While the description of resistance to quinolones is almost as old as these antimicrobial agents themselves, transferable mechanisms of quinolone resistance (TMQR) remained absent from the scenario for more than 36 years, appearing first as sporadic events and afterward as epidemics. In 1998, the first TMQR was soundly described, that is, QnrA.
KEYWORDS: CrpP, OqxAB, PMQR, QepA, quinolones, TMQR, aac(6)Ib-cr, antibiotic resistance, plasmid-mediated quinolone resistance, qnr
SUMMARY
While the description of resistance to quinolones is almost as old as these antimicrobial agents themselves, transferable mechanisms of quinolone resistance (TMQR) remained absent from the scenario for more than 36 years, appearing first as sporadic events and afterward as epidemics. In 1998, the first TMQR was soundly described, that is, QnrA. The presence of QnrA was almost anecdotal for years, but in the middle of the first decade of the 21st century, there was an explosion of TMQR descriptions, which definitively changed the epidemiology of quinolone resistance. Currently, 3 different clinically relevant mechanisms of quinolone resistance are encoded within mobile elements: (i) target protection, which is mediated by 7 different families of Qnr (QnrA, QnrB, QnrC, QnrD, QnrE, QnrS, and QnrVC), which overall account for more than 100 recognized alleles; (ii) antibiotic efflux, which is mediated by 2 main transferable efflux pumps (QepA and OqxAB), which together account for more than 30 alleles, and a series of other efflux pumps (e.g., QacBIII), which at present have been sporadically described; and (iii) antibiotic modification, which is mediated by the enzymes AAC(6′)Ib-cr, from which different alleles have been claimed, as well as CrpP, a newly described phosphorylase.
INTRODUCTION
Research on transferable mechanisms of quinolone resistance (TMQR) (see “Is it correct to use the term plasmid-mediated quinolone resistance?” below, for an explanation about the use of this term and acronym) is almost as old as quinolones themselves (1–3). Nonetheless, despite some sporadic unconfirmed descriptions of TMQR (2–6) and the presence of transferable mechanisms of resistance related to a slowdown of the bacterial duplication time associated with a 2.2-kb region of the plasmid pKM101 containing the korB, traL, korA, and traM genes (7), TMQR remained undetected. In most of the early studies in which quinolone resistance transfer was claimed, transconjugants were selected with nalidixic acid, which possesses a relatively high frequency of mutation (8, 9). Furthermore, some studies used low MICs to select nalidixic acid-resistant transconjugants as nalidixic acid resistant (e.g., 8 μg/ml in a study by Jonsson [2]). These findings suggest that the selection of spontaneous mutants rather than the presence of true transconjugants is the most feasible scenario. This led to a debate about the feasibility of TMQR development (10, 11). Indeed, some studies on TMQR were further reanalyzed, and it was proposed that plasmids act as a mutator factor able to induce the development of nalidixic acid resistance (12), and the presence of spontaneous quinolone target mutations (10) showed a lack of transfer of nalidixic acid resistance (13).
Thus, for 36 years after the first nalidixic acid description (14), TMQR remained a unicorn or a vanishing hitchhiker, a myth or urban legend, until 1998, when the presence of TMQR was first unequivocally demonstrated (15).
Quinolones
Quinolones are synthetic products that were first synthetized in 1949 (16). Thereafter, a high number of derivatives and related substances were developed, some of which showed antibacterial properties. Although some quinolone derivative molecules were patented in the late 1950s, it is largely considered that the quinolone era began in 1962, with the synthesis of nalidixic acid (14, 17–19). The first clinical trial reports on the use of nalidixic acid are from 1963 (20–23). Subsequently, nalidixic acid was introduced into clinical practice as early as 1964 (18, 24, 25) albeit limited to the treatment of urinary tract infections (26). Despite this limitation, this agent has played a role in the treatment of other infections, such as those of the gastrointestinal tract, especially in some developing areas (27–30). In subsequent years, the quinolone family grew, and some of its members were introduced into the antibacterial clinical armamentarium; these include oxolinic acid (31), piromidic acid (32), cinoxacin (33), and pipemidic acid (34), among others. Although first proposed in 1960 (17), the next step in the history of quinolones was the addition of a fluorine atom, which opened the door to the fluoroquinolone era. Norfloxacin is considered the starting point of this era (35), despite the fluorine atom first being present in another quinolone introduced into clinical practice, flumequine (36). This fluorine atom was thereafter maintained in almost all the quinolones introduced into clinical practice, largely expanding their bacterial spectra and levels of activity (37). Nonetheless, some recent quinolones, a few of which were introduced into clinical practice only a few years ago (e.g., garenoxacin, nemonoxacin, and ozenoxacin), lack the fluorine atom substituent at position 6, although they may present a fluorine atom(s) in other positions (for instance, the garenoxacin molecule possesses an OCHF2 group in position 8), usually referred to as “desfluoroquinolones” or “nonfluorinated quinolones” (38–42). Finally, some recently developed molecules present antibiotic hybrid characteristics, such as cadazolid, a quinoxolidinone (fluoroquinolone-oxazolidone hybrid molecule) that has been investigated to treat Clostridium difficile infections (43) (Fig. 1).
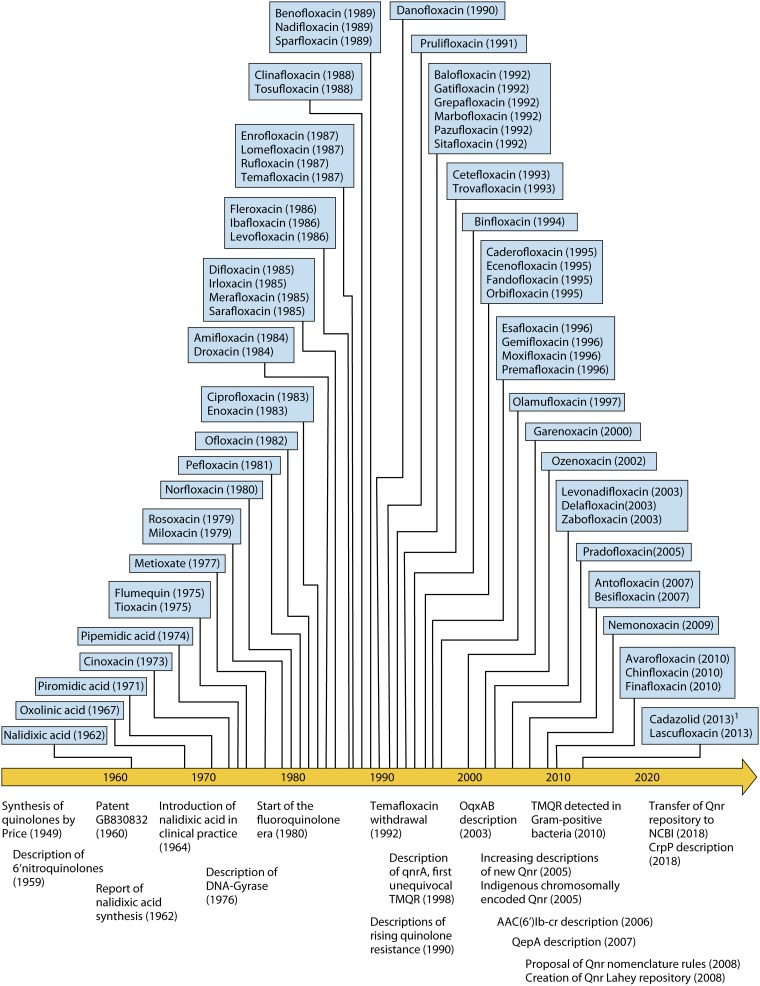
FIG 1.
The chronology of the quinolones (1949 to the present). The top side of the temporal line indicates the years of discovery/synthesis of a series of representative quinolones. In all cases, the year of the most ancient report found in the literature is reported. Note that although several thousand quinolones have been synthetized and their anti-infective potential has been explored, only a selection is presented. Furthermore, no quinolone without a “specific” name has been included (because in the vast majority of cases, their development was discontinued), leading to a lower number of quinolones from ∼2010 onward, as most are in the first stages of development (for instance, DS-8587 or KPI-10 [both of which are described in articles from 2013 {452, 453}]). The bottom side of the temporal line indicates a series of milestones in the history of quinolones. 1, fluoroquinolone-oxazolidone hybrid molecule (43).
In summary, since the beginning of the quinolone era, more than 10,000 quinolones have been synthetized, and their activities and properties have been explored (25, 44, 45), with more than 40 being approved for either human or veterinary applications. Currently, 21 quinolones remain in human use in the European Union and/or the United States (Fig. 2), while several others are used in other countries; for instance, balofloxacin has been approved for human use in South Korea and India (46, 47).
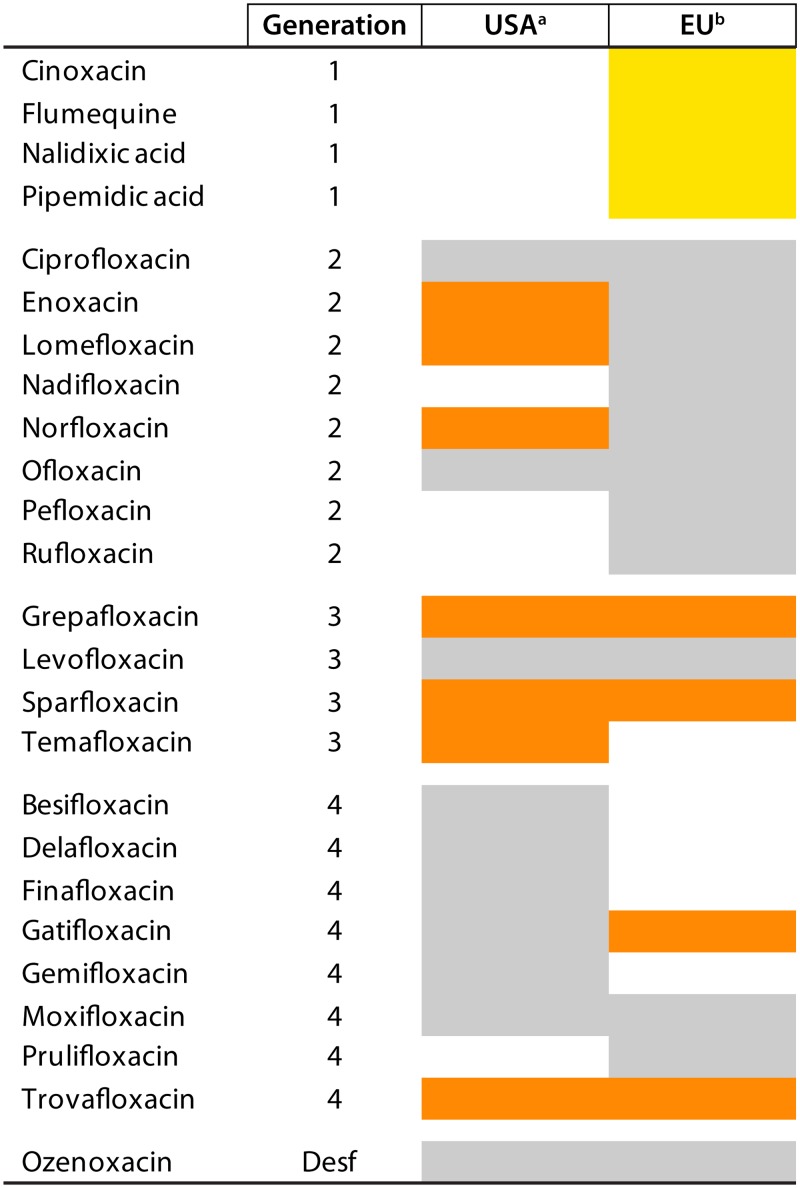
FIG 2.
Quinolones in use in human therapeutics in the United States and the European Union. In gray are quinolones currently (as of March 2019) used in human health. In orange are quinolones that have been discontinued (marked only when this information has been found). In yellow are quinolones proposed for withdrawal from use on March 2019 (https://www.ema.europa.eu/en/documents/referral/quinolone-fluoroquinolone-article-31-referral-annex-i_en.pdf). Note that discontinuation may be related to adverse events (e.g., trovafloxacin) or to economical and market reasons. Some of these antibiotics (or other quinolones that have not been approved or are in the investigational phase) may be considered in special circumstances as last-resort treatment (454). Note that in all the cases, the data listed refer only to the United States and the European Union (including the United Kingdom at the time of writing). The introduction or current or past use/nonuse of these or other quinolones in other geographical areas may not be inferred by this figure. a, extracted from https://www.accessdata.fda.gov/scripts/cder/ob/index.cfm; b, in use in at least one European Union member country (including the United Kingdom at the time of writing) (https://www.ema.europa.eu/documents/referral/quinolone-fluoroquinolone-article-31-referral-annex-i_en.pdf). G, generation; Desf, desfluoroquinolone.
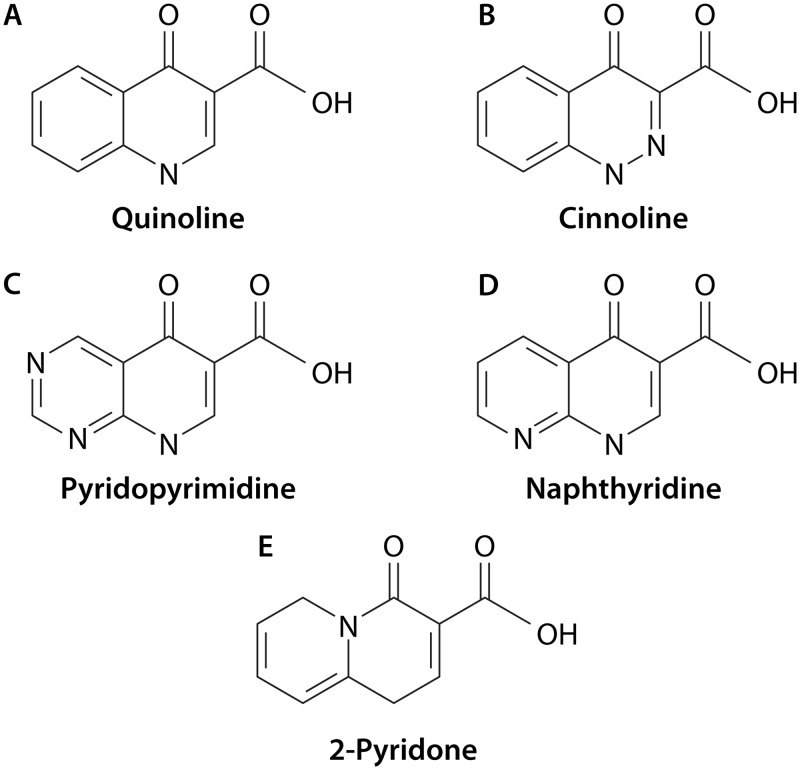
Despite the fact that the classification of quinolones, or other antibacterial agents, in “generations” is imprecise and subject to different interpretations, the quinolones have traditionally been classified into 4 generations based on their spectrum of activity (48, 49) (see Table 1 for definitions used in this article). In addition, the quinolone ring may be subdivided into 4 subclasses (cinnolines, naphthyridines, pyridopyrimidines, and quinolines) related to the position of the nitrogen atoms within the molecule. Of these, the quinoline subclass has by far the largest number of quinolones that have been introduced into clinical/veterinary practice. Furthermore, other structurally related molecules, such as 2-pyridones, quinazoline-2,4-diones, and isothiazoloquinolones, also have antibacterial activity. Of these, 2-pyridones have probably been the most extensively studied. Although usually classified as a different antibiotic family, the 2-pyridones might be structurally considered a fifth quinolone subclass. In fact, they differ from quinolones only in the position of the N atom, which is located at the ring juncture, and the subsequent loss of a double bond (50) (Table 1 and Fig. 3). Nevertheless, to date, no 2-pyridone has been introduced into clinical practice.
TABLE 1.
General classification of the main quinolonesa
| Structural class | Quinolonesb
|
||||
|---|---|---|---|---|---|
| 1st generation | Fluoroquinolones |
Desfluoroquinolones | |||
| 2nd generation | 3rd generation | 4th generation | |||
| Naphthyridines | Nalidixic acid (V) | Enoxacin | Ecenofloxacin | Gemifloxacin | |
| Esafloxacin | Tosufloxacin | Trovafloxacin | |||
| Oxoenoxacin | Zabofloxacin | ||||
| Cinnolines | Cinoxacin | ||||
| Pyridopyrimidines | Piromidic acid | ||||
| Pipemidic acid | |||||
| Quinolines | Droxacin | Amifloxacin | Balofloxacin | Antofloxacin | Nemonoxacin |
| Flumequine (V)c | Benofloxacin (V) | Cetefloxacin | Avarofloxacin | Ozenoxacin | |
| Metioxated | Ciprofloxacin (V) | Chinfloxacin | Besifloxacine | Garenoxacin | |
| Miloxacin | Enrofloxacin (V) | Difloxacin (V) | Caderofloxacin | Piroxacin | |
| Oxolinic acid (V) | Fandofloxacin | Danofloxacin (V) | Clinafloxacin | ||
| Rosoxacin | Fleroxacin | Grepafloxacine | Delafloxacin | ||
| Tioxacin | Lomefloxacin | Ibafloxacin (V) | Finafloxacin | ||
| Merafloxacin | Levofloxacinf | Gatifloxacin | |||
| Nadifloxacin | Marbofloxacin (V) | Lascufloxacin | |||
| Norfloxacin (V) | Orbifloxacin (V) | Levonadifloxacinf | |||
| Ofloxacin (V) | Olamufloxacin | Moxifloxacin | |||
| Pefloxacin | Pazufloxacin | Premafloxacin | |||
| Pirfloxacin | Pradofloxacin (V) | Prulifloxacin | |||
| Rufloxacin | Sarafloxacin (V) | Sitafloxacine | |||
| Sparfloxacin | |||||
| Temafloxacine | |||||
| 2-Pyridonesg | |||||
V, frequent use (past or present) for animal health (livestock and/or companion animals). Note that the list of quinolones used in veterinary health include quinolones with extended use for human health, such as ciprofloxacin, norfloxacin, and ofloxacin, some of which are included in the list of essential medicines (http://www.who.int/medicines/publications/essentialmedicines/20th_EML2017_FINAL_amendedAug2017.pdf?ua=1). Most of the listed quinolones have not been introduced into either human or veterinary medicine. Note that some quinolones may be found in the literature under different names. For instance, avarofloxacin, benofloxacin, caderofloxacin, enoxacin, nadifloxacin, pirfloxacin, rosoxacin, and tioxic acid may also be found as acorafloxacin, vebufloxacin, cadrofloxacin, enofloxacin, jinofloxacin, irloxacin, acrosoxacin, and tioxacin, respectively.
Note that in some classifications, several quinolones may be considered to belong to another generation. The criteria followed in the present scheme are as follows. First generation indicates quinolones presenting activity against some Gram-negative microorganisms (e.g., not against P. aeruginosa), almost all of which are nonfluorinated and with a limited spectrum of clinical indications. Second generation indicates quinolones with an expanded spectrum of activity (includes most Gram-negative microorganisms, e.g., P. aeruginosa, and some Gram-positive organisms), with expanded indications (which may include systemic infections), and marked with the stable introduction of a fluor atom in position 6. Third generation is similar to second generation, with expanded Gram-positive coverage; members of this class may possess activity against some atypical pathogens. Fourth generation is similar to third generation but with activity against anaerobic microorganisms. Desfluoroquinolones are new quinolones in which the fluor atom in position 6 has been removed, usually not included in the fourth-generation scheme.
Carries a fluor atom in position 6.
Metioxate has almost the same structure as tioxic acid but with COOH in position 3 modified by the presence of 4-methylpiperidine (https://pubchem.ncbi.nlm.nih.gov/search/#query=Metioxate). Nonetheless, it is usually classified as a first-generation quinolone.
Presence of a chloride substituent in position 8.
Those with the prefix “levo” (i.e., levofloxacin and levonadifloxacin) are isomers of previously described quinolones which exhibit enhanced activity.
At present, no 2-pyridone has been introduced as an antibacterial agent in clinical practice. Examples of this antibacterial agent group are ABT-719 (50) and KRQ-10018 (459). The 2-pyridone ABT-719 should not be confused with modimelanotide, which also receives the same ABT-719 code. Modimelanotide is an unrelated molecule developed later and designed to prevent acute kidney injury (460).
FIG 3.
General structure and subclasses of quinolones. Quinolones have a bicyclic structure. At present, 4 quinolone subclasses, differing in the positions and numbers of the nitrogen atoms present in the basal bicyclic structure, have been developed and introduced into clinical and/or veterinary settings (A to D). In these 4 quinolone subclasses, the atom numeration is usually described using the quinoline subclass as a general model (A). Position 1 is considered the N atom, and the subsequent positions are numbered anticlockwise. Note that no atoms present in the ring junctions are numbered. Fluoroquinolones present a fluoridine as a substituent in position 6. The radicals present in positions 1 and 7 are critical for quinolone-target interactions. Thus, it has been proposed that radical 1 interacts with amino acid 83 of GyrA (numeration of E. coli) or its equivalent in ParC by means of Van der Waals forces, while radical 7 interacts with amino acid 87 by charge attraction (455). (A) Quinoline. This molecule presents a nitrogen atom only in position 1 (e.g., oxolinic acid, ciprofloxacin, and norfloxacin). (B) Cinnoline. This molecule presents nitrogen atoms in positions 1 and 2 (e.g., cinoxacin). (C) Pyridopyrimidine. The molecule presents nitrogen atoms in positions 1, 6, and 8 (e.g., pipemidic acid). (D) Naphthyridine. This molecule presents nitrogen atoms in positions 1 and 8 (e.g., nalidixic acid and trovafloxacin). (E) 2-Pyridone. This molecule is usually not considered a member of the quinolone antibiotic family. The atom numeration of this molecule differs from those in panels A to D (the nitrogen atom in the upper ring juncture is numbered as atom 5).
Utility of quinolones.
Quinolones have been used to treat a great variety of bacterial infections by either Gram-positive or Gram-negative microorganisms, including intracellular pathogens (37, 51–56). Quinolones have also been used or proposed for prophylactic treatment of specific at-risk populations, such as patients with cirrhosis or neoplasms, and in posttransplant or presurgery/postsurgery patients (57–62). Nonetheless, in several cases, the benefits have been controversial or unsatisfactory, and the addition of new factors, such as increasing levels of antibiotic resistance, has led to the restriction or avoidance of some of these uses (60, 63–65).
The utility of quinolones has also been explored in the treatment of parasitic infections such as malaria, toxoplasmosis, and leishmaniosis; fungal infections (e.g., Candida albicans or Aspergillus fumigatus); or viral infections such as those by BK polyomavirus, rhinovirus, and hepatitis C virus (HCV) (66–75). Meanwhile, the structurally related quinolone molecules elvitegravir and ivacaftor are currently in use as an integrase inhibitor in HIV therapy and in the treatment of cystic fibrosis, respectively (76–78). Moreover, in addition to the antineoplastic potential of some established quinolones such as gemifloxacin (79), new quinolone derivatives are being explored as specific antineoplastic agents (71, 80).
Beyond human uses, fluoroquinolones have also been included in the veterinary armamentarium, in livestock as a growth promoter (an application which is currently forbidden in different countries, including those of the European Union) (81–83), and as a prophylactic agent (84, 85) or in the treatment of infections (81, 85). Indeed, at the end of the 20th century, quinolones ranked among the antibiotics most widely used worldwide, with ciprofloxacin being considered the antibacterial agent most frequently used (81).
Adverse events.
To our knowledge, adverse events were described in early clinical trials of nalidixic acid (20, 21), and the first warning as to the possible occurrence of adverse events related to the use of quinolones was reported as far back as 1965 (4). Since then, several adverse effects related to the use of quinolones have been described, including blood disorders, central nervous system events (dizziness, sleep disorders, and seizures, among others), gastrointestinal disturbances and C. difficile-associated diarrhea, myasthenia gravis exacerbations, peripheral neuropathy, phototoxicity, rashes, and torsade de pointes, among others (25, 45, 86, 87). Of note, adverse events are related to specific quinolone substituents. Thus, halogen atoms at position 8, such as a chloro atom in clinafloxacin or sitafloxacin or a fluor atom in sparfloxacin, have been involved in phototoxicity reactions (45, 88–90). Interestingly, several adverse events have been related to ethnic background (89). In this sense, ethnic differences in phototoxicity reactions have led to the introduction of sitafloxacin into human clinical practice in several Asian countries, such as Japan (https://www.pmda.go.jp/files/000152974.pdf) and Thailand (91), while remaining absent from the antibiotic armamentariums of the European Union, the United States, and other countries.
The most commonly known adverse events are considered to be antibiotic class related and include arthralgias, cartilage affectations, tendinitis, and tendon ruptures. Due to known teratogenic and mutagenic effects, the use of quinolones has classically been avoided in pregnant women and children (88, 92). It is of note that different reports have reanalyzed the use of quinolones in children, including neonates, and they have been considered to be safe and beneficial in specific circumstances, such as severe or life-threating infections by quinolone-susceptible microorganisms; furthermore, secondary effects were considered reversible (88, 93–96). In this line, several fluoroquinolones have been introduced in established pediatric antibacterial armamentariums, such as tosufloxacin in Japan for treating respiratory infections (97, 98). Meanwhile, a recent meta-analysis and systematic review on the use of quinolones in pregnant women highlighted the safety of using fluoroquinolones during the first trimester of pregnancy (99).
There have been recent warnings regarding the possible development of aortic aneurysms and dissection in patients at risk as antibiotic-class-related severe adverse events (100, 101). In this line, mice challenged with a high-fat diet and a low-dose angiotensin infusion exposed to ciprofloxacin were more prone to developing aortic destruction and aneurysms (102). Other unexpected severe adverse events are shown in Table 2.
TABLE 2.
Quinolone-related severe adverse eventsa
| Severe adverse event(s)b | Relevant quinolone(s)c |
|---|---|
| Aortic destruction and aneurysmsd | FQd |
| Cardiovascular events | GRX, MXF, SPX |
| Dysglycemia | CIP, CLX, GAT, LVX |
| Hemolysis and HUS | TMX |
| Hepatotoxicity | TMX, TVA |
| Nephritis | TSX |
| Phototoxicity | CLX, FLE, LOM, PFX, SPX, STX |
| Renal failure | TMX |
| Thrombocytopenia | SPX, TMX, TSX |
CIP, ciprofloxacin; CLX, clinafloxacin; FLE, fleroxacin; FQ, fluoroquinolones; GAT, gatifloxacin; GRX, grepafloxacin; LOM, lomefloxacin; LVX, levofloxacin; MXF, moxifloxacin; PFX, pefloxacin; SPX, sparfloxacin; STX, sitafloxacin; TMX, temafloxacin; TSX, tosufloxacin; TVA, trovafloxacin; HUS, hemolytic-uremic syndrome.
This is a nonexhaustive list. Only especially severe adverse events are shown, which in several cases have resulted in patient death (45, 87, 88, 90, 101, 461). In several cases, these findings have led to the discontinuation of research, withdrawal from clinical practice, and strong restrictions to specific nonsystemic applications (such as topical or ophthalmic), to serious life- or limb-threatening infections, or to last-resort applications, such as compassionate use of several of these quinolones (45, 100, 454, 462, 463). Note that these regulations may differ among different countries. In addition, ethnic background may play a role in the frequency of these adverse events (89). For information about systemic quinolones in use in the European Union and the United States, see Fig. 2.
Most relevant quinolones involved in the specific adverse event. Note that other quinolones may also be able to cause similar effects in specific cases or circumstances.
Immediate quinolone-induced hypersensitivity, mediated by quinolone-specific IgE (103), has been on the rise in the last years, with quinolones currently likely ranking second to β-lactams as the antimicrobial agents most frequently involved in allergic reactions (104), with an especially high incidence among patients treated with moxifloxacin (105–107). The severity of these reactions ranges from anaphylaxis and urticaria to life-threatening anaphylactic shock (87, 105), with a few fatal cases being reported in the literature (108).
Evolution of Quinolone Resistance over Time
Although early studies described the presence of nalidixic acid-resistant clinical isolates, highlighting the feasibility of selecting nalidixic acid resistance during treatment (20, 21, 109), up until the 1980s, reports of quinolone resistance were unusual (1, 2, 4, 110–112). In the 1990s, descriptions of quinolone-resistant microorganisms rapidly increased (111, 113–119), in parallel with the exponential growth of quinolone use (120, 121). In fact, at the end of the 20th century in specific geographical areas, some microorganisms, such as Campylobacter spp., presented extremely high percentages of fluoroquinolone resistance, which in some cases were >80% (111, 122), and therapeutic failure and/or the development of resistance during quinolone treatment was increasingly reported (123–127).
The risks of the rising levels of antimicrobial resistance worldwide have been increasingly discussed and highlighted (128–130), with subsequent proposals for the implementation of different actions (128, 131, 132). Nevertheless, in the present century, the continuously rising global trend toward the isolation of pathogenic or nonpathogenic quinolone-resistant microorganisms has remained unaltered (55, 113, 133–140) and has expanded to regions with limited access to antibiotic agents (141–143). In fact, the use of quinolones has been compromised in different areas because of the high percentages of quinolone resistance among specific pathogens (134, 144–147).
Quinolone Resistance Mechanisms
The first studies determining the molecular mechanisms involved in the development of quinolone resistance showed the important role of point mutations in the genes encoding the different topoisomerase type II subunits (GyrA and GyrB for DNA gyrase and ParC and ParE for topoisomerase IV). In addition, the role of increased efflux activity or permeability alterations, both resulting in decreased cytoplasm quinolone concentrations, was also observed (37) although frequently misconsidered (148). Other less frequent chromosomal mechanisms of quinolone resistance have been described, such as an increase in resistance levels related to lower expression levels of GrlA/GrlB (ParC/ParE) (149).
Although classically not considered mechanisms of resistance, bacteria may also display life strategies that make it difficult for quinolones to access their targets and thereby allow microorganisms to survive in the presence of quinolones. The best example of this is the development of biofilms, in which the extracellular matrix diminishes the interaction of quinolones with microorganisms (150, 151). Microorganisms can also use quiescence as a way to survive in the presence of quinolones (151), probably because these agents need the presence of biological processes involving the activity of topoisomerases to be active. In this line, a curious phenomenon of paradoxical bacterial survival has been described in the presence of extremely high levels of quinolones, which modify the bactericidal action of quinolones to a bacteriostatic effect. It has been proposed that this is related to the blocking of bacterial processes leading to a quiescent state (152).
As mentioned above, the search for TMQR was unfruitful for years, until 1998, when Martínez-Martínez et al. (15) described an increase in quinolone resistance related to the presence of an unknown determinant present in a Klebsiella pneumoniae plasmid which was related to the presence of a Qnr determinant (see QNR, below). Thereafter, AAC(6′)Ib-cr, a variant of an aminoglycoside acetylase able to modify some quinolones, was described in 2006 (153). Until recently, this enzyme remained the only quinolone-inactivating enzyme, but in 2018, a Pseudomonas aeruginosa phosphorylase, called CrpP, was first described (154). Finally, in 2003, a transferable mechanism able to confer olaquindox resistance to Escherichia coli (155) was first detected, being characterized as a resistance-nodulation-division (RND) efflux system and called OqxAB (156). Its ability to extrude some quinolones was established in 2007 (157), almost in parallel with the description of QepA (158, 159), another transferable efflux pump. Further studies have led to the description of other transferable efflux pumps able to confer resistance to quinolones (see Transferable Efflux Pumps, below).
GENERAL OVERVIEW OF TMQR
Classically, the relevance of TMQR of Gram-negative and Gram-positive microorganisms is not equivalent (160). Despite the description of the exchange of genes between Gram-negative and Gram-positive microorganisms (161, 162) and the spread of antibiotic resistance determinants among them (161–165), prior to 2016, only a few specific Gram-positive TMQR, such as QacBIII, had been detected in Staphylococcus aureus (166). Nonetheless, PCR studies performed in the last years have shown the presence of other TMQR, such as qnrA, qnrB, qnrD, oqxAB, or aac(6′)Ib-cr, in Gram-positive microorganisms (167–169). Unfortunately, to our knowledge, of these genes, only oqxA and oqxB have been fully validated by DNA sequencing (169).
Nomenclature of the Transferable Mechanisms of Quinolone Resistance
The term “qnr” was successfully introduced in the first article in the field to refer to the first plasmid-mediated mechanism of quinolone resistance described (15). Although the meaning of “qnr” was not explicitly explained in the text (15), it has been largely considered an acronym of “quinolone resistance.” Similarly, although not abbreviated, Martínez-Martínez et al. (15) also used the term “plasmid-mediated quinolone resistance,” which was adopted in most subsequent works in the field, although to the best of my knowledge, the commonly used acronym “PMQR” was introduced in 2006, after the description of AAC(6′)Ib-cr (170). Further chromosomal descriptions of transferable qnr or other transferable genes involved in quinolone resistance have led to proposal of other genetic nomenclatures (see “Is it correct to use the term plasmid-mediated quinolone resistance?,” below).
The introduction of a letter to define the exact qnr gene was first introduced in 2005, when a new Qnr exhibiting only 59% amino acid identity with the only known Qnr at that time (currently QnrA1) was detected in Shigella flexneri 2b in Japan; this new Qnr was named “QnrS” because of the microorganisms from which it was recovered (171). Thereafter, the qnr gene, which was first isolated in 1998, was rapidly reported as qnrA to avoid confusion (172–174). Although qnrA2 was described in 2004 (GenBank accession number AY675584), the introduction of allele numeration was also developed in 2005, with the reporting of a series of different qnrA gene variants (171).
The subsequent names QnrB, QnrC, and QnrD were adopted in alphabetical order for gene name assignation. Meanwhile, QnrVC maintains the first name proposed, related to its presence in Vibrio cholerae (175). Regarding QnrE, the last transferable Qnr family described (in 2017), both the alphabetical order and the original bacterial source (Enterobacter spp.) coincided at the moment to propose a specific name (176).
Regarding transferable efflux pumps, the term OqxAB was proposed in 2004 because the first description of this transferable efflux pump was made during a study aimed at determining the presence of olaquindox resistance mechanisms (155, 156). Similar to other TMQR, allelic numeration was proposed after the description of amino acid variants in both OqxA and OqxB (177).
Later, in 2007, another efflux pump able to extrude quinolones was isolated and named “qepA” for “quinolone efflux pump” (158, 159). Nonetheless, in the related GenBank submission (GenBank accession number EF150886), Périchon et al. (159) used the unsuccessful term “pef” (derived from “plasmid efflux pump”) to refer to this mechanism of resistance. Then, in 2008, after the description of a new qepA gene differing in 2 amino acid positions, a number was introduced to refer to qepA variants differing in one or a few amino acid residues (178, 179).
The term “qac” was established in the 1980s to refer to a series of Gram-positive efflux pumps able to extrude quaternary ammonium compounds (180), and it is used in the nomenclature of efflux pumps belonging to different families, such as the RND (e.g., QacA and QacB) and small multidrug resistance (SMR) (e.g., QacG, QacH, and QacJ) families (181). Similar to what is described for Qnr, different letters differentiate different genes (e.g., qacA and qacB, etc.). Despite being analyzed in multiple studies, the role of qac determinants in resistance to quinolones has been established only recently; in the same study, roman numerals were introduced to differentiate QacB alleles (166).
Regarding inactivating enzymes, the term AAC(6′)Ib-cr was derived from the nomenclature of aminoglycoside-modified enzymes, because this gene derives from that encoding the aminoglycoside acetyltransferase AAC(6′)Ib (for an explanation of the meaning of the term, see the revision of Ramírez and Tolmasky [182]); the “cr” at the end represents “ciprofloxacin resistance” (153). Finally, CrpP, the most recently described TMQR, was named as such for ciprofloxacin resistance protein, plasmid encoded (154).
Is it correct to use the term plasmid-mediated quinolone resistance?
Classically, mobilizable genes leading to the development of quinolone resistance have been collectively named PMQR genes in order to highlight their nonchromosomal nature (15, 170). Nonetheless, plasmids may exchange material with other chromosomal or plasmidic elements, and therefore, the plasmid content may be fully or partially integrated within the bacterial chromosome (183, 184). The integration of mobilizable antibiotic resistance genes within the bacterial chromosome has been largely described. Thus, many transferable resistance genes, including those encoding β-lactamases, aminoglycoside-modifying enzymes, or trimethoprim resistance determinants, have been detected in the chromosomes of different microorganisms (141, 185–196). With regard to TMQR, this finding has also been described. Thus, the aac(6′)Ib-cr gene (197, 198) and the qepA1 gene (e.g., GenBank accession number NZ_CP019051) have been described in the chromosome of E. coli, while oqxAB, indigenous to Klebsiella spp. (160, 199–201), has been detected in the chromosome of Salmonella enterica serovar Derby (202). Similarly, different transferable qnr determinants, such as qnrA, qnrB, qnrS, and qnrVC, have been found to be inserted within the chromosomes of nonindigenous microorganisms, such as E. coli, Acinetobacter baumannii, and Pseudomonas putida (195, 203–206).
Therefore, the term PMQR may not be completely correct and might lead to a misunderstanding and erroneous interpretations of the genetic locations of the quinolone resistance determinants. In this sense, some authors have introduced alternative nomenclatures to refer to these genes, such as quinolone resistance determinants (QRDs), quinolone resistance genes (QRGs), transferable mechanisms of quinolone Resistance (TMQR), or transferable quinolone resistance determinants (TQRDs) (160, 196, 207, 208). The acronym TMQR is used throughout this review.
TMQR misidentification.
Unfortunately, misidentification of genes is a relevant scientific problem and involves all study fields. This problem can lead to erroneous result interpretation, and when stated in the literature or in gene sequence databases, a series of bona fide misanalyzed or subsequent erroneously interpreted data is generated. In this sense, GenBank is a powerful tool in which millions of DNA sequences are deposited. However, this database may have misidentified or erroneous sequences, which compromises the utility of the tool (209).
Regarding TMQR, the clearest example of misidentification is related to the exponential growth which qnr scientific literature has undergone in the last years. This phenomenon has favored the publication of partial qnr sequences to which allele numeration has been “assigned” or, more seriously, full qnr sequences which have been erroneously assigned to either an allele or a qnr family (209, 210). The possibility of posting sequences in GenBank without validation (see “Qnr Classification,” below), in order to be verified and correctly named and numbered, contributes to the perpetuation and amplification of nomenclature errors because they are included in the most relevant gene database worldwide (209, 210). Indeed, upon analyzing 1,657 Qnr sequences recorded in GenBank (209), it was observed that 340 (20.5%) sequences presented a major error. These errors included 105 Qnr sequences introduced in GenBank as “PipB2,” a type III effector protein which has been associated with the formation of vesicles (211); 145 sequences classified within an erroneous Qnr family; 16 sequences with an erroneous allele assignation; 24 partial sequences with an allele assignation; and 50 sequences with a nonnormative initial ATG codon assigned (209). Moreover, 449 (27.1%) of the sequences were only partially or not identified, and 9 unreported transferable alleles were detected, which were later allele numbered within GenBank records (209). Another finding is the nondetection of qnr genes when sequences are submitted to GenBank, leading to “hidden” qnr genes which can be detected only after a direct DNA BLAST search (210). This may lead to unnoticed mistakes in the discussion of results obtained in field studies.
Although this problem is less frequent because of the lower number of variants, it also affects the remaining TMQR. Thus, different qepA variants are recorded in GenBank as “qac” variants, while a series of GenBank-recorded qepA variants remain undescribed (see “QepA,” below) (179). Regarding OqxAB, it is of note that a series of allelic variants of OqxA and OqxB, which are either transferable or indigenous to K. pneumoniae, have received further numeration; for instance, “OqxB20” and “OqxB29” were first reported in the same article (212).
In an effort to minimize these problems, GenBank has developed “RefSeq” to name proteins as consistently and as correctly as possible (for instance, a search for qnrA alleles may be performed at https://www.ncbi.nlm.nih.gov/pathogens/isolates#/refgene/gene_family:qnrA; for other determinants, all that is needed is a change from “qnrA” to the desired gene). Nonetheless, it is the responsibility of all researchers to facilitate this effort, providing the most correct and normative nomenclature, submitting newly described sequences to the respective repositories, when available, in order to be numbered in a rational manner, and providing these data in GenBank submissions, manuscripts, and presentations.
The most serious problem with AAC(6′)Ib-cr is the assertion of new allelic variants in the development of quinolone resistance. Since a single amino acid change may alter the functionality of an antibiotic-modifying enzyme by amplifying, limiting, or modifying its spectrum of activity, it is important to determine the effect of any new allele on different quinolones prior to asserting its role as a quinolone resistance determinant.
Changing Resistance Paradigms
There are reports of exceptional microorganisms, such as Stenotrophomonas maltophilia, in which chromosomal target mutations have no relevance in the acquisition of nalidixic acid or fluoroquinolone resistance. In these microorganisms, quinolone resistance is mainly related to highly potent efflux pumps acting concomitantly with factors such as the presence of native chromosomal Qnr (213–216). Nonetheless, efflux pump overexpression by itself does not normally lead to full resistance to quinolones. Therefore, the presence of target mutations has even been reported to be the most relevant quinolone resistance mechanism in both Gram-positive and Gram-negative microorganisms (37). In the latter microorganisms, it has been established that the activity of old quinolones, such as nalidixic acid, is highly affected by these mutations, with the resistance breakpoint being surpassed in the presence of a single target mutation (37, 217–219). Thus, the bacterial phenotype of resistance to nalidixic acid and susceptibility (or diminished susceptibility) to fluoroquinolones has frequently been described and associated with the presence of at least one target mutation (37, 115, 118, 119, 217, 218), being considered a risk factor for the development of full resistance to fluoroquinolones (219, 220). These findings have led to the modification of several CLSI quinolone resistance breakpoints in 2016 (221, 222). In fact, in several reports, the use of nalidixic acid was suggested as a predictor of fluoroquinolone resistance (219, 223). Along this line, natural resistance to nalidixic acid and at least diminished susceptibility to fluoroquinolones related to the presence of a specific wild-type GyrA amino acid in position 83 and/or position 87 (E. coli numeration) have been described in microorganisms such as Bartonella spp. and Brevundimonas spp., among others (224–226).
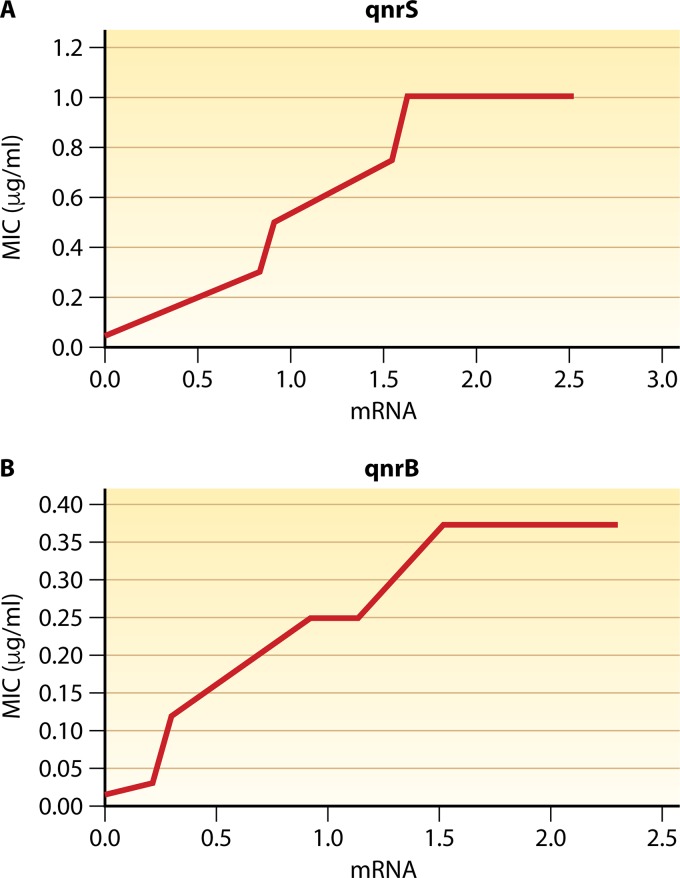
The eruption of TMQR has altered this scenario, increasing the isolation of quinolone-resistant microorganisms in the absence of target mutations and descriptions of microorganisms exhibiting the unusual phenotype of nalidixic acid susceptibility and ciprofloxacin resistance. Thus, the presence of more than one TMQR may increase the final MIC for resistance breakpoints even in the absence of quinolone target mutations (153, 227–231, 471). Furthermore, this finding has also been reported in isolates in which only one TMQR was identified or introduced (15, 228, 231–233). For instance, a recent swine isolate of Salmonella enterica serovar Rissen, without target mutations and in which the only TMQR detected was QnrVC4, showed MICs of nalidixic acid, norfloxacin, and ciprofloxacin of 32 μg/ml, 1 μg/ml, and 0.5 μg/ml, respectively (231). In this line, it has been shown that the levels of quinolone resistance produced by QnrB and QnrS are directly related to their levels of expression. Thus, it was observed that the final MICs of ciprofloxacin were increased when QnrB1, QnrS1, or derived mutants with impaired functionality were cloned in expression vectors under IPTG (isopropyl-β-d-thiogalactopyranoside) induction (234, 235). Furthermore, a study by Garoff et al. (236) in which qnrB and qnrS (no allele was specified) were cloned alone into E. coli (strain MG1655) and expressed under the control of different promoters showed that the MIC of ciprofloxacin increased according to gene expression levels until reaching plateaus of 0.375 and 1 μg/ml for qnrB and qnrS, respectively, representing increases of 25 and 66.6 times the original MIC for MG1655 (0.015 μg/ml) (Fig. 4). Although in this study, qnrB and qnrS were cloned as a single resistance determinant, it is important to again highlight that the final MICs are the result of multiple phenomena, therefore also being related to intrinsic bacterial factors such as cell wall permeability or intrinsic efflux pump activity. In microorganisms such as Acinetobacter spp. or P. aeruginosa, low membrane permeability and the extrusion of quinolones by several powerful efflux pumps strongly affect the intrinsic MICs of quinolones. Thus, intrinsic ciprofloxacin resistance ranges from 0.125 to 1 μg/ml (>8 times that of E. coli MG1655) to 0.25 to 4 μg/ml (>16 times that of E. coli MG1655) in the cases of A. baumannii and P. aeruginosa, respectively (37, 237–239). Thus, it could be predicted that similar qnrB or qnrS expression levels in these microorganisms lead to the detection of higher final MICs.
FIG 4.
Effect of expression levels of qnrS and qnrB on final MICs. In both panels, the mRNA levels are relative to those of the control genes hcaT, idnT, and cysG and are expressed in log10 units (based on data from reference 236). (A) qnrS; (B) qnrB.
Prior to the boom of TMQR, another atypical scenario was the presence of the unusual phenotype of nalidixic acid susceptibility and ciprofloxacin (fluoroquinolone) resistance or diminished susceptibility. Previously, this had been reported only in specific microorganisms such as the above-mentioned S. maltophilia, in which this phenotype is especially frequent, probably due to the unusual mechanisms involving the development of quinolone resistance (214, 216). In addition, this phenotype is also shown in a few E. coli clinical isolates, with decreased quinolone uptake associated with the highly unusual GyrA mutation D82G (240, 241) and in in vitro-constructed E. coli mutants carrying the GyrA substitution G81D (242). This has also been observed in Campylobacter jejuni and Neisseria gonorrhoeae, for which no analysis of the mechanisms of resistance has been reported (117, 243), or in C. jejuni presenting the GyrA substitution T86A, thereby suggesting the presence of an unidentified factor (244). Nonetheless, this phenotype may be mediated by the concomitant presence of TMQR that have an effect on specific quinolones, such as AAC(6′)Ib-cr or the QepA-like efflux pumps, affecting ciprofloxacin activity but being unable to affect that of nalidixic acid (160). Furthermore, the concomitant presence of any of these TMQR with others affecting all quinolone MICs even at different levels, such as Qnr determinants, also leads to the same scenario (133, 245). In this way, in 2012, two K. pneumoniae isolates without DNA gyrase or topoisomerase IV amino acid substitutions but carrying the aac(6′)-Ib-cr, qnrB4, and qnrS2 genes and showing resistance to ciprofloxacin but susceptibility to nalidixic acid were described (245). Similarly, the transfer of aac(6′)Ib-cr and qnrS1 from a clinical isolate of Salmonella enterica serovar Typhimurium to a competent E. coli isolate resulted in a transconjugant exhibiting an MIC of ciprofloxacin of 2 μg/ml (133).
Clinical Relevance of TMQR
As mentioned above, the levels of resistance conferred by TMQR are low and usually do not surpass the established quinolone resistance breakpoints. Therefore, while it might be considered that TMQR play an accessory and secondary role in the development of quinolone resistance, several factors should be taken into account (Table 3).
TABLE 3.
Essential data on TMQR
| Datum |
|---|
| Epidemiologya |
| Usually encoded and spread together with other antibiotic resistance determinantsb |
| Possible presence of more than one TMQR in the same genetic structure or microorganisms |
| Horizontal dissemination of transposons, genomic islands, phages |
| Vertical dissemination (indigenous presence, integration within chromosome) |
| Effect on quinolone MIC |
| Additive effect → possibility of full resistance to FQ |
| Multiple copies, ↑ expression → ↑MIC |
| In some cases, a maximum MIC can be reached, irrespective of TMQR expression levels |
| Affects all quinolones (Qnr) |
| Affects specific quinolones [AAC(6′)Ib-cr, CrpP, QepA, OqxAB, other efflux pumps] |
| Single amino acid substitutions, ↑ or ↓ final MICs or modify substrate profile |
| Facilitation of acquisition of further mutations leading to increasing levels of quinolone resistance |
| No effect on increasing no. of quinolone target mutations |
| Possible effect on MICs of unrelated agents (e.g., novobiocin, tigecycline, or colistin) |
The effect on clinical outcome is a negative impact (pending confirmation).
This finding results in coselection of antibiotic resistance.
Thus, the effect of TMQR on final MICs is always additive to that conferred by other transferable or chromosome-encoded mechanisms of quinolone resistance (160); expression levels may modulate the effect of TMQR on the final MIC (236), and no factor limits the number of identical or different TMQR that may be present in a single microorganism, encoded or not within the same genetic structure (138, 197, 229, 245–254) (Tables 3 and 4); for instance, 3 copies of the qnrB6 gene, 2 copies of the aac(6′)Ib-cr gene, and 1 qnrB pseudogene are carried on the E. coli plasmid pAMSH1 (GenBank accession number CP030940) (Table 4). In the same sense, Vinué et al. (229) described an E. coli isolate carrying the aac(6′)Ib-cr, qnrA1, qepA1, and oqxAB genes. Similarly, a recent study showed the presence of up to 5 qnrA1 copies within a plasmid selected under ciprofloxacin pressure (251).
TABLE 4.
Epidemiology of coresistance involving TMQRa
| GSc | Inc type | Size (kb) | Antibiotic resistance determinantsb
|
Reference(s) or GenBank accession no. | |
|---|---|---|---|---|---|
| TMQR | Other [AMG/β-lactam (βL/ESBL/CBP/pAmpC)/CHL/CST/FOF/MC/RIF/SUL/TET/TMP/other] | ||||
| pHS1387 | 48 | aac(6′)Ib-crd | aadA16/(—/—/—/—)/—/—/—/—/arr3/sul1Δ/—/dfrA27/— | 464, 465 | |
| pC1865 | 220 | aac(6′)Ib-cr | aadA2/(blaTEM-1, blaOXA1e /blaCTX-M-15/—/—)/—/—/—/—/—/sul1/—/dfrA12/— | 197 | |
| pA3T | HI2 | 253 | aac(6′)Ib-cr, oqxAB | aph(4)-Ia/(blaOXA-1/blaCTX-M-14/—/—)/catB3, floR/—/fosA3/—/arr3/sul1, sul2/—/dfrA12/— | 254 |
| pD90-1 | 222.5 | aac(6′)Ib-cr, oqxAB | aac(3)-IV, aadA5, armA, strA, strB/(blaTEM-1b, blaOXA-1/blaCTX-M-6/—/—)/catB3, floR/—/fosA3/—/arr3/sul1, sul2/—/dfrA17/— | 252 | |
| pHSH2 | 68 | aac(6′)Ib-cr, qnrA1f | —/(blaOXA-1/—/—/—)/catB3/—/—/—/arr3/sul1/—/—/—/— | 253 | |
| pHE96 | 70 | aac(6′)Ib-cr, qnrA3g | aadA16Δ/(blaOXA-1/—/—/—)/catB3e/—/—/—/arr3/sul1/—/dfrA27Δ/— | 246 | |
| pNDM-MAR | H | 267 | aac(6′)Ib-cr, qnrB1 | —/(blaOXA-1/blaCTX-M-15/blaNDM-1/—)/cat, catB3/—/—/—/—/—/—/—/— | 250 |
| ND | aac(6′)Ib-cr, qnrB4h | —/(blaOXA-1/—/—/blaDHA-1)/catB3/—/—/—/arr3/sul1/—/—/— | 245 | ||
| pC2367 | R | 195 | aac(6′)Ib-cr, qnrB6i | aadA16/(—/—/—/—)/—/—/—/—/arr3/sul1/—/dfrA27/— | 197 |
| pAMSH1j | 257.4 | aac(6′)Ib-cr, qnrB6, qnrBΔ | aac(3)IId, aph(3′)Ib, aph(6)Id/(blaTEM-1/blaCTX-M-55/—/—)/floR/—/—/mph(A)/arr3/sul1, sul2/tet(A)/dfrA27/— | CP030940 | |
| ND | aac(6′)Ib-cr, qnrB10k | —/(blaOXA-1/—/—/—)/catB3/—/—/—/arr3/sul1/—/—/— | 248 | ||
| p19051-IMP | HI5 | 316.8 | aac(6′)Ib-cr,l qnrB52, qnrS1 | aacC2, aadA2, armA, strA, strB/(blaTEM-1B/blaCTX-M-3, blaSHV-12/blaIMP-4/—)/—/—/fosA3/mph(E), msr(E)/arr3/sul1/—/dfrA12/tmrBm | 247 |
| pUM505 | I | 123 | crpP | —/(—/—/—/—)/—/—/—/—/—/—/—/—/— | 154 |
| pHNSHP45-2 | H12 | 251.5 | oqxAB | aac(3)-IV, aadA1, aadA2, aph(3′)-Ia/(—/blaCTX-M-14/—/—)/cmlA, floR/mcr-1/fosA3/—/—/sul1, sul2, sul3/—/dfrA12/— | 278 |
| pOLA52 | X1 | 52 | oqxAB | —/(blaTEM-1/—/—/—)/—/—/—/—/—/—/—/—/— | 200 |
| pHPA | FII | 70 | qepA1 | rmtB/(blaTEM-1/blaCTX-M-12/—/—)/—/—/—/mph(A)/—/—/—/—/— | 158 |
| pIP1206 | FI | 168 | qepA1 | aadA4, rmtB/(blaTEM-1/—/—/—)/catA1/—/—/—/—/—/tet(A)/dfrA17/— | 286 |
| pMG252n | 185.6 | qnrA1 | aadA2, aadB, aph(3)II/(blaCARB-2/—/—/blaFOX-5)/catB3, catB11, cmlA1, mdtL/—/—/mph(E), msr(E)/—/sul1/—/—/dfrA19 | 15, 251, 284, 317 | |
| SGI-1Vo | 42.9 | qnrA1 | aac(6′)Ib, aadB/(—/blaVEB-6/—/—)/—/—/—/—/—/sul1/tet(A)Δ/dfrA1/— | 277 | |
| pJIBE401p | L/M | >150 | qnrB2 | aac(6′)Ib4/(—/—/blaIMP-4, blaOXA-73/—)/catB3c/—/—/mph(A)/—/sul1/—/—/— | 279 |
| pENVA | H | 253 | qnrB4 | aacC2, aadA/(blaTEM-1/blaCTX-M-15/—/blaDHA-1)/—/—/—/—/—/sul1/tet(A)/dfrA15/— | 276 |
| pECY6-7 | ColE | 2.7 | qnrB19 | —/(—/—/—/—)/—/—/—/—/—/—/—/—/— | 466 |
| pLRM24 | 80 | qnrB19 | aac(6′)Ib, aadA1/(blaTEM-1/—/blaKPC-3/—)/—/—/—/—/—/—/—/—/— | 283 | |
| Chromq | qnrB62 | aac(6′)II/(—/—/blaVIM-2/—)/—/—/—/—/—/sul1/—/— | 275 | ||
| pHS10 | 120 | qnrC | —/(—/—/—/—)/—/—/—/—/—/—/—/—/— | 340 | |
| p2007057 | 4.27 | qnrD1 | —/(—/—/—/—)/—/—/—/—/—/—/—/—/— | 344 | |
| pKP41M | M1 | 70 | qnrE1 | aac(6′)-Ib, aadA1/(blaTEM-1, blaOXA-9/blaCTX-M-8/—/—)/—/—/—/—/—/—/—/—/— | 273 |
| pGN26-KPC | X6 | 46.3 | qnrS1 | —/(blaTEM-1Δ/—/blaKPC-2/—)/—/—/—/—/—/—/—/—/— | 280 |
| pK245 | 98 | qnrS1 | aacC2, strA, strB/(blaLAP-2/blaSHV-2/—/—)/cat2/—/—/—/—/—/tet(D)/dfrA14/— | 272 | |
| pUR19829-KPC21 | Q2 | 12.7 | qnrS2 | —/(—/—/blaKPC-21/—)/—/—/—/—/—/—/—/—/— | 281 |
| pKAZ3 | A/C | 148 | qnrVC1 | —/(—/blaVEB-9/—/—)/—/—/—/—/—/—/tet(A)′, tet(C)/dfrA1, dfrA23/— | 274 |
| pKAZ4r | A/C | 167 | qnrVC1 | aadA1e (blaOXA-10/blaSHV-12/—/—)/—/—/—/—/arr3/sul1/—/—/— | 274 |
| pVAS3-1 | A/C | 175 | qnrVC4 | aac(6′)I, aadA1, aadA2, aph(3′)I, strA, strB/(blaOXA-10/—/—/blaCMY-2)/cmlA1, cmlA5, floR/—/—/mph(A)/—/—/tet(A)/dfrA14/— | 285 |
TMQR are shown in alphabetical order and thereafter by allelic variant. Prior or posterior presence in the table does not preclude a prior or posterior identification or a specific position within the reported genetic structure. For the exact gene position within the structure, see the references provided. In all cases, the final effect of the genes reported should be considered within the bacterial genetic background. Furthermore, the presence of additional mutations in the chromosome or other genes involved in antibiotic resistance cannot be ruled out. When the TMQR are carried within an integron environment, information about the full integron gene content is reported. GS, genetic structure; Inc type, plasmid incompatibility group; TMQR, transferable mechanisms of quinolone resistance; AMG, aminoglycosides; βL, β-lactam (not including third- or fourth-generation cephalosporins, carbapenems, or monobactams); ESBL, extended-spectrum β-lactamases; CBP, carbapenems; pAmpC, plasmid AmpC; CHL, chloramphenicol; MB, monobactams; MC, macrolides; RIF, rifampin and derivatives; SUL, sulfonamides; TET, tetracyclines; TMP, trimethoprim; Δ, pseudogene (presence of an internal stop codon or deletion leading to a frameshift) with an impaired ability to confer antibiotic resistance (in these cases, the gene is reported for epidemiological purposes); ND, not determined; 3′-CS, 3′ conserved segment.
Only antibiotic resistance genes in the reported structure and therefore spreading concomitantly. Note that the presence of a specific antibiotic resistance-encoding gene is not synonymous with phenotypic resistance; the effect (e.g., TMQR) may be only a modest increase in MICs, not reaching the established resistance breakpoint, or gene expression may be downregulated or impaired. Furthermore, in several cases, such as specific TMQR, aminoglycoside-modifying enzymes, or transferable mechanisms of macrolide resistance, the resistance conferred by the reported mechanisms may not be extended to all antibiotic class family members. The number of described genetic structures carrying TMQR alone or in association with other antibiotic resistance genes is enormous and growing on an almost daily basis. Only a series of representative genetic structures are described here. When a TMQR is reported to be present within a plasmid but no exact plasmid name is provided, the name of the bacterial strain is used as the plasmid name, and the genetic structure name is highlighted in boldface type.
In several cases, the whole sequence or full content data of the reported genetic structure are not available, with the subsequent possible presence of unnoticed antibiotic resistance genes; for instance, although not reported when fully sequenced (GenBank accession number MK638972), in the plasmid pMG252, which started the TMQR era, 2 other β-lactamases with isoelectric points of 7.0 and 7.6 were detected, in addition to β-lactamases with isoelectric points of 5.6 and 7.2, and further identified as blaCARB-2 and blaFOX-5 and reported in the table (15).
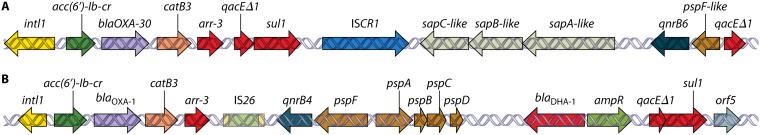
aac(6′)Ib-cr is harbored within an ∼4.8-kb integron {In1021 [aac(6′)Ib-cr-arr3-dfrA27-aadA16-IS15-sul1Δ-IS1]}.
Also reported as blaOXA-30 or blaOXA-1/30.
Both TMQR are harbored within an ∼15-kb complex class 1 integron {In37 [aac(6′)Ib-cr-blaOXA-1-catB3-arr3 qacEΔ1-sul1-ISCR1-qnrA1-ampR-qacEΔ1-sul1]}.
The qnrA3 gene is located after a structure (qacEΔ1-sul1-ISCR1-qnrA3) suggesting the presence of an undescribed complex class 1 integron, while aac(6′)Ib-cr is placed upstream of qnrA3 just after the location of IS26 and tnpA within In640 [aac(6′)Ib-cr-blaOXA-1-arr3-dfrA27Δ-aadA16Δ-qacEΔ1].
Integron identical to In37 but carrying a qnrB4 allele instead of qnrA1 and an additional blaDHA-1 gene just upstream from the qnrB gene.
Integron similar to In1021 but with a different 3′-CS region leading to a complex class 1 integron. A qnrB6 gene is located in the additional variable region [aac(6′)Ib-cr-arr3-dfrA27-aadA16-qacEΔ1-sul1-ISCR1-qnrB6-qacEΔ1 sul1].
The aac(6′)Ib-cr and qnrB6 genes are harbored within a complex integron {In1229-like [aac(6′)Ib-cr-arr3-dfrA27-qacEΔ1-sul1-ISCR1-ORF (putative oxidoreductase)-qnrB6-pspFΔ-qacEΔ1-sul1]}. This integron is present twice in the plasmid. An additional qnrB6 gene within a structure resembling that of the end of the complex integron is also present.
Integron identical to In37 but carrying a qnrB10 allele instead of qnrA1.
The aac(6′)Ib-cr gene is harbored within a class 1 integron {In1231 [aac(6′)Ib-cr-arr3]}.
TmrB confers resistance to tunicamycin, an antibiotic produced by members of the genus Streptomyces (e.g., Streptomyces lysosuperificus) which targets the cell wall synthesis of Gram-positive bacteria. Toxicity for eukaryotic cells has precluded its clinical use (467).
qnrA1 is harbored within a complex class 1 integron (In580-like [blaCARB-2-aadA2-cmlA1c-catB11-qacEΔ1-sul1-ISCR1-qnrA1-qacEΔ1-sul1-ISCR1-dfrA19]). Of note, in the original article, pMG252 was reported to have a size of ∼56 kb (15).
Genomic island.
The TMQR is present within a complex class 1 integron environment {In585-like [blaIMP-4-qacG2-aac(6′)Ib4-catB3-qacEΔ1-ISCR1-sapA-orf2-qnrB2-pspF-qacEΔ1-sul1]}.
The TMQR is present within a complex class 1 integron environment {In1184 [blaVIM-2-aac(6′)II-gucD-qacEΔ1-sul1-ISCR1-pspF-qnrB62-qacEΔ1-sul1]}.
The qnrVC1 gene is harbored in a class 1 integron (In1214 [arr3-qnrVC1-blaOXA-10-aadA1e-qacEΔ1-sul1]).
Furthermore, the presence of decreased susceptibility to quinolones has also been shown to be a risk factor for the development of full quinolone resistance (219). In this sense, early studies showed a scenario in which the presence of qnr genes increased by >100 times the selection of quinolone-resistant mutants in the absence of a general mutator effect (15). Nonetheless, a study published in 2019 showed that qnrB has a general mutator effect in both the presence and the absence of ciprofloxacin and also showed that QnrB interacts with DnaA, subsequently regulating the formation of the DnaA-oriC complex and leading to the upregulation of genes near oriC. This results in DNA replication stress, which in turn favors both an increase in the number of plasmids, independently of Qnr-topoisomerase interactions, and higher mutation rates (255). In the last years, the effect of TMQR on the further selection of quinolone resistance mechanisms and bacterial survival in the presence of lethal concentrations of quinolones has been analyzed (256–258). Indeed, Cesaro et al. showed that the selection of quinolone-resistant microorganisms does not differ in the presence or absence of qnrA1, qnrA3, qnrB1, or qnrS1 determinants and that the presence of these qnr determinants increases quinolone mutant prevention concentrations (MPCs). They also highlighted an unexpected finding in which the presence of qnr determinants results in a significantly (P < 0.0001) lower number of quinolone target mutations (256). Thus, in the genetic environment of E. coli J53 and KL16, while 65/329 (20%) of the mutants selected in the presence of qnr carried at least 1 gyrA mutation, the number of gyrA mutations increased up to 94/119 (79%) among the mutants selected from qnr-free E. coli (256). Similar studies with qnrA1 by Goto et al. showed increases in the MPC and a lack of association with the selection of target mutations (257). Furthermore, upon extending this scenario to other TMQR, such as qepA and aac(6′)Ib-cr (258), it was suggested that the presence of TMQR favors the selection of a series of chromosomal mutations outside the classical quinolone targets that are able to act in an additive or cooperative manner, each providing lower levels of resistance but concomitantly achieving high levels of quinolone resistance (258). In this sense, Vinué et al. (258) showed that under quinolone pressure, the presence of qnrA favors the overexpression of different efflux pumps such as AcrAB-TolC, AcrEF-TolC, MdtEF, and MdtK (also known as YdhE), which, in most cases, correlates with the presence of mutations in different regulator genes like marR, soxR, and evgA. Furthermore, most of these mutants also presented decreased expression of the porins OmpC and OmpF as well as a series of mutations in apparently unrelated genes whose possible role in the development of low levels of quinolone resistance remains to be elucidated (258). In addition, a series of mutations in genes involved in the inner part of the core oligosaccharide of lipopolysaccharide (LPS) biosynthesis, which were correlated with a parallel increase in novobiocin susceptibility, were also detected in several of these mutants (258). Deficient LPS expression has been involved in increased susceptibility to the most hydrophobic quinolones, such as nalidixic acid and flumequine, without affecting those that are more hydrophilic, such as ciprofloxacin and norfloxacin (241, 259). Furthermore, while unexplored in these mutants, these alterations in LPS may also play a role in the final susceptibility levels to especially relevant and last-resort antimicrobial agents, either increasing or decreasing the final MICs. For instance, a slight increase in colistin susceptibility has been observed in E. coli carrying mutations affecting core LPS biosynthesis, while an inverse effect has been observed with tigecycline (260, 261).
Finally, while the relationships between the presence of TMQR and longer hospitalization as well as other clinical parameters, including final patient outcomes, remain to be fully elucidated, different authors have suggested that the presence of the qnr genes is associated with a trend toward long hospital stays and increased 30-day mortality (262, 263). In support of this, the presence of TMQR has also been associated with the poorest clinical response and fatal outcomes in murine models of pneumonia and urinary tract infection (264, 265).
Therefore, the presence of TMQR is a true risk factor for the development of quinolone resistance/survival in the presence of high quinolone concentrations even in the absence of target mutations. Furthermore, their presence may have a direct impact on the susceptibility levels of unrelated agents by alterations in the expression levels of specific genes and might have a direct impact on patient management.
The above-mentioned scenarios suggest the appropriateness of including these genes in routine clinical laboratory procedures, even in the absence of reliable data on their impact on patient management. This goal is of special relevance for the most sensitive hospital settings, such as intensive care units, in which commensal microorganisms also play a key role as antibiotic resistance gene reservoirs (266). Nonetheless, while different high-throughput approaches, such as DNA microarray technology (267), whole-genome sequencing (268), or matrix-assisted laser desorption ionization–time of flight (MALDI TOF) mass spectrometry (269), that are able to be automated and scalable to large numbers of clinical samples might be implemented (or adapted) for routine detection of TMQR, most of the currently used methodologies, such as different in-house multiplex PCR approaches (270, 271), have been conceived for research purposes or to be used in settings with a low or moderate number of samples for processing.
Molecular Epidemiology of TMQR
TMQR have been largely related to different mobile elements, including plasmids, transposons, and genomic islands, as well as to the presence of other specific antibiotic resistance determinants that are cocarried within the same genetic structures (Table 4). Thus, this concomitant carriage within the same genetic structure of extended-spectrum β-lactamase (ESBL) genes such as blaCTX-M-2, blaCTX-M-6, blaCTX-M-8, blaCTX-M-14, blaCTX-M-15, blaSHV-2, blaSHV-12, blaVEB-6, and blaVEB-9 (158, 178, 247, 252, 254, 272–278); carbapenemase genes like blaKPC-2, blaKPC-3, blaKPC-21, blaNDM-1, blaOXA-72, and blaVIM-2 (247, 279–283); plasmid AmpC (pAmpC) genes such as blaCMY-2, blaDHA-1, and blaFOX-5 (245, 276, 284, 285); 16S rRNA methylase genes such as rmtB (158, 286); or other mechanisms of resistance to unrelated agents is a common fact (Table 4). This fact might underlie the dissemination of difficult-to-treat pathogenic microorganisms (262, 287). It has also been considered an underlying reason for the role of previous fluoroquinolone exposure as an independent risk factor for the development of infections by microorganisms possessing unrelated specific antibiotic determinants such as blaKPC (288). In this sense, different studies have observed a link between the presence of blaCTX-M-15, which ranks among the most widely distributed and described ESBLs, and that of aac(6′)Ib-cr (185, 197, 289, 290). For instance, Pitout et al. found that 34 out of 63 E. coli strains with the aac(6′)Ib-cr gene also carried the blaCTX-M-15 gene (289). Accordingly, the neighboring presence of both genes within the same plasmid has been reported (197, 250). It should be mentioned that other studies, such as those of Brahmi et al. and Moremi et al., did not find this association (291, 292), probably suggesting the presence of local differences in the spread of microorganisms/plasmids related to factors such as specific antimicrobial pressure and geographical factors.
TMQR are frequently encoded within plasmids belonging to different incompatibility groups and with heterogeneous sizes (Table 4), although surrounding structures are often similar, suggesting the presence of a limited number of mobilizations of ancestral chromosomal origin followed by a series of transpositions, recombinations, deletions, insertions, and every other genetic material arrangement leading to the currently observed variety. In this sense, the boom of next-generation sequencing approaches has increased the availability of data related to the presence and distribution of TMQR. Further molecular epidemiological data are presented in the specific sections devoted to each TMQR.
Qnr
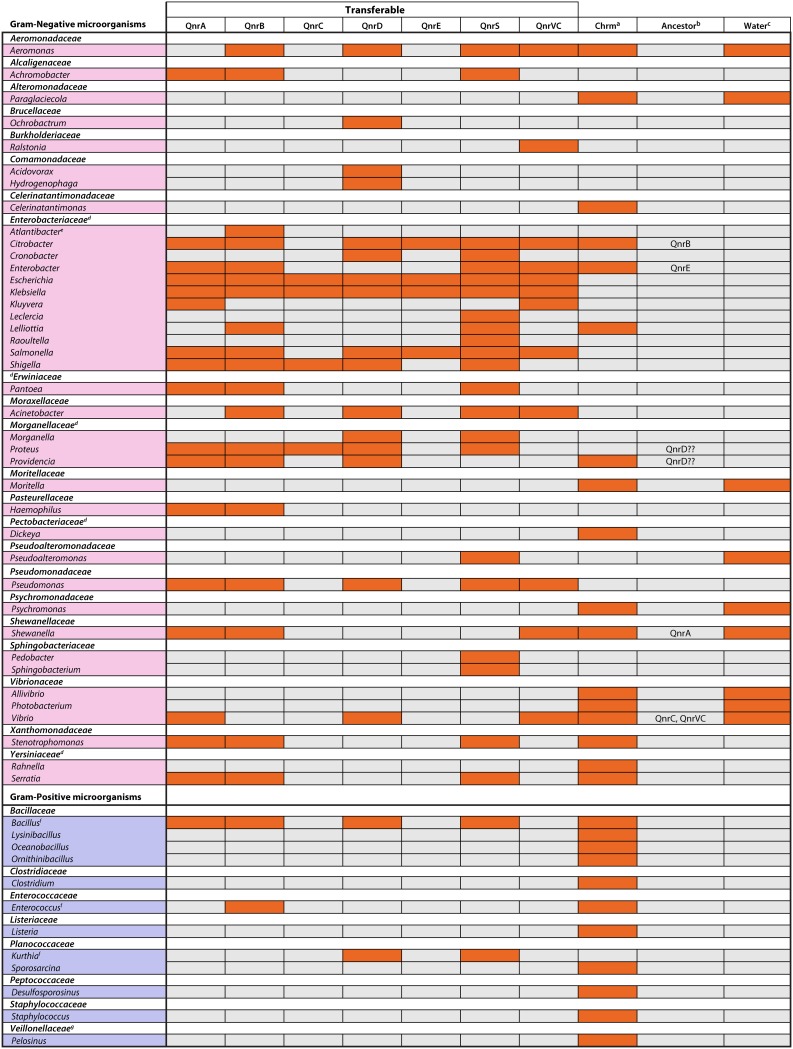
At the beginning of 1998, Martínez-Martínez and colleagues (15) described the presence of a TMQR within the plasmid pMG252 in a K. pneumoniae strain isolated in Alabama in 1994, which was named “qnr” (see “Nomenclature of the Transferable Mechanisms of Quinolone Resistance,” above). The transfer of this plasmid led to increases in the quinolone MICs of 8- to 64-fold irrespective of the initial MICs. The effect was dissimilar among the different quinolones tested, being maximum with nalidixic acid and minimum with clinafloxacin (15). Thus, when the plasmid was transferred to E. coli J53, the nalidixic acid susceptibility changed from susceptible (4 μg/ml) to resistant (32 μg/ml). Moreover, the authors showed that microorganisms carrying the qnr determinant were more prone to developing full resistance to fluoroquinolones (15). Subsequently, different studies were designed to determine the true prevalence and relevance of this mechanism of resistance, demonstrating its rare and low prevalence. Thus, this mechanism was found in only 6 (1 E. coli, 4 K. pneumoniae, and 1 Klebsiella species isolates) out of 420 Gram-negative microorganisms tested (338 clinical isolates from 19 countries and a series of laboratory strains carrying different plasmids), with all 6 having been collected in 1994 in Alabama (13). In the subsequent years, this qnr determinant as well as new qnr variants (Table 5) were increasingly detected worldwide in different microorganisms, mainly Enterobacteriaceae (138, 293–297) and a few isolates of Aeromonadaceae (297–300), Moraxellaceae (301) Pseudomonadaceae (206, 302), and Vibrionaceae (175, 303, 304), among others (Fig. 5).
TABLE 5.
Overall view of TMQRa
| Mechanism | TMQRc | No. of allelesb
|
Size (bp) | Yrf | Presence of integronsg | Ancestorsh | |
|---|---|---|---|---|---|---|---|
| Litd | RefSeqe | ||||||
| Target protection | QnrA | 8 | 8i | 218 | 1998 | Y | Shewanella spp. |
| QnrB | 88j | 81 | 214 | 2006 | Y | Citrobacter spp. | |
| QnrC | 1 | 1 | 221 | 2009 | Vibrio spp. | ||
| QnrD | 3 | 3 | 214 | 2009 | Morganellaceae? | ||
| QnrE | 1 | 2 | 214 | 2017 | Enterobacter spp. | ||
| QnrS | 9 | 14k | 218 | 2005 | Vibrio spp. | ||
| QnrVCl | 9 | 7 | 218 | 2008 | Y | Vibrio spp. | |
| Efflux system | QepA | 10 | 10 | 511m | 2007 | Comamonadaceae | |
| OqxAB | 14/28 | 5/7 | 391/1,050n | 2003 | Klebsiella spp.o | ||
| QacA | 1 | 2p | |||||
| QacB | 1q | 2p | 2010 | ||||
| pRSB101 | 2004 | ||||||
| Antibiotic modification | AAC(6′)Ib-cr | >5 | 7 | 2006 | Y | ||
| CrpP | >37r | 1 | 2018 | Pseudomonadaceae? | |||
| Slow growth | pKM101 | ||||||
TMQR, transferable mechanisms of quinolone resistance; Lit, data present in the literature; RefSeq, data present as reference sequence data in GenBank.
Only alleles confirmed or proposed to be involved in the development of quinolone resistance.
Only confirmed TMQR. When a TMQR was related to the presence of a specific plasmid but no gene-specific nomenclature is available, the name of the plasmid is indicated.
Based on GenBank and bibliographic searches. Regarding Qnr, only those included in the Lahey database (formerly at http://www.lahey.org/qnrStudies/) as of 31 December 2018 are shown. This database is no longer available. Other unnamed or erroneously assigned alleles may be found in GenBank (209).
Based on a RefSeq search (https://www.ncbi.nlm.nih.gov/pathogens/isolates#/refgene/gene_family:XXX, where XXX is the name of the gene) (updated on 12 April 2019).
Publication of the first allele of the family that has been considered a TMQR, irrespective of the time at which the ability to confer quinolone resistance was demonstrated. Note that previous conference presentations may have been made and that these presentations may be included in reviews reported prior to the reported data.
Y indicates that all or several alleles were detected within the integron environment.
Established or proposed original chromosomal sources.
While considered at the Lahey website, QnrA8 is not included in RefSeq. Of note, the only description of this gene was in the S. algae chromosome; therefore, transferability has not been demonstrated (331).
QnrB89 has not been included (a high number of amino acid differences with established QnrB alleles has been reported [formerly at http://www.lahey.org/qnrStudies/], and no data on the exact sequence are provided at either the Lahey website or GenBank).
QnrS3 has not been included in RefSeq. Of note, the reported sequence lacks the initial amino acid (356).
While QnrVC8 and QnrVC9 were considered at the Lahey website, they are not included in RefSeq. It is of note that in the only description of these genes, they were located within Vibrio species chromosomes; therefore, transferability has not been demonstrated (363).
The proposed QepA8 protein has a 2-amino-acid insertion leading to a final size of 513 amino acids.
Standard size of OqxA and OqxB, respectively. Note that transferable OqxA and OqxB presenting amino acid insertions have been detected (see Tables 11 and 12).
Klebsiella pneumoniae and Klebsiella aerogenes (see “OqxAB”).
No data about the ability of one of these alleles to extrude quinolones have been reported.
Only the QacBIII allele has been associated with the ability to extrude quinolones.
The presence of more than 37 closely related alleles with identity levels of >90% has been highlighted (414). Nonetheless, the effect on ciprofloxacin has been established for only one allele, with the others remaining to be studied.
FIG 5.
Main genera in which the presence of qnr genes has been described. This is a nonexhaustive list; the possible presence of nonreported qnr genes in genera presented in the figure or the presence of qnr genes in genera not presented in the figure should be taken into account. a, genera in which indigenous qnr-like genes have been described (note that when a qnr gene is present in the chromosome of a nonindigenous microorganism, it is reported as a TMQR); b, proposed original source of specific transferable qnr; c, microorganisms related to water environments; d, classically classified together within the family Enterobacteriaceae and classified here in separate genera according to the proposal of Adeolu et al. (347); e, new genus proposed in 2016, including Escherichia hermannii and Salmonella subterranea (456) (at the time of writing of this review, the genus “Atlantibacter” was not present in the List of Prokaryotic Names with Standing in Nomenclature [http://www.bacterio.net/index.html]); f, in none of these Gram-positive organisms are confirmatory sequences for qnrA, qnrB, qnrD, and qnrS available; g, Gram stain variable and phylogenetically closely related to Clostridium spp.
Thereafter, some studies were designed using older bacterial collections in order to detect the presence of these genes in isolates recovered prior to 1994. Thus, the older transferable qnr gene detected belonged to the qnrB family, being identified in one K. pneumoniae strain isolated in 1988 in Cordoba, Argentina (305). In the same study, another qnrB gene carried by a Citrobacter freundii isolate from Brooklyn, NY, was detected, which had also been isolated in 1988 (305). Nonetheless, in the absence of specific analysis to determine its genetic environment, and since subsequent studies have established the possible origin of qnrB genes in the chromosome of Citrobacter spp. (306), it is uncertain whether this gene was present within a transferable structure or was an intrinsic resident gene (see “QnrB,” below). In fact, the presence of qnrB60 and a qnrB pseudogene has been described in the C. freundii collection strains ATCC 6879 and ATCC 8090, respectively (307), both of which were isolated in the late 1920s or early 1930s (308, 309).
It was observed that Qnr also confers slight protection against 2-pyridones, quinazoline-2,4-diones (both of which are structurally closely related to quinolones), and spiropyrimidinetriones but has no protective effect against other topoisomerase type II-targeting molecules such as aminocoumarins (coumermycin A1, novobiocin, and simocyclinone D8), gyramide A, microcin B17, pyrazolopyridones, or tricyclic pyrimidoindoles (310).
Although the presence of Gram-positive chromosomally encoded Qnr has been described (311), all the currently known transferable Qnr families derive from Gram-negative ancestors. Therefore, the presence of qnr-related genes has been described in the chromosomes of different Gram-negative microorganisms, mostly related to water environments, such as members of the Shewanellaceae and Vibrionaceae families as well as in other microorganisms belonging to other bacterial families, like the Enterobacteriaceae and Xanthomonadaceae, among others (152, 172, 174, 312, 313) (Fig. 5). In light of these data, different ancestors for current transferable qnr genes have been proposed (see from “QnrA” to “QnrVC,” below).
In addition to plasmids and transposons, the presence of unexpected qnr transmission pathways has been described, including the presence of qnr genes within genomic islands (277), which may be conjugatively transferred under specific circumstances in the presence of helper plasmids, similar to what occurs with the so-called Salmonella genomic island 1 (SGI1) (314). Thus, in 2011, a Proteus mirabilis isolate carrying a new variant of SGI1 (SGI1-V) was described, in which, together with the aacA4 [encoding an AAC(6′)I enzyme, but no exact allelic variant is indicated], aadB, dfrA1, and blaVEB-6 genes, a qnrA1 gene was present (277). The potential role of bacteriophages in qnr dissemination has also been reported (315, 316). Thus, by analyzing the role of phages in the dissemination of antibiotic resistance genes, the presence of qnrA was detected in 18 bacteriophage samples recovered from human wastewater as well as in 18 water samples from a strongly anthropogenically impacted river but in only 19 out of 28 samples from animal wastewater (315). The same study included qnrS, which was detected in only 7, 4, and 1 of the above-mentioned human wastewater, river water, and animal wastewater bacteriophage samples, respectively, but with even higher densities than qnrA (315).
Qnr Classification
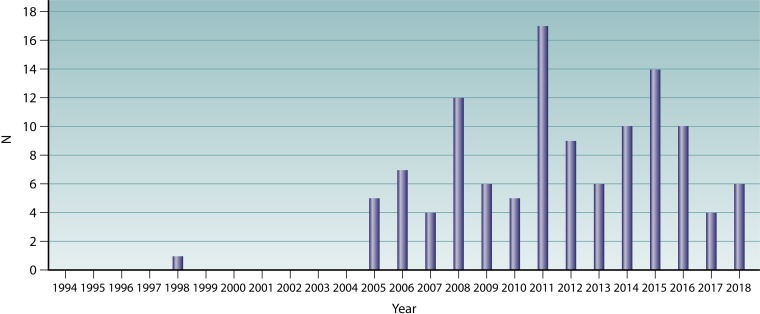
In the first years after the description of qnrA1 in 1998, literature on qnr genes was scarce, being mostly addressed to describe the presence and prevalence of qnrA1 in different geographical areas (13, 253, 317) or to advance the knowledge of the mode of action of qnr (318). Nonetheless, in the mid-2000s (Fig. 6), the presence of new qnr genes/alleles in GenBank and published reports on qnr suddenly increased. This led to the presence of different qnr genes with the same name and the subsequent increasing chaos (319). Therefore, in 2008, a series of rules was implemented to unify the criteria to define a new qnr allele or gene (319) (Table 6). Thus, the Qnr proteins were classified within families (genes) and subdivided into alleles based on single or multiple amino acid differences among them (319).
FIG 6.
Description of new qnr alleles (1998 to 2017). Only transferable genes/alleles with a standard name in the nomenclature as of 31 December 2018 according to the Lahey website (formerly at http://www.lahey.org/qnrStudies/) are shown. The reporting year has been considered following the next-priority order. (A) Date of oldest publication by the describing authors. Note that in several cases, the paper by the original authors might have been published several years after the original inclusion in GenBank, and therefore, data regarding these alleles may be present in other precedent articles. For example, QnrB6 was included in GenBank in 2006, being considered when the QnrB nomenclature was normalized and reorganized (319), but the oldest article found by the describing authors was published in 2009 (457). (B) In the absence of publication data by the describing authors, the oldest article published by any author was considered. (C) Presence of the allele in meeting presentations by (i) describing authors and (ii) other authors. (D) When neither the published article nor meeting presentation was found, reporting data have been annotated by the year of the GenBank record. Note that the presence of the allele in GenBank may precede the time of article publication. In addition, several sequences might be added to the repository at a later time. Note that the absence of an identified citing article/communication does not preclude the absence of related publications or communications. (E) In the absence of all above-described data, personal communication was considered if recorded at the Lahey website (e.g., from qnrB84 to qnrB87). qnrE1 was proposed as a new gene in 2017 (176), although qnrB88 was submitted to GenBank in 2016 and also first reported (a meeting presentation) in 2016 (352).
TABLE 6.
Characteristics defining a new Qnr allele/gene
| New Qnr element | Characteristic(s) |
|||
|---|---|---|---|---|
| Requisiteb | Discardc | Optionald | Not considered | |
| Allelea | Natural source | Synthetic origin | Increase in MICe | Promoter alterations |
| Full-length sequence | Partial sequence | Silent mutations | ||
| Amino acid change | ||||
| Identity of >70% (same ancestor)f | ||||
| Geneg | Identity of ≤70% (same ancestor)f | No effect on MIC | Promoter alterations | |
| Increase in MICe | Silent mutations | |||
Within a defined qnr gene. Allele numeration will be assigned after submission to the qnr repository (https://www.ncbi.nlm.nih.gov/pathogens/submit-beta-lactamase). Similarly, prior to defining a new qnr gene, it needs to be submitted to the qnr repository. Note that the website manager may not perform continuous surveillance of the thousands of new articles published monthly or the innumerable new sequences added to GenBank. Researchers need to act responsibly and ask for an allele assignation prior to any publication and should not pirate or appropriate an allele number in an irresponsible manner, which only contributes to increasing confusion and a lack of trust in publishing results. Similarly, it would be of great interest for journals to ask authors about previous submission to an internet repository to use the numeration provider by the curator, in order to reinforce the use of correct nomenclature.
All these characteristics are needed to classify a new qnr allele/gene.
Characteristics invalidating the description of a new qnr allele/gene.
Desirable but not essential.
Effect on any quinolone MIC.
Transferable Qnr genes are named “Qnr” followed by a letter (e.g., QnrA). Chromosomal Qnr should be named with the initials of the microorganism followed by “Qnr” (e.g., accordingly, qnr from Photobacterium profundum should be named Ppqnr for the DNA and PpQnr for the protein). In addition, it was proposed that if a chromosomal Qnr has at least 70% identity to one of the established transferable qnr families, it may be named according to transferable or chromosomal name rules (e.g., SaQnrA3, where Sa represents Shewanella algae). Usually, the latter consideration is applied only for proposed original bacterial sources of established transferable qnr families.
The qnr nomenclature rules published in 2008 (319) considered the presence of either a DNA or amino acid identity difference of ≥30% as a requisite to define a new gene. While this has been considered a general rule, in 2017 qnrB88 was renamed qnrE1 because its original source (Enterobacter instead of Citrobacter) was taken into account (176), classifying a new gene irrespective of having an amino acid identity of ∼85% with established QnrB alleles, including QnrB1.
Silent mutations are not considered to determine the presence of new qnr alleles (319). Nonetheless, the presence of silent mutations may be of epidemiological and evolutive interest because they may play a role in the diversification of qnr, since further codon alterations may result in new variants. In this sense, it is of note that in vitro studies have shown that amino acid changes may alter the levels of resistance conferred by different Qnr proteins (see “In vitro mutations,” below). Furthermore, the effect of silent differences in DNA sequences in qnr genes on mRNA stability or on the efficiency of translation to the final protein related to codon usage or other phenomena, which may lead to different levels of Qnr proteins in the bacterial cytoplasm, with a subsequent effect on final MICs, remains unexplored. In this sense, it has been observed that increased levels of expression of qnrB and qnrS may be related to higher MICs of ciprofloxacin (236) (Fig. 4).
Identity criteria (<70% identity) were established to determine the different Qnr families. Accordingly, at present, 7 families of transferable qnr genes have been described (qnrA, qnrB, qnrC, qnrD, qnrE, qnrS, and qnrVC) (see “QnrE,” below, for levels of identity between QnrE and QnrB). Nonetheless, while a series of closely clustered qnr alleles can be found and can even be easily classified within a common family, a series of gradually divergent alleles, which act as a bridge between different clusters, may obscure the Qnr family borders. These divergent alleles are the result of natural qnr sequence divergences within the ancestral host, followed by more than one independent mobilization phenomenon, the natural evolution of transferable qnr genes within different hosts, and the description of transferable qnr alleles/genes with a different, but closely phylogenetically related, original source (e.g., QnrB and QnrE). All these forces are not exclusive, and indeed, all act in a simultaneous manner.
A repository website used to number and order new qnr families and alleles (the Lahey database, formerly at https://www.lahey.org/qnrStudies/) was developed, including more than 100 different qnr alleles (Table 5). Similar to what occurred on July 2015 with the repository of β-lactamases (https://www.lahey.org/Studies/), in 2018, this repository was transferred to the NCBI for Qnr nomenclature maintenance and future gene/allele assignations, and the original Lahey database is no longer online. Thus, β-lactamase, qnr, as well as mcr gene/allele assignations are currently centralized at (https://www.ncbi.nlm.nih.gov/pathogens/submit-beta-lactamase). Meanwhile, to my knowledge, no database has compiled the chromosomal qnr genes, except for chromosomal qnr genes present in the bacterial chromosome of S. maltophilia described at a website (http://www.icms.qmul.ac.uk/centres/immunologyandinfectiousdisease/Smqnr%20Web%20v2.htm) that seems to have disappeared (I was unable to find this website) but existed at the beginning of the 2010s (320).
Qnr Families
Although the presence of Gram-positive chromosomally encoded Qnr proteins has been described (311, 321), all the currently known transferable Qnr families derive from Gram-negative ancestors (see from “QnrA” to “Chromosomal Qnr,” below).
As mentioned above (see “Nomenclature of the Transferable Mechanisms of Quinolone Resistance”), the names of transferable Qnr determinants have been assigned in a mixed manner. Thus, some follow an alphabetical order according to the first description, while others have been named based on the microorganisms from which they were first recovered, irrespective of the ordinal moment and the true prevalence in this microorganism. The next 7 subsections are strictly ordered alphabetically. Therefore, the order in which the different families are presented does not preclude the order described in time.
QnrA.
As mentioned above, the qnrA gene, which encodes a 218-amino-acid protein, was the first well-established TMQR (15). Further studies determined that this qnr gene is located in an integron-like environment (318). The integron containing qnrA1 was fully sequenced in 2007, confirming the presence of qnrA1 in a complex class 1 integron downstream from the first qacEΔ1-sul1, between 2 ISCR1 elements (284). The same study also highlighted the presence of qnrA1 in other complex integrons (284). The association between ISCR1 and qnrA1 has been largely reported, although in most cases, only the first ISCR1 is present (253). Furthermore, other genetic structures lacking the presence of ISCR1 have also been described (322). The presence of qnrA between 2 ISCR1 elements may facilitate spontaneous (or induced under quinolone pressure) qnrA gene duplication, with the subsequent effect on the final MIC of quinolones (251). Thus, when the in vitro-selected pMG252 plasmid-derived mutant pMG252A was cloned into an E. coli J53 background, the qnrA1 expression level and the MIC of ciprofloxacin rose 2.2- and 8-fold, respectively, higher than those related to the presence of pMG252 (258). Recent reanalysis and sequencing of pMG252A showed that these findings were due to the presence of 4 additional qnrA1 copies after the original ISCR1-qnrA-qacEΔ1-sul1-ISCR1, following the scheme qnrA-qacEΔ1-sul1-ISCR1 (251). Of note, this phenomenon might be extended to other TMQR placed between 2 insertion sequences. For reading of literature published before 2006, note that ISCR1 is usually referred to as orf513 or orf341. Additionally, the term “CR” for “common region” may also be present (196, 323–325).
Although variants of qnrA1 containing silent mutations were detected in 2003 (253), until 2004, no other qnrA allele (qnrA2) was described (GenBank accession number AY675584). Nevertheless, to my knowledge, there has been no published report by sequence authors, with the first report including QnrA2 data, together with those of QnrA3, QnrA4, and QnrA5, being published in 2005 (172).
Currently, 8 qnrA alleles have been described in accordance with nomenclature rules, mainly described within complex integron-like genetic environments (246, 284, 325–328), while a search of GenBank detected other new transferable QnrA alleles together with 2 new chromosomal Shewanella algae qnr variants (see the next paragraph). Of note, QnrA8 recorded previously in Lahey's repository is not in the RefSeq list, probably because no description within a plasmid or other transferable genetic structure has been made.
Shewanella species has been proposed as the chromosomal ancestor of this Qnr family. Thus, QnrA1 (329) and QnrA2 (GenBank accession number BAF95541) have been detected in the chromosome of Shewanella putrefaciens, while QnrA2, QnrA3, Qnr4, QnrA5, and QnrA8 have been identified in that of S. algae (172, 330, 331). QnrA7 has also been detected in S. algae both in the chromosome (GenBank accession number CP018456) and within a 33-kb plasmid (332). A GenBank search using QnrA1 (GenBank accession number AAL60061) also detected other QnrA alleles within the chromosome of S. algae, including the sequences under GenBank accession numbers WP_044735234 (333) and WP_045283443 (334).
QnrB.
The first qnrB gene was described within the pMG298 plasmid in a South Indian K. pneumoniae isolate (335), being the third transferable qnr family described. In the same study, the second qnrB allele (qnrB2) was detected in isolates from the United States when designing primers to determine the prevalence of qnrB in different microorganisms (335). Since then, the number of new qnrB alleles has been continuously rising, with a total of 87 unique sequences with an assigned allele numeration in the Lahey database as of 31 December 2018 and, as remaining qnr genes, an uncertain number of unnoticed alleles (209). Similar to what has been described for the qnrA genes, qnrB may be located within a complex integron environment associated with ISCR1 or other insertion sequences like IS26 (245, 275, 336) (Fig. 7). In addition, other dissemination pathways, such as transposons like Tn2012 (formed by ISEcp1C and qnrB19) or the so-called KQ element (for KPC and Qnr), have been described, in which the acquisition of qnrB19 has been suggested to be related to ISEcp1-like mobilization (283, 336, 337).
FIG 7.
Example of a complex integron containing qnr determinants. In both panels, the first variable region is identical. Note that in addition to the qnr determinants, other TMQR such as aac(6′)Ib-cr may be also present. (A) Complex integron containing a typical ISCR1 element (note that in older studies, ISCR1 is referred to as orf513 or orf341) after the classical 3′-end region of class 1 integrons (qacEΔ1-sulA) and just before the qnrB6 determinant environment (between psp and sap clusters) present in the second variable region. (Adapted from reference 458 with permission.) (B) Complex integron in which ISCR1 has been replaced by an IS26 element. Note that the environment of qnrB4 slightly differs from that in panel A (the sap cluster has been lost). (Adapted from reference 245 with permission from Elsevier.)
Although in 2005, two different sizes for QnrB1 (226 amino acids long) and QnrB2 (214 amino acids long) were described (335), in 2008, consensus was achieved regarding the use of the initial codon present in QnrB2 as the initial QnrB family ATG because of its commonness to all the QnrB proteins described (319). Thus, caution is needed when analyzing QnrB in order to ensure the use of the correct size and amino acid numeration.
In 2004, the presence of a qnrB gene was detected during marine metagenomic studies, leading to the proposal that an unknown marine microorganism may be the original source of QnrB (338, 339). Nonetheless, taking into account the high prevalence and diversity of QnrB, together with the lack of surrounding mobile elements in Citrobacter spp., members of the C. freundii complex are considered to be the original source of QnrB (306).
Within the genomes of the C. freundii complex (as well as in most of the transferable QnrB proteins detected), the genetic environment of qnrB is highly conserved, being located between two gene clusters, the psp (phage shock protein) cluster downstream and the sap (sensitivity to antimicrobial peptides) cluster upstream (306, 307). These clusters are contiguously present in the chromosomes of other Enterobacteriaceae, including other Citrobacter spp. such as Citrobacter koseri and Citrobacter rodentium, in which no qnrB is present. In view of these findings, it was proposed that a common ancestor of the C. freundii complex had acquired an exogenous qnrB-like gene between the psp and sap clusters, which was thereafter maintained or partially deleted, leading to the presence of pseudogenes (307). Further mobilization phenomena led to the transfer of qnrB to plasmids, which were then acquired by other microorganisms, resulting in the current widespread scenario.
Although not demonstrated, it seems evident that this spread from a member of the Enterobacteriaceae family would strongly favor the wide dissemination of this qnr family within Enterobacteriaceae, which are probably the microorganisms most commonly isolated and studied, leading to their current status as the most frequently described (and numerous) qnr family worldwide.
QnrC.
To date, only one allele of QnrC has been described, first detected in a plasmid (pHS10) from P. mirabilis (340). This Qnr is slightly larger than the remaining transferable Qnr proteins, being 221 amino acids long, similar to the lengths of some chromosomal Qnr proteins from different Vibrio spp. Accordingly, the origin of QnrC has been proposed to be among Vibrionaceae (340). Indeed, different VpQnr and VrQnr proteins (chromosomal Qnr of Vibrio parahaemolyticus and Vibrio rumoiensis, respectively) present amino acid identity levels ranging from 94 to 97% (e.g., GenBank accession numbers ODZ33109.1 and OEF25096). Furthermore, a chromosomal Qnr was found (GenBank accession number WP_105901077.1) in a recently sequenced Vibrio gangliei isolate (GenBank accession number NZ_PPSN01000001), differing in 9 bp compared to QnrC and resulting in 1 amino acid change, thereby presenting DNA and amino acid identities of 98.6% and 99.5%, respectively. This origin within Vibrionaceae produces identity levels of QnrC with some members of the QnrVC family of more than 70% (210) and highlights the recent suggestion that the ancestral microorganism should be taken into account to define new transferable qnr families (176).
To date, QnrC seems to be infrequent, although it has been described in different genera, including the above-mentioned genus Proteus (340) as well as in Escherichia (341), Klebsiella (342), and Shigella (343) (Fig. 5).
QnrD.
QnrD was first described as being encoded in a small plasmid of 4.3 kb (p2007057), which was isolated from 4 S. enterica strains belonging to serovar Bovismorbificans (3 strains) and serovar Kentucky (1 strain), isolated in China in 2006 or 2007 (344). QnrD is 214 amino acids long. In contrast to other Qnr proteins, when cloned into E. coli DH10B, the first studies showed a null or very limited effect on the MIC of nalidixic acid (only a 2-fold increase, from 2 to 4 μg/ml) and a higher impact on final ciprofloxacin MICs, which increased from 0.002 to 0.06 μg/ml (increase of 32-fold) (344, 345). Nonetheless, further studies on the cloning of QnrD in E. coli TOP10 showed slightly higher increases in the MIC from 2 to 8/16 μg/ml for nalidixic acid (346).
At present, 3 alleles have been formally described, while other QnrD family members are present in GenBank (e.g., GenBank accession numbers WP_084978381 and WP_108479726). QnrD has been detected in a variety of microorganisms (Fig. 5), including Ochrobactrum anthropi (GenBank accession numbers CCV01662.1 and CCU60984.1, among others) belonging to Brucellaceae, in which Qnr proteins have been scarcely detected. Nonetheless, QnrD proteins have been described as being especially prevalent in members of the genera Proteus and Providencia, belonging to the Proteeae tribe (currently proposed to be reclassified as the new family Morganellaceae [347]), encoded within small (∼2.6- to ∼5.2-kb) nonconjugative plasmids of an undescribed incompatibility group, usually linked to an open reading frame (ORF) (ORF-2) of unknown function (345, 346, 348–351). Thus, while Guillard et al. detected QnrD in 7 out of 332 (2.1%) Morganellaceae isolates (349), in more-recent studies, the presence of qnrD was observed in 40 out of 203 (19.7%) Morganellaceae isolates (348) and in 19 out of 24 (79.2%) Proteus species isolates (351). Considering these findings, it has been proposed that the origin of QnrD lies within an as-yet-unidentified member of this bacterial family, and different Proteus spp., Providencia spp., and Morganella spp. may play a role as intermediate reservoirs from which QnrD has expanded to other microorganisms (346, 348, 349). However, as yet, this remains to be demonstrated.
Although there is no information on the specific microorganisms used as QnrD donors, the above-mentioned role of phages in the dissemination of QnrD has been experimentally observed by the transduction of qnrD into E. coli JM109 (348).
QnrD is one of the few TMQR that has been described among Gram-positive microorganisms. Thus, in a study aimed at determining the presence of antibiotic-resistant microorganisms in treated and untreated river water, qnrD was detected in Bacillus spp. and Kurthia spp. (167). Unfortunately, no attempt to confirm these data by DNA sequencing and to determine either the exact genetic environment or the exact allelic variant of qnrD was performed, and no further data are available.
QnrE.
In the middle of 2017, a K. pneumoniae strain isolated in 2007 in Argentina exhibiting low-level quinolone resistance, wild-type GyrA, and negative results when TMQR were sought was reported. Further analysis showed the presence of a 645-bp ORF carried within an IncM1 transferable plasmid (pKp1130; GenBank accession number KY073238) (176). This ORF showed an average identity of 75% with members of the QnrB family, and a literature search showed 100% identity with qnrB88, which was isolated in Brazil from a K. pneumoniae plasmid (pKp145-11b; GenBank accession number KX118608) (352). Thereafter, another K. pneumoniae isolate carrying this gene within an IncM1 plasmid (pKP41M) as well as 3 S. enterica isolates belonging to serotypes Enteritidis, Infantis, and Newport also carrying qnrE1 within an IncM1 plasmid were again described in Brazil (273, 353). A further search of GenBank showed the presence of qnrE1 within S. Typhimurium (e.g., GenBank accession number KYE08263), C. freundii (e.g., GenBank accession number PUU65120), and other K. pneumoniae isolates. Moreover, a variant with only 2 amino acid differences currently recorded as QnrE2 in GenBank (GenBank accession number WP_078207746.1) is present within an E. coli plasmid (pEC422_1; GenBank accession number CP018961) as well as (although initially recorded as QnrS1) a whole-genome sequence of K. pneumoniae (GenBank accession number UJVU01000046).
Although, according to the rules of qnr gene nomenclature, this gene should be considered a member of the qnrB family (Table 6), QnrE has been classified as the first member of a new family (qnrE). An in-depth analysis showed high identity with the chromosomal qnr gene of Enterobacter spp., also suggesting that ISEcp1 is responsible for gene mobilization (176) and therefore has a different ancestral origin than other qnrB genes. Notwithstanding, this finding highlights the need to use the concept of gene family with caution as well as to advance toward the introduction of the concept of original bacterial chromosomal source to nomenclature rules. Indeed, there are numerous genomic data for different Enterobacter spp., including an Enterobacter kobei Qnr protein (GenBank accession number OTW32454) with 100% amino acid identity with QnrE1. In addition, a series of closely related qnr genes (even with >85% identity in either DNA base pairs or amino acids) are also present in other bacterial genomes, especially within the genus Serratia, such as Serratia plymuthica (e.g., GenBank accession number AGP44145) and other Serratia spp. (e.g., GenBank accession number OKP25762), as well as in the genomes of other Enterobacteriaceae such as Lelliottia spp. (e.g., GenBank accession number ASV55492) and Buttiauxella spp. (e.g., GenBank accession number KFC78735).
Curiously, at present, all transferable qnrE genes (those present outside a potential indigenous bacterial source) for which the geographical source is available (18 out of 20 GenBank-recorded sequences) have been described in South America, within Citrobacter spp. from Argentina, Klebsiella spp. from Argentina and Brazil, Salmonella spp. from Brazil, and E. coli from Ecuador. It is likely that upon routine screening of this gene, the number of microorganisms carrying QnrE variants, alleles described, and geographical locations would be greatly increased.
QnrS.
QnrS was first described in 2006 as being encoded within a conjugative plasmid of 47 kb (pAH0376) of an S. flexneri 2b strain isolated in Japan in 2003 (171). Studies performed in subsequent years showed a low, albeit increasing, prevalence of QnrS and a wide geographical distribution. Thus, QnrS was described in microorganisms such as S. enterica, E. coli, and Enterobacter cloacae, among others (328, 354). Since then, up to 9 QnrS alleles have been described and numbered in a previous Internet repository (https://www.lahey.org/qnrStudies), and at least 5 other alleles (QnrS10 to QnrS15) are present in GenBank (209), being described in a great variety of microorganisms and environments (133, 203, 209, 245, 297, 300, 355). Of note, QnrS3 is not numbered in RefSeq, probably because the reported sequence lacks the initial amino acid (356).
The mobilization and spread of qnrS alleles have been mediated by structures such as insertion sequences such as IS2, IS26, and ISEc12 (a member of the IS21 family) (272, 354, 357, 358). Additionally, qnrS has also been described as occurring near, but not within, Tn3-like structures carrying blaTEM-1 (171, 354). In this sense, Kehrenberg et al. described the plasmid pINF5 from Salmonella enterica serovar Infantis which, after a Tn3-like structure, carried a defective ISEc12 element followed by a qnrS2 gene, an internal segment of the CS12 fimbrial gene cluster of E. coli, and a defective IS26 element (354).
The origin of QnrS has been proposed to be among aquatic microorganisms. Thus, in 2007, the presence of chromosomal Qnr was observed in Vibrio splendidus, with identity levels of 83.1 to 83.9% with QnrS1 and 87.1 to 87.6% with QnrS2 (the 2 QnrS proteins described at that time) (312). Thereafter, Vibrio species sequences presenting higher identity levels have been detected. For instance, 97% and 95.5 to 96% amino acid identities with chromosomal Qnr from Vibrio mytili (GenBank accession number KIN11186.1) and Vibrio parahaemolyticus (GenBank accession numbers WP_029802054.1 and WP_029823919.1), respectively, have been observed. Furthermore, in a V. parahaemolyticus isolate from Malaysia, a Qnr protein (GenBank accession number KKF68274.1) has been detected, located between transposases having 100% amino acid identity with QnrS1. Despite being classified as “genomic,” the size of the containing DNA sequence (2,178 bp long) allows the possible presence of this qnr gene within a plasmid environment.
QnrVC.
Although qnr genes were described within the genomes of Vibrionaceae as early as 2005 (171, 174), the presence of a new transferable qnr family within an integron environment in V. cholerae was not shown until 2008 (175). This transferable Qnr variant was subsequently detected in other Vibrionaceae (303, 304, 359, 360) and in other bacterial families such as Aeromonadaceae (361) and Moraxellaceae (362), even within plasmids or in integron environments. Despite these evidences, the presence of this transferable qnr family was not proposed and subsequently included in the Lahey database until 2013 (210).
Nine different QnrVC alleles (QnrVC1 and QnrVC3 to QnrVC10), which have a length of 218 amino acids, have been classified by the Lahey website. Of these, at present, QnrVC8 and QnrVC9 are not included as numbered QnrVC variants in RefSeq, being in fact present within the Vibrio species chromosome (363). Note that qnrVC2 is not considered in the list because of the presence of 3 base insertions and 1 deletion (361) leading to a frameshift and premature stop, erasing its functionality. Similar to QnrB, another possible initial codon may be present 13 amino acids before the initial ATG codon (e.g., GenBank accession number APA29731).
These alleles have been described worldwide in plasmid and integron environments, either in the first variable region of a classical class 1 integron or downstream from ISCR1 within a complex class 1 integron (274, 364). In this sense, it is worth mentioning that qnrVC may be included in a gene cassette with an attC site (175, 231, 361). In contrast to classical integron gene cassettes, qnrVC gene cassettes may possess their own promoter, which allows them to surpass the decreased levels of expression related to a delayed position within the overall integron or to the absence of the strong promoter P2 (231).
It is of interest that QnrVC may be subdivided into two main groups, which are composed of (i) QnrVC1, QnrVC3, QnrVC6, QnrVC10, and one P. aeruginosa sequence erroneously classified as QnrVC7 (GenBank accession number AWT08553), in which the more divergent allele is QnrVC3, differing by 4 and 5 amino acids with respect to QnrVC1, QnrVC6, and QnrVC10 and QnrVCAWT08553, respectively, and therefore with identity levels of 97 to 99.5%, and (ii) QnrVC4, QnrVC5, and QnrVC7, in which the maximum difference is 3 amino acids between QnrVC5 and QnrVC7. The identity levels of these alleles are around 98.5 to 99.5%. QnrVC1-like and QnrVC4-like proteins differ by a minimum of 40 amino acids, with a maximum identity of 81.6%.
At present, QnrVC has been detected in additional genera within the Enterobacteriaceae (E. coli, Enterobacter spp. Citrobacter spp., Klebsiella spp., and Salmonella spp.) and Pseudomonadaceae (P. aeruginosa and P. putida), among others (206, 231, 249, 302, 350, 364–366) (Fig. 5).
Chromosomal Qnr.
The presence of chromosomal qnr genes was first observed in 2005 in Photobacterium profundum and Vibrio vulnificus (171). The same year, the chromosomal qnr gene of a strain of V. parahaemolyticus was cloned in a pUC118-derived plasmid and introduced into an E. coli isolate (174). The wild-type cloned gene does not affect the MIC of quinolones, but a mutant cloned gene with a single amino acid change, C115Y, accidentally introduced during PCR procedures showed 8- to 16-fold increases in the MICs of nalidixic acid, ciprofloxacin, and levofloxacin. Afterwards, the effect of this point mutation was confirmed by mutagenesis studies (174).
In subsequent years, a great number of chromosomally encoded Qnr-related proteins have been detected, in both Gram-negative and Gram-positive microorganisms, and their ability to confer resistance to quinolones has been confirmed in most cases (215, 367). Thus, the 2008 Qnr nomenclature rules, in order to differentiate a plasmid-encoded from a chromosomally encoded Qnr protein, proposed that the initials of the microorganism be added before chromosomal Qnr (e.g., EfsQnr for chromosomal Qnr of Enterococcus faecalis) (Table 6).
Among microorganisms possessing chromosomal Qnr, a high heterogeneity of chromosome-encoded Qnr proteins of S. maltophilia was observed. Thus, 11 different alleles were sequenced when SmQnr was detected (368). This finding was thereafter confirmed (369, 370), and as mentioned above (see “Qnr Classification”), the development of a website repository to bring order to the SmQnr nomenclature was proposed (320). The number of SmQnr alleles has increased enormously in the last years, with ∼150 different SmQnr alleles being detected in a GenBank search performed at the time of this review.
Although indigenous chromosomal Qnr proteins have been detected in microorganisms, such as in the above-mentioned E. faecalis and C. freundii (306, 311, 321), a high number has been found in microorganisms from water environments (367). In fact, as indicated when the different Qnr families are presented in this review, 4 out of 7 of the currently established transferable Qnr families (QnrA, QnrC, QnrS, and QnrVC) are derived from chromosomes of water-related microorganisms (367).
Qnr Structure
Qnr proteins are dimeric proteins belonging to the pentapeptide repeat protein (PRP) family, which encompasses thousands of proteins present in more than 1,500 eukaryotic or prokaryotic organisms (371, 372).Thus, the PRP family includes MfpA-like proteins, a chromosome-encoded Mycobacterium species protein group able to confer slight protection against the action of quinolones (373, 374), and McbG from E. coli, which is involved in resistance to microcin B17, a natural topoisomerase II inhibitor (375). This protein family is characterized by a tandem repeat of 5 amino acids, in which the residue located in a central position is indicated as “i.” Those on the right are indicated by adding 1 or 2 (i+1, i+2), and those on the left are indicated by subtracting 1 or 2 (i−1, i−2), leading to the structure i−2, i−1, i, i+1, i+2 (372). The typical PRP motif follows the amino acid scheme i−2 (S, T, A, or V), i−1 (D or N), i (L or F), i+1 (S, T, or R), i+2 (G) (372).
Although data are inferred from the limited number of Qnr crystallography studies, it has been observed that the Qnr dimer has a rodlike form in which each monomeric structure folds into a right-handed quadrilateral β-helix (376, 377). In this quadrilateral, the PRP amino acid in position i interacts with the amino acid i−2, stabilizing the structure; both residues i and i−2 are oriented toward the internal quadrilateral surface, while the remaining residues are oriented toward the exterior, resulting in a usual anionic surface (378). This quadrilateral structure has 9 square repeating units, called coils, numbered from 0 to 9, and these coils are stacked on each other, leading to a 4-faced protein (372). Thus, each coil has 4 faces numbered from 1 to 4, and 5 amino acids forming a PRP motif are presented on each face (372, 376). Most of the coils present a type II turn, which clusters toward the C-terminal region, while a few (7 in QnrB1) have a type IV turn, being located on faces 2 and 4 toward the N-terminal region (376). Of these coils, the first (coil 0) is capped in some Qnr proteins, such as QnrB1, due to the presence of the noncanonical PRP residues E8 and E18 at the i−2 position (376). The last (coil 9) is also capped in all the Qnr proteins analyzed (376, 379) because of the presence in the protein C terminus of the dimerization module formed by the structure strand (β)/helix (α)/strand (β) (376, 377, 380). In this structure, the orientation of each subunit is flexible thanks to the small, ∼730-Å2/subunit, and hydrophobic contact area (376).
In transferable as well as in several chromosomally encoded Qnr proteins, the quadrilateral structure is broken by 2 outward-projecting loops, named loop A and loop B, present between the second and third faces of coil 2 and between the fourth face of coil 4 and the first face of coil 5, respectively. These loops account for 8 amino acids (loop A) (from amino acid 46 to amino acid 53) and 12 amino acids (loop B) (from amino acid 102 to amino acid 113) (371, 376, 377) (Fig. 8). These loops are essential for the protective action of Qnr. Thus, in QnrB1 and AhQnr (the Qnr-like protein encoded in the chromosome of Aeromonas hydrophila), it has been shown that a deletion of loop A produces a strong decrease in the protective action of Qnr, while a deletion of loop B (or of both loops) leads to a full loss of Qnr protection (376, 377). In this line, point amino acid changes within loops may also lead to deleterious effects on protective activity (see “In vitro mutations,” below) (Table 7). Nonetheless, it has been observed that EfsQnr (chromosomal Qnr of E. faecalis) has no loop, retaining its protective action (381). This finding has been related to a series of peculiarities in the N-terminal region, in which the first 26 amino acids are not part of the quadrilateral β-helix (381). Thus, the first 18 amino acids of EfsQnr are extended over the β-helix, with which they interact, while amino acids 19 to 26 transverse the diagonal of the β-helix (381). Of note, the deletion of the first 17 amino acids strongly affects the protective action of EfsQnr (381).
FIG 8.
Structure of Qnr. (A) Sequence of QnrB1. The sequence is presented under 4 columns, one for each of the four faces of the right-handed quadrilateral β-helix. At the top are the faces (each one marked with a different color; note that the colors are maintained in the 3 panels) as well as the naming convention for each PRP residue. The positions of loop A and loop B are marked (*, loop A; **, loop B). The sequences of both loops are at the bottom of the panel. Highlighted in salmon is the N-terminal α-helix. (B) Structures of 4 Qnr proteins, QnrB1, MfpA (PRP from Mycobacterium spp.), AlbG (PRP produced by Xanthomonas albilineans), and EfsQnr (chromosomal Qnr of Enterococcus faecalis). MfpA is able to confer slight resistance to quinolones, while AlbG results in resistance to albicin (a natural topoisomerase inhibitor). (C) QnrB1 dimeric rodlike structure. The amino acid positions of loop A and loop B are indicated. A diamond shows the molecular 2-fold symmetry. Type II turn-containing faces are shown as spheres, and type IV-containing faces are shown as strands. (Reproduced from reference 376 with permission of the publisher.)
TABLE 7.
Effect of specific amino acid substitutions and deletions at loops A and B on Qnr protection levelsa
| Position(s) within TR | Change(s) in indicated Qnr causing a decrease in protection |
|||
|---|---|---|---|---|
| QnrA1 | QnrB1 | QnrC1 | QnrS1 | |
| Loop A | ΔA41–G56 | |||
| ΔI49–E55 | ||||
| ΔS51–G56 | ||||
| Y46A | ||||
| ΔY46–Q51 | ||||
| S50A | ||||
| ΔG56/D | ΔG53A/D/E | ΔG56D/E | ||
| Loop B | N103A | |||
| M104A | ||||
| ΔM104–S113 | ||||
| I105A | V108A | |||
| ΔT106 | S109A | |||
| ΔT106–T107 | ||||
| ΔT107 | ||||
| ΔR108 | ||||
| ΔT106–R108 | ||||
| ΔT107-R108 | ||||
| M112A | ||||
| W110A | Y113A | |||
| F114D | F111A/D | F114A | ||
| C115Y | C112A/Y | C115A/Y | ||
| R58H + R87H + C112Yb + N178Y | ||||
| S116D | S113A/D | S116P | S116A | |
| Loops A + B | ΔY46–Q51 + ΔM104–S113 | |||
Amino acid substitutions are listed from top to bottom in the direction from the N to C termini. Changes in the same row are analogous to each other. The amino acids highlighted in boldface type are those leading to a full loss of Qnr activity and a subsequent lack of an effect on the final quinolone MICs. Note that cloning in different vectors may slightly affect the final full or partial loss of activity. The amino acid change is reported only when there is a positive or negative effect of at least 4-fold on the MIC of the quinolones described. Note that different studies have determined the Qnr effect on different quinolones. Loop A, amino acids 46 to 53 (QnrB) and amino acids 49 to 56 (QnrA, QnrC, QnrS, and QnrVC); loop B, amino acids 102 to 113 (QnrB); TR, tandem repeat; Δ, deletion (based on data from references 234, 235, 376, and 391–394).
The amino acid change C112Y is located in loop B.
Original Qnr Function
It has been suggested that the original Qnr function is similar to that of GyrI (a protein also able to confer slight protection against quinolones). GyrI has been related to bacterial protection against natural topoisomerase II inhibitors such as CcdB and ParE (not to be confounded with topoisomerase IV subunit B). Both proteins belong to addiction systems involved in plasmid maintenance (382). Nonetheless, further studies have described the inability of Qnr to protect DNA gyrase from the action of these toxins and the lack of an effect of these toxins on the quinolone protection conferred by Qnr or on quinolone susceptibility levels (383).
Other authors have considered the possible involvement of several Qnr proteins, such as QnrB and QnrD, in the bacterial response to DNA damage (384). This proposal considers the presence of a lexA box in the Qnr promoter region of these genes (but not in that of other Qnr variants), which results in the expression of Qnr in a LexA/RecA-dependent manner under SOS system control (385, 386).
Another proposal focused on the original Qnr function in the water-related environment of several Qnr ancestral microorganisms. Thus, it has been observed that qnrA is overexpressed with cold shocks, suggesting an adaptive role favoring the survival of bacteria in water environments at low temperatures by regulating negative DNA supercoiling through the stabilization or modulation of the activity of DNA gyrase or topoisomerase type III (330). In this sense, it should be mentioned that most Shewanella spp., the ancestral source of QnrA, are psychrotolerant and are able to grow at temperatures of <5°C (387).
Nonetheless, despite these suggestions, in the absence of definitive studies, the original function of Qnr remains unknown.
Mechanisms of Qnr Action
The exact manner as to how Qnr interacts with DNA, type II topoisomerases, and/or quinolones to confer quinolone protection remains controversial, and further studies are needed to establish the definitive mode of action of Qnr.
Initial studies to determine the mode of action of Qnr showed that Qnr minimizes the action of quinolones by interacting with type II topoisomerases (388, 389). Furthermore, it was observed that this interaction did not need either DNA or quinolones, suggesting the presence of different possible mechanistic explanations (388, 389). Thus, an alteration of the quinolone-binding pocket conformation was considered, which hinders further quinolone-target interactions and leads to a reduction of the presence of the cleavage complex among quinolones, DNA, and DNA gyrase/topoisomerase IV (388, 389). Other options have also been considered, such as the destabilization of these cleavage complexes, which subsequently allows DNA replication, or the reduction of the interactions between DNA and DNA gyrase, leading to a lower number of replication forks (388, 389).
In order to answer these questions, Xiong et al. (377) analyzed the structure of AhQnr in silico, and its interactions with DNA gyrase led to the proposal of an interaction model in which Qnr is located within the DNA gyrase structure interacting by electrostatic charges through faces 1 and 2. In this model, loops A and B interact with the topoisomerase-primase (TOPRIM) domain of GyrB and the GyrA “tower,” respectively, although a specific orientation of the GyrA and GyrB subunits is needed (377). The authors propose that this specific GyrA/GyrB orientation is favored when quinolones interact with topoisomerase and subsequently allows Qnr to interact with and disrupt the complex between quinolones and topoisomerase (377), and the latter recovers its catalytic activity.
At around the same time, working with a crystallographic QnrB1 model, Vetting et al. (376) proposed a model in which Qnr acts in a posterior step, when the DNA-poison-topoisomerase is cleaved, destabilizing the cleavage complex and restoring DNA replication.
Subsequent studies analyzing the effect of point mutations of either loop A or loop B found no correlation between the Qnr-topoisomerase interaction and final protection levels, proposing that the loops are not involved in direct interactions with topoisomerase but rather are involved in proper Qnr positioning in gyrase, blocking the access of quinolones. Thus, when the Qnr-gyrase complex interacts with DNA, Qnr is removed, and the enzyme becomes catalytic (390).
In vitro mutations.
As mentioned above, the first evidence of the effect of point mutations on quinolone protection levels conferred by Qnr determinants was observed in 2005 due to a PCR accident which produced an amino acid change, C115Y, during cloning of a V. parahaemolyticus chromosomal Qnr determinant (174). Further studies analyzing the effect of the same amino acid change in QnrA1 and QnrS1 showed a different scenario, in which despite both mutated Qnr proteins exhibiting a certain degree of protection, this was lower than that for parental Qnr (391). Along the same line, when this mutation was generated in the equivalent position of QnrB1 (C112), a full loss of Qnr activity was observed (392) (Table 8).
TABLE 8.
Effect of specific amino acid deletions at PRP units on Qnr protection levelsa
| Deletion in indicated Qnr causing a decrease in protection |
|---|
| QnrA1 |
| ΔD2–Q10 |
| ΔD2–S21 |
| ΔL187–D218 |
| ΔL207–D218 |
| QnrB1 |
| ΔM1–F15 |
| ΔM1–I20 |
| M205–G214 |
| ΔI210–G214 |
| ΔV212–G214 |
| QnrC1 |
| ΔI11–S20 |
| ΔN77–G96 |
| ΔC137–K156 |
In all cases, these deletions lead to a full loss of Qnr activity and a subsequent lack of an effect on the final quinolone MICs. Note that different studies have determined the Qnr effect on different quinolones. Deletions within loop A or B are reported in Table 7. Deletions of single amino acids are reported in Table 9 (based on data from references 234, 235, 376, and 391–394).
Similar differences in final quinolone MICs were observed when different S. maltophilia chromosomal Qnr alleles were cloned (368). Thus, SmQnr5 does not affect the MICs of norfloxacin, ciprofloxacin, sparfloxacin, or gatifloxacin but rather has an almost imperceptible effect (≤2-fold) on the MICs of nalidixic acid, levofloxacin, and moxifloxacin. Meanwhile, except in the case of nalidixic acid, the effect of SmQnr6 leads to >5-fold increases, reaching increases of up to >62-fold for the MICs of sparfloxacin (from <0.002 to 0.125 μg/ml) (368).
Rodríguez-Martínez et al. (392) analyzed the effects of different amino acid codon changes by random and directed mutagenesis. Although most of the amino acid variations detected led to decreased or null Qnr protection, the results showed that in QnrS1, the amino acid change D185Y increases the MICs of ciprofloxacin and moxifloxacin in E. coli 4-fold (both from 0.125 to 0.5 μg/ml), and increases of 2-fold were observed when norfloxacin, ofloxacin, and levofloxacin were tested (392). In the same study, it was also observed that the effect of the same substitution led to different final results when introduced into different Qnr families. Thus, the D185Y amino acid change had no effect on the final MICs when introduced into QnrA, while it negatively impacted the Qnr protective role when introduced into QnrB (392). Furthermore, while the deletion of Gly56 in QnrA results in a reduction of protective levels and the substitution G56D produces a full lack of QnrA activity, the same modifications lead to a different scenario in QnrS, in which the deletion of G56 results in a loss of Qnr activity, and the substitution G56D was able to confer slight protection. Meanwhile, in QnrB, both modifications inactivated Qnr (392). It is of note that G56 (QnrA and QnrS) or the equivalent amino acid G53 (QnrB) is located in loop A (Table 7).
In fact, a series of studies attempted to determine the impact of substitutions on the Qnr loops or on specific positions within pentapeptides. Along these lines, as mentioned above, the loss of loop A, loop B, or both loops has a negative effect on QnrB activity. In addition, it has been proposed that amino acid substitutions for polar amino acids in position i, or for amino acids with bulky side chains in position i+2 or i−2, destabilize the right-handed quadrilateral β-helix structure, having a negative impacting on Qnr activity (393, 394) (Tables 8 and 9).
TABLE 9.
Effect of single amino acid substitutions at PRP units on Qnr protection levelsa
| Position within TR | Change(s) in the indicated Qnr causing the indicated change in protection |
||||||
|---|---|---|---|---|---|---|---|
| QnrA1 (Neg) | QnrB1 (Neg) | QnrC1 (Neg) | QnrS1 |
QnrVC7 |
|||
| Neg | Pos | Neg | Pos | ||||
| i | F13S | ||||||
| i | F25A | ||||||
| i+2 | T27I | ||||||
| i | F30A | ||||||
| i−2 | A36L/D | ||||||
| i | L38P | L35A | L38R | ||||
| i | F40A | ||||||
| i−2 | C43A | ||||||
| i | F45A | ||||||
| i−2 | S62D/G/L | ||||||
| i | F66A | ||||||
| i−2 | C72Y | C69A/Y | C72Y | C72Y | |||
| i | F76A | ||||||
| 1−2 | A82I/D/G/L/T | ||||||
| i−2 | C92Y | C89Y | C92Y | ||||
| i+2 | G96D | G93D | G96D | ||||
| i−2 | A97Y | ||||||
| i | F96A | ||||||
| i−2 | A102D/G/L/T | ||||||
| i | F101A | ||||||
| i−2 | A114C/V | A117C/V | |||||
| i | I116A | ||||||
| i−1 | N120A | ||||||
| i | L121A | ||||||
| i+2 | Y123A | ||||||
| i−1 | N125A | ||||||
| i−2 | Q132D | Q132L | |||||
| i+1 | E132A | ||||||
| i | L136A | ||||||
| i−2 | N139A | ||||||
| i+2 | G148A | ||||||
| i−2 | T152D/G/L | T152A | |||||
| i−1 | S153P | ||||||
| i−2 | S154A | ||||||
| i−1 | D155A | ||||||
| i | L159D | L156A/D | L159D | ||||
| i+1 | S157A | ||||||
| i−2 | G159A | ||||||
| i | F161A | ||||||
| i | W166A | ||||||
| i−2 | C172D/G | C172L | |||||
| i | L176A | ||||||
| i | L181A | ||||||
| i+1 | D182Y | D185Y | |||||
| i−2 | L184A | ||||||
| i+1 | D188V | ||||||
| i+2 | G193A | ||||||
| dm | G209A | ||||||
| N27I + A91V + K195R | |||||||
| Q132L + T152A | |||||||
| T152A + C172L | |||||||
| F114Y + G196S | |||||||
| G7R + G98D | |||||||
| V6A + D95Y | |||||||
Amino acid substitutions are listed from top to bottom in the direction from the N to C termini. When more than one amino acid change is reported simultaneously, they are placed at the bottom. Changes in the same row are analogous to each other. The amino acids highlighted in boldface type are those leading to a full loss of Qnr activity and a subsequent lack of an effect on the final quinolone MICs. Note that cloning in different vectors may slightly affect the final full or partial loss of activity. The amino acid change is reported only when there is a positive or negative effect of at least 4-fold on the MIC of quinolones described. Note that different studies have determined the Qnr effects on different quinolones. No data obtained by IPTG induction have been considered because they represent the effect of higher Qnr expression levels but not the intrinsic Qnr activity. TR, tandem repeat; dm, dimerization module; Neg, protective levels conferred by Qnr decrease; Pos, protective levels conferred by Qnr increase (based on data from references 234, 235, 376, and 391–394).
The effect of amino acid changes on different levels of quinolone protection has also been observed among different wild variants of Qnr belonging to the same family. Thus, QnrVC7 confers lower levels of protection than QnrVC5 or QnrVC6 (394). This finding has been related to the wild-type presence of A152 in QnrVC5 and QnrVC6.
QUINOLONE MODIFICATION
The first evidence of quinolone biodegradation was provided in the mid-1990s when Martens and colleagues demonstrated the ability of some wood-rotting fungi to degrade enrofloxacin (395). Further studies focused on the ability of Gloeophyllum striatum to degrade enrofloxacin have proposed up to 4 possible degradation pathways, including oxidative decarboxylation, defluorination, hydroxylation at C-8, or oxidation of the piperazinyl moiety (396), all showing different hydroxyl radical quinolone attack points (396). This model was thereafter expanded to other basidiomycetes, including Cyathus stercoreus, a coprophilous fungus, and quinolones such as ciprofloxacin (397). In addition, it showed the ability of other fungi (i.e., Xylaria longipes) to transform danofloxacin into danofloxacin N-oxide, a metabolite with substantially less antibacterial activity (398).
Further analyses of the ability of soil microorganisms to degrade quinolones suggest the potential of different fungi (e.g., Candida spp.) and also of Mycobacterium spp. and Pseudomonas spp. to fully degrade the danofloxacin piperazine ring (399). On the other hand, the same study also described the ability of several microorganisms, including Pseudomonas spp. and Mycobacterium spp., to metabolize danofloxacin to N-desmethyl danofloxacin (399), a metabolite which retains antibacterial activity.
It is obvious that the intrinsic basal quinolone MIC is silently influenced in bacteria that are able to inactivate or modify quinolones by any of these routes. Nonetheless, to date, none of these mechanisms has been detected in a transferable genetic element.
AAC(6′)Ib-cr
In 2003, during a study focused on analyzing a series of plasmids carrying qnrA, a few plasmids conferring unusually high levels of ciprofloxacin resistance (∼4-fold higher than usual) were detected (253). The authors suggested that differences in qnrA expression levels might explain these differences but highlighted that the modulatory effect on final qnr expression of unnoticed plasmid factors or the presence of unknown TMQR might also be a reason (253). Further analyses resulted in the description of the first bacterial enzyme able to selectively modify several fluoroquinolones, such as ciprofloxacin and levofloxacin, increasing bacterial resistance levels (153, 170). Currently, this enzyme has been largely described in different microorganisms from various environments, probably being the most common and frequent TMQR. In addition to its presence in microorganisms from human samples, a recent report has shown a high prevalence of AAC(6′)Ib-cr in animal and environmental samples (400).
The aac(6′)Ib-cr gene is frequently detected as a cassette of class 1 integrons or associated with mobile elements such as IS26 (197, 245, 401, 402). Among these, In37-like integrons of around 3.6 kb are largely detected in K. pneumoniae and have disseminated to other bacterial species, including other Enterobacteriaceae and Aeromonadaceae. In this integron, aac(6)-Ib-cr is placed in the first position, thereafter being followed by three other gene cassettes: blaOXA-1, catB3, and arr3 (403–405). The different In37 variants may in addition possess other antimicrobial resistance genes after the first variable region, becoming complex integrons. Thus, the presence of qnrA1 or qnrB alleles (for instance, qnrB4 or qnrB10), which may also be linked to a blaDHA-1 gene, has also been reported in In37-like structures (245, 249, 253).
Differences have been observed in the final MICs conferred by AAC(6′)Ib-cr. Thus, in addition to possible differences related to additional amino acid substitutions (see below), upon analyzing 2 Klebsiella aerogenes (formerly Enterobacter aerogenes) isolates, Ruiz et al. (402) showed differences in final ciprofloxacin MICs associated with differences in AAC(6′)Ib-cr expression levels. It was proposed that these differences were related to the presence of a 12-bp deletion between IS26 and the initial ATG codon, which displaces the −10 box in one of the K. aerogenes isolates, leading to an almost complete absence of expression of aac(6′)Ib-cr (402). It should be mentioned that the AAC(6′)Ib-like enzymes have a great diversity in size (170, 406, 407); therefore, this text follows the numeration provided by Robicsek et al. (170), but it should be noted that other numerations may be considered in different reports (407, 408).
The originally described AAC(6′)Ib-cr protein [see “AAC(6′)Ib-cr subtypes,” below] was reported as a protein of 172 amino acids in length belonging to the AAC(6′)Ib group (253), but the initial ATG codon was corrected early on and was reported as a protein of 199 amino acids in length. AAC(6′)Ib-cr is characterized by 2 amino-acid-specific substitutions, W102R and D179Y, which are located in two external enzyme loops (153, 407, 409). Furthermore, at the time of its description, it also presented its own N-terminal region with 12 unique amino acids. Currently, this original enzyme is present in GenBank as AAC(6′)Ib-cr5 (GenBank accession number WP_001749987). Subsequent sequencing has shown the presence of several AAC(6′)Ib-cr variants with high heterogeneity in size, mainly related to the enlarged N-terminal region [see “AAC(6′)Ib-cr subtypes,” below]. The above-mentioned amino acid modifications confer to the enzyme the ability to acetylate quinolones possessing an unsubstituted piperazinyl group, such as ciprofloxacin and norfloxacin, but not other quinolones such as levofloxacin and moxifloxacin (153). In addition to the above-mentioned substrates, a later study showed AAC(6′)Ib-cr (at least the original allelic variant described by Robicsek et al.) activity over tosufloxacin (410). Mutagenesis analyses focused on both single amino acid changes showed that when these are absent, the enzyme loses its ability to confer quinolone resistance, while in the absence of W102R, D179Y also confers slight protection against quinolones but lower than that of wild-type AAC(6′)Ib-cr (153).
Similar to AAC(6′)Ib, acetyl coenzyme A acts as an acetyl donor (410), while a series of differences regarding specific AAC(6′)Ib-cr substrates have been reported. Of these, the most evident is a modest decrease [∼10-fold with respect wild-type AAC(6′)Ib] in the ability to acetylate kanamycin and the inability to acetylate neomycin B (410). In fact, despite being characterized for its ability to acetylate some fluoroquinolones, AAC(6′)Ib-cr acetylates kanamycin more efficiently (catalytic efficiency of kanamycin acetylation ∼50-fold higher than that of fluoroquinolones) (410). In addition, it is interesting that the specific optimum substrate pHs for AAC(6′)Ib-cr also differ, being 6.1 to acetylate aminoglycosides and 7.7 to act on fluoroquinolones (410).
When AAC(6′)Ib-cr acts on aminoglycosides, the interactions are based on the presence of numerous hydrogen-bonding interactions, while it has been suggested that the interactions with quinolones are related to stacking interactions. In this sense, the model of interaction between AAC(6′)Ib-cr and quinolones proposed by Vetting et al. (410) considers that the amino nitrogen of the piperazinyl group located 2.7 Å from Asp115 interacts with acetyl coenzyme A, playing a role as a proton acceptor during catalysis. In this model, the quinolones establish stacking interactions with the β6/β7 and α1′-α2 loops of AAC(6′)Ib-cr (410). Within the α1′-α2 loop, Trp49 directly interacts with quinolones, and the Gly50-Ala54 region interacts with the pyridinone ring of the quinolones through van der Waals forces. Tyr179 located on the β6/β7 loop can interact with the quinolone ring due to the establishment of attractive, noncovalent interactions (π-stacking interactions) of the p-orbitals of Tyr179 and quinolone rings. In this model, Trp49 plays a relevant role in the stabilization of the structure. Meanwhile, Arg102 establishes interactions with Tyr179 leading to the stabilization of the optimal interaction position of Tyr179 (410).
Maurice et al. (407) proposed an alternative interaction model. While this model considered a similar role in stacking interactions for Tyr179, it proposed that Arg102 directly interacts with the keto or carboxy groups of fluoroquinolones through the development of a hydrogen bond (407).
AAC(6′)Ib-cr subtypes.
As mentioned above, 2 amino acid substitutions are sufficient to confer a new ability to AAC(6′)Ib-cr. This is of special relevance because it highlights the fact that small changes may modify the enzyme substrates. In this sense, different AAC(6′)Ib-cr variants have been shown to confer quinolone resistance, or at least this role has been strongly suggested (406, 409). Nonetheless, a series of studies have not addressed this question and have mainly described the presence of AAC(6′)Ib-cr by means of approaches such as specific PCR followed by restriction fragment length polymorphism (RFLP) analysis (138, 411), while other studies report full sequences presenting the 2 above-mentioned specific amino acids, as well as other amino acid differences, even in the absence of specific studies to determine enzyme activity. While these are valid approaches, they have the same limitations as any other antibiotic resistance gene PCR detection method or new allele descriptions in the absence of activity determinations. Indeed, a series of enzymes have been reported to be able to confer quinolone resistance in the absence of definitive data.
On the other hand, different AAC(6′)Ib variants may possess the unnoticed ability to confer slight resistance to specific quinolones. In this sense, many AAC(6′)Ib-cr sequences currently present in GenBank possess W102R and/or D179Y, differing mainly in the N-terminal region or presenting a few amino acid differences. Therefore, systematic cloning and evaluation of these enzymes could lead to a series of relevant modifications in the current knowledge about this mechanism of quinolone resistance. In this sense, Kim et al. (406) detected a variant of AAC(6′)Ib-cr able to acetylate gemifloxacin and zabofloxacin. This AAC(6′)Ib-cr variant was characterized to present Y179 plus L117 (406), with both amino acids being present in the enzyme described by Robicsek et al. (170) but lacking the amino acid change W102R (406). Table 10 shows a series of AAC(6′)Ib-cr variants which have shown the ability to acetylate quinolones.
TABLE 10.
AAC(6′)Ib-cr variants with demonstrated quinolone-acetylating activitya
| GenBank accession no. | IDb | Size (no. of amino acids) | Amino acid positionc | Quinolone(s)d | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WP_001749987 | cr5 | 199 | M1 | S2 | N3 | A4 | K5 | N20 | R102 | L117 | Y179 | D198 | CIP, NOR, TSX | 170 |
| EUM72348 | 202 | I | Q | H | F | Q | D | V | CIP | 406 | ||||
| —e | ? | ? | ? | ? | ? | ? | G | ? | CIP | 406 | ||||
| EUM72320 | 199 | W | CIP, GMX, ZBX | 406 | ||||||||||
| WP_015059932 | cr4 | 225 | T | P | G | N | D | T | CIP, NOR | 409 | ||||
CIP, ciprofloxacin; GMX, gemifloxacin; NOR, norfloxacin; TSX, tosufloxacin; ZBX, zabofloxacin. Amino acids considered relevant for the acetylation of quinolones are highlighted in boldface type. Note that possible enhanced or diminished activity as well as expanded or contracted substrate profile alterations might be related to nonhighlighted amino acid differences. The absence of a specific quinolone does not preclude the inability of the enzyme to inactive it.
In the description present in GenBank, note that the first described AAC(6′)Ib-cr enzyme (170) is numbered “cr5.”
Following the numeration provided by Robicsek et al. (170), note that the different sizes of acetylases may lead to different numeric positions in any variant. For instance, the reported amino acid substitution “N20T” under GenBank accession number WP_015059932 is located in the equivalent position 46. When longer, amino acid positions prior to M1 under GenBank accession number WP_001749987 have not been analyzed.
Quinolones which might be acetylated.
No GenBank accession number is provided in the article by Kim et al. (406), and after a search in GenBank using the sequence under accession number WP_001749987 and introducing a glycine in position 179, no concordant enzyme was found among the first 100 results.
Other Quinolone Modification Enzymes
The description of AAC(6′)Ib-cr demonstrates the possible effects of bacterial enzymes able to modify antimicrobial agents on quinolones. In this regard, a recent study reported the presence of a plasmid-encoded phosphorylase able to inactivate ciprofloxacin, called CrpP (154). This enzyme of 65 amino acids in length was detected in P. aeruginosa, encoded within a conjugative plasmid (pUM505) of 123 kbp (154) belonging to the IncI1 group, which was isolated in the early 1980s during a study designed to determine the presence of resistance to heavy metals (412). Thereafter, in 2011, a further analysis of this plasmid showed its ability to increase the MIC of ciprofloxacin in P. aeruginosa, although no TMQR known at that time was detected, thereby suggesting the presence of an unknown TMQR encoded within this plasmid (413). This finding was confirmed in 2018 upon the identification of the crpP gene (154).
CrpP has 40% identity with an internal 42-amino-acid region of the aminoglycoside phosphotransferase (APH) from Mycobacterium smegmatis and, despite not having the catalytic APH enzyme motifs, presents Gly7 and Ile26, two conserved residues in APH enzymes which are involved in catalysis and in ATP binding, respectively. Of note, the concomitant presence of G7 and Ile26 was detected only in 63 of the >1,000 closely related (identity levels of ≥90%) amino acid sequences present in GenBank (see below) (414). CrpP seems to be able to confer a slight increase in the MIC of ciprofloxacin (from 0.008 to 0.06 μg/ml) when cloned into pUC_20 and introduced into an E. coli J53 environment but had no effect on the remaining quinolones tested (levofloxacin, moxifloxacin, nalidixic acid, and norfloxacin). Interestingly, upon comparing the effects of CrpP when cloned into pUC_20 and when the native pUM505 plasmid was transferred to another P. aeruginosa isolate, the effect of the native plasmid was greater and extended to norfloxacin and moxifloxacin, suggesting the presence of another unknown quinolone resistance mechanism (154). Further analysis using the above-mentioned quinolones as substrates showed that CrpP is able to phosphorylate norfloxacin but with an even lower efficiency than that of ciprofloxacin (with a Vmax/Km ratio approximately 5-fold lower than that for ciprofloxacin) (154). Due to the novelty of the CrpP description (to my knowledge, at the time of writing of this review, it has been reported in only four articles), there is practically no knowledge of the extension of this mechanism. Nonetheless, more than a thousand CrpP-like proteins grouped into more than 37 different alleles, possessing one or a few amino acid changes, are present in GenBank (414). Interestingly, all these sequences except for one uncharacterized 102-amino-acid-long protein of A. baumannii (GenBank accession number SST11961) belong to members of the Pseudomonadaceae (414). Most of these sequences were reported within the P. aeruginosa genomic islands PAPI-1 and PAGI-5 (414), which are able to disseminate through conjugation (415). These genomic islands have evolved from the ancestral conjugative plasmid pKLC102, which has been proposed to have an environmental origin (416). These finding might suggest that the origin of CrpP is from plant-related bacterial communities, perhaps belonging to the Pseudomonadaceae (414).
At the beginning of 2019, Chávez-Jacobo et al. (417) explored the presence of crpP-like genes in 77 ESBL-carrying Enterobacteriaceae and in a historical collection of 66 E. coli J53-2 isolates containing plasmids transferred from ESBL-carrying clinical isolates which were collected between 1988 and 2012. The authors detected 9 samples (4 clinical isolates and 5 plasmids from the historical collection) carrying a series of related crpP genes. Of these, the original sources were an E. coli isolate in 5 cases and a K. pneumoniae isolate in the remaining 4 cases (417). Further analysis of 5 of these plasmids (2 from E. coli and 3 from K. pneumoniae) showed that the first 2 encoded products had protein identity levels ranging from 10.1% to 43.7% with original CrpP, while the identity levels between the 3 CrpP-like proteins from K. pneumoniae were higher, ranging between 57.1% and 65.7% (417). An analysis of the specific effects of 3 of these CrpP-like proteins on the ciprofloxacin MICs showed increases equivalent to those of the original CrpP protein (417). Nonetheless, neither data on the presence of quinolone target selection nor those on the nalidixic acid MIC were provided, allowing the possible selection of spontaneous mutants. In this sense, an in silico analysis of these crpP-like sequences showed the presence of base insertions/deletions that result in frameshifts (414). Pending further confirmation, current data suggest the existence of a family of genes with the ability to slightly affect the MICs of specific quinolones.
It should be taken into account that the authors proposed that a carboxyl group present at position 3 of the ciprofloxacin molecule binds with ATP to be thereafter phosphorylated and degraded (154). This radical is common to all quinolones (except metioxate) (Fig. 3); therefore, it should be considered that CrpP may present activity against untested quinolones, and as indicated above, the presence of CrpP modifications may lead to alterations of either expansion or constraint in the substrate pattern as well as the final activity levels. In this sense, further antibiotic spectrum determination of detected CrpP variants will be of relevance.
TRANSFERABLE EFFLUX PUMPS
Although transferable oqxAB genes were first described in 2004 (156), the ability of transferable efflux pumps to extrude quinolones from bacteria to the environment was demonstrated in 2007 (157–159).
Most of the studies in the field are focused on the detection of 2 different efflux pump groups (QepA and OqxAB). In addition, other scarcely characterized efflux pumps such as QacA and QacB or those encoded within the plasmid pRSB101 are also able to pump out quinolones from bacteria.
OqxAB
In 2003, Sørensen et al. (155) were the first to describe the presence of a plasmid (pOLA52)-borne mechanism of olaquindox resistance in E. coli isolated from swine. Further studies to determine the exact mechanism of olaquindox resistance encoded within pOLA52 showed its efflux pump nature, detecting 2 ORFs, encoding a 391- and a 1,050-amino-acid-long protein, which were named OqxA and OqxB (here, these 2 specific alleles are referred to as OqxA1 and OqxB1). OqxA and OqxB present a certain degree of identity, especially at the amino acid level, with inner membrane (OqxA) and periplasmic (OqxB) components of previously described RND efflux pumps, such as MexEF of Xanthomonas axonopodis, MexXY of P. aeruginosa, and AcrAB of E. coli (156). The RND efflux pumps need a third component to be active, that is, an outer membrane protein (OMP). Hansen et al. (156) showed that in an E. coli genetic background, TolC plays this role, and therefore, TolC-like proteins presumably play this role in other Enterobacteriaceae. Nonetheless, the role of OqxAB as a quinolone resistance mechanism was not established until 2007, when its ability to extrude different quinolones, such as nalidixic acid, ciprofloxacin, and norfloxacin, together with a series of unrelated antibacterial agents and toxins was observed (157). Similar to what has been described for other RND efflux pumps, such as AcrAB-like efflux pumps in Enterobacteriaceae or MexAB-like efflux pumps in Pseudomonadaceae (239, 418, 419), it has been suggested that Phe-Arg-β-naphtylamide may inhibit OqxAB (420).
Further studies aimed at characterizing pOLA52 showed a plasmid size of 51,602 bp, classified as belonging to the IncX1 incompatibility group (200). Norman et al. (200) also showed that together with a putative regulator gene (later proposing the name OqxR), OqxAB proteins were flanked by IS26, thus being a composite transposon, demonstrating the presence of 100% (oqxA) and 99% (oqxB) identities with the components of a K. pneumoniae putative efflux pump, which was thereafter considered the OqxAB original bacterial source (199–201). Posterior analyses of the role of OqxR have shown that it may be involved in the downregulation of OqxAB expression levels. Thus, upon analyzing the expression levels of K. pneumoniae chromosomal oqxA and oqxB genes, it was shown that the presence of the specific OqxR amino acid changes reported below correlates with higher levels of expression of these genes, which revert when a plasmid carrying wild-type OqxR is used to complement the bacteria (421, 422):
F6S
V102G
Q11L, D95E, and V113I
N38T, D95E, V113I, and H159L
Δ73–77
Δ88–94
where Δ indicates a deletion. An association between these mutations and the final levels of OqxAB expression has been proposed (421, 422). Note that when there is more than one amino acid change, the presence of simple polymorphisms needs to be considered. To my knowledge, there is no study on the role of OqxR in transferable oqxAB expression levels. Nonetheless, transferable oqxAB regulation mediated by chromosomal regulatory genes such as ramA and rarA has been shown in S. Typhimurium (201, 420). This finding correlates with the final MICs of the antibacterial agents tested. Therefore, the specific chromosomal genetic background plays a crucial role in the final levels of expression of oqxAB and, subsequently, in its contribution to the final antimicrobial resistance levels. This finding can likely be extrapolated to other TMQR.
Although the presence of differences in the sequence of K. pneumoniae chromosome-encoded OqxAB has been described (tens of different chromosomal OqxA and OqxB alleles are recorded in GenBank), most of the studies on transferable OqxAB address only the presence/absence of OqxAB. In 2012, the presence of new transferable allelic variants of OqxA and OqxB were described, differing in 1 or 2 amino acids from the original OqxA and OqxB (thereafter named OqxA1 and OqxB1, respectively) (177). Furthermore, an in silico analysis of the presence of new transferable OqxA/OqxB variants in GenBank showed the presence of at least 14 new OqxA and 28 OqxB previously unnoticed variants, which had clearly evolved from K. pneumoniae chromosomal OqxAB, as well as the dissemination of these variants through a series of microorganisms (Tables 11 and 12). While most of these alterations are probably polymorphisms that do not affect the final OqxAB activities, the possible involvement of activity levels and/or the expansion/contraction of substrate profiles cannot be ruled out. In any case, the effect of these modifications on the final activity of OqxAB remains to be elucidated. Interestingly, K. pneumoniae is not the only member of the Enterobacteriaceae carrying an OqxAB-like efflux pump. Closely related efflux pumps have been described as being indigenous to other microorganisms such as other Klebsiella spp. or Enterobacter spp. (201). In this regard, the above-mentioned analysis also showed the presence in GenBank of an E. coli genome (strain TUM18641; GenBank accession number NZ_BGTY01000006) carrying OqxA (GenBank accession number WP_087857967) and OqxB (GenBank accession number WP_113374178) with 30 and 29 amino acid differences from OqxA1 and OqxB1, respectively, but closely related to OqxAB of K. aerogenes; thus, OqxA presents a maximum identity of 100% with that of K. aerogenes strain CRK0055 (GenBank accession number RNT35078), and OqxB, with only 2 amino acid differences, presents an identity of 99.8% with OqxB of K. aerogenes strains AR_0431 and GN04835 (GenBank accession number WP_047066491). Therefore, the unnoticed dissemination of another variant of OqxAB with an undefined effect on quinolones (or other antibiotic agents) has been suggested. This finding, especially if this new OqxAB-like protein expands to other E. coli isolates or other microorganisms, opens the door to suggesting an alternative name for this new variant, as occurred with QnrE.
TABLE 11.
Amino acid changes described in transferable OqxAa
| Yrc | GenBank accession no.b |
% identityd | Amino acid difference | Gram-negative genus or generae | Referencef | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DNA | Amino acid | |||||||||||||||||
| 2007 | EU370913g | AAP43109 | 100.0 | G8 | G16 | M19 | S30 | A113 | R116 | I159 | S183 | A199 | Q244 | R338 | D339 | ins | Enterobacter, Escherichia, Salmonella, Shigella | 156 |
| 2012 | NG_050420 | WP_063865358 | 99.7 | V | Escherichia, Raoultella, Salmonella, Enterococcus | 177 | ||||||||||||
| 2016 | NZ_LVMD01000084 | WP_063501637 | 99.7 | S | Escherichia | |||||||||||||
| 2016 | KT727030 | AMW92379 | 99.7 | R | Enterococcus | |||||||||||||
| 2018 | CP029215 | AWJ07841 | 99.7 | E | Escherichia | |||||||||||||
| 2018 | CP026492 | AUY47426 | 99.7 | I | Escherichia | |||||||||||||
| 2018 | KT716392 | AMW92376 | 99.7 | H | Enterococcus | |||||||||||||
| 2016 | FLWH01000008 | SBY67709 | 99.5 | E | V | Escherichia | ||||||||||||
| 2017 | NZ_MKFO01000039 | WP_089069082 | 99.5 | E | V | Escherichia | ||||||||||||
| 2017 | MOGH01000057 | OJM31094 | 99.5 | V | R | Escherichia | 56 | |||||||||||
| 2018 | JAMW01000061 | OXY63391 | 99.5 | K | V | Salmonella | ||||||||||||
| 2018 | MG028668 | AWG41968 | 99.5 | V | T | Enterobacter | ||||||||||||
| 2018 | MG028669 | AWG41969 | 99.5 | V | C | Enterobacter | ||||||||||||
| 2018 | PTMN01000089 | PPW02434 | 99.2 | 41PAP | Escherichia | |||||||||||||
Gram-positive microorganisms are highlighted in boldface type. No allelic numeration has been included to avoid confusion with a series of K. pneumoniae chromosomal OqxA proteins that are present in GenBank with an assigned allelic numeration. Note that these variants may be present in other genera and that other OqxA variants may remain undetected. ins, insertion.
In several cases, multiple valid DNA/amino acid sequences may be found in GenBank.
Date of first published report. If no published report has been found, the data for the older GenBank record are reported.
The minimum degree of identity considered was 99%. A series of OqxAB efflux pumps with lower levels of identity are present in GenBank, including a transferable oqxAB gene probably derived from an ancestral K. aerogenes isolate (GenBank accession number NZ_BGTY01000006) (see “OqxAB”).
In all cases, it is considered that any OqxA reported in a microorganism other than Klebsiella spp. belongs to a transferable gene, irrespective of chromosomal/plasmid location or the absence of specific genetic environment data. The presence of transferable OqxA in Klebsiella spp. is not reported here.
First published report.
First OqxA described as a transferable determinant.
TABLE 12.
Amino acid changes described in transferable OqxBa
| Yrc | GenBank accession no.b |
% identityd | Amino acid difference | Gram-negative genus or generae | Referencef | ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DNA | Amino acid | ||||||||||||||||||||||||||||||||
| 2007 | EU370913g | WP_000347934 | 100.0 | P102 | L138 | G148 | V217 | M313 | Q325 | A432 | L284 | H434 | G463 | A493 | G540 | G544 | G547 | A551 | V612 | V645 | A650 | L672 | A699 | Q778 | D749 | Y783 | E788 | Q799 | V839 | T1050 | ins | Enterobacter, Escherichia, Salmonella, Shigella | 156 |
| 2012 | JF912901 | AEM45928 | 99.9 | I | Escherichia | 177 | |||||||||||||||||||||||||||
| 2017 | KX518744 | APO16513 | 99.9 | I | Escherichia | 468 | |||||||||||||||||||||||||||
| 2018 | NZ_NRYU01000105 | WP_113449358 | 99.9 | L | Escherichia | ||||||||||||||||||||||||||||
| 2018 | NZ_PJHN01000068 | WP_112910686 | 99.9 | Q | Escherichia | 469 | |||||||||||||||||||||||||||
| 2018 | NZ_PJGF01000097 | WP_112889165 | 99.9 | K | Escherichia | 469 | |||||||||||||||||||||||||||
| 2017 | MOIC01000152 | OJO61665 | 99.9 | E | Escherichia | 470 | |||||||||||||||||||||||||||
| 2016 | FLWH01000008 | SBY67683 | 99.9 | N | Escherichia | ||||||||||||||||||||||||||||
| 2018 | NZ_NRXP01000002 | WP_113402128 | 99.9 | S | Escherichia | ||||||||||||||||||||||||||||
| 2018 | NZ_PQHV01000042 | WP_105906775 | 99.9 | E | Escherichia | ||||||||||||||||||||||||||||
| 2018 | KT716391 | AMW92372 | 99.9 | Y | Escherichia, Salmonella, Enterococcus | 169 | |||||||||||||||||||||||||||
| 2017 | NZ_NGVB01000050 | WP_087525850 | 99.9 | F | Escherichia | ||||||||||||||||||||||||||||
| 2017 | NZ_FZIQ01000054 | WP_096216538 | 99.9 | L | Escherichia | ||||||||||||||||||||||||||||
| 2012 | JF912902 | AEM45930 | 99.8 | Y | I | Escherichia, Raoultella, Salmonella | 177 | ||||||||||||||||||||||||||
| 2018 | PTNE01000065 | PPW37064 | 99.8 | Q | E | Escherichia | |||||||||||||||||||||||||||
| 2016 | KT334335 | ALI92905 | 99.8 | H | Q | Salmonella | |||||||||||||||||||||||||||
| 2016 | KT727030 | AMW92380 | 99.8 | L | Y | Enterococcus | |||||||||||||||||||||||||||
| 2016 | NZ_KQ089493 | WP_048299610 | 99.8 | N | E | Escherichia | |||||||||||||||||||||||||||
| 2016 | MOIX01000077 | OJP65617 | 99.8 | E | E | Escherichia | |||||||||||||||||||||||||||
| 2018 | NZ_NSAZ01000061 | WP_113455638 | 99.8 | Y | E | Escherichia | 469 | ||||||||||||||||||||||||||
| 2015 | KF414089 | AGZ20250 | 99.7 | Y | I | I | Escherichia | 212 | |||||||||||||||||||||||||
| 2016 | AGZ20250 | AMW92377 | 99.7 | Y | S | E | Enterococcus | ||||||||||||||||||||||||||
| 2018 | PTML01000068 | PPX08745 | 99.7 | 411XVD | Escherichia | ||||||||||||||||||||||||||||
| 2018 | PTML01000068 | PPV65433 | 99.6 | 411XVVD | Escherichia | ||||||||||||||||||||||||||||
| 2015 | KF414080 | AGZ20241 | 99.6 | N | S | E | F | Escherichia | 212 | ||||||||||||||||||||||||
| 2016 | LUIP01000142 | KYE44549 | 99.5 | K | S | R | E | N | Salmonella | ||||||||||||||||||||||||
| 2011 | FN811184 | CBL62366 | 99.5 | K | S | S | R | E | Salmonella | ||||||||||||||||||||||||
| 2018 | NZ_BGKO01000006 | WP_109956222 | 99.3 | N | K | S | V | G | F | I | Escherichia | ||||||||||||||||||||||
Gram-positive microorganisms are highlighted in boldface type. No allelic numeration has been included to avoid confusion with a series of K. pneumoniae chromosomal OqxB proteins which are present in GenBank with an assigned allelic numeration. Note that these variants may be present in other genera and that other OqxB variants may remain undetected. ins, insertion.
In several cases, multiple valid DNA/amino acid sequences may be found in GenBank.
Date of the first published report. If no published report has been found, the data for the older GenBank record are reported.
The minimum degree of identity considered was 99%. A series of OqxAB efflux pumps with lower levels of identity are present in GenBank, including a transferable oqxAB gene probably derived from an ancestral K. aerogenes isolate (GenBank accession number NZ_BGTY01000006) (see “OqxAB”).
In all cases, it is considered that any OqxB reported in a microorganism other than Klebsiella spp. belongs to a transferable gene, irrespective of chromosomal/plasmid location or the absence of specific genetic environment data. The presence of transferable OqxB in Klebsiella spp. is not reported here.
First published report.
First OqxB described as a transferable determinant.
Internal partial genomic integrations or recombinations of transferable genetic structures leading to the full or partial loss of oqxA or oqxB may explain the studies in which only one of these genes is detected in nonindigenous microorganisms (341, 423). In addition, the presence of transferable OqxAB not derived from the K. pneumoniae genome may also contribute to explaining this phenomenon.
Initial studies were developed using samples of veterinary origin and thereafter in animal-exposed human populations (e.g., farmworkers), suggesting a relevant role of livestock management in the selection and spread of OqxAB-carrying isolates (155, 230). Nonetheless, despite the relevance of the farm environment, transferable OqxAB proteins have currently also been described in samples from healthy and sick people as well as from food samples, wild animals, and environmental samples (202, 424–426). Furthermore, the range of plasmids in which oqxAB has been reported has expanded, including plasmids classified within the IncF or IncHI2 group, among others (354, 425, 427).
In 2018, the presence of OqxAB in Gram-positive microorganisms was first reported. A study performed in China showed the presence of this efflux pump in E. faecalis in a genetic structure flanked by IS26 and linked to an incomplete aph(3′)-IIa gene (169). Interestingly, these samples were from a pig farm.
QepA
To date, 4 QepA variants have been described in the literature. The first qepA gene was described at almost the same time by 2 independent French and Japanese groups in 2007 (158, 159). A new QepA variant, QepA2, differing in 2 amino acid codons, was described in 2008 (178). Finally, QepA3 and QepA4 were described in 2015 and 2017, respectively (428–430). Nonetheless, an analysis of GenBank records showed that at least 6 other new complete qepA genes have been fully sequenced as well as the presence of additional partial QepA sequences exhibiting their own specific amino acid codon changes (179). All 6 uncharacterized QepA proteins were detected in members of the Enterobacteriaceae, thereby being acquired from an external source, irrespective of their exact genetic environment (chromosome or plasmid) (179). Similar to Qnr (and, in fact, with a long series of genes), a centralized QepA repository is needed to bring order to the nomenclature.
Similar to other TMQR, the mobilization of qepA has been related to the presence of different insertion sequences, such as IS26 and ISCR3C (158, 175, 430). The qepA4 gene has been detected in an atypical class 1 integron preceded by a defective dfrB4 gene and followed by an ISCR3 element (428).
QepA is a proton-dependent efflux pump which varies in size, presenting 14 transmembrane segments (14-TMS) which belong to the major facilitator superfamily (MFS) (159). An extrusion product pattern has been established in QepA1 and QepA2, including more-hydrophilic quinolones such as ciprofloxacin and norfloxacin but with a marginal or null ability to extrude hydrophobic quinolones such as ofloxacin levofloxacin, moxifloxacin, and nalidixic acid (159, 178, 431). Furthermore, these variants are unable to pump out nonquinolone agents such as erythromycin, chloramphenicol, rifampin, and tetracycline (159, 178). No data on pumping substrates of the remaining QepA variants are available. In this regard, it should be mentioned that despite having identities of ≥90% among the 10 QepA variants, these slight differences include insertions and deletions (179). Thus, while the 4 reported QepA variants have 511 amino acids, a deletion of 5 amino acids (Δ432SAALP), just after the 13th transmembrane section, and an insertion of 2 amino acids (Ins357LG) within the 11th transmembrane transept have been detected in QepA GenBank records (179). These insertions and deletions may affect the final QepA structure and may therefore have a major impact on either the expansion or the restriction of efflux pump substrate profiles as well as on substrate affinities, which may lead to different efficiencies of pumping out, with a direct impact on the final effect on antimicrobial resistance levels.
The first phylogenetic analysis proposed that the origin of QepA was within the Actinomycetales order (158). Furthermore, a possible ancestor within the actinomycetes also agrees with the G+C content of QepA (∼72%) (158). Nonetheless, the addition of new genomic data in GenBank has shown that the higher levels of identity with the currently described or putative Actinomycetales efflux pumps are ∼40%, while the identities of QepA with a series of 512 putative efflux pumps of the Comamonadaceae family (i.e., Pseudorhodoferax spp.) reach values of ∼80%. Despite the scarce data in this respect, the G+C content of the Pseudorhodoferax genus has been reported to be ∼68 to 70%, thereby agreeing with that of QepA. These findings suggested the possibility of an alternative origin within the Comamonadaceae family (179). In agreement with this alternative origin of QepA, in a study by Yamane et al., the chromosomal efflux pump with higher levels of identity with QepA1 in fact belonged to a member of the Comamonadaceae (i.e., Polaromonas spp.) (158).
QacA and QacB
Although described in the 1980s (180), the ability of the MFS 14-TMS proton-dependent efflux pumps QacA and QacBIII (an allelic variant of QacB) to extrude quinolones from bacteria was established only in 2010 (166).
The Qac determinants are mechanisms of resistance to antiseptic and disinfectant monovalent and divalent cations and are widely disseminated in S. aureus (432). Thus, in a study designed to analyze the variability of Qac determinants, an allelic variant of QacB, named QacBIII, was observed, possessing 4 amino acid differences from the originally described QacB (I26V, I167L, A184V, and A320E). Of these, the presence of E320 in transmembrane segment 10 was directly related to 4-fold increases in the MICs of norfloxacin and ciprofloxacin when cloned into S. aureus RDN1 within pTZN10; further in vitro introduction of A320 in QacBIII returned the MICs to parental levels (166). In the same study, it was observed that upon cloning QacA, the MICs of ciprofloxacin and norfloxacin increased slightly (2-fold), which was suggested to be related to the presence of aspartic acid at position 323 (166). Interestingly, D332 has also been related to the increased activity of QacA over divalent cations (433). This study showed a high prevalence of QacBIII (20 isolates, representing 80% of the isolates analyzed). Nonetheless, a recent study in Japan showed a slight decrease in the prevalence of QacBIII in methicillin-resistant S. aureus (MRSA) isolates from the period from 2010 to 2011 to the period from 2014 to 2015 (13.3% to 5.5%) (434). Furthermore, the authors highlighted that 80.7% of QacBIII-positive isolates belonged to staphylococcal cassette chromosome mec (SCCmec) type II; of these, the multilocus sequence typing (MLST) pattern was established in eight cases, with all belonging to sequence type 764 (ST764) (434). Another study in Spain of 159 S. aureus clinical isolates failed to detect the QacBIII variant (in fact, they detected only 1 isolate carrying QacB) (435). The diversity in the prevalence of QacB might reflect local differences and suggests the need to expand the studies on the presence of QacBIII to new geographical areas. Unfortunately, no other article related to the presence of QacBIII or about the role of QacBIII (or other QacA/B variants) has been found.
Other Transferable Efflux Pumps
A series of usually unconsidered efflux pumps with the ability or the potential to extrude quinolones from bacteria has been described.
Thus, among other plasmids recovered from a wastewater treatment plant bacterial community, Szczepanowski et al. characterized a plasmid called pRSB101 (436). This plasmid of 47,829 bp does not fit within any described incompatibility group. pRSB101 carries different antibiotic resistance determinants, including a mixed-type tripartite multidrug efflux pump. When cloned into E. coli, increases from 80 to 550 μg/ml and from 0.5 to 1.25 μg/ml were observed for the MICs of nalidixic acid and norfloxacin, respectively (436).
This efflux pump possesses 2 components belonging to the RND family. These components include a TetR/AcrR family transcriptional regulator showing 40 % identity with a putative regulator of Geobacter sulfurreducens (GenBank accession number NP_952005). Downstream from this regulator gene, an RND membrane fusion protein is encoded, possessing 45% identity with that of G. sulfurreducens (GenBank accession number NP_952003). These components are followed by a permease and by an ATPase (probably involved in providing energy for active transportation through ATP hydrolysis) belonging to the ATP-binding cassette (ABC) efflux pump family, also showing a high degree of identity (59% and 45%, respectively) with components of the same efflux system of G. sulfurreducens. No OMP-encoding gene was present in pRSB101, and therefore, the authors proposed that the system required a chromosomally encoded OMP to be functional (436).
The role of other plasmid-encoded efflux pumps in the development of quinolone resistance remains to be elucidated. For instance, in 2013, a MexAB-OprD-like efflux pump was described, located within 2 IS1 elements within an IncH1 plasmid (pNDM-CIT) from a multidrug-resistant C. freundii isolate for which no further data are available (437).
TMQR AND THE FUTURE
Antibiotic resistance mechanisms are as old as antibiotic production (438) and may be traced back to ancient samples from hundreds or thousands of years ago (439, 440). In some cases, these mechanisms have been related to adaptative mechanisms of antibiotic producer microorganisms to avoid self-killing or of other neighboring microorganisms to increase survival (441–444). Other mechanisms, such as TMQR, are recent adaptative functions of ancient molecules, possessing another original physiological function, in response to the presence of new nonnatural toxic agents related to human action, such as quinolones, a fully synthetic antibacterial agent family.
The first scientific studies on antibacterial agents date from the mid- to late 19th century (445–448), and the introduction of antibacterial agents into clinical practice to fight infectious diseases dates from the late 19th century and the early 1900s (449, 450). Nonetheless, the effect of these items on the ecology of bacteria was minimal if any. However, the changes in the microbial world induced by the pressure exerted after the clinical introduction of the current antibacterial agents in the 1930s (451) became exorbitant and absolutely immeasurable. The success of penicillin and subsequent antibiotics against infectious microorganisms led to an imbalance in the bacterial population, favoring the selection of microorganisms possessing antibiotic resistance mechanisms (Fig. 9).
FIG 9.
Selection of antibiotic resistance. (A) In the preantibiotic era, the presence of antibiotic-resistant microorganisms was rare. The introduction of antibiotics led to killing of antibiotic-susceptible bacterial populations; the selection of the minority antibiotic-resistant populations, which were present in the preantibiotic era; the dispersion of plasmid-encoded mechanisms of resistance; as well as the generation of new resistant strains (by selecting chromosomal mutations). This phenomenon led to the current high levels of antibiotic resistance. (B) Antibiotic resistance may be stable, with a minimal or null effect on bacterial fitness, thus being easily fixed in the bacterial population. The antibiotic-resistant microorganisms tend to be present even in the absence of antibiotic pressure. Antibiotic resistance may also be unstable, affecting bacterial fitness; the absence of antibiotic pressure tends to dilute in the general bacterial population. Nonetheless, antibiotic resistance may act in an indirect manner by other antibacterial agents favoring the fixing of genetic structures carrying multiple mechanisms of antibiotic resistance. Compensatory mutations are mutations which have no direct effect on antibiotic resistance but lead to the recovery of lost fitness. When selected, the microorganisms act as those with stable resistance.
TMQR have evolved from a series of genes with an as-yet-unknown physiological function, with their unexpected effect on the final MICs of quinolones benefitting their spread outside their original environment to the whole world. In this scenario, and fueled by the direct, continuous growth selection pressure exerted by quinolones in human, veterinary, and environment settings, further steps in the history of TMQR will include their installation in new genetic structures and their spread to new microorganisms, including a greater presence in Gram-positive microorganisms, as has recently been described (167–169). More efficient variants that are able to expand or modify their quinolone affinity or to confer higher levels of quinolone resistance will be selected. The description of new transferable Qnr families derived from new chromosomal Qnr ancestors is also foreseen, including derivatives from chromosomal Qnr of Gram-positive microorganisms. Thus, the incessant continuous description of new variants of the currently established TMQR, such as Qnr, QepA, AAC(6′)Ib-cr, and OqxAB, will advance in the future. Furthermore, a dribble of TMQR belonging to new families, such as the recently described CrpP (154), will also probably be described. In this sense, the potential of amino acid changes in the current antibiotic-modifying enzymes (or in other physiological acetylases or phosphorylases or other enzymes able to modify molecules) is a fertile and practically unexplored field.
CONCLUSION
Within a little over 20 years, TMQR have evolved from being a scientific, almost undetectable rarity to a worldwide epidemic. This finding has been made possible by human attitudes. Thus, quinolone use, similar to that of all antibacterial agents, has become one of absolute overuse and misuse; that is, the use of quinolones has expanded beyond the treatment of bacterial infectious diseases to a series of unnecessary roles, such as livestock growth promotion. This, in addition to the high levels of use in veterinary settings as a prophylactic agent, the uncontrolled access to antibacterial agents in a long series of countries, the high rate of misprescription, self-medication, erroneous posology, and early treatment discontinuation, has led to the current scenario of quinolone resistance that, as with all other antibacterial agents, has squandered the treasure that antimicrobials are.
The direct, continuous, and growing selection pressure exerted by antibiotics in human and microbiota commensal members and pathogenic microorganisms is incessant, favoring the survival of microorganisms which carry antibiotic resistance determinants such as Qnr or other TMQR. These microorganisms that possess adequate antibiotic resistance genes are the winners of the race involving the survival of bacteria in the antibiotic era (Fig. 9).
It is undeniable that TMQR will remain with us. At present, not even the strictest antibiotic control policies will return the presence of TMQR determinants to the level of the prequinolone era, but these measures are necessary to control and limit their continuous spread.
ACKNOWLEDGMENTS
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
I thank Donna Pringle for language correction.
Biography

Joaquim Ruiz, M.S.C., Ph.D., is a microbiologist who graduated from the Science Faculty of the Universitat Autònoma de Barcelona. He has developed his scientific career at the Hospital Clinic of Barcelona, being Associate Research Professor at the Barcelona Institute for Global Health and Visiting Lecturer at the Universidad Peruana Cayetano Heredia in Lima, Peru. At present, he is an External Researcher at the Universidad Continental in Lima, Peru. He has worked in the area of antibiotic resistance, being especially interested in the study of quinolone resistance mechanisms and spread since 1993. In recent years, he has focused his research on the knowledge of the dynamics of the development of antibiotic resistance and dissemination in different environments and on the study of infectious diseases in low- and middle-income countries, mainly diarrheagenic and bacteremic pathogens affecting children. Since 2008, he has also been leading a research line devoted to the understanding and knowledge of Carrion’s disease.
REFERENCES
- 1.Burman LG. 1977. Apparent absence of transferable resistance to nalidixic acid in pathogenic Gram-negative bacteria. J Antimicrob Chemother 3:509–516. doi: 10.1093/jac/3.5.509. [DOI] [PubMed] [Google Scholar]
- 2.Jonsson M. 1973. Antibiotic resistance and R factors in Gram-negative bacteria isolated in a hospital for infectious diseases. III. The effect of antibacterial treatment on the incidence of R factor-mediated antibiotic resistance. Scand J Infect Dis 5:41–47. doi: 10.3109/inf.1973.5.issue-1.08. [DOI] [PubMed] [Google Scholar]
- 3.Lebek G. 1967. Medizinische aspekte der infektiösen antibiotika-resistenz gram-negativer darmbakterien. Pathol Microbiol 30:1015–1036. doi: 10.1159/000161739. [DOI] [PubMed] [Google Scholar]
- 4.Babcock GF, Berryhill DL, Marsh DH. 1973. R-factors of Escherichia coli from dressed beef and humans. Appl Microbiol 25:21–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Munshi MH, Sack DA, Haider K, Ahmed ZU, Rahaman MM, Morshed MG. 1987. Plasmid-mediated resistance to nalidixic acid in Shigella dysenteriae type 1. Lancet ii:419–421. doi: 10.1016/s0140-6736(87)90957-3. [DOI] [PubMed] [Google Scholar]
- 6.Panhotra BR, Desai B, Sharma PL. 1985. Nalidixic-acid-resistant Shigella dysenteriae I. Lancet i:763. doi: 10.1016/s0140-6736(85)91310-8. [DOI] [PubMed] [Google Scholar]
- 7.Clerch B, Rivera E, Llagostera M. 1996. Identification of a pKM101 region which confers a slow growth rate and interferes with susceptibility to quinolone in Escherichia coli AB1157. J Bacteriol 178:5568–5572. doi: 10.1128/jb.178.19.5568-5572.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanders CC, Sanders WE Jr, Goering RV, Werner V. 1984. Selection of multiple antibiotic resistance by quinolones, beta-lactams, and aminoglycosides with special reference to cross-resistance between unrelated drug classes. Antimicrob Agents Chemother 26:797–801. doi: 10.1128/aac.26.6.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith JT. 1986. Frequency and expression of mutational resistance to the 4-quinolone antibacterials. Scand J Infect Dis Suppl 49:115–123. [PubMed] [Google Scholar]
- 10.Courvalin P. 1990. Plasmid-mediated 4-quinolone resistance: a real or apparent absence? Antimicrob Agents Chemother 34:681–684. doi: 10.1128/aac.34.5.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crumplin GC. 1987. Plasmid-mediated resistance to nalidixic acid and new 4-quinolones. Lancet ii:854–855. doi: 10.1016/s0140-6736(87)91039-7. [DOI] [PubMed] [Google Scholar]
- 12.Ashraf MM, Ahmed ZU, Sack DA. 1991. Unusual association of a plasmid with nalidixic acid resistance in an epidemic strain of Shigella dysenteriae type 1 from Asia. Can J Microbiol 37:59–63. doi: 10.1139/m91-009. [DOI] [PubMed] [Google Scholar]
- 13.Jacoby GA, Chow N, Waites KB. 2003. Prevalence of plasmid-mediated quinolone resistance. Antimicrob Agents Chemother 47:559–562. doi: 10.1128/AAC.47.2.559-562.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lesher GY, Froelich EJ, Gruett MD, Bailey JH, Brundage RP. 1962. 1,8-Naphthyridine derivatives: a new class of chemotherapy agents. J Med Chem 5:1063–1068. doi: 10.1021/jm01240a021. [DOI] [PubMed] [Google Scholar]
- 15.Martínez-Martínez L, Pascual A, Jacoby GA. 1998. Quinolone resistance from a transferable plasmid. Lancet 351:797–799. doi: 10.1016/S0140-6736(97)07322-4. [DOI] [PubMed] [Google Scholar]
- 16.Price JR. 1949. Alkaloids of the Australian Rutaceae: Melicope fareana. IV. Some reactions of I-methyl-y-quinolone-3-carboxylic acid, a degradation product of the alkaloids. Aust J Sci Res 2:272–281. doi: 10.1071/CH9490272. [DOI] [Google Scholar]
- 17.Barton N, Crowther AF, Hepworth W, Richardson ND, Driver GW. 1960. New quinolones and therapeutic compositions containing them. British patent 830832.
- 18.Bisacchi GS. 2015. Origins of the quinolone class of antibacterials: an expanded “discovery story.” J Med Chem 58:4874–4882. doi: 10.1021/jm501881c. [DOI] [PubMed] [Google Scholar]
- 19.Hepworth W. 1959. Manufacture of 1-methyl-6-nitro-4-quinolone-3-carboxylic acid. British patent 820295.
- 20.Carroll G. 1963. Neggram (nalidixic acid): a new antimicrobial chemotherapeutic agent. J Urol 90:476–478. doi: 10.1016/s0022-5347(17)64441-0. [DOI] [PubMed] [Google Scholar]
- 21.Lishman IV, Swinney J. 1963. Studies of a new antibacterial agent, nalidixic acid. Br J Urol 35:116–121. doi: 10.1111/j.1464-410X.1963.tb02602.x. [DOI] [Google Scholar]
- 22.Jameson RM, Swinney J. 1963. A clinical trial of the treatment of gram-negative urinary infections with nalidixic acid. Br J Urol 35:122–124. doi: 10.1111/j.1464-410X.1963.tb02603.x. [DOI] [PubMed] [Google Scholar]
- 23.Slade DA. 1963. A clinical trial of nalidixic acid (Negram, WIN. 18,320) in thirty-three patients with urinary infections treated in general practice. Br J Urol 35:125–128. doi: 10.1111/j.1464-410X.1963.tb02604.x. [DOI] [PubMed] [Google Scholar]
- 24.Cahal DA. 1965. Nalidixic acid—request for prompt reporting of suspected adverse reactions. Br Med J i:130. [PMC free article] [PubMed] [Google Scholar]
- 25.Tillotson GS. 1996. Quinolones: structure-activity relationships and future predictions. J Med Microbiol 44:320–324. doi: 10.1099/00222615-44-5-320. [DOI] [PubMed] [Google Scholar]
- 26.Emmerson AM, Jones AM. 2003. The quinolones: decades of development and use. J Antimicrob Chemother 51(Suppl S1):13–20. doi: 10.1093/jac/dkg208. [DOI] [PubMed] [Google Scholar]
- 27.Heyman SN, Ginosar Y, Shapiro M, Kluger Y, Marx N, Maayan S. 1997. Diarrheal epidemics among Rwandan refugees in 1994: management and outcome in a field hospital. J Clin Gastroenterol 25:595–601. doi: 10.1097/00004836-199712000-00009. [DOI] [PubMed] [Google Scholar]
- 28.Mandomando IM, Macete EV, Ruiz J, Sanz S, Abacassamo F, Vallès X, Sacarlal J, Navia MM, Vila J, Alonso PL, Gascon J. 2007. Etiology of diarrhea in children younger than 5 years of age admitted in a rural hospital of southern Mozambique. Am J Trop Med Hyg 76:522–527. doi: 10.4269/ajtmh.2007.76.522. [DOI] [PubMed] [Google Scholar]
- 29.Rogerie F, Ott D, Vandepitte J, Verbist L, Lemmens P, Habiyaremye I. 1986. Comparison of norfloxacin and nalidixic acid for treatment of dysentery caused by Shigella dysenteriae type 1 in adults. Antimicrob Agents Chemother 29:883–886. doi: 10.1128/aac.29.5.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salam MA, Bennish ML. 1988. Therapy for shigellosis. I. Randomized, double-blind trial of nalidixic acid in childhood shigellosis. J Pediatr 113:901–907. doi: 10.1016/S0022-3476(88)80029-5. [DOI] [PubMed] [Google Scholar]
- 31.Turner FJ, Ringel SM, Martin JF, Storino PJ, Daly JM, Schwartz BS. 1967. Oxolinic acid, a new synthetic antimicrobial agent. I. In vitro and in vivo activity. Antimicrob Agents Chemother 7:475–479. [PubMed] [Google Scholar]
- 32.Shimizu M, Nakamura S, Takase Y. 1970. Piromidic acid, a new antibacterial agent: antibacterial properties. Antimicrob Agents Chemother 10:117–122. [PubMed] [Google Scholar]
- 33.Wick WE, Preston DA, White WA, Gordee RS. 1973. Compound 64716, a new synthetic antibacterial agent. Antimicrob Agents Chemother 4:415–420. doi: 10.1128/aac.4.4.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Lajudie P, Roquet F, Reynier M, Adamowicz P. 1974. Etude d’un nouvel antibactérien de synthesé: l’acyde éthyl-8 oxo-5 pipérazinyl-2 dihydro-5,8 pyrido [2,3-d]pyrimidine-6 carboxylique (ácide pipemidique). C R Acad Sci Hebd Seances Acad Sci D 279:1931–1934. [PubMed] [Google Scholar]
- 35.Ito A, Hirai K, Inoue M, Koga H, Suzue S, Irikura T, Mitsuhashi S. 1980. In vitro antibacterial activity of AM-715, a new nalidixic acid analog. Antimicrob Agents Chemother 17:103–108. doi: 10.1128/aac.17.2.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stilwell G, Holmes K, Turck M. 1975. In vitro evaluation of a new quinolone antibacterial. Antimicrob Agents Chemother 7:483–485. doi: 10.1128/aac.7.4.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruiz J. 2003. Mechanisms of resistance to quinolones: target alterations, decreased accumulation and DNA gyrase protection. J Antimicrob Chemother 51:1109–1117. doi: 10.1093/jac/dkg222. [DOI] [PubMed] [Google Scholar]
- 38.Fung-Tomc JC, Minassian B, Kolek B, Huczko E, Aleksunes L, Stickle T, Washo T, Gradelski E, Valera L, Bonner DP. 2000. Antibacterial spectrum of a novel des-fluoro(6) quinolone, BMS-284756. Antimicrob Agents Chemother 44:3351–3356. doi: 10.1128/AAC.44.12.3351-3356.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gropper S, Albareda N, Chelius K, Kruger D, Mitha I, Vahed Y, Gani M, García-Alonso F, Ozenoxacin in Impetigo Trial Investigators Group. 2014. Ozenoxacin 1% cream in the treatment of impetigo: a multicenter, randomized, placebo- and retapamulin-controlled clinical trial. Future Microbiol 9:1013–1023. doi: 10.2217/fmb.14.78. [DOI] [PubMed] [Google Scholar]
- 40.Poole RM. 2014. Nemonoxacin: first global approval. Drugs 74:1445–1453. doi: 10.1007/s40265-014-0270-0. [DOI] [PubMed] [Google Scholar]
- 41.Roychoudhury S, Ledoussal B. 2002. Non-fluorinated quinolones (NFQs): new antibacterials with unique properties against quinolone-resistant gram-positive pathogens. Curr Drug Targets Infect Disord 2:51–65. doi: 10.2174/1568005024605891. [DOI] [PubMed] [Google Scholar]
- 42.Wiles JA, Bradbury BJ, Pucci MJ. 2010. New quinolone antibiotics: a survey of the literature from 2005 to 2010. Expert Opin Ther Pat 20:1295–1319. doi: 10.1517/13543776.2010.505922. [DOI] [PubMed] [Google Scholar]
- 43.Slayton ET, Hay AS, Babcock CK, Long TE. 2016. New antibiotics in clinical trials for Clostridium difficile. Expert Rev Anti Infect Ther 14:789–800. doi: 10.1080/14787210.2016.1211931. [DOI] [PubMed] [Google Scholar]
- 44.Klopman G, Wang S, Jacobs MR, Bajaksouzian S, Edmonds K, Ellner JJ. 1993. Anti-Mycobacterium avium activity of quinolones: in vitro activities. Antimicrob Agents Chemother 37:1799–1806. doi: 10.1128/aac.37.9.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mandell L, Tillotson G. 2002. Safety of fluoroquinolones: an update. Can J Infect Dis 13:54–61. doi: 10.1155/2002/864789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alksne L. 2003. Balofloxacin Choongwae. Curr Opin Investig Drugs 4:224–229. [PubMed] [Google Scholar]
- 47.Panda BK, Pawar A, Patel C, Marne SR, Singh P. 2012. Balofloxacin (Q35), new fluoroquinolone as an anti-infective: a systematic review. Am J Pharmtech Res 2:287–303. [Google Scholar]
- 48.King DE, Malone R, Lilley SH. 2000. New classification and update on the quinolone antibiotics. Am Fam Physician 61:2741–2748. [PubMed] [Google Scholar]
- 49.Naber K, Adam D. 1998. Classification of fluoroquinolones. Int J Antimicrob Agents 10:255–257. doi: 10.1016/S0924-8579(98)00059-4. [DOI] [PubMed] [Google Scholar]
- 50.Flamm RK, Vojtko C, Chu DT, Li Q, Beyer J, Hensey D, Ramer N, Clement JJ, Tanaka SK. 1995. In vitro evaluation of ABT-719, a novel DNA gyrase inhibitor. Antimicrob Agents Chemother 39:964–970. doi: 10.1128/aac.39.4.964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alonso D, Muñoz J, Ruiz J, Carmona F, Nadal A, Gascón J. 2007. Salmonella ovarian abscess following travel diarrhoea episode. Arch Gynecol Obstet 276:551–553. doi: 10.1007/s00404-007-0380-y. [DOI] [PubMed] [Google Scholar]
- 52.Horcajada JP, Vila J, Moreno-Martínez A, Ruiz J, Martínez JA, Sánchez M, Soriano E, Mensa J. 2002. Molecular epidemiology and evolution of resistance to quinolones in Escherichia coli after prolonged administration of ciprofloxacin in patients with prostatitis. J Antimicrob Chemother 49:55–59. doi: 10.1093/jac/49.1.55. [DOI] [PubMed] [Google Scholar]
- 53.Kohlhoff SA, Hammerschlag MR. 2015. Treatment of chlamydial infections: 2014 update. Expert Opin Pharmacother 16:205–212. doi: 10.1517/14656566.2015.999041. [DOI] [PubMed] [Google Scholar]
- 54.Mogle BT, Steele JM, Thomas SJ, Bohan KH, Kufel WD. 2018. Clinical review of delafloxacin: a novel anionic fluoroquinolone. J Antimicrob Chemother 73:1439–1451. doi: 10.1093/jac/dkx543. [DOI] [PubMed] [Google Scholar]
- 55.Ruiz J, Marco F, Oliveira I, Vila J, Gascón J. 2007. Trends in antimicrobial resistance in Campylobacter spp. causing traveler’s diarrhea. APMIS 115:218–224. doi: 10.1111/j.1600-0463.2007.apm_567.x. [DOI] [PubMed] [Google Scholar]
- 56.Wang J, Xu H, Wang D, Li M. 2017. Comparison of pathogen eradication rate and safety of anti-bacterial agents for bronchitis: a network meta-analysis. J Cell Biochem 118:3171–3183. doi: 10.1002/jcb.25951. [DOI] [PubMed] [Google Scholar]
- 57.Copeland V, McLaughlin M, Trifilio S. 2018. Ciprofloxacin vs levofloxacin for prophylaxis during hematopoietic stem-cell transplantation. Clin Transplant 32:e13145. doi: 10.1111/ctr.13145. [DOI] [PubMed] [Google Scholar]
- 58.Freifeld AG, Bow EJ, Sepkowitz KA, Boeckh MJ, Ito JI, Mullen CA, Raad II, Rolston KV, Young JA, Wingard JR. 2011. Clinical practice guideline for the use of antimicrobial agents in neutropenic patients with cancer: 2010 update by the Infectious Diseases Society of America. Clin Infect Dis 52:e56–e93. doi: 10.1093/cid/cir073. [DOI] [PubMed] [Google Scholar]
- 59.Haripriya A, Chang DF, Ravindran RD. 2017. Endophthalmitis reduction with intracameral moxifloxacin prophylaxis: analysis of 600 000 surgeries. Ophthalmology 124:768–775. doi: 10.1016/j.ophtha.2017.01.026. [DOI] [PubMed] [Google Scholar]
- 60.Gustinetti G, Mikulska M. 2016. Bloodstream infections in neutropenic cancer patients: a practical update. Virulence 7:280–297. doi: 10.1080/21505594.2016.1156821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Humar A, Gill J, Johnston O, Fergusson D, House AA, Lebel L, Cockfield S, Kim SJ, Zaltzman J, Cantarovich M, Karpinski M, Ramsay T, Knoll GA. 2013. Quinolone prophylaxis for the prevention of BK virus infection in kidney transplantation: study protocol for a randomized controlled trial. Trials 14:185. doi: 10.1186/1745-6215-14-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thevenot T, Degand T, Grelat N, Elkrief L, Christol C, Moreau R, Henrion J, Cadranel JF, Sheppard F, Bureau C, di Martino V, Pauwels A, National Association of General Hospital Hepatogastroenterologists. 2013. A French national survey on the use of antibiotic prophylaxis in cirrhotic patients. Liver Int 33:389–397. doi: 10.1111/liv.12093. [DOI] [PubMed] [Google Scholar]
- 63.Knoll G, Humar A, Fergusson D, Johnston O, House AA, Kim SJ, Ramsay T, Chassé M, Pang X, Zaltzman J, Cockfield S, Cantarovich M, Karpinski M, Lebel L, Gill JS. 2014. Levofloxacin for BK virus prophylaxis following kidney transplantation: a randomized clinical trial. JAMA 312:2106–2114. doi: 10.1001/jama.2014.14721. [DOI] [PubMed] [Google Scholar]
- 64.Mikulska M, Cordonnier C. 2018. Fluoroquinolone prophylaxis during neutropenia: what can we expect nowadays? Clin Microbiol Infect 24:678–679. doi: 10.1016/j.cmi.2018.02.031. [DOI] [PubMed] [Google Scholar]
- 65.Song TR, Rao ZS, Qiu Y, Liu JP, Huang ZL, Wang XD, Lin T. 2016. Fluoroquinolone prophylaxis in preventing BK polyomavirus infection after renal transplant: a systematic review and meta-analysis. Kaohsiung J Med Sci 32:152–159. doi: 10.1016/j.kjms.2016.01.004. [DOI] [PubMed] [Google Scholar]
- 66.Beteck RM, Smit FJ, Haynes RK, N’Da DD. 2014. Recent progress in the development of anti-malarial quinolones. Malar J 13:339. doi: 10.1186/1475-2875-13-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dalhoff A. 2015. Antiviral, antifungal, and antiparasitic activities of fluoroquinolones optimized for treatment of bacterial infections: a puzzling paradox or a logical consequence of their mode of action? Eur J Clin Microbiol Infect Dis 34:661–668. doi: 10.1007/s10096-014-2296-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gaillard T, Madamet M, Tsombeng FF, Dormoi J, Pradines B. 2016. Antibiotics in malaria therapy: which antibiotics except tetracyclines and macrolides may be used against malaria? Malar J 15:556. doi: 10.1186/s12936-016-1613-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Montazeri M, Sharif M, Sarvi S, Mehrzadi S, Ahmadpour E, Daryani A. 2017. A systematic review of in vitro and in vivo activities of anti-Toxoplasma drugs and compounds (2006-2016). Front Microbiol 8:25. doi: 10.3389/fmicb.2017.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Romero IC, Saravia NG, Walker J. 2005. Selective action of fluoroquinolones against intracellular amastigotes of Leishmania (Viannia) panamensis in vitro. J Parasitol 91:1474–1479. doi: 10.1645/GE-3489.1. [DOI] [PubMed] [Google Scholar]
- 71.Leyva-Ramos S, Hernández-López H. 2017. Fluoroquinolonas: perspectivas no antibacterianas. Rev Esp Quimioter 30:1–8. [PubMed] [Google Scholar]
- 72.Stergiopoulou T, Meletiadis J, Sein T, Papaioannidou P, Tsiouris I, Roilides E, Walsh TJ. 2009. Comparative pharmacodynamic interaction analysis between ciprofloxacin, moxifloxacin and levofloxacin and antifungal agents against Candida albicans and Aspergillus fumigatus. J Antimicrob Chemother 63:343–348. doi: 10.1093/jac/dkn473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jeffers-Francis LK, Burger-Calderon R, Webster-Cyriaque J. 2015. Effect of leflunomide, cidofovir and ciprofloxacin on replication of BKPyV in a salivary gland in vitro culture system. Antiviral Res 118:46–55. doi: 10.1016/j.antiviral.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Khan IA, Siddiqui S, Rehmani S, Kazmi SU, Ali SH. 2012. Fluoroquinolones inhibit HCV by targeting its helicase. Antivir Ther 17:467–476. doi: 10.3851/IMP1937. [DOI] [PubMed] [Google Scholar]
- 75.Yamaya M, Nishimura H, Hatachi Y, Yasuda H, Deng X, Sasaki T, Mizuta K, Kubo H, Nagatomi R. 2012. Levofloxacin inhibits rhinovirus infection in primary cultures of human tracheal epithelial cells. Antimicrob Agents Chemother 56:4052–4061. doi: 10.1128/AAC.00259-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dryden C, Wilkinson J, Young D, Brooker RJ, Scottish Paediatric Cystic Fibrosis Managed Clinical Network. 2018. The impact of 12 months treatment with ivacaftor on Scottish paediatric patients with cystic fibrosis with the G551D mutation: a review. Arch Dis Child 103:68–70. doi: 10.1136/archdischild-2015-310420. [DOI] [PubMed] [Google Scholar]
- 77.Psichogiou M, Poulakou G, Basoulis D, Paraskevis D, Markogiannakis A, Daikos GL. 2017. Recent advances in antiretroviral agents: potent integrase inhibitors. Curr Pharm Des 23:2552–2567. doi: 10.2174/1381612823666170329142059. [DOI] [PubMed] [Google Scholar]
- 78.Sato M, Motomura T, Aramaki H, Matsuda T, Yamashita M, Ito Y, Kawakami H, Matsuzaki Y, Watanabe W, Yamataka K, Ikeda S, Kodama E, Matsuoka M, Shinkai H. 2006. Novel HIV-1 integrase inhibitors derived from quinolone antibiotics. J Med Chem 49:1506–1508. doi: 10.1021/jm0600139. [DOI] [PubMed] [Google Scholar]
- 79.Kan JY, Hsu YL, Chen YH, Chen TC, Wang JY, Kuo PL. 2013. Gemifloxacin, a fluoroquinolone antimicrobial drug, inhibits migration and invasion of human colon cancer cells. Biomed Res Int 2013:159786. doi: 10.1155/2013/159786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sayar H, Bashardoust P. 2017. Therapies for acute myeloid leukemia: vosaroxin. Onco Targets Ther 10:3957–3963. doi: 10.2147/OTT.S121477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Acar JF, Goldstein FW. 1997. Trends in bacterial resistance to fluoroquinolones. Clin Infect Dis 24(Suppl 1):S67–S73. doi: 10.1093/clinids/24.Supplement_1.S67. [DOI] [PubMed] [Google Scholar]
- 82.Castanon JIR. 2007. History of the use of antibiotic as growth promoters in European poultry feeds. Poult Sci 86:2466–2471. doi: 10.3382/ps.2007-00249. [DOI] [PubMed] [Google Scholar]
- 83.Anonymous. 2005. Ban on antibiotics as growth promoters in animal feed enters into effect, European Commission—IP/05/1687. European Commission, Brussels, Belgium: http://europa.eu/rapid/press-release_IP-05-1687_en.htm. [Google Scholar]
- 84.Om C, McLaws ML. 2016. Antibiotics: practice and opinions of Cambodian commercial farmers, animal feed retailers and veterinarians. Antimicrob Resist Infect Control 5:42. doi: 10.1186/s13756-016-0147-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Trouchon T, Lefebvre S. 2016. A review of enrofloxacin for veterinary use. Open J Vet Med 6:40–58. doi: 10.4236/ojvm.2016.62006. [DOI] [Google Scholar]
- 86.De Sarro A, De Sarro G. 2001. Adverse reactions to fluoroquinolones. An overview on mechanistic aspects. Curr Med Chem 8:371–384. doi: 10.2174/0929867013373435. [DOI] [PubMed] [Google Scholar]
- 87.Owens RC Jr, Ambrose PG. 2005. Antimicrobial safety: focus on fluoroquinolones. Clin Infect Dis 41(Suppl 2):S144–S157. doi: 10.1086/428055. [DOI] [PubMed] [Google Scholar]
- 88.Grady RW. 2005. Systemic quinolone antibiotics in children: a review of the use and safety. Expert Opin Drug Saf 4:623–630. doi: 10.1517/14740338.4.4.623. [DOI] [PubMed] [Google Scholar]
- 89.Dawe RS, Ibbotson SH, Sanderson JB, Thomson EM, Ferguson J. 2003. A randomized controlled trial (volunteer study) of sitafloxacin, enoxacin, levofloxacin and sparfloxacin phototoxicity. Br J Dermatol 149:1232–1241. doi: 10.1111/j.1365-2133.2003.05582.x. [DOI] [PubMed] [Google Scholar]
- 90.Rubinstein E. 2001. History of quinolones and their side effects. Chemotherapy 47(Suppl 3):3–8. doi: 10.1159/000057838. [DOI] [PubMed] [Google Scholar]
- 91.Thamlikitkul V, Tiengrim S. 2014. In vitro susceptibility test of sitafloxacin against resistant gram-negative bacilli isolated from Thai patients by disk diffusion method. J Med Assoc Thai 97(Suppl 3):S7–S12. [PubMed] [Google Scholar]
- 92.Lipsky BA, Baker CA. 1999. Fluoroquinolone toxicity profiles: a review focusing on newer agents. Clin Infect Dis 28:352–364. doi: 10.1086/515104. [DOI] [PubMed] [Google Scholar]
- 93.Kaguelidou F, Turner MA, Choonara I, Jacqz-Aigrain E. 2011. Ciprofloxacin use in neonates: a systematic review of the literature. Pediatr Infect Dis J 30:e29–e37. doi: 10.1097/INF.0b013e3181fe353d. [DOI] [PubMed] [Google Scholar]
- 94.Okoye NV, Oyawole MR, Uzochukwu PU, Oyetunde OO. 2013. Review of ciprofloxacin use in children. Nig Q J Hosp Med 23:43–47. [PubMed] [Google Scholar]
- 95.Patel K, Goldman JL. 2016. Safety concerns surrounding quinolone use in children. J Clin Pharmacol 56:1060–1075. doi: 10.1002/jcph.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Principi N, Esposito S. 2015. Appropriate use of fluoroquinolones in children. Int J Antimicrob Agents 45:341–346. doi: 10.1016/j.ijantimicag.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 97.Ouchi K, Sunakawa K. 2014. Effect of new oral antimicrobial agents in outpatient treatment of pneumonia in children. Jpn J Antibiot 67:157–166. [PubMed] [Google Scholar]
- 98.Tanaka E, Hara N, Wajima T, Ochiai S, Seyama S, Shirai A, Shibata M, Shiro H, Natsume Y, Noguchi N. 2019. Emergence of Haemophilus influenzae with low susceptibility to quinolones and persistence in tosufloxacin treatment. J Glob Antimicrob Resist 18:104–108. doi: 10.1016/j.jgar.2019.01.017. [DOI] [PubMed] [Google Scholar]
- 99.Ziv A, Masarwa R, Perlman A, Ziv D, Matok I. 2018. Pregnancy outcomes following exposure to quinolone antibiotics—a systematic-review and meta-analysis. Pharm Res 35:109. doi: 10.1007/s11095-018-2383-8. [DOI] [PubMed] [Google Scholar]
- 100.Frankel WC, Trautner BW, Spiegelman A, Grigoryan L, LeMaire SA. 2019. Patients at risk for aortic rupture often exposed to fluoroquinolones during hospitalization. Antimicrob Agents Chemother 63:e01712-18. doi: 10.1128/AAC.01712-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pasternak B, Inghammar M, Svanström H. 2018. Fluoroquinolone use and risk of aortic aneurysm and dissection: nationwide cohort study. BMJ 360:k678. doi: 10.1136/bmj.k678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.LeMaire SA, Zhang L, Luo W, Ren P, Azares AR, Wang Y, Zhang C, Coselli JS, Shen YH. 2018. Effect of ciprofloxacin on susceptibility to aortic dissection and rupture in mice. JAMA Surg 153:e181804. doi: 10.1001/jamasurg.2018.1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Manfredi M, Severino M, Testi S, Macchia D, Ermini G, Pichler WJ, Campi P. 2004. Detection of specific IgE to quinolones. J Allergy Clin Immunol 113:155–160. doi: 10.1016/j.jaci.2003.09.035. [DOI] [PubMed] [Google Scholar]
- 104.Doña I, Blanca-López N, Torres MJ, García-Campos J, García-Núñez I, Gómez F, Salas M, Rondón C, Canto MG, Blanca M. 2012. Drug hypersensitivity reactions: response patterns, drug involved, and temporal variations in a large series of patients. J Investig Allergol Clin Immunol 22:363–371. doi: 10.18176/jiaci.0078. [DOI] [PubMed] [Google Scholar]
- 105.Aranda A, Mayorga C, Ariza A, Doña I, Rosado A, Blanca-Lopez N, Andreu I, Torres MJ. 2011. In vitro evaluation of IgE-mediated hypersensitivity reactions to quinolones. Allergy 66:247–254. doi: 10.1111/j.1398-9995.2010.02460.x. [DOI] [PubMed] [Google Scholar]
- 106.Blanca-López N, Ariza A, Doña I, Mayorga C, Montañez MI, Garcia-Campos J, Gomez F, Rondón C, Blanca M, Torres MJ. 2013. Hypersensitivity reactions to fluoroquinolones: analysis of the factors involved. Clin Exp Allergy 43:560–567. doi: 10.1111/cea.12099. [DOI] [PubMed] [Google Scholar]
- 107.Sachs B, Riegel S, Seebeck J, Beier R, Schichler D, Barger A, Merk HF, Erdmann S. 2006. Fluoroquinolone-associated anaphylaxis in spontaneous adverse drug reaction reports in Germany: differences in reporting rates between individual fluoroquinolones and occurrence after first-ever use. Drug Saf 229:1087–1100. doi: 10.2165/00002018-200629110-00008. [DOI] [PubMed] [Google Scholar]
- 108.Peters B, Pinching AJ. 1989. Fatal anaphylaxis associated with ciprofloxacin in a patient with AIDS related complex. BMJ 298:605. doi: 10.1136/bmj.298.6673.605-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jespersen HG, Stenderup A. 1965. Treatment of urinary tract infections in the aged with nalidixic acid. Gerontol Clin 7:303–310. doi: 10.1159/000244913. [DOI] [PubMed] [Google Scholar]
- 110.Barry AL, Jones RN, Thornsberry C, Ayers LW, Gerlach EH, Sommers HM. 1984. Antibacterial activities of ciprofloxacin, norfloxacin, oxolinic acid, cinoxacin, and nalidixic acid. Antimicrob Agents Chemother 25:633–637. doi: 10.1128/aac.25.5.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hoge CW, Gambel JM, Srijan A, Pitarangsi C, Echeverria P. 1998. Trends in antibiotic resistance among diarrheal pathogens isolated in Thailand over 15 years. Clin Infect Dis 26:341–345. doi: 10.1086/516303. [DOI] [PubMed] [Google Scholar]
- 112.Rudrik JT, Cavalieri SJ, Britt EM. 1984. In vitro activities of enoxacin and 17 other antimicrobial agents against multiply resistant, gram-negative bacteria. Antimicrob Agents Chemother 26:97–100. doi: 10.1128/aac.26.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dalhoff A. 2012. Global fluoroquinolone resistance epidemiology and implications for clinical use. Interdiscip Perspect Infect Dis 2012:976273. doi: 10.1155/2012/976273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Jones RN. 1996. Impact of changing pathogens and antimicrobial susceptibility patterns in the treatment of serious infections in hospitalized patients. Am J Med 100(6A):3S–12S. doi: 10.1016/s0002-9343(96)00102-7. [DOI] [PubMed] [Google Scholar]
- 115.Navia MM, Ruiz J, Ribera A, Jiménez de Anta MT, Vila J. 1999. Analysis of the mechanisms of quinolone resistance in clinical isolates of Citrobacter freundii. J Antimicrob Chemother 44:743–748. doi: 10.1093/jac/44.6.743. [DOI] [PubMed] [Google Scholar]
- 116.Rautelin H, Renkonen OV, Kosunen TU. 1991. Emergence of fluoroquinolone resistance in Campylobacter jejuni and Campylobacter coli in subjects from Finland. Antimicrob Agents Chemother 35:2065–2069. doi: 10.1128/aac.35.10.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Reina J, Ros MJ, Muñoz-López A. 1996. Aislamiento de una cepa de Campylobacter jejuni con resistencia a las fluoroquinolonas y sensibilidad al ácido nalídixico. Enferm Infecc Microbiol Clin 14:124–125. [PubMed] [Google Scholar]
- 118.Ruiz J, Castro D, Goñi P, Santamaria JA, Borrego JJ, Vila J. 1997. Analysis of the mechanism of quinolone-resistance in nalidixic acid-resistant clinical isolates of Salmonella serotype Typhimurium. J Med Microbiol 46:623–628. doi: 10.1099/00222615-46-7-623. [DOI] [PubMed] [Google Scholar]
- 119.Ruiz J, Marco F, Goñi P, Gallardo F, Mensa J, Trilla A, Jiménez de Anta T, Vila J. 1995. High frequency of mutations at codon 83 of the gyrA gene of quinolone-resistant clinical isolates of Escherichia coli. J Antimicrob Chemother 36:737–738. doi: 10.1093/jac/36.4.737. [DOI] [PubMed] [Google Scholar]
- 120.Endtz HP, Ruijs GJ, van Klingeren B, Jansen WH, van der Reyden T, Mouton RP. 1991. Quinolone resistance in Campylobacter isolated from man and poultry following the introduction of fluoroquinolones in veterinary medicine. J Antimicrob Chemother 27:199–208. doi: 10.1093/jac/27.2.199. [DOI] [PubMed] [Google Scholar]
- 121.Mirelis B, Miro E, Navarro F, Ogalla CA, Bonal J, Prats G. 1993. Increased resistance to quinolone in Catalonia, Spain. Diagn Microbiol Infect Dis 16:137–139. doi: 10.1016/0732-8893(93)90009-V. [DOI] [PubMed] [Google Scholar]
- 122.Ruiz J, Goñi P, Marco F, Gallardo F, Mirelis B, Jiménez de Anta T, Vila J. 1998. Increased resistance to quinolones in Campylobacter jejuni: a genetic analysis of gyrA gene mutations in quinolone-resistant clinical isolates. Microbiol Immunol 42:223–226. doi: 10.1111/j.1348-0421.1998.tb02274.x. [DOI] [PubMed] [Google Scholar]
- 123.Deguchi T, Saito I, Tanaka M, Sato K, Deguchi K, Yasuda M, Nakano M, Nishino Y, Kanematsu E, Ozeki S, Kawada Y. 1997. Fluoroquinolone treatment failure in gonorrhea. Emergence of a Neisseria gonorrhoeae strain with enhanced resistance to fluoroquinolones. Sex Transm Dis 24:247–250. doi: 10.1097/00007435-199705000-00002. [DOI] [PubMed] [Google Scholar]
- 124.Le Lostec Z, Fegueux S, Jouve P, Cheron M, Mornet P, Boisivon A. 1997. Reduced susceptibility to quinolones in Salmonella typhi acquired in Europe: a clinical failure of treatment. Clin Microbiol Infect 3:576–577. doi: 10.1111/j.1469-0691.1997.tb00313.x. [DOI] [PubMed] [Google Scholar]
- 125.Vasallo FJ, Martín-Rabadán P, Alcalá L, García-Lechuz JM, Rodríguez-Créixems M, Bouza E. 1998. Failure of ciprofloxacin therapy for invasive nontyphoidal salmonellosis. Clin Infect Dis 126:535–536. doi: 10.1086/517087. [DOI] [PubMed] [Google Scholar]
- 126.Vila J, Olmos L, Ballesteros J, Vázquez JA, Giménez MJ, Marco F, Aguilar L. 1997. Development of in-vivo resistance after quinolone treatment of gonococcal urethritis. J Antimicrob Chemother 39:841. doi: 10.1093/jac/39.6.841a. [DOI] [PubMed] [Google Scholar]
- 127.Vila J, Ruiz J, Sanchez F, Navarro F, Mirelis B, Jiménez de Anta MT, Prats G. 1999. Increase in quinolone resistance in a Haemophilus influenzae strain isolated from a patient with recurrent respiratory infections treated with ofloxacin. Antimicrob Agents Chemother 43:161–162. doi: 10.1128/AAC.43.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Llor C, Bjerrum L. 2014. Antimicrobial resistance: risk associated with antibiotic overuse and initiatives to reduce the problem. Ther Adv Drug Saf 5:229–241. doi: 10.1177/2042098614554919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ruiz J, Pons MJ. 2013. Prevention of travellers’ diarrhoea: where and who? Lancet Infect Dis 13:911–912. doi: 10.1016/S1473-3099(13)70243-3. [DOI] [PubMed] [Google Scholar]
- 130.US Centers for Diseases Control and Prevention. 2013. Antibiotic resistance threats in the United States, 2013. US Department of Health and Human Services, Centers for Disease Control and Prevention, Atlanta, GA. [Google Scholar]
- 131.Gavaldà L, Soriano AM, Cámara J, Gasull R, Arch O, Ferrer M, Shaw E, Granada RM, Dominguez MA, Pujol M. 2016. Control of endemic extensively drug-resistant Acinetobacter baumannii with a cohorting policy and cleaning procedures based on the 1 room, 1 wipe approach. Am J Infect Control 44:520–524. doi: 10.1016/j.ajic.2015.11.036. [DOI] [PubMed] [Google Scholar]
- 132.Saam M, Huttner B, Harbarth S. 2017. Evaluation of antibiotic awareness campaigns. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 133.de Toro M, Rojo-Bezares B, Vinué L, Undabeitia E, Torres C, Sáenz Y. 2010. In vivo selection of aac(6′)-Ib-cr and mutations in the gyrA gene in a clinical qnrS1-positive Salmonella enterica serovar Typhimurium DT104B strain recovered after fluoroquinolone treatment. J Antimicrob Chemother 65:1945–1949. doi: 10.1093/jac/dkq262. [DOI] [PubMed] [Google Scholar]
- 134.Gao L, Lyu Y, Li Y. 2017. Trends in drug resistance of Acinetobacter baumannii over a 10-year period: nationwide data from the China surveillance of antimicrobial resistance program. Chin Med J 130:659–664. doi: 10.4103/0366-6999.201601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Horna G, Quezada K, Ramos S, Mosqueda N, Rubio M, Guerra H, Ruiz J. 2019. Specific type IV pili groups in clinical isolates of Pseudomonas aeruginosa. Int Microbiol 22:131–141. doi: 10.1007/s10123-018-00035-3. [DOI] [PubMed] [Google Scholar]
- 136.Mendez Arancibia E, Pitart C, Ruiz J, Marco F, Gascón J, Vila J. 2009. Evolution of antimicrobial resistance in enteroaggregative Escherichia coli and enterotoxigenic Escherichia coli causing traveller’s diarrhoea. J Antimicrob Chemother 64:343–347. doi: 10.1093/jac/dkp178. [DOI] [PubMed] [Google Scholar]
- 137.Pons MJ, Gomes C, Martínez-Puchol S, Ruiz L, Mensa L, Vila J, Gascón J, Ruiz J. 2013. Antimicrobial resistance in Shigella spp. causing traveller’s diarrhoea (1995-2010): a retrospective analysis. Travel Med Infect Dis 5:315–319. doi: 10.1016/j.tmaid.2013.06.010. [DOI] [PubMed] [Google Scholar]
- 138.Pons MJ, Mosquito S, Gomes C, del Valle LJ, Ochoa TJ, Ruiz J. 2014. Analysis of quinolone-resistance in commensal and diarrheagenic Escherichia coli isolates from infants in Lima, Peru. Trans R Soc Trop Med Hyg 108:22–28. doi: 10.1093/trstmh/trt106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Salazar de Vegas EZ, Nieves B, Ruiz M, Ruíz J, Vila J, Araque M, Velázco E. 2007. Molecular epidemiology and characterization of resistance mechanisms to various antimicrobial agents in Acinetobacter baumannii isolated in Mérida, Venezuela. Med Sci Monit 13:BR89–BR94. [PubMed] [Google Scholar]
- 140.WHO Western Pacific Gonococcal Antimicrobial Surveillance Programme. 2006. Surveillance of antibiotic resistance in Neisseria gonorrhoeae in the WHO Western Pacific Region, 2005. Commun Dis Intell Q Rep 30:430–433. [PubMed] [Google Scholar]
- 141.Guiral E, Pons MJ, Vubil D, Marí M, Sigauque B, Soto SM, Alonso PL, Ruiz J, Vila J, Mandomando I. 2018. Epidemiology and molecular characterization of multidrug-resistant Escherichia coli isolates harboring blaCTX-M group 1 extended-spectrum β-lactamases causing bacteremia and urinary tract infection in Manhiça, Mozambique. Infect Drug Resist 11:927–936. doi: 10.2147/IDR.S153601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Pallecchi L, Bartoloni A, Riccobono E, Fernandez C, Mantella A, Magnelli D, Mannini D, Strohmeyer M, Bartalesi F, Rodriguez H, Gotuzzo E, Rossolini GM. 2012. Quinolone resistance in absence of selective pressure: the experience of a very remote community in the Amazon forest. PLoS Negl Trop Dis 6:e1790. doi: 10.1371/journal.pntd.0001790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Pons MJ, Vubil D, Guiral E, Jaintilal D, Fraile O, Soto SM, Sigauque B, Nhampossa T, Aide P, Alonso PL, Vila J, Mandomando I, Ruiz J. 2015. Characterisation of extended spectrum β-lactamases among Klebsiella pneumoniae isolates causing bacteraemia and urinary tract infection in Mozambique. J Glob Antimicrob Resist 3:19–25. doi: 10.1016/j.jgar.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 144.Ahmed A, Bashir T. 2015. Trends in quinolone resistance among common urinary tract isolates over three years. J Coll Physicians Surg Pak 25:543–545. [PubMed] [Google Scholar]
- 145.Lahra MM, Australian Gonococcal Surveillance Programme. 2015. Australian Gonococcal Surveillance Programme annual report, 2014. Commun Dis Intell Q Rep 39:E347–E354. [PubMed] [Google Scholar]
- 146.Lluque A, Riveros M, Prada A, Ochoa TJ, Ruiz J. 2017. Campylobacter spp. from a Peruvian pediatric cohort. Scientifica (Cairo) 2017:7848926. doi: 10.1155/2017/7848926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Palma N, Pons MJ, Gomes C, Mateu J, Riveros M, García W, Jacobs J, García C, Ochoa TJ, Ruiz J. 2017. Resistance to quinolones, cephalosporins and macrolides in Escherichia coli causing bacteremia in Peruvian children. J Glob Antimicrob Resist 11:28–33. doi: 10.1016/j.jgar.2017.06.011. [DOI] [PubMed] [Google Scholar]
- 148.Sáenz Y, Ruiz J, Zarazaga M, Teixidó M, Torres C, Vila J. 2004. Effect of the efflux pump inhibitor Phe-Arg-β-naphthylamide on the MIC values of the quinolones, tetracycline and chloramphenicol, in Escherichia coli isolates of different origin. J Antimicrob Chemother 53:544–545. doi: 10.1093/jac/dkh117. [DOI] [PubMed] [Google Scholar]
- 149.Ince D, Hooper DC. 2003. Quinolone resistance due to reduced target enzyme expression. J Bacteriol 185:6883–6892. doi: 10.1128/JB.185.23.6883-6892.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Abdi-Ali A, Mohammadi-Mehr M, Agha Alaei Y. 2006. Bactericidal activity of various antibiotics against biofilm-producing Pseudomonas aeruginosa. Int J Antimicrob Agents 27:196–200. doi: 10.1016/j.ijantimicag.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 151.Blango MG, Mulvey MA. 2010. Persistence of uropathogenic Escherichia coli in the face of multiple antibiotics. Antimicrob Agents Chemother 54:1855–1863. doi: 10.1128/AAC.00014-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Crumplin GC, Smith JT. 1975. Nalidixic acid: an antibacterial paradox. Antimicrob Agents Chemother 8:251–261. doi: 10.1128/aac.8.3.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Robicsek A, Strahilevitz J, Jacoby GA, Macielag M, Abbanat D, Park CH, Bush K, Hooper DC. 2006. Fluoroquinolone-modifying enzyme: a new adaptation of a common aminoglycoside acetyltransferase. Nat Med 12:83–88. doi: 10.1038/nm1347. [DOI] [PubMed] [Google Scholar]
- 154.Chávez-Jacobo VM, Hernández-Ramírez KC, Romo-Rodríguez P, Pérez-Gallardo RV, Campos-García J, Gutiérrez-Corona JF, García-Merinos JP, Meza-Carmen V, Silva-Sánchez J, Ramírez-Díaz MI. 2018. CrpP is a novel ciprofloxacin-modifying enzyme encoded by the Pseudomonas aeruginosa pUM505 plasmid. Antimicrob Agents Chemother 62:e02629-17. doi: 10.1128/AAC.02629-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Sørensen AH, Hansen LH, Johannesen E, Sørensen SJ. 2003. Conjugative plasmid conferring resistance to olaquindox. Antimicrob Agents Chemother 47:798–799. doi: 10.1128/AAC.47.2.798-799.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Hansen LH, Johannesen E, Burmølle M, Sørensen AH, Sørensen SJ. 2004. Plasmid-encoded multidrug efflux pump conferring resistance to olaquindox in Escherichia coli. Antimicrob Agents Chemother 48:3332–3337. doi: 10.1128/AAC.48.9.3332-3337.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Hansen LH, Jensen LB, Sørensen HI, Sørensen SJ. 2007. Substrate specificity of the OqxAB multidrug resistance pump in Escherichia coli and selected enteric bacteria. J Antimicrob Chemother 60:145–147. doi: 10.1093/jac/dkm167. [DOI] [PubMed] [Google Scholar]
- 158.Yamane K, Wachino J, Suzuki S, Kimura K, Shibata N, Kato H, Shibayama K, Konda T, Arakawa Y. 2007. New plasmid-mediated fluoroquinolone efflux pump, QepA, found in an Escherichia coli clinical isolate. Antimicrob Agents Chemother 51:3354–3360. doi: 10.1128/AAC.00339-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Périchon B, Courvalin P, Galimand M. 2007. Transferable resistance to aminoglycosides by methylation of G1405 in 16S rRNA and to hydrophilic fluoroquinolones by QepA-mediated efflux in Escherichia coli. Antimicrob Agents Chemother 51:2464–2469. doi: 10.1128/AAC.00143-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ruiz J, Pons MJ, Gomes C. 2012. Transferable mechanisms of quinolone resistance. Int J Antimicrob Agents 40:196–203. doi: 10.1016/j.ijantimicag.2012.02.011. [DOI] [PubMed] [Google Scholar]
- 161.Aviv G, Rahav G, Gal-Mor O. 2016. Horizontal transfer of the Salmonella enterica serovar Infantis resistance and virulence plasmid pESI to the gut microbiota of warm-blooded hosts. mBio 7:e01395-16. doi: 10.1128/mBio.01395-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Courvalin P. 1994. Transfer of antibiotic resistance genes between gram-positive and gram-negative bacteria. Antimicrob Agents Chemother 38:1447–1451. doi: 10.1128/aac.38.7.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Alonso A, Sanchez P, Martínez JL. 2000. Stenotrophomonas maltophilia D457R contains a cluster of genes from gram-positive bacteria involved in antibiotic and heavy metal resistance. Antimicrob Agents Chemother 44:1778–1782. doi: 10.1128/AAC.44.7.1778-1782.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Ribera A, Ruiz J, Vila J. 2003. Presence of the Tet M determinant in a clinical isolate of Acinetobacter baumannii. Antimicrob Agents Chemother 47:2310–2312. doi: 10.1128/AAC.47.7.2310-2312.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Gomes C, Martínez-Puchol S, Palma N, Horna G, Ruiz-Roldán L, Pons MJ, Ruiz J. 2017. Macrolide resistance mechanisms in Enterobacteriaceae: focus in azithromycin. Crit Rev Microbiol 43:1–30. doi: 10.3109/1040841X.2015.1136261. [DOI] [PubMed] [Google Scholar]
- 166.Nakaminami H, Noguchi N, Sasatsu M. 2010. Fluoroquinolone efflux by the plasmid-mediated multidrug efflux pump QacB variant QacBIII in Staphylococcus aureus. Antimicrob Agents Chemother 54:4107–4111. doi: 10.1128/AAC.01065-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Osińska A, Harnisz M, Korzeniewska E. 2016. Prevalence of plasmid-mediated multidrug resistance determinants in fluoroquinolone-resistant bacteria isolated from sewage and surface water. Environ Sci Pollut Res Int 23:10818–10831. doi: 10.1007/s11356-016-6221-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Laport MS, Pontes PVM, Dos Santos DS, Santos-Gandelman JDF, Muricy G, Bauwens M, Giambiagi-deMarval M, George I. 2016. Antibiotic resistance genes detected in the marine sponge Petromica citrina from Brazilian coast. Braz J Microbiol 47:617–620. doi: 10.1016/j.bjm.2016.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Yuan L, Zhai YJ, Wu H, Sun HR, He ZP, Wang YB, Pan YS, Kuang NN, Hu GZ. 2018. Identification and prevalence of RND family multidrug efflux pump oqxAB genes in Enterococci isolates from swine manure in China. J Med Microbiol 67:733–739. doi: 10.1099/jmm.0.000736. [DOI] [PubMed] [Google Scholar]
- 170.Robicsek A, Jacoby GA, Hooper DC. 2006. The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect Dis 6:629–640. doi: 10.1016/S1473-3099(06)70599-0. [DOI] [PubMed] [Google Scholar]
- 171.Hata M, Suzuki M, Matsumoto M, Takahashi M, Sato K, Ibe S, Sakae K. 2005. Cloning of a novel gene for quinolone resistance from a transferable plasmid in Shigella flexneri 2b. Antimicrob Agents Chemother 49:801–803. doi: 10.1128/AAC.49.2.801-803.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Poirel L, Rodríguez-Martínez JM, Mammeri H, Liard A, Nordmann P. 2005. Origin of plasmid-mediated quinolone resistance determinant QnrA. Antimicrob Agents Chemother 49:3523–3525. doi: 10.1128/AAC.49.8.3523-3525.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Robicsek A, Sahm DF, Strahilevitz J, Jacoby GA, Hooper DC. 2005. Broader distribution of plasmid-mediated quinolone resistance in the United States. Antimicrob Agents Chemother 49:3001–3003. doi: 10.1128/AAC.49.7.3001-3003.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Saga T, Kaku M, Onodera Y, Yamachika S, Sato K, Takase H. 2005. Vibrio parahaemolyticus chromosomal qnr homologue VPA0095: demonstration by transformation with a mutated gene of its potential to reduce quinolone susceptibility in Escherichia coli. Antimicrob Agents Chemother 49:2144–2145. doi: 10.1128/AAC.49.5.2144-2145.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Fonseca EL, Dos Santos Freitas F, Vieira VV, Vicente ACP. 2008. New qnr gene cassettes associated with superintegron repeats in Vibrio cholerae 01. Emerg Infect Dis 14:1129–1131. doi: 10.3201/eid1407.080132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Albornoz E, Tijet N, De Belder D, Gomez S, Martino F, Corso A, Melano RG, Petroni A. 2017. qnrE1, a member of a new family of plasmid-located quinolone resistance genes, originated from the chromosome of Enterobacter species. Antimicrob Agents Chemother 61:e02555-16. doi: 10.1128/AAC.02555-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Yuan J, Xu X, Guo Q, Zhao X, Ye X, Guo Y, Wang M. 2012. Prevalence of the oqxAB gene complex in Klebsiella pneumoniae and Escherichia coli clinical isolates. J Antimicrob Chemother 67:1655–1659. doi: 10.1093/jac/dks086. [DOI] [PubMed] [Google Scholar]
- 178.Cattoir V, Poirel L, Nordmann P. 2008. Plasmid-mediated quinolone resistance pump QepA2 in an Escherichia coli isolate from France. Antimicrob Agents Chemother 52:3801–3804. doi: 10.1128/AAC.00638-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ruiz J. 2018. In silico analysis of transferable QepA variants and related chromosomal efflux pumps. Rev Esp Quimioter 31:537–541. [PMC free article] [PubMed] [Google Scholar]
- 180.Lyon BR, Skurray R. 1987. Antimicrobial resistance of Staphylococcus aureus: genetic basis. Microbiol Rev 51:88–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Andersen JL, He GX, Kakarla P, KC R, Kumar S, Lakra WS, Mukherjee MM, Ranaweera I, Shrestha U, Tran T, Varela MF. 2015. Multidrug efflux pumps from Enterobacteriaceae, Vibrio cholerae and Staphylococcus aureus bacterial food pathogens. Int J Environ Res Public Health 12:1487–1547. doi: 10.3390/ijerph120201487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Ramírez MS, Tolmasky ME. 2010. Aminoglycoside modifying enzymes. Drug Resist Updat 13:151–171. doi: 10.1016/j.drup.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Foster TJ, Howe TG, Richmond KM. 1975. Translocation of the tetracycline resistance determinant from R100-1 to the Escherichia coli K-12 chromosome. J Bacteriol 124:1153–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Rychlik I, Gregorova D, Hradecka H. 2006. Distribution and function of plasmids in Salmonella enterica. Vet Microbiol 112:1–10. doi: 10.1016/j.vetmic.2005.10.030. [DOI] [PubMed] [Google Scholar]
- 185.Coelho A, González-López JJ, Miró E, Alonso-Tarrés C, Mirelis B, Larrosa MN, Bartolomé RM, Andreu A, Navarro F, Johnson JR, Prats G. 2010. Characterisation of the CTX-M-15-encoding gene in Klebsiella pneumoniae strains from the Barcelona metropolitan area: plasmid diversity and chromosomal integration. Int J Antimicrob Agents 36:73–78. doi: 10.1016/j.ijantimicag.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 186.Fabre L, Delauné A, Espié E, Nygard K, Pardos M, Polomack L, Guesnier F, Galimand M, Lassen J, Weill FX. 2009. Chromosomal integration of the extended-spectrum β-lactamase gene blaCTX-M-15 in Salmonella enterica serotype Concord isolates from internationally adopted children. Antimicrob Agents Chemother 53:1808–1816. doi: 10.1128/AAC.00451-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Ferreira JC, Penha Filho RAC, Andrade LN, Berchieri A Jr, Darini ALC. 2014. Detection of chromosomal blaCTX-M-2 in diverse Escherichia coli isolates from healthy broiler chickens. Clin Microbiol Infect 20:O623–O626. doi: 10.1111/1469-0691.12531. [DOI] [PubMed] [Google Scholar]
- 188.Hamamoto K, Ueda S, Toyosato T, Yamamoto Y, Hirai I. 2016. High prevalence of chromosomal blaCTX-M-14 in Escherichia coli isolates possessing blaCTX-M-14. Antimicrob Agents Chemother 60:2582–2584. doi: 10.1128/AAC.00108-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Harada S, Ishii Y, Saga T, Kouyama Y, Tateda K, Yamaguchi K. 2012. Chromosomal integration and location on IncT plasmids of the blaCTX-M-2 gene in Proteus mirabilis clinical isolates. Antimicrob Agents Chemother 56:1093–1096. doi: 10.1128/AAC.00258-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Hirai I, Fukui N, Taguchi M, Yamauchi K, Nakamura T, Okano S, Yamamoto Y. 2013. Detection of chromosomal blaCTX-M-15 in Escherichia coli O25b-B2-ST131 isolates from the Kinki region of Japan. Int J Antimicrob Agents 42:500–506. doi: 10.1016/j.ijantimicag.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 191.Huang W, Wang G, Sebra R, Zhuge J, Yin C, Aguero-Rosenfeld ME, Schuetz AN, Dimitrova N, Fallon JT. 2017. Emergence and evolution of multidrug-resistant Klebsiella pneumoniae with both blaKPC and blaCTX-M integrated in the chromosome. Antimicrob Agents Chemother 61:e00076-17. doi: 10.1128/AAC.00076-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Mahrouki S, Belhadj O, Chihi H, Mohamed BM, Celenza G, Amicosante G, Perilli M. 2012. Chromosomal blaCTX-M-15 associated with ISEcp1 in Proteus mirabilis and Morganella morganii isolated at the Military Hospital of Tunis, Tunisia. J Med Microbiol 61(Part 9):1286–1289. doi: 10.1099/jmm.0.039487-0. [DOI] [PubMed] [Google Scholar]
- 193.Castanheira M, Deshpande LM, DiPersio JR, Kang J, Weinstein MP, Jones RN. 2009. First descriptions of blaKPC in Raoultella spp. (R. planticola and R. ornithinolytica): report from the SENTRY Antimicrobial Surveillance Program. J Clin Microbiol 47:4129–4130. doi: 10.1128/JCM.01502-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Villegas MV, Lolans K, Correa A, Kattan JN, Lopez JA, Quinn JP, Colombian Nosocomial Resistance Study Group. 2007. First identification of Pseudomonas aeruginosa isolates producing a KPC-type carbapenem-hydrolyzing β-lactamase. Antimicrob Agents Chemother 51:1553–1555. doi: 10.1128/AAC.01405-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Martínez T, Vázquez GJ, Aquino EE, Martínez I, Robledo IE. 2014. ISEcp1-mediated transposition of blaKPC into the chromosome of a clinical isolate of Acinetobacter baumannii from Puerto Rico. J Med Microbiol 63(Part 12):1644–1648. doi: 10.1099/jmm.0.080721-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Toleman MA, Bennett PM, Walsh TR. 2006. Common regions e.g. orf513 and antibiotic resistance: IS91-like elements evolving complex class 1 integrons. J Antimicrob Chemother 58:1–6. doi: 10.1093/jac/dkl204. [DOI] [PubMed] [Google Scholar]
- 197.Ruiz E, Sáenz Y, Zarazaga M, Rocha-Gracia R, Martínez-Martínez L, Arlet G, Torres C. 2012. qnr, aac(6′)-Ib-cr and qepA genes in Escherichia coli and Klebsiella spp.: genetic environments and plasmid and chromosomal location. J Antimicrob Chemother 67:886–897. doi: 10.1093/jac/dkr548. [DOI] [PubMed] [Google Scholar]
- 198.Musumeci R, Rausa M, Giovannoni R, Cialdella A, Bramati S, Sibra B, Giltri G, Viganò F, Cocuzza CE. 2012. Prevalence of plasmid-mediated quinolone resistance genes in uropathogenic Escherichia coli isolated in a teaching hospital of northern Italy. Microb Drug Resist 18:33–41. doi: 10.1089/mdr.2010.0146. [DOI] [PubMed] [Google Scholar]
- 199.Kim HB, Wang M, Park CH, Kim EC, Jacoby GA, Hooper DC. 2009. oqxAB encoding a multidrug efflux pump in human clinical isolates of Enterobacteriaceae. Antimicrob Agents Chemother 53:3582–3584. doi: 10.1128/AAC.01574-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Norman A, Hansen LH, She Q, Sørensen SJ. 2008. Nucleotide sequence of pOLA52: a conjugative IncX1 plasmid from Escherichia coli which enables biofilm formation and multidrug efflux. Plasmid 60:59–74. doi: 10.1016/j.plasmid.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 201.Wong MHY, Chan EWC, Chen S. 2015. Evolution and dissemination of OqxAB-like efflux pumps, an emerging quinolone resistance determinant among members of Enterobacteriaceae. Antimicrob Agents Chemother 59:3290–3297. doi: 10.1128/AAC.00310-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Wong MHY, Chen S. 2013. First detection of oqxAB in Salmonella spp. isolated from food. Antimicrob Agents Chemother 57:658–660. doi: 10.1128/AAC.01144-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Cavaco LM, Hansen DS, Friis-Møller A, Aarestrup FM, Hasman H, Frimodt-Møller N. 2007. First detection of plasmid-mediated quinolone resistance (qnrA and qnrS) in Escherichia coli strains isolated from humans in Scandinavia. J Antimicrob Chemother 59:804–805. doi: 10.1093/jac/dkl554. [DOI] [PubMed] [Google Scholar]
- 204.Tomova A, Ivanova L, Buschmann AH, Godfrey HP, Cabello FC. 2018. Plasmid-mediated quinolone resistance (PMQR) genes and class 1 integrons in quinolone-resistant marine bacteria and clinical isolates of Escherichia coli from an aquacultural area. Microb Ecol 75:104–112. doi: 10.1007/s00248-017-1016-9. [DOI] [PubMed] [Google Scholar]
- 205.Veldman K, Kant A, Dierikx C, van Essen-Zandbergen A, Wit B, Mevius D. 2014. Enterobacteriaceae resistant to third-generation cephalosporins and quinolones in fresh culinary herbs imported from Southeast Asia. Int J Food Microbiol 177:72–77. doi: 10.1016/j.ijfoodmicro.2014.02.014. [DOI] [PubMed] [Google Scholar]
- 206.Chen D, Yang L, Peters BM, Liu J, Li L, Li B, Xu Z, Shirtliff ME. 2019. Complete sequence of a novel multidrug-resistant Pseudomonas putida strain carrying two copies of qnrVC6. Microb Drug Resist 25:1–7. doi: 10.1089/mdr.2018.0104. [DOI] [PubMed] [Google Scholar]
- 207.Pérez-Moreno MO, Picó-Plana E, de Toro M, Grande-Armas J, Quiles-Fortuny V, Pons MJ, Gomes C, Sáenz Y, Torres C, Ruiz J. 2013. β-Lactamases, transferable quinolone resistance determinants, and class 1 integron-mediated antimicrobial resistance in human clinical Salmonella enterica isolates of non-Typhimurium serotypes. Int J Med Microbiol 303:25–31. doi: 10.1016/j.ijmm.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 208.Zhao Z, Lan F, Liu M, Chen W, Huang L, Lin Q, Li B. 2017. Evaluation of automated systems for aminoglycosides and fluoroquinolones susceptibility testing for carbapenem-resistant Enterobacteriaceae. Antimicrob Resist Infect Control 6:77. doi: 10.1186/s13756-017-0235-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Ruiz J. 2018. Analysis of the presence of erroneous Qnr sequences in GenBank. J Antimicrob Chemother 73:1213–1216. doi: 10.1093/jac/dky025. [DOI] [PubMed] [Google Scholar]
- 210.Pons MJ, Gomes C, Ruiz J. 2013. QnrVC a new transferable Qnr-like family. Enferm Infecc Microbiol Clin 31:191–192. doi: 10.1016/j.eimc.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 211.Leone P, Méresse S. 2011. Kinesin regulation by Salmonella. Virulence 2:63–66. doi: 10.4161/viru.2.1.14603. [DOI] [PubMed] [Google Scholar]
- 212.Zhao L, Zhang J, Zheng B, Wei Z, Shen P, Li S, Li L, Xiao Y. 2015. Molecular epidemiology and genetic diversity of fluoroquinolone-resistant Escherichia coli isolates from patients with community-onset infections in 30 Chinese county hospitals. J Clin Microbiol 53:766–770. doi: 10.1128/JCM.02594-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.García-León G, Ruiz de Alegría Puig C, García de la Fuente C, Martínez-Martínez L, Martínez JL, Sánchez MB. 2015. High-level quinolone resistance is associated with the overexpression of smeVWX in Stenotrophomonas maltophilia clinical isolates. Clin Microbiol Infect 21:464–467. doi: 10.1016/j.cmi.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 214.Ribera A, Doménech-Sanchez A, Ruiz J, Benedi VJ, Jimenez de Anta MT, Vila J. 2002. Mutations in gyrA and parC QRDRs are not relevant for quinolone resistance in epidemiological unrelated Stenotrophomonas maltophilia clinical isolates. Microb Drug Resist 8:245–252. doi: 10.1089/10766290260469499. [DOI] [PubMed] [Google Scholar]
- 215.Sánchez MB, Martínez JL. 2010. SmQnr contributes to intrinsic resistance to quinolones in Stenotrophomonas maltophilia. Antimicrob Agents Chemother 54:580–581. doi: 10.1128/AAC.00496-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Valdezate S, Vindel A, Saéz-Nieto JA, Baquero F, Cantón R. 2005. Preservation of topoisomerase genetic sequences during in vivo and in vitro development of high-level resistance to ciprofloxacin in isogenic Stenotrophomonas maltophilia strains. J Antimicrob Chemother 56:220–223. doi: 10.1093/jac/dki182. [DOI] [PubMed] [Google Scholar]
- 217.Cabrera R, Ruiz J, Marco F, Oliveira I, Arroyo M, Aladueña A, Usera MA, Jiménez De Anta MT, Gascón J, Vila J. 2004. Mechanism of resistance to several antimicrobial agents in Salmonella clinical isolates causing traveler’s diarrhea. Antimicrob Agents Chemother 48:3934–3939. doi: 10.1128/AAC.48.10.3934-3939.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Capilla S, Ruiz J, Goñi P, Castillo J, Rubio MC, Jiménez de Anta MT, Gómez-Lus R, Vila J. 2004. Characterization of the molecular mechanisms of quinolone-resistance in Yersinia enterocolitica O:3 clinical isolates. J Antimicrob Chemother 53:1068–1071. doi: 10.1093/jac/dkh225. [DOI] [PubMed] [Google Scholar]
- 219.Ruiz J, Gómez J, Navia MM, Ribera A, Sierra JM, Marco F, Mensa J, Vila J. 2002. High prevalence of nalidixic acid resistant, ciprofloxacin susceptible phenotype among clinical isolates of Escherichia coli and other Enterobacteriaceae. Diagn Microbiol Infect Dis 42:257–261. doi: 10.1016/S0732-8893(01)00357-1. [DOI] [PubMed] [Google Scholar]
- 220.Wain J, Hoa NT, Chinh NT, Vinh H, Everett MJ, Diep TS, Day NP, Solomon T, White NJ, Piddock LJ, Parry CM. 1997. Quinolone-resistant Salmonella typhi in Viet Nam: molecular basis of resistance and clinical response to treatment. Clin Infect Dis 25:1404–1410. doi: 10.1086/516128. [DOI] [PubMed] [Google Scholar]
- 221.CLSI. 2015. Performance standards for antimicrobial susceptibility testing; twenty-fifth informational supplement. Document M100-S25, 25th ed CLSI, Wayne, PA. [Google Scholar]
- 222.CLSI. 2016. Performance standards for antimicrobial susceptibility testing. Document M100S, 26th ed CLSI, Wayne, PA. [Google Scholar]
- 223.Hakanen A, Kotilainen P, Jalava J, Siitonen A, Huovinen P. 1999. Detection of decreased fluoroquinolone susceptibility in salmonellas and validation of nalidixic acid screening test. J Clin Microbiol 37:3572–3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Angelakis E, Biswas S, Taylor C, Raoult D, Rolain JM. 2008. Heterogeneity of susceptibility to fluoroquinolones in Bartonella isolates from Australia reveals a natural mutation in gyrA. J Antimicrob Chemother 61:1252–1255. doi: 10.1093/jac/dkn094. [DOI] [PubMed] [Google Scholar]
- 225.del Valle LJ, Flores L, Vargas M, García-de-la-Guarda R, Quispe RL, Ibañez ZB, Alvarado D, Ramírez P, Ruiz J. 2010. Bartonella bacilliformis, endemic pathogen of the Andean region, is intrinsically resistant to quinolones. Int J Infect Dis 14:e506–e510. doi: 10.1016/j.ijid.2009.07.025. [DOI] [PubMed] [Google Scholar]
- 226.Han XY, Andrade RA. 2005. Brevundimonas diminuta infections and its resistance to fluoroquinolones. J Antimicrob Chemother 55:853–859. doi: 10.1093/jac/dki139. [DOI] [PubMed] [Google Scholar]
- 227.Sato T, Yokota S, Uchida I, Okubo T, Ishihara K, Fujii N, Tamura Y. 2011. A fluoroquinolone-resistant Escherichia coli clinical isolate without quinolone resistance-determining region mutations found in Japan. Antimicrob Agents Chemother 55:3964–3965. doi: 10.1128/AAC.00532-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Vien LTM, Baker S, Thao LTP, Tu LTP, Thuy CT, Nga TTT, Hoang NVM, Campbell JI, Yen LM, Hieu NT, Chau NVV, Farrar J, Schultsz C. 2009. High prevalence of plasmid-mediated quinolone resistance determinants in commensal members of the Enterobacteriaceae in Ho Chi Minh City, Vietnam. J Med Microbiol 58:1585–1592. doi: 10.1099/jmm.0.010033-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Vinué L, Hooper DC, Jacoby GA. 2018. Chromosomal mutations that accompany qnr in clinical isolates of Escherichia coli. Int J Antimicrob Agents 51:479–483. doi: 10.1016/j.ijantimicag.2018.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Zhao J, Chen Z, Chen S, Deng Y, Liu Y, Tian W, Huang X, Wu C, Sun Y, Sun Y, Zeng Z, Liu JH. 2010. Prevalence and dissemination of oqxAB in Escherichia coli isolates from animals, farmworkers, and the environment. Antimicrob Agents Chemother 54:4219–4224. doi: 10.1128/AAC.00139-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Luk-In S, Pulsrikarn C, Bangtrakulnonth A, Chatsuwan T, Kulwichit W. 2017. Occurrence of a novel class 1 integron harboring qnrVC4 in Salmonella Rissen. Diagn Microbiol Infect Dis 88:282–286. doi: 10.1016/j.diagmicrobio.2017.03.016. [DOI] [PubMed] [Google Scholar]
- 232.Galani I, Souli M, Mitchell N, Chryssouli Z, Giamarellou H. 2010. Presence of plasmid-mediated quinolone resistance in Klebsiella pneumoniae and Escherichia coli isolates possessing blaVIM-1 in Greece. Int J Antimicrob Agents 36:252–254. doi: 10.1016/j.ijantimicag.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 233.Karczmarczyk M, Martins M, McCusker M, Mattar S, Amaral L, Leonard N, Aarestrup FM, Fanning S. 2010. Characterization of antimicrobial resistance in Salmonella enterica food and animal isolates from Colombia: identification of a qnrB19-mediated quinolone resistance marker in two novel serovars. FEMS Microbiol Lett 313:10–19. doi: 10.1111/j.1574-6968.2010.02119.x. [DOI] [PubMed] [Google Scholar]
- 234.Jacoby GA, Corcoran MA, Mills DM, Griffin CM, Hooper DC. 2013. Mutational analysis of quinolone resistance protein QnrB1. Antimicrob Agents Chemother 57:5733–5736. doi: 10.1128/AAC.01533-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Tavío MM, Jacoby GA, Hooper DC. 2014. QnrS1 structure-activity relationships. J Antimicrob Chemother 69:2102–2109. doi: 10.1093/jac/dku102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Garoff L, Yadav K, Hughes D. 2018. Increased expression of Qnr is sufficient to confer clinical resistance to ciprofloxacin in Escherichia coli. J Antimicrob Chemother 73:348–352. doi: 10.1093/jac/dkx375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Akasaka T, Tanaka M, Yamaguchi A, Sato K. 2001. Type II topoisomerase mutations in fluoroquinolone-resistant clinical strains of Pseudomonas aeruginosa isolated in 1998 and 1999: role of target enzyme in mechanism of fluoroquinolone resistance. Antimicrob Agents Chemother 45:2263–2268. doi: 10.1128/AAC.45.8.2263-2268.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Bruchmann S, Dötsch A, Nouri B, Chaberny IF, Häussler S. 2013. Quantitative contributions of target alteration and decreased drug accumulation to Pseudomonas aeruginosa fluoroquinolone resistance. Antimicrob Agents Chemother 57:1361–1368. doi: 10.1128/AAC.01581-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Li XZ, Plésiat P, Nikaido H. 2015. The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin Microbiol Rev 28:337–418. doi: 10.1128/CMR.00117-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Cambau E, Bordon F, Collatz E, Gutmann L. 1993. Novel gyrA point mutation in a strain of Escherichia coli resistant to fluoroquinolones but not to nalidixic acid. Antimicrob Agents Chemother 37:1247–1252. doi: 10.1128/AAC.37.6.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Moniot-Ville N, Guibert J, Moreau N, Acar JF, Collatz E, Gutmann L. 1991. Mechanisms of quinolone resistance in a clinical isolate of Escherichia coli highly resistant to fluoroquinolones but susceptible to nalidixic acid. Antimicrob Agents Chemother 35:519–523. doi: 10.1128/AAC.35.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Truong QC, Nguyen Van JC, Shlaes D, Gutmann L, Moreau NJ. 1997. A novel, double mutation in DNA gyrase A of Escherichia coli conferring resistance to quinolone antibiotics. Antimicrob Agents Chemother 41:85–90. doi: 10.1128/AAC.41.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Ragunathan PL, Ison C, Livermore DM. 2005. Nalidixic acid-susceptible, ciprofloxacin-resistant Neisseria gonorrhoeae strain in the UK. J Antimicrob Chemother 56:437. doi: 10.1093/jac/dki201. [DOI] [PubMed] [Google Scholar]
- 244.Bachoual R, Ouabdesselam S, Mory F, Lascols C, Soussy CJ, Tankovic J. 2001. Single or double mutational alterations of gyrA associated with fluoroquinolone resistance in Campylobacter jejuni and Campylobacter coli. Microb Drug Resist 7:257–261. doi: 10.1089/10766290152652800. [DOI] [PubMed] [Google Scholar]
- 245.Pérez-Moreno MO, Estepa V, Sáenz Y, Cortell-Ortolá M, Fort-Gallifa I, Ruiz J, Torres C. 2012. Intrahospitalary dissemination of Klebsiella pneumoniae carrying blaDHA-1 and qnrB4 genes within a novel complex class 1 integron. Diagn Microbiol Infect Dis 73:210–211. doi: 10.1016/j.diagmicrobio.2012.02.011. [DOI] [PubMed] [Google Scholar]
- 246.Lascols C, Podglajen I, Verdet C, Gautier V, Gutmann L, Soussy CJ, Collatz E, Cambau E. 2008. A plasmid-borne Shewanella algae gene, qnrA3, and its possible transfer in vivo between Kluyvera ascorbata and Klebsiella pneumoniae. J Bacteriol 190:5217–5223. doi: 10.1128/JB.00243-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Liang Q, Jiang X, Hu L, Yin Z, Gao B, Zhao Y, Yang W, Yang H, Tong Y, Li W, Jiang L, Zhou D. 2019. Sequencing and genomic diversity analysis of IncHI5 plasmids. Front Microbiol 9:3318. doi: 10.3389/fmicb.2018.03318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Quiroga MP, Andres P, Petroni A, Soler Bistué AJC, Guerriero L, Vargas LJ, Zorreguieta A, Tokumoto M, Quiroga C, Tolmasky ME, Galas M, Centrón D. 2007. Complex class 1 integrons with diverse variable regions, including aac(6′)-Ib-cr, and a novel allele, qnrB10, associated with ISCR1 in clinical enterobacterial isolates from Argentina. Antimicrob Agents Chemother 51:4466–4470. doi: 10.1128/AAC.00726-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Tamariz J, Llanos C, Seas C, Montenegro P, Lagos J, Fernandes MR, Cerdeira L, Lincopan N. 2018. Draft genome sequence of the first New Delhi metallo-β-lactamase (NDM-1)-producing Escherichia coli strain isolated in Peru. Genome Announc 6(13):e00199-18. doi: 10.1128/genomeA.00199-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Villa L, Poirel L, Nordmann P, Carta C, Carattoli A. 2012. Complete sequencing of an IncH plasmid carrying the blaNDM-1, blaCTX-M-15 and qnrB1 genes. J Antimicrob Chemother 67:1645–1650. doi: 10.1093/jac/dks114. [DOI] [PubMed] [Google Scholar]
- 251.Vinué L, Sater MRA, Herriott I, Huntley MH, Jacoby GA, Hooper DC. 3 June 2019. Multiple copies of qnrA1 on an Inc A/C2 plasmid explain enhanced quinolone resistance in an Escherichia coli mutant. Antimicrob Agents Chemother doi: 10.1128/AAC.00718-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Wang J, Li X, Li J, Hurley D, Bai X, Yu Z, Cao Y, Wall E, Fanning S, Bai L. 2017. Complete genetic analysis of a Salmonella enterica serovar Indiana isolate accompanying four plasmids carrying mcr-1, ESBL and other resistance genes in China. Vet Microbiol 210:142–146. doi: 10.1016/j.vetmic.2017.08.024. [DOI] [PubMed] [Google Scholar]
- 253.Wang M, Tran JH, Jacoby GA, Zhang Y, Wang F, Hooper DC. 2003. Plasmid-mediated quinolone resistance in clinical isolates of Escherichia coli from Shanghai, China. Antimicrob Agents Chemother 47:2242–2248. doi: 10.1128/AAC.47.7.2242-2248.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Wong MHY, Chan EWC, Xie L, Li R, Chen S. 2016. IncHI2 plasmids are the key vectors responsible for oqxAB transmission among Salmonella species. Antimicrob Agents Chemother 60:6911–6915. doi: 10.1128/AAC.01555-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Li X, Zhang Y, Zhou X, Hu X, Zhou Y, Liu D, Maxwell A, Mi K. 2019. The plasmid-borne quinolone resistance protein QnrB, a novel DnaA-binding protein, increases the bacterial mutation rate by triggering DNA replication stress. Mol Microbiol 111:1529–1543. doi: 10.1111/mmi.14235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Cesaro A, Bettoni RR, Lascols C, Mérens A, Soussy CJ, Cambau E. 2008. Low selection of topoisomerase mutants from strains of Escherichia coli harbouring plasmid-borne qnr genes. J Antimicrob Chemother 61:1007–1015. doi: 10.1093/jac/dkn077. [DOI] [PubMed] [Google Scholar]
- 257.Goto K, Kawamura K, Arakawa Y. 2015. Contribution of QnrA, a plasmid-mediated quinolone resistance peptide, to survival of Escherichia coli exposed to a lethal ciprofloxacin concentration. Jpn J Infect Dis 68:196–202. doi: 10.7883/yoken.JJID.2014.153. [DOI] [PubMed] [Google Scholar]
- 258.Vinué L, Corcoran MA, Hooper DC, Jacoby GA. 2016. Mutations that enhance the ciprofloxacin resistance of Escherichia coli with qnrA1. Antimicrob Agents Chemother 60:1537–1545. doi: 10.1128/AAC.02167-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Hirai K, Aoyama H, Irikura T, Iyobe S, Mitsuhashi S. 1986. Difference in susceptibility to quinolones of outer membrane mutants of Salmonella typhimurium and Escherichia coli. Antimicrob Agents Chemother 29:535–538. doi: 10.1128/AAC.29.3.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Ebbensgaard A, Mordhorst H, Aarestrup FM, Hansen EB. 2018. The role of outer membrane proteins and lipopolysaccharides for the sensitivity of Escherichia coli to antimicrobial peptides. Front Microbiol 9:2153. doi: 10.3389/fmicb.2018.02153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Linkevicius M, Sandegren L, Andersson DI. 2013. Mechanisms and fitness costs of tigecycline resistance in Escherichia coli. J Antimicrob Chemother 68:2809–2819. doi: 10.1093/jac/dkt263. [DOI] [PubMed] [Google Scholar]
- 262.Lee CC, Lui G, Ip M, Ling TK, Lee N. 2012. Frequent detection of plasmid-mediated quinolone resistance (qnr) genes in multidrug-resistant Enterobacteriaceae blood isolates, Hong Kong. Eur J Clin Microbiol Infect Dis 31:3183–3189. doi: 10.1007/s10096-012-1683-x. [DOI] [PubMed] [Google Scholar]
- 263.Liao CH, Hsueh PR, Jacoby GA, Hooper DC. 2013. Risk factors and clinical characteristics of patients with qnr-positive Klebsiella pneumoniae bacteraemia. J Antimicrob Chemother 68:2907–2914. doi: 10.1093/jac/dkt295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Domínguez-Herrera J, Velasco C, Docobo-Pérez F, Rodríguez-Martínez JM, López-Rojas R, Briales A, Pichardo C, Díaz-de-Alba P, Rodríguez-Baño J, Pascual A, Pachón J. 2013. Impact of qnrA1, qnrB1 and qnrS1 on the efficacy of ciprofloxacin and levofloxacin in an experimental pneumonia model caused by Escherichia coli with or without the GyrA mutation Ser83Leu. J Antimicrob Chemother 68:1609–1615. doi: 10.1093/jac/dkt063. [DOI] [PubMed] [Google Scholar]
- 265.Jakobsen L, Cattoir V, Jensen KS, Hammerum AM, Nordmann P, Frimodt-Møller N. 2012. Impact of low-level fluoroquinolone resistance genes qnrA1, qnrB19 and qnrS1 on ciprofloxacin treatment of isogenic Escherichia coli strains in a murine urinary tract infection model. J Antimicrob Chemother 67:2438–2444. doi: 10.1093/jac/dks224. [DOI] [PubMed] [Google Scholar]
- 266.Ruiz J, Pons MJ. 2018. In an inhospitable ICU, not even antibiotic cycling or mixing are the solutions. Ann Infect 2:2. doi: 10.21037/aoi.2018.07.01. [DOI] [Google Scholar]
- 267.Card R, Zhang J, Das P, Cook C, Woodford N, Anjum MF. 2013. Evaluation of an expanded microarray for detecting antibiotic resistance genes in a broad range of Gram-negative bacterial pathogens. Antimicrob Agents Chemother 57:458–465. doi: 10.1128/AAC.01223-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Rossen JWA, Friedrich AW, Moran-Gilad J, ESCMID Study Group for Genomic and Molecular Diagnostics. 2018. Practical issues in implementing whole-genome-sequencing in routine diagnostic microbiology. Clin Microbiol Infect 24:355–360. doi: 10.1016/j.cmi.2017.11.001. [DOI] [PubMed] [Google Scholar]
- 269.Schubert S, Kostrzewa M. 2017. MALDI-TOF MS in the microbiology laboratory: current trends. Curr Issues Mol Biol 23:17–20. doi: 10.21775/cimb.023.017. [DOI] [PubMed] [Google Scholar]
- 270.Cattoir V, Poirel L, Rotimi V, Soussy CJ, Nordmann P. 2007. Multiplex PCR for detection of plasmid-mediated quinolone resistance qnr genes in ESBL-producing enterobacterial isolates. J Antimicrob Chemother 60:394–397. doi: 10.1093/jac/dkm204. [DOI] [PubMed] [Google Scholar]
- 271.Ciesielczuk H, Hornsey M, Choi V, Woodford N, Wareham DW. 2013. Development and evaluation of a multiplex PCR for eight plasmid-mediated quinolone-resistance determinants. J Med Microbiol 62(Part 12):1823–1827. doi: 10.1099/jmm.0.064428-0. [DOI] [PubMed] [Google Scholar]
- 272.Chen YT, Shu HY, Li LH, Liao TL, Wu KM, Shiau YR, Yan JJ, Su IJ, Tsai SF, Lauderdale TL. 2006. Complete nucleotide sequence of pK245, a 98-kilobase plasmid conferring quinolone resistance and extended-spectrum-β-lactamase activity in a clinical Klebsiella pneumoniae isolate. Antimicrob Agents Chemother 50:3861–3866. doi: 10.1128/AAC.00456-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Cunha MPV, Davies YM, Cerdeira L, Dropa M, Lincopan N, Knöbl T. 2017. Complete DNA sequence of an IncM1 plasmid bearing the novel qnrE1 plasmid-mediated quinolone resistance variant and blaCTX-M-8 from Klebsiella pneumoniae sequence type 147. Antimicrob Agents Chemother 61:e00592-17. doi: 10.1128/AAC.00592-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Flach CF, Johnning A, Nilsson I, Smalla K, Kristiansson E, Larsson DG. 2015. Isolation of novel IncA/C and IncN fluoroquinolone resistance plasmids from an antibiotic-polluted lake. J Antimicrob Chemother 70:2709–2717. doi: 10.1093/jac/dkv167. [DOI] [PubMed] [Google Scholar]
- 275.Lee JJ, Kim MN, Park KS, Lee JH, Karim AM, Park M, Kim JH, Lee SH. 2016. Complex class 1 integron carrying qnrB62 and blaVIM-2 in a Citrobacter freundii clinical isolate. Antimicrob Agents Chemother 60:6937–6940. doi: 10.1128/AAC.00614-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Schlüter A, Nordmann P, Bonnin RA, Millemann Y, Eikmeyer FG, Wibberg D, Pühler A, Poirel L. 2014. IncH-type plasmid harboring blaCTX-M-15, blaDHA-1, and qnrB4 genes recovered from animal isolates. Antimicrob Agents Chemother 58:3768–3773. doi: 10.1128/AAC.02695-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Siebor E, Neuwirth C. 2011. The new variant of Salmonella genomic island 1 (SGI1-V) from a Proteus mirabilis French clinical isolate harbours blaVEB-6 and qnrA1 in the multiple antibiotic resistance region. J Antimicrob Chemother 66:2513–2520. doi: 10.1093/jac/dkr335. [DOI] [PubMed] [Google Scholar]
- 278.Zhi C, Lv L, Yu L-F, Doi Y, Liu J-H. 2016. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis 16:292–293. doi: 10.1016/S1473-3099(16)00063-3. [DOI] [PubMed] [Google Scholar]
- 279.Espedido BA, Partridge SR, Iredell JR. 2008. blaIMP-4 in different genetic contexts in Enterobacteriaceae isolates from Australia. Antimicrob Agents Chemother 52:2984–2987. doi: 10.1128/AAC.01634-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Li B, Feng J, Zhan Z, Yin Z, Jiang Q, Wei P, Chen X, Gao B, Hou J, Mao P, Wu W, Chen W, Tong Y, Wang J, Li B, Zhou D. 2018. Dissemination of KPC-2-encoding IncX6 plasmids among multiple Enterobacteriaceae species in a single Chinese hospital. Front Microbiol 9:478. doi: 10.3389/fmicb.2018.00478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Manageiro V, Romão R, Moura IB, Sampaio DA, Vieira L, Ferreira E, Network EuSCAPE-Portugal, Caniça M. 2018. Molecular epidemiology and risk factors of carbapenemase-producing Enterobacteriaceae isolates in Portuguese hospitals: results from European survey on carbapenemase-producing Enterobacteriaceae (EuSCAPE). Front Microbiol 9:2834. doi: 10.3389/fmicb.2018.02834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Mendes RE, Bell JM, Turnidge JD, Yang Q, Yu Y, Sun Z, Jones RN. 2008. Carbapenem-resistant isolates of Klebsiella pneumoniae in China and detection of a conjugative plasmid (blaKPC-2 plus qnrB4) and a blaIMP-4 gene. Antimicrob Agents Chemother 52:798–799. doi: 10.1128/AAC.01185-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Rice LB, Carias LL, Hutton RA, Rudin SD, Endimiani A, Bonomo RA. 2008. The KQ element, a complex genetic region conferring transferable resistance to carbapenems, aminoglycosides, and fluoroquinolones in Klebsiella pneumoniae. Antimicrob Agents Chemother 52:3427–3429. doi: 10.1128/AAC.00493-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Rodríguez-Martínez JM, Velasco C, García I, Cano ME, Martínez-Martínez L, Pascual A. 2007. Characterisation of integrons containing the plasmid-mediated quinolone resistance gene qnrA1 in Klebsiella pneumoniae. Int J Antimicrob Agents 29:705–709. doi: 10.1016/j.ijantimicag.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 285.Ye L, Li R, Lin D, Zhou Y, Fu A, Ding Q, Chan EW, Yao W, Chen S. 2016. Characterization of an IncA/C multidrug resistance plasmid in Vibrio alginolyticus. Antimicrob Agents Chemother 60:3232–3235. doi: 10.1128/AAC.00300-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Périchon B, Bogaerts P, Lambert T, Frangeul L, Courvalin P, Galimand M. 2008. Sequence of conjugative plasmid pIP1206 mediating resistance to aminoglycosides by 16S rRNA methylation and to hydrophilic fluoroquinolones by efflux. Antimicrob Agents Chemother 52:2581–2592. doi: 10.1128/AAC.01540-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Jeong HS, Bae IK, Shin JH, Jung HJ, Kim SH, Lee JY, Oh SH, Kim HR, Chang CL, Kho WG, Lee JN. 2011. Prevalence of plasmid-mediated quinolone resistance and its association with extended-spectrum beta-lactamase and AmpC beta-lactamase in Enterobacteriaceae. Korean J Lab Med 31:257–264. doi: 10.3343/kjlm.2011.31.4.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Tuon FF, Rocha JL, Toledo P, Arend LN, Dias CH, Leite TM, Penteado-Filho SR, Pilonetto M, Zavascki AP. 2012. Risk factors for KPC-producing Klebsiella pneumoniae bacteremia. Braz J Infect Dis 16:416–419. doi: 10.1016/j.bjid.2012.08.006. [DOI] [PubMed] [Google Scholar]
- 289.Pitout JD, Wei Y, Church DL, Gregson DB. 2008. Surveillance for plasmid-mediated quinolone resistance determinants in Enterobacteriaceae within the Calgary Health Region, Canada: the emergence of aac(6′)-Ib-cr. J Antimicrob Chemother 61:999–1002. doi: 10.1093/jac/dkn068. [DOI] [PubMed] [Google Scholar]
- 290.Piekarska K, Zacharczuk K, Wołkowicz T, Wolaniuk N, Rzeczkowska M, Gierczyński R. 2019. Emergence of Enterobacteriaceae co-producing CTX-M-15, ArmA and PMQR in Poland. Adv Clin Exp Med 28:249–254. doi: 10.17219/acem/94165. [DOI] [PubMed] [Google Scholar]
- 291.Brahmi S, Touati A, Dunyach-Remy C, Sotto A, Pantel A, Lavigne JP. 2018. High prevalence of extended-spectrum β-lactamase-producing Enterobacteriaceae in wild fish from the Mediterranean sea in Algeria. Microb Drug Resist 24:290–298. doi: 10.1089/mdr.2017.0149. [DOI] [PubMed] [Google Scholar]
- 292.Moremi N, Manda EV, Falgenhauer L, Ghosh H, Imirzalioglu C, Matee M, Chakraborty T, Mshana SE. 2016. Predominance of CTX-M-15 among ESBL producers from environment and fish gut from the shores of Lake Victoria in Mwanza, Tanzania. Front Microbiol 7:1862. doi: 10.3389/fmicb.2016.01862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Argente M, Miró E, Martí C, Vilamala A, Alonso-Tarrés C, Ballester F, Calderón A, Gallés C, Gasós A, Mirelis B, Morta M, Olsina M, Sauca G, Sierra M, Rivera A, Navarro F. 2019. Molecular characterization of OXA-48 carbapenemase-producing Klebsiella pneumoniae strains after a carbapenem resistance increase in Catalonia. Enferm Infecc Microbiol Clin 37:82–88. doi: 10.1016/j.eimc.2018.02.003. [DOI] [PubMed] [Google Scholar]
- 294.Riveros M, Riccobbono E, Durand D, Mosquito S, Ruiz J, Gotuzzo E, Rossolini GM, Ochoa TJ, Pallecchi L. 2012. Plasmid-mediated quinolone resistance genes in enteroaggregative Escherichia coli from infants in Lima, Perú. Int J Antimicrob Agents 39:540–542. doi: 10.1016/j.ijantimicag.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 295.Rodríguez-Martínez JM, Poirel L, Pascual A, Nordmann P. 2006. Plasmid-mediated quinolone resistance in Australia. Microb Drug Resist 12:99–102. doi: 10.1089/mdr.2006.12.99. [DOI] [PubMed] [Google Scholar]
- 296.Veldman K, Cavaco LM, Mevius D, Battisti A, Franco A, Botteldoorn N, Bruneau M, Perrin-Guyomard A, Cerny T, De Frutos Escobar C, Guerra B, Schroeter A, Gutierrez M, Hopkins K, Myllyniemi AL, Sunde M, Wasyl D, Aarestrup FM. 2011. International collaborative study on the occurrence of plasmid-mediated quinolone resistance in Salmonella enterica and Escherichia coli isolated from animals, humans, food and the environment in 13 European countries. J Antimicrob Chemother 66:1278–1286. doi: 10.1093/jac/dkr084. [DOI] [PubMed] [Google Scholar]
- 297.Wen Y, Pu X, Zheng W, Hu G. 2016. High prevalence of plasmid-mediated quinolone resistance and IncQ plasmids carrying qnrS2 gene in bacteria from rivers near hospitals and aquaculture in China. PLoS One 11:e0159418. doi: 10.1371/journal.pone.0159418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Cattoir V, Poirel L, Aubert C, Soussy CJ, Nordmann P. 2008. Unexpected occurrence of plasmid-mediated quinolone resistance determinants in environmental Aeromonas spp. Emerg Infect Dis 14:231–237. doi: 10.3201/eid1402.070677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Chenia HY. 2016. Prevalence and characterization of plasmid-mediated quinolone resistance genes in Aeromonas spp. isolated from South African freshwater fish. Int J Food Microbiol 231:26–32. doi: 10.1016/j.ijfoodmicro.2016.04.030. [DOI] [PubMed] [Google Scholar]
- 300.Sánchez-Céspedes J, Blasco MD, Martí S, Alba V, Alcalde E, Esteve C, Vila J. 2008. Plasmid-mediated QnrS2 determinant from a clinical Aeromonas veronii isolate. Antimicrob Agents Chemother 52:2990–2991. doi: 10.1128/AAC.00287-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Islam MA, Islam M, Hasan R, Hossain MI, Nabi A, Rahman M, Goessens WHF, Endtz HP, Boehm AB, Faruque SM. 2017. Environmental spread of New Delhi metallo-β-lactamase-1-producing multidrug-resistant bacteria in Dhaka, Bangladesh. Appl Environ Microbiol 83:e00793-17. doi: 10.1128/AEM.00793-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Belotti PT, Thabet L, Laffargue A, André C, Coulange-Mayonnove L, Arpin C, Messadi A, M’Zali F, Quentin C, Dubois V. 2015. Description of an original integron encompassing blaVIM-2, qnrVC1 and genes encoding bacterial group II intron proteins in Pseudomonas aeruginosa. J Antimicrob Chemother 70:2237–2240. doi: 10.1093/jac/dkv103. [DOI] [PubMed] [Google Scholar]
- 303.Kumar P, Thomas S. 2011. Presence of qnrVC3 gene cassette in SXT and class 1 integrons of Vibrio cholerae. Int J Antimicrob Agents 37:280–281. doi: 10.1016/j.ijantimicag.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 304.Singh R, Rajpara N, Tak J, Patel A, Mohanty P, Vinothkumar K, Chowdhury G, Ramamurthy T, Ghosh A, Bhardwaj AK. 2012. Clinical isolates of Vibrio fluvialis from Kolkata, India, obtained during 2006: plasmids, qnr and mutation in gyrase A as mechanisms of multidrug resistance. J Med Microbiol 61:369–374. doi: 10.1099/jmm.0.037226-0. [DOI] [PubMed] [Google Scholar]
- 305.Jacoby GA, Gacharna N, Black TA, Miller GH, Hooper DC. 2009. Temporal appearance of plasmid-mediated quinolone resistance genes. Antimicrob Agents Chemother 53:1665–1666. doi: 10.1128/AAC.01447-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Jacoby GA, Griffin CM, Hooper DC. 2011. Citrobacter spp. as a source of qnrB alleles. Antimicrob Agents Chemother 55:4979–4984. doi: 10.1128/AAC.05187-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Saga T, Sabtcheva S, Mitsutake K, Ishii Y, Tateda K, Yamaguchi K, Kaku M. 2013. Characterization of qnrB-like genes in Citrobacter species of the American Type Culture Collection. Antimicrob Agents Chemother 57:2863–2866. doi: 10.1128/AAC.02396-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Sarles WB, Hammer BW. 1933. Species of Escherichia-Aerobacter organisms responsible for some defects in dairy products. J Bacteriol 25:461–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Werkman CH, Gillen GF. 1932. Bacteria producing trimethylene glycol. J Bacteriol 23:167–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Jacoby GA, Corcoran MA, Hooper DC. 2015. Protective effect of Qnr on agents other than quinolones that target DNA gyrase. Antimicrob Agents Chemother 59:6689–6695. doi: 10.1128/AAC.01292-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Rodríguez-Martínez JM, Velasco C, Briales A, García I, Conejo MC, Pascual A. 2008. Qnr-like pentapeptide repeat proteins in Gram-positive bacteria. J Antimicrob Chemother 61:1240–1243. doi: 10.1093/jac/dkn115. [DOI] [PubMed] [Google Scholar]
- 312.Cattoir V, Poirel L, Mazel D, Soussy CJ, Nordmann P. 2007. Vibrio splendidus as the source of plasmid-mediated QnrS-like quinolone resistance determinants. Antimicrob Agents Chemother 51:2650–2651. doi: 10.1128/AAC.00070-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Poirel L, Liard A, Rodriguez-Martinez J-M, Nordmann P. 2005. Vibrionaceae as a possible source of Qnr-like quinolone resistance determinants. J Antimicrob Chemother 56:1118–1121. doi: 10.1093/jac/dki371. [DOI] [PubMed] [Google Scholar]
- 314.Doublet B, Boyd D, Mulvey MR, Cloeckaert A. 2005. The Salmonella genomic island 1 is an integrative mobilizable element. Mol Microbiol 55:1911–1924. doi: 10.1111/j.1365-2958.2005.04520.x. [DOI] [PubMed] [Google Scholar]
- 315.Colomer-Lluch M, Jofre J, Muniesa M. 2014. Quinolone resistance genes (qnrA and qnrS) in bacteriophage particles from wastewater samples and the effect of inducing agents on packaged antibiotic resistance genes. J Antimicrob Chemother 69:1265–1274. doi: 10.1093/jac/dkt528. [DOI] [PubMed] [Google Scholar]
- 316.Marti E, Variatza E, Balcázar JL. 2014. Bacteriophages as a reservoir of extended-spectrum β-lactamase and fluoroquinolone resistance genes in the environment. Clin Microbiol Infect 20:O456–O459. doi: 10.1111/1469-0691.12446. [DOI] [PubMed] [Google Scholar]
- 317.Rodríguez-Martínez JM, Pascual A, García I, Martínez-Martínez L. 2003. Detection of the plasmid-mediated quinolone resistance determinant qnr among clinical isolates of Klebsiella pneumoniae producing AmpC-type β-lactamase. J Antimicrob Chemother 52:703–706. doi: 10.1093/jac/dkg388. [DOI] [PubMed] [Google Scholar]
- 318.Tran JH, Jacoby GA. 2002. Mechanism of plasmid-mediated quinolone resistance. Proc Natl Acad Sci U S A 99:5638–5642. doi: 10.1073/pnas.082092899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Jacoby G, Cattoir V, Hooper D, Martínez-Martínez L, Nordmann P, Pascual A, Poirel L, Wang M. 2008. qnr gene nomenclature. Antimicrob Agents Chemother 52:2297–2299. doi: 10.1128/AAC.00147-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Wareham DW, Gordon NC, Shimizu K. 2011. Two new variants of and creation of a repository for Stenotrophomonas maltophilia quinolone protection protein (Smqnr) genes. Int J Antimicrob Agents 37:89–90. doi: 10.1016/j.ijantimicag.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 321.Arsène S, Leclercq R. 2007. Role of a qnr-like gene in the intrinsic resistance of Enterococcus faecalis to fluoroquinolones. Antimicrob Agents Chemother 51:3254–3258. doi: 10.1128/AAC.00274-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Park YJ, Yu JK, Lee S, Oh EJ, Woo GJ. 2007. Prevalence and diversity of qnr alleles in AmpC-producing Enterobacter cloacae, Enterobacter aerogenes, Citrobacter freundii and Serratia marcescens: a multicentre study from Korea. J Antimicrob Chemother 60:868–871. doi: 10.1093/jac/dkm266. [DOI] [PubMed] [Google Scholar]
- 323.Fluit AC, Schmitz FJ. 2004. Resistance integrons and super-integrons. Clin Microbiol Infect 10:272–288. doi: 10.1111/j.1198-743X.2004.00858.x. [DOI] [PubMed] [Google Scholar]
- 324.Toleman MA, Bennett PM, Walsh TR. 2006. ISCR elements: novel gene-capturing systems of the 21st century? Microbiol Mol Biol Rev 70:296–316. doi: 10.1128/MMBR.00048-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Cambau E, Lascols C, Sougakoff W, Bébéar C, Bonnet R, Cavallo JD, Gutmann L, Ploy MC, Jarlier V, Soussy CJ, Robert J. 2006. Occurrence of qnrA-positive clinical isolates in French teaching hospitals during 2002-2005. Clin Microbiol Infect 12:1013–1020. doi: 10.1111/j.1469-0691.2006.01529.x. [DOI] [PubMed] [Google Scholar]
- 326.Mahrouki S, Perilli M, Bourouis A, Chihi H, Ferjani M, Ben Moussa M, Amicosante G, Belhadj O. 2013. Prevalence of quinolone resistance determinant qnrA6 among broad- and extended-spectrum beta-lactam-resistant Proteus mirabilis and Morganella morganii clinical isolates with sul1-type class 1 integron association in a Tunisian hospital. Scand J Infect Dis 45:600–605. doi: 10.3109/00365548.2013.795657. [DOI] [PubMed] [Google Scholar]
- 327.Mammeri H, Van De Loo M, Poirel L, Martinez-Martinez L, Nordmann P. 2005. Emergence of plasmid-mediated quinolone resistance in Escherichia coli in Europe. Antimicrob Agents Chemother 49:71–76. doi: 10.1128/AAC.49.1.71-76.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Poirel L, Leviandier C, Nordmann P. 2006. Prevalence and genetic analysis of plasmid-mediated quinolone resistance determinants QnrA and QnrS in Enterobacteriaceae isolates from a French university hospital. Antimicrob Agents Chemother 50:3992–3997. doi: 10.1128/AAC.00597-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Potter RF, D’Souza AW, Wallace MA, Shupe A, Patel S, Gul D, Kwon JH, Andleeb S, Burnham CA, Dantas G. 2017. Draft genome sequence of the blaOXA-436- and blaNDM-1-harboring Shewanella putrefaciens SA70 isolate. Genome Announc 5(29):e00644-17. doi: 10.1128/genomeA.00644-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Kim HB, Park CH, Gavin M, Jacoby GA, Hooper DC. 2011. Cold shock induces qnrA expression in Shewanella algae. Antimicrob Agents Chemother 55:414–416. doi: 10.1128/AAC.00991-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Melvold JA, Wyrsch ER, McKinnon J, Roy Chowdhury P, Charles IG, Djordjevic SP. 2017. Identification of a novel qnrA allele, qnrA8, in environmental Shewanella algae. J Antimicrob Chemother 72:2949–2952. doi: 10.1093/jac/dkx226. [DOI] [PubMed] [Google Scholar]
- 332.Zhou MM, Tu HX, Zhou TL, Fei JX, Li C, Zhao Y, Bao Q. 2010. Detection of a new qnrA7 genotypes in Shewanella algae. Chin J Microbiol Immunol 30:593–596. [Google Scholar]
- 333.Cimmino T, Olaitan AO, Rolain JM. 2016. Whole genome sequence to decipher the resistome of Shewanella algae, a multidrug-resistant bacterium responsible for pneumonia, Marseille, France. Expert Rev Anti Infect Ther 14:269–275. doi: 10.1586/14787210.2016.1106936. [DOI] [PubMed] [Google Scholar]
- 334.Aigle A, Michotey V, Bonin P. 2015. Draft-genome sequence of Shewanella algae strain C6G3. Stand Genomic Sci 10:43. doi: 10.1186/s40793-015-0022-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Jacoby GA, Walsh KE, Mills DM, Walker VJ, Oh H, Robicsek A, Hooper DC. 2006. qnrB, another plasmid-mediated gene for quinolone resistance. Antimicrob Agents Chemother 50:1178–1182. doi: 10.1128/AAC.50.4.1178-1182.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Richter SN, Frasson I, Bergo C, Manganelli R, Cavallaro A, Palù G. 2010. Characterisation of qnr plasmid-mediated quinolone resistance in Enterobacteriaceae from Italy: association of the qnrB19 allele with the integron element ISCR1 in Escherichia coli. Int J Antimicrob Agents 35:578–583. doi: 10.1016/j.ijantimicag.2010.02.015. [DOI] [PubMed] [Google Scholar]
- 337.Cattoir V, Nordmann P, Silva-Sanchez J, Espinal P, Poirel L. 2008. ISEcp1-mediated transposition of qnrB-like gene in Escherichia coli. Antimicrob Agents Chemother 52:2929–2932. doi: 10.1128/AAC.00349-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W, Fouts DE, Levy S, Knap AH, Lomas MW, Nealson K, White O, Peterson J, Hoffman J, Parsons R, Baden-Tillson H, Pfannkoch C, Rogers Y-H, Smith HO. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 339.Sánchez MB, Hernández A, Rodríguez-Martínez JM, Martínez-Martínez L, Martínez JL. 2008. Predictive analysis of transmissible quinolone resistance indicates Stenotrophomonas maltophilia as a potential source of a novel family of Qnr determinants. BMC Microbiol 8:148. doi: 10.1186/1471-2180-8-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Wang M, Guo Q, Xu X, Wang X, Ye X, Wu S, Hooper DC, Wang M. 2009. New plasmid-mediated quinolone resistance gene, qnrC, found in a clinical isolate of Proteus mirabilis. Antimicrob Agents Chemother 53:1892–1897. doi: 10.1128/AAC.01400-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Azargun R, Sadeghi MR, Barhaghi MHS, Kafil HS, Yeganeh F, Oskouee MA, Ghotaslou R. 2018. The prevalence of plasmid-mediated quinolone resistance and ESBL-production in Enterobacteriaceae isolated from urinary tract infections. Infect Drug Resist 11:1007–1014. doi: 10.2147/IDR.S160720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.El-Badawy MF, Tawakol WM, El-Far SW, Maghrabi IA, Al-Ghamdi SA, Mansy MS, Ashour MS, Shohayeb MM. 2017. Molecular identification of aminoglycoside-modifying enzymes and plasmid-mediated quinolone resistance genes among Klebsiella pneumoniae clinical isolates recovered from Egyptian patients. Int J Microbiol 2017:8050432. doi: 10.1155/2017/8050432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Das A, Natarajan M, Mandal J. 2016. The emergence of quinolone resistant Shigella sonnei, Pondicherry, India. PLoS One 11:e0160290. doi: 10.1371/journal.pone.0160290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Cavaco LM, Hasman H, Xia S, Aarestrup FM. 2009. qnrD, a novel gene conferring transferable quinolone resistance in Salmonella enterica serovar Kentucky and Bovismorbificans strains of human origin. Antimicrob Agents Chemother 53:603–608. doi: 10.1128/AAC.00997-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Guillard T, Cambau E, Neuwirth C, Nenninger T, Mbadi A, Brasme L, Vernet-Garnier V, Bajolet O, de Champs C. 2012. Description of a 2,683-base-pair plasmid containing qnrD in two Providencia rettgeri isolates. Antimicrob Agents Chemother 56:565–568. doi: 10.1128/AAC.00081-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Zhang S, Sun J, Liao XP, Hu QJ, Liu BT, Fang LX, Deng H, Ma J, Xiao X, Zhu HQ, Liu YH. 2013. Prevalence and plasmid characterization of the qnrD determinant in Enterobacteriaceae isolated from animals, retail meat products, and humans. Microb Drug Resist 19:331–335. doi: 10.1089/mdr.2012.0146. [DOI] [PubMed] [Google Scholar]
- 347.Adeolu M, Alnajar S, Naushad S, Gupta RS. 2016. Genome-based phylogeny and taxonomy of the ‘Enterobacteriales’: proposal for Enterobacterales ord. nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. nov., Pectobacteriaceae fam. nov., Yersiniaceae fam. nov., Hafniaceae fam. nov., Morganellaceae fam. nov., and Budviciaceae fam. nov. Int J Syst Evol Microbiol 66:5575–5599. doi: 10.1099/ijsem.0.001485. [DOI] [PubMed] [Google Scholar]
- 348.Chen L, Zhang Y, Du J, Zhang X, Li M, Chen H, Yu X, Sun Y, Zhou T. 2018. Description and plasmid characterization of the qnrD determinant in Proteeae in Wenzhou, Southern China. J Microbiol Immunol Infect 51:115–122. doi: 10.1016/j.jmii.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 349.Guillard T, Grillon A, de Champs C, Cartier C, Madoux J, Berçot B, Lebreil AL, Lozniewski A, Riahi J, Vernet-Garnier V, Cambau E. 2014. Mobile insertion cassette elements found in small non-transmissible plasmids in Proteeae may explain qnrD mobilization. PLoS One 9:e87801. doi: 10.1371/journal.pone.0087801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Kraychete GB, Campana EH, Picão RC, Bonelli RR. 2019. qnrD-harboring plasmids in Providencia spp. recovered from food and environmental Brazilian sources. Sci Total Environ 646:1290–1292. doi: 10.1016/j.scitotenv.2018.07.378. [DOI] [PubMed] [Google Scholar]
- 351.Pathirana HNKSP, Shin G-W, Wimalasena SHMP, De Silva BCJ, Hossain S, Heo G-J. 2018. Prevalence and characterization of quinolone resistance genes in Proteus species isolated from pet turtles. J Exot Pet Med 27:67–73. doi: 10.1053/j.jepm.2017.10.026. [DOI] [Google Scholar]
- 352.Bueno MFC, Francisco GR, Cerdeira LT, Ienne S, Souza TA, Lincopan N, Garcia DO. 2016. Sequenciamento completo de plasmídeo que carrega blaCTX-M-8 de Klebsiella pneumoniae multirresistente, abstr 8573. Abstr 52nd Congr Soc Bras Med Trop.
- 353.Soares FB, Camargo CH, Cunha MPV, de Almeida EA, Bertani AMJ, Carvalho E, de Paiva JB, Fernandes SA, Tiba-Casas MR. 2019. Co-occurrence of qnrE1 and blaCTX-M-8 in IncM1 transferable plasmids contributing to MDR in different Salmonella serotypes. J Antimicrob Chemother 74:1155–1156. doi: 10.1093/jac/dky516. [DOI] [PubMed] [Google Scholar]
- 354.Kehrenberg C, Friederichs S, de Jong A, Michael GB, Schwarz S. 2006. Identification of the plasmid-borne quinolone resistance gene qnrS in Salmonella enterica serovar Infantis. J Antimicrob Chemother 58:18–22. doi: 10.1093/jac/dkl213. [DOI] [PubMed] [Google Scholar]
- 355.Lavilla S, González-López JJ, Sabaté M, García-Fernández A, Larrosa MN, Bartolomé RM, Carattoli A, Prats G. 2008. Prevalence of qnr genes among extended-spectrum β-lactamase-producing enterobacterial isolates in Barcelona, Spain. J Antimicrob Chemother 61:291–295. doi: 10.1093/jac/dkm448. [DOI] [PubMed] [Google Scholar]
- 356.Yue L, Jiang HX, Liao XP, Liu JH, Li SJ, Chen XY, Chen CX, Lü DH, Liu YH. 2008. Prevalence of plasmid-mediated quinolone resistance qnr genes in poultry and swine clinical isolates of Escherichia coli. Vet Microbiol 132:414–420. doi: 10.1016/j.vetmic.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 357.Hu FP, Xu XG, Zhu DM, Wang MG. 2008. Coexistence of qnrB4 and qnrS1 in a clinical strain of Klebsiella pneumoniae. Acta Pharmacol Sin 29:320–324. doi: 10.1111/j.1745-7254.2008.00757.x. [DOI] [PubMed] [Google Scholar]
- 358.Poirel L, Cattoir V, Soares A, Soussy CJ, Nordmann P. 2007. Novel Ambler class A β-lactamase LAP-1 and its association with the plasmid-mediated quinolone resistance determinant QnrS1. Antimicrob Agents Chemother 51:631–637. doi: 10.1128/AAC.01082-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Kim HB, Wang M, Ahmed S, Park CH, LaRocque RC, Faruque ASG, Salam MA, Khan WA, Qadri F, Calderwood SB, Jacoby GA, Hooper DC. 2010. Transferable quinolone resistance in Vibrio cholerae. Antimicrob Agents Chemother 54:799–803. doi: 10.1128/AAC.01045-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Rajpara N, Patel A, Tiwari N, Bahuguna J, Antony A, Choudhury I, Ghosh A, Jain R, Ghosh A, Bhardwaj AK. 2009. Mechanism of drug resistance in a clinical isolate of Vibrio fluvialis: involvement of multiple plasmids and integrons. Int J Antimicrob Agents 34:220–222. doi: 10.1016/j.ijantimicag.2009.03.020. [DOI] [PubMed] [Google Scholar]
- 361.Xia R, Guo X, Zhang Y, Xu H. 2010. qnrVC-like gene located in a novel complex class 1 integron harboring the ISCR1 element in an Aeromonas punctata strain from an aquatic environment in Shandong province, China. Antimicrob Agents Chemother 54:3471–3474. doi: 10.1128/AAC.01668-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Wu K, Wang F, Sun J, Wang Q, Chen Q, Yu S, Rui Y. 2012. Class 1 integron gene cassettes in multidrug-resistant Gram-negative bacteria in southern China. Int J Antimicrob Agents 40:264–267. doi: 10.1016/j.ijantimicag.2012.05.017. [DOI] [PubMed] [Google Scholar]
- 363.Zhang Y, Zheng Z, Chan EW, Dong N, Xia X, Chen S. 2018. Molecular characterization of qnrVC genes and their novel alleles in Vibrio spp. isolated from food products in China. Antimicrob Agents Chemother 62:e00529-18. doi: 10.1128/AAC.00529-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Bado I, Ezdra RP, Cordeiro N, Outeda M, Caiata L, García-Fulgueiras V, Seija V, Vignoli R. 2018. Detection of qnrVC6, within a new genetic context, in a NDM-1-producing Citrobacter freundii clinical isolate from Uruguay. J Glob Antimicrob Resist 14:95–98. doi: 10.1016/j.jgar.2018.02.023. [DOI] [PubMed] [Google Scholar]
- 365.Janvier F, Otto MP, Jové T, Mille A, Contargyris C, Meaudre E, Brisou P, Plésiat P, Jeannot K. 2017. A case of multiple contamination with methylase ArmA-producing pathogens. J Antimicrob Chemother 72:618–620. doi: 10.1093/jac/dkw418. [DOI] [PubMed] [Google Scholar]
- 366.Tacão M, Moura A, Correia A, Henriques I. 2014. Co-resistance to different classes of antibiotics among ESBL-producers from aquatic systems. Water Res 48:100–107. doi: 10.1016/j.watres.2013.09.021. [DOI] [PubMed] [Google Scholar]
- 367.Jacoby GA, Hooper DC. 2013. Phylogenetic analysis of chromosomally determined qnr and related proteins. Antimicrob Agents Chemother 57:1930–1934. doi: 10.1128/AAC.02080-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Shimizu K, Kikuchi K, Sasaki T, Takahashi N, Ohtsuka M, Ono Y, Hiramatsu K. 2008. Smqnr, a new chromosome-carried quinolone resistance gene in Stenotrophomonas maltophilia. Antimicrob Agents Chemother 52:3823–3825. doi: 10.1128/AAC.00026-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Gracia-Paez JI, Ferraz JR, Silva IA, Rossi F, Levin AS, Costa SF. 2013. Smqnr variants in clinical isolates of Stenotrophomonas maltophilia in Brazil. Rev Inst Med Trop Sao Paulo 55:417–420. doi: 10.1590/S0036-46652013000600008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Kanamori H, Yano H, Tanouchi A, Kakuta R, Endo S, Ichimura S, Ogawa M, Shimojima M, Inomata S, Ozawa D, Aoyagi T, Weber DJ, Kaku M. 2015. Prevalence of Smqnr and plasmid-mediated quinolone resistance determinants in clinical isolates of Stenotrophomonas maltophilia from Japan: novel variants of Smqnr. New Microbes New Infect 7:8–14. doi: 10.1016/j.nmni.2015.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Shah S, Heddle JG. 2014. Squaring up to DNA: pentapeptide repeat proteins and DNA mimicry. Appl Microbiol Biotechnol 98:9545–9560. doi: 10.1007/s00253-014-6151-3. [DOI] [PubMed] [Google Scholar]
- 372.Vetting MW, Hegde SS, Fajardo JE, Fiser A, Roderick SL, Takiff HE, Blanchard JS. 2006. Pentapeptide repeat proteins. Biochemistry 45:1–10. doi: 10.1021/bi052130w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Hegde SS, Vetting MW, Roderick SL, Mitchenall LA, Maxwell A, Takiff HE, Blanchard JS. 2005. A fluoroquinolone resistance protein from Mycobacterium tuberculosis that mimics DNA. Science 308:1480–1483. doi: 10.1126/science.1110699. [DOI] [PubMed] [Google Scholar]
- 374.Montero C, Mateu G, Rodríguez R, Takiff H. 2001. Intrinsic resistance of Mycobacterium smegmatis to fluoroquinolones may be influenced by new pentapeptide protein MfpA. Antimicrob Agents Chemother 45:3387–3392. doi: 10.1128/AAC.45.12.3387-3392.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Garrido MC, Herrero M, Kolter R, Moreno F. 1988. The export of the DNA replication inhibitor microcin B17 provides immunity for the host cell. EMBO J 7:1853–1862. doi: 10.1002/j.1460-2075.1988.tb03018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Vetting MW, Hegde SS, Wang M, Jacoby GA, Hooper DC, Blanchard JS. 2011. Structure of QnrB1, a plasmid-mediated fluoroquinolone resistance factor. J Biol Chem 286:25265–25273. doi: 10.1074/jbc.M111.226936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Xiong X, Bromley EH, Oelschlaeger P, Woolfson DN, Spencer J. 2011. Structural insights into quinolone antibiotic resistance mediated by pentapeptide repeat proteins: conserved surface loops direct the activity of a Qnr protein from a gram-negative bacterium. Nucleic Acids Res 39:3917–3927. doi: 10.1093/nar/gkq1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Hooper DC, Jacoby GA. 2016. Topoisomerase inhibitors: fluoroquinolone mechanisms of action and resistance. Cold Spring Harb Perspect Med 6:a025320. doi: 10.1101/cshperspect.a025320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Park KS, Lee JH, Jeong DU, Lee JJ, Wu X, Jeong BC, Kang CM, Lee SH. 2011. Determination of pentapeptide repeat units in Qnr proteins by the structure-based alignment approach. Antimicrob Agents Chemother 55:4475–4478. doi: 10.1128/AAC.00041-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Vetting MW, Hegde SS, Zhang Y, Blanchard JS. 2011. Pentapeptide-repeat proteins that act as topoisomerase poison resistance factors have a common dimer interface. Acta Crystallogr Sect F Struct Biol Cryst Commun 67:296–302. doi: 10.1107/S1744309110053315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Hegde SS, Vetting MW, Mitchenall LA, Maxwell A, Blanchard JS. 2011. Structural and biochemical analysis of the pentapeptide repeat protein EfsQnr, a potent DNA gyrase inhibitor. Antimicrob Agents Chemother 55:110–117. doi: 10.1128/AAC.01158-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Ellington MJ, Woodford N. 2006. Fluoroquinolone resistance and plasmid addiction systems: self-imposed selection pressure? J Antimicrob Chemother 57:1026–1029. doi: 10.1093/jac/dkl110. [DOI] [PubMed] [Google Scholar]
- 383.Kwak YG, Jacoby GA, Hooper DC. 2015. Effect of Qnr on plasmid gyrase toxins CcdB and ParE. Antimicrob Agents Chemother 59:5078–5079. doi: 10.1128/AAC.00524-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Rodríguez-Martínez JM, Machuca J, Cano ME, Calvo J, Martínez-Martínez L, Pascual A. 2016. Plasmid-mediated quinolone resistance: two decades on. Drug Resist Updat 29:13–29. doi: 10.1016/j.drup.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 385.Da Re S, Garnier F, Guérin E, Campoy S, Denis F, Ploy MC. 2009. The SOS response promotes qnrB quinolone-resistance determinant expression. EMBO Rep 10:929–933. doi: 10.1038/embor.2009.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Wang M, Jacoby GA, Mills DM, Hooper DC. 2009. SOS regulation of qnrB expression. Antimicrob Agents Chemother 53:821–823. doi: 10.1128/AAC.00132-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Hau HH, Gralnick JA. 2007. Ecology and biotechnology of the genus Shewanella. Annu Rev Microbiol 61:237–258. doi: 10.1146/annurev.micro.61.080706.093257. [DOI] [PubMed] [Google Scholar]
- 388.Tran JH, Jacoby GA, Hooper DC. 2005. Interaction of the plasmid-encoded quinolone resistance protein Qnr with Escherichia coli DNA gyrase. Antimicrob Agents Chemother 49:118–125. doi: 10.1128/AAC.49.1.118-125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Tran JH, Jacoby GA, Hooper DC. 2005. Interaction of the plasmid-encoded quinolone resistance protein QnrA with Escherichia coli topoisomerase IV. Antimicrob Agents Chemother 49:3050–3052. doi: 10.1128/AAC.49.7.3050-3052.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Kim ES, Chen C, Braun M, Kim HY, Okumura R, Wang Y, Jacoby GA, Hooper DC. 2015. Interactions between QnrB, QnrB mutants, and DNA gyrase. Antimicrob Agents Chemother 59:5413–5419. doi: 10.1128/AAC.00771-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Cattoir V, Poirel L, Nordmann P. 2007. In-vitro mutagenesis of qnrA and qnrS genes and quinolone resistance in Escherichia coli. Clin Microbiol Infect 13:940–943. doi: 10.1111/j.1469-0691.2007.01778.x. [DOI] [PubMed] [Google Scholar]
- 392.Rodríguez-Martínez JM, Briales A, Velasco C, Conejo MC, Martínez-Martínez L, Pascual A. 2009. Mutational analysis of quinolone resistance in the plasmid-encoded pentapeptide repeat proteins QnrA, QnrB and QnrS. J Antimicrob Chemother 63:1128–1134. doi: 10.1093/jac/dkp111. [DOI] [PubMed] [Google Scholar]
- 393.Guo Q, Weng J, Xu X, Wang M, Wang X, Ye X, Wang W, Wang M. 2010. A mutational analysis and molecular dynamics simulation of quinolone resistance proteins QnrA1 and QnrC from Proteus mirabilis. BMC Struct Biol 10:33. doi: 10.1186/1472-6807-10-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Po KH, Chan EW, Chen S. 2016. Mutational analysis of quinolone resistance protein QnrVC7 provides novel insights into the structure-activity relationship of Qnr proteins. Antimicrob Agents Chemother 60:1939–1942. doi: 10.1128/AAC.01805-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Martens R, Wetzstein HG, Zadrazil F, Capelari M, Hoffmann P, Schmeer N. 1996. Degradation of the fluoroquinolone enrofloxacin by wood-rotting fungi. Appl Environ Microbiol 62:4206–4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Wetzstein HG, Schmeer N, Karl W. 1997. Degradation of the fluoroquinolone enrofloxacin by the brown rot fungus Gloeophyllum striatum: identification of metabolites. Appl Environ Microbiol 63:4272–4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Wetzstein HG, Stadler M, Tichy HV, Dalhoff A, Karl W. 1999. Degradation of ciprofloxacin by basidiomycetes and identification of metabolites generated by the brown rot fungus Gloeophyllum striatum. Appl Environ Microbiol 65:1556–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Rusch M, Kauschat A, Spielmeyer A, Römpp A, Hausmann H, Zorn H, Hamscher G. 2015. Biotransformation of the antibiotic danofloxacin by Xylaria longipes leads to an efficient reduction of its antibacterial activity. J Agric Food Chem 63:6897–6904. doi: 10.1021/acs.jafc.5b02343. [DOI] [PubMed] [Google Scholar]
- 399.Chen Y, Rosazza JP, Reese CP, Chang HY, Nowakowski MA, Kiplinger JP. 1997. Microbial models of soil metabolism: biotransformations of danofloxacin. J Ind Microbiol Biotechnol 19:378–384. doi: 10.1038/sj.jim.2900409. [DOI] [PubMed] [Google Scholar]
- 400.Kim DW, Thawng CN, Lee K, Cha CJ. 2018. Revisiting polymorphic diversity of aminoglycoside N-acetyltransferase AAC(6′)-Ib based on bacterial genomes of human, animal, and environmental origins. Front Microbiol 9:1831. doi: 10.3389/fmicb.2018.01831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Albornoz E, Lucero C, Romero G, Quiroga MP, Rapoport M, Guerriero L, Andres P, Rodriguez C, Galas M, Centrón D, Corso A, Petroni A. 2017. Prevalence of plasmid-mediated quinolone resistance genes in clinical Enterobacteria from Argentina. Microb Drug Resist 23:177–187. doi: 10.1089/mdr.2016.0033. [DOI] [PubMed] [Google Scholar]
- 402.Ruiz E, Ocampo-Sosa AA, Alcoba-Flórez J, Román E, Arlet G, Torres C, Martínez-Martínez L. 2012. Changes in ciprofloxacin resistance levels in Enterobacter aerogenes isolates associated with variable expression of the aac(6′)-Ib-cr gene. Antimicrob Agents Chemother 56:1097–1100. doi: 10.1128/AAC.05074-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Chang CY, Fang YT, Tsai SM, Chang LL, Yu WL. 2009. Characterization of class 1 integrons and gene cassettes in clinical isolates of Klebsiella pneumoniae from Taiwan. Diagn Microbiol Infect Dis 65:214–216. doi: 10.1016/j.diagmicrobio.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 404.Moura A, Pereira C, Henriques I, Correia A. 2012. Novel gene cassettes and integrons in antibiotic-resistant bacteria isolated from urban wastewaters. Res Microbiol 163:92–100. doi: 10.1016/j.resmic.2011.10.010. [DOI] [PubMed] [Google Scholar]
- 405.Pérez-Moreno MO, Centelles-Serrano MJ, Cortell-Ortolá M, Fort-Gallifa I, Ruiz J, Llovet-Lombarte MI, Picó-Plana E, Jardí-Baiges AM. 2011. Molecular epidemiology and resistance mechanisms involved in reduced susceptibility to amoxicillin/clavulanic acid in Klebsiella pneumoniae isolates from a chronic care centre. Int J Antimicrob Agents 37:462–466. doi: 10.1016/j.ijantimicag.2010.12.010. [DOI] [PubMed] [Google Scholar]
- 406.Kim DW, Thawng CN, Choi JH, Lee K, Cha CJ. 2018. Polymorphism of antibiotic-inactivating enzyme driven by ecology expands the environmental resistome. ISME J 12:267–276. doi: 10.1038/ismej.2017.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Maurice F, Broutin I, Podglajen I, Benas P, Collatz E, Dardel F. 2008. Enzyme structural plasticity and the emergence of broad-spectrum antibiotic resistance. EMBO Rep 9:344–349. doi: 10.1038/embor.2008.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Wang D, Wang H, Qi Y, Liang Y, Zhang J, Yu L. 2012. Characteristics of Klebsiella pneumoniae harboring QnrB32, Aac(6′)-Ib-cr, GyrA and CTX-M-22 genes. Folia Histochem Cytobiol 50:68–74. doi: 10.2478/18698. [DOI] [PubMed] [Google Scholar]
- 409.de Toro M, Rodríguez I, Rojo-Bezares B, Helmuth R, Torres C, Guerra B, Sáenz Y. 2013. pMdT1, a small ColE1-like plasmid mobilizing a new variant of the aac(6′)-Ib-cr gene in Salmonella enterica serovar Typhimurium. J Antimicrob Chemother 68:1277–1280. doi: 10.1093/jac/dkt001. [DOI] [PubMed] [Google Scholar]
- 410.Vetting MW, Park CH, Hegde SS, Jacoby GA, Hooper DC, Blanchard JS. 2008. Mechanistic and structural analysis of aminoglycoside N-acetyltransferase AAC(6)-Ib and its bifunctional, fluoroquinolone-active AAC(6)-Ib-cr variant. Biochemistry 47:9825–9835. doi: 10.1021/bi800664x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Jones GL, Warren RE, Skidmore SJ, Davies VA, Gibreel T, Upton M. 2008. Prevalence and distribution of plasmid-mediated quinolone resistance genes in clinical isolates of Escherichia coli lacking extended-spectrum beta-lactamases. J Antimicrob Chemother 62:1245–1251. doi: 10.1093/jac/dkn406. [DOI] [PubMed] [Google Scholar]
- 412.Cervantes-Vega C, Chávez J, Córdova NA, de la Mora P, Amador Velasco J. 1986. Resistance to metals by Pseudomonas aeruginosa clinical isolates. Microbios 48:159–163. [PubMed] [Google Scholar]
- 413.Ramírez-Díaz MI, Díaz-Magaña A, Meza-Carmen V, Johnstone L, Cervantes C, Rensing C. 2011. Nucleotide sequence of Pseudomonas aeruginosa conjugative plasmid pUM505 containing virulence and heavy-metal resistance genes. Plasmid 66:7–18. doi: 10.1016/j.plasmid.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 414.Ruiz J. 23 July 2019. CrpP, a passenger or a hidden stowaway in the Pseudomonas aeruginosa genome? J Antimicrob Chemother doi: 10.1093/jac/dkz316. [DOI] [PubMed] [Google Scholar]
- 415.Kung VL, Ozer EA, Hauser AR. 2010. The accessory genome of Pseudomonas aeruginosa. Microbiol Mol Biol Rev 74:621–641. doi: 10.1128/MMBR.00027-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Klockgether J, Reva O, Larbig K, Tümmler B. 2004. Sequence analysis of the mobile genome island pKLC102 of Pseudomonas aeruginosa C. J Bacteriol 186:518–534. doi: 10.1128/JB.186.2.518-534.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417.Chávez-Jacobo VM, Hernández-Ramírez KC, Silva-Sánchez J, Garza-Ramos U, Barrios-Camacho H, Ortiz-Alvarado R, Cervantes C, Meza-Carmen V, Ramírez-Díaz MI. 2019. Prevalence of the crpP gene conferring decreased ciprofloxacin susceptibility in enterobacterial clinical isolates from Mexican hospitals. J Antimicrob Chemother 74:1253–1259. doi: 10.1093/jac/dky562. [DOI] [PubMed] [Google Scholar]
- 418.Horna G, López M, Guerra H, Saénz Y, Ruiz J. 2018. Interplay between MexAB-OprM and MexEF-OprN in clinical isolates of Pseudomonas aeruginosa. Sci Rep 8:16463. doi: 10.1038/s41598-018-34694-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Lomovskaya O, Warren MS, Lee A, Galazzo J, Fronko R, Lee M, Blais J, Cho D, Chamberland S, Renau T, Leger R, Hecker S, Watkins W, Hoshino K, Ishida H, Lee VJ. 2001. Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: novel agents for combination therapy. Antimicrob Agents Chemother 45:105–116. doi: 10.1128/AAC.45.1.105-116.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Chen Y, Hu D, Zhang Q, Liao XP, Liu YH, Sun J. 2017. Efflux pump overexpression contributes to tigecycline heteroresistance in Salmonella enterica serovar Typhimurium. Front Cell Infect Microbiol 7:37. doi: 10.3389/fcimb.2017.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421.Bialek-Davenet S, Lavigne JP, Guyot K, Mayer N, Tournebize R, Brisse S, Leflon-Guibout V, Nicolas-Chanoine MH. 2015. Differential contribution of AcrAB and OqxAB efflux pumps to multidrug resistance and virulence in Klebsiella pneumoniae. J Antimicrob Chemother 70:81–88. doi: 10.1093/jac/dku340. [DOI] [PubMed] [Google Scholar]
- 422.Veleba M, Higgins PG, Gonzalez G, Seifert H, Schneiders T. 2012. Characterization of RarA, a novel AraC family multidrug resistance regulator in Klebsiella pneumoniae. Antimicrob Agents Chemother 56:4450–4458. doi: 10.1128/AAC.00456-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423.Furlan JPR, Sanchez DG, Gallo IFL, Stehling EG. 2019. Characterization of acquired antimicrobial resistance genes in environmental Stenotrophomonas maltophilia isolates from Brazil. Microb Drug Resist 25:475–479. doi: 10.1089/mdr.2018.0216. [DOI] [PubMed] [Google Scholar]
- 424.Chen X, Zhang W, Pan W, Yin J, Pan Z, Gao S, Jiao X. 2012. Prevalence of qnr, aac(6′)-Ib-cr, qepA, and oqxAB in Escherichia coli isolates from humans, animals, and the environment. Antimicrob Agents Chemother 56:3423–3427. doi: 10.1128/AAC.06191-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425.Dotto G, Giacomelli M, Grilli G, Ferrazzi V, Carattoli A, Fortini D, Piccirillo A. 2014. High prevalence of oqxAB in Escherichia coli isolates from domestic and wild lagomorphs in Italy. Microb Drug Resist 20:118–123. doi: 10.1089/mdr.2013.0141. [DOI] [PubMed] [Google Scholar]
- 426.Janecko N, Halova D, Jamborova I, Papousek I, Masarikova M, Dolejska M, Literak I. 2018. Occurrence of plasmid-mediated quinolone resistance genes in Escherichia coli and Klebsiella spp. recovered from Corvus brachyrhynchos and Corvus corax roosting in Canada. Lett Appl Microbiol 67:130–135. doi: 10.1111/lam.12993. [DOI] [PubMed] [Google Scholar]
- 427.Li L, Liao X, Yang Y, Sun J, Li L, Liu B, Yang S, Ma J, Li X, Zhang Q, Liu Y. 2013. Spread of oqxAB in Salmonella enterica serotype Typhimurium predominantly by IncHI2 plasmids. J Antimicrob Chemother 68:2263–2268. doi: 10.1093/jac/dkt209. [DOI] [PubMed] [Google Scholar]
- 428.Manageiro V, Félix D, Jones-Dias D, Sampaio DA, Vieira L, Sancho L, Ferreira E, Caniça M. 2017. Genetic background and expression of the new qepA4 gene variant recovered in clinical TEM-1- and CMY-2-producing Escherichia coli. Front Microbiol 8:1899. doi: 10.3389/fmicb.2017.01899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 429.Rahman Z, Islam A, Rashid MU, Johura FT, Monira S, Watanabe H, Ahmed N, Camilli A, Alam M. 2017. Existence of a novel qepA variant in quinolone resistant Escherichia coli from aquatic habitats of Bangladesh. Gut Pathog 9:58. doi: 10.1186/s13099-017-0207-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430.Wang D, Huang X, Chen J, Mou Y, Li H, Yang L. 2015. Characterization of genetic structures of the QepA3 gene in clinical isolates of Enterobacteriaceae. Front Microbiol 6:1147. doi: 10.3389/fmicb.2015.01147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Machuca J, Briales A, Díaz-de-Alba P, Martínez-Martínez L, Pascual Á, Rodríguez-Martínez JM. 2015. Effect of the efflux pump QepA2 combined with chromosomally mediated mechanisms on quinolone resistance and bacterial fitness in Escherichia coli. J Antimicrob Chemother 70:2524–2527. doi: 10.1093/jac/dkv144. [DOI] [PubMed] [Google Scholar]
- 432.Littlejohn TG, Paulsen IT, Gillespie MT, Tennent JM, Midgley M, Jones IG, Purewal AS, Skurray RA. 1992. Substrate specificity and energetics of antiseptic and disinfectant resistance in Staphylococcus aureus. FEMS Microbiol Lett 74:259–265. doi: 10.1111/j.1574-6968.1992.tb05376.x. [DOI] [PubMed] [Google Scholar]
- 433.Paulsen IT, Brown MH, Littlejohn TG, Mitchell BA, Skurray RA. 1996. Multidrug resistance proteins QacA and QacB from Staphylococcus aureus: membrane topology and identification of residues involved in substrate specificity. Proc Natl Acad Sci U S A 93:3630–3635. doi: 10.1073/pnas.93.8.3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Miyajima E, Harada D, Nakaminami H, Kitamura Y, Tamura T, Kawakubo T, Noguchi N. 9 April 2019. Decreased prevalence of qacA-positive methicillin-resistant Staphylococcus aureus in hospitalized patients in Tokyo, Japan. Microb Drug Resist doi: 10.1089/mdr.2018.0351. [DOI] [PubMed] [Google Scholar]
- 435.Infante-Martínez V, Rodríguez-Martínez JM, Velasco C, Pascual A. 2012. Ausencia del sistema de expulsión de quinolonas mediado por plásmido QacBIII en aislados bacteriémicos de Staphylococcus aureus en el área hospitalaria Virgen Macarena de Sevilla. Enferm Infecc Microbiol Clin 30:51. doi: 10.1016/j.eimc.2011.07.019. [DOI] [PubMed] [Google Scholar]
- 436.Szczepanowski R, Krahn I, Linke B, Goesmann A, Pühler A, Schlüter A. 2004. Antibiotic multiresistance plasmid pRSB101 isolated from a wastewater treatment plant is related to plasmids residing in phytopathogenic bacteria and carries eight different resistance determinants including a multidrug transport system. Microbiology 150:3613–3630. doi: 10.1099/mic.0.27317-0. [DOI] [PubMed] [Google Scholar]
- 437.Dolejska M, Villa L, Poirel L, Nordmann P, Carattoli A. 2013. Complete sequencing of an IncHI1 plasmid encoding the carbapenemase NDM-1, the ArmA 16S RNA methylase and a resistance-nodulation-cell division/multidrug efflux pump. J Antimicrob Chemother 68:34–39. doi: 10.1093/jac/dks357. [DOI] [PubMed] [Google Scholar]
- 438.Abraham EP, Chain E. 1940. An enzyme from bacteria able to destroy penicillin. Nature 146:837. doi: 10.1038/146837a0. [DOI] [PubMed] [Google Scholar]
- 439.Appelt S, Fancello L, Le Bailly M, Raoult D, Drancourt M, Desnues C. 2014. Viruses in a 14th-century coprolite. Appl Environ Microbiol 80:2648–2655. doi: 10.1128/AEM.03242-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Petrova M, Gorlenko Z, Mindlin S. 2011. Tn5045, a novel integron-containing antibiotic and chromate resistance transposon isolated from a permafrost bacterium. Res Microbiol 162:337–345. doi: 10.1016/j.resmic.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 441.Hall BG, Barlow M. 2004. Evolution of the serine beta-lactamases: past, present and future. Drug Resist Updat 7:111–123. doi: 10.1016/j.drup.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 442.Mosher RH, Camp DJ, Yang K, Brown MP, Shaw WV, Vining LC. 1995. Inactivation of chloramphenicol by O-phosphorylation. A novel resistance mechanism in Streptomyces venezuelae ISP5230, a chloramphenicol producer. J Biol Chem 270:27000–27006. doi: 10.1074/jbc.270.45.27000. [DOI] [PubMed] [Google Scholar]
- 443.Uchiyama H, Weisblum B. 1985. N-Methyl transferase of Streptomyces erythraeus that confers resistance to the macrolide-lincosamide-streptogramin B antibiotics: amino acid sequence and its homology to cognate R-factor enzymes from pathogenic bacilli and cocci. Gene 38:103–110. doi: 10.1016/0378-1119(85)90208-2. [DOI] [PubMed] [Google Scholar]
- 444.Van Goethem MW, Pierneef R, Bezuidt OKI, Van De Peer Y, Cowan DA, Makhalanyane TP. 2018. A reservoir of ‘historical’ antibiotic resistance genes in remote pristine Antarctic soils. Microbiome 6:40. doi: 10.1186/s40168-018-0424-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445.Duchesne E. 1897. Contribution à l’etude de la concurrence vitale chez les microorganismes. Antagonisme entre les moisissures et le microbes. PhD thesis Faculté de Médecine et de Pharmacie de Lyon, Lyon, France. [Google Scholar]
- 446.Garre C. 1887. Ueber antagonisten unter den Bakterien. Korrespondenzbl Schweiz Aerzte 17:385–392. [Google Scholar]
- 447.Ruiz J. 2005. La prehistoria de los antimicrobianos, p 41–44. In Barriobero Neiva JI, Soldevilla Armas A (ed), Un breve viaje por la ciencia. Universidad de La Rioja, Logroño, Spain. [Google Scholar]
- 448.Tiberio V. 1895. Sugli estratti di alcune muffe. An Ig Sper 5:91–103. [Google Scholar]
- 449.Emmerich R, Löw O. 1899. Bakteriolytische enzyme als ursache der erworbenen immunität und die heilung von infectionskrankheiten durch dieselben. Z Hyg Infektionskr 31:1–56. [Google Scholar]
- 450.Jones HW. 1911. Report of a series of cases of syphilis treated by Ehrlich’s arsenobenzol at Walter Reed General Hospital. District of Columbia. Boston Med Surg J 164:381–383. doi: 10.1056/NEJM191103161641103. [DOI] [Google Scholar]
- 451.Domagk G. 1935. Ein beitrag zur chemotherapie der bakteriellen infektionen. Dtsch Med Wochenschr 61:250–253. doi: 10.1055/s-0028-1129486. [DOI] [Google Scholar]
- 452.Higuchi S, Onodera Y, Chiba M, Hoshino K, Gotoh N. 2013. Potent in vitro antibacterial activity of DS-8587, a novel broad-spectrum quinolone, against Acinetobacter baumannii. Antimicrob Agents Chemother 57:1978–1981. doi: 10.1128/AAC.02374-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453.Pucci MJ, Bush K. 2013. Investigational antimicrobial agents of 2013. Clin Microbiol Rev 26:792–821. doi: 10.1128/CMR.00033-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 454.Balwan A, Nicolau DP, Wungwattana M, Zuckerman JB, Waters V. 2016. Clinafloxacin for treatment of Burkholderia cenocepacia infection in a cystic fibrosis patient. Antimicrob Agents Chemother 60:1–5. doi: 10.1128/AAC.01428-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 455.Madurga S, Sánchez-Céspedes J, Belda I, Vila J, Giralt E. 2008. Mechanism of binding of fluoroquinolones to the quinolone resistance-determining region of DNA gyrase: towards an understanding of the molecular basis of quinolone resistance. Chembiochem 9:2081–2086. doi: 10.1002/cbic.200800041. [DOI] [PubMed] [Google Scholar]
- 456.Hata H, Natori T, Mizuno T, Kanazawa I, Eldesouky I, Hayashi M, Miyata M, Fukunaga H, Ohji S, Hosoyama A, Aono E, Yamazoe A, Tsuchikane K, Fujita N, Ezaki T. 2016. Phylogenetics of family Enterobacteriaceae and proposal to reclassify Escherichia hermannii and Salmonella subterranea as Atlantibacter hermannii and Atlantibacter subterranea gen. nov., comb. nov. Microbiol Immunol 60:303–311. doi: 10.1111/1348-0421.12374. [DOI] [PubMed] [Google Scholar]
- 457.Ma X, Lü X, Chen H, Gao Y, Mu L, Yu R, Du R, Niu F. 2009. Study on qnrB-mediated quinolone resistance among Enterobacteriaceae species. Chin J Antibiot 2:114–119. [Google Scholar]
- 458.Ma J, Zeng Z, Chen Z, Xu X, Wang X, Deng Y, Lü D, Huang L, Zhang Y, Liu J, Wang M. 2009. High prevalence of plasmid-mediated quinolone resistance determinants qnr, aac(6′)-Ib-cr, and qepA among ceftiofur-resistant Enterobacteriaceae isolates from companion and food-producing animals. Antimicrob Agents Chemother 53:519–524. doi: 10.1128/AAC.00886-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.Lenaerts AJ, Bitting C, Woolhiser L, Gruppo V, Marietta KS, Johnson CM, Orme IM. 2008. Evaluation of a 2-pyridone, KRQ-10018, against Mycobacterium tuberculosis in vitro and in vivo. Antimicrob Agents Chemother 52:1513–1515. doi: 10.1128/AAC.00897-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 460.McCullough PA, Bennett-Guerrero E, Chawla LS, Beaver T, Mehta RL, Molitoris BA, Eldred A, Ball G, Lee HJ, Houser MT, Khan S. 2016. ABT-719 for the prevention of acute kidney injury in patients undergoing high-risk cardiac surgery: a randomized phase 2b clinical trial. J Am Heart Assoc 5:e003549. doi: 10.1161/JAHA.116.003549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Park-Wyllie LY, Juurlink DN, Kopp A, Shah BR, Stukel TA, Stumpo C, Dresser L, Low DE, Mamdani MM. 2006. Outpatient gatifloxacin therapy and dysglycemia in older adults. N Engl J Med 354:1352–1361. doi: 10.1056/NEJMoa055191. [DOI] [PubMed] [Google Scholar]
- 462.Blum MD, Graham DJ, McCloskey CA. 1994. Temafloxacin syndrome: review of 95 cases. Clin Infect Dis 18:946–950. doi: 10.1093/clinids/18.6.946. [DOI] [PubMed] [Google Scholar]
- 463.Crowell EL, Koduri VA, Groat RS, Lee DA. 2017. Cost comparison of commonly used postoperative topical ophthalmic antibiotics. J Cataract Refract Surg 43:1322–1327. doi: 10.1016/j.jcrs.2017.07.028. [DOI] [PubMed] [Google Scholar]
- 464.Wang D, Yang J, Fang M, He W, Zhang Y, Liu C, Zhou D. 2017. Characterization of the novel In1059 harbouring VIM gene cassette. Antimicrob Resist Infect Control 6:50. doi: 10.1186/s13756-017-0204-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 465.Wei Q, Jiang X, Yang Z, Chen N, Chen X, Li G, Lu Y. 2009. dfrA27, a new integron-associated trimethoprim resistance gene from Escherichia coli. J Antimicrob Chemother 63:405–406. doi: 10.1093/jac/dkn474. [DOI] [PubMed] [Google Scholar]
- 466.Pallecchi L, Riccobono E, Sennati S, Mantella A, Bartalesi F, Trigoso C, Gotuzzo E, Bartoloni A, Rossolini GM. 2010. Characterization of small ColE-like plasmids mediating widespread dissemination of the qnrB19 gene in commensal enterobacteria. Antimicrob Agents Chemother 54:678–682. doi: 10.1128/AAC.01160-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 467.Hering J, Dunevall E, Ek M, Brändén G. 2018. Structural basis for selective inhibition of antibacterial target MraY, a membrane-bound enzyme involved in peptidoglycan synthesis. Drug Discov Today 23:1426–1435. doi: 10.1016/j.drudis.2018.05.020. [DOI] [PubMed] [Google Scholar]
- 468.Li R, Xie M, Lv J, Wai-Chi Chan E, Chen S. 2017. Complete genetic analysis of plasmids carrying mcr-1 and other resistance genes in an Escherichia coli isolate of animal origin. J Antimicrob Chemother 72:696–699. doi: 10.1093/jac/dkw509. [DOI] [PubMed] [Google Scholar]
- 469.Shen Y, Zhou H, Xu J, Wang Y, Zhang Q, Walsh TR, Shao B, Wu C, Hu Y, Yang L, Shen Z, Wu Z, Sun Q, Ou Y, Wang Y, Wang S, Wu Y, Cai C, Li J, Shen J, Zhang R, Wang Y. 2018. Anthropogenic and environmental factors associated with high incidence of mcr-1 carriage in humans across China. Nat Microbiol 3:1054–1062. doi: 10.1038/s41564-018-0205-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 470.Wang Y, Zhang R, Li J, Wu Z, Yin W, Schwarz S, Tyrrell JM, Zheng Y, Wang S, Shen Z, Liu Z, Liu J, Lei L, Li M, Zhang Q, Wu C, Zhang Q, Wu Y, Walsh TR, Shen J. 2017. Comprehensive resistome analysis reveals the prevalence of NDM and MCR-1 in Chinese poultry production. Nat Microbiol 2:16260. doi: 10.1038/nmicrobiol.2016.260. [DOI] [PubMed] [Google Scholar]
- 471.Lascols C, Robert J, Cattoir V, Bébéar C, Cavallo JD, Podglajen I, Ploy MC, Bonnet R, Soussy CJ, Cambau E. 2007. Type II topoisomerase mutations in clinical isolates of Enterobacter cloacae and other enterobacterial species harbouring the qnrA gene. Int J Antimicrob Agents 29:402–409. doi: 10.1016/j.ijantimicag.2006.11.008. [DOI] [PubMed] [Google Scholar]