Abstract
Recent studies indicate that neuroprotection afforded by brain-derived neurotrophic factor (BDNF) is mediated by extracellular signal-regulated kinase (ERK) and phosphatidylinositol-3 kinase (PI3K). However, the mechanisms by which ERK and PI3K exert neuroprotection are not completely understood. Because ERK1/2 and PI3K both stimulate serum response element (SRE)-mediated gene expression, and serum response factor (SRF) is indispensable for SRE-mediated transcription, we investigated whether SRF contributes to ERK1/2 and PI3K neuroprotection. To accomplish this goal, we used an established experimental paradigm in which BDNF protects postnatal cortical neurons against both trophic deprivation and camptothecin-induced DNA damage. BDNF protection against camptothecin is mediated primarily by ERK1/2 activation, whereas its protection against trophic deprivation is mainly through stimulation of the PI3K pathway (Hetman et al., 1999). Here we demonstrate that expression of a wild-type SRF is sufficient to protect postnatal cortical neurons against camptothecin or trophic deprivation. Expression of a dominant-negative SRF partially reversed BDNF neuroprotection against both apoptotic insults. Moreover, the dominant-negative SRF inhibited neuroprotection against trophic withdrawal afforded by expression of a constitutive active PI3K. In addition, protection against camptothecin by expression of constitutive active mitogen-activated protein kinase kinase 1, an upstream kinase that activates ERK1/2, was also blocked by expression of the dominant-negative SRF. These data suggest that SRF is both necessary and sufficient for BDNF neuroprotection of cortical neurons against trophic deprivation and DNA damage. Our data provide a direct demonstration of a biological function of SRF in neurons and a novel downstream neuroprotective mechanism common to both ERK1/2 and PI3K pathways.
Keywords: SRF, SRE, BDNF, PI3K, ERK1/2, neuronal survival, apoptosis, cortical neuron, CNS, transcription
Introduction
Neurotrophins are important for the development and maintenance of the nervous system and regulate several aspects of neuronal function, including differentiation and survival (Segal and Greenberg, 1996; Kaplan and Miller, 2000). For example, brain-derived neurotrophic factor (BDNF) activates extracellular signal-regulated kinase 1/2 (ERK1/2), ERK5, and phosphatidylinositol-3 kinase (PI3K) pathways in cortical neurons to promote neuronal survival (Hetman et al., 1999; Liu et al., 2003). However, the relative contribution of each pathway to neuronal survival depends on the specific type of cellular injury and the developmental stage of the neurons. For instance, ERK5 signaling is critical for BDNF neuroprotection against trophic deprivation in developing embryonic but not postnatal mature neurons (Liu et al., 2003; Shalizi et al., 2003). Although the ERK1/2 pathway plays a major role in BDNF neuroprotection against DNA damage, the PI3K pathway is the dominant trophic-promoted survival mechanism in postnatal cortical neurons (Hetman et al., 1999).
The mechanisms downstream of ERK1/2 activation that lead to neuronal survival are not completely defined. ERK1/2 activates p90rsk, which in turn phosphorylates and inactivates the proapoptotic protein BAD (Bonni et al., 1999). Alternatively, activation of p90rsk and subsequent phosphorylation of cAMP response element-binding protein (CREB) as well as increased CREB-mediated gene expression constitute a potential mechanism (Bonni et al., 1999; Riccio et al., 1999). ERK1/2 also protects cortical neurons against glycogen synthase kinase 3β (GSK3β; Hetman et al., 2002).
Activation of the PI3K signal transduction system may promote survival through several mechanisms, including phosphorylation and inhibition of GSK3β, BAD, or caspase 9 (Cross et al., 1995; Datta et al., 1997; del Peso et al., 1997; Cardone et al., 1998; Pap and Cooper, 1998; Crowder and Freeman, 2000; Hetman et al., 2000, 2002). The PI3K/Akt pathway can also transduce its neuroprotective signal via transcription-dependent mechanisms. For example, Akt inhibits fork-head transcription factor (FKHRL1)-mediated Fas ligand transcription, thereby suppressing apoptosis in cerebellar granule neurons (Brunet et al., 1999).
In addition to the “passive” inhibition of proapoptotic gene transcriptions, PI3K/Akt signaling may also promote neuroprotection via “active” induction of antiapoptotic genes. We recently discovered that PI3K activates serum response element (SRE)- and serum response factor (SRF)-dependent transcription (Poser et al., 2000). SRF is a ubiquitous transcription factor belonging to the MADS transcription factor family (Treisman, 1994). It binds to the cis-acting SRE as a dimer and is obligatory for the activation of SRE-mediated gene expression (Norman et al., 1988). SRE is important for mitogen-stimulated transcription of c-fos and many immediate early genes (Treisman, 1995). Because SRE/SRF-mediated transcription is required for PI3K-regualted cell proliferation in PC12 and NIH3TC cells (Poser et al., 2000), we were interested in determining whether SRE/SRF-dependent transcription plays a role in PI3K-promoted neuroprotection.
In addition to SRF, SRE can also interact with other transcription factors, including the ternary complex factor (TCF), a family of Ets domain-containing transcription factors, including Elk-1, Sap-1a, Sap-1b, and Sap-2/Net/ERP (Rao et al., 1989; Shaw et al., 1989; Hipskind et al., 1991; Dalton and Treisman, 1992; Giovane et al., 1994; Treisman, 1994). TCFs form a ternary complex composed of an SRF dimer and a single molecule of TCF (Mueller and Nordheim, 1991; Shaw, 1992; Treisman, 1992). Phosphorylation of TCFs by members of the MAP kinase family including ERK1/2 is critical for its ability to stimulate transcription in response to serum or growth factor addition (Gille et al., 1992; Janknecht et al., 1993; Marais et al., 1993; Hipskind et al., 1994; Gille et al., 1995; Miranti et al., 1995; Price et al., 1995; Xia et al., 1996; Davis, 2000; Johnson and Lapadat, 2002). Because SRF is obligatory for SRE-mediated transcription, and TCFs interact with SRE only when SRF is already bound, we investigated the possibility that SRE/SRF-mediated transcription is also critical for neuroprotection downstream of the ERK1/2 signaling pathway.
In this study, we report that BDNF activates SRE-mediated transcription in cortical neurons in an SRF-dependent manner. By transfecting cortical neurons with various dominant negative and constitutive active constructs of the pathways, we demonstrate a critical role for SRF in neuroprotection downstream of ERK1/2 and PI3K signaling.
Materials and Methods
Materials. The following plasmids have been described: UB6-lacZ, cloning vector CGN, wild-type and dominant-negative SRF, 5×SRE-luc reporter, constitutive active PI3 kinase catalytic subunit (p110*), a dominant negative PI3K (dn p85), a constitutive active mitogen-activated protein kinase kinase (MKK)1 (Hetman et al., 1999; Poser et al., 2000), pSVα1 for the α-globin gene (Shyu et al., 1989), pAF42.SRE.wt and pAF42.SRE.mut2 (Rivera et al., 1990), and pAF42.SRE.pm1 (Misra et al., 1994). Camptothecin and cytosine-β-d-arabinofuranoside were purchased from Sigma (St. Louis, MO). The specific kinase inhibitors LY294002 and PD98059 were purchased from Calbiochem (San Diego, CA). BDNF and LipofectAMINE 2000 were purchased from Invitrogen (Carlsbad, CA).
Cell culture. Primary cortical neurons were prepared from newborn Sprague Dawley rats as described (Xia et al., 1996). Briefly, Postnatal day 0 cortical neurons were dissociated and cultured at a density of 0.5 × 106 per well in 24-well plates for luciferase (luc) reporter gene assay, 2 × 106 per plate in 35 mm plates with coverslips for apoptosis assay, or 3 × 106 per plate in 60 mm plates for RNase protection assay. All plates and coverslips were coated with poly-d-lysine and laminin. The neurons were cultured in basal medium Eagle (BME) supplemented with 10% heat-inactivated bovine calf serum, 35 mm glucose, 1 mm l-glutamine, 100 U/ml penicillin, and 0.1 mg/ml streptomycin. Cytosine-β-d-arabinofuranoside (2.5 μm) was added at day 2 in vitro (DIV 2) after plating to prevent the proliferation of non-neuronal cells.
Luciferase reporter gene assays. At DIV 3, cortical neurons plated on 24-well plates were transfected with LipofectAMINE 2000 per the manufacturer's instructions with slight modifications. Briefly, 1 μg of total DNA was diluted in 50 μl of OptiMEM medium. LipofectAMINE (2 μl) was combined with 50 μl of OptiMEM and added drop-wise to the DNA in OptiMEM. Conditioned media were removed and saved. Five hundred microliters of BME supplemented with 35 mm glucose, 1 mm l-glutamine, 100 U/ml penicillin, and 0.1 mg/ml streptomycin were added to each well. One hundred microliters of the LipofectAMINE-DNA mix was added drop-wise to each well. The cells were then incubated at 37°C for 2 hr. The transfection media were removed, and the saved conditioned media were added back. At 48-60 hr after transfection, cells were switched to serum-free media to reduce background. Cells were treated with 10 ng/ml BDNF or vehicle control where indicated. Luciferase and β-galactosidase activities were assayed 6 hr after switching to serum-free media as described (Impey et al., 1996). The reporter gene luciferase activity was normalized to β-galactosidase activity and expressed as percent SRE-luciferase activity relative to control. There is no observable difference in the response of a cell to transcriptional regulation monitored by luciferase assay during this time frame (48-60 hr after transfection).
Transient transfection of primary cortical neurons and cell death assays. Neurons were transiently transfected at DIV 3-4 as described using a calcium-phosphate coprecipitation protocol (Xia et al., 1996; Hetman et al., 1999). Neurons were cotransfected with an expression vector encoding β-galactosidase (UB6-lacZ) as a marker for transfected cells. Two days after transfection, cells were treated with camptothecin or trophic withdrawal for 24 hr, and apoptosis was scored by nuclear fragmentation and condensation as described (Hetman et al., 1999, 2000; Namgung and Xia, 2000; Figueroa-Masot et al., 2001). To ensure unbiased counting, slides were coded, and apoptosis was scored without knowledge of the treatment. Statistical analysis of the data was performed using one-way ANOVA.
RNA isolation and RNase protection analysis. Neurons were cultured and transfected as described (Xia et al., 1996). Cells were stimulated with BDNF for 1 hr at 48-60 hr after transfection. RNA was then isolated, and RNase protection was performed as described (Chomczynski and Sacchi, 1987; Sheng et al., 1988; Xia et al., 1996). There is no observable difference in the response of a cell to transcriptional regulation during this time frame (48-60 hr after transfection). In each experiment, the levels of expression of the transfected human c-fos gene (fosh), the endogenous rat c-fos gene (fosr), and the transfected α-globin gene were determined using an RNase protection assay (Sheng et al., 1988). The correctly initiated and spliced human c-fos gene transcript protects a 296-nucleotide fragment of the c-fos probe, whereas the endogenous rat c-fos mRNA protects a 65-nucleotide fragment. The correctly initiated human α-globin gene transcript protects a 133-nucleotide fragment of the globin probe. The endogenous rat c-fos mRNA was used as a control for BDNF stimulation and RNase protection, whereas the cotransfected human α-globin gene was used as a control to normalize transfection.
Trophic deprivation. Trophic deprivation was performed at DIV 5 or 2 d after transfection as described (Hetman et al., 2000). Briefly, conditioned media were removed and saved. The cells were washed twice with serum-free BME medium and then placed in serum-free BME medium in the presence of 10 μm MK801, an NMDA receptor blocker (-serum), or in the saved conditioned media as a control (+serum).
Mitotracker staining to assay for cell viability. Cortical neurons were incubated with 10 nm Mitotracker Red CMXRos (Molecular Probes, Eugene, OR) at 37°C for 30 min to stain mitochondria. Cells were washed in PBS and fixed with 3.7% formaldehyde at 37°C for 15 min. The cells were then stained with Hoechst dye 33342 as previously described (Hetman et al., 1999).
Western analysis. Western blot analysis of p-Akt and the transfected SRF was performed as described (Hetman et al., 1999). The polyclonal anti-p-Akt antibody (p-Ser 473) was from Cell Signaling (Beverly, WA); the anti-SRF antibody was from Santa Cruz Biotechnology (Santa Cruz, CA), and the monoclonal anti-β-actin antibody was from Sigma. Using this anti-SRF antibody and two other SRF antibodies, we were unable to detect endogenous SRF; therefore, we cannot directly compare the level of the expressed SRF with that of the endogenous SRF. This is probably because of the low level of endogenous SRF expression, which is below the detection limit of available antibodies. This is not an uncommon phenomenon for transcription factors, which are generally expressed at very low levels under normal conditions.
Other treatments. Where indicated, BDNF was added at the final concentration of 10 ng/ml; LY294002 was added at 30 μm; PD98059 was added at 50 μm; and camptothecin was added at 5 μm. Equal volumes of DMSO were added to control samples as a vehicle control for LY294002, PD98059, and camptothecin.
Results
BDNF stimulates SRE/SRF-mediated transcription via PI3K activation in cortical neurons
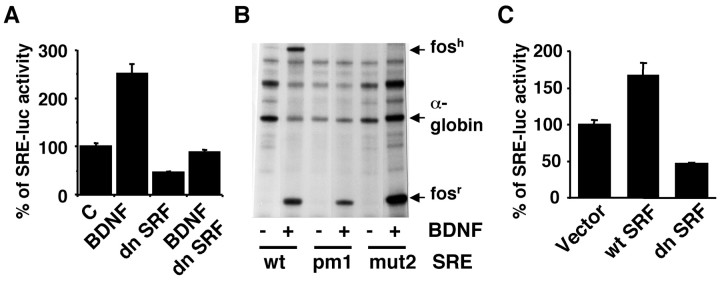
To determine whether growth factor stimulation of SRE/SRF-mediated transcription in neurons from the CNS requires PI3K activation, we transiently transfected cortical neurons cultured from newborn rats with a luc reporter gene construct driven by five repeats of SRE (5×SRE-luciferase; Poser et al., 2000). All luciferase activities were assayed under serum-free conditions to reduce background. Addition of 10 ng/ml BDNF stimulated SRE-luciferase activity (Fig. 1A). To ascertain whether SRF is required for BDNF stimulation of SRE-mediated transcription, cortical neurons were cotransfected with a dominant-negative SRF expression vector. This dominant-negative SRF has a deletion in the C-terminal activation domain of SRF (Δ339-508). Thus, the mutant SRF can still interact with endogenous wild-type SRF and can form dimers that do not activate transcription (Johansen and Prywes, 1993). Expression of the dominant negative SRF prevented BDNF from activating SRE (Fig. 1A), suggesting that SRF is obligatory for BDNF stimulation of the SRE reporter gene in cortical neurons.
Figure 1.
An obligatory role of SRF in BDNF stimulation of SRE-mediated transcription in cortical neurons. A, BDNF stimulation of SRE-mediated transcription is blocked by coexpression of a dn SRF. Cortical neurons plated in 24-well plates were transfected using LipofectAMINE 2000 with 0.6 μg of DNA per well of a 5×SRE-driven luc reporter and 0.3 μg of DNA per well of a dn SRF or vector control to measure SRE-mediated transcription. At 60 hr after transfection, cells were placed in serum-free media and treated with 10 ng/ml BDNF or vehicle control (C) where indicated. Reporter gene activity was measured 6 hr later. B, Effects of mutations within the SRE that interfere with the binding of SRF on SRE-mediated transcription. Cortical neurons (3 × 106 cells per 60 mm plate) were transfected using a calcium phosphate coprecipitation method with 1 μg of pAF42.SRE.wt or constructs with mutations in the SRE that disrupt SRF binding (pAF42.SRE.pm1 and pAF42.SRE.mut2). A α-globin gene was cotransfected as a control for transfection efficiency and equal loading. Forty eight to 60 hr after transfection, the cells were treated for 1 hr with vehicle control (-) or BDNF (+). Similar results were obtained from three independent experiments. The transcription from the endogenous fos gene (fosr) serves as a control for BDNF stimulation and RNase protection. C, Expression of a wild-type SRF but not the vector control or the dominant negative SRF, is sufficient to induce SRE-mediated transcription even under trophic withdrawal conditions. Cortical neurons were transfected with a wt or dn SRF or vector control (0.6 μg of DNA per well) as described in A. Data (A, C) are from three independent experiments of quadruplicate determinants. Error bars indicate SEM.
Further evidence supporting a role for SRF in BDNF stimulation of SRE-mediated transcription was provided by an experiment using the SRE promoter element of a native gene, c-fos (Fig. 1B). Various human fosh reporter genes were transfected into cortical neurons. The plasmid pAF42.SRE.wt (wild-type SRE) contains a single copy of the wild-type human c-fos SRE inserted 42 bp 5′ of the transcription initiation site in the plasmid pAF42. pAF42 contains a copy of the human c-fos gene in which the promoter has been deleted of all known transcription regulatory sequences 5′ of the TATAA box (Rivera et al., 1990). Three days after transfection, cells were stimulated with BDNF for 1 hr. Transcription from the pAF42.SRE.wt reporter was efficiently induced by BDNF (Fig. 1 B, fosh). In contrast, mutations in the SRE that disrupt the binding of SRF to the SRE (SRE.pm1 or SRE.mut2; Rivera et al., 1990; Misra et al., 1994) abolished BDNF induction of transcription through the SRE (Fig. 1B). This experiment established that SRF is necessary for BDNF-stimulation of SRE transcription. Is SRF sufficient for SRE-mediated transcription? Expression of a wild-type but not the dominant-negative SRF induced SRE-mediated transcription even under trophic withdrawal conditions (Fig. 1C), suggesting a sufficiency of SRF in SRE-mediated transcription in cortical neurons.
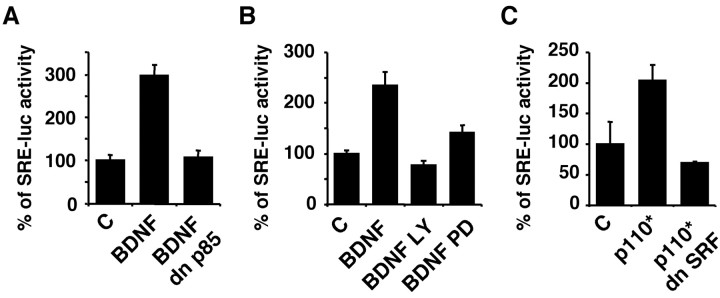
To determine whether the PI3K pathway mediates BDNF stimulation of SRE transcription, cortical neurons were transfected with a dominant negative form of the p85 subunit to selectively inhibit PI3K (Dhand et al., 1994). Expression of the dominant interfering p85 blocked BDNF stimulation of SRE-mediated transcription (Fig. 2A). Furthermore, treatment with a PI3K inhibitor, LY294002, also prevented BDNF stimulation of SRE-mediated transcription (Fig. 2B). These data demonstrate that PI3K activity is required for BDNF stimulation of SRE-mediated transcription in cortical neurons.
Figure 2.
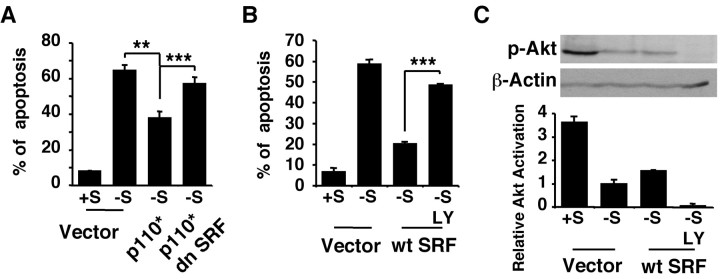
BDNF stimulation of SRE/SRF-mediated transcription in cortical neurons requires PI3K activity. Cortical neurons plated in 24-well plates were transfected with a 5×SRE-driven luciferase reporter to measure SRE-mediated transcription as described in Figure 1A. A, BDNF (10 ng/ml) stimulation of SRE-mediated transcription is blocked by coexpression of a dominant negative PI3K (dn p85). B, BDNF stimulation of SRE-mediated transcription is blocked by LY294002 (LY, 30 μm), a PI3K inhibitor, and by PD98059 (PD, 50 μm), an MKK1 inhibitor that inhibits ERK1/2 activation. C, Expression of a constitutive active PI3K (p110*) stimulates SRE-mediated transcription, which is blocked by coexpression of a dominant negative SRF. Data are from three independent experiments of quadruplicate determinants. Error bars indicate SEM.
Is SRF critical for PI3K stimulation of SRE-mediated transcription? To address this issue, cortical neurons were transfected with a constitutive active (p110*) catalytic subunit of PI3K (Hu et al., 1995) with or without cotransfection of a dominant-negative SRF (Fig. 2C). Expression of p110* stimulated transcription initiated from SRE, which was blocked by the dominant negative SRF. Together, these data suggest that PI3K is key to BDNF stimulation of SRE/SRF-mediated transcription in cortical neurons.
Expression of a wild-type SRF is sufficient to protect cortical neurons from trophic deprivation-induced apoptosis
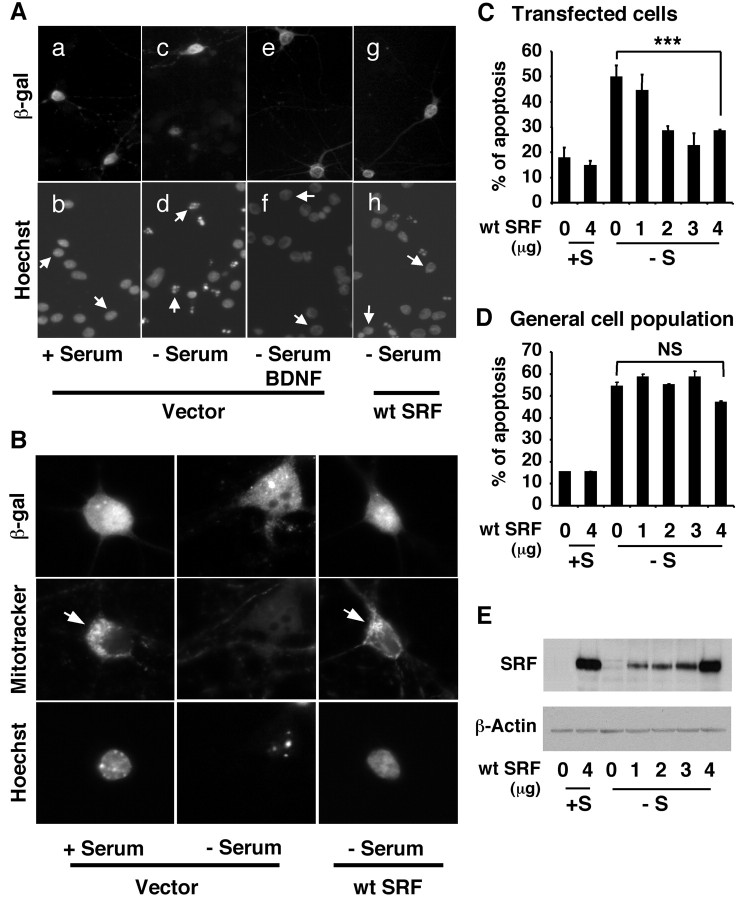
We previously established that BDNF protects postnatal cortical neurons against trophic deprivation (Hetman et al., 1999). To test the hypothesis that SRE/SRF-mediated transcription plays a role in BDNF neuroprotection, we asked whether expression of a wild-type SRF protects against trophic withdrawal. Cortical neurons were transiently transfected with an expression vector for wild-type SRF or its cloning vector as a control (Fig. 3). Two days after transfection, cells were treated with trophic withdrawal to induce apoptosis, as previously described (Hetman et al., 1999). Transfected cells were identified by immunostaining of the cotransfected marker protein β-galactosidase (Fig. 3A,B, top panels). Apoptotic cells were identified after Hoechst staining of the nuclei (Fig. 3A,B, bottom panels). Under normal conditions (Fig. 3A, a, b), most of the transfected cells were healthy with uniformly stained nuclei. After 24 hr trophic withdrawal, many of the vector control transfected cells have fragmented and condensed nuclei, indicative of apoptosis (Fig. 3A, c, d). Treatment with 10 ng/ml BDNF prevented this apoptosis (Fig. 3A, e, f). Likewise, expression of the wild-type SRF rescued cells from trophic withdrawal (Fig. 3A, g, h). To ascertain that cells expressing SRF are not only prevented from undergoing nuclear fragmentation and condensation but also viable, we evaluated neuronal viability by staining the cells with a mitotracker, CMXRos. This mitotracker is sensitive to mitochondria membrane potential; thus, this staining can be used as an indicator of cell viability. In live cells, mitochondria membrane potential is maintained, and the mitotracker is retained in the mitochondria, manifesting a punctate staining pattern. In contrast, when mitochondria membrane potential is lost, the mitotracker cannot be retained in the mitochondria, and the staining is diffuse throughout the cytoplasm, indicative of nonviable cells. Like cells in the presence of serum, cells transfected with a wild-type SRF showed punctate mitotracker staining and uniformly stained nuclear morphology even under trophic deprivation conditions (Fig. 3B), indicating that they have maintained their mitochondria membrane potential and are viable. However, vector control-transfected cells under trophic deprivation conditions exhibited diffuse mitotracker staining and fragmented nuclear morphology, indicating that they are nonviable.
Figure 3.
SRF protects cortical neurons from trophic withdrawal. A, Representative photomicrographs of cortical neurons. Primary cortical neurons were transfected with vector control or an expression vector for wt SRF (4 μg). In all cases, a β-galactosidase expression vector was A, Representative photomicrographs of cortical neurons. Primary cortical neurons (-S), serum-containing conditioned medium (+S), or 10 ng/ml BDNF as indicated. Twenty-four hours later, cells were fixed and immunostained with an antibody against β-galactosidase to identify transfected cells (top panels). Hoechst 33342 staining was used to visualize nuclear morphology (bottom panels). Arrows point to the nuclei of transfected cells. The arrow-pointed nuclei in d are fragmented and condensed, characteristic of apoptosis, whereas those in panels b, f, and h are evenly stained, indicating healthy cells. B, Cells expressing SRF maintain viability after serum withdrawal. Cortical neurons were transfected with vector control or wt SRF (4 μg) and subjected to serum deprivation (-S) for 24 hr as in A. The cells were then incubated with 10 nm Mitotracker Red CMXRos to stain mitochondria (middle panels). Thirty minutes later, the cells were fixed and stained with an antibody against β-galactosidase to identify transfected cells (top panels) and with Hoechst dye 33342 to visualize nuclear morphology (bottom panels). Note that the cell transfected with the wt SRF has evenly stained nuclei and exhibits a punctate staining pattern for the mitotracker (arrow), indicating that it is nonapoptotic and viable. In contrast, the control transfected cell has typical apoptotic nuclear morphology and diffuse mitotracker staining, indicating that it is apoptotic and nonviable. C, D, SRF protects cortical neurons from trophic withdrawal in a dose dependent manner. Cortical neurons were transfected at DIV 3 with varying concentrations of a wt SRF DNA (0-4 μg). Cells were also cotransfected with a plasmid DNA encoding β-galactosidase as a marker for transfection. The CGN vector DNA was used as a supplement so that all plates have an equal amount of DNA. Two days after transfection, cells were washed twice with serum-free medium and then placed in serum-free medium (-S) or serum-containing conditioned medium (+S). Apoptosis in the transfected cell population (β-galactosidase-stained neurons; C) and the general cell population (D) was scored 24 hr later. Results are averages of five independent experiments ± SEM; ***p ≤ 0.001(ANOVA). NS, Not statistically significant. E, Western analysis for transfected SRF. Cortical neurons were transfected and treated as in C and D. Cell lysates were prepared 2 d after transfection, and 25 μg of total protein was used for Western blotting against SRF. Western analysis of β-actin was used as a loading control. The transfected SRF is expressed in a dose-dependent manner.
The neuroprotection by SRF was dose-dependent and statistically significant (Fig. 3C; p ≤ 0.001). Cells transfected with the vector control showed a significant increase in apoptosis after trophic withdrawal. Although 1 μg of SRF DNA had little effect, cells transfected with 2-4 μg SRF DNA showed a substantial reduction in apoptosis. The protective effect of SRF was maximal when cells were transfected with 3-4 μg of SRF DNA. To control for transfection-associated neurotoxicity, apoptosis was also quantitated in the general (all) cell population (Fig. 3D). Because the transfection efficiency is low (<5%), apoptosis in the general cell population should closely reflect that in the nontransfected cell population. Trophic withdrawal induced similar levels (∼50%) of apoptosis in vector control-transfected cells and in the general population (Fig. 3C,D), suggesting that transient transfection per se did not have a nonspecific effect on neuronal apoptosis in transfected cells. Furthermore, the dose-dependent neuroprotection correlated well with the dose-dependent expression of SRF (Fig. 3E). Together with data shown in Figure 1C, these results suggest that expression of a wild-type SRF is sufficient to activate SRE-mediated gene expression and to protect cortical neurons from apoptosis during trophic deprivation.
SRF contributes to BDNF and PI3K neuroprotection against trophic withdrawal
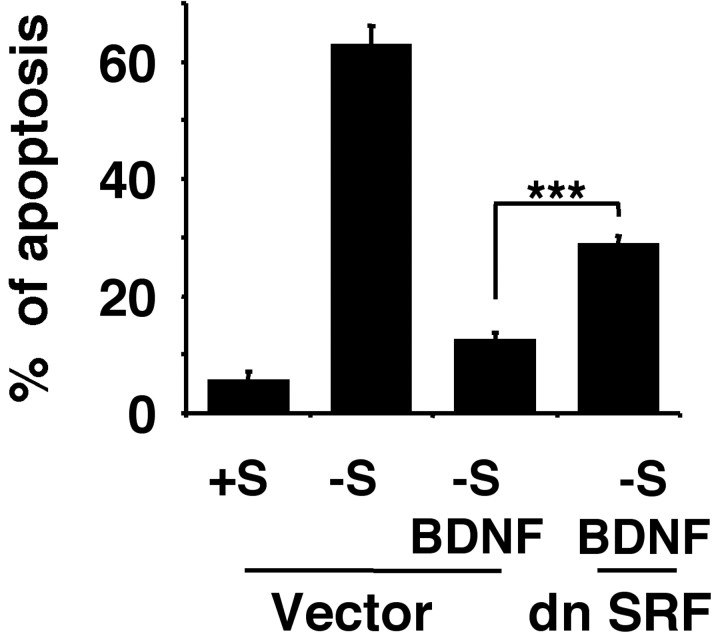
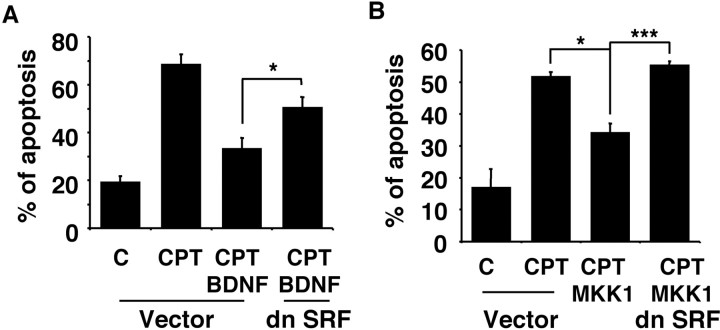
To determine whether SRE/SRF-mediated transcription is important for the neuroprotective effect of BDNF, cortical neurons were transfected with a vector control or 4 μg of the dominant-negative SRF DNA. Two days after transfection, cells were subjected to trophic withdrawal in the presence or absence of BDNF, and apoptosis in transfected cells was scored 24 hr later (Fig. 4). As shown earlier, BDNF protected cortical neurons from trophic withdrawal. However, expression of a dominant negative SRF, which interferes with SRE-mediated gene expression, significantly inhibited the protective effect of BDNF (p ≤ 0.001). These data suggest that SRE/SRF-mediated transcription plays a critical role in BDNF neuroprotection against trophic deprivation. The incomplete inhibition of neuroprotection afforded by the dominant negative SRF suggests that several pathways may contribute to BDNF neuroprotection.
Figure 4.
SRF is necessary for full neuroprotection provided by BDNF against trophic deprivation. Cortical neurons were transfected with control vector (4 μg) or dn SRF (4 μg) and treated as described in Figure 3. BDNF was added at 10 ng/ml where indicated. Results are averages of three independent experiments ± SEM; ***p ≤ 0.001(ANOVA).
BDNF neuroprotection against trophic deprivation is primarily mediated through PI3K signaling (Hetman et al., 1999). To determine whether PI3K neuroprotection is mediated through SRF, we transiently transfected cortical neurons with a constitutive active PI3K (p110*; Hu et al., 1995) with or without cotransfection with a dominant negative SRF (Fig. 5). Expression of p110* partially protected cortical neurons from trophic deprivation (Fig. 5A; p ≤ 0.01), consistent with our previous report (Hetman et al., 1999). Coexpression of a dominant negative SRF significantly reversed the protection afforded by p110* (p ≤ 0.001), placing SRF downstream from PI3K in the BDNF neuroprotection signaling pathway.
Figure 5.
Role of PI3K signaling in SRF neuroprotection against trophic deprivation. Cortical neurons were transfected with 4 μg of DNA of a control vector, a dn SRF, or a constitutive active PI3K (p110*) where indicated and treated as described in Figure 3. A, Neuroprotection against trophic deprivation provided by expression of a constitutive active PI3K is reversed by coexpression of a dn SRF, suggesting that SRF contributes to PI3K neuroprotection. B, Neuroprotection afforded by expression of the wild-type SRF is completely reversed by 30 μm LY294002 treatment (LY), suggesting that the activity of the endogenous PI3K is necessary for SRF neuroprotection. C, Anti-phospho-Akt (p-Akt) Western analysis demonstrating complete inhibition of endogenous PI3K activity in the presence of 30 μm LY294002 (LY). Relative levels of p-Akt were normalized to the loading control β-actin. Results are averages of three independent experiments ± SEM; **p ≤ 0.01; ***p ≤ 0.001 (ANOVA).
We also examined the effect of endogenous PI3K activity on the neuroprotective capacity of SRF. Cortical neurons were transfected with wild-type SRF or control vector and subjected to trophic withdrawal with or without LY294002 treatment (Fig. 5B,C). In the presence of serum, there is a considerable level of basal PI3K activity assessed by Akt phosphorylation (Fig. 5C). This endogenous PI3K activity is dramatically reduced on serum withdrawal and completely inhibited by LY294002 (Fig. 5C), consistent with our previous report (Hetman et al., 2000). Inhibition of endogenous PI3K activity by LY294002 almost completely reversed the protective activity of SRF against trophic withdrawal (Fig. 5B; p ≤ 0.001). This suggests that the neuroprotective activity of the transfected wild-type SRF is regulated by upstream endogenous PI3K activity. Together with data shown in Figure 2B, these data implicate BDNF-PI3K-SRE/SRF-mediated transcription as a neuroprotective pathway against trophic deprivation.
Expression of wild-type SRF protects cortical neurons against DNA damage
SRE-mediated transcription in CNS neurons is also regulated by the ERK1/2 pathway via phosphorylation of TCFs (Xia et al., 1996). Indeed, PD98059, an inhibitor for MKK1/2, inhibited BDNF stimulation of SRE-mediated transcription in cortical neurons (Fig. 2B). Because SRF is obligatory for SRE-mediated transcription in all cells, SRF function is also essential for ERK1/2 activation of SRE-mediated transcription (Treisman, 1995; Xia et al., 1996). Thus, we examined whether SRE/SRF-mediated gene expression also plays a role in neuronal survival directed by ERK1/2 signaling. We used our previously established experimental paradigm in which BDNF protects cortical neurons against camptothecin in an ERK1/2-dependent manner (Hetman et al., 1999). Camptothecin is an anticancer drug, and apoptosis induced by camptothecin has been used as a model to study neuronal apoptosis induced by DNA damage (Morris and Geller, 1996; Park et al., 1997; Hetman et al., 1999).
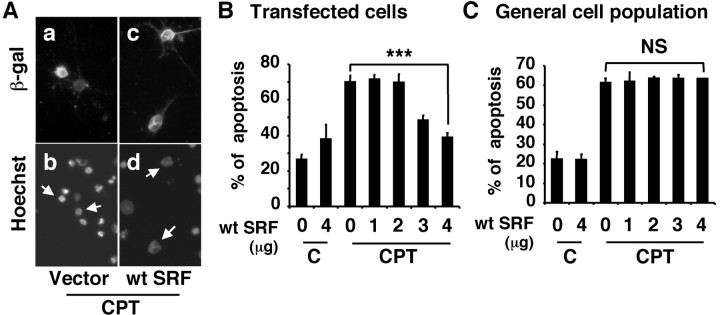
To test the hypothesis that ERK1/2 induction of SRE/SRF-mediated transcription plays a role in BDNF neuroprotection, we determined whether expression of wild-type SRF protects neurons against camptothecin. Cortical neurons were transiently transfected with varying amounts of a wild-type SRF DNA or its vector control (Fig. 6). Two days after transfection, cells were treated with 5 μm camptothecin to induce apoptosis, as previously described (Hetman et al., 1999). Expression of SRF protected cortical neurons from camptothecin-induced apoptosis in a dose-dependent manner (Fig. 6B). Camptothecin induced similar levels (∼60-70%) of apoptosis in vector control-transfected cells and in the general population (Fig. 6B,C), suggesting that transient transfection per se did not have a nonspecific effect on neuronal apoptosis in transfected cells. These results suggest that SRE/SRF-mediated gene expression is sufficient to counteract DNA damage-induced apoptosis in cortical neurons.
Figure 6.
Expression of a wild-type SRF is sufficient to protect cortical neurons against camptothecin, a DNA-damaging agent. A, Representative photomicrographs of cortical neurons. Primary cortical neurons were transfected with vector control or an expression vector for wild-type SRF (4 μg). In all cases, a β-galactosidase expression vector was cotransfected as a marker for transfection. Two days after transfection, cells were treated with 5 μm camptothecin (CPT). Twenty-four hours later, cells were fixed and immunostained with an antibody against β-galactosidase to identify transfected cells (a, c). Hoechst 33342 staining was used to visualize nuclear morphology (b, d). Arrows point to the nuclei of transfected cells. The arrow-pointed nuclei in b are apoptotic, whereas those in d are healthy. B, C, Quantitation of SRF neuroprotection against camptothecin. Cortical neurons were transfected with varying concentrations of a wt SRF (0-4 μg). The CGN vector DNA was used as a supplement so that all plates have equal amount of DNA. Two days after transfection, cells were treated with 5 μm camptothecin or vehicle control (C), and apoptosis in transfected cells (B) and in the general cell population (C) was scored 24 hr later. Results are averages of three independent experiments ± SEM; ***p ≤ 0.001(ANOVA). NS, Not statistically significant.
BDNF protection against camptothecin is mediated by SRF
We next asked whether SRF is necessary for the neuroprotection of BDNF against camptothecin. Cortical neurons were transfected with 4 μg of DNA encoding a dominant-negative SRF or vector control (Fig. 7A). Two days after transfection, cells were treated with 5 μm camptothecin in the presence or absence of 10 ng/ml BDNF as indicated. Apoptosis in transfected cells was scored 24 hr later. Consistent with our previous report (Hetman et al., 1999), BDNF protected cortical neurons from camptothecin-induced apoptosis (Fig. 7A). Expression of the dominant-negative SRF partially inhibited the neuroprotective effect of BDNF (p ≤ 0.05). Partial inhibition of neuroprotection afforded by dominant-negative SRF suggests that several mechanisms may contribute to BDNF neuroprotection.
Figure 7.
BDNF protection against camptothecin is mediated by SRF. A, Expression of a dominant negative SRF reduces BDNF neuroprotection against camptothecin. B, Expression of a dominant negative SRF reduces neuroprotection of a constitutive active MKK1 against camptothecin. Cortical neurons were transfected with control vector (4 μg), a dn SRF (4 μg), or a constitutive active MKK1 (4 μg), a kinase that phosphorylates and activates ERK1/2. Cells were treated with vehicle control (C) or 5 μm camptothecin (CPT) as described in Figure 6. BDNF was added at 10 ng/ml at the time of camptothecin treatment. Results are averages of three independent experiments ± SEM; *p ≤ 0.05; ***p ≤ 0.001(ANOVA).
BDNF neuroprotection against camptothecin is mediated primarily by activation of the ERK1/2 signaling pathway (Hetman et al., 1999). Does ERK1/2 activate SRE/SRF-mediated transcription to counteract camptothecin toxicity? We addressed this question using dominant negative SRF to prevent ERK1/2 activation of SRE-mediated transcription. Cortical neurons were cotransfected with a constitutive active MKK1 to active ERK1/2 and a dominant negative SRF to block SRF function (Fig. 7B). Expression of a constitutive active MKK1 provided protection against camptothecin (p ≤ 0.05). Coexpression of a dominant-negative SRF completely reversed this neuroprotection (p ≤ 0.001). These data suggest that SRF-mediated transcription is necessary for BDNF protection against camptothecin, a process mediated through ERK1/2 signaling.
Discussion
The objective of this study was to define the role of SRE/SRF-mediated gene expression for neuronal survival. SRF is a ubiquitously expressed transcription factor that plays a role in muscle differentiation, cellular growth, and cardiac development (Zhang et al., 2001; Chen et al., 2002; Miano, 2003). Furthermore, SRF is widely expressed in the CNS both during development and in adults (Stringer et al., 2002). Because SRE/SRF-mediated transcription is regulated by glutamate and calcium in neurons, it has been implicated in regulating synaptic activity (Xia et al., 1996; Johnson et al., 1997). However, a biological function of SRF in neurons has not been directly demonstrated. Here we show that SRF promotes neuronal survival, providing the first description of a biological function for SRF in CNS neurons.
Expression of a wild-type SRF was sufficient to protect cortical neurons against trophic deprivation and camptothecin treatment. In contrast, expression of a dominant negative SRF blocked BDNF and PI3K neuroprotection against trophic withdrawal. It also prevented BDNF and ERK1/2 neuroprotection against the DNA-damaging agent camptothecin. Our data suggest that SRE/SRF-mediated transcription is critical for BDNF neuroprotection mediated through the PI3K and ERK1/2 signaling pathways.
It is widely accepted that growth factor stimulation of SRE-mediated transcription depends on ERK1/2 activation and subsequent phosphorylation of TCFs (Gille et al., 1992; Janknecht et al., 1993; Marais et al., 1993; Hipskind et al., 1994; Gille et al., 1995; Miranti et al., 1995; Price et al., 1995). Indeed, BDNF stimulation of SRE-mediated transcription is reduced by PD98059 inhibition of ERK1/2 signaling. Interestingly, we report here that BDNF stimulates SRE-mediated gene expression in cortical neurons in an SRF- and PI3K-dependent manner. Furthermore, PI3K induction of SRE-mediated transcription required SRF. These results suggest a critical role for PI3K in BDNF stimulation of SRE-mediated transcription in CNS neurons. Thus, neurotrophin stimulation of SRE/SRF-mediated transcription requires both PI3K and ERK1/2 signal transduction systems.
Although the mechanism by which SRF contributes to neuroprotection is unknown, induction of prosurvival genes through SRE is a likely explanation. SRE is found in a variety of genes, including the immediate early genes c-fos, fosB, junB, egr-1 and -2, nurr1, and nur77, as well as actins and myosins (Chai and Tarnawski, 2002). For example, SRE was originally identified in the promoter of the immediate early gene c-fos, whose expression is regulated by SRF binding to SRE (Treisman, 1992, 1995). Interestingly, it was recently reported that c-fos plays a critical role in neuronal survival after kainite treatment by regulating the expression of BDNF and glutamate receptor 6 (Zhang et al., 2002). Another antiapoptotic protein that may be regulated through SRE is MCL-1, a member of the Bcl-2 family that protects cell lines from DNA damage and other cytotoxic drugs (Yang et al., 1996; Zhan et al., 1997; Townsend et al., 1999; Akgul et al., 2000).
We observed that transfected SRF exerted a dose-dependent protection against trophic withdrawal. Because the effect of SRF depends on its interaction with much lower amounts of endogenous SRE promoter element, one might expect that any amount of transfected SRF would be more than enough to complex with endogenous SRE. However, only a wild-type SRF was transfected into neurons in this study. The transcriptional activity and neuroprotective effect of both the endogenous and transfected wild-type SRF depend on its activation by the upstream PI3K pathway. In the presence of serum, there is a considerable level of basal PI3K activity that is sufficient to activate the endogenous SRF or the transfected wild-type SRF. In fact, in the presence of serum, expression of 4 μg of SRF does not further decrease the basal level of apoptosis compared with vector control transfected cells (Fig. 3C). This suggests that when there is relatively high level of PI3K activity in the presence of serum; there is no dose-dependent effect of SRF; and the endogenous level of SRF is sufficient to provide the neuroprotection. However, on serum withdrawal, the endogenous PI3K activity is dramatically reduced, and this is likely to yield a much less efficient activation of the downstream SRF so that SRF binding to and activating the “much lower amounts of the endogenous SRE” is far from saturated. With an increasing amount of SRF transfected and expressed, it may facilitate SRF activation by increasing substrate SRF concentration. This may yield more functionally active SRF, thus the observed SRF dose-response protection. LY294002 completely inhibited the remaining endogenous PI3K activity in the absence of serum, thus preventing the activation of the transfected wild-type SRF. This may explain why LY294002 abolishes neuroprotection of expressed SRF.
Several mechanisms downstream from PI3K have been implicated in executing the prosurvival effect of PI3K against trophic withdrawal, including inhibition of GSK3β and phosphorylation and inactivation of BAD (Datta et al., 1997; del Peso et al., 1997), caspase 9 (Cardone et al., 1998), and FKHRL1 (Brunet et al., 1999). Data presented here suggest SRE/SRF-mediated transcription as another neuroprotective mechanism downstream from PI3K.
ERK1/2 neuroprotection against trophic withdrawal in cerebellar granule neurons and sympathetic ganglion neurons has been attributed to activation of CREB or phosphorylation and inactivation of BAD (Bonni et al., 1999; Riccio et al., 1999). However, downstream mechanisms mediating ERK1/2 neuroprotection against camptothecin have not been identified. Our data suggest that ERK1/2 protects cortical neurons against camptothecin, at least in part, by activating SRE/SRF-mediated transcription. Furthermore, BDNF protects neurons from trophic withdrawal- and camptothecin-induced apoptosis by actively inducing SRE/SRF-mediated transcription, providing a downstream neuroprotective mechanism common to both PI3K and ERK1/2 pathways.
In summary, we describe here a novel mechanism whereby SRE/SRF-mediated transcription plays an important role in neuroprotection against both trophic deprivation and camptothecin-induced DNA damage. This is, to our knowledge, the first direct demonstration of a biological function for SRF in CNS neurons and identifies SRF as a pivotal downstream target for ERK1/2- and PI3K-mediated neuroprotection.
Footnotes
This work was supported by Burroughs Wellcome Fund for New Investigator Award in Toxicology Grant APP 3010 (Z.X.), National Institutes of Health (NIH) Grants NS44069 and AG19193 (Z.X.), and NIH National Institute of Environmental Health Sciences Environmental Pathology/Toxicology Postdoctoral Training Grant 5T32ES07032 (S.H.C.). We thank Kathryn Vandemark for technical support and members of the Xia laboratory for critical reading of this manuscript.
Correspondence should be addressed to Zhengui Xia, Department of Environmental and Occupational Health Sciences, Box 357234, University of Washington, Health Sciences Building, Room F561C, Seattle, WA 98195-7234. E-mail: zxia@u.washington.edu.
Copyright © 2004 Society for Neuroscience 0270-6474/04/242277-09$15.00/0
References
- Akgul C, Turner PC, White MR, Edwards SW (2000) Functional analysis of the human MCL-1 gene. Cell Mol Life Sci 57: 684-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonni A, Brunet A, West AE, Datta SR, Takasu MA, Greenberg ME (1999) Cell survival promoted by the Ras-MAPK signaling pathway by transcription-dependent and -independent mechanisms. Science 286: 1358-1362. [DOI] [PubMed] [Google Scholar]
- Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, Anderson MJ, Arden KC, Blenis J, Greenberg ME (1999) Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 96: 857-868. [DOI] [PubMed] [Google Scholar]
- Cardone MH, Roy N, Stennicke HR, Salvesen GS, Franke TF, Stanbridge E, Frisch S, Reed JC (1998) Regulation of cell death protease caspase-9 by phosphorylation. Science 282: 1318-1321. [DOI] [PubMed] [Google Scholar]
- Chai J, Tarnawski AS (2002) Serum response factor: discovery, biochemistry, biological roles and implications for tissue injury healing. J Physiol Pharmacol 53: 147-157. [PubMed] [Google Scholar]
- Chen F, Kook H, Milewski R, Gitler AD, Lu MM, Li J, Nazarian R, Schnepp R, Jen K, Biben C, Runke G, Mackay JP, Novotny J, Schwartz RJ, Harvey RP, Mullins MC, Epstein JA (2002) Hop is an unusual homeobox gene that modulates cardiac development. Cell 110: 713-723. [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162: 156-159. [DOI] [PubMed] [Google Scholar]
- Cross DA, Alessi DR, Cohen P, Andjelkovich M, Hemmings BA (1995) Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature 378: 785-789. [DOI] [PubMed] [Google Scholar]
- Crowder RJ, Freeman RS (2000) Glycogen synthase kinase-3beta activity is critical for neuronal death caused by inhibiting phosphatidylinositol 3-kinase or akt but not for death caused by nerve growth factor withdrawal. J Biol Chem 275: 34266-34271. [DOI] [PubMed] [Google Scholar]
- Dalton S, Treisman R (1992) Characterization of SAP-1, a protein recruited by serum response factor to the c-fos serum response element. Cell 68: 597-612. [DOI] [PubMed] [Google Scholar]
- Datta SR, Dudek H, Tao X, Masters S, Fu HA, Gotoh Y, Greenberg ME (1997) Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell 91: 231-241. [DOI] [PubMed] [Google Scholar]
- Davis RJ (2000) Signal transduction by the JNK group of MAP kinases. Cell 103: 239-252. [DOI] [PubMed] [Google Scholar]
- del Peso L, Gonzalez-Garcia M, Page C, Herrera R, Nunez G (1997) Interleukin-3-induced phosphorylation of BAD through the protein kinase Akt. Science 278: 687-689. [DOI] [PubMed] [Google Scholar]
- Dhand R, Hara K, Hiles I, Bax B, Gout I, Panayotou G, Fry MJ, Yonezawa K, Kasuga M, Waterfield MD (1994) PI 3-kinase: structural and functional analysis of intersubunit interactions. EMBO J 13: 511-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa-Masot XA, Hetman M, Higgins MJ, Kokot N, Xia Z (2001) Taxol induces apoptosis in cortical neurons by a mechanism independent of Bcl-2 phosphorylation. J Neurosci 21: 4657-4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gille R, Sharrocks AD, Shaw PE (1992) Phosphorylation of transcription factor p62TCF by MAP kinase stimulates ternary complex formation at the c-fos promoter. Nature 358: 414-417. [DOI] [PubMed] [Google Scholar]
- Gille R, Kortenjann M, Thomae O, Moomaw C, Slaughter C, Cobb MH, Shaw PE (1995) ERK phosphorylation potentiates Elk-1-mediated ternary complex formation and transactivation. EMBO J 14: 951-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovane A, Pintzas A, Maira S-M, Sobieszczuk P, Wasylyk B (1994) Net, a new ets transcription factor that is activated by Ras. Genes Dev 8: 1502-1513. [DOI] [PubMed] [Google Scholar]
- Hetman M, Kanning K, Cavanaugh JE, Xia Z (1999) Neuroprotection by brain-derived neurotrophic factor is mediated by extracellular-signal-regulated kinase and phosphatidylinositol-3 kinase. J Biol Chem 274: 22569-22580. [DOI] [PubMed] [Google Scholar]
- Hetman M, Cavanaugh JE, Kimelman D, Xia Z (2000) Role of glycogen synthase kinase-3β in neuronal apoptosis induced by trophic withdrawal. J Neurosci 20: 2567-2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetman M, Hsuan SL, Habas A, Higgins MJ, Xia Z (2002) ERK1/2 antagonizes glycogen synthase kinase-3β-induced apoptosis in cortical neurons. J Biol Chem 277: 49577-49584. [DOI] [PubMed] [Google Scholar]
- Hipskind RA, Rao VN, Mueller CG, Reddy ESP, Nordheim A (1991) Ets-related protein Elk-1 is homologous to the c-fos regulatory factor p62TCF Nature 354: 531-534. [DOI] [PubMed] [Google Scholar]
- Hipskind RA, Büscher D, Nordheim A, Baccarini M (1994) Ras/MAP kinase-dependent and -independent signaling pathways target distinct ternary complex factors. Genes Dev 8: 1803-1816. [DOI] [PubMed] [Google Scholar]
- Hu Q, Klippel A, Muslin AJ, Fantl WJ, Williams LT (1995) Ras-dependent induction of cellular responses by constitutively active phosphatidylinositol-3 kinase. Science 268: 100-102. [DOI] [PubMed] [Google Scholar]
- Impey S, Mark M, Villacres EC, Poser S, Chavkin C, Storm DR (1996) Induction of CRE-mediated gene expression by stimuli that generate long-lasting LTP in area CA1 of the hippocampus. Neuron 16: 973-982. [DOI] [PubMed] [Google Scholar]
- Janknecht R, Ernst WH, Pingoud V, Nordheim A (1993) Activation of ternary complex factor Elk-1 by MAP kinase. EMBO J 12: 5097-5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen FE, Prywes R (1993) Identification of transcriptional activation and inhibitory domains in serum response factor (SRF) by using GAL4-SRF constructs. Mol Cell Biol 13: 4640-4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CM, Hill CS, Chawla S, Treisman R, Bading H (1997) Calcium controls gene expression via three distinct pathways that can function independently of the Ras/mitogen-activated protein kinases (ERKs) signaling cascade. J Neurosci 17: 6189-6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson GL, Lapadat R (2002) Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science 298: 1911-1912. [DOI] [PubMed] [Google Scholar]
- Kaplan DR, Miller FD (2000) Neurotrophin signal transduction in the nervous system. Curr Opin Neurobiol 10: 381-391. [DOI] [PubMed] [Google Scholar]
- Liu L, Cavanaugh JE, Wang Y, Sakagami H, Mao Z, Xia Z (2003) ERK5 activation of MEF2-mediated gene expression plays a critical role in BDNF-promoted survival of developing but not mature cortical neurons. Proc Natl Acad Sci USA 100: 8532-8537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marais R, Wynne J, Treisman R (1993) The SRF accessory protein Elk-1 contains a growth factor-regulated transcriptional activation domain. Cell 73: 381-393. [DOI] [PubMed] [Google Scholar]
- Miano JM (2003) Serum response factor: toggling between disparate programs of gene expression. J Mol Cell Cardiol 35: 577-593. [DOI] [PubMed] [Google Scholar]
- Miranti CK, Ginty DD, Huang G, Chatila T, Greenberg ME (1995) Calcium activates serum response factor-dependent transcription by a Ras- and Elk-1-independent mechanism that involves a Ca2+/calmodulin-dependent kinase. Mol Cell Biol 15: 3672-3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra RP, Bonni A, Miranti CK, Rivera VM, Sheng M, Greenberg ME (1994) Calcium entry through L-type voltage-sensitive calcium channels can activate transcription via the serum response factor. J Biol Chem 269: 25483-25493. [PubMed] [Google Scholar]
- Morris EJ, Geller HM (1996) Induction of neuronal apoptosis by camptothecin, an inhibitor of DNA topoisomerase-I: evidence for cell cycle-independent toxicity. J Cell Biol 134: 757-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller CGF, Nordheim A (1991) A protein domain conserved between yeast MCM1 and human SRF directs ternary complex formation. EMBO J 10: 4219-4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namgung U, Xia Z (2000) Arsenite-induced apoptosis in cortical neurons is mediated by c-Jun N-terminal protein kinase 3 and p38 mitogen-activated protein kinase. J Neurosci 20: 6442-6451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman C, Runswick M, Pollock R, Treisman R (1988) Isolation and properties of cDNA clones encoding SRF, a transcription factor that binds to the c-fos serum response element. Cell 55: 989-1003. [DOI] [PubMed] [Google Scholar]
- Pap M, Cooper GM (1998) Role of glycogen synthase kinase-3 in the phosphatidylinositol 3-kinase/Akt cell survival pathway. J Biol Chem 273: 19929-19932. [DOI] [PubMed] [Google Scholar]
- Park DS, Morris EJ, Greene LA, Geller HM (1997) G1/S cell cycle blockers and inhibitors of cyclin-dependent kinases suppress camptothecin-induced neuronal apoptosis. J Neurosci 17: 1256-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poser S, Impey K, Trinh K, Xia Z, Storm DR (2000) SRF-dependent gene expression is required for PI3-kinase-regulated cell proliferation. EMBO J 19: 4955-4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MA, Rogers AE, Treisman R (1995) Comparative analysis of the ternary complex factors Elk-1, SAP-1a and SAP-2 (ERP/NET). EMBO J 14: 2589-2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao VN, Huebner K, Isobe M, Ar-Rushdie A, Croce CM, Reddy ES (1989) elk, tissue-specific ets-related genes on chromosomes X and 14 near translocation breakpoints. Science 244: 66-70. [DOI] [PubMed] [Google Scholar]
- Riccio A, Ahn S, Davenport CM, Blendy JA, Ginty DD (1999) Mediation by a CREB family transcription factor of NGF-dependent survival of sympathetic neurons. Science 286: 2358-2361. [DOI] [PubMed] [Google Scholar]
- Rivera VM, Sheng M, Greenberg ME (1990) The inner core of the serum response element mediates both the rapid induction and subsequent repression of c-fos transcription following serum stimulation. Genes Dev 4: 255-268. [DOI] [PubMed] [Google Scholar]
- Segal RA, Greenberg ME (1996) Intracellular signaling pathways activated by neurotrophic factors. Annu Rev Neurosci 19: 463-489. [DOI] [PubMed] [Google Scholar]
- Shalizi A, Lehtinen M, Gaudilliere B, Donovan N, Han J, Konishi Y, Bonni A (2003) Characterization of a neurotrophin signaling mechanism that mediates neuron survival in a temporally specific pattern. J Neurosci 23: 7326-7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw PE (1992) Ternary complex formation over the c-fos serum response element: p62TCF exhibits dual component specificity with contacts to DNA and an extended structure in the DNA-binding domain of p67SRF. EMBO J 11: 3011-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw PE, Shroter H, Nordheim A (1989) The ability of a ternary complex to form over the serum response element correlates with serum inducibility of the c-fos promoter. Cell 56: 563-572. [DOI] [PubMed] [Google Scholar]
- Sheng M, Dougan ST, McFadden G, Greenberg ME (1988) Calcium and growth factor pathways of c-fos transcriptional activation require distinct upstream regulatory sequences. Mol Cell Biol 8: 2787-2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyu A-B, Greenberg ME, Belasco JG (1989) The c-fos transcript is targeted for rapid decay by two distinct mRNA degradation pathways. Genes Dev 3: 60-72. [DOI] [PubMed] [Google Scholar]
- Stringer JL, Belaguli NS, Iyer D, Schwartz RJ, Balasubramanyam A (2002) Developmental expression of serum response factor in the rat central nervous system. Brain Res Dev Brain Res 138: 81-86. [DOI] [PubMed] [Google Scholar]
- Townsend KJ, Zhou P, Qian L, Bieszczad CK, Lowrey CH, Yen A, Craig RW (1999) Regulation of MCL1 through a serum response factor/Elk-1-mediated mechanism links expression of a viability-promoting member of the BCL2 family to the induction of hematopoietic cell differentiation. J Biol Chem 274: 1801-1813. [DOI] [PubMed] [Google Scholar]
- Treisman R (1992) The serum response element. Trends Biochem Sci 17: 423-426. [DOI] [PubMed] [Google Scholar]
- Treisman R (1994) Ternary complex factors: growth factor regulated transcriptional activators. Curr Opinion Genet Dev 4: 96-101. [DOI] [PubMed] [Google Scholar]
- Treisman R (1995) Journey to the surface of the cell: Fos regulation and the SRE. EMBO J 14: 4905-4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Z, Dudek H, Miranti CK, Greenberg ME (1996) Calcium influx via the NMDA receptor induces immediate early gene transcription by a MAP kinase/ERK-dependent mechanism. J Neurosci 16: 5425-5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Buchan HL, Townsend KJ, Craig RW (1996) MCL-1, a member of the BLC-2 family, is induced rapidly in response to signals for cell differentiation or death, but not to signals for cell proliferation. J Cell Physiol 166: 523-536. [DOI] [PubMed] [Google Scholar]
- Zhan Q, Bieszczad CK, Bae I, Fornace Jr AJ, Craig RW (1997) Induction of BCL2 family member MCL1 as an early response to DNA damage. Oncogene 14: 1031-1039. [DOI] [PubMed] [Google Scholar]
- Zhang J, Zhang D, McQuade JS, Behbehani M, Tsien JZ, Xu M (2002) c-fos regulates neuronal excitability and survival. Nat Genet 30: 416-420. [DOI] [PubMed] [Google Scholar]
- Zhang X, Chai J, Azhar G, Sheridan P, Borras AM, Furr MC, Khrapko K, Lawitts J, Misra RP, Wei JY (2001) Early postnatal cardiac changes and premature death in transgenic mice overexpressing a mutant form of serum response factor. J Biol Chem 276: 40033-40040. [DOI] [PubMed] [Google Scholar]