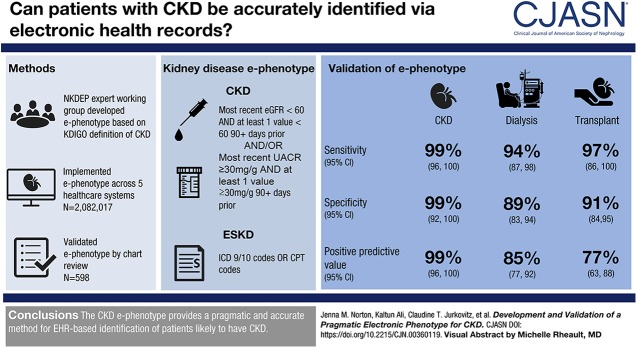
Visual Abstract
Keywords: adult; humans; renal dialysis; creatinine; outpatients; chronic kidney failure; chronic renal insufficiency; glomerular filtration rate; EGFR protein, human; epidermal growth factor; receptor, epidermal growth factor; albumins; phenotype
Abstract
Background and objectives
Poor identification of individuals with CKD is a major barrier to research and appropriate clinical management of the disease. We aimed to develop and validate a pragmatic electronic (e-) phenotype to identify patients likely to have CKD.
Design, setting, participants, & measurements
The e-phenotype was developed by an expert working group and implemented among adults receiving in- or outpatient care at five healthcare organizations. To determine urine albumin (UA) dipstick cutoffs for CKD to enable use in the e-phenotype when lacking urine albumin-to-creatinine ratio (UACR), we compared same day UACR and UA results at four sites. A sample of patients, spanning no CKD to ESKD, was randomly selected at four sites for validation via blinded chart review.
Results
The CKD e-phenotype was defined as most recent eGFR <60 ml/min per 1.73 m2 with at least one value <60 ml/min per 1.73 m2 >90 days prior and/or a UACR of ≥30 mg/g in the most recent test with at least one positive value >90 days prior. Dialysis and transplant were identified using diagnosis codes. In absence of UACR, a sensitive CKD definition would consider negative UA results as normal to mildly increased (KDIGO A1), trace to 1+ as moderately increased (KDIGO A2), and ≥2+ as severely increased (KDIGO A3). Sensitivity, specificity, and diagnostic accuracy of the CKD e-phenotype were 99%, 99%, and 98%, respectively. For dialysis sensitivity was 94% and specificity was 89%. For transplant, sensitivity was 97% and specificity was 91%.
Conclusions
The CKD e-phenotype provides a pragmatic and accurate method for EHR-based identification of patients likely to have CKD.
Introduction
CKD is associated with increased morbidity, mortality, and health care costs (1) as well as decreased quality of life (2). An estimated 30 million American adults—nearly 15% of the population—have CKD, and millions more are at risk for the disease (3). CKD is progressive, often resulting in ESKD or death from cardiovascular disease (1). Treatments exist to slow progression and manage complications of CKD; yet, many with the disease do not receive such treatments (4). For example, in 2015, serum creatinine, lipids levels, and albuminuria were assessed in only about one third of Medicare recipients with CKD, and just under two thirds were prescribed recommended renin-angiotensin-aldosterone system blockers (4). Limited progress has been made in reducing the burden of CKD in the United States, despite published clinical guidelines and efforts to raise awareness of the disease and improve patient care.
A major barrier to appropriate CKD management is delayed identification of individuals with the disease. National estimates indicate that only 8% of people with CKD are aware of their condition (5). Although recent studies suggest that questions used to evaluate CKD awareness in national samples may underestimate actual awareness, such studies still find low CKD awareness at about 30%–40% (6). Because CKD is often asymptomatic in early stages, it frequently goes undiagnosed until the disease is very advanced. Therefore, diagnostic codes are inadequate to identify individuals with CKD for population management, surveillance, and research (7,8).
An electronic (e-) CKD phenotype using data widely available in the electronic health record (EHR) could facilitate identification of patients likely to have CKD (9). CKD is typically detected by objective laboratory data, including serum creatinine used to estimate the GFR (eGFR) and urine albumin-to-creatinine ratio (UACR) to detect albuminuria (9,10). Although e-phenotypes for CKD exist (8), they are complex and require substantial information technology (IT) infrastructure and support that many health systems lack. The National Kidney Disease Education Program (NKDEP) established a working group to develop a pragmatic CKD e-phenotype to help health care organizations, providers, and researchers identify patients likely to have CKD to facilitate population health management, surveillance, and research. The NKDEP CKD e-Phenotype working group functions under the NKDEP Health IT working group, which was established in October 2012 to “enable and support the widespread interoperability of data related to kidney health among health care software applications to optimize CKD detection and management.” This manuscript describes the development, implementation, and validation of the pragmatic CKD e-phenotype across multiple health systems.
Materials and Methods
Development of the NKDEP CKD e-Phenotype
The NKDEP CKD e-phenotype was iteratively developed by the working group over a series of conference calls. The working group aimed to keep the phenotype as simple as possible to enable broad implementation. The steps in the development of the e-phenotype were to (1) determine a definition of CKD, (2) identify clinical variables integral to the definition, (3) identify Logical Observation Identifiers Names and Codes (LOINC) for each clinical variable, and (4) identify billing and procedure codes for important related diagnoses (e.g., kidney transplantation and dialysis).
Determining Cutoffs for Urine Albumin
Prior experience with EHR data suggested that UACR would only be available in a small proportion of patients, but urinalyses (urine albumin [UA]) would be more widely available. However, the appropriate cutoffs for determining CKD on the basis of UA have not been well established. Therefore, in order to determine UA results corresponding with the accepted definition of CKD on the basis of UACR, four sites—the Cleveland Clinic, Columbia University, the University of Minnesota, and the Veteran’s Health Administration—identified patients with simultaneous UACR and UA results using data available from the EHR. The distribution of UACR results was compared across UA result categories at each site. In secondary analyses, three of the sites also identified simultaneous urine protein-to-creatinine ratio (UPCR) and UA results and compared the distribution of UPCR results across UA result categories.
Phenotype Implementation and Validation
After developing the NKDEP e-phenotype, the working group validated the implementation feasibility and precision of the e-phenotype, including an electronic validation and a manual chart review to determine its sensitivity, specificity, and accuracy. The e-phenotype was implemented among adult patients (ages ≥18 years old) receiving in- or outpatient care at five health care organizations using EHR data. Unique health systems with varying population demographics, distinct EHR systems, and diverse levels of health IT infrastructure were selected to mitigate selection bias and enhance generalizability of findings. Assumptions and considerations for implementing the NKDEP e-phenotype varied across sites (Table 1).
Table 1.
Assumptions and considerations for electronic phenotype implementation by site
| Site | Determination of eGFR | Race Assumption | Personnel Involved | Estimated Time Investment | Challenges/Barriers | Facilitators |
|---|---|---|---|---|---|---|
| Christiana Care | From laboratory (multiple equations) | All patients had a race, and GFR was checked against race | One nephrologist | Total: 223 h | LOINC not used, requiring crosswalk to internal codes | None |
| One information technologist | Communication: 13 | Separate EHRs used for in- and outpatient care during the study period; required matching and merging multiple warehouses and data marts and pulls directly from the EHRs to produce the dataset | ||||
| One student | Data extraction: 79 | |||||
| One resident | Data merging and cleaning: 131 | |||||
| Columbia University | Recalculated from serum creatinine (CKD-EPI) | Assumed nonblack if race not available | One nephrologist | Total: 20 h | LOINC not used, requiring crosswalk to internal codes; however, crosswalk to internal codes existed | Existing clinical data warehouse; eMERGE site with adequate infrastructure and IT support |
| One informaticist | Communication: 5 | |||||
| One research coordinator | Data extraction: 10 | |||||
| Data cleaning: 5 | ||||||
| University of Minnesota | Recalculated from serum creatinine (MDRD) | Assumed nonblack if race not available | One nephrologist | Total: 44 h | eGFR was less available than serum creatinine | Leveraged data extraction completed for an ongoing project |
| One IT person | Communication: 4 | |||||
| Data extraction: 20 | ||||||
| Data cleaning: 20 | ||||||
| UCSF | From laboratory (CKD-EPI) | Assumed nonblack if race not available | One nephrologist | Total: 49 h | LOINC not used; no existing crosswalk to Clarity codes | Leveraged personal relationships |
| One informaticist | Communication: 5 | |||||
| One IT person | Data extraction: 20 | |||||
| One statistician | Data cleaning: 24 | |||||
| University of Utah | From laboratory (multiple equations) | Assumed nonblack if race not available | One informaticist | Total: 40 h | Identifying relevant laboratory codes as LOINC was not universally available | None |
| Communication: 0 | ||||||
| Data extraction: 10 | ||||||
| Data cleaning: 10 | ||||||
| Data analysis: 20 |
LOINC, Logical Observation Identifiers Names and Codes; EHR, electronic health record; CKD-EPI, Chronic Kidney Disease Epidemiology Collaboration; eMERGE, Electronic Medical Records and Genomics; IT, information technology; MDRD, Modification of Diet in Renal Disease; UCSF, University of California, San Francisco.
Patients were considered active if they had any of the following in the last 18 months: vital signs, laboratory value, clinic visit, or hospitalization. Patients were excluded if they had died before the data extraction date. Each site collected the following information from the EHR and calculated means and SDs or proportions (as appropriate): age, sex, race (black versus nonblack), eGFR, UACR, UPCR, UA, transplant status, and ESKD status. For each of the laboratory values, the sites determined the number of results per patient and whether a value that would qualify a patient as having CKD (e.g., eGFR<60 ml/min per 1.73 m2 or UACR≥30 mg/g) existed at least 90 days before the most recent value. The NKDEP e-phenotype was used to identify patients for the manual validation at four of the five sites. Patients were randomly selected for manual validation across stages of CKD to reduce spectrum bias, wherein the phenotype may be more likely to capture individuals with advanced disease. Patient selection targeted (1) seven to ten patients within each of CKD stages 3A, 3B, 4, and 5; (2) five to ten patients who had received a transplant; (3) five to ten patients on hemodialysis or peritoneal dialysis; and (4) 20 patients without phenotyped CKD; however, numbers varied across sites. Reviewers were blind to the e-phenotype—except for five “no CKD” charts reviewed at the University of California, San Francisco (UCSF) due to a protocol error. At the University of Minnesota and UCSF, two reviewers assessed each chart, and a third reviewer adjudicated disagreements. At Columbia University and Christiana Care Health System (Christiana Care), one reviewer performed all chart reviews. Reviewers recorded the following EHR data: (1) most recent eGFR, UACR, UPCR, and UA before data extraction; (2) dialysis status; (3) transplant status; and (4) race. Sensitivity, specificity, and positive and negative predictive values were calculated using binom.test in R version 3.4.1.
The study was approved by the institutional review board at each site.
Results
The NKDEP e-Phenotype
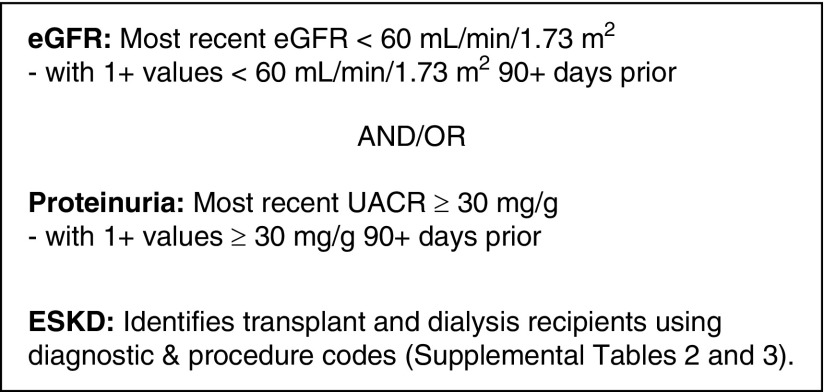
The working group adhered to its guiding principle of simplicity by focusing primarily on CKD. However, the group acknowledges that organizations may expand on the NKDEP e-phenotype to include important CKD-related data, such as relevant conditions (e.g., hypertension and diabetes), medications, and interventions (e.g., vascular access and nephrology consults). The working group defined the CKD e-phenotype as follows: most recent eGFR <60 ml/min per 1.73 m2 with at least one value <60 ml/min per 1.73 m2 >90 days prior and/or proteinuria presenting as a UACR≥30 mg/g in the most recent test with at least one positive value >90 days prior (Figure 1). The NKDEP e-phenotype uses LOINC to identify laboratory values from the EHR (Supplemental Table 1). To determine eGFR, implementers may need to make assumptions about race when race data are unavailable in the EHR, which often is the case. The NKDEP e-phenotype will be less sensitive and more specific if patients are assumed to be black, and it will be more sensitive and less specific if patients are assumed to be nonblack. Similarly, because UACR is frequently unavailable in the EHR, the e-phenotype allows sites to use the most recent result among all proteinuria measures (UACR, UPCR, or UA) for more specific results or any positive result among the most recent UACR, UPCR, or UA for more sensitive results. To facilitate implementation at sites with a variety of technical capabilities, the e-phenotype allows for implementers to use eGFR as reported by the laboratory (regardless of eGFR equation used). Given the small differences between performance of the Modification of Diet in Renal Disease and the Kidney Disease Epidemiology Collaboration (CKD-EPI) estimating equations relative to measured GFR—especially for individuals with an eGFR≥60 ml/min per 1.73 m2—any differences in e-phenotype from using one equation or the other are unlikely to be clinically meaningful (11,12). However, for more consistent results, implementors may choose to recalculate the eGFR from the reported serum creatinine value using the CKD-EPI equation (11). The NKDEP e-phenotype also identifies transplant or dialysis recipients using billing/encounter data using International Classification of Diseases (ICD), Ninth or Tenth Revision and Current Procedural Terminology codes (Supplemental Tables 2 and 3).
Figure 1.
The NKDEP CKD e-phenotype identifies patients likely to have CKD based on laboratory measures. UACR, urine albumin-to-creatinine ratio.
UA Cutoffs
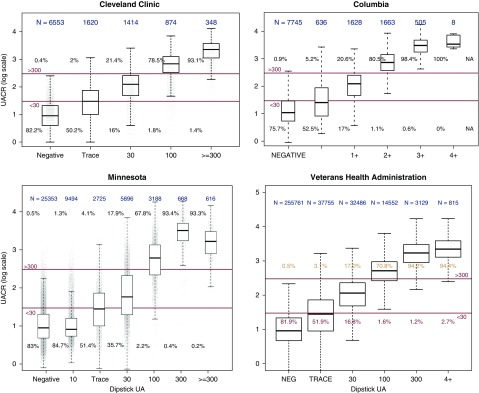
Four sites identified patients with same-day UACR and UA results: University of Minnesota (n=47,940), Cleveland Clinic (n=10,809), Columbia University (n=12,185), and the Veterans Health Administration (n=344,498) (Figure 2). Across the sites, the majority of patients with negative UA result had a same-day UACR <30 mg/g (76%–83%). Approximately one half (50%–53%) of patients with a trace UA result had a UACR<30 mg/g. The majority of patients with a 1+ UA result had a UACR≥30 mg/g (65%–84%), and virtually all patients with 2+, 3+, or 4+ UA results had a UACR≥30 mg/g (97%+). On the basis of these findings, the working group concluded that—in the absence of a UACR result—a sensitive definition of CKD would consider a negative UA result as normal to mildly increased (Kidney Disease Improving Global Outcomes [KDIGO] A1 category), UA results in the trace to 1+ range as moderately increased (KDIGO A2 category), and UA results of 2+ or greater as severely increased (KDIGO A3 category). However, shifting the UA cutoffs to include negative/trace as A1 would yield more specific results. Three sites assessed values from patients with same-day UPCR/UACR and UPCR/UA laboratory results: University of Minnesota (n=9193 and 37,865, respectively), Cleveland Clinic (n=539 and 6403, respectively), and Columbia University (n=4642 and 29,438, respectively). Results are shown in Supplemental Figures 1 and 2.
Figure 2.
Urine albumin-to-creatinine ratio correlates with same-day urine albumin results. The majority of patients with negative UA result had a same-day UACR <30 mg/g (76%–83%). Approximately one half (50%–53%) of patients with a trace UA result had a UACR <30 mg/g. The majority of patients with a 1+ UA result had a UACR ≥30 mg/g (65%–84%), and virtually all patients with 2+, 3+, or 4+ UA results had a UACR ≥30 mg/g (97%+).
Population Characteristics
Table 2 shows the characteristics of the total implementation population from all five sites overall and across eGFR levels. Supplemental Tables 4–8 show the population characteristics by site. The implementation population totaled 2,082,017 patients. Average age was 50±19 years old. Of these patients, 58% were women, 9% were black, 55% had at least one eGFR, and 39% had a proteinuria measurement. The proportion of patients with proteinuria measurements varied across sites from 20% to 52% and increased as eGFR decreased.
Table 2.
Characteristics of the implementation population
| Variable | Total Population | Group 1: No eGFR | Group 2: eGFR≥60 | Group 3: eGFR 45–59 | Group 4: eGFR 30–44 | Group 5: eGFR 15–29 | Group 6: eGFR<15 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | %/SD | N | %/SD | N | %/SD | N | %/SD | N | %/SD | N | %/SD | N | %/SD | |
| No. | 2,082,017 | 940,223 | 1,001,213 | 84,348 | 34,045 | 13,009 | 9170 | |||||||
| Age, yr | 50 | 19 | 45 | 18 | 51 | 17 | 70 | 14 | 74 | 14 | 71 | 15 | 62 | 16 |
| Women | 1,209,280 | 58% | 549,534 | 58% | 581,798 | 58% | 47,886 | 57% | 19,154 | 56% | 6854 | 53% | 4049 | 44% |
| Black | 181,742 | 9% | 65,481 | 7% | 104,267 | 10% | 5567 | 7% | 2768 | 8% | 1603 | 12% | 2055 | 22% |
| No. eGFR measurements | 6 | 17 | N/A | N/A | 7 | 15 | 16 | 29 | 24 | 44 | 31 | 54 | 35 | 68 |
| Days to most recent eGFR | 604 | 816 | N/A | N/A | 649 | 870 | 456 | 704 | 355 | 561 | 355 | 550 | 482 | 691 |
| Prior eGFR <60 | 178,099 | 9% | N/A | N/A | 79,812 | 8% | 52,214 | 62% | 27,905 | 82% | 10,960 | 84% | 7199 | 79% |
| Prior GFR <60 90+ d prior | 143,487 | 7% | N/A | N/A | 63,949 | 6% | 42,343 | 50% | 22,389 | 66% | 8941 | 69% | 5864 | 64% |
| Any UACR, UPCR, or UA | 811,462 | 39% | 128,244 | 14% | 585,936 | 59% | 56,305 | 67% | 24,789 | 73% | 9877 | 76% | 6305 | 69% |
| Any UACR | 140,256 | 7% | 2251 | 0.2% | 102,858 | 10% | 18,758 | 22% | 10,224 | 30% | 4073 | 31% | 2090 | 23% |
| No. UACR measurements | 2.0 | 2.9 | 0.9 | 1.2 | 2.0 | 2.6 | 2.5 | 3.2 | 2.7 | 3.6 | 2.9 | 4.2 | 2.3 | 3.9 |
| Most recent UACR, mg/g | ||||||||||||||
| <30 | 101,137 | 72% | 1773 | 79% | 81,015 | 79% | 12,139 | 65% | 5033 | 49% | 1030 | 25% | 146 | 7% |
| 30–300 | 28,637 | 20% | 357 | 16% | 18,105 | 13% | 4963 | 27% | 3420 | 34% | 1380 | 34% | 412 | 20% |
| >300 | 10,482 | 8% | 121 | 5% | 3738 | 3% | 1656 | 9% | 1771 | 17% | 1663 | 41% | 1532 | 73% |
| Any UPCR | 68,131 | 3% | 1910 | 0.2% | 41,646 | 4% | 9245 | 11% | 7658 | 23% | 4673 | 36% | 2998 | 33% |
| No. UPCR measurements | 2.3 | 5.2 | 1.1 | 3.0 | 1.9 | 4.3 | 3.0 | 6.4 | 3.5 | 7.7 | 3.7 | 6.6 | 3.6 | 7.3 |
| Most recent UPCR, mg/g | ||||||||||||||
| <150 | 30,932 | 45% | 838 | 44% | 21,755 | 52% | 4600 | 50% | 2820 | 34% | 780 | 17% | 138 | 5% |
| 150–500 | 21,952 | 32% | 638 | 33% | 14,027 | 34% | 2907 | 31% | 2639 | 35% | 1398 | 30% | 343 | 11% |
| >500 | 15,247 | 22% | 434 | 23% | 5864 | 14% | 1738 | 19% | 2199 | 29% | 2495 | 53% | 2517 | 84% |
| Any UA | 754,379 | 36% | 125,792 | 13% | 541,761 | 54% | 50,852 | 60% | 21,967 | 65% | 8461 | 65% | 5541 | 60% |
| No. UA measurements | 2.8 | 5.2 | 1.4 | 2.6 | 3.3 | 5.4 | 4.2 | 7.0 | 5.1 | 8.3 | 5.7 | 9.6 | 5.8 | 11.5 |
| Most recent UA | ||||||||||||||
| Negative | 589,612 | 78% | 97,753 | 78% | 436,605 | 81% | 37,424 | 74% | 13,717 | 62% | 3516 | 42% | 594 | 11% |
| Trace to 30 (trace, 1+) | 118,870 | 16% | 20,951 | 17% | 80,438 | 15% | 9226 | 18% | 4977 | 23% | 2206 | 26% | 1071 | 19% |
| 100 to >300 (2+, 3+, 4+) | 45,608 | 6% | 7072 | 6% | 24,545 | 5% | 4174 | 8% | 3244 | 15% | 2712 | 32% | 3860 | 70% |
| Dialysis | 26,766 | 1% | 4562 | 0.5% | 5696 | 0.6% | 3518 | 4% | 3258 | 10% | 3059 | 24% | 6672 | 73% |
| Transplant | 12,765 | 0.6% | 810 | 0.1% | 4818 | 0.5% | 2699 | 3% | 2166 | 6% | 1182 | 9% | 1089 | 12% |
N/A, not applicable; UACR, urine albumin-to-creatinine ratio; UPCR, urine protein-to-creatinine ratio; UA, urine albumin.
Supplemental Table 9 shows population characteristics from the four validation sites overall and across eGFR levels. The validation population totaled 1,680,334 patients, with similar characteristics to the implementation population (average age =50±19 years old, 59% women, 10% black, 60% with at least one eGFR, and 41% with a proteinuria measurement).
Implementation
All sites successfully implemented the NKDEP e-phenotype. The time investment for implementation varied considerably across sites, ranging from 20 to 223 hours (Table 1). However, four sites completed implementation in <50 hours. The considerable time investment by site 5 involved efforts to match and merge data across two distinct EHR systems used for in- and outpatient care. Time investment depended on existing infrastructure, personnel involved, use of data standards (e.g., LOINC), and IT support. Both nephrologist input and informaticist (or other information technologist) input were instrumental to efficient implementation. Both were involved at four of five sites. The site lacking nephrologist input—the University of Utah—received nephrology guidance from the working group.
Validation
Table 3 shows results of the NKDEP e-phenotype validation for CKD, dialysis, and transplantation. For CKD, 207 charts were reviewed: 71 at Christiana Care, 60 at Columbia University, 58 at the University of Minnesota, and 18 at UCSF. Sensitivity for identification of patients with CKD was 99%, with a 95% confidence interval (95% CI) of 96% to 100%. Three sites achieved perfect sensitivity. The remaining site achieved 93% sensitivity (95% CI, 66% to 100%). Specificity was 99% (95% CI, 92% to 100%). Three sites achieved perfect specificity. The remaining site achieved 96% specificity (95% CI, 79% to 100%). Table 4 shows the adjudication for CKD stage overall and across the validation sites. Of 207 charts analyzed, 202 were correctly categorized by CKD stage, suggesting a diagnostic accuracy of 98%. The five misclassifications resulted from (1) outside laboratory values not captured by the e-phenotype, (2) delayed transfer of a laboratory result to the corporate data warehouse, (3) an eGFR value just at the cutoff (14.8 by e-phenotype versus 15 by chart review), (4) a case of IgA nephropathy where laboratory values did not indicate CKD, and (5) a case where the most recent recorded eGFR for a patient on dialysis was 39 ml/min per 1.73 m2.
Table 3.
Accuracy of the e-phenotype for CKD, dialysis, and transplant across validation sites
| Population | True Positive | False Negative | False Positive | True Negative | Total | Sensitivity (95% CI) | Specificity (95% CI) | Positive Predictive Value (95% CI) | Negative Predictive Value (95% CI) |
|---|---|---|---|---|---|---|---|---|---|
| CKD | |||||||||
| Total | 138 | 1 | 1 | 67 | 207 | 99% (96% to 100%) | 99% (92% to 100%) | 99% (96% to 100%) | 99% (92% to 100%) |
| University of Minnesota | 34 | 0 | 1 | 23 | 58 | 100% (90% to 100%) | 96% (79% to 100%) | 97% (85% to 100%) | 100% (85% to 100%) |
| Christiana Care | 51 | 0 | 0 | 20 | 71 | 100% (93% to 100%) | 100% (83% to 100%) | 100% (93% to 100%) | 100% (83% to 100%) |
| Columbia University | 40 | 0 | 0 | 20 | 60 | 100% (91% to 100%) | 100% (83% to 100%) | 100% (91% to 100%) | 100% (83% to 100%) |
| UCSF | 13 | 1 | 0 | 4 | 18 | 93% (66% to 100%) | 100% (40% to 100%) | 100% (75% to 100%) | 80% (28% to 100%) |
| Dialysis | |||||||||
| Total | 88 | 6 | 15 | 122 | 231 | 94% (87% to 98%) | 89% (83% to 94%) | 85% (77% to 92%) | 95% (90% to 98%) |
| University of Minnesota | 44 | 1 | 1 | 12 | 58 | 98% (88% to 100%) | 92% (64% to 100%) | 98% (88% to 100%) | 92% (64% to 100%) |
| Christiana Care | 14 | 2 | 3 | 53 | 72 | 88% (62% to 98%) | 95% (85% to 99%) | 82% (57% to 96%) | 96% (87% to 100%) |
| Columbia University | 19 | 3 | 5 | 53 | 80 | 86% (65% to 97%) | 91% (81% to 97%) | 79% (58% to 93%) | 95% (85% to 99%) |
| UCSF | 11 | 0 | 6 | 4 | 21 | 100% (72% to 100%) | 40% (12% to 74%) | 65% (38% to 86%) | 100% (40% to 100%) |
| Transplant | |||||||||
| Total | 37 | 1 | 11 | 111 | 160 | 97% (86% to 100%) | 91% (84% to 95%) | 77% (63% to 88%) | 99% (95% to 100%) |
| University of Minnesota | 10 | 0 | 1 | 47 | 58 | 100% (69% to 100%) | 98% (89% to 100%) | 91% (59% to 100%) | 100% (93% to 100%) |
| Christiana Care | 7 | 0 | 1 | 64 | 72 | 100% (59% to 100%) | 98% (92% to 100%) | 88% (47% to 100%) | 100% (94% to 100%) |
| Columbia University | 16 | 1 | 6 | 57 | 80 | 94% (71% to 100%) | 91% (80% to 96%) | 73% (50% to 89%) | 98% (91% to 100%) |
| UCSF | 4 | 0 | 3 | 3 | 10 | 100% (40% to 100%) | 50% (12% to 88%) | 57% (18% to 90%) | 100% (29% to 100%) |
Sensitivity, specificity, positive predictive value, and negative predictive value were calculated using binom.test in R. 95% CI, 95% confidence interval; UCSF, University of California, San Francisco.
Table 4.
Cumulative adjudication by CKD stage on the basis of eGFR overall and across sites
| Electronic Phenotype | Adjudicated | ||||
|---|---|---|---|---|---|
| No CKD | Stage 1/2 | Stage 3 | Stage 4 | Stage 5 | |
| No CKD | 67 (23, 20, 20, 4) | 1 (0, 0, 0, 1) | |||
| Stage 1/2 | 35 (10, 12, 10, 3) | ||||
| Stage 3 | 27 (8, 6, 9, 4) | 1 (1, 0, 0, 0) | 1 (0, 0, 1, 0) | ||
| Stage 4 | 1 (1, 0, 0, 0) | 32 (6, 13, 10, 3) | |||
| Stage 5 | 1 (1, 0, 0, 0) | 41 (8, 20, 10, 3) | |||
Results are displayed as the four-site total (University of Minnesota; Christiana Care; Columbia University; University of California, San Francisco).
The two approaches to defining proteinuria were evaluated at the University of Minnesota. Using the sensitive definition (any positive result from among the last UACR, last UPCR, or last UA), 34,271 patients were identified as having CKD. Of these, 26,897 patients were positive on the last available UACR, UPCR, or UA. The 7374 patients identified by the sensitive definition were more likely to have mild to moderate increased proteinuria.
To evaluate the NKDEP e-phenotype for identification of patients on dialysis, 231 charts were reviewed: 80 at Columbia University, 72 at Christiana Care, 58 at the University of Minnesota, and 21 at UCSF. Sensitivity was 94% (95% CI, 87% to 98%) overall, ranging from 86% (95% CI, 65% to 97%) to 100% (95% CI, 72% to 100%). Overall specificity was 89% (95% CI, 83% to 94%), ranging from 40% (95% CI, 12% to 74%) to 95% (95% CI, 85% to 99%).
To evaluate the NKDEP e-phenotype for identification of patients with transplants, 160 charts were reviewed: 80 at Columbia University, 72 at Christiana Care, 58 at the University of Minnesota, and ten at UCSF. Sensitivity was 97% (95% CI, 86% to 100%), ranging from 94% (95% CI, 71% to 100%) to 100% (95% CI, 69% to 100%). Specificity was 91% (95% CI, 84% to 95%), ranging from 50% (95% CI, 12% to 88%) to 98% (95% CI, 92% to 100%).
Discussion
Identification of patients likely to have CKD using laboratory data available in the EHR provides an opportunity to facilitate quality improvement, disease surveillance, health systems research, and clinical trial recruitment. To advance automatic, EHR-based identification of patients with CKD, the NKDEP CKD e-phenotype working group developed a laboratory value–based e-phenotype, implemented the e-phenotype across five sites with diverse informatics infrastructure and capabilities, and validated the e-phenotype at four sites. The NKDEP CKD e-phenotype performed well in identifying patients with CKD, yielding a sensitivity and specificity of 99%. The ICD code–based identification of maintenance dialysis and transplant recipients had slightly lower accuracy: at 94% sensitivity and 89% specificity for dialysis and 97% sensitivity and 91% specificity for transplant.
The performance of the NKDEP e-phenotype for CKD was slightly better than an algorithm developed as part of the Electronic Medical Records and Genomics (eMERGE) Network (sensitivity of 93% and specificity of 96%) (8). Key differences between the two phenotypes—regarding the criteria specified both for identifying CKD and in the validation populations—should be noted. Whereas the NKDEP e-phenotype relied on laboratory values only, the eMERGE algorithm used diagnostic codes, procedure codes, laboratory results, and physician observation reports to identify patients with CKD. The eMERGE algorithm was designed to include only those with hypertensive and diabetic kidney disease, excluding those with primary glomerular diseases and other potential secondary CKD, such as sickle cell and HIV-associated nephropathy. The eMERGE algorithm also does not include proteinuria in the definition. Additionally, the eMERGE group did not calculate sensitivity and specificity on the basis of the entire active clinical population at each site but rather, on the specified patient and control populations, which were characterized by specific exclusion criteria. Selection of the NKDEP e-phenotype versus the eMERGE algorithm will depend on the intended purpose. The NKDEP e-phenotype was specifically designed for ease of implementation to identify a group of patients likely to have CKD; the eMERGE algorithm was designed to identify patients with hypertensive or diabetic kidney disease for research.
The NKDEP CKD e-phenotype has limitations. It relies on availability and accuracy of EHR data, including eGFR, UACR (or UA/UPCR), and race. Among the study’s implementation patient sample, only about 55% had an eGFR, and <7% had a UACR in the EHR (Table 2). Of those with at least one eGFR indicative of CKD, the proportion with no prior eGFR <60 ml/min per 1.73 m2 was high: ranging from 50% in those with a most recent eGFR of 45 ml/min per 1.73 m2 to 60%–31% in those with a most recent eGFR of 15–29 ml/min per 1.73 m2 (Table 2). Furthermore, spot UA measurements used in calculating the UACR have not been standardized, and results from commercially available UA measurement procedures showed positive and negative biases in the range of 40% compared with an isotope dilution mass spectrometry procedure (13). The NKDEP e-phenotype allows use of UA in place of UACR, but UA was only available in the EHR for 36% of patients at implementation sites (Table 2). The NKDEP e-phenotype uses both inpatient and outpatient laboratory values and may misclassify some patients with AKI as having CKD. Race is often absent in the EHR, requiring assumptions to implement the NKDEP e-phenotype. Sensitivity and specificity of the NKDEP e-phenotype may vary depending on the degree of missingness of these data elements in the EHR. In addition, the e-phenotype relies on use of LOINC to identify laboratory data from the EHR. LOINC is widely but not universally implemented, and some sites may need to crosswalk an internal coding system to LOINC to implement the e-phenotype, which will increase time for implementation, as was the case for several sites in this study. Finally, despite the intended pragmatic nature of the e-phenotype and its successful implementation across five unique sites, implementation time varied significantly across sites, with one site requiring significant time and resource investment due to use of different EHRs for in- and outpatient settings.
Despite these limitations, the NKDEP CKD e-phenotype advances existing methods for identifying patients with CKD from the EHR. The NKDEP e-phenotype performed better compared with previous reported use of diagnosis codes in identifying patients with CKD (7). Additionally, the NKDEP e-phenotype has unique characteristics that distinguish it from the eMERGE e-phenotype (8)—including use of proteinuria measures and less complex requirements—which may make it more appropriate in certain settings, particularly small health centers with less robust informatics infrastructure.
Key strengths and limitations of the study design should also be noted. The study was implemented and validated at numerous sites with distinct patient populations and diverse informatics capabilities. However, the sites were primarily large, research-based institutions, which may not reflect implementation barriers and facilitators or level of accuracy that may occur in smaller settings. The overall validation population was large, approaching 1.7 million patients, with record reviews of >200 patients. Yet, the number of record reviews varied across sites and was small for certain sites. In addition, a protocol deviation leading to a nonblinded review of some patient charts may have affected results. However, this deviation affected only five charts: <3% of the charts assessed. Furthermore, lack of laboratory data for some patients in the EHR may subject the results of the sensitivity and specificity analyses to what may be best described as confounding by indication; patients with multiple eGFR and UA laboratory results are more likely to be detected by the e-phenotype and identified as having CKD in the manual chart review. This may lead to overestimation of the sensitivity of the e-phenotype in identifying patients with CKD, because neither the e-phenotype nor the manual chart review would detect those with only a single or fewer eGFR and/or UA measures.
Individuals with CKD cannot be accurately identified from the EHR using diagnosis codes alone. Using laboratory data available in the EHR to automate identification of patients with CKD may facilitate population health management, surveillance, and research. The NKDEP CKD e-phenotype provides a pragmatic and accurate method for EHR-based identification of patients likely to have CKD.
Disclosures
Dr. Navaneethan reports personal fees from Bayer, Boehringer Ingelheim, and Tricida outside the submitted work. Ms. Ali, Dr. Drawz, Dr. Jurkovitz, Dr. Kawamoto, Dr. Kiryluk, Dr. Narva, Ms. Norton, Dr. Park, and Dr. Shang have nothing to disclose.
Funding
The manual validation was supported by a contract from the National Kidney Disease Education Program. Dr. Jurkovitz reports partial support from Institutional Development Awards from National Institute of General Medical Sciences, National Institutes of Health grants U54-GM104941 and P20 GM103446, and contract funding from National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health. Dr. Kiryluk and Dr. Shang report partial support from National Institute of Diabetes and Digestive and Kidney Diseases Columbia Kidney Precision Medicine Project grant UG3DK114926 and National Human Genome Research Institute Electronic Medical Records and Genomics Consortium grant U01HG8680. Dr. Navaneethan reports receiving a grant from Keryx. Dr. Park reports partial support from National Center for Advancing Translational Sciences, National Institutes of Health University of California, San Francisco Clinical & Translational Science Institute grant UL1 TR001872.
Supplementary Material
Acknowledgments
The authors acknowledge the students, residents, and staff who supported this work across the implementation sites: Susana Arrigain, Yuliya Dochupailo, J. Thomas Laughery, Devin Mckelvey, Karla Mehl, Alexa Meinhardt, Tania Moody, Daniel Murphy, Joseph Nally, and Jesse Schold.
Dr. Navaneethan is an employee of the US Department of Veterans Affairs (VA). The interpretation and reporting of VA data are the responsibility of the authors and in no way should be seen as official policy or interpretation of the VA or the US Government. The contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
The authors also acknowledge members of the National Kidney Disease Education Program (NKDEP) CKD e-phenotype working group who are not listed as authors on this manuscript but who contributed to the development of the NKDEP CKD e-phenotype: Khaled Abdel-Kader, Michelle Denburg, Kevin Fowler, Chester Fox, W. Ed Hammond, Jamie S. Hirsch, Clement J. McDonald, Sumit Mohan, Nilka Rios Burrows, and Lipika Samal.
Footnotes
Published online ahead of print. Publication date available at www.cjasn.org.
See related editorial, “An Electronic CKD Phenotype: A Step Forward in Improving Kidney Care,” on pages 1277–1279.
Supplemental Material
This article contains the following supplemental material online at http://cjasn.asnjournals.org/lookup/suppl/doi:10.2215/CJN.00360119/-/DCSupplemental.
Supplemental Table 1. eGFR and UACR LOINC used in the NKDEP e-phenotype for CKD.
Supplemental Table 2. Dialysis codes used in the NKDEP e-phenotype for ESKD.
Supplemental Table 3. Transplant codes used in the NKDEP e-phenotype for ESKD.
Supplemental Table 4. Characteristics of the Christiana Care Health System population.
Supplemental Table 5. Characteristics of the Columbia University population.
Supplemental Table 6. Characteristics of the University of California, San Francisco population.
Supplemental Table 7. Characteristics of the University of Minnesota population.
Supplemental Table 8. Characteristics of the University of Utah population.
Supplemental Table 9. Characteristics of the validation population.
Supplemental Figure 1. Correlation between same-day UACR and UPCR results at three sites.
Supplemental Figure 2. Correlation between same-day UPCR and UA results at three sites.
References
- 1.United States Renal Data System : USRDS Annual Data Report: Epidemiology of Kidney Disease in the United States, Bethesda, MD, National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases, 2017 [Google Scholar]
- 2.Abdel-Kader K, Unruh ML, Weisbord SD: Symptom burden, depression, and quality of life in chronic and end-stage kidney disease. Clin J Am Soc Nephrol 4: 1057–1064, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention : National Chronic Kidney Disease Fact Sheet, 2017, Atlanta, GA, US Department of Health and Human Services, Centers for Disease Control and Prevention, 2017 [Google Scholar]
- 4.Healthy People 2020 : Chronic Kidney Disease Objectives, Washington, DC, US Department of Health and Human Services, Office of Disease Prevention and Health Promotion; Available at: https://www.healthypeople.gov/2020/topics-objectives/topic/chronic-kidney-disease/objectives. Accessed March 13, 2019 [Google Scholar]
- 5.Centers for Disease Control and Prevention: Chronic Kidney Disease Surveillance System—United States, Atlanta, GA, Centers for Disease Control and Prevention, 2018 [Google Scholar]
- 6.Tuot DS, Zhu Y, Velasquez A, Espinoza J, Mendez CD, Banerjee T, Hsu CY, Powe NR: Variation in patients’ awareness of CKD according to how they are asked. Clin J Am Soc Nephrol 11: 1566–1573, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grams ME, Plantinga LC, Hedgeman E, Saran R, Myers GL, Williams DE, Powe NR; CDC CKD Surveillance Team : Validation of CKD and related conditions in existing data sets: A systematic review. Am J Kidney Dis 57: 44–54, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nadkarni GN, Gottesman O, Linneman JG, Chase H, Berg RL, Farouk S, Nadukuru R, Lotay V, Ellis S, Hripcsak G, Peissig P, Weng C, Bottinger EP: Development and validation of an electronic phenotyping algorithm for chronic kidney disease. AMIA Annu Symp Proc 2014: 907–916, 2014 [PMC free article] [PubMed] [Google Scholar]
- 9.Drawz PE, Archdeacon P, McDonald CJ, Powe NR, Smith KA, Norton J, Williams DE, Patel UD, Narva A: CKD as a model for improving chronic disease care through electronic health records. Clin J Am Soc Nephrol 10: 1488–1499, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levin A, Stevens PE, Bilous RW, Coresh J, De Francisco ALM, De Jong PE, Griffith KE, Hemmelgarn BR, Iseki K, Lamb EJ, Levey AS, Riella MC, Shlipak MG, Wang H, White CT, Winearls CG: Kidney disease: Improving Global Outcomes (KDIGO) CKD work group : KDIGO 2012 clinical practice guideline for the evaluation and management of chronic kidney disease. Kidney Int Suppl 3: 1–150, 2013 [Google Scholar]
- 11.Levey AS, Stevens LA, Schmid CH, Zhang YL, Castro AF 3rd, Feldman HI, Kusek JW, Eggers P, Van Lente F, Greene T, Coresh J; CKD-EPI (Chronic Kidney Disease Epidemiology Collaboration) : A new equation to estimate glomerular filtration rate. Ann Intern Med 150: 604–612, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller WG, Myers GL, Ashwood ER, Killeen AA, Wang E, Thienpont LM, Siekmann L: Creatinine measurement: State of the art in accuracy and interlaboratory harmonization. Arch Pathol Lab Med 129: 297–304, 2005 [DOI] [PubMed] [Google Scholar]
- 13.Bachmann LM, Nilsson G, Bruns DE, McQueen MJ, Lieske JC, Zakowski JJ, Miller WG: State of the art for measurement of urine albumin: Comparison of routine measurement procedures to isotope dilution tandem mass spectrometry. Clin Chem 60: 471–480, 2014 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.