Abstract
The p75 neurotrophin receptor (p75NTR) regulates neuronal survival, apoptosis, and growth. Recent studies have reported that disruption of Exon IV produces a null mouse lacking all p75NTR gene products (p75NTRExonIV-/-), whereas mice lacking p75NTR Exon III (p75NTRExonIII-/-) maintain expression of an alternatively spliced form of p75NTR (s-p75NTR). Here, we report that p75NTRExonIV-/- mice express a p75NTR gene product that encodes a truncated protein containing the extracellular stalk region together with the entire transmembrane and intracellular domains. The gene product is initiated from a cryptic Kozak consensus/initiator ATG sequence within a region of Exon IV located 3′ to the pGK-Neo insertion site. Overexpression of this fragment in heterologous cells results in activation of Jun kinase and induces Pro-caspase-3 cleavage, indicating that it activates p75NTR signaling cascades. These results indicate that aspects of the p75NTRExonIV-/- phenotype may reflect a gain-of-function mutation rather than loss of p75NTR function.
Keywords: apoptosis, knock-out, neurotrophin, signal transduction, jun kinase, Trk
Introduction
The p75 neurotrophin receptor (p75NTR) is a receptor for the neurotrophins, a family of growth factors that includes nerve growth factor (NGF), brain-derived neurotrophic factor (BDNF), neurotrophin-3 (NT-3), and neurotrophin-4/5 (NT-4/5). Along with the tyrosine kinase (Trk) receptors, p75NTR mediates the diverse functions of the neurotrophins. The Trk receptors show neurotrophin binding specificity and are generally associated with regulation of neuronal growth, differentiation, and survival. p75NTR binds all neurotrophins with approximately equal affinity, and its functions include enhancing Trk receptor signaling and initiating autonomous signaling cascades that regulate growth, survival, and apoptosis (Barrett, 2000; Roux and Barker, 2002). Recent studies indicate that p75NTR is a component of a receptor complex that mediates growth inhibition in response to myelin-based inhibitory proteins (McKerracher and Winton, 2002; Kaplan and Miller, 2003).
To address the in vivo role of p75NTR, Lee and colleagues (Lee et al., 1992) constructed a p75NTR knock-out mouse in which Exon III of the p75NTR locus was targeted for deletion (p75NTRExonIII-/-). This mouse exhibited profound deficits in the peripheral nervous system in vivo, and neonatal sympathetic and embryonic sensory neurons derived from these animals showed reduced sensitivity to NGF and displayed deficits in developmental and injury-induced apoptosis (Davies et al., 1993; Lee et al., 1994). Recent studies, however, have indicated that the p75NTR locus produces an alternatively spliced isoform of p75NTR (s-p75NTR) that lacks Exon III. This transcript, which is apparently present in both wild-type and p75NTRExonIII-/- mice, produces a protein product that lacks the portion of the extracellular domain responsible for neurotrophin binding (von Schack et al., 2001). To produce animals completely deficient in p75NTR expression, von Schack and colleagues (2001) targeted the fourth exon of the p75NTR gene to generate p75NTRExonIV-/- mice. These animals produce neither full-length p75NTR nor s-p75NTR and exhibit severe nervous system and vascular system defects. Approximately 40% of p75NTRExonIV-/- mice do not survive beyond the perinatal period.
To initiate a characterization of the biochemical basis for phenotypic differences between p75NTRExonIII-/- and p75NTRExonIV-/- mice, we directly compared p75NTR gene products expressed in the two mouse lines. From this, we identified a p75NTR gene product that is selectively expressed in p75NTRExonIV-/- mice. This p75NTR fragment has an apparent molecular weight of 26 kDa, is not observed in wild-type animals, shows moderate expression in heterozygotes, and is most highly expressed in p75NTRExonIV-/- animals. RT-PCR analysis revealed that the protein expression pattern correlates with the aberrant expression of a p75NTR mRNA that is initiated 3′ to the inserted pGK-neo cassette. The resulting protein encodes a portion of the extracellular stalk region and the entire transmembrane and intracellular domains and is likely produced through utilization of an enhancer sequence within the pGK-Neo cassette inserted into Exon IV. When overexpressed, the p75NTRExonIV gene product activates p75NTR signaling cascades that result in Jun kinase phosphorylation and Pro-caspase-3 cleavage. The signaling properties of this p75NTR fragment suggest that some of the phenotypes observed within the p75NTRExonIV-/- mouse may be caused by gain-of-function mutations.
Materials and Methods
Materials. Cell culture reagents were purchased from BioWhittaker (Walkersville, MD) unless indicated otherwise. αP1 (Majdan et al., 1997) and αp75NTR (Promega, Madison, WI; catalog #G3231) antibodies are directed against the rat and human p75NTR intracellular domains, respectively. REX antibody, directed against the p75NTR extracellular domain, was a kind gift from Lou Reichardt (University of California San Francisco). Polyclonal antibodies directed against NRH 2 (neuotrophin receptor homolog 2) were produced in rabbits using a KLH-conjugated NRH 2 intracellular domain peptide (KARTVELGDPDRDQR); this antibody detects human, rat, and mouse PLAIDD and shows no cross-reactivity to p75NTR (data not shown). The Jun N-terminal kinase (JNK) antibody (C-17; catalog #sc-474) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA), and the Bcl-2 antibody (catalog #6620) was purchased from BD Biosciences. Cleaved caspase-3 (catalog #9661), phospho-Thr183/Tyr185 JNK (catalog #9255), phospho-Thr69/71 activating transcription factor 2 (ATF-2) (catalog #9225S), ATF-2 (cat #9222), and IκB (cat #9242) specific antibodies were purchased from Cell Signaling Technology.
Animal colonies and genotyping. Mice bearing the p75NTRExonIII mutation (Lee et al., 1992) were obtained from the The Jackson Laboratory (Bar Harbor, ME), interbred on a C57BL/6 background via heterozygous matings, and genotyped by PCR using primers directed to Intron II (p75-IntII, 5′-CGATGCTCCTATGGCTACTA), Intron III (p75IntIII, 5′-CCTCGCATTCGGCGTCAGCC), and the pGK-Neo cassette (pGK, 5′-GGGAACTTCCTGACTAGGGG). Mice bearing the p75NTRExonIV mutation (von Schack et al., 2001) were maintained on a C57BL/6 background and genotyped as described (von Schack et al., 2001).
Tissue processing and immunoblotting. Tissues were lysed, assayed for total protein, and immunoblotted as described previously (Roux et al., 2001). Blocking and antibody incubations of immunoblots were performed in Tris-buffered saline/Tween 20 (10 mm Tris, pH 7.4, 150 mm NaCl, and 0.2% Tween 20) supplemented with 5% (w/v) dried skim milk powder, except for those involving phospho-specific antibodies that were performed in Tris-buffered saline/Tween supplemented with 2% (w/v) bovine serum albumin.
Reverse transcription-PCR analysis. mRNA was isolated from P1 brains using an RNEasy Mini kit according to the manufacturer's instructions (Qiagen, Hilden, Germany). cDNA was generated using the Qiagen Omniscript RT kit and random hexamers as primers. PCR was performed for 30 cycles (94°C for 1 min, 58°C for 1 min, 72°C for 1 min) using the following primers (300 nm, final concentration): β-actin sense, 5′-CACCACTTTCTACAATGAGC; β-actin antisense, 5′-CGGTCAGGATCTTCATGAGG; p75NTR-intracellular domain (ICD) sense, 5′-CCAGCAGACCCACACACAGACTG; p75NTR-ICD antisense, 5′-CCCTACACAGAGATGCTCGGTTC; p75NTR-Exon IV (791-813) sense, 5′-CACCACCTCCAGAGCGAGACCTCATAG; p75NTR Exon VI (1467-1448) antisense, 5′-GAACATCAGCGGTCGGAATG.
PCR was performed using primers directed against Exon I and Exon III for a total of 30 cycles (94°C for 1 min, 4 cycles of 68°C for 1 min, 8 cycles of 68°C for 1 min ramping down by 1°C each cycle, 18 cycles of 60°C for 1 min, extension for each cycle was 72°C for 1 min) using the following primers: p75NTR-Exon I sense, 5′-AGCACCATCGGTCCGCAG; p75NTR Exon III antisense, 5′-TCATCTGAGTATGTGCCCTCTGG.
Twenty microliters of the reaction products were separated on a 1% agarose gel, and PCR products were visualized under UV light.
Mammalian expression vector construction, cell culture, and transfection. RT-PCR reaction was performed on mRNA isolated from P1 p75NTRExonIV brains using primers directed against mouse p75NTR Exon IV and Exon IV to amplify nucleotides 791-1467 of p75NTR cDNA (GenBank Index 23468246). The PCR reaction product was cloned, sequenced (Bio S&T, Montreal, Quebec), and subcloned into the GATEWAY pDEST12.2 mammalian expression as per the manufacturer's instructions (Invitrogen, Gaithersburg, MD). PC12 or 293T cells (350,000) plated onto a six-well plate were transfected using LipofectAMINE 2000 (Invitrogen) according to the manufacturer's instructions and were subsequently lysed in Laemmli sample buffer and analyzed by SDS-PAGE and immunoblotting.
Subcellular fractionation. P1 brain tissues were lysed in homogenization buffer [20 mm HEPES, pH 7.4, 1 mm EDTA, 255 mm sucrose, protease inhibitor mixture (Roche, Hertforshire, UK; catalog #1836153)] and then centrifuged at 600 × g for 5 min. The supernatant was centrifuged at 10,000 × g, and the pellet obtained was designated heavy membrane. The supernatant was centrifuged at 100,000 × g, the pellet obtained was designated light membrane, and the supernatant was designated cytosol.
Immunocytochemistry. COS-7 cells (25,000) were transfected as above, fixed in 2% PFA, blocked in 2% donkey serum and 0.2% Triton X-100 in PBS, and incubated with polyclonal antibodies directed against p75NTR (αP1, 1:1000) and nucleolin (1:1000). Cells were washed and incubated in Cy3-labeled anti-rabbit IgG (1:1000; Jackson ImmunoResearch, West Grove, PA), FITC-labeled anti-mouse IgG (1:1000; The Jackson Laboratory), and Hoescht 33258 (1:10,000; Molecular Probes, Eugene, OR). For embryo section staining, 20 μm frozen sections were fixed in 4% PFA, pH 7.0, permeabilized (0.1% Triton X-100, 0.1% sodium citrate in water), and then blocked in 5% donkey serum, 3% BSA, 1% glycine, and 0.4% Triton X-100 in PBS. Sections were incubated in αP1 (5 μg/ml), REX (1:250), or block buffer alone, washed, incubated in donkey anti-rabbit Cy3 (1:1000) and Hoescht 33258, washed, and mounted.
Results
A fragment of p75NTR is expressed in p75NTRExonIV-/- mice
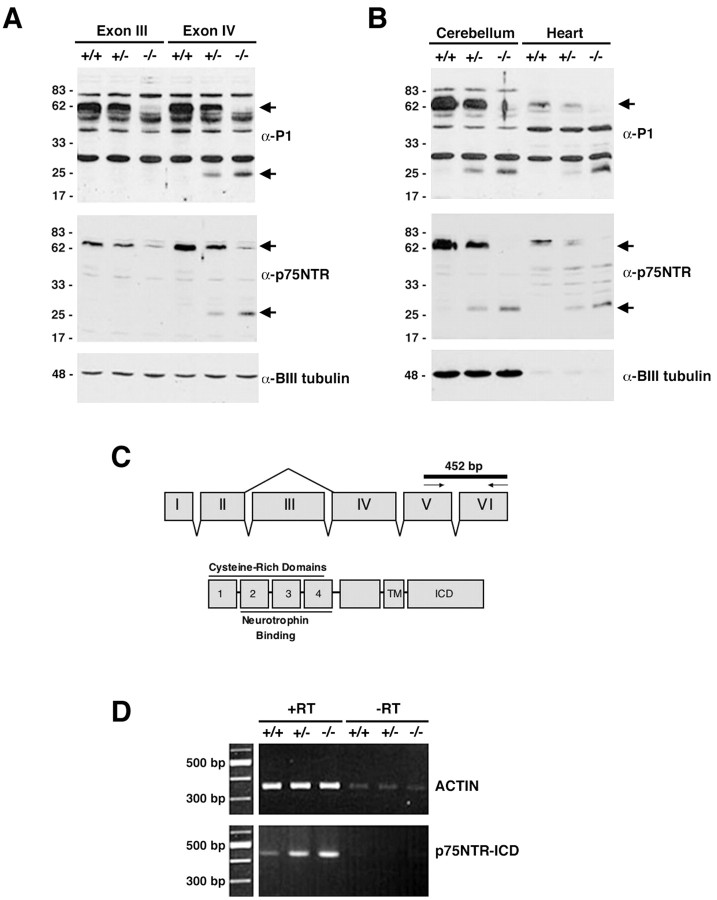
Expression analysis of p75NTR in mice bearing mutations within Exon III or Exon IV of the p75NTR locus was performed. As expected, P1 brain lysates of p75NTRExonIV-/- mice contained no full-length p75NTR (Fig. 1A, lane 3), and P1 brain lysates from p75NTRExonIV-/- mice also lacked full-length p75NTR (Fig 1A, lane 6). Surprisingly, we found that mice bearing either one or two copies of the p75NTRExonIV mutation contained a 26 kDa protein that was immunoreactive with two distinct antibodies directed against the p75NTR intracellular domain (Fig. 1A, lanes 5, 6). This 26 kDa band was not present in wild-type mice, nor was it detected in p75NTRExonIII mice (Fig. 1A, lanes 1-4). The genotype-specific expression profile of the 26 kDa band was observed in all tissues from P1 p75NTRExonIV animals examined, including cerebellum and heart (Fig. 1B, lanes 2, 3 and lanes 5, 6). We have been unable to identify the alternatively spliced s-p75NTR protein in p75NTRExonIII lysates from several tissues, suggesting either that it is not produced in vivo or that it is produced at low levels that are below our detection limit (Fig. 1A) (data not shown).
Figure 1.
A p75NTR fragment is expressed in p75NTRExonIV-/- mice. A, P1 brain lysates from p75NTRExonIII and p75NTRExonIV wild-type (+/+), heterozygous (+/-), and null (-/-) littermates were separated on a 15% SDS-PAGE gel, followed by immunoblotting with αP1 (top panel) or αp75NTR (middle panel), distinct polyclonal antibodies directed against the p75NTR-ICD (see Materials and Methods). Arrowheads indicate full-length p75NTR and a ∼26 kDa band present in lysates from mice bearing the p75NTRExonIV mutant allele. Blots were reprobed for βIII-tubulin to verify equal loading. B, P1 cerebellar and heart lysates from p75NTRExonIV wild-type (+/+), heterozygous (+/-), and null (-/-) littermates were prepared and subjected to SDS-PAGE and immunoblotting as described above. C, Schematic diagram showing the genomic structure of the mouse p75NTR locus and corresponding protein domains. D, RNA was isolated from P1 brain lysates of p75NTRExonIV wild-type (+/+), heterozygous (+/-), and null (-/-) littermates. cDNA was prepared in the presence (+RT) and absence (-RT) of reverse transcriptase, followed by RT-PCR using primers for β-actin and for p75NTR-ICD. Arrows in C indicate location of primers used for this analysis. Experiments in A, B, and D were repeated three times with identical results.
A p75NTR homolog termed NRH2 has been identified recently (Frankowski et al., 2002; Kanning et al., 2003; Wang et al., 2003). The intracellular domain of NRH2 is similar to that of p75NTR, and it is possible that antibodies directed against p75NTR may cross-react with NHR2 and that the 26 kDa protein observed in p75NTRExonIV mice reflects a compensatory increase in levels of an NRH2 gene product. To specifically determine whether NRH2 levels are altered in p75NTRExonIV mice and to learn whether the 26 kDa protein observed in p75NTRExonIV mice is an NRH2 gene product, we prepared anti-peptide antibodies against NRH2 and used these to probe brain lysates derived from wild-type, heterozygote, and homozygote p75NTRExonIII and p75NTRExonIV-/- mice. Supplementary Figure 1 (available at www.jneurosci.org) shows that NRH2 levels are not altered in p75NTRExonIII or p75NTRExonIV-/- mice, indicating that the 26 kDa protein produced in p75NTRExonIV mice is a p75NTR gene product.
To determine whether the increase in the 26 kDa protein reflects an increase in transcription of p75NTR, RT-PCR was performed on P1 brain cDNAs derived from p75NTRExonIV wild-type, heterozygous, and null littermates, using primers directed to Exon V and Exon VI of the p75NTR locus (which encode the portion of the intracellular domain indicated in Fig. 1C). This fragment of the p75NTR mRNA was transcribed at higher levels in mice bearing the p75NTRExonIV mutant allele (Fig. 1D) than in wild-type littermates. Collectively, these data indicate that mRNA encoding a truncated p75NTR gene product is produced and translated to protein in p75NTRExonIV-/- mice.
Transcription of the transmembrane and intracellular domains of p75NTR in p75NTRExonIV-/- mice is the result of pGK-Neo cassette insertion
To generate the p75NTRExonIV-/- mice, von Schack et al. (2001) inserted a pGK-Neo cassette into an AatII restriction site within Exon IV of mouse p75NTR. Several studies indicate that the retention of the selectable marker (a hybrid gene consisting of the phosphoglycerate kinase I promoter driving the neomycin phosphotransferase gene) in targeted loci of “knock-out” mice can cause unexpected phenotypes attributable to effects on flanking genes (Olson et al., 1996; Pham et al., 1996), and we reasoned that pGK-Neo may lead to aberrant expression of 3′ gene products in the p75NTR locus. To determine whether the inserted pGK-Neo cassette affected the transcription of p75NTR in p75NTRExonIV-/- mice, we designed primers to amplify p75NTR gene products produced upstream and downstream of the pGK-Neo cassette (Fig. 2A) and performed RT-PCR on P1 brain mRNA from p75NTRExonIV wild-type, heterozygous, and null littermates. Figure 2A shows that levels of p75NTR mRNA derived from a portion of the p75NTR locus located 5′ to the pGK-Neo insertion site were significantly reduced in brains from P1 mice bearing the p75NTRExonIV mutation, likely because of mRNA instability resulting from absence of a polyA tail on the 3′ end of mRNAs driven off the endogenous p75NTR promoter. In contrast, levels of p75NTR mRNA derived from a portion of the gene located 3′ to the pGK-neo insertion site was significantly enhanced, suggesting that enhancer sequences within the pGK-Neo cassette are responsible for transcription of 3′ fragments of the p75NTR gene in p75NTRExonIV-/- mice.
Figure 2.
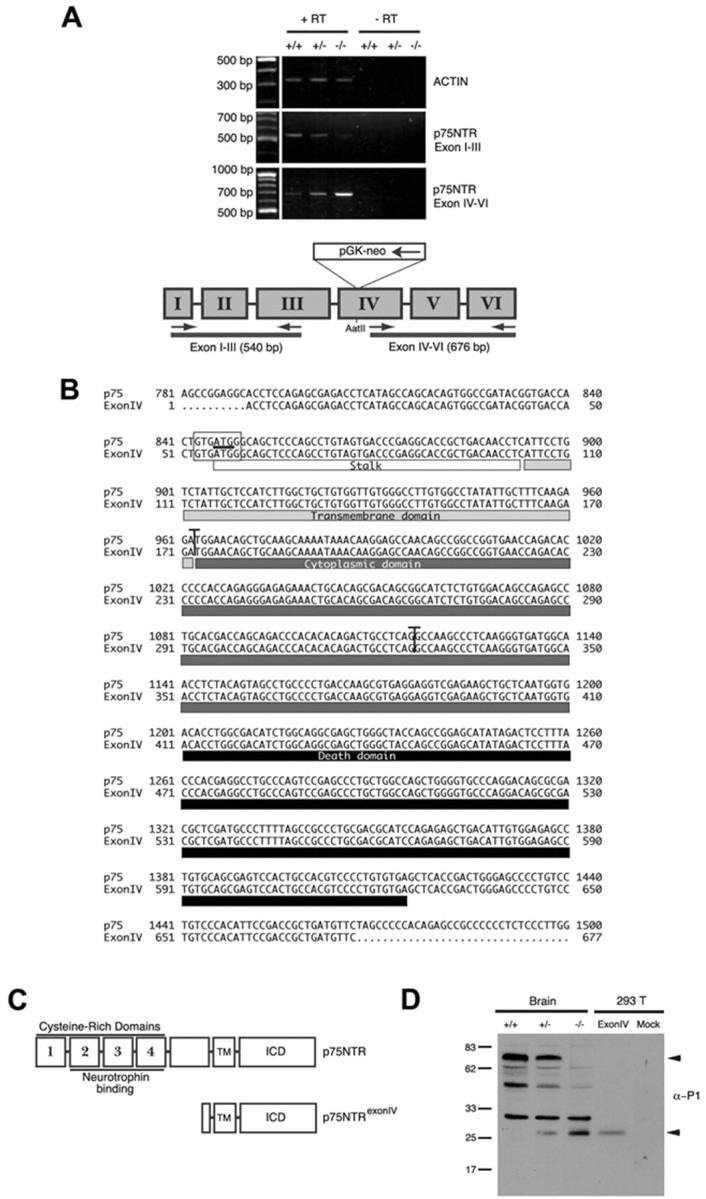
Transcription of p75NTR fragment in p75NTRExonIV-/- mice is the result of pGK-Neo cassette insertion into the p75NTRExonIV targeting vector. A, mRNA from p75NTRExonIV wild-type (+/+), heterozygous (+/-), and null (-/-) littermates was subjected to RT-PCR using primers that target regions of p75NTR upstream or downstream of the pGK-Neo cassette insertion in the p75NTRExonIV targeting vector, as indicated in the schematic diagram. B, Alignment of mouse p75NTR cDNA sequence with p75NTR transcript cloned from p75NTRExonIV-/- mice. The putative start codon is underlined, and the putative Kozak sequence is boxed. Vertical lines indicate Exon IV-V and Exon V-VI junctions, and boxes underneath the sequence denote protein domains. C, Schematic diagram showing the protein domains of mouse p75NTR and p75NTRExonIV proteins. D, P1 brain lysates from p75NTRExonIV wild-type (+/+), heterozygous (+/-), and null (-/-) littermates and cellular lysates from 293T cells transiently transfected with plasmids encoding p75NTRExonIV (ExonIV) or the parental vector (Mock) were immunoblotted using αP1. Arrowheads indicate full-length p75NTR (top) and p75NTRExonIV (bottom) products. Experiments in A and D were repeated three times with identical results.
We used RACE (rapid amplification of cDNA ends)-PCR to clone the fragment of p75NTR expressed in the p75NTRExonIV-/-, but this was not successful because of the presence of a strong reverse transcription stop within the cDNA that invariably produced truncated products (data not shown). We therefore scanned the sequence 3′ to the pGK-neo insertion site to identify in-frame ATG codons that were flanked by potential cryptic Kozak consensus sequences. One candidate was identified (nt 846-848) (Fig. 2B), and RT-PCR was used to determine whether cDNA containing this putative start site could be directly cloned from P1 brains of p75NTRExonIV-/- mice. Figure 2B shows the sequence of the 676 bp cDNA that was cloned from the p75NTRExonIV-/- mice. The predicted protein encoded by this fragment is a 187 amino acid fragment of p75NTR containing a small portion of the extracellular stalk together with the entire transmembrane and intracellular domains of the receptor (Fig. 2C). To confirm that this cDNA could direct protein expression, the p75NTRExonIV-/- cDNA was subcloned into a mammalian expression vector and transfected into human embryonic kidney 293 cells. Figure 2D shows that the p75NTRExonIV-/- cDNA directed expression of a p75NTR fragment that co-migrates with the protein product identified within p75NTRExonIV-/- brain lysates (Fig. 2C).
The 26 kDa p75NTRExonIV protein is membrane-associated and activates p75NTR signaling cascades
To assess the cellular distribution of the p75NTRExonIV fragment, immunocytochemistry was performed on embryonic day 13 thoracic spinal cords derived from p75NTRExonIV null and wild-type littermates using REX, which is directed against the p75NTR extracellular domain, and αP1, which is directed against the p75NTR intracellular domain. Supplementary Figure 2 (available at www.jneurosci.org) shows that the immunostaining patterns obtained with REX and αP1 are essentially identical on sections from wild-type animals, with prominent staining of spinal motoneurons and dorsal root ganglia sensory neurons. REX immunostaining is lost in p75NTRExonIV null mice, whereas αP1 immunoreactivity is detectable, albeit at considerably lower levels than in wild-type mice. αP1 staining in the null mice appears relatively homogeneous, with slightly higher staining in motoneurons and dorsal root ganglia.
The p75NTR fragment produced in the p75NTRExonIV-/- mouse lacks the signal sequence necessary for membrane insertion, but retains the hydrophobic transmembrane domain. To determine whether the p75NTRExonIV product is membrane-associated, P1 brains from p75NTRExonIV heterozygote and null littermates were subjected to subcellular fractionation. Figure 3A shows that the 26 kDa protein is lacking in cytosolic fractions but is enriched in heavy and light membrane fractions, similar to the distribution of full-length p75NTR (Fig. 3A). To further characterize the subcellular localization of the p75NTRExonIV product, COS-7 cells were transiently transfected with the p75NTRExonIV-/- cDNA, and immunocytochemistry was performed using αP1 and antibodies directed against nucleolin, a nuclear protein. Figure 3B shows that p75NTRExonIV protein is excluded from the nucleus but is detected at the plasma membrane and within the cytosol, where it is presumably associated with intracellular membranes. Together these results indicate that the p75NTRExonIV fragment is a membrane-associated protein.
Figure 3.

p75NTRExonIV is membrane-associated and induces p75NTR signaling events. A, P1 brains from p75NTRExonIV heterozygote (Exon IV +/-) and null (Exon IV -/-) littermates were subjected to subcellular fractionation and analyzed by immunoblot. C, Cytosol; M1, heavy membrane; M2, light membrane fraction. B, COS-7 cells were transfected with plasmids encoding p75NTRExonIV or the p75NTR intracellular domain tagged with a myristoylation sequence (Roux et al., 2001) and immunostained with αP1 and α-nucleolin. Left-hand panels show p75NTR staining, middle panels show nucleolin staining, and right-hand panels show merged images. C, PC12 cells were transfected with control plasmid or plasmid expressing p75NTRExonIV in the presence or absence of plasmid expressing Bcl-2. Staurosporine treatment was used as a positive control for caspase cleavage. Cellular lysates were analyzed by immunoblot to detect p75NTR, Bcl-2, and active caspase-3, as indicated. IκBα levels were analyzed to confirm equal loading between lanes. D, PC12 cells were left untransfected or were transfected with 2 μg of a control plasmid (pcDNA3), or expression plasmids encoding p75NTRExonIV alone or p75NTRExonIV and Bcl-2 and levels of phospho-Thr183/Tyr185-JNK and total JNK, phospho-Thr69/71-ATF-2, and total ATF-2 were assessed by immunoblotting. Experiments in A-C were repeated three times with identical results.
We have shown previously that the p75NTR intracellular domain can initiate p75NTR signaling events that range from activation of Akt to induction of apoptosis (Majdan et al., 1997; Roux et al., 2001) and therefore set out to determine whether the p75NTRExonIV protein exhibits a similar range of activities. Figure 3C shows that expression of the p75NTRExonIV protein in PC12 cells induces Pro-caspase-3 cleavage, which is attenuated by coexpression of Bcl-2, indicating that the p75NTRExonIV protein can activate the intrinsic death pathway. Consistent with this, Figure 3D shows that expression of the p75NTRExonIV protein in PC12 cells induced phosphorylation of JNK and ATF-2, a JNK target (van Dam et al., 1995; Eilers et al., 2001). Together, these data show that overexpression of the p75NTRExonIV protein results in activation of p75NTR signaling cascades that result in caspase cleavage.
Discussion
Our results show that p75NTRExonIV-/- mice express a fragment of the p75NTR protein that contains a portion of the extracellular domain and the transmembrane and intracellular domains. The genotype-specific expression profile of this fragment is consistent with the hypothesis that the pGK-neo cassette inserted into Exon IV of the p75NTR gene is responsible for this expression pattern. The PGK-1 promoter and enhancer sequence functions in an orientation and a position-independent manner (McBurney et al., 1991), and several groups have demonstrated that retention of the pGK-Neo cassette in targeted loci can profoundly influence the expression of surrounding genes upstream and downstream from the cassette (Olson et al., 1996; Pham et al., 1996). Our results suggest that the pGK-Neo placed within Exon IV of p75NTR results in aberrant transcription of the adjacent 3′ portion of the p75NTR gene, which results in production of the p75NTRExonIV protein product because of the presence of a cryptic Kozak consensus sequence in exon 4. The resulting protein contains the entire p75NTR transmembrane and intracellular domain and is associated with membrane. p75NTR has recently been shown to be cleaved by α- and γ-secretase, and the released intracellular fragment may accumulate in the nucleus (Jung et al., 2003; Kanning et al., 2003). We have not observed nuclear accumulation of p75NTR gene products in transfected cells, but it is possible that the p75NTRExonIV protein product is a substrate for secretase activity in vivo.
Initial characterization of the p75NTRExonIV-/- mice revealed severe peripheral neuron loss as well as dilation and rupture of large blood vessels (von Schack et al., 2001). We have demonstrated that the p75NTRExonIV protein is highly expressed in heart and brain lysates of P1 p75NTRExonIV-/- mice and have shown that the p75NTRExonIV protein product activates signaling cascades previously ascribed to p75NTR, including JNK activation and caspase-3 cleavage (Harrington et al., 2002; Troy et al., 2002). Our previous studies have shown that expression of the p75NTR intracellular domain can similarly activate p75NTR signaling cascades and thus induce apoptosis of peripheral and central neurons in vivo (Majdan et al., 1997). The numerous activities of p75NTR are highly dependent on cell context (Roux and Barker, 2002), however, and it is likely that the in vivo consequences of p75NTRExonIV protein expression will be complex and vary between tissues.
Although p75NTR was the founding member of the TNF receptor superfamily and was the first identified neurotrophin receptor, its physiological function has not been completely elucidated. Recent findings indicate roles that include promotion of survival pathways, induction of apoptosis, effects on cell cycle and differentiation, and facilitation or inhibition of growth (Barrett, 2000; McKerracher and Winton, 2002; Roux and Barker, 2002; Kaplan and Miller, 2003). The use of genetically modified mice that have altered p75NTR expression has provided important insights into the biological function of the receptor. Judicious application of the lines produced to date, together with the creation of new transgenic strains in which p75NTR deletion is accomplished in a regulated manner, is certain to enhance our knowledge of the physiological functions of this receptor.
Footnotes
C.E.P. was supported by a scholarship from the Natural Sciences and Engineering Research Council of Canada, and P.A.B. is an Investigator of the Canadian Institutes of Health Research. This work was supported by grants from the Canadian Institutes of Health Research. We gratefully acknowledge the technical assistance of Genevieve Dorval. Wayne Sossin provided helpful comments on this manuscript. The p75NTRExonIV-/- mice were a kind gift from Georg Dechant (University of Innsbruck).
Correspondence should be addressed to Philip A. Barker, Montreal Neurological Institute, McGill University, 3801 University Avenue, Montreal, Quebec, Canada, H3A 2B4. E-mail: phil.barker@mcgill.ca.
Copyright © 2004 Society for Neuroscience 0270-6474/04/241917-07$15.00/0
References
- Barrett GL (2000) The p75 neurotrophin receptor and neuronal apoptosis. Prog Neurobiol 61: 205-229. [DOI] [PubMed] [Google Scholar]
- Davies AM, Lee KF, Jaenisch R (1993) p75-deficient trigeminal sensory neurons have an altered response to NGF but not to other neurotrophins. Neuron 11: 565-574. [DOI] [PubMed] [Google Scholar]
- Eilers A, Whitfield J, Shah B, Spadoni C, Desmond H, Ham J (2001) Direct inhibition of c-Jun N-terminal kinase in sympathetic neurones prevents c-jun promoter activation and NGF withdrawal-induced death. J Neurochem 76: 1439-1454. [DOI] [PubMed] [Google Scholar]
- Frankowski H, Castro-Obregon S, del Rio G, Rao RV, Bredesen DE (2002) PLAIDD, a type II death domain protein that interacts with p75 neurotrophin receptor. Neuromol Med 1: 153-170. [DOI] [PubMed] [Google Scholar]
- Harrington AW, Kim JY, Yoon SO (2002) Activation of Rac GTPase by p75 is necessary for c-jun N-terminal kinase-mediated apoptosis. J Neurosci 22: 156-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung KM, Tan S, Landman N, Petrova K, Murray S, Lewis R, Kim PK, Kim DS, Ryu SH, Chao MV, Kim TW (2003) Regulated intramembrane proteolysis of the p75 neurotrophin receptor modulates its association with the TrkA receptor. J Biol Chem 278: 42161-42169. [DOI] [PubMed] [Google Scholar]
- Kanning KC, Hudson M, Amieux PS, Wiley JC, Bothwell M, Schecterson LC (2003) Proteolytic processing of the p75 neurotrophin receptor and two homologs generates C-terminal fragments with signaling capability. J Neurosci 23: 5425-5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan DR, Miller FD (2003) Axon growth inhibition: signals from the p75 neurotrophin receptor. Nat Neurosci 6: 435-436. [DOI] [PubMed] [Google Scholar]
- Lee K-F, Li E, Huber J, Landis SC, Sharpe AH, Chao MV, Jaenisch R (1992) Targeted mutation of the gene encoding the low affinity NGF receptor leads to deficits in the peripheral sensory nervous system. Cell 69: 737-749. [DOI] [PubMed] [Google Scholar]
- Lee K-F, Davies AM, Jaenisch R (1994) p75-deficient embryonic dorsal root sensory and neonatal sympathetic neurons display a decreased sensitivity to NGF. Development 120: 1027-1033. [DOI] [PubMed] [Google Scholar]
- Majdan M, Lachance C, Gloster A, Aloyz R, Zeindler C, Bamji S, Bhakar A, Belliveau D, Fawcett J, Miller FD, Barker PA (1997) Transgenic mice expressing the intracellular domain of the p75 neurotrophin receptor undergo neuronal apoptosis. J Neurosci 17: 6988-6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBurney MW, Sutherland LC, Adra CN, Leclair B, Rudnicki MA, Jardine K (1991) The mouse Pgk-1 gene promoter contains an upstream activator sequence. Nucleic Acids Res 19: 5755-5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKerracher L, Winton MJ (2002) Nogo on the go. Neuron 36: 345-348. [DOI] [PubMed] [Google Scholar]
- Olson EN, Arnold HH, Rigby PW, Wold BJ (1996) Know your neighbors: three phenotypes in null mutants of the myogenic bHLH gene MRF4. Cell 85: 1-4. [DOI] [PubMed] [Google Scholar]
- Pham CT, MacIvor DM, Hug BA, Heusel JW, Ley TJ (1996) Long-range disruption of gene expression by a selectable marker cassette. Proc Natl Acad Sci USA 93: 13090-13095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux PP, Barker PA (2002) Neurotrophin signaling through the p75 neurotrophin receptor. Prog Neurobiol 67: 203-233. [DOI] [PubMed] [Google Scholar]
- Roux PP, Bhakar AL, Kennedy TE, Barker PA (2001) The p75 neurotrophin receptor activates Akt (protein kinase B) through a phosphatidylinositol 3-kinase-dependent pathway. J Biol Chem 276: 23097-23104. [DOI] [PubMed] [Google Scholar]
- Troy CM, Friedman JE, Friedman WJ (2002) Mechanisms of p75-mediated death of hippocampal neurons: role of caspases. J Biol Chem 3: 34295-34302. [DOI] [PubMed] [Google Scholar]
- van Dam H, Wilhelm D, Herr I, Steffen A, Herrlich P, Angel P (1995) ATF-2 is preferentially activated by stress-activated protein kinases to mediate c-jun induction in response to genotoxic agents. EMBO J 14: 1798-1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Schack D, Casademunt E, Schweigreiter R, Meyer M, Bibel M, Dechant G (2001) Complete ablation of the neurotrophin receptor p75NTR causes defects both in the nervous and the vascular system. Nat Neurosci 4: 977-978. [DOI] [PubMed] [Google Scholar]
- Wang X, Shao Z, Zetoune FS, Zeidler MG, Gowrishankar K, Vincenz C (2003) NRADD, a novel membrane protein with a death domain involved in mediating apoptosis in response to ER stress. Cell Death Differ 10: 580-591. [DOI] [PubMed] [Google Scholar]