Abstract
Flip (i) and flop (o) alternatively spliced variants of the four glutamate AMPA receptor subunits (GluR1-4) are differentially expressed in the CNS and can display distinct rates of desensitization that contribute to the heterogeneity of native AMPA receptor-dependent synaptic responses. In the present study, we initially compared the kinetics of desensitization in response to fast application of glutamate (1 mm) for the eight different homomeric recombinant human AMPA receptors (hGluR1-4i and o) heterologously expressed in mammalian cells. Consistent with previous reports on recombinant rat AMPA receptors, the time constants of desensitization between human GluR1i and GluR1o receptors were the same, whereas the flip isoforms for GluR2-4 receptors exhibited significantly slower rates of desensitization compared with the flop isoforms. To identify the molecular determinants responsible for these functional differences, the effects of exchanging amino acid residues in the flip-flop cassette of GluR2i and GluR2o were investigated. Three amino acid residues in the flip-flop region (Thr765, Pro766, and Ser775 in flip and Asn765, Ala766, and Asn775 in flop) were identified that contribute to splice-variant differences in the rate of desensitization. Recent structural data show that these three residues are located on helix J, which forms part of the intradimer interface of AMPA receptor ligand-binding cores, and that the stability of this interface may regulate desensitization. The present results suggest that these three residues may confer differences in flip and flop receptor desensitization rates by directly and/or indirectly influencing the stability of the interface between adjacent subunits.
Keywords: AMPA, desensitization, glutamate, alternative splicing, flip, flop
Introduction
Ionotropic glutamate AMPA receptors are expressed ubiquitously throughout the CNS and mediate the majority of rapid excitatory neurotransmission. These receptors are tetrameric complexes assembled from subunits encoded by four different genes [glutamate AMPA receptor subunits 1-4 (GluR1-4)] (Hollmann et al., 1989) yielding functional homomeric and heteromeric receptors. Recent work on the assembly of AMPA receptors provides evidence that certain subunit stoichiometries are favored; however, the rules governing assembly still remain essentially unknown (Mansour et al., 2001; Brorson et al., 2004). Additional molecular diversity comes from alternative splicing of RNA for each subunit in a region that encodes a 38 amino acid sequence in the extracellular ligand-binding core (LBC), giving rise to two splice variants, flip (i) and flop (o) (Sommer et al., 1990). The amino acid sequence in the flip-flop region differs by <10 residues for each isoform of GluR1-4. Although the flip-flop region does not appear to significantly influence the sensitivity to glutamate or the rate of deactivation of splice variants, for GluR2-4, the two isoforms display markedly different desensitization kinetics (Sommer et al., 1990; Mosbacher et al., 1994; Koike et al., 2000; Krampfl et al., 2001). Homomeric GluR2-4i receptors exhibit slower rates of desensitization and faster recovery from desensitization than GluR2-4o receptors (Mosbacher et al., 1994; Koike et al., 2000). Flip and flop isoforms are differentially expressed in a cell-specific manner in the CNS (Geiger et al., 1995) that has been proposed to contribute to the functional heterogeneity of AMPA receptor-dependent synaptic transmission (Jonas and Sakmann, 1992).
Crystallography studies of the GluR2o LBC have provided insights into the role of the flip-flop region in desensitization. Evidence indicates that LBC protomers can form dimers and that stabilization of the intradimer interface is correlated with the rate of desensitization (Sun et al., 2002). The majority of the flip-flop region is contained within the LBC, and residues within this region make intradimer contacts (Armstrong and Gouaux, 2000). As such, the molecular differences between splice variants may govern their rates of desensitization by directly and/or indirectly influencing the stability of the intradimer interface. As a first step toward testing this hypothesis, the present studies have investigated the critical residues responsible for differences in desensitization rates of flip and flop receptors by systematically exchanging amino acids in the flip-flop cassette on recombinant human homomeric GluR2i and GluR2o desensitization kinetics.
Materials and Methods
Molecular biology. Production of stable cell lines was performed as described previously (Quirk and Nisenbaum, 2003). Briefly, human embryonic kidney 293 (HEK293) cells were transfected with the mammalian expression vectors pcDNA3.1 or pRc-CMV containing a GluR subunit gene. Transfected cells were selected on the basis of G418 resistance, and expression of genes was confirmed by reverse transcription-PCR. The stable GluR3o cell line was established in a similar manner using adenovirus (12)-transformed Syrian hamster kidney cells. Production of mutations was performed as described previously (Quirk and Nisenbaum, 2003). Briefly, stable HEK293 cell lines were generated using the Flp-In system (Invitrogen, San Diego, CA). Cell lines were established on the basis of hygromycin B (100 mg/ml; Invitrogen) resistance. Site-directed mutagenesis was performed using the QuikChange mutagenesis kit (Stratagene, La Jolla, CA) using complementary primers. Presence of the desired mutant and verification of the absences of unwanted substitutions were confirmed by direct sequencing of the complete coding region.
Electrophysiological recordings from HEK293 cells. Glutamate (1 mm)-evoked currents were recorded from small isolated cells 1-3 d after plating. In the case of transient transfections, recordings were performed on isolated green fluorescent protein-positive cells 30-54 hr after transfection. Electrodes were fire polished and filled with a solution containing (in mm): 165 N-methyl-d-glucamine, 40 HEPES, 4 MgCl2, 3.6 Na2ATP, 0.35 GTP, 3.5 BAPTA, pH 7.2, osmolarity = 280 mOsm/l with phosphocreatine. The extracellular recording solution contained (in mm): 120 NaCl, 1 MgCl2, 10 HEPES, 5 BaCl2, 20 CsCl, pH 7.4, osmolarity = 300 mOsm/l with glucose. After placing the recording electrode into the bath, offset potentials were corrected, and electrode resistances were 2-5 MΩ. Voltage-clamp recordings were made while the membrane potential of cells was held at -80 mV, and currents were filtered at 5 kHz and digitized at 20 kHz. Series resistance was compensated (65-80%) and monitored periodically. Glutamate (1 mm) was rapidly applied to small cells via a double-barreled glass theta tube with a final diameter of ∼100 μm. Theta-tube position was shifted using a piezoelectric actuator having a charging time of 0.3 msec. Onset-to-offset time of junction potential measurements (0.1 and 1.0 m NaCl) were ∼700 μsec. Solutions were delivered to the theta tube using a gravity-induced flow method. All experiments were conducted at room temperature.
The rate of onset of desensitization was evaluated by fitting the decay of responses in the presence of glutamate from 75-95% of the peak current to steady state with a single exponential function to determine the time constant of desensitization (τDES). Statistical differences between τDES and the time constant of recovery from desensitization (τREC) for splice variants and/or subunits were determined using an ANOVA followed by Tukey's multiple post hoc comparison tests.
Results
Recombinant human homomeric AMPA receptors display different rates of desensitization
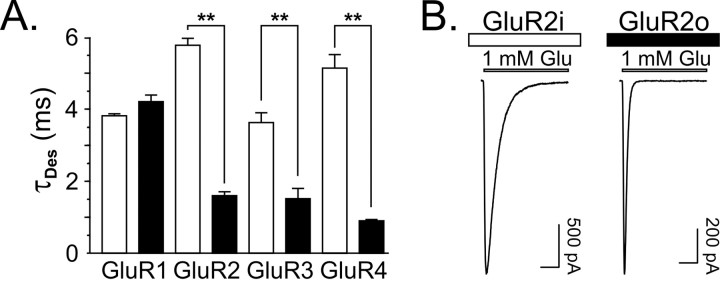
Consistent with results from previous studies of rat homomeric AMPA receptors (Mosbacher et al., 1994; Koike et al., 2000), the rates of desensitization of recombinant human homomeric AMPA receptors differed between splice variants for GluR2, GluR3, and GluR4 receptors. Specifically, desensitization rates of GluR2-4i were significantly faster than GluR2-4o receptors (Table 1, Fig. 1A,B). In contrast, the desensitization rates for GluR1i and GluR1o were not different (Table 1). These data show that the relative differences in the rate of desensitization for flip and flop isoforms are not species specific. Nonetheless, absolute differences between species for particular subunit-isoform combinations may exist. For example, the desensitization time constants for human GluR1i (3.8 msec) and GluR1o (4.2 msec) (measured from whole cells; present study) are ∼50% longer than rat GluR1i (2.5 msec) and GluR1o (3.2 msec) (measured from outside-out patches) (Partin et al., 1996). These disparities likely result from differences in amino acid sequence between species peripheral to the splice variant region and/or differences in recording techniques between studies.
Table 1.
Desensitization time constants
|
Receptor |
Residues exchanged |
τDES (msec) |
|---|---|---|
| GluR1i | - | 3.8 ± 0.05 (12) |
| GluR1o | - | 4.2 ± 0.2 (10) |
| GluR2i | - | 5.8 ± 0.2 (20) |
| GluR2o | - | 1.6 ± 0.1 (16) |
| GluR3i | - | 3.6 ± 0.3 (10) |
| GluR3o | - | 1.5 ± 0.1 (7) |
| GluR4i | - | 5.1 ± 0.4 (16) |
| GluR4o | - | 0.9 ± 0.03 (14) |
| GluR2io,i,i | T765N, P766A | 2.7 ± 0.1 (15) |
| GluR2ii,o,i | S775N | 4.7 ± 0.2 (16) |
| GluR2iV779L | V779L | 5.1 ± 0.2 (13) |
| GluR2ii,i,o | A796S, K797G, D798G, S799G, G800D | 5.8 ± 0.2 (23) |
| GluR2io,o,i | T765N, P766A, S775N | 1.8 ± 0.1 (18) |
| GluR2iT765N | T765N | 3.6 ± 0.1 (18) |
| GluR2iP766A | P766A | 3.6 ± 0.2 (15) |
| GluR2io,i,o | T765N, P766A, K797G, D798G, S799G, G800D | 2.5 ± 0.1 (18) |
| GluR2io,i,V779L,i | T765N, P766A, V779L | 2.1 ± 0.2 (15) |
| GluR2oi,o,o | N765T, A766P | 4.2 ± 0.2 (15) |
| GluR2oo,i,o | N775S | 2.3 ± 0.1 (13) |
| GluR2oi,i,o
|
N765T, A766P, N775S |
5.7 ± 0.2 (20) |
τDES was estimated from single exponential fits as described in Materials and Methods. Numbers in parentheses indicate the number of cells.
Figure 1.
Desensitization kinetics for human homomeric AMPA receptors. A, Bar graph of the time constants of desensitization (mean ± SEM). Open bars represent flip isoform, and closed bars represent flop isoform. Asterisks indicate that for GluR2-4, the τDES for the flop isoform is significantly faster than for the flip isoform (p < 0.001). B, Representative traces of GluR2i and GluR2o. Calibration bar, 10 msec.
Substitution of three complementary flop-specific residues in GluR2i confers GluR2o kinetics of desensitization
To determine the molecular determinants that underlie differences in desensitization kinetics between isoforms, a mutational strategy to exchange residues within the flip-flop module of GluR2 was conducted. Alignment of the amino acid sequences for the two isoforms of GluR2 reveals that differences in the flip-flop domain are clustered in three different regions (Fig. 2A). Region 1 is at the N-terminal border of the alternatively spliced domain and consists of a Thr765-Pro766 in GluR2i and an Asn765-Ala766 in GluR2o subunits. Region 2 is 10 amino acids downstream and contains a Ser775 in GluR2i and an Asn775 in GluR2o subunits. Region 3 is located at the C-terminal boundary of the flip-flop domain and is comprised of five residues, Ala796-Lys797-Asp798-Ser799-Gly800 in GluR2i and Ser796-Gly797-Gly798-Gly799-Asp800 in GluR2o subunits. In addition, there is a relatively conservative difference of a Val779 in GluR2i to a Leu779 in GluR2o. In the present report, the exchange of regions 1, 2, and 3 in GluR2i and GluR2o are identified as a subscripted “i” or “o” in the mutated receptors (Partin et al., 1996; Quirk and Nisenbaum, 2003). For example, an exchange of the two residues in region 1 at the N-terminal border in GluR2i for the residues in GluR2o is indicated as GluR2io,i,i. Exchanges of single residues, such as the V/L779 site, also are subscripted; for example, GluR2iV779L indicates that Val779 in GluR2i was exchanged for Leu779 in GluR2o.
Figure 2.
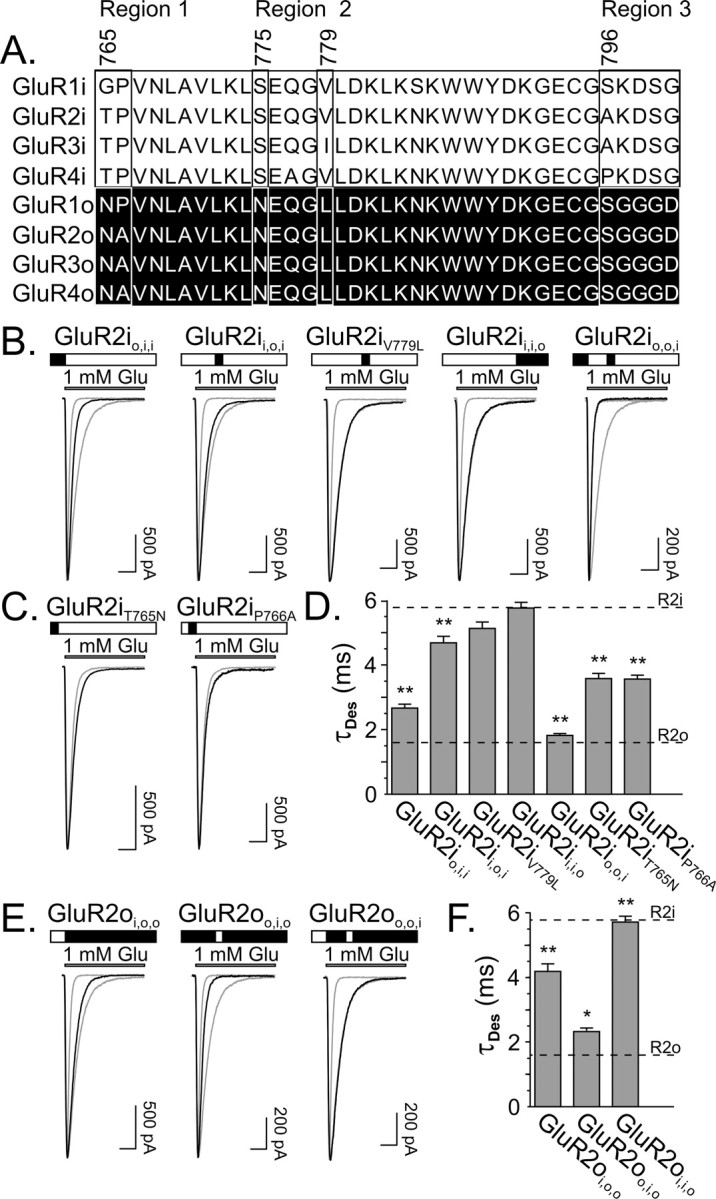
Desensitization kinetics are sensitive to the exchange of three residues in GluR2i and GluR2o. A, Sequence alignment of the flip and flop domains for GluR1-4. Numbering is based on GluR2 (accession number NP_000817), showing four regions of heterogeneity (regions 1, 2, 3, and Val/Leu779 residue). B, Representative traces from mutant GluR2i with exchanges of region 1 (GluR2io,i,i), region 2 (GluR2ii,o,i), V/L residue (GluR2iV779L), region 3 (GluR2ii,i,o), and region 1 and 2 (GluR2io,o,i). Traces from GluR2i and GluR2o are superimposed in gray for comparison. Above each trace, open rectangles indicate GluR2i residues, and filled rectangles indicate GluR2o residues. C, Representative traces from GluR2iT765N and GluR2iP765A. Trace from GluR2io,i,i is superimposed in gray for comparison. D, Bar graph of the τDES (mean ± SEM) for mutant GluR2i. Asterisks indicate mutant receptors that have faster τDES than GluR2i (p < 0.001). The mean τDES for GluR2i (R2i) and GluR2o (R2o) wild-type receptors are indicated for comparison. E, Representative traces from GluR2oi,o,o, GluR2oo,i,o, or GluR2oi,i,o. Traces from GluR2i and GluR2o are superimposed in gray for comparison. Calibration bar, 10 msec. F, Bar graph of the τDES (mean ± SEM) for mutant GluR2o. Asterisks indicate mutant receptors having slower τDES than GluR2o (*p < 0.05; **p < 0.001). The mean τDES for GluR2I and GluR2o wild-type receptors are indicated for comparison.
Initial experiments focused on exchange of individual regions in GluR2i for the corresponding regions in GluR2o. Results showed that substitution of region 1 (T765N, P766A; GluR2io,i,i) and region 2 (S775N; GluR2ii,o,i), but not region 3 (A796S, K797G, D798G, S799G, G800D; GluR2ii,i,o), alone in GluR2i increased the rate of desensitization (Table 1, Fig. 2B,D). However, neither substitution of region 1 nor region 2 alone in GluR2i was sufficient to decrease the time constant of desensitization to levels equivalent to GluR2o. Indeed, only when region 1 and region 2 were exchanged in combination was the rate of desensitization identical to GluR2o (Table 1, Fig. 2B,D).
Because region 1 contains two residues (Thr765-Pro766 in GluR2i and Asn765-Ala766 in GluR2o), it was possible that exchange of only one of these residues in GluR2i was required to increase the rate of desensitization. Furthermore, the facts that GluR1i and GluR1o have similar rates of desensitization (Fig. 1A) and both of these isoforms contain a Pro as the second residue in region 1 (Gly-Pro in GluR1i and Asn-Pro in GluR1o) suggested that substitution of Asn765 for Thr765 in region 1 of GluR2i might account for the increase in rate of desensitization associated with mutating this entire region. Results showed that although this substitution (GluR2iT765N) did increase the rate of desensitization, exchange of Pro for Ala (GluR2iP766A) had a similar effect (Table 1, Fig. 2C,D). However, neither exchange alone decreased the desensitization time constant to levels equivalent to that produced by exchange of the two amino acids in combination (GluR2io,i,i), indicating that both residues contribute equally to effects of region 1 mutation in GluR2i.
Besides the three regional sequence differences between isoforms of GluR2, there is an additional difference at residue 779 (Val in GluR2I; Leu in GluR2o). Although relatively conservative, the V/L residue recently has been shown to be critical for the activity of an allosteric modulator that binds at the dimer interface (Quirk and Nisenbaum, 2003; D. Timm, personal communication), suggesting a possible role for this residue in the differences in GluR2i and GluR2o desensitization rates. In addition, although exchange of region 1 in combination with region 2 in GluR2i was sufficient to confer the GluR2o rate of desensitization, the possibility that exchange of region 1 in combination with either the V/L site or region 3 could produce similar effects was explored. Results showed that mutation of the V/L site did not alter the rate of desensitization of GluR2i (Table 1, Fig. 2B,D). In addition, substitution of region 1 in combination with either L779 (GluR2io,i,V779L,i) or region 3 (GluR2io,i,o) in GluR2i did not increase the rate of desensitization compared with mutation of region 1 alone (GluR2io,i,i) (Table 1). Collectively, these data indicate that region 1 and region 2 are necessary and sufficient to account for the differences between the desensitization rates of GluR2i and GluR2o.
Substitution of the three complementary flip-specific residues in GluR2o confers GluR2i kinetics of desensitization
To provide further confirmation of the critical role of region 1 and region 2 in the differences in desensitization kinetics between AMPA receptor splice variants, a complementary mutational analysis was conducted with GluR2o. Analogous to results described above, individual substitutions of region 1 (N765T, A766P; GluR2oi,o,o) or region 2 (N775S; GluR2oo,i,o) in GluR2o attenuated the rate of desensitization (Table 1, Fig. 2E,F). However, neither mutation of region 1 nor region 2 alone was sufficient to confer GluR2i desensitization kinetics. Only exchange of region 1 and region 2 in combination (GluR2oi,i,o) reduced the rate of desensitization equivalent to GluR2i (Table 1, Fig. 2E,F).
The same three residues are responsible for splice-variant differences in kinetics of recovery from desensitization
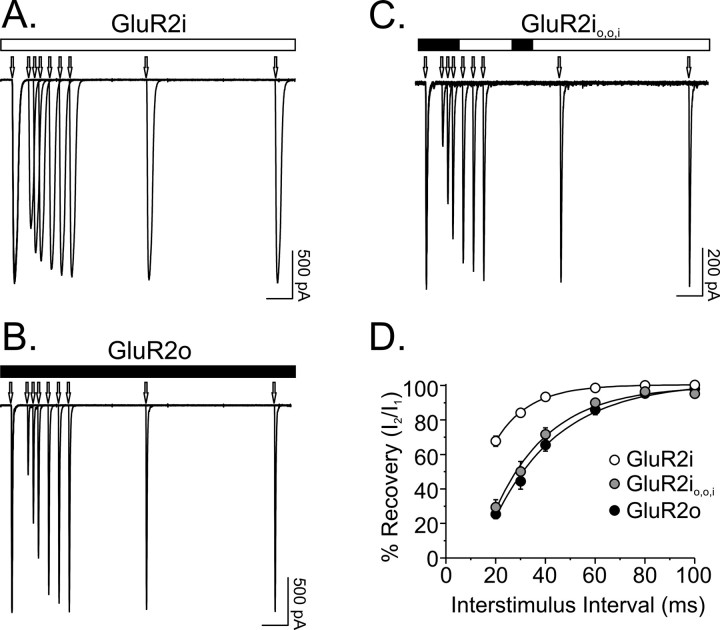
Previous studies of rat GluR2 flip and flop isoforms have demonstrated that the more rapid desensitization of GluR2o is associated with a slower time constant of recovery from desensitization than that for GluR2i (Koike et al., 2000). Similar results were found in the present studies of human receptors, such that recovery kinetics for GluR2o (τREC = 26.5 ± 2.4 msec; n = 5) were approximately twofold slower than for GluR2i (τREC = 14.1 ± 1.0 msec; n = 7) (Fig. 3A,B,D). These differences were not attributable to differences in the editing status of the R/G764 residue at the N-terminal boundary of the flip-flop domain that has previously been shown to influence kinetics of recovery from desensitization (Lomeli et al., 1994), because both GluR2i and GluR2o contained the edited residue. To determine whether the amino acids responsible for splice-variant differences in rate of desensitization also accounted for differences in recovery kinetics, the effects of mutating region 1 and region 2 of GluR2i individually and in combination were evaluated. Exchange of region 1 (T765N, P766A) or region 2 (S775N) had minimal to modest effects on the recovery kinetics of GluR2i (GluR2io,i,i: τREC = 15.8 ± 1.7 msec, n = 11; GluR2ii,o,i: τREC = 17.9 ± 2.8 msec, n = 8). However, substitution of both regions in combination yielded recovery kinetics equivalent to GluR2o (GluR2io,o,i: τREC = 21.3 ± 1.8 msec; n = 16) (Fig. 3C,D). Thus, the residues that account for differences in desensitization kinetics between flip and flop receptors also are responsible for the distinct kinetics of recovery from desensitization for the two isoforms.
Figure 3.
Recovery from desensitization kinetics are sensitive to the exchange of three residues in GluR2i. A-C, Recovery from desensitization was evaluated by delivering pairs of 10 msec pulses of 1 mm glutamate separated by interstimulus intervals ranging from 20 to 500 msec. Traces represent superimposed responses from cells expressing GluR2i, GluR2o, and mutant GluR2i with exchanges of region 1 and 2 (GluR2io,o,i). Arrows denote on set of glutamate pulses. Calibration bar, 50 msec. D, The time course of recovery from desensitization was determined for each cell by measuring the amplitude of the second response (I2) relative to the first response (I1) of a pair of pulses and plotting these values as a function of interstimulus interval. The τREC for each cell was determined by fitting these recovery plots with a single exponential function. Statistically significant differences were found between τREC for GluR2i, GluR2o, and GluR2io,o,i (F(2,27) = 7.2; p < 0.005). Post hoc comparisons showed that although the τREC for GluR2io,o,i was not different from the τREC for GluR2o (p > 0.05), both differed from the τREC for GluR2i (p < 0.05).
Discussion
Molecular determinants of differences in desensitization kinetics of flip and flop receptors
Desensitization occurs in nearly all ligand-gated ion channels and involves entry of the channel into a nonconducting state despite the continued presence of agonist. For ionotropic glutamate AMPA receptors, desensitization proceeds relatively rapidly and can govern the amplitude, kinetics, and frequency of excitatory synaptic transmission (Jones and Westbrook, 1996). Modulation of the desensitization process of AMPA receptors occurs naturally through the expression of flip and flop alternatively spliced variants of GluR2-4. Consistent with previous reports (Mosbacher et al., 1994; Koike et al., 2000), the present studies show that GluR2-4 composed of the flip isoform desensitize more slowly than corresponding GluR2-4 of the flop isoform. Mutagenic analysis of GluR2 isoforms reveals that the differences in desensitization kinetics are determined by the identity of only three amino acid residues in the flip-flop module: GluR2i: Thr765, Pro766, Ser775 and GluR2o: Asn765, Ala766, Asn775. Because all three residues identified in GluR2 are found in the equivalent positions in GluR3 and GluR4, these amino acids most likely also are responsible for differences in isoform desensitization rates in these subunits. However, inspection of the sequence alignments in the flip-flop domain for GluR3 shows that the residue equivalent to Val779 in GluR2i is an Ile in GluR3i (Leu in GluR3o). A similar examination for GluR4 reveals two differences at positions equivalent to Gln777 and Ala796 in GluR2i between GluR4i (Ala and Pro) and GluR4o (Gln and Ser) (Fig. 2A). These additional differences also may influence the rate of desensitization in GluR3 and GluR4 splice variants.
In contrast to splice-variant differences in desensitization kinetics for GluR2-4, GluR1 isoforms display similar rates of desensitization despite expressing different residues in two of the three positions (765 and 775) that contribute to differences in desensitization rates of GluR2 isoforms (Fig. 2A). The absence of isoform differences in GluR1 may be attributable to the presence of Gly in GluR1i compared with Thr765 in GluR2-4i and/or the expression of Pro in both splice variants of GluR1 compared with Pro766 in GluR2i and Ala766 in GluR2o. Alternatively, sequence differences between GluR1 and GluR2-4 peripheral to the flip-flop domain may preclude GluR1 splice-variant differences in desensitization rates.
Differences in the rate of desensitization for homomeric AMPA receptors independent of splice variant also were observed (Fig. 1A) (Mosbacher et al., 1994). For example, the rate of desensitization for GluR3i is significantly faster than the rates for flip isoforms of GluR2 and GluR4 subunits. Likewise, desensitization of GluR4o proceeds more rapidly than GluR1-3o (Table 1). These kinetic differences most likely arise from residues that differ between the four different subunits outside the flip-flop domain. Indeed, comparison of the sequences for GluR2i and GluR3i in the splice variant region shows that the only difference is the Val/Ile779 residue, which had no impact on desensitization rates when mutated in GluR2i in the present studies. A similar comparison between GluR4o and GluR2o reveals no differences in sequence in the flip-flop domain, indicating that the other residues must confer these distinct desensitization rates. Additional support for this conclusion comes from chimera studies using GluR1o and GluR3o that have identified a tyrosine residue outside of the flip-flop region that contributes to the differences in the rate of desensitization for these subunits (Banke et al., 2001).
Implications for molecular determinants of desensitization of kainate receptors
Previous investigations of the molecular determinants of desensitization of kainate receptors have shown that mutation of residues corresponding to those in region 1 and region 2 of GluR2-4 modifies the desensitization kinetics of homomeric GluR6 (Fleck et al., 2003). Alignment of the amino acid sequences for human GluR2-4 with the rat GluR6 shows that the Thr/Asn 765, Pro/Ala766, Ser/Asn775 residues in GluR2-4i/o are equivalent to Asp, Lys, and Gln in GluR6. Exchange of Asp in GluR6 for either Thr or Gly (corresponding residue in GluR1i) markedly increased the rate of desensitization compared with wild-type GluR6 (Fleck et al., 2003). The kinetics of GluR6 desensitization also were modified after exchange of Gln for Ser, such that this mutation attenuated desensitization nearly threefold (Partin et al., 1995; Partin, 2001). In conjunction with the present results, these data suggest that although they are not identical (Stern-Bach et al., 1998), considerable homology exists between the molecular determinants of kainate and AMPA receptor desensitization (Fleck et al., 2003).
Structural implications
Recent structure-function studies combining crystallographic and electrophysiological analyses have provided insights into the mechanism that governs AMPA receptor desensitization (Sun et al., 2002). Evidence indicates that the stability of the dimer interface between adjacent LBCs is a critical structural feature that regulates the gating of AMPA receptors. According to the model of Sun et al. (2002), desensitization involves destabilization of the intradimer interface. In the crystal structure of the GluR2o LBC, the three key residues comprising regions 1 (Thr/Asn 765, Pro/Ala766) and 2 (Ser/Asn775) in the flip-flop module that govern differences in splice-variant desensitization and recovery kinetics are located on helix J at the dimer interface (Armstrong and Gouaux, 2000; Sun et al., 2002). Consistent with the absence of an influence of region 3, these residues are not contained within the crystal structure and do not appear to contribute to the dimer interface. Of the three determinant residues, Asn775 has been shown to contribute directly to stabilization of the dimer in the activated state by forming an intermolecular hydrogen bond with the backbone carbonyl of Ser729 from the adjacent protomer (Armstrong and Gouaux, 2000; Sun et al., 2002). Additional support for a critical role for this residue comes from mutational studies in GluR2i demonstrating that exchange of Ser for Asp leads to a marked increase in the rate of desensitization with the prediction that this would also contribute to the associated destabilization of the dimer interface (Sun et al., 2002); similar results were found for GluR1i (Partin et al., 1996). Viewed in the context of the present results, these data suggest that the faster desensitization rate of GluR2o is attributable in part to a less stable dimer interface conferred by Asn775 compared with Ser775 in GluR2i.
An additional contribution to dimerization comes from an intermolecular hydrophobic cluster containing residues from helix J (Armstrong and Gouaux, 2000). To date, only the GluR2o LBC has been crystallized, so the extent (if any) to which these intermolecular bonds differ in wild-type GluR2i remains unknown. However, mutation of residues in this intermolecular hydrophobic cluster can alter the stability of the dimer and rate of desensitization (Stern-Bach et al., 1998; Sun et al., 2002; Horning and Mayer, 2004). Although Asn765 and Ala766 in GluR2o are not directly involved in this hydrophobic cluster, the present results demonstrating their role in governing desensitization and recovery kinetics suggest that the identity of these residues may indirectly influence the stability of this hydrophobic cluster and/or other protomer-protomer interactions (e.g., Asn775-Ser729). A more definitive description of the precise mechanisms by which these three residues contribute to differences in splice-variant desensitization-recovery kinetics will await future biochemical and structural experiments investigating the effects of residue exchanges in this region of GluR2o (and GluR2i) on dimer stability.
Footnotes
We thank Brian Eastwood, David Timm, and Dennis Zimmerman for their insightful contributions to this manuscript.
Correspondence should be addressed to Dr. Eric S. Nisenbaum, Eli Lilly and Company, Lilly Corporate Center, Indianapolis, IN 46285. E-mail: ESN@lilly.com.
Copyright © 2004 Society for Neuroscience 0270-6474/04/2411416-05$15.00/0
References
- Armstrong N, Gouaux E (2000) Mechanisms for activation and antagonism of an AMPA-sensitive glutamate receptor: crystal structures of the GluR2 ligand binding core. Neuron 28: 165-181. [DOI] [PubMed] [Google Scholar]
- Banke TG, Greenwood JR, Christensen JK, Liljefors T, Traynelis SF, Schousboe A, Pickering DS (2001) Identification of amino acid residues in GluR1 responsible for ligand binding and desensitization. J Neurosci 21: 3052-3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brorson JR, Li D, Suzuki T (2004) Selective expression of heteromeric AMPA receptors driven by flip-flop differences. J Neurosci 24: 3461-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleck MW, Cornell E, Mah SJ (2003) Amino-acid residues involved in glutamate receptor 6 kainate receptor gating and desensitization. J Neurosci 23: 1219-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger JR, Melcher T, Koh DS, Sakmann B, Seeburg P, Monyer H (1995) Relative abundance of subunit mRNAs determines gating and Ca2+ permeability of AMPA receptors in principal neurons and interneurons in rat CNS. Neuron 15: 193-204. [DOI] [PubMed] [Google Scholar]
- Hollmann M, O'Shea-Greenfield A, Rogers SW, Heinemann S (1989) Cloning and functional expression of a member of the glutamate receptor family. Nature 342: 643-648. [DOI] [PubMed] [Google Scholar]
- Horning MS, Mayer ML (2004) Regulation of AMPA receptor gating by ligand binding core dimers. Neuron 41: 379-388. [DOI] [PubMed] [Google Scholar]
- Jonas P, Sakmann B (1992) Glutamate receptor channels in isolated patches from CA1 and CA3 pyramidal cells of rat hippocampal slices. J Physiol (Lond) 455: 143-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MV, Westbrook GL (1996) The impact of receptor desensitization on fast synaptic transmission. Trends Neurosci 19: 96-101. [DOI] [PubMed] [Google Scholar]
- Koike M, Tsukada S, Tsuzuki K, Kijima H, Ozawa S (2000) Regulation of kinetic properties of GluR2 AMPA receptor channels by alternative splicing. J Neurosci 20: 2166-2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krampfl K, Schlesinger F, Wolfes H, Dengler R, Bufler J (2001) Functional diversity of recombinant human AMPA type glutamate receptors: possible implications for selective vulnerability of motor neurons. J Neurol Sci 191: 19-23. [DOI] [PubMed] [Google Scholar]
- Lomeli H, Moschbacher J, Melcher T, Hoger T, Geiger JR, Kuner T, Monyer H, Higuchi M, Bach A, Seeburg PH (1994) Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science 266: 1709-1713. [DOI] [PubMed] [Google Scholar]
- Mansour M, Nagarajan N, Nehring RB, Clements JD, Rosenmund C (2001) Heteromeric AMPA receptors assemble with a preferred subunit stoichiometry and spatial arrangement. Neuron 32: 841-853. [DOI] [PubMed] [Google Scholar]
- Mosbacher J, Schoepfer R, Monyer H, Burnashev N, Seeburg PH, Ruppersberg JP (1994) A molecular determinant for submillisecond desensitization in glutamate receptors. Science 266: 1059-1062. [DOI] [PubMed] [Google Scholar]
- Partin KM (2001) Domain interactions regulating AMPA receptor desensitization. J Neurosci 21: 1939-1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partin KM, Bowie D, Mayer ML (1995) Structural determinants of allosteric regulation in alternatively spliced AMPA receptors. Neuron 14: 833-843. [DOI] [PubMed] [Google Scholar]
- Partin KM, Fleck MW, Mayer ML (1996) AMPA receptor flip/flop mutants affecting deactivation, desensitization, and modulation by cyclothiazide, aniracetam, and thiocyanate. J Neurosci 16: 6634-6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quirk JC, Nisenbaum ES (2003) Multiple molecular determinants for allosteric modulation of alternatively spliced AMPA receptors. J Neurosci 23: 10953-10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer B, Keinanen K, Verdoorn TA, Wisden W, Burnashev N, Herb A, Kohler M, Takagi T, Sakmann B, Seeburg PH (1990) Flip and flop: a cell-specific functional switch in glutamate-operated channels of the CNS. Science 249: 1580-1585. [DOI] [PubMed] [Google Scholar]
- Stern-Bach Y, Russo S, Neuman M, Rosenmund C (1998) A point mutation in the glutamate binding site blocks desensitization of AMPA receptors. Neuron 21: 907-918. [DOI] [PubMed] [Google Scholar]
- Sun Y, Olson R, Horning M, Armstrong N, Mayer M, Gouaux E (2002) Mechanism of glutamate receptor desensitization. Nature 417: 245-253. [DOI] [PubMed] [Google Scholar]