Abstract
A variation in the BDNF gene (val66met) affects the function of BDNF in neurons, predicts variation in human memory, and is associated with several neurological and psychiatric disorders. Here, we show that, in magnetic resonance imaging scans of a large sample of normal individuals, this polymorphism affects the anatomy of the hippocampus and prefrontal cortex, identifying a genetic mechanism of variation in brain morphology related to learning and memory.
Keywords: Alzheimer, Down syndrome, gene, Huntington, memory, morphometry, neurotrophic, schizophrenia, BDNF, voxel-based, VBM, imaging genomics
Introduction
Brain-derived neurotrophic factor (BDNF) modulates hippocampal plasticity and hippocampal-dependent learning and memory in physiologic models and in animals (Lu and Gottschalk, 2000). A common polymorphism in the BDNF gene (val66met) in the 5′ signal domain has been shown to affect intracellular packaging and regulated secretion of BDNF (Egan et al., 2003; Chen et al., 2004) and also human hippocampal function and episodic memory (Egan et al., 2003; Hariri et al., 2003). Cultured hippocampal neurons transfected with met-BDNF show reduced depolarization-induced secretion and fail to localize BDNF to secretory granules and dendritic processes (Egan et al., 2003; Chen et al., 2004). Normal individuals with met alleles have poorer episodic memory performance and reduced hippocampal physiologic engagement during memory studied with functional magnetic resonance imaging (fMRI) (Egan et al., 2003; Hariri et al., 2003). The role of BDNF in human learning and memory may be related to modulation of synaptic transmission and plasticity, particularly long-term potentiation (LTP) (Figurov et al., 1996; Huang et al., 1999; Lee et al., 2004), but also to its importance in mediating long-term developmental phenomena, such as neuronal survival, migration, and differentiation and also activity-dependent refinement of synaptic architecture (Huang et al., 1999; Gorski et al., 2003; Baquet et al., 2004; Hua and Smith, 2004). Thus, the effects of the val/met-BDNF polymorphism during memory processing in humans may reflect rapid, context-dependent plasticity in the hippocampal formation and it may reflect a trait characteristic related to hippocampal development and morphology.
To investigate whether this polymorphism impacts gross morphological changes in the hippocampus, we analyzed highresolution anatomical magnetic resonance images of 111 normal healthy volunteers with optimized voxel-based morphometry (VBM), a sophisticated, fully automated morphological imaging technique (Ashburner and Friston, 2000; Good et al., 2001). We hypothesized that, consistent with the cellular and clinical effects of the BDNF val66met polymorphism and the role of BDNF in cortical development, met allele carriers would have reduced hippocampal gray matter volume.
Materials and Methods
Demographics. All of the subjects included in val/met-BDNF comparisons and subjects being used for customized template creation were culled from a larger population after careful screening with extensive historical and interview procedures (Egan et al., 2001). All of the subjects gave written informed consent and participated in the study according to the guidelines of the National Institute of Mental Health Institutional Review Board.
Template creation was based on a sample of 214 subjects recruited as “normal volunteers” with available and visually inspected anatomical MRI scans. Most subjects were Caucasian (n = 163); other ethnicities included African-American (n = 26), Hispanic (n = 14), Asian (n = 8), and Native American (n = 1). The range of the sample was 18-60 years of age (mean, 34.2 ± SD 9.7).
From these 214 subjects, the sample for the val/met-BDNF comparison was drawn. The BDNF genotyping procedure was described previously (Egan et al., 2003). To minimize confounding environmental and ethnicity effects, we included only Caucasian subjects of European ancestry (n = 111) who were free of any lifetime history of psychiatric or neurological illness, of psychiatric treatment, or of drug or alcohol abuse (basic demographic data can be seen in the supplemental material, available at www.jneurosci.org). The individuals in the BDNF genotype groups were also genotyped at 100 unlinked single-nucleotide polymorphisms and showed no significant variations in frequency in alleles at any of these loci, including several that have been associated with variation in cortical function (e.g., catechol-O-methyltransferase, serotonin transporter promotor polymorphism, glutamate receptor metatropic 3, apolipoprotein e4).
Image processing. Three-dimensional structural MRI was performed on a 1.5 tesla GE (Milwaukee, WI) scanner using a T1-weighted SPGR sequence (repetition time, 24 msec; echo time, 5 msec; excitations, 1; flip angle, 45°; matrix size, 256 × 256; field of view, 24 × 24 cm), with 124 sagittal slices at a thickness of 1.5 mm and an in-plane resolution of 0.94 × 0.94 mm. Data were analyzed on a Linux workstation (Red Hat 8.0) using MATLAB 6.51SP1 (Math-Works, Natick, MA) and SPM2 (http://www.fil.ion.ucl.ac.uk/spm/; Wellcome Department of Imaging Neuroscience, London, UK). Additional imaging software packages used were Analyses of Functional NeuroImages (AFNI) (Cox, 1996), Surface Mapping with AFNI (SUMA) (Saad et al., 2004), and Nonparametric nonuniform intensity normalization (N3) (Sled et al., 1998).
All of the scans (n = 214) first underwent inhomogeneity correction, followed by “Winsorizing” (Yuen, 1971) (spatial noise filter available in AFNI; for details, see supplemental material, available at www.jneurosci.org). Optimal inhomogeneity correction was achieved by using an automated histogram-based algorithm to choose between the corrections made by either AFNI or N3 based on the following measures: entropy, peak height, overlap between gray and white matter curves, and ratio of gray-to-white matter peak.
We used an iterative procedure to create customized templates optimally suited for our sample. In the first step, preliminary anatomical, gray and white matter and CSF templates were created by segmentation and spatial normalization of the structural image and the tissue compartment maps to the SPM2 T1 template, followed by smoothing with an 8 mm full-width at half-maximum (FWHM) isotropic Gaussian kernel. Images were then reprocessed in the same way but using the preliminary templates. This resulted in the final set of templates, which were again smoothed with an 8 mm FWHM kernel.
Images of subjects included in our genetic analysis were processed using optimized VBM in SPM2 as described in detail previously (Ashburner and Friston, 2000; Good et al., 2001; Ashburner et al., 2003). Briefly, scans (in native space) were segmented into gray matter images, followed by a series of fully automated morphological operations for removing unconnected nonbrain voxels from the segmented images. The extracted segmented images were normalized to the gray matter template, and deformation parameters were applied to the original images, followed by a second segmentation step in stereotactic space. To correct for volume changes by the normalization transformation, images were modulated by multiplication with the determinant of the Jacobian of the spatial normalization function. An integration of modulated gray matter voxel values resulted in estimates for total gray matter volume. The normalized segmented images were smoothed using a 12 mm FWHM isotropic Gaussian kernel.
Statistics. The normalized and smoothed gray matter images were analyzed using statistical parametric mapping with the framework of the General Linear Model (Friston et al., 1995) in SPM2. Effects of val/val-BDNF and met-BDNF on gray matter were examined by using an analysis of covariance model. The following covariates were included in the model to control for confounds: (1) total gray matter volume, accounting for global differences of gray matter volume across subjects; (2) orthogonalized first- and second-order polynomial expansions of age to remove linear and nonlinear effects of age (Buchel et al., 1996) (a third-order polynomial expansion did not contribute to our age effect model of our sample and was therefore deleted from our design matrix) (Buchel et al., 1998); and (3) gender.
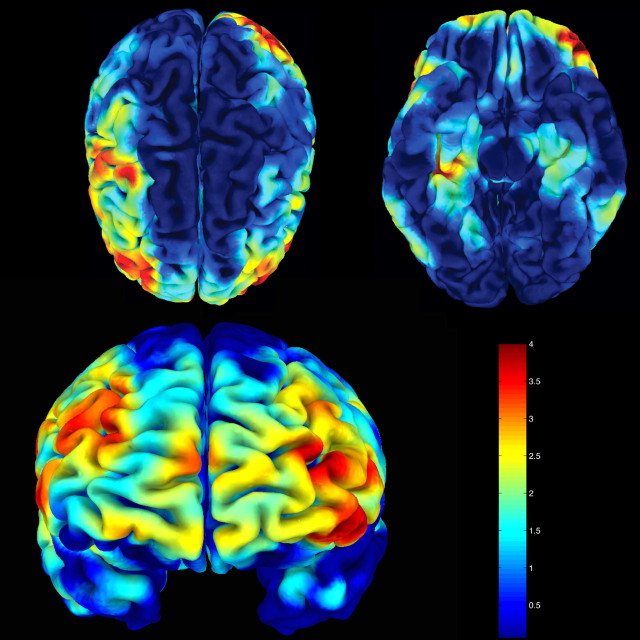
A hypothesis-driven regions of interest (ROIs) approach was used to investigate the hippocampus using an ROI from the Wake Forest University PickAtlas (Maldjian et al., 2003). Hippocampal gray matter volume change was assessed statistically using a two-tailed t contrast with a significance level set to 0.05 (corrected for multiple comparisons within the ROI). Uncorrected exploratory full-brain statistics were also performed with two-tailed t contrasts at a significance level set to 0.001. Anatomical labels of reported coordinates [transformed from Montreal Neurological Institute (MNI) to Talairach space] of peak clusters were retrieved from the Talairach Daemon database (Lancaster et al., 2000) within a 5 mm cubical search range (supplemental material, available at www.jneurosci.org). Figure 3 was created by converting t-scores into a map of z-scores using AFNI software. The resulting map was warped from MNI into Talairach space. Maximal z-scores within the cortex were projected onto a single-subject brain surface (part of the SUMA distribution) and displayed with SUMA.
Figure 3.
Statistical gray matter maps of the entire brain showing volume reductions of met-BDNF carriers in comparison to val/val-BDNF have been transformed from MNI space in Talairach space and converted to z-scores. Maximal z-scores within cortical regions have been projected onto a brain surface. The figure shows that the met-BDNF group has significantly reduced cortical volume (red and yellow areas) compared with val/val-BDNF carriers being found predominantly at the lateral convexity of the frontal lobe. Peak differences (red) are also found within dorsolateral prefrontal cortical areas, which are related to memory function.
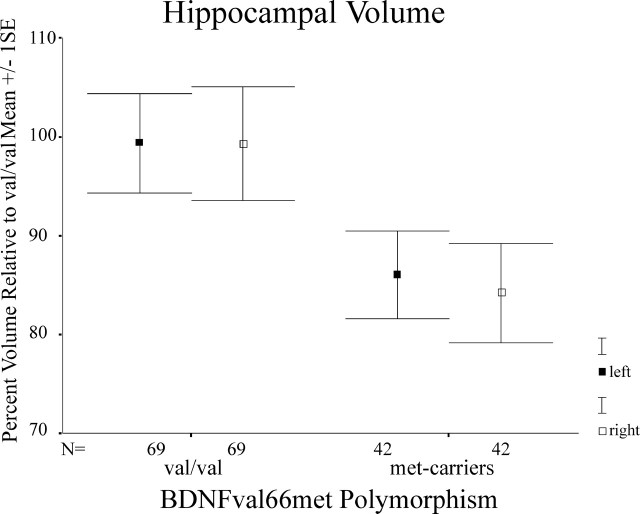
For Figure 2, data adjusted for confounding covariates within the peak clusters for the val/val-BDNF versus the met-BDNF comparison were exported from both hippocampal ROIs into SPSS 11.0 (SPSS, Chicago, IL) for Mac OS X (Apple Computers, Cupertino, CA). Data were converted into percentage volume relative to val/val-BDNF (see Fig. 2).
Figure 2.
Mean differences (±SEM) in hippocampal volume reduction in met-BDNF carriers relative to val/val-BDNF subjects within regions of statistical significance (p = 0.05) as shown in Figure 1. Measures for hippocampal volume have been extracted from the hippocampal cluster and transformed in percentage volume relative to val/val-BDNF while controlling for confounding variables such as age, gender, and total gray matter volume.
Results
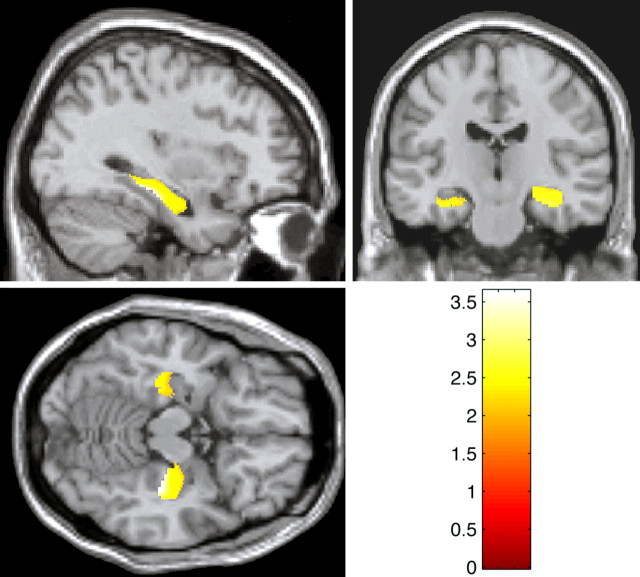
Consistent with our initial hypothesis, we found bilateral reductions of hippocampal gray matter volumes (right, p < 0.001; t = 3.41; left, p = 0.013; t = 2.24) in met-BDNF carriers compared with val/val-BDNF subjects (Fig. 1). It is noteworthy that these differences were age and gender independent and substantial in terms of volume differences in both significant hippocampal clusters (Fig. 2).
Figure 1.
Statistical maps of t-transformed hippocampal volume differences derived by optimized voxel-based morphometry in met relative to val/val-BDNF carriers thresholded at p = 0.05 (corrected) in coronal, sagittal, and axial views, showing bilateral significant hippocampal volume reduction in met-BDNF carriers.
Our hypothesis-driven analysis focused on the hippocampal formation because previous work highlighted the effects of val/met-BDNF on hippocampal function. However, BDNF is expressed abundantly throughout the human brain, particularly in areas such as the prefrontal cortex (Baquet et al., 2004), in which it has been implicated in various psychiatric and neurological diseases (Zuccato et al., 2001; Bimonte-Nelson et al., 2003; Weickert et al., 2003; Baquet et al., 2004). Therefore, we performed an additional analysis of the effects of BDNF val66met on gray matter anatomy throughout the entire brain. We found that, compared with val/val individuals, met-BDNF carriers exhibited additional loci of reduced gray matter volumes, predominately in the lateral convexity of the frontal lobes with peak values encompassing the dorsolateral prefrontal cortex bilaterally (Fig. 3) (supplemental material, available at www.jneurosci.org).
Discussion
Our data indicate that a functional variation at the val66met locus in the 5′ prodomain of BDNF impacts cortical morphology in normal humans. Specifically, met-BDNF carriers have substantial relative decreases in hippocampal volume, which are gender and age independent, suggesting that these changes occurred before adulthood. Our data are consistent with the hypothesis that reduced memory function of met-BDNF carriers might, at least in part, be expressed by fixed changes of synaptic and cellular plasticity and not exclusively by an effect of rapid changes of neural transmission during memorization. The mechanism of this structural effect presumably relates to abnormal regulated secretion of met-BDNF alleles, which alters activity-dependent processes of cortical development and plasticity. We also found that val/met-BDNF has effects on the volume of gray matter in the cerebral neocortex of normal humans, and again met-BDNF is associated with volume reductions, primarily in the lateral convexity of the prefrontal cortex. Analogous to the finding of reduced hippocampal volume, the neocortical results also suggest that met-BDNF leads to stable changes in prefrontal anatomy before adulthood and might create an anatomical substrate related to variation in the function of distributed memory networks (Egan et al., 2003). Recent work in prefrontally targeted BDNF knock-out mice has shown analogous reductions in cortical volume during adulthood (Baquet et al., 2004).
It is interesting that the anatomical effects of this functional variation in BDNF are most apparent in hippocampal formation and in prefrontal cortex, two brain regions that show especially abundant expression of BDNF and that are central to lifelong neuroplastic adaptations related to learning and memory (Bimonte-Nelson et al., 2003; Egan et al., 2003; Hariri et al., 2003; Weickert et al., 2003; Baquet et al., 2004). Moreover, expression of BDNF in these brain regions has been implicated in neurodevelopmental and neurodegenerative disorders, including Alzheimer's disease, Huntington's disease, Down's syndrome, and schizophrenia (Zuccato et al., 2001; Bimonte-Nelson et al., 2003; Weickert et al., 2003; Baquet et al., 2004). Thus, the val/met-BDNF polymorphism may be a modifying genetic factor in the expression of a number of normal and abnormal brain conditions, in part because of its effects on the development and plasticity of these critical brain systems.
Footnotes
This work was supported by the National Institute of Mental Health Intramural Research Program. We thank A. Goldman and P. Fisher for technical assistance.
Correspondence should be addressed to Dr. Daniel R. Weinberger, Genes, Cognition, and Psychosis Program, National Institute of Mental Health, Intramural Research Program, National Institutes of Health, Room 4S-235, 10 Center Drive, Bethesda, MD 20892. E-mail: weinberd@intra.nimh.nih.gov.
Copyright © 2004 Society for Neuroscience 0270-6474/04/2410099-04$15.00/0
References
- Ashburner J, Friston KJ (2000) Voxel-based morphometry—the methods. NeuroImage 11: 805-821. [DOI] [PubMed] [Google Scholar]
- Ashburner J, Csernansky JG, Davatzikos C, Fox NC, Frisoni GB, Thompson PM (2003) Computer-assisted imaging to assess brain structure in healthy and diseased brains. Lancet Neurol 2: 79-88. [DOI] [PubMed] [Google Scholar]
- Baquet ZC, Gorski JA, Jones KR (2004) Early striatal dendrite deficits followed by neuron loss with advanced age in the absence of anterograde cortical brain-derived neurotrophic factor. J Neurosci 24: 4250-4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bimonte-Nelson HA, Hunter CL, Nelson ME, Granholm AC (2003) Frontal cortex BDNF levels correlate with working memory in an animal model of Down syndrome. Behav Brain Res 139: 47-57. [DOI] [PubMed] [Google Scholar]
- Buchel C, Wise RJS, Mummery CJ, Poline J-B, Friston KJ (1996) Nonlinear regression in parametric activation studies. NeuroImage 4: 60-66. [DOI] [PubMed] [Google Scholar]
- Buchel C, Holmes AP, Rees G, Friston KJ (1998) Characterizing stimulus-response functions using nonlinear regressors in parametric fMRI experiments. NeuroImage 8: 140-148. [DOI] [PubMed] [Google Scholar]
- Chen ZY, Patel PD, Sant G, Meng CX, Teng KK, Hempstead BL, Lee FS (2004) Variant brain-derived neurotrophic factor (BDNF) (Met66) alters the intracellular trafficking and activity-dependent secretion of wild-type BDNF in neurosecretory cells and cortical neurons. J Neurosci 24: 4401-4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox RW (1996) AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput Biomed Res 29: 162-173. [DOI] [PubMed] [Google Scholar]
- Egan MF, Goldberg TE, Kolachana BS, Callicott JH, Mazzanti CM, Straub RE, Goldman D, Weinberger DR (2001) Effect of COMT Val108/158 Met genotype on frontal lobe function and risk for schizophrenia. Proc Natl Acad Sci USA 98: 6917-6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan MF, Kojima M, Callicott JH, Goldberg TE, Kolachana BS, Bertolino A, Zaitsev E, Gold B, Goldman D, Dean M, Lu B, Weinberger DR (2003) The BDNF val66met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell 112: 257-269. [DOI] [PubMed] [Google Scholar]
- Figurov A, Pozzo-Miller LD, Olafsson P, Wang T, Lu B (1996) Regulation of synaptic responses to high-frequency stimulation and LTP by neurotrophins in the hippocampus. Nature 381: 706-709. [DOI] [PubMed] [Google Scholar]
- Friston KJ, Holmes AP, Worsley KJ, Poline JP, Frith CD, Frackowiak RSJ (1995) Statistical parametric maps in functional imaging: a general linear approach. Hum Brain Mapp 2: 189-210. [Google Scholar]
- Good CD, Johnsrude IS, Ashburner J, Henson RN, Friston KJ, Frackowiak RS (2001) A voxel-based morphometric study of ageing in 465 normal adult human brains. NeuroImage 14: 21-36. [DOI] [PubMed] [Google Scholar]
- Gorski JA, Zeiler SR, Tamowski S, Jones KR (2003) Brain-derived neurotrophic factor is required for the maintenance of cortical dendrites. J Neurosci 23: 6856-6865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariri AR, Goldberg TE, Mattay VS, Kolachana BS, Callicott JH, Egan MF, Weinberger DR (2003) Brain-derived neurotrophic factor val66met polymorphism affects human memory-related hippocampal activity and predicts memory performance. J Neurosci 23: 6690-6694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua JY, Smith SJ (2004) Neural activity and the dynamics of central nervous system development. Nat Neurosci 7: 327-332. [DOI] [PubMed] [Google Scholar]
- Huang ZJ, Kirkwood A, Pizzorusso T, Porciatti V, Morales B, Bear MF, Maffei L, Tonegawa S (1999) BDNF regulates the maturation of inhibition and the critical period of plasticity in mouse visual cortex. Cell 98: 739-755. [DOI] [PubMed] [Google Scholar]
- Lancaster JL, Woldorff MG, Parsons LM, Liotti M, Freitas CS, Rainey L, Kochunov PV, Nickerson D, Mikiten SA, Fox PT (2000) Automated Talairach atlas labels for functional brain mapping. Hum Brain Mapp 10: 120-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JL, Everitt BJ, Thomas KL (2004) Independent cellular processes for hippocampal memory consolidation and reconsolidation. Science 304: 839-843. [DOI] [PubMed] [Google Scholar]
- Lu B, Gottschalk W (2000) Modulation of hippocampal synaptic transmission and plasticity by neurotrophins. Prog Brain Res 128: 231-241. [DOI] [PubMed] [Google Scholar]
- Maldjian JA, Laurienti PJ, Kraft RA, Burdette JH (2003) An automated method for neuroanatomic and cytoarchitectonic atlas-based interrogation of fMRI data sets. NeuroImage 19: 1233-1239. [DOI] [PubMed] [Google Scholar]
- Saad ZS, Reynolds RC, Argall B, Japee S, Cox RW (2004) SUMA: an interface for surface-based intra- and inter-subject analysis with AFNI. Paper presented at the IEEE international symposium on biomedical imaging, Arlington, VA, April 15-18, pp 1510-1513.
- Sled JG, Zijdenbos AP, Evans AC (1998) A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imaging 17: 87-97. [DOI] [PubMed] [Google Scholar]
- Weickert CS, Hyde TM, Lipska BK, Herman MM, Weinberger DR, Kleinman JE (2003) Reduced brain-derived neurotrophic factor in prefrontal cortex of patients with schizophrenia. Mol Psychiatry 8: 592-610. [DOI] [PubMed] [Google Scholar]
- Yuen KK (1971) A note on Winsorized t. Appl Stat 20: 297-304. [Google Scholar]
- Zuccato C, Ciammola A, Rigamonti D, Leavitt BR, Goffredo D, Conti L, MacDonald ME, Friedlander RM, Silani V, Hayden MR, Timmusk T, Sipione S, Cattaneo E (2001) Loss of huntingtin-mediated BDNF gene transcription in Huntington's disease. Science 293: 493-498. [DOI] [PubMed] [Google Scholar]