peroxidase-conjugated anti-mouse secondary antibody and ECL blotting reagents (Amersham Biosciences, Arlington Heights, IL) for detection. Blots were stripped following manufacturer protocols and reprobed with a monoclonal antibody against dynamin (BD Biosciences, San Jose, CA) to control for loading.
Olfactory conditioning. Olfactory conditioning was performed following the protocols outlined by Pascual and Preat (2001) and Isabel et al. (2004).
Results
Isolation of dCREB2 isoforms
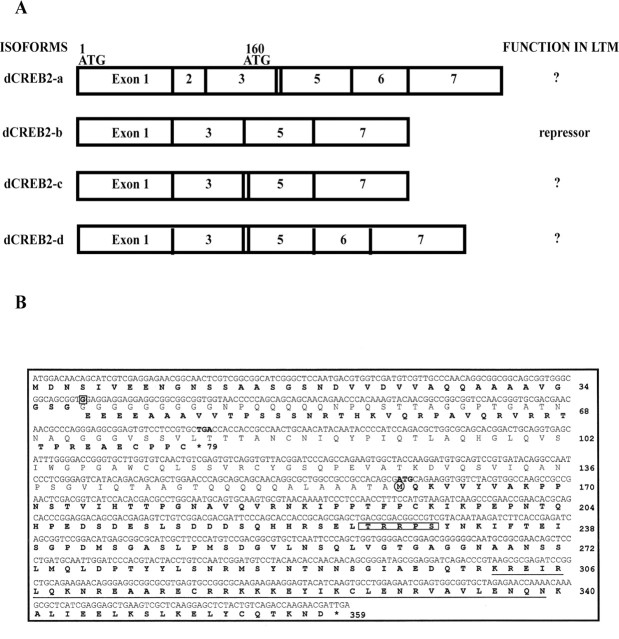
The previously reported organization of alternatively spliced variants of the dCREB2 gene is illustrated in Figure 1A. Primers spanning the 5′ and 3′ untranslated and coding regions of the dCREB2 gene were used to amplify dCREB2 messages by PCR from a λZAPII cDNA library of high complexity made from Drosophila heads. PCR products of 0.8 and 1.1 kb were observed on DNA agarose gels (data not shown) and subcloned into plasmid vectors. One hundred of these clones were screened for the presence of exon 2, which is present only in dCREB2-a, and in a smaller variant dCREB2-q (data not shown). The dCREB2-q variant was excluded from the cloned PCR products because of its small size. None of the 100 clones surveyed contained exon 2. Only clones representing the dCREB2-b, -c, and -d isoforms were identified based on the presence or absence of exons 2, 4, and 6. Thus, the RNAs represented by these analyses include only those coding for dCREB2-b, -c, and -d.
Figure 1.
Drosophila dCREB2 isoforms. A, Schematic representation of four different dCREB2 isoforms. B, DNA and protein sequence of the predicted full-length open reading frame and hs-dCREB2-a open reading frame in transgenes present in lines C28 and C30. A deletion of a single nucleotide, g (boxed), at amino acid position 38 produces a shift in reading frame to a new stop codon (bold) at amino acid position 79. The DNA sequence illustrated is the sequence with revisions (see Results) except for this single nucleotide g, which is preserved from the original sequence to illustrate the effect of its deletion on the open reading frame. An alternative and downstream start codon is highlighted in bold at position 160. The hypothetical and truncated version predicted by this open reading frame is shown in bold. The PKA-dependent phosphorylation recognition site is boxed and the bZIP domain is underlined.
dCREB2-a in transgenic lines C28 and C30
Two independent hs-dCREB2-a transgenic lines, named C28 and C30, were previously reported to enhance LTM (Yin et al., 1995a). We used genomic DNA from these lines to isolate the dCREB-a coding cassette for subsequent experiments. A PCR primer in the hsp70 promoter region of the P-element carrying the dCREB2-a transgene was used as a forward primer, and one in the 3′ end of the dCREB2-a gene was used as a reverse primer to ensure specific amplification of the transgenic form of dCREB2-a. PCR products were subcloned and sequenced. Surprisingly, we discovered that the sequences of the C28 and C30 dCREB2-a cassettes differed from the published sequence of the hs-dCREB2-a isoform (Yin et al., 1995a) at four sites. One base substitution (g→a) caused the change of glycine→serine at amino acid position 36. At amino acid position 47, the triplet ggt coding for glycine was missing. A single nucleotide difference (t→c) produced no amino acid change at tyrosine 353. The most significant difference mapped to amino acid position 38, where we found that a single g nucleotide was missing. This deletion shifts the reading frame at amino acid 38, leading to a stop codon at position 79. To exclude the unlikely possibility that a spontaneous mutation at this position had occurred in the transgenic lines of our own stock collection, we isolated and sequenced the dCREB2-a transgene from independent C28 and C30 fly stocks that were maintained in three other laboratories. The same differences were present in all of the lines tested. Inverse PCR (iPCR) was used to isolate genomic regions flanking the transgenes in the C28 and C30 lines. Sequence analysis of the iPCR clones revealed that these insertions mapped to the X and third chromosomes, respectively, therefore representing two independent isolates. This eliminated the possibility that either the C28 or C30 stocks had become mislabeled as the other since their construction.
The first open reading frame in the hs-CREB2-a transgene therefore predicts a peptide product encoding 78 amino acids, of which the first 37 match the previously reported protein sequence of dCREB2-a. It is possible but unlikely, however, that a full-length form of CREB2-a might be expressed as a result of reading frame shifts during translation or of nonsense codon suppression coupled with a reading frame shift during translation. Alternatively, translation might initiate at a downstream, but in-frame, start codon to produce a truncated molecule. The next available start codon within the transgene resides at amino acid 160. If used in vivo, this translational start predicts a putative protein product of 199 amino acids with a mass of 22 kDa (Fig. 1B). This hypothetical molecule retains the PKA-dependent phosphorylation site and the C-terminal basic region with the leucine zipper domain. These data indicate, therefore, that the open reading frame originally assigned as dCREB2-a cannot be expressed as a protein product in the C28 and C30 transgenics without invoking rare processes such as translation-coupled frameshifting and nonsense codon suppression.
dCREB2 expression in control and transgenic flies
We performed RT-PCR for the C28 and C30 lines in parallel with the hs-dCREB2-b repressor line 17-2 (Yin et al., 1994) and control flies Canton-S (CS), w1118 (wCS10), and ry506 (ry) to examine the induced and uninduced expression of dCREB2 RNAs. As controls, we synthesized PCR products with the same primer sets on plasmids carrying the dCREB2-a, -b, and -d isoforms. We observed that the control flies expressed three distinct but faint bands that by size comparison corresponded to dCREB2 isoforms -a, -b, and -d. The dCREB2 isoform-c may also be present but cannot be discriminated from dCREB2-b because only 12 base pairs of exon 4 distinguish these two isoforms. The hs-dCREB2-b and hs-dCREB2-a transgenes were both induced a few-fold by heat shock (Fig. 2). We also confirmed the presence and organization of dCREB2-a RNA by showing that primers specific to exons 2 and 6, exons that are both present in dCREB2-a, produced a RT-PCR product of a size predicted to represent the dCREB-a RNA in control flies. This product was sequenced to confirm the organization of dCREB-a illustrated in Figure 1A. These data, therefore, show that at least three isoforms of the dCREB2 gene are expressed in the heads of control flies. However, no information about the neurons in which they are expressed can be gathered from these data.
Figure 2.
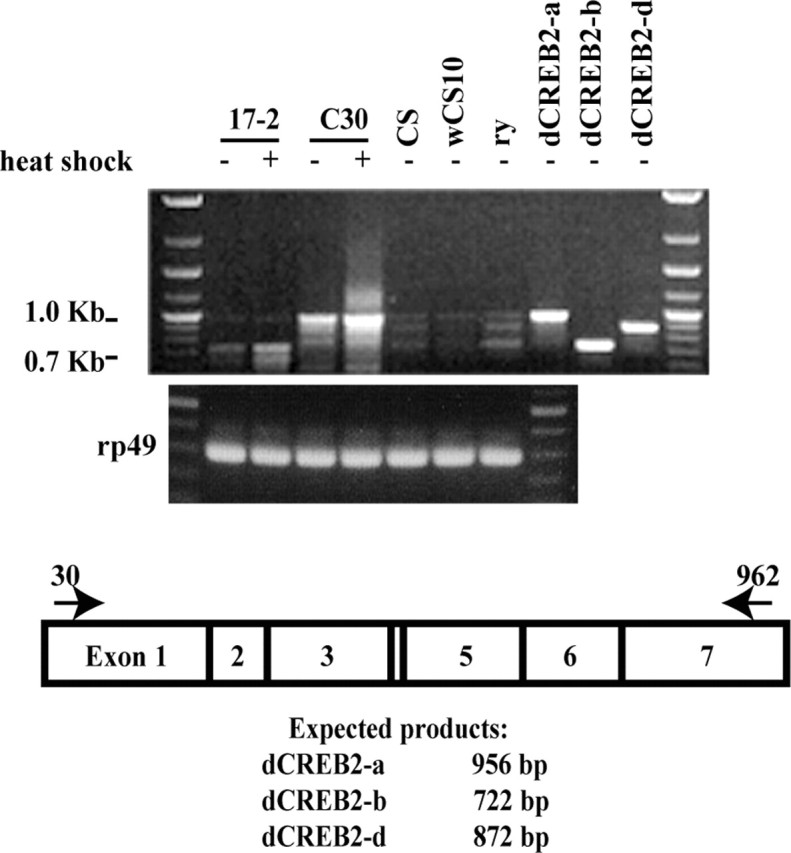
RT-PCR analysis of transgenic and control flies. RT-PCR was performed using total RNA isolated from heads from the hs-dCREB2-b line 17-2, hs-dCREB2-a line C30, and control lines CS, wCS10, and ry. Plasmids containing the dCREB2-a, -b, and -d isoforms were used as PCR templates to synthesize size standards using the same primers used for the RT-PCR. All flies were raised at 22°C, collected within 2 d from eclosion, and kept overnight at 25°C. Those that were treated with heat shock were transferred to empty food vials and placed in a 37°C water bath for 30 min. The flies were rested in food vials at 25°C for 3 hr after heat shock and then RNA was isolated. Heat shock treatment induced synthesis of hs-dCREB2-b and hs-dCREB2-a RNAs in flies carrying the respective transgenes. The level of the three detectable CREB splice variants in the control genotypes is very low. A schematic of the CREB gene exons with the location of the primers used is shown at the bottom of the figure along with the size expected for the PCR product of each CREB variant.
Western analysis of dCREB2 expression
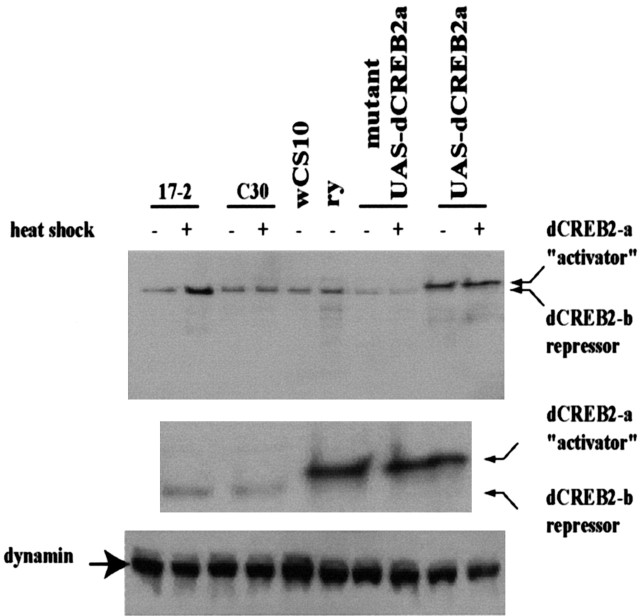
We used a monoclonal antibody raised against the dCREB2-b protein (Belvin et al., 1999) to analyze dCREB2 protein expression in extracts isolated from fly heads (Fig. 3). The dCREB2-b protein was detected as a band on Western blots that migrated at 33,500 Da [predicted molecular weight (MW), 31,000]. The expression level of the repressor isoform in hs-CREB2-b line 17-2 increased several-fold with heat shock, as predicted (Fig. 3). A protein with the same mobility whose expression did not increase with heat shock was detected in the hs-dCREB2-a lines C28 and C30 (data shown only for C30) and in extracts of control flies wCS10 and ry (Fig. 3). We were unable to detect a protein band corresponding to the predicted molecular weight of the dCREB2-a isoform potentially initiated at either Met1 or Met160 (Fig. 1) of the hs-dCREB2-a transgene, despite the fact that dCREB2-b shares all sequences with dCREB2-a. To determine whether the anti-dCREB2-b antibody recognizesepitopes found in the full-length form of dCREB2-a, we assayed protein extracts from flies expressing the dCREB2-a full-length protein from a transgene that was corrected for the base deletion that leads to the translational frameshift (predicted MW 39,500 Da). A protein band of 38,500 Da was detected in these flies, although only very modest increases in expression have been observed using the heat-shock GAL4 (hs-GAL4) driver (Fig. 3). This genotype was also used as a control to mark the mobility position of any dCREB2-a protein of full length that may arise from the mutant hs-dCREB2-a transgene in C30 by nonsense codon suppression and/or reading-frame shifts during translation. We found that in the hs-dCREB2-a line C30, as well as in a mutant UAS-dCREB2-a version, the only detectable product corresponded to the endogenous dCREB2-b protein. These results indicate that the anti-dCREB2-b antibody recognizes epitopes present on a full-length dCREB2-a protein, and that the full-length dCREB-a protein is not detectable in the C28 (data not shown) or C30 lines, nor in the mutant UAS-dCREB2-a line (Fig. 3).
Figure 3.
Western analysis of dCREB2 isoforms. Total protein was prepared from 10 fly heads from line 17-2 expressing the hs-dCREB2-b transgene, C30 expressing the hs-dCREB-a mutant transgene, control lines wCS10 and ry, and UAS-CREB2-a lines containing the dCREB2-a mutant cassette and the dCREB2-a full-length isoform (line T7.1), expressed from a hs-GAL4 element. The equivalent of two heads was loaded in each well. The blot was probed with a 1:20 dilution of a mouse monoclonal antibody raised against a bacterially expressed full-length dCREB2-b protein. The middle panel is a magnified image of the last four lanes of the top panel. This better illustrates the size difference of dCREB2-a and dCREB-b. To control for loading, the blot was stripped and reprobed with a 1:100 dilution of a mouse monoclonal anti-dynamin antibody (bottom).
Memory formation in dCREB2 transgenic flies
We used olfactory classical conditioning to assay the LTM of flies carrying transgenes with mutant and corrected dCREB2-a open reading frames, as well as flies carrying the dCREB2-b repressor. In the first set of experiments, we trained flies with a spaced conditioning protocol consisting of five training sessions (5×) with 15 min rest intervals between training sessions. Training with multiple but spaced sessions induces LTM persisting for several days (Tully et al., 1994; Pascual and Preat, 2001). Conditioned C30 flies carrying the dCREB2-a mutant transgene and CS control flies were tested for LTM 4 d after training. We found no significant difference between the two genotypes confirming that control and C30 flies without heat shock were able to form normal LTM persisting at least 4 d after spaced training (Fig. 4A). A second conditioning protocol consisted of a single training trial (1×) also followed by testing 4 d after conditioning. Overexpression of the hs-dCREB2-a transgene in line C30 produced by heat shock 3 hr before training was previously reported to eliminate the requirement for multiple, spaced training sessions for the establishment of maximal LTM (Yin et al., 1995a). Two separate experiments were performed in attempt to replicate these previous results. The results of the two separate experiments were equivalent and the pooled results are illustrated in Figure 4A. Induction of the mutant dCREB2-a transgene 3 hr before 1× training (Yin et al., 1995a) failed to elevate memory performance to levels produced by 5× training. Indeed, the performance of C30 flies after 1× training was not significantly different from any other group given 1× training. To ensure that the dCREB2-a mutant transgene was actually induced by heat shock, we removed a subset of animals after heat shock at the time they would normally have been trained and analyzed dCREB2 expression by RT-PCR analysis. The hs-dCREB2-a animals showed the expected increases in dCREB2-a mRNAs after heat shock (Fig. 4B).
Figure 4.
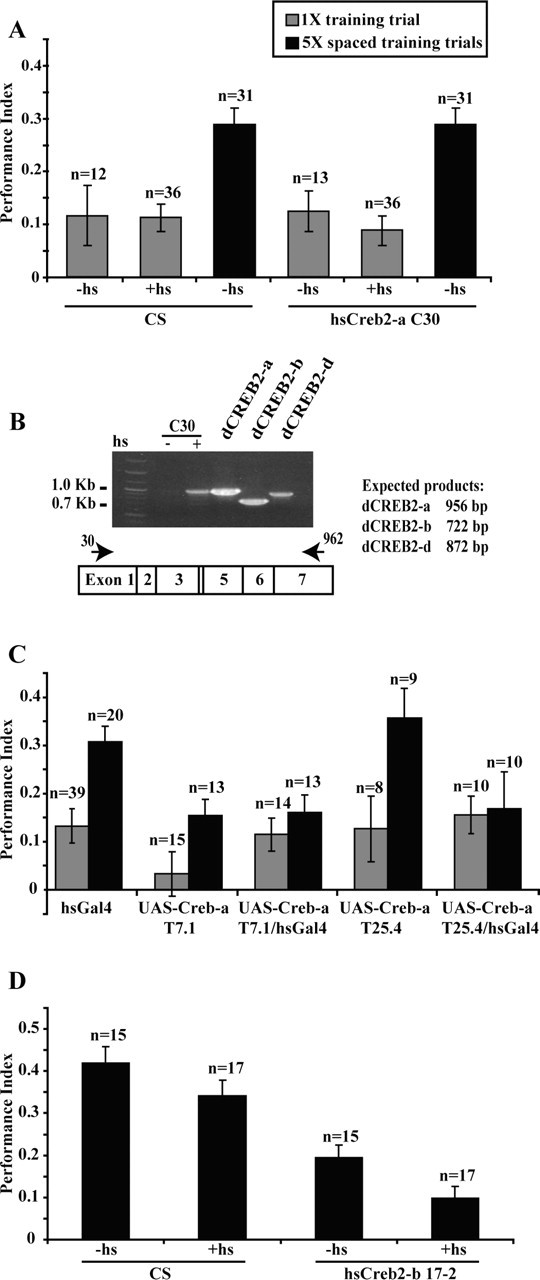
Induction of hs-dCREB2-a and of UAS-dCREB2-a has no effect on memory performance. A, Performance indices are shown for flies that were conditioned 3 hr after heat shock (+hs) or without heat shock (-hs) using a spaced training protocol consisting of five training trials with 15 min rest intervals or a single training trial. Tests were performed 4 d after training. Performance indexes were calculated as described previously (Pascual and Preat, 2001). ANOVA with Fisher's post hoc tests was used to make several important comparisons: the 5× CS (-hs) group was significantly different from both the 1× CS (-hs) (p = 0.0009) and 1× CS (+hs) (p < 0.0001) groups. The 5× hs-dCREB2-a C30 (-hs) group was significantly different from both the 1× hs-dCREB2-a C30 (-hs) (p = 0.0032) and 1× hs-dCREB2-a C30 (+hs) (p = 0.0028) groups. No significant differences were found between the 5× CS (-hs) and 5× hs-dCREB2-a C30 (-hs) groups (p = 0.2708). No significant differences were found between any of the groups provided with 1× training. These comparisons illustrate the significant effect of 5× spaced over 1× training and the lack of an effect of dCREB2-a induction in the hs-dCREB2-a C30 line on LTM. B, RT-PCR analyses of the transgenic flies used for training (hs-dCREB2-a C30) in A showed the heat shock-dependent induction of the transgene. Plasmids containing the dCREB2-a, -b, and -d isoforms were used as templates for PCR to generate size standards. C, Behavioral analysis of UAS-dCREB2-alines T7.1 and T25.4 containing the corrected dCREB2-a open reading frame and driven by hs-GAL4 showed no improvement in memory performance after 1× training compared with the wild-type control. ANOVA with Fisher's post hoc tests was used to make several comparisons. Most importantly, the 1× UAS-dCREB2-a T7.1 and 1× UAS-dCREB2-a T7.1-hsGAL4 groups were not significantly different (p = 0.2237), and the 1× UAS-dCREB2-a T25.4 and 1× UAS-dCREB2-a T25.4-hsGAL4 groups were not significantly different (p = 0.7323). The 5× hsGAL4 group was significantly different from the 1× hsGAL4 group (p = 0.0005), and the 5× UAS-dCREB2-a T25.4 group was significantly different to the 1× UAS-dCREB2-a T25.4 group (p = 0.0095). No significant differences were found between the 1× and 5× trained groups for UAS-dCREB2-a T7.1, UAS-dCREB2-a T7.1-hsGAL4, and UAS-dCREB-a T25.4-hsGAL4. The reason for failing to observe the effect of 5× spaced training for the latter three groups is unclear. D, Twenty-four hour memory after 5× spaced training with and without heat shock was measured for wild-type flies (CS) and the dCREB2-b repressor line 17-2. ANOVA with Fisher's post hoc tests was used to make several important comparisons; no significant differences were found between the CS (-hs) and CS (+hs) groups (p = 0.1064). The hs-dCREB2-b 17-2 (-hs) group was significantly different from both the CS (-hs) (p < 0.0001) and CS (+hs) (p < 0.0001) groups, showing the effect of basal expression of the transgene. The hs-dCREB2-b 17-2 (+hs) group was significantly different from all groups but most importantly from the hs-dCREB2-b 17-2 (-hs) group (p = 0.0449), showing the effect of induced expression of the transgene. Error bars indicate SEM.
To test whether LTM after a single training trial would be enhanced in flies expressing a corrected dCREB2-a open reading frame, we used the hs-GAL4 driver to drive the expression of two independent UAS-dCREB2-a transgenic lines, T7.1 and T25.4, each corrected for the original mutation. Unexpectedly, however, we discovered that heat shock-induced overexpression of the corrected dCREB2-a transgene produced a slow lethality, despite the marginal induction of the protein with heat shock (Fig. 3), at least in head extracts. Four days after a 30 min heat shock at 37°C, >50% of the flies were dead, with lethality reaching 99% by day 7.
Although the lethal phenotype indicated that the corrected dCREB2-a transgenes had biological function, this compromised the use of heat shock with flies containing these transgenes because of the probable physiological deterioration during the slow death and the unequivocal need to use healthy flies for behavior analyses. However, Western analysis on flies carrying the corrected transgene indicated a very significant basal level of expression in the absence of heat shock, well above the undetectable level in control flies (Fig. 3, data shown for T7.1). We therefore trained and tested these flies without heat shock, reasoning that the basal level of expression of the corrected dCREB2-a protein was well above control levels. We tested flies carrying UAS-dCREB2-a T7.1 (corrected), UAS-dCREB2-a T25.4 (corrected), both corrected transgenes in combination with hs-GAL4, and hs-GAL4 as a control. The anticipated effect of 5× spaced training over 1× training on LTM was observed for hs-GAL4 and UAS-dCREB2-a T25.4. The UAS-dCREB2-a T7.1-hs-GAL4, UAS-dCREB2-a T7.1-CS, and UAS-dCREB2-T25.4-hs-GAL4 flies, however, did not show the expected effect of 5× spaced training. Indeed, 5× spaced training of these flies produced 4 d memory that was not significantly different from that produced with 1× training, and both, in turn, were not significantly different from the 1× memory scores for the other lines tested (Fig. 4C). Therefore, overexpression of the corrected dCREB2-a protein failed to improve memory when tested at 4 d (Fig. 4C).
We also tested the hs-CREB2-b repressor line 17-2 (Yin et al., 1994) to determine whether expression of this molecule impairs LTM. We unexpectedly observed a reduction in 24 hr memory of nonheat-shocked 17-2 flies relative to CS. We also observed an additional significant reduction in LTM with heat shock. Therefore, overexpression of the dCREB2-b repressor did impair LTM.
Discussion
We provide evidence here that the hs-dCREB2-a transgene originally reported to enhance LTM carries a deletion of one base coding for amino acid 38, and that this mutation produces a reading-frame shift with the consequent formation of a stop codon at position 79. We also provide evidence that an antibody capable of detecting the predicted dCREB2-a protein fails to detect an expressed product in the hs-dCREB2-a transgenic flies. We additionally assayed the hs-dCREB2-a transgenic animals before and after heat shock in an attempt to reproduce the enhancement in LTM that has been previously reported. These experiments failed to reveal improvement of LTM in the hs-dCREB2 flies relative to the control flies. Finally, we overexpressed a corrected dCREB2-a protein using the GAL4-UAS system. This overexpression also failed to produce memory enhancement after a single conditioning trial.
It is difficult to reconcile our observations with those of the original report. However, there are three possible, but unlikely and partial, explanations. First, it is possible that there exists in the hs-dCREB2-a transgenic animals a low level of nonsense codon suppression and/or reading-frame shifts during translation that would allow the nonsense codon in the hs-dCREB2-a mRNA to be suppressed or eliminated by frameshifts during translation. This hypothesis predicts the existence of the full-length hs-CREB2-a protein after heat shock in the transgenic animals. We failed to detect this molecule by Western blotting, either because it is not there or because it is too rare to be detected. We also failed to find any improvement in LTM using flies overexpressing a corrected version of dCREB2-a. Second, it is possible that an ATG downstream from the first codon in the long open reading frame is used to initiate translation. Inspection of the sequence revealed that there is a second ATG in frame with the original reading frame that predicts a protein with 199 amino acids. This hypothetical and truncated dCREB2-a product retains the PKA phosphorylation site and the bZIP domains. We cannot exclude the possibility that this shorter protein product, which is predicted to be 22,000 Da and missing 45% of the N-terminal sequences, is expressed from the mutated hs-dCREB2-a transgene. However, we failed to find any evidence for its existence. Furthermore, it seems unlikely that the truncation of nearly half of the protein would result in a molecule with the same function as the full-length dCREB2-a transcription factor. It is possible, nonetheless, that this hypothetical mini-dCREB2-a molecule has some alternative function, perhaps through interactions with other members of the ATF family of transcription factors to produce memory enhancement. Finally, there is the possibility that unknown factors influence the behavioral assays such that the hs-dCREB2-a protein enhances memory under some circumstances but not others. Behavior is very sensitive to unknown variables and one of these could be held accountable for the discrepancy in the behavioral results.
A recent study by Balschun et al. (2003) questioned the role of CREB in hippocampal synaptic plasticity and hippocampal-dependent LTM after failing to demonstrate impairments of hippocampal long-term synaptic plasticity and LTM in mice lacking all CREB isoforms in the brain. In light of this report using the mouse and of our findings with Drosophila, it is difficult to accept the ratiometric model of dCREB2-a and dCREB2-b function as a molecular switch for LTM. A simpler model would use only the dCREB2-b function as an LTM suppressor, whose suppressing functions are alleviated under the appropriate training conditions. Clearly, the role of CREB in the formation of LTM in Drosophila is far from being understood.
Footnotes
This research was supported by National Institutes of Health Grant NS19904, the R. P. Doherty-Welch Chair in Science to R.L.D., and a Human Frontier Science Program grant to T.P.
Correspondence should be addressed to Ronald L. Davis, Department of Molecular and Cellular Biology, Baylor College of Medicine, Houston, TX 77030. E-mail: rdavis@bcm.tmc.edu.
Copyright © 2004 Society for Neuroscience 0270-6474/04/248823-06$15.00/0
B.P. and G.I. contributed equally to this manuscript.
References
- Balschun D, Wolfer DP, Grass P, Mantamadiotis T, Welzl H, Schütz G, Frey JU, Lipp HP (2003) Does cAMP response element-binding protein have a pivotal role in hippocampal synaptic plasticity and hippocampus-dependent memory? J Neurosci 23: 6304-6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belvin MP, Yin JC (1997) Drosophila learning and memory: recent progress and new approaches. Bioessays 19: 1083-1089. [DOI] [PubMed] [Google Scholar]
- Belvin MP, Zhou H, Yin JC (1999) The Drosophila dCREB2 gene affects the circadian clock. Neuron 22: 777-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RL (1996) Physiology and biochemistry of Drosophila learning mutants. Physiol Rev 76: 299-317. [DOI] [PubMed] [Google Scholar]
- Dubnau J, Chiang AS, Grady L, Barditch J, Gossweiler S, McNeil J, Smith P, Buldoc F, Scott R, Certa U, Broger C, Tully T (2003) The staufen/pumilio pathway is involved in Drosophila long-term memory. Curr Biol 13: 286-296. [DOI] [PubMed] [Google Scholar]
- Friedrich MJ (2000) Reseach with Drosophila provides clues to enhancing human memory. JAMA 284: 2857-2858. [DOI] [PubMed] [Google Scholar]
- Isabel G, Pascual A, Preat T (2004) Exclusive consolidated memory phases in Drosophila Science 304: 1024-1027. [DOI] [PubMed] [Google Scholar]
- Pascual A, Preat T (2001) Localization of long-term memory within the Drosophila mushroom body. Science 294: 1115-1117. [DOI] [PubMed] [Google Scholar]
- Roman G, Davis RL (2001) Molecular biology and anatomy of Drosophila olfactory associative learning. Bioessays 23: 571-581. [DOI] [PubMed] [Google Scholar]
- Roman G, He J, Davis RL (1999) New series of Drosophila expression vectors suitable for behavioral rescue. Biotechniques 27: 54-56. [DOI] [PubMed] [Google Scholar]
- Scott R, Bourtchuladze R, Gossweiler S, Dubnau J, Tully T (2002) CREB and the discovery of cognitive enhancers. J Mol Neurosci 19: 171-177. [DOI] [PubMed] [Google Scholar]
- Tully T (1997) Regulation of gene expression and its role in long-term memory and synaptic plasticity. Proc Natl Acad Sci USA 94: 4239-4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tully T, Preat T, Boynton SC, Del Vecchio M (1994) Genetic dissection of consolidated memory in Drosophila Cell 79: 35-47. [DOI] [PubMed] [Google Scholar]
- Tully T, Bourtchouladze R, Scott R, Tallman J (2003) Targeting the CREB pathway for memory enhancers. Nat Rev Drug Discov 2: 267-277. [DOI] [PubMed] [Google Scholar]
- Usui T, Smolik SM, Goodman RH (1993) Isolation of Drosophila CREB-B: a novel CRE-binding protein. DNA Cell Biol 12: 589-595. [DOI] [PubMed] [Google Scholar]
- Yin JC, Tully T (1996) CREB and the formation of long-term memory. Curr Opin Neurobiol 6: 264-268. [DOI] [PubMed] [Google Scholar]
- Yin JC, Wallach JS, Del Vecchio M, Wilder EL, Zhou H, Quinn WG, Tully T (1994) Induction of a dominant negative CREB transgene specifically blocks long-term memory in Drosophila Cell 79: 49-58. [DOI] [PubMed] [Google Scholar]
- Yin JC, Del Vecchio M, Zhou H, Tully T (1995a) CREB as a memory modulator: induced expression of a dCREB2 activator isoform enhances long-term memory in Drosophila Cell 81: 107-115. [DOI] [PubMed] [Google Scholar]
- Yin JC, Wallach JS, Wilder EL, Klingensmith J, Dang D, Perrimon N, Zhou H, Tully T, Quinn WG (1995b) A Drosophila CREB/CREM homolog encodes multiple isoforms, including a cyclic AMP-dependent protein kinase-responsive transcriptional activator and antagonist. Mol Cell Biol 15: 5123-5130. [DOI] [PMC free article] [PubMed] [Google Scholar]