Abstract
CNS lesions induce production of ECM molecules that inhibit axon regeneration. One major inhibitory family is the chondroitin sulfate proteoglycans (CSPGs). Reduction of their glycosaminoglycan (GAG) chains with chondroitinase ABC leads to increased axon regeneration that does not extend well past the lesion. Chondroitinase ABC, however, is unable to completely digest the GAG chains from the protein core, leaving an inhibitory “stub” carbohydrate behind. We used a newly designed DNA enzyme, which targets the mRNA of a critical enzyme that initiates glycosylation of the protein backbone of PGs, xylosyltransferase-1. DNA enzyme administration to TGF-β-stimulated astrocytes in culture reduced specific GAG chains. The same DNA enzyme applied to the injured spinal cord led to a strong reduction of the GAG chains in the lesion penumbra and allowed axons to regenerate around the core of the lesion. Our experiments demonstrate the critical role of PGs, and particularly those in the penumbra, in causing regeneration failure in the adult spinal cord.
Keywords: proteoglycan, extracellular matrix, dorsal root ganglion, antisense, reactive astrocytes, spinal cord
Introduction
After many different types of injuries to the brain and spinal cord that open the blood-brain barrier (Fitch et al., 1999), reactive astrocytes (Menet et al., 2003) and other non-neuronal cells (Chen et al., 2002; Jones et al., 2002; Pasterkamp et al., 1999) produce various growth inhibitory molecules. Chemorepulsive keratan sulfate proteoglycans (Jones and Tuszynski, 2002) semaphorin 3A (Song et al., 1998), ephrins such as EphB2 (Bundesen et al., 2003), and slit (Brose et al., 1999) are largely confined to the lesion core. In the lesion penumbra where mesenchymal elements are lacking, various inhibitory chondroitin sulfate proteoglycans (CSPGs) are most prevalent (Snow et al., 1990; McKeon et al., 1995; Davies et al., 1999). All of these inhibitory molecules, in concert, are believed to constitute an impenetrable barrier to the passage of severed axons either through or around CNS lesions. The enzymatic digestion of just one of these components, the inhibitory glycosaminoglycan (GAG) chains of extracellular matrix PGs (McKeon et al., 1995; Bradbury et al., 2002; Pizzorusso et al., 2002; Tropea et al., 2003), is one of several key strategies that can be used to increase regeneration or sprouting of axons.
The primary enzyme that has shown efficacy in decreasing PG-mediated inhibition is bacterial chondroitinase ABC that cleaves the GAG side chains on CS-containing PGs (Bradbury et al., 2002). The use of chondroitinase ABC, however, has one major disadvantage, which is its inability to completely digest the GAG chains from the protein core. The incomplete cleavage leaves the so-called carbohydrate “stubs” behind (Caterson et al., 1985). These undigested side-chain segments, although less potently inhibitory than the complete GAG, nonetheless can reduce the capacity of axons to regenerate (Lemons et al., 2004). Therefore, an improved reagent might be one that has the ability to more completely interrupt glycosylation of the PG core protein. Targeted disruption of the enzymes that assemble the GAG chains to the protein core is one potential solution to this problem. In this paper we show that local administration of a DNA enzyme against the GAG-chain initiating enzyme, xylosyltransferase-1 (XT-1) (Goetting et al., 2000), can greatly reduce the presence of GAG chains, which normally create an inhibitory penumbra around the epicenter of spinal cord stab lesions. The reduction of fully glycosylated PGs allows for the regeneration of microtransplanted adult sensory axons around and well past the central core in our lesion model.
Materials and Methods
Preparation of primary astrocyte cultures. Astrocytes were prepared from cerebral cortices of newborn Sprague Dawley rats and cultured for 14 d in DMEM-F12 (Invitrogen, Gaithersburg, MD). The astrocytes were stimulated with TGF-β (Sigma, St. Louis, MO) at a concentration of 2 ng/μl to increase CSPG expression (Asher et al., 2000). Some cultures were also treated with 8 μm DNA enzyme or control DNA enzyme (DNA mb) for 2 d followed by fixing and immunostaining.
Immunohistochemistry: CSPG, green fluorescent protein, and GFAP. The primary astrocyte cultures were fixed, and the spinal cords were perfused with 4% paraformaldehyde (PFA). The cultures and horizontal vibratome sections through the cord (70 μm) were incubated at 37°C for 30 min followed by rinsing in PBS followed by incubation with the first antibody, the mouse monoclonal CS-56 IgM antibody (1:200; Sigma), polyclonal rabbit anti-green fluorescent protein (GFP) antibody (1:1000; Molecular Probes, Eugene, OR), or an antibody against mouse GFAP (1:500; Sigma) with 5% normal goat serum (Invitrogen) diluted in PBS and 0.1% bovine serum albumin (Sigma) overnight at 4°C on a shaker. The coverslips or cord sections were rinsed again in PBS and incubated on a shaker for 2 hr at room temperature with the second antibody, a biotin-labeled goat anti-mouse IgM (1:200; Chemicon, Temecula, CA). This was followed by rinsing the coverslips or sections in PBS and incubation for 2 hr at room temperature with Texas Red-labeled streptavidin (1:200; Molecular Probes) or an Alexa 488-labeled goat anti-rabbit or anti-mouse IgG (1:500; Molecular Probes) for the cord sections. For the primary astrocytes, Oregon Green-labeled streptavidin (1:200; Molecular Probes) was used. The tissues or cultures were then rinsed and mounted in Citifluor (Ted Pella).
To quantify the decrease in GAG staining after DNA enzyme treatment against XT-1 compared with mixed base-treated astrocyte cultures, five coverslips of each group were stained with the CS-56 antibody. The center of each coverslip was chosen grossly without first looking through the microscope objective. All fields chosen in this “blind” manner were photographed under identical conditions, and their pixel intensities were measured, averaged, and compared by Tina 2.09 (Raytest). For the statistical evaluation, the ANOVA t test was used.
Dot blot. Six wells of astrocytes for each group [DNA enzyme treated and control (mb) DNA enzyme treated; each 8 μm] were collected and treated in the same manner as described in Grimpe et al. (2002). Equal amounts of protein, measured by the bicinchoninic acid assay (Pierce, Rockford, IL), were diluted in Tris-buffered saline (TBS) at a concentration of 1:100, and 200 μl were loaded in the dot blot apparatus (Bio-Rad, Hercules, CA) on a nitrocellulose membrane (Bio-Rad) in a triple experiment. The membrane was incubated with the CS-56 antibody (1:500) overnight at 4°C and stained with the second antibody conjugated with horseradish peroxidase (HRP) (1:1000; Chemicon). This step was followed by an ECL reaction (Amersham Biosciences, Arlington Heights, IL). The reaction was terminated and the dot blot was developed on Hyperfilm (Amersham) after 1 min. The film was quantified by Aida v3.21 (Raytest). For further evaluation, Excel (Microsoft) was used. The dot blot was reused by incubation in stripping buffer (100 mm DTT, 2% SDS, 62.5 mm Tris-HCl, pH 6.7) at 50°C two times for 30 min, reblocked, and restained with a polyclonal rabbit anti-GFAP antibody (Accurate) in a 1:1000 dilution overnight at 4°C on a shaker. The next day the blot was rinsed three times in TBS-T (T = Tween at 0.05%) and incubated with the second HRP-conjugated antibody in a 1:1000 dilution for 2 hr at room temperature. This was followed by rinsing the dot blot three times for 30 min in TBS-T. For statistical evaluation we used the ANOVA t test.
Design and biotinylation of the DNA enzyme against XT-1. The DNA enzyme was designed as end-capped phosphorothioated oligodeoxyribonucleotides (ODN-PSs) with two nucleotides on each end. They were obtained from MWG Biotech. The sequence corresponded to the 3′ end of the xylosyltransferase-1 mRNA sequence (accession no. RNO295748) (Goetting et al., 2000) and included the following nucleotides: DNAXT-1as2: TGG GGG GAC TTG GGC TAG CTA CAA CGA GAC CTT G; control DNAXT-1mb2: ACG AGT CAG GAA CAT CGA TCG GGA AGT CCC ATG C. The DNA enzyme has no homology to other mammalian sequences registered in the GenBank databases of the National Institutes of Health (Altschult et al., 1997). For the catalytic digestion of the targeted mRNA, the DNA enzyme contained a loop structure (Santoro and Joyce, 1997). The control DNA enzyme was designed not to bind to any mRNA and serves as a control for the presence of exogenous single-stranded DNA in a cell. The DNA enzyme was biotinylated at the 3′ end by MWG Biotech. Labeled DNA enzyme diffusion was evaluated in a separate group of three lesioned animals. After 7 d, spinal cord sections were stained with streptavidin-labeled Texas Red (1:200; Molecular Probes) overnight on a shaker followed by rinsing in PBS and mounting in Citifluor (Ted Pella). Evaluation of enzyme diffusion in the spinal cord tissue was measured on an Orthoplan 2 (Leitz) microscope. The same biotinylated DNA enzyme was used in our astrocyte cultures to monitor DNA enzyme uptake. The cultures were processed as described above.
Microtransplantation of dorsal root ganglia into the spinal cord. The adult dorsal root ganglia (DRG) were harvested from 2-month-old mice that expressed GFP under the control of the β-actin promoter (Okabe et al., 1997) using the technique described by Davies et al. (1999). Briefly, the dorsal columns of the spinal cord were exposed between the C4 and C5 vertebrae, and a small incision was made in the dura. A glass capillary with an outer diameter of ∼70-90 μm was inserted 1 mm into the fasciculus gracilis and via a Picospritzer (General Valve, Fairfield, NJ); ∼1 × 103 DRG in 0.5-0.9 μl of buffer was slowly injected. A dual-stab lesion was placed between the C5 and C6 vertebrae by inserting a 25 gauge needle two times into fasciculus gracilis and two times into fasciculus cuneatus ∼1 mm deep. Additionally, an intrathecal catheter (PE 10; Becton Dickinson, Sparks, MD) was placed 1.9 cm into the subarachnoidal space of the spinal cord via the alanto-occipital membrane. The tubing was filled with the DNA enzyme against the XT-1 or control DNA enzyme and connected to an osmotic minipump via PE 60 tubing. The pumping rate was 0.5 μl/hr with a concentration of the DNA enzyme at 10 μg/μl over a period of 7 d. After that time the animals were killed and perfused with 4% PFA, and the spinal cords were embedded into 5% agarose/5% gelatin.
Quantification of axon regeneration: numbers of animals. In total, we used 18 DNA enzyme-treated, 6 control (mb) DNA enzyme-treated, and 12 untreated animals for our experiments. Because of variability in placement (p) of the cannula tip and successful engraftment and axonal regeneration of the DRG neurons (r), for quantification we selected only those animals in which the tubing was correctly positioned and regeneration of any axons to the vicinity of the lesion was achieved (see Fig. 2). This left a total of 6 DNA-enzyme treated (excluding 12 of 18-3p/r, -6p, -3r), 3 control (mb) DNA enzyme-treated animals (excluding 3 of 6-2p, -1r), and 6 untreated (excluding 6 of 12-6r) for our quantification.
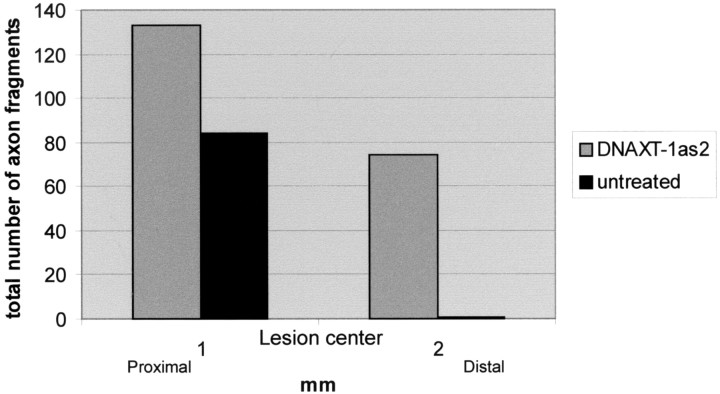
Figure 2.
The histogram shows the numbers of microtransplanted DRG axon fragments proximal and distal to the lesion in DNA enzyme-treated and untreated spinal cords.
For the quantification shown in Figure 2 and because the great number of sections exceeded our capacity for reconstructive confocal microscopy (∼9 lengthy sections per animal), the number of discrete GFP+ axon fragments in each spinal cord section of all 12 experimental animals (6 DNA enzyme-treated and 6 untreated animals) was documented with the use of a Leica Orthoplan 2 microscope. An “axon fragment” was defined as any discrete piece of an axon that was stained continuously throughout the focal plane of the section. We examined an area that spanned a distance of 1 mm rostral and 2 mm caudal to the center of the lesion.
RNA isolation and PCR. Twenty-four wells, each with 100,000 stimulated astrocytes, were treated for 3 d with the DNA enzyme against XT-1 at a concentration of 8 μm. The same number of wells was used for the untreated group. Additionally, crush-lesioned spinal cords were treated for 3 d with the DNA enzyme and control (mb) DNA enzyme, and the lesion site was punched out with a 3 mm diameter Uni-Punch (Premier Medical Products). The astrocytes as well as the spinal cord tissues were lysed and homogenized in Trizol (Invitrogen). Total RNA was prepared according to the supplier's protocol. Two micrograms of total astrocyte RNA and 0.6 μg of total spinal cord RNA were reverse transcribed into cDNA (Table 1) using the GenAmp RNA PCR Core Kit (PerkinElmer Life Sciences, Emeryville, CA) in accordance with the manufacturer's instructions. All RT-PCRs were analyzed on 1.0% agarose gels, stained with ethidium bromide, and photographed with a Polaroid camera (Kodak,Rochester, NY).
Table 1.
PCR conditions for XT-1, GAPDH, and cyclophilin
|
Name |
Sequence |
Length of end product |
Reference |
|---|---|---|---|
| XT-1 | XT1s1 | 316bp | Goetting et al. (2000) |
| 5′-CTTCATCCGCCTGGGTCTTCG-3′ | |||
| XT1as1 | |||
| 5′-GTAGTGTGTGAATTCCGCAGTGG-3′ | |||
| nested XT1s2 | |||
| 5′-GGGCTGAGTCATCGCTACACACG-3′ | |||
| nested XT1as2 | |||
| 5′-CGATGACATTGACAGGATCCACC-3′ | |||
| GAPDH | GAPDHs | 78bp | Tajima et al. (1999) |
| 5′-GAACATCATCCCTGCATACA-3′ | |||
| GAPDHas | |||
| 5′-CCAGTGAGCTTCCCGTTCA-3′ | |||
| Cyclophilin | cyclo as | 65 bp | Danielson et al. (1988) |
| 5′-TGTGCCAGGGTGGTGACTT-3′ | |||
| cyclo as | |||
|
|
5′-TTTCTCTCCGTAGATGGACTT-3′ |
|
|
Southern blot analysis. The oligodeoxynucleotide XT-1 Southern blot probe (5′-CTG ACC GAT TCC AGG GCT TTC TGA TCA AGC ACC ATG TGA C-3′) (Goetting et al., 2000) was obtained from MWG Biotech. This oligodeoxynucleotide probe was digoxigenin labeled according to the manufacture's protocol (Boehringer Mannheim, Indianapolis, IN). The PCR products were electrophoretically separated on a 1.0% agarose gel. The gel was denatured for 15 min in 3 m NaCl, 0.4 m NaOH followed by a 15 min incubation in transfer solution (3 m NaCl, 8 mm NaOH) and blotted onto Hybond N+ (Amersham Biosciences) as described previously (Sambrook et al., 1989). The following day the membrane was blocked and treated according to Engler-Blum et al. (1993). As a detection reagent, CDP-Star (Boehringer Mannheim) was used. The blots were exposed to Amersham Biosciences Hyperfilm for 1-2 sec for the astrocyte preparation and 10 sec for the spinal cord preparation.
Results
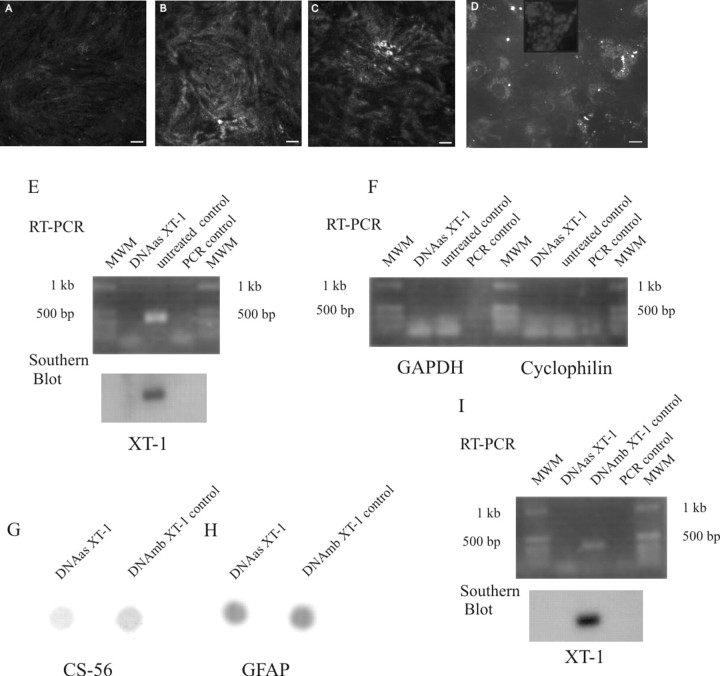
To investigate the functionality of our DNA enzyme against rat XT-1 and a nonfunctional control (mb) DNA enzyme, we used TGF-β to stimulate PG upregulation in reactive astrocytes (Asher et al., 2000). We observed after staining for extracellular PGs (i.e., no Triton added) with the CS-56 antibody, which recognizes the GAG chains on CSPGs, a 2.6-fold (p > 0.0088; see Materials and Methods for details) decrease in staining intensity in the DNA enzyme-treated cultures versus controls (Fig. 1A-C). With the use of a biotin-labeled DNA enzyme, we were able to monitor enzyme uptake within the glia (Fig. 1D). In a nested RT-PCR experiment we show that the mRNA of XT-1 in DNA enzyme-treated astrocyte cultures is strongly reduced compared with untreated cultures (Fig. 1E). To confirm the specificity of our DNA enzyme, an RT-PCR of the aforementioned mRNA for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and cyclophilin was performed that showed equal amounts of the mRNAs for these “housekeeping” genes (Fig. 1F). A dot-blot experiment with the CS-56 antibody showed a DNA enzyme-mediated reduction in the GAG chains of PGs (Fig. 1G). This reduction was 1.5-fold (p > 0.005) compared with control DNA enzyme-treated astrocyte cultures. To document that equal amounts of total protein were loaded, the nitrocellulose membrane was stripped and restained with an anti-GFAP antibody, which resulted in comparable staining (Fig. 1H). The reason for the inability to see more complete reduction of the PG level in the dot-blot assay is likely attributable to the fact that the astrocytes accumulated intracellular PGs before the treatment started and these were released along with extracellular PGs during the dot-blot procedure.
Figure 1.
Confocal photomicrographs of TGF-β-stimulated primary astrocyte cultures treated with the DNA enzyme (A) or control DNA enzyme (B) or left untreated (C) and stained with the CS-56 antibody. Uptake of the biotin-streptavidin-labeled DNA enzyme can be seen in the astrocytes (D). Scale bars, 10 μm. E, F, RT-PCR-Southern blot of primary astrocyte cultures for XT-1 (E) and RT-PCR for GAPDH and cyclophilin (F) of the same mRNA. G, Dot blot of primary astrocyte cultures treated with the DNA enzyme or control DNA enzyme and stained with the CS-56 antibody. H, Restaining of the same dot blot with GFAP. I, RT-PCR-Southern blot for the XT-1 in a DNA enzyme- and control DNA enzyme-treated spinal cord.
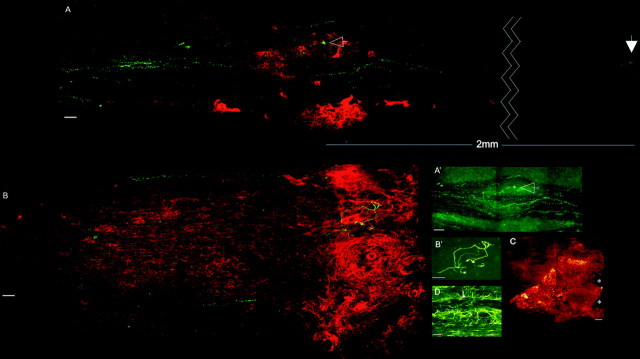
Microtransplantation techniques, which allow for an unambiguous identification of regenerating adult axons even within an incompletely lesioned white matter tract, have demonstrated the critical role of the lesion environment in regeneration failure (Davies et al., 1999). We repeated these experiments, and, additionally, the lesion was treated via infusion of the DNA enzyme at a concentration that was 117 times higher at the cannula tip than the concentration that was used in vitro. One group of control animals was treated with the control DNA enzyme and another control group was untreated. After 7 d the animals were killed, and spinal cord sections were stained for PGs with the CS-56 antibody as well as GFP to visualize the transplanted DRGs and their regenerating axons (see Fig. 3A,B). In both control groups, the rapidly regenerating axons failed to navigate beyond the confines of the lesion. Once they approached the lesion penumbra, which contains the lowest concentration of PGs (Fig. 3B), axons seemed trapped and were rarely able to leave this territory. Rather, just as axons behave in a PG step gradient in vitro (Snow and Letourneau, 1992), they preferred a path up the increasing concentration gradient of inhibitory matrix that brought them deeper into the lesion. Because the trajectory of the growth cone is documented by the shape of the axon, one gets the impression that fibers within the lesion environment have the capacity to struggle for a time, growing in contorted paths that end inevitably in the formation of dystrophic clubs (Fig. 3B,B′). In the six untreated animals, we observed 84 DRG axon fragments rostral to the lesion but just one axon fragment caudal to the lesion (Fig. 2).
Figure 3.
Confocal photomicrographs of treated and untreated spinal cords. Red/green (A) and green (A′) channels show the green DRGs and their axons passing the distinct borders of the lesion cores (red CS-56 staining) in a DNA enzyme-treated spinal cord. Axons that enter the lesion core become dystrophic (A, A′, arrows). Red/green (B) and green (B′) channels show an untreated spinal cord with the green DRG and red CS-56 staining. Note the struggling axon in the heart of the lesion. Scale bar, 50 μm. C, Diffusion of a labeled DNA enzyme in vivo 7 d after treatment. Scale bar, 100 μm. D, GFAP-stained section of the lesion area in a different animal. Note the disrupted astrocyte alignment in the core and borders of the lesion (L) and the aligned astrocytes (arrows) in the region where axons regenerate.
In the DNA enzyme-treated spinal cords the regeneration phenotype was quite different. In six treated spinal cords, 133 axon fragments could be identified 1 mm rostral to the lesion center and 74 were found well caudal to the lesion (Fig. 2). In all of these animals the PG penumbra was variably absent. Instead, in five of these six animals a more discrete (i.e., with a sharply delineated outer edge) core of PG staining was visible very close to the original lesion site. One animal showed a complete reduction of PG staining (data not shown). In all of these animals the regenerating axons grew straight toward the lesion; however, unlike their behavior in controls, the regenerating fibers were able to maneuver very tightly around the entirety of the outside edge of the lesion core or between the paired lesions and beyond, up to distances of a maximum of 2 mm in the dorsal columns. Axons that did penetrate the lesion core formed dystrophic end balls and were unable to pass (Fig. 3A,A′). As fibers navigated around the lesion, they appeared to use regions of cord tissue in which the longitudinal geometry of the intrafascicular astroglia was relatively undisturbed (Fig. 3D).
A biotinylated DNA enzyme was used to assess its range of diffusion from the tip of the cannula into the cord tissue. After 7 d, labeling extended ∼1 mm in all directions away from the epicenter (Fig. 3C). The XT-1 mRNA decrease within the lesion penumbra was also demonstrated by an RT-PCR Southern blot of treated versus control animals after 3 d (Fig. 1I).
Discussion
Our goal in these experiments was to test the hypothesis that the upregulation of GAG-bearing PGs after spinal cord injury plays a critical role in causing sensory axon regeneration failure. To achieve this we have used a novel DNA enzyme technology (Grimpe et al., 2002) to disrupt a critical enzyme in the synthetic pathway of all CS-bearing PGs. Although we could demonstrate a marked reduction of CSPGs within the lesion penumbra, those CSPGs within the core of the lesion remained at much higher levels. The reasons for incomplete reduction of PG upregulation in the lesion core could be attributable to the compensatory activity of XT-2, a second enzyme that is involved with GAG-chain synthesis that may be associated with various cell types that populate the center of stab lesions (Goetting et al., 2000). Another possibility is that the stimulus for PG upregulation in the lesion core may be extremely high, thereby necessitating even greater concentrations of the DNA enzyme. It is likely that inhibitory myelin components or fibroblast-associated inhibitory molecules that are not direct targets of the DNA-enzyme may still be present in the heart of the lesion and add to its inhospitality to the regenerating fibers (Brose et al., 1999; Pasterkamp et al., 1999; Bundesen et al., 2003). The remarkable behavior, however, of the regenerating axons growing in intimate association with the perimeter of the central lesion suggests that if these molecules help to prevent regeneration in the lesion core, they must act via highly local contact-inhibitory mechanisms. Most importantly, comparison of the different behaviors of the regenerating fibers in DNA enzyme-treated versus control animals suggests that it is the geometric distribution of PG in a gradient that reaches out well beyond the center of the lesion that is especially critical in regeneration failure. Uncovering the mechanisms by which proteoglycans in the penumbra transform the growth cone into a state that appears incapable of freeing itself from the lesion environment should be a major target of future research. Finally, it is important to comment on the interesting biocatalyst activity of DNA enzymes. These new reagents, which have been used primarily in cancer and AIDS research, are relatively simple to design and have greater flexibility for selection of specific cleavage sites than ribozymes, and their synthesis is relatively inexpensive. They can provide important tools for future use in spinal cord injury research.
Footnotes
This work was supported by National Institute of Neurological Disorders and Stroke Grant NS 25713, The Christopher Reeve Paralysis Foundation, The Daniel Heumann Fund, and the Brumagin Memorial Fund. We thank Albert Ries for his outstanding help in protein chemistry and photographic quantification. We also thank Prof. Paul Jones (Department of Epidemiology and Biostatistics, Case Western Reserve University) for his help in the statistical evaluation procedures.
Correspondence should be addressed to Barbara Grimpe, Case Western Reserve University, School of Medicine, Department of Neurosciences, 10900 Euclid Avenue, Cleveland, OH 44106. E-mail: bxg15@po.cwru.edu.
Copyright © 2004 Society for Neuroscience 0270-6474/04/241393-05$15.00/0
References
- Altschult SF, Madden TL, Schaeffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher RA, Morgenstern DA, Fidler PS, Adcock KH, Oohira A, Braistead JE, Levine JM, Margolis RU, Rogers JH, Fawcett JW (2000) Neurocan is upregulated in injured brain and in cytokine-treated astrocytes. J Neurosci 20: 2427-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury EJ, Moon LD, Popat RJ, King VR, Bennett GS, Patel PN, Fawcett JW, McMahon SB (2002) Chondroitinase ABC promotes functional recovery after spinal cord injury. Nature 416: 636-640. [DOI] [PubMed] [Google Scholar]
- Brose K, Bland KS, Wang KH, Arnott D, Henzel W, Goodman CS, Tessier-Lavigne M, Kidd T (1999) Slit proteins bind Robo receptors and have an evolutionarily conserved role in repulsive axon guidance. Cell 96: 795-806. [DOI] [PubMed] [Google Scholar]
- Bundesen LQ, Scheel TA, Bregman BS, Kromer LF (2003) Ephrin-B2 and EphB2 regulation of astrocyte-meningeal fibroblast interactions in response to spinal cord lesions in adult rats. J Neurosci 23: 7789-7800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caterson B, Christner JE, Baker JR, Couchman JR (1985) Production and characterization directed against connective tissue proteoglycans. Fed Proc 44: 386-393. [PubMed] [Google Scholar]
- Chen ZJ, Negra M, Levine A, Ughrin Y, Levine JM (2002) Oligodendrocyte precursor cells: reactive cells that inhibit axon growth and regeneration. J Neurocytol 31: 481-495. [DOI] [PubMed] [Google Scholar]
- Danielson PE, Forss-Petter S, Brow MA, Calavetta L, Douglass J, Milner RJ, Sutcliffe JG (1988) p1B15: a cDNA clone of the rat mRNA encoding cyclophilin. DNA 7: 261-267. [DOI] [PubMed] [Google Scholar]
- Davies SJ, Goucher DR, Doller C, Silver J (1999) Robust regeneration of adult sensory axons in degenerating white matter of the adult rat spinal cord. J Neurosci 19: 5710-5822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler-Blum G, Meier M, Frank J, Miller GA (1993) Reduction of background problems in nonradioactive Northern and Southern blot analysis enables higher sensitivity than 32P-based hybridization. Anal Biochem 210: 235-244. [DOI] [PubMed] [Google Scholar]
- Fitch MT, Doller C, Combs CK, Landreth GE, Silver J (1999) Cellular and molecular mechanisms of glial scarring and progressive cavitation in vivo and in vitro: analysis of inflammation-induced secondary injury after CNS trauma. J Neurosci 19: 8182-8192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetting C, Kuhn J, Zahn R, Brinkmann T, Kleesiek K (2000) Molecular cloning and expression of human UDP-d-xylose: proteoglycan core protein beta-d-xylosyltransferase and its first isoform XT-I. J Mol Biol 304: 517-528. [DOI] [PubMed] [Google Scholar]
- Grimpe B, Dong S, Doller C, Temple K, Malouf AT, Silver J (2002) The critical role of basement membrane-independent laminin gamma 1 chain during axon regeneration in the CNS. J Neurosci 22: 3144-3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones LL, Tuszynski MH (2002) Spinal cord injury elicits expression of keratan sulfate proteoglycans by macrophages, reactive microglia, and oligodendrocyte progenitors. J Neurosci 22: 4611-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemons ML, Sandy JD, Anderson DK, Howland DR (2004) Intact aggrecan and chondroitin sulfate depleted aggrecan core glycoprotein inhibit axon growth in the adult rat spinal cord. Exp Neurol 184: 981-990. [DOI] [PubMed] [Google Scholar]
- McKeon RJ, Hoke A, Silver J (1995) Injury-induced proteoglycans inhibit the potential for laminin-mediated axon growth on astrocytic scars. Exp Neurol 136: 32-43. [DOI] [PubMed] [Google Scholar]
- Menet V, Prieto M, Privat A, Gimenez y Ribotta M (2003) Axonal plasticity and functional recovery after spinal cord injury in mice deficient in both glial fibrillary acidic protein and vimentin genes. Proc Natl Acad Sci USA 100: 8999-9004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe M, Ikawa M, Kominami K, Nakanishi T, Nishimune Y (1997) “Green mice” as a source of ubiquitous green cells. FEBS Lett 407: 313-319. [DOI] [PubMed] [Google Scholar]
- Pasterkamp RJ, Giger RJ, Ruitenberg MJ, Holtmaat AJ, De Wit J, De Winter F, Verhaagen J (1999) Expression of the gene encoding the chemorepellent semaphoring III is induced in the fibroblast component of neural scar tissue formed following injuries of adult but not neonatal CNS. Mol Cell Neurosci 13: 143-166. [DOI] [PubMed] [Google Scholar]
- Pizzorusso T, Medini P, Berardi N, Chierzi S, Fawcett JW, Maffei L (2002) Reactivation of ocular dominance plasticity in the adult visual cortex. Science 298: 1248-1251. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory.
- Santoro SW, Joyce GF (1997) A general purpose RNA-cleaving DNA-enzyme. Proc Natl Acad Sci USA 94: 4262-4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow DM, Letourneau PC (1992) Neurite outgrowth on a step gradient of chondroitin sulfate proteoglycan (CS-PG). J Neurobiol 23: 322-336. [DOI] [PubMed] [Google Scholar]
- Snow DM, Lemmon V, Carrino DA, Caplan AI, Silver J (1990) Sulfated proteoglycans in astroglial barriers inhibit neurite outgrowth in vitro. Exp Neurol 109: 111-130. [DOI] [PubMed] [Google Scholar]
- Song H, Ming G, He Z, Lehmann M, McKerracher L, Tessier-Lavigne M, Poo M (1998) Conversion of neuronal growth cone responses from repulsion to attraction by cyclic nucleotides. Science 281: 1515-1518. [DOI] [PubMed] [Google Scholar]
- Tajima H, Tsuchiya K, Yamada M, Kondo K, Katsube N, Ishitani R (1999) Over-expression of GAPDH induces apoptosis in COS-7 cells transfected with cloned GAPDH cDNAs. NeuroReport 10: 2029-2033. [DOI] [PubMed] [Google Scholar]
- Tropea D, Caleo M, Maffei L (2003) Synergistic effects of brain-derived neurotrophic factor and chondroitinase ABC on retinal fiber sprouting after denervation of the superior colliculus in adult rats. J Neurosci 23: 7034-7044. [DOI] [PMC free article] [PubMed] [Google Scholar]