Abstract
The identification of the chemical structure of an odorant by the vertebrate olfactory system is thought to occur through the combinatorial activity from multiple receptors, each tuned to recognize different chemical features. What are the molecular determinants underlying the selectivity of individual odorant receptors for their cognate ligands? To address this question, we performed molecular modeling and site-directed mutagenesis on the ligand-binding region of two orthologous amino acid odorant receptors belonging to the “C family” of G-protein-coupled receptors in goldfish and zebrafish. We identified the critical ligand-receptor interactions that afford ligand binding as well as selectivity for different amino acids. Moreover, predictions regarding binding pocket structure allowed us to alter, in a predictable manner, the receptor preferences for different ligands. These results reveal how this class of odorant receptor has evolved to accommodate ligands of varying chemical structure and further illuminate the molecular principles underlying ligand recognition and selectivity in this family of chemosensory receptors.
Keywords: olfaction, olfactory, odorant receptor, G-protein-coupled receptor, ligand binding, signal transduction
Introduction
The perception and discrimination of thousands of different odorants by the vertebrate olfactory system results from the activation of specific odorant receptors expressed by olfactory neurons in the nose (Firestein, 2001; Mombaerts, 2004). Olfactory receptors in vertebrates comprise three different families of G-protein-coupled receptors (GPCRs): the OR receptor gene family, the largest family (∼1000 functional members in some mammalian species); the V1R receptors (∼150 members); and the V2R receptors, with ∼50-100 members (Mombaerts, 2004). The V2R receptors belong to the “C family” of GPCRs, which includes the calcium sensing receptor (CaSR), metabotropic glutamate (mGlu) receptors, GABAB receptors, and T1R taste receptors (Parmentier et al., 2002; Pin et al., 2003). A hallmark of this receptor family is a long extracellular N-terminal domain (NTD) that comprises the ligand-binding site (Pin et al., 2003).
Using an expression-cloning strategy, we previously identified a goldfish V2R-like or C family olfactory receptor responsive to amino acid ligands (Speca et al., 1999), which are potent odorants for fish and used as olfactory feeding cues (Hara, 1994; Sorensen and Caprio, 1998). This receptor (receptor 5.24) is broadly tuned to detect all 20 naturally occurring amino acids, with a preference for the basic amino acids arginine and lysine (Speca et al., 1999). These observations are consistent with the idea that olfactory discrimination is accomplished through the combinatorial activity of an array of broadly tuned odorant receptors (Malnic et al., 1999; Araneda et al., 2000; Uchida et al., 2000).
The identification of the activating ligands for receptor 5.24 provides a means to understand the principles governing the molecular receptive field properties of a vertebrate odorant receptor. For example, it is of great interest to elucidate what features of the receptor molecule are responsible for determining ligand specificity. Are certain regions of the ligand-binding pocket tuned to interact with particular chemical moieties? What aspects of the ligand-receptor interaction allow for the broad tuning profiles for certain ligands within a class of compounds (e.g., different types of amino acids)? The homology of receptor 5.24 to other C family GPCRs allows us to address these questions. Extensive structure-function information, including crystal structures of the ligand-binding NTD of the mGlu1 receptor (Kunishima et al., 2000; Tsuchiya et al., 2002), is known regarding the ligand-receptor interactions in the larger group of C family GPCRs (Pin et al., 2003).
In this study, we describe an analysis of the residues involved in the interactions between receptor 5.24 and amino acid agonists. Comparative modeling of receptor 5.24 based on the mGlu1 NTD crystal structure was used to predict the residues that contribute to the binding of amino acid ligands; these predictions were then tested in functional assays. Our results reveal residues that mediate ligand binding and influence selectivity for specific amino acid side chains. In addition, a comparison of goldfish receptor 5.24 with its zebrafish ortholog highlights the importance of specific binding pocket sites as determinants of ligand selectivity. From this analysis, we propose a general model of ligand recognition for amino acid odorant receptors.
Materials and Methods
Homology modeling of receptor NTDs. A sequence alignment of receptor NTDs was generated according to Bessis et al. (2000) and further refined using predicted (goldfish receptor 5.24 and zebrafish receptor ZO6) and known [mGlu1 NTD; Protein Data Bank (PDB) code 1ewk] secondary structures. Homology models for the closed form of the ligand binding domains from wild-type and mutant receptors were generated by the automated comparative modeling tool MODELER 7.00 (DS Modeling 1.1; Accelrys, San Diego, CA) as described previously (Bertrand et al., 2002). Models were generated by using the coordinates of the mGlu1 NTD closed form bound with glutamate (PDB code 1ewk:A) and based on the sequence alignment described in supplemental Figure S1 (available at www.jneurosci.org as supplemental material). The structural quality of the models was assessed according to the MODELER probability density functions as well as Profiles-3D analysis (DS Modeling 1.1; Accelrys). The selected models were also used for docking.
Docking of arginine and glutamate ligands in the ligand-binding domain models. Assuming that the glycine moiety of the ligand would bind in the same manner as that of glutamate in 1ewk:A (Bertrand et al., 2002), the obtained protein-ligand complex was submitted to energy minimization while tethering the Cα trace. This was performed using the CHARMm 29b1 calculation engine (Brooks et al., 1983) with the consistent force field (DS Modeling version 1.1; Accelrys). CHARMm and the consistent force field were also used to perform 500 psec of molecular dynamics at 298 K. Once the system was equilibrated, the coordinates of snapshots collected over a period of 20 psec were averaged and submitted again to energy minimization (Bertrand et al., 2002). These procedures were performed for wild-type receptors 5.24 and ZO6 as well as for the E47L, E47K, D388A, and M389K receptor 5.24 mutants.
High-throughput docking of other amino acid ligands and model scoring. The model of arginine docked into the wild-type receptor 5.24 ligand binding domain was used for further docking of different amino acids, using LigandFit (Venkatachalam et al., 2003) (DS Modeling version 1.2; Accelrys). In such a process, the protein is kept rigid while the ligands undergo Monte Carlo conformational searching. For each ligand, 20 poses were generated, clustered, and selected according to their binding mode.
As a preliminary test of the structural validity of our model, we performed a computational docking-scoring experiment of ligands for which we have previous measurements of ligand-receptor affinities (Speca et al., 1999). Our expectation is that the rank order of ligand affinities should only be predicted by a reliable molecular model. Conformations of all 20 naturally occurring amino acids were docked in the three-dimensional model of the binding pocket using a docking engine and subsequently ranked with the Jain scoring function (Jain, 1996). Indeed, calculated binding scores correlate well with measured binding affinities (supplemental material, available at www.jneurosci.org), indicating that this scoring function allows us to predict the rank orders with remarkable accuracy. The correlation between observed and predicted binding affinities supports, in general terms, the validity of our homology model of the receptor 5.24 NTD.
Receptor expression plasmids and site-directed mutagenesis. A receptor 5.24::eGFP (enhanced green fluorescent protein) expression plasmid was constructed by PCR fusion of the complete cDNA sequence of goldfish receptor 5.24 (Speca et al., 1999) with the full-length sequence of eGFP. The 3′ primer used to generate this PCR product contained an Asc I site replacing the stop codon, placing it in frame with another engineered Asc I site immediately upstream of the eGFP start codon. This introduced two amino acids (Gly-Glu) into the fusion junction between receptor 5.24 and eGFP. The entire fusion construct was cloned into pCDNA3 (Invitrogen, Carlsbad, CA).
For the zebrafish receptor ZO6, the goldfish receptor 5.24 cDNA insert was used to screen a zebrafish cDNA library (Barth et al., 1997), resulting in the isolation of a 1.1 kb partial 3′ cDNA sequence, ZO6A. ZO6A, in turn, was used to screen a zebrafish genomic bacterial artificial chromosome library (Barth et al., 1997) to identify the full protein coding region of this receptor gene. We then isolated the full protein coding sequence by reverse transcription (RT)-PCR on zebrafish olfactory RNA, using oligonucleotide primers spanning the initiator methionine and terminator codons. The receptor ZO6 RT-PCR product (GenBank accession number AY770492) was subsequently subcloned into a eukaryotic expression plasmid (pEGFP-N1, with the eGFP coding sequence deleted; Clontech, Palo Alto, CA) containing the cytomegalovirus immediate early promoter. The receptor ZO6 RT-PCR product and corresponding genomic sequence are 99.8% identical at the nucleotide level and 98.7% identical at the amino acid level (with 100% amino acid identity in the NTD sequence) (supplemental material, available at www.jneurosci.org) (T. Alioto and J. Ngai, unpublished results).
Site-directed mutagenesis was conducted using the QuickChange mutagenesis kit (Stratagene, La Jolla, CA) according to the protocols of the manufacturer. Mutants were constructed on receptor 5.24::GFP or receptor ZO6 backgrounds. All mutations were confirmed by DNA sequencing.
Calcium imaging of human embryonic kidney 293 cells expressing wild-type and mutant receptors. Human embryonic kidney (HEK) 293 cells were maintained in DMEM supplemented with 10% FBS and glutamine. Typically, cells were transiently transfected with 2 μg of receptor expression plasmid using Lipofectamine 2000 (Invitrogen). Twenty-four hours after transfection, samples were replated at a density of 1 × 105/ml on poly-d-lysine-coated coverslips. Transfected samples were allowed to attach overnight and were subsequently loaded with the calcium-sensitive dye fura-2 AM (Molecular Probes, Eugene, OR) at a concentration of 5 μm for 30 min at 37°C. After washout and recovery for 30 min at room temperature, cells were imaged on a Nikon (Tokyo, Japan) Diaphot inverted microscope fitted with a 20×/0.75 numerical aperture objective. For receptor 5.24 constructs, the field of cells was first examined for GFP fluorescence to select regions to analyze. Cells were perfused with a continuous flow of buffer, with or without different concentrations of ligand. “CIB” buffer (Caterina et al., 1997) was used in all loading and perfusion solutions. Cells were illuminated alternately at 340 or 380 nm, and fluorescence emission was monitored at 510 nm. The F340/F380 ratio was used to measure relative intracellular calcium levels. Images were captured with a Sutter Instruments (Novato, CA) CCD camera and quantitated using Axon Imaging Workstation imaging software (version AIW 4.1; Axon Instruments, Union City, CA), and data were analyzed using the Origin analysis software. We typically obtained data from 50-80 cells per concentration point; responses were measured at multiple concentrations of ligand (l-amino acids or derivatives) and individually normalized to the saturating response for each cell. Representative traces and the dose-response curve are shown in supplemental Figure S3 (available at www.jneurosci.org as supplemental material). In this cell-based functional assay, wild-type receptor 5.24 exhibits saturation at ∼2-3 μm arginine and half maximal activation (EC50) at ∼0.8-0.9 μm (supplemental material, available at www.jneurosci.org) (Tables 1, 2), which is 10-fold higher than the Kd of arginine (80 nm) as determined by radiolabeled ligand binding (Speca et al., 1999). The most likely explanation for this disparity is the nonlinearity of the measured calcium response (Hill coefficients, 1.9 vs 0.95 for ligand binding) (Speca et al., 1999), which lies far downstream of the initial receptor activation event and involves several amplified steps. Nonetheless, the rank-order potency for selected ligands in these calcium-imaging assays is similar to the rank order determined by direct ligand binding (most potent or highest affinity in both cases: arginine/lysine > citrulline/ornithine > glutamate) (data not shown).
Table 1.
Effects of receptor 5.24 proximal pocket mutations on arginine potency
|
Construct |
Arginine EC50 (μm) |
|---|---|
| WT 5.24 | 0.91 ± 0.041 n = 5 |
| S152A | 39 ± 2.8 n = 5 |
| T175A | 769 ± 64 n = 2 |
| Y223A | 100 ± 1.6 n = 2 |
| Y223H | 177 n = 1 |
| Y223F | >500 n = 2 |
| D309L |
>>1000 n = 2 |
Arginine EC50 values were generated for receptor 5.24 wild-type (WT) and mutant constructs as described in Materials and Methods. Values were derived from separate transfection and imaging experiments. Where indicated, mean EC50 values are shown, followed by the SE (n = number of independent determinations).
Table 2.
Effects of receptor 5.24 distal pocket mutations on potencies of amino acid ligands
|
|
Arginine EC50 (μm) |
Lysine EC50 (μm) |
Glutamate EC50 (μm) |
Aspartate EC50 (μm) |
Glycine EC50 (μm) |
Selectivity (arginine/glutamate) |
Fold-change selectivity (mut/wt)−1 |
|---|---|---|---|---|---|---|---|
| 5.24WT | 0.91 ± 0.041 | 1.9 ± 0.44 | 60 ± 7.4 | 1478 ± 108 | 22 ± 3.2 | 66 | |
| n = 5 | n = 5 | n = 10 | n = 3 | n = 5 | |||
| E47L | 4.4 ± 0.2 | 3.4 ± 0.33 | 37 ± 2.1 | 1724 ± 27 | 50 ± 4.7 | 8.4 | 8X |
| n = 4 | n = 3 | n = 4 | n = 3 | n = 3 | |||
| E47K | 11 ± 1.5 | 9.4 ± 2.3 | 95 ± 6.3 | 1928 ± 572 | 114 ± 2.9 | 8.6 | 8X |
| n = 6 | n = 2 | n = 4 | n = 2 | n = 4 | |||
| E47K/Y72K | 15 ± 1.6 | 9.3 ± 0.79 | 36 ± 4.1 | 560 ± 54 | 101 ± 12 | 2.4 | 28X |
| n = 4 | n = 3 | n = 9 | n = 4 | n = 5 | |||
| D388A | 4.2 ± 0.3 | 8.5 ± 0.9 | 21 ± 4.2 | 1637 ± 292 | 28 ± 5.0 | 5.0 | 13X |
| n = 3 | n = 2 | n = 3 | n = 4 | n = 3 | |||
| D388K | 5.6 ± 1.1 | 9.4 ± 4.4 | 23 ± 5.7 | 1483 ± 228 | 46 ± 2.3 | 4.1 | 16X |
| n = 2 | n = 2 | n = 2 | n = 3 | n = 3 | |||
| E47L/D388A | 13 ± 0.9 | ND | 37 ± 5.0 | 1352 ± 151 | 52 ± 3.8 | 2.8 | 24X |
| n = 4 | n = 4 | n = 2 | n = 3 | ||||
| M389K | 22 ± 2.2 | ND | 3 ± 0.67 | ND | 52 ± 16 | 0.14 | 471X |
|
|
n = 3 |
|
n = 3 |
|
n = 6 |
|
|
EC50 values were generated for each ligand (top) and receptor construct (left) as described in Materials and Methods. Each value was derived from the mean of multiple transfection and imaging experiments (mean ± SE; n, number of independent determinations). ND, Not determined. For selectivity of arginine versus glutamate (EC50 E/EC50 R), a value of >1 indicates a preference for arginine over glutamate, whereas a value of <1 indicates a preference for glutamate over arginine. Fold-change selectivity compares arginine/glutamate selectivity relative to wild-type ratio. WT, Wild type.
To control for the effects of mutations on processes other than the ligand-receptor interaction itself, for example, the coupling of ligand binding to receptor activation (Colquhoun, 1998) or the effects of receptor density on the extent of downstream signaling (Hermans et al., 1999), we compared the peak responses for wild-type receptor 5.24 and ZO6 and selected mutants activated by various ligands. If the introduced mutations cause large reductions in receptor expression or other effects on receptor structure and/or activation, we would expect to see large differences in peak response values when comparing wild-type receptors with their respective mutants. Even in the most severe cases (e.g., the receptor 5.24 S152A or Y223A mutants, which cause ∼40- to 100-fold reductions in apparent ligand potency) (Table 1), peak responses are indistinguishable from wild-type levels (supplemental material, available at www.jneurosci.org). We also used the level of eGFP fluorescence from the receptor 5.24::eGFP fusions as an indicator for receptor expression from the different receptor 5.24 constructs. For wild-type receptor 5.24, we determined that an eightfold difference in eGFP fluorescence resulted in less than a twofold difference in the EC50 value (data not shown). Because most of our mutant constructs displayed fluorescence comparable with wild-type levels, this suggests that differences in receptor expression have only a very minor effect on measured EC50 values.
Results
Homology modeling of the receptor 5.24 ligand-binding domain
Modeling proteins based on other known protein structures constitute a powerful means to predict structure-function relationships for a protein for which direct structural data are lacking. For example, key features of glutamate binding to the mGlu1 receptor ligand-binding pocket were identified by modeling the NTD of this receptor to the bacterial periplasmic leucine-isoleucinevaline binding protein (LIVBP) (O'Hara et al., 1993). Similar predictions of receptor structures and their interactions with specific ligands have also been made for other mGlu receptor subtypes as well as for the GABAB receptor and ionotropic glutamate receptors (Armstrong et al., 1998; Bessis et al., 2000; Galvez et al., 2000; Parmentier et al., 2000). Indeed, many of these early predictions were confirmed through the determination of the crystal structures for the metabotropic and ionotropic glutamate receptor ligand binding domains (Armstrong et al., 1998; Kunishima et al., 2000). The homology between the NTD of receptor 5.24 and mGlu receptors (Speca et al., 1999) therefore enables the generation of a structural model on which to base direct functional studies.
To identify residues critical for ligand binding, we began by constructing a three-dimensional model of the receptor 5.24 NTD followed by the docking of cognate ligands within the predicted binding pocket. This model was based on an alignment of receptor 5.24 with mGlu1a (supplemental material, available at www.jneurosci.org) and generated by comparative modeling using the closed, ligand-bound form of mGlu1a (1ewk:A) as a template (Bessis et al., 2000; Bertrand et al., 2002) (see Materials and Methods). The most potent agonist, arginine, was docked according to a molecular mechanics-dynamics protocol allowing protein side chain and ligand flexibility as described previously (Bertrand et al., 2002). The results are shown in Figure 1. As with other C family GPCRs, the receptor 5.24 NTD adopts a bilobate “clamshell” conformation, connected by a flexible hinge (Fig. 1A). Ligand interactions with the inner surfaces of lobes 1 and 2 are thought to stabilize a closed conformation of the clamshell and lead to subsequent activation (Kunishima et al., 2000; Bessis et al., 2002; Parmentier et al., 2002; Tsuchiya et al., 2002). An inspection of the core binding residues reveals numerous potential contacts with ligand (Fig. 1B). These contact residues can be sorted into two groups, which we refer to as the “proximal” and “distal” binding pockets. The proximal pocket residues are predicted to bind the glycine moiety of the amino acid ligand (i.e., the α-carboxyl together with the α-amino group and α-proton). We have shown previously that a common “signature” motif that interacts with the glycine moiety can be found among all amino acid binding proteins in the periplasmic binding protein-like I family (SCOP classification, http://scop.mrc-lmb.cam.ac.uk/scop/data/scop.b.d.bbg.b.A.html) (Bertrand et al., 2002). Among these are the bacterial leucine binding protein (LBP), LIVBP, eight subtypes of mGlu receptors, CaSR, and T1R taste receptors as well as receptor 5.24 (Bertrand et al., 2002; Pin et al., 2003). Side chains of five residues (S152, T175, D195, Y223, and D309 in receptor 5.24) and a backbone carbonyl (A173) constitute this motif, which is highlighted in the sequence alignment of supplemental Figure S1 (available at www.jneurosci.org as supplemental material) (Fig. 1B).
Figure 1.

Homology model of receptor 5.24 based on the crystal structure of mGlu1. A, Ribbon diagram of receptor 5.24 extracellular domain from amino acids 28-484. Lobe 1 is depicted in cyan, whereas lobe 2 is shaded magenta. The flexible hinge region is indicated in yellow. The structure is shown in the ligand-bound state in a closed conformation, with an l-arginine molecule present in the binding cleft. The α-carbon of the ligand is toward the left, with the side chain extending toward the right. B, Structural diagram of the proposed binding pocket of receptor 5.24 binding l-arginine. Amino acids proposed to interact with an arginine ligand are indicated by the residue number. Ligand contacts through hydrogen bonds (or ionic interactions in the case of E47 and D388) are depicted by green dashed lines, whereas inter-residue hydrogen bonds are shown as red dashed lines. Side chains of residues S111, S150, E47, and D388 may interact with the ligand via bridging water molecules. Residues D195 (hinge) and K283 have been omitted for clarity. The color scheme and general orientation of the receptor are the same as in A. On the ligand, hydrogens are shown in white, carbons are shown in green, nitrogens are shown in blue, and oxygens are shown in red.
The distal pocket residues are predicted to coordinate the interactions between the R group side chain and receptor of the ligand. When we examined the distal pocket residues, no common motif was found with any C family GPCR. However, most residues that interact with ligands are localized in homologous loops at the interface of the two lobes (supplemental material, available at www.jneurosci.org). In the case of receptor 5.24, two types of distal pocket residues can be identified: those that make hydrophobic contacts with the methylene groups of the arginine ligand and those that make polarionic interactions with the distal guanidinium moiety. The hydrophobic ligand environment is provided by the methyl and methylene groups of S151, A173, K283, and M389 (for clarity, K283 is not shown in Fig. 1B) (Fig. 2). The polar and ionic interactions predicted to contribute to the binding of the distal portion of the ligand side chain (Fig. 1B, right, dashed green lines) are mediated by two clusters of serines (S111, S150, and S151 in lobe 1; S284 and S285 in lobe 2), E47, D388, Y72, and N310. Most of these interactions are likely to be provided by hydrogen bonding, although some may be bridged by water molecules. Interestingly, two ionic interactions (with the carboxylate groups of E47 and D388) are predicted to coordinate the positively charged guanidinium group. Several residues predicted to contact the ligand may also play structural roles in the receptor itself by forming hydrogen bond networks that stabilize the overall tertiary structure (Fig. 1B, red dashed lines). In addition, it should be noted that the activation of C family GPCRs involves favorable interactions not only between the ligand and protein but also between the two lobes of NTD of the receptor during closure, which leads to an optimal activated state (Kunishima et al., 2000; Bertrand et al., 2002).
Figure 2.
Model for arginine binding in receptor 5.24: contribution of residues in the proximal, middle-distal, and far-distal binding pockets. A model of the receptor 5.24 binding pocket bound with l-arginine is shown. This view shows polar interactions with the α-amino and α-carboxylate groups of the ligand in the proximal pocket (S152, T175, Y223, and D309), possible hydrophobic interactions with the backbone of the side chain of the ligand in the middle-distal pocket (S151, A173, K283, and M389), and polar-ionic interactions with the distal guanidinium of the ligand (S111, S150, E47, Y72, D388, S284, S285, and N310). The color scheme is the same as in Figure 1 B, except that residues proposed to form a hydrophobic ring in the middle-distal pocket are highlighted in yellow.
Proximal binding pocket residues contact all amino acids via their common glycine moieties
Previous studies have identified a conserved signature motif in amino acid binding proteins that interacts with the glycine moiety common to all amino acids (Bertrand et al., 2002). These signature residues are also present in our homology model of receptor 5.24 (supplemental material, available at www.jneurosci.org) (Fig. 1). We therefore engineered single amino acid substitutions at these positions and tested the mutant receptors to confirm the roles of these residues in ligand binding and receptor activation. Because binding and closure of the two lobes of the NTD clamshell around the ligand are closely linked to receptor activation in C family GPCRs (Parmentier et al., 2002), we decided to use a functional assay to assess the effects of any mutations on receptor activation, which reflects both ligand binding and clamshell closure. The ligand activation properties of wild-type and mutant receptors were therefore tested by expressing them in HEK293 cells and measuring calcium mobilization in response to ligand application. In this assay, the EC50 value for arginine in the wild-type receptor is 0.9 μm (see Materials and Methods) (Table 1). Mutations at any one of the signature motif residues described above have dramatic effects on the EC50 value for arginine (Table 1). T175 and D309 were confirmed to be particularly important, because alanine substitution at these positions resulted in complete abrogation of activation (∼850-fold to >>1000-fold increase in EC50). Other mutations (S152A, Y223A) also caused large effects on arginine activation (EC50, ∼40-100 μm). These observations are in agreement with previous studies of C family GPCRs, including receptor 5.24 (O'Hara et al., 1993; Hampson et al., 1999; Kunishima et al., 2000; Malherbe et al., 2001; Bertrand et al., 2002; Kuang et al., 2003), which showed that the residues comprising the conserved signature binding motif subserve interactions with the α-carboxylate (via S152 and T175) and α-amino (via T175, Y223, and D309) groups of the amino acid ligand (Fig. 1B).
Distal binding pocket residues confer specificity for basic amino acid ligands
From our homology model (Fig. 1B), residues in the distal binding pocket coordinate binding via contacts with the side chain of the ligand. For example, the positively charged guanidinium moiety of arginine is predicted to interact with the receptor via a network of hydrogen bonds made with a number of residues and water molecules. Perturbation at any one site in the distal pocket therefore might not be expected to have profound effects on ligand potency. Indeed, introduction of single mutations at some sites within the distal pocket either has only modest effects on arginine potency (e.g., S150A EC50, 2.6 μm; S111A EC50, 3.4 μm; compared with wild-type EC50, 0.91 μm) (data not shown) or has no appreciable effect at all (S284A EC50, 1.2 μm; Y72A EC50, 0.89 μm) (data not shown). These observations are consistent with the observation that receptor 5.24 can accommodate amino acid ligands of widely varying structure.
Because receptor 5.24 shows a preference for basic amino acids, we postulated that negatively charged residues must contribute to a local negative electrostatic environment of the distal binding pocket, thus providing a favorable environment for the positively charged side chain of arginine or lysine. We therefore surveyed our structural model for acidic residues close enough to interact with the extended side chain of arginine docked in the binding pocket. Our model predicts that favorable ionic interactions could be provided by E47 and D388, with Y72 contributing stabilizing interactions (Fig. 3A). At physiological pH, the distal carboxylate groups of E47 and D388 should be ionized and therefore contribute negative charge to the local environment of the binding pocket. In this scenario, the positive charge of guanidinium moiety of arginine (or distal amino group of lysine) is distributed between D388 and E47. Note that the predicted distances between the carboxylates of these residues and guanidinium of the ligand are on the order of ∼5 Å, probably too far for strong ionic interactions. This suggests that the proposed ionic interactions may be mediated by bridging water molecules. Y72 is predicted to interact directly with the ligand through hydrogen bonding and may also function to position E47 and K74 (which may form a salt bridge with D388) in the optimal conformation for ligand binding through hydrophobic stacking with the aliphatic side chains of these latter residues.
Figure 3.
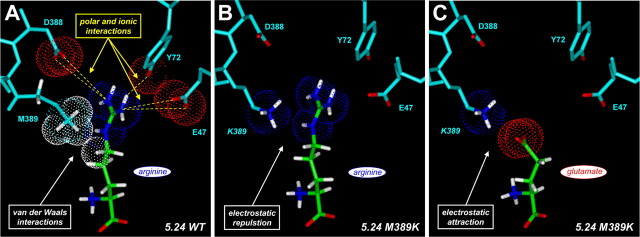
Methionine-389 is a critical determinant of ligand selectivity in receptor 5.24. A, Schematic view of selected distal pocket residues (M389, D388, Y72, and E47) involved in contacting the guanidinium moiety of an arginine ligand in the wild-type (WT) receptor. The orientation of this “top-down” view is from above lobe 1 looking down onto the bound ligand, with the proximal pocket toward the bottom and the distal pocket toward the top of the figure. Receptor residues are shaded cyan, with oxygens shown in red. On the ligand, hydrogens are shown in white, carbons are shown in green, nitrogens are shown in blue, and oxygens are shown in red. Proposed intermolecular interactions and distances are indicated with dashed green lines. M389 faces toward the binding pocket, possibly making van der Waals contacts with the n-aliphatic side chain of the docked arginine (Fig. 4 A). B, Predicted interaction of K389 in the receptor 5.24 M389K mutant with bound glutamate. In this model of the mutant binding pocket, the amino group of the side chain of K389 (nitrogen atom shown in blue) can make a direct ionic interaction with the distal carboxylate of the bound glutamate (distances indicated). C, Representative dose-response curves for wild-type and M389K receptor 5.24. HEK293 cells expressing wild-type or mutant receptor were exposed to arginine (blue lines) or glutamate (red lines), and receptor activation was measured by calcium imaging (see Materials and Methods). Note that the wild-type receptor prefers arginine to glutamate, whereas this selectivity is inverted in the M389K mutant. EC50 values for the curves shown in this panel are as follows. Wild type: 0.82 μm arginine, 29.6 μm glutamate; M389K mutant: 24.9 μm arginine, 2.1 μm glutamate.
To test the roles of E47 and D388 in coordinating the positively charged R groups of basic ligands by receptor 5.24, we made substitutions at these positions and examined their effects after arginine activation. Interestingly, we found that the neutralizing mutations E47L and D388A have only modest (approximately fourfold) effects on arginine and lysine potencies (Table 2). Neutralization of both charges in the E47L/D388A double mutant has a cumulative effect, with an ∼14-fold reduction in arginine potency compared with the wild-type receptor (Table 2). Consistent with these results, long-chain neutral amino acids (e.g., cysteine, leucine, isoleucine, methionine, and glutamine) and the amino acid analog citrulline (which is structurally very similar to arginine except that a neutral urethane moiety replaces the charged guanidinium group) exhibit only 5-fold to 10-fold lower affinities for the receptor than arginine or lysine (Speca et al., 1999). These latter observations therefore establish the maximum possible effect that neutralizing ionic interactions (caused by either site-directed mutagenesis or alterations in ligand structure) can have on the interaction between receptor and ligand.
Substitutions at E47 and D388 are only mildly charge dependent, because charge-reversing substitutions at these sites have only a minor additional effect: arginine potency is decreased by just 33% in the D388K mutant compared with the D388A construct, whereas E47K shows a 2.5-fold decrease compared with E47L (Table 2). However, it is important to note that the charge dependence of the E47K mutation on arginine and lysine potencies may be overestimated because of a structural effect of the mutation on receptor activation. This is indicated by an approximately fivefold effect of this mutation on glycine potency (Table 2), a ligand that is only expected to interact with the receptor via proximal pocket interactions. (The potential confounds introduced by structural effects are addressed and accounted for in the analysis described in the Discussion.)
We ascribe the modest effects of both charge-neutralizing and charge-reversing substitutions at D388 and E47 to two principal factors. First, the ionic interactions provided by these residues are weakened because of the long distances that they span (∼5Å) and are probably screened by water molecules. Just as water molecules can screen favorable interactions between attractive ionic pairs, they can also shield unfavorable interactions between repulsive ionic pairs. We would therefore expect that neutralization (or reversal) of these ionic interactions, either by chemical alterations in the ligand or by substitutions at these residues, might have fairly modest effects on ligand potency. Second, there are probably compensatory changes between the binding pocket side chains and the ligand in the receptor 5.24 mutants, such that the ligand adopts altered conformations, resulting in the guanidinium group moving closer to the remaining acidic side chain in the distal pocket (supplemental material, available at www.jneurosci.org). For the charge-reversing E47K or D388K substitutions, in the presence of a positively charged ligand, the side chains of K388 or K47 probably adopt alternate conformations by swinging away from the binding pocket (supplemental material, available at www.jneurosci.org).
Selectivity for basic versus acidic amino acid ligands is reduced in distal binding pocket mutants
From our structural model, we predict that substitutions at E47, D388, and Y72 would alter the proposed polar interactions between ligand and receptor and therefore should have effects on the selectivity of the receptor for basic versus acidic amino acids. To test this idea, we determined the potencies of multiple amino acids in activating receptors with single or double mutations at these positions (Table 2). The ligands tested include arginine and lysine (basic), glutamate and aspartate (acidic), and glycine (neutral; no side chain). The ratios of EC50 values for glutamate:arginine were calculated for each receptor construct (Table 2). It is important to note that whatever effects a given mutation might have on the structure or activation of the receptor (i.e., not directly affecting ligand-receptor affinity) will be canceled out in this ratiometric calculation. We then used these ratios as indicators of receptor selectivity for basic versus acidic amino acid ligands.
As shown in Table 2, wild-type receptor 5.24 exhibits a 66-fold preference for arginine over glutamate ([EC50 glutamate]: [EC50 arginine] = 66:1). Mutations of the above-mentioned distal pocket residues result in significant effects on ligand selectivity, with arginine:glutamate selectivity ratios reduced to ∼8-9:1 for the E47L and E47K mutants, ∼4-5:1 for the D388A and D388K mutants, and ∼3:1 for the E47L/D388A double mutant (Table 2). The similar behaviors of charge-neutralizing and charge-reversing mutations (i.e., E47L and E47K; D388A and D388K) could be explained by the ability of the positively charged side chains to adopt alternate rotameric conformations, possibly facing away from the binding pocket and therefore reducing the effects of the charge-reversing substitutions on repelling basic ligands (supplemental material, available at www.jneurosci.org).
Although our structural model predicts an interaction between the hydroxyl group of Y72 and guanidinium of the arginine ligand, the Y72A mutation results in no appreciable change in the potency of arginine (EC50, 0.83 μm) (data not shown) or arginine:glutamate selectivity (52:1; data not shown). However, arginine:glutamate selectivity is reduced to ∼2:1 in the E47K/Y72K double mutant (compared with ∼9:1 for E47K), consistent with the prediction that Y72 plays a role in the distal binding pocket.
Overall, these results are corroborated by the effects of the distal pocket mutations on arginine:aspartate, lysine:glutamate, and lysine:aspartate selectivities (calculations not shown) (Table 2), which mirror the effects on arginine:glutamate selectivity. In summary, our data confirm the participation of E47, D388, and possibly also Y72 not only in coordinating the positive charges of the arginine and lysine R groups but also in determining the selectivity of the receptor for basic versus acidic amino acid ligands.
A recent study by Kuang et al. (2003) also identified D388 as a residue important in receptor 5.24 for coordinating the positively charged ϵ-amino group of bound lysine. Unlike the present model, the model in the study by Kuang et al. (2003) predicts the close apposition of this functional group with the γ-carboxylate (<3 Å) of D388 but no interaction with E47. In addition, these authors observed a 26-fold reduction in lysine potency in this mutant, much higher than the approximately fourfold effect we observed in our assay (Table 2). Although the reasons for these discrepancies are presently unclear, our data demonstrate important roles for both D388 and E47 in ligand binding-receptor activation (as well as selectivity), with effects of the mutations in accord with previous structure-activity data (Speca et al., 1999).
Methionine-389 in the receptor 5.24 distal binding pocket is a major determinant of selectivity
The results presented thus far indicate that polar interactions between the positively charged R groups of arginine or lysine and D388, Y72, and E47 play important roles in ligand binding and selectivity in receptor 5.24. We next wanted to determine whether other residues might contribute to selectivity in the distal binding pocket of this receptor. To this end, we asked whether there are common positions in the distal binding pockets of other amino acid binding proteins that contact the R group of the cognate ligand of each receptor. If so, this would instruct our search for other potential binding sites in the receptor 5.24 distal pocket. We therefore examined the alignment of selected amino acid binding proteins (supplemental material, available at www.jneurosci.org) and identified one position, the equivalent to K409 in mGlu1 and M389 in receptor 5.24, that fulfilled this criterion. In mGlu1, K409 makes a direct ionic interaction with the distal carboxylate of the bound glutamate and is required for ligand binding (Kunishima et al., 2000; Rosemond et al., 2002); this lysine is strictly conserved in all mGlu receptor isoforms (Bertrand et al., 2002). In the bacterial LBP and LIVBP, the amino acids in the equivalent position (Y276 and F276, respectively) (supplemental material, available at www.jneurosci.org) participate in van der Waals interactions with the hydrophobic R group of the bound ligand (Sack et al., 1989; Magnusson et al., 2004). Similarly, molecular modeling and site-directed mutagenesis of the GABAB1 receptor implicate E465, which aligns with K409 in mGlu1, in making an ionic interaction with the distal amino group of the bound GABA ligand (Kniazeff et al., 2002). We therefore investigated the potential role of M389 in receptor 5.24 in ligand binding.
Our structural model predicts that the side chain of M389 faces toward the binding pocket, possibly making hydrophobic contacts with the δ-CH2 of the R group of the preferred basic ligand (Figs. 3A, 4A). This may maintain the long n-aliphatic side chain of arginine and lysine in an extended conformation, such that the distal guanidinium or amino group of the ligand can interact with D388, Y72, and E47 in the “far-distal” pocket. Whereas M389 provides a permissive environment for basic amino acid binding, we predict that substitution of a lysine at this position (M389K) would create an environment nonpermissive for arginine binding because of electrostatic repulsion between the side chain of the mutant lysine and the positively charged R group of the basic ligand (Fig. 4B). Moreover, modeling of the M389K mutant suggests that the altered binding pocket should be favorable for glutamate binding because of a direct ionic interaction between the amino group of the mutant K389 side chain and γ-carboxylate of the glutamate ligand (Figs. 3B, 4C).
Figure 4.
Models of receptor-ligand interactions in wild-type (WT) and M389K mutant receptor 5.24. Proposed hydrophobic and electrostatic interactions between distal binding pocket residues and docked ligands are shown for wild-type and M389K mutant receptor 5.24. The top-down orientation of these views is the same as in Figure 3, A and B. Van der Waals spheres for selected atoms are displayed (white, hydrogen; blue, nitrogen; red, oxygen). A, For arginine docked in the wild-type binding pocket, M389 makes van der Waals contacts with the methylene backbone of the n-aliphatic side chain of the ligand. This may serve to hold the side chain of the ligand in an extended conformation, such that the distal guanidinium group can participate in favorable polar and ionic interactions with D388, Y72, and E47 in the far-distal pocket. B, In the M389K mutant, electrostatic repulsion occurs between the distal amino group of K389 and the guanidinium of the arginine ligand. This unfavorable interaction is shown here for illustrative purposes, although in reality the repulsion probably prevents binding in this conformation. C, The lysine residue introduced into the binding pocket of the M389K mutant is positioned to form a favorable electrostatic interaction (a salt bridge) with the distal carboxylate of the glutamate ligand. Consistent with this model, the M389K mutant shows a marked increase in selectivity for glutamate over arginine.
To test these predictions, we created the M389K mutant and measured the potencies of selected ligands. Consistent with our model, the M389K mutation causes an ∼24-fold reduction in arginine potency, whereas glutamate potency is increased by 20-fold (Fig. 3C, Table 2). Thus, the mutant receptor is more selective for glutamate than for arginine (arginine:glutamate selectivity ratio, ∼1:7), a complete inversion in tuning for basic versus acidic ligands representing a 471-fold change in selectivity (Table 2). The dramatic reversal of arginine:glutamate selectivity in the M389K mutant is consistent with our prediction that the 389 side chain is positioned to contact the backbone of the R group of the preferred ligand in the “middle-distal” binding pocket. The preference for glutamate of the receptor 5.24 M389K mutant, which introduces a lysine at the equivalent position of the critical K409 residue in the distal binding pocket of mGlu1, underscores the structural similarities between receptor 5.24 and the metabotropic glutamate receptors. More generally, these results are in accord with similar structure-function and x-ray crystallographic studies, which highlight the importance of the equivalent position in coordinating the distal R group functions of the ligand in other amino acid binding proteins (Sack et al., 1989; Kunishima et al., 2000; Bertrand et al., 2002; Kniazeff et al., 2002; Pin et al., 2003; Magnusson et al., 2004). In the case of receptor 5.24, M389 functions together with other sites in the far-distal binding pocket to determine ligand selectivity.
Different selectivity of the zebrafish ortholog of receptor 5.24 can be attributed to a single amino acid substitution
We wanted to determine whether the preference of receptor 5.24 for basic amino acids and the role of M389 in determining this preference were in fact selected for during evolution. We therefore compared the amino acid sequence and ligand activation properties of the zebrafish ortholog of goldfish receptor 5.24, receptor ZO6. Of the ∼60 C family olfactory receptor genes identified from an exhaustive search of the zebrafish genome database (http://www-.sanger.ac.uk/Projects/D_rerio/), receptor ZO6 is the most closely related to goldfish receptor 5.24 (Alioto and Ngai, unpublished results), sharing 73% amino acid identity over the entire protein coding sequence. An alignment of receptor ZO6 and receptor 5.24 NTDs reveals that almost all of the binding pocket residues identified in the present study are in fact conserved between these two receptors (supplemental material, available at www.jneurosci.org). However, one notable exception occurs at the position equivalent to M389 in receptor 5.24, which is a lysine (K386) in receptor ZO6. Molecular modeling of the receptor ZO6 binding pocket predicts that K386 can form a direct and favorable ionic interaction with bound glutamate (Fig. 5A). Based on the observed selectivities of wild-type and M389K receptor 5.24 proteins, we further predict that receptor ZO6 should prefer glutamate over basic amino acid ligands.
Figure 5.

Zebrafish receptor ZO6 is a glutamate receptor and can be rationally retuned by the K386M mutation. A, View of the receptor ZO6 binding pocket looking down onto glutamate docked in the ligand-binding pocket from the perspective of lobe 1. Distal pocket residues K386, D385, Y70, and E45 (corresponding, respectively, to M389, D388, Y72, and E47 in receptor 5.24) are highlighted. The amino group of the K386 side chain can make a direct ionic interaction with the distal carboxylate of the bound glutamate (distances indicated). B, Representative dose-response curves for wild-type (WT) and K386M receptor ZO6. HEK293 cells expressing wild-type or mutant receptor were exposed to arginine (blue lines) or glutamate (red lines), and receptor activation was measured by calcium imaging (see Materials and Methods). Note that the wild-type zebrafish receptor prefers glutamate to arginine, whereas the K386M mutant is rendered nonselective for glutamate and arginine. EC50 values for the curves shown in this panel are as follows. Wild type: 671 μm arginine, 7.1 μm glutamate; K386M mutant: 80.8 μm arginine, 126 μm glutamate.
To test these predictions, we expressed receptor ZO6 in HEK293 cells and assessed its activation by selected amino acid ligands. In support of our molecular model, receptor ZO6 shows a strong preference for glutamate over arginine (glutamate EC50, 7.1 μm, vs arginine EC50, ∼1 mm) (Table 3). Similar to receptor 5.24, receptor ZO6 appears to be somewhat broadly tuned for amino acids, with the neutral amino acids serine and glutamine exhibiting only threefold to fourfold lower potencies than glutamate (Table 3). We next asked whether mutation of K386 in receptor ZO6 to a methionine (K386M) would diminish the preference of this receptor for glutamate. Indeed, the receptor ZO6 K386M mutant is rendered essentially nonselective for arginine and glutamate, with EC50 values of ∼100 and ∼130 μm, respectively (Fig. 5B, Table 3). These observations are consistent with the alterations in selectivity observed in the complementary receptor 5.24 M389K mutant. Together, our results on the two orthologous receptors provide compelling experimental validation for the notion that the M389/K386 position is a critical determinant of selectivity for basic versus acidic amino acid ligands.
Table 3.
Amino acid selectivities of wild-type and mutant zebrafish receptor Z06
|
|
Arginine EC50 (μm) |
Serine EC50 (μm) |
Glutamine EC50 (μm) |
Glutamate EC50 (μm) |
Aspartate EC50 (μm) |
Glycine EC50 (μm) |
Selectivity (arginine/glutamate) |
Fold-change selectivity (mut/wt) |
|---|---|---|---|---|---|---|---|---|
| Z06WT | 1059 ± 151 | 22 ± 4.9 | 27 ± 1.1 | 7.1 ± 0.6 | >> 1000 | 1129 ± 78 | 0.0067 | |
| n = 4 | n = 2 | n = 2 | n = 4 | n = 1 | n = 2 | |||
| K389M | 101 ± 17 | 48 ± 17 | 44 ± 2.6 | 129 ± 17 | >> 1000 | 526 ± 23 | 1.3 | 194× |
|
|
n = 4 |
n = 2 |
n = 3 |
n = 4 |
n = 1 |
n = 4 |
|
|
EC50 values were generated for each ligand (top) and receptor construct (left) as described in Materials and Methods. Where indicated, each value was derived from the mean of multiple transfection and imaging experiments (mean ± SE; n, number of independent determinations). For selectivity of arginine versus glutamate (EC50 E/EC50 R), a value of >1 indicates a preference for arginine over glutamate, whereas a value of <1 indicates a preference for glutamate over arginine. Fold-change selectivity compares arginine/glutamate selectivity relative to wild-type ratio. WT, Wild type.
Discussion
A structural basis for ligand recognition by a C family odorant receptor: implications for the molecular mechanisms underlying olfactory coding
Through the use of molecular modeling, we identified a number of key residues involved in ligand binding and selectivity in the receptor 5.24 binding pocket. Site-directed mutagenesis and receptor activity assays confirm many aspects of our model, as highlighted by our ability to effect a predictable and marked alteration of ligand selectivity in specific distal binding pocket mutations (Figs. 3, 5; Tables 2, 3). However, it should be noted that not all mutants had entirely predictable effects, underscoring the limitations of this structure-function approach. Although our molecular model can be iteratively refined with data on additional mutants and ligands, full validation will likely require a more definitive approach (e.g., the solving of a crystal structure for the receptor 5.24 NTD). Nonetheless, we believe that the present study provides a useful framework for considering ligand-protein interactions in this receptor. With these caveats in mind, based on our structure-function studies as well as our previous binding data (Speca et al., 1999), we propose a general model for recognition of amino acids by receptor 5.24. In our model, proximal residues in lobe 1 and lobe 2 coordinate the glycine moiety (common to all amino acids) via both ionic and polar interactions. The physical requirements at this region are extremely stringent, because disruption of any one of these interactions, whether by mutation of the receptor (Table 1) (Kuang et al., 2003) or by chemically altering the ligand around the α-carbon center (Speca et al., 1999), severely inhibits ligand-dependent activation.
Whereas binding to all amino acids requires strict coordination by residues in the proximal binding pocket, selectivity for different ligands is determined by the distal binding pocket, where some of the requirements are predicted to be less stringent for binding and activation. It is here that the side chain of the ligand dictates the formation of either greater or fewer favored contacts, thus allowing the receptor to become stabilized in the active (closed) conformation at lesser or greater concentrations of ligands, respectively. The side chain of the ligand is likely kept in an extended state through the presence of a hydrophobic “ring” in the middle-distal portion of the binding pocket (Fig. 2). In particular, we identified one residue, M389 in receptor 5.24, as playing a critical role in this region, probably by making hydrophobic contacts with the backbone of the n-aliphatic side chain of the ligand (Fig. 4A). The negative electrostatic environment provided by E47 and D388 in the far-distal pocket further narrows this specificity for basic side chains. Thus, a number of residues in the distal binding pocket work in concert to coordinate binding of the preferred side chains of the ligands (Figs. 2, 4A).
The relaxed stringency of the far-distal binding pocket interactions (as evidenced by the absence of any killing mutations in this region) is consistent with the observation that receptor 5.24 can bind to amino acids of widely varying structure (Speca et al., 1999). In addition, it should be noted that for shorter ligands bearing polar or ionic functional groups, water molecules can either screen unfavorable interactions or bridge favorable ones. The promiscuity of receptor 5.24 for amino acids of varying structure (Speca et al., 1999) and tolerance of the far-distal pocket residues to mutations is in contrast to the specificity of mGlu receptors for their cognate ligands, which are chemically highly restricted (Naples and Hampson, 2001). This difference is likely attributable to the greater reliance on direct ionic interactions in the mGlu distal binding pocket for binding glutamate (Kunishima et al., 2000; Malherbe et al., 2001; Rosemond et al., 2002; Sato et al., 2003) compared with receptor 5.24, which appears instead to use longer distance, and therefore weaker, ionic interactions as well as a network of mostly uncharged polar residues to coordinate the side chain of the ligand (Fig. 2). The reason for this difference may lie in the biological function of receptor 5.24 as an odorant receptor. Unlike glutamate receptors, which initiate synaptic transmission in a neurotransmitter-specific manner, odorant receptors are responsible for querying the environment for a myriad of chemical signatures. The utility of a receptor for sensing odorants is arguably higher if the stringency of its selectivity is reduced so that it can respond to multiple, albeit chemically similar structures. Discrimination by the system as a whole is then afforded by the combinatorial activation of multiple receptors, each with a broad odorant tuning profile. Thus, the evolution of relatively promiscuous odorant receptors has probably allowed the olfactory system to recognize a diversity of chemicals far greater than the number of odorant receptors encoded in the genome. Our analysis of the receptor 5.24 binding pocket provides an explanation for how this odorant receptor has evolved to recognize multiple ligands.
Interestingly, the T1R1 subunit of the heteromeric T1R1/T1R3 taste receptor (another C family chemosensory receptor) can recognize multiple amino acid ligands (Nelson et al., 2002). Presumably, the broad ligand tuning properties of this receptor also reflect some tolerance in the ligand-receptor interaction in the T1R distal binding pocket. Extension of the present structure-function approach to other C family olfactory and taste receptors should facilitate a more general understanding of the molecular principles underlying ligand selectivity in these chemosensory receptors.
Divergent ligand selectivities in orthologous goldfish and zebrafish receptors
Curiously, goldfish receptor 5.24 and its ortholog from zebrafish, receptor ZO6, have very different ligand specificities. Goldfish receptor 5.24 shows a clear preference for basic versus acidic amino acids, whereas zebrafish receptor ZO6 exhibits the opposite tuning. Consistent with our structure-function analysis of the receptor 5.24 distal binding pocket, our studies identified a difference at one amino acid position (M389 in receptor 5.24 and K386 in receptor ZO6) as a major contributor to these contrasting ligand selectivities. As shown with goldfish and zebrafish receptors both wild-type and mutant at this position, a methionine is permissive for n-aliphatic and basic amino acid binding, whereas a lysine creates a highly unfavorable electrostatic environment for basic amino acids in the binding pocket and instead can form a favorable ionic interaction with glutamate.
Both goldfish and zebrafish can detect a broad spectrum of amino acids through their olfactory systems (Michel and Lubomudrov, 1995; Friedrich and Korsching, 1997; Sorensen and Caprio, 1998). It is therefore unclear what evolutionary pressures may have driven the selection of such different ligand selectivities of these orthologous receptors. Nonetheless, our comparison of goldfish receptor 5.24 and zebrafish receptor ZO6 underscores the importance of M389 (goldfish)/K386 (zebrafish) in determining ligand selectivity and moreover indicates that this position was selected for during the evolution of these receptors.
Footnotes
This work was supported by grants from the National Institute on Deafness and Other Communication Disorders and the Human Frontiers Science Program (J.N.), the Centre National de la Recherche Scientifique/Institut National de la Santé et de la Recherche Médicale Molécules et Cibles Thérapeutiques program and the Fondation de France, comité Parkinson (F.A.), and by a genomics training grant from the National Institutes of Health (P.L.). We thank M. Donlan (Accelrys), E. VanName, and S. DeMaria for providing technical assistance and receptor activation data; T. Machen for the use of his fluorescence imaging microscope; and A.-S. Bessis, R. Kramer, E. Isacoff, T. Machen, and K. Scott for helpful discussions throughout the course of this study.
Correspondence should be addressed to John Ngai, Department of Molecular and Cell Biology, 269 Life Sciences Addition-3200, University of California, Berkeley, CA 94720-3200. E-mail: jngai@socrates.berkeley.edu.
P. Luu's present address: Department of Psychiatry and Behavioral Sciences, Stanford University School of Medicine, Stanford, CA 94304.
J. Fan's present address: Cytokinetics, South San Francisco, CA 94080.
Copyright © 2004 Society for Neuroscience 0270-6474/04/2410128-10$15.00/0
References
- Araneda RC, Kini AD, Firestein S (2000) The molecular receptive range of an odorant receptor. Nat Neurosci 3: 1248-1255. [DOI] [PubMed] [Google Scholar]
- Armstrong N, Sun Y, Chen GQ, Gouaux E (1998) Structure of a glutamate-receptor ligand-binding core in complex with kainate. Nature 395: 913-917. [DOI] [PubMed] [Google Scholar]
- Barth AL, Dugas JC, Ngai J (1997) Noncoordinate expression of odorant receptor genes tightly linked in the zebrafish genome. Neuron 19: 359-369. [DOI] [PubMed] [Google Scholar]
- Bertrand HO, Bessis AS, Pin JP, Acher FC (2002) Common and selective molecular determinants involved in metabotropic glutamate receptor agonist activity. J Med Chem 45: 3171-3183. [DOI] [PubMed] [Google Scholar]
- Bessis AS, Bertrand HO, Galvez T, De Colle C, Pin JP, Acher F (2000) Three-dimensional model of the extracellular domain of the type 4a metabotropic glutamate receptor: new insights into the activation process. Protein Sci 9: 2200-2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessis AS, Rondard P, Gaven F, Brabet I, Triballeau N, Prezeau L, Acher F, Pin JP (2002) Closure of the Venus flytrap module of mGlu8 receptor and the activation process: insights from mutations converting antagonists into agonists. Proc Natl Acad Sci USA 99: 11097-11102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M (1983) CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J Comput Chem 4: 187-217. [Google Scholar]
- Caterina MJ, Schumacher MA, Tominaga M, Rosen TA, Levine JD, Julius D (1997) The capsaicin receptor: a heat-activated ion channel in the pain pathway. Nature 389: 816-824. [DOI] [PubMed] [Google Scholar]
- Colquhoun D (1998) Binding, gating, affinity and efficacy: the interpretation of structure-activity relationships for agonists and of the effects of mutating receptors. Br J Pharmacol 125: 924-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firestein S (2001) How the olfactory system makes sense of scents. Nature 413: 211-218. [DOI] [PubMed] [Google Scholar]
- Friedrich RW, Korsching SI (1997) Combinatorial and chemotopic odorant coding in the zebrafish olfactory bulb visualized by optical imaging. Neuron 18: 737-752. [DOI] [PubMed] [Google Scholar]
- Galvez T, Prezeau L, Milioti G, Franek M, Joly C, Froestl W, Bettler B, Bertrand HO, Blahos J, Pin JP (2000) Mapping the agonist-binding site of GABAB type 1 subunit sheds light on the activation process of GABAB receptors. J Biol Chem 275: 41166-41174. [DOI] [PubMed] [Google Scholar]
- Hampson DR, Huang XP, Pekhletski R, Peltekova V, Hornby G, Thomsen C, Thogersen H (1999) Probing the ligand-binding domain of the mGluR4 subtype of metabotropic glutamate receptor. J Biol Chem 274: 33488-33495. [DOI] [PubMed] [Google Scholar]
- Hara TJ (1994) Olfaction and gustation in fish: an overview. Acta Physiol Scand 152: 207-217. [DOI] [PubMed] [Google Scholar]
- Hermans E, Challiss RA, Nahorski SR (1999) Effects of varying the expression level of recombinant human mGlu1alpha receptors on the pharmacological properties of agonists and antagonists. Br J Pharmacol 126: 873-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain AN (1996) Scoring noncovalent protein-ligand interactions: a continuous differentiable function tuned to compute binding affinities. J Comput Aided Mol Des 10: 427-440. [DOI] [PubMed] [Google Scholar]
- Kniazeff J, Galvez T, Labesse G, Pin JP (2002) No ligand binding in the GB2 subunit of the GABAB receptor is required for activation and allosteric interaction between the subunits. J Neurosci 22: 7352-7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang D, Yao Y, Wang M, Pattabiraman N, Kotra LP, Hampson DR (2003) Molecular similarities in the ligand binding pockets of an odorant receptor and the metabotropic glutamate receptors. J Biol Chem 278: 42551-42559. [DOI] [PubMed] [Google Scholar]
- Kunishima N, Shimada Y, Tsuji Y, Sato T, Yamamoto M, Kumasaka T, Nakanishi S, Jingami H, Morikawa K (2000) Structural basis of glutamate recognition by a dimeric metabotropic glutamate receptor. Nature 407: 971-977. [DOI] [PubMed] [Google Scholar]
- Magnusson U, Salopek-Sondi B, Luck LA, Mowbray SL (2004) X-ray structures of the leucine-binding protein illustrate conformational changes and the basis of ligand specificity. J Biol Chem 279: 8747-8752. [DOI] [PubMed] [Google Scholar]
- Malherbe P, Knoflach F, Broger C, Ohresser S, Kratzeisen C, Adam G, Stadler H, Kemp JA, Mutel V (2001) Identification of essential residues involved in the glutamate binding pocket of the group II metabotropic glutamate receptor. Mol Pharmacol 60: 944-954. [DOI] [PubMed] [Google Scholar]
- Malnic B, Hirono J, Sato T, Buck LB (1999) Combinatorial receptor codes for odors. Cell 96: 713-723. [DOI] [PubMed] [Google Scholar]
- Michel WC, Lubomudrov LM (1995) Specificity and sensitivity of the olfactory organ of the zebrafish, Danio rerio J Comp Physiol [A] 177: 191-199. [DOI] [PubMed] [Google Scholar]
- Mombaerts P (2004) Genes and ligands for odorant, vomeronasal and taste receptors. Nat Rev Neurosci 5: 263-278. [DOI] [PubMed] [Google Scholar]
- Naples MA, Hampson DR (2001) Pharmacological profiles of the metabotropic glutamate receptor ligands. Neuropharmacology 40: 170-177. [DOI] [PubMed] [Google Scholar]
- Nelson G, Chandrashekar J, Hoon MA, Feng L, Zhao G, Ryba NJ, Zuker CS (2002) An amino-acid taste receptor. Nature 416: 199-202. [DOI] [PubMed] [Google Scholar]
- O'Hara PJ, Sheppard PO, Thogersen H, Venezia D, Haldeman BA, McGrane V, Houamed KM, Thomsen C, Gilbert TL, Mulvihill ER (1993) The ligand-binding domain in metabotropic glutamate receptors is related to bacterial periplasmic binding proteins. Neuron 11: 41-52. [DOI] [PubMed] [Google Scholar]
- Parmentier ML, Galvez T, Acher F, Peyre B, Pellicciari R, Grau Y, Bockaert J, Pin JP (2000) Conservation of the ligand recognition site of metabotropic glutamate receptors during evolution. Neuropharmacology 39: 1119-1131. [DOI] [PubMed] [Google Scholar]
- Parmentier ML, Prezeau L, Bockaert J, Pin JP (2002) A model for the functioning of family 3 GPCRs. Trends Pharmacol Sci 23: 268-274. [DOI] [PubMed] [Google Scholar]
- Pin JP, Galvez T, Prezeau L (2003) Evolution, structure and activation mechanism of family 3/C G-protein coupled receptors. Pharmacol Ther 98: 325-354. [DOI] [PubMed] [Google Scholar]
- Rosemond E, Peltekova V, Naples M, Thogersen H, Hampson DR (2002) Molecular determinants of high affinity binding to group III metabotropic glutamate receptors. J Biol Chem 277: 7333-7340. [DOI] [PubMed] [Google Scholar]
- Sack JS, Saper MA, Quiocho FA (1989) Periplasmic binding protein structure and function. Refined X-ray structures of the leucine/isoleucine/valine-binding protein and its complex with leucine. J Mol Biol 206: 171-191. [DOI] [PubMed] [Google Scholar]
- Sato T, Shimada Y, Nagasawa N, Nakanishi S, Jingami H (2003) Amino acid mutagenesis of the ligand binding site and the dimer interface of the metabotropic glutamate receptor 1: identification of crucial residues for setting the activated state. J Biol Chem 278: 4314-4321. [DOI] [PubMed] [Google Scholar]
- Sorensen PW, Caprio JC (1998) Chemoreception. In: The physiology of fishes, Ed 2 (Evans DH, ed), pp 375-405. Boca Raton, FL: CRC.
- Speca DJ, Lin DM, Sorensen PW, Isacoff EY, Ngai J, Dittman AH (1999) Functional identification of a goldfish odorant receptor. Neuron 23: 487-498. [DOI] [PubMed] [Google Scholar]
- Tsuchiya D, Kunishima N, Kamiya N, Jingami H, Morikawa K (2002) Structural views of the ligand-binding cores of a metabotropic glutamate receptor complexed with an antagonist and both glutamate and Gd3+. Proc Natl Acad Sci USA 99: 2660-2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida N, Takahashi YK, Tanifuji M, Mori K (2000) Odor maps in the mammalian olfactory bulb: domain organization and odorant structural features. Nat Neurosci 3: 1035-1043. [DOI] [PubMed] [Google Scholar]
- Venkatachalam CM, Jiang X, Oldfield T, Waldman M (2003) LigandFit: a novel method for the shape-directed rapid docking of ligands to protein active sites. J Mol Graph Model 21: 289-307. [DOI] [PubMed] [Google Scholar]