Abstract
Genomic analyses of cancer have identified recurrent point mutations in the RNA splicing factor–encoding genes SF3B1, U2AF1, and SRSF2 that confer an alteration of function1–6. Cancer cells bearing these mutations are preferentially dependent on wild-type (WT) spliceosome function7–11, but clinically relevant means to therapeutically target the spliceosome do not currently exist. Here we describe an orally available modulator of the SF3b complex, H3B-8800, which potently and preferentially kills spliceosome-mutant epithelial and hematologic tumor cells. These killing effects of H3B-8800 are due to its direct interaction with the SF3b complex, as evidenced by loss of H3B-8800 activity in drug-resistant cells bearing mutations in genes encoding SF3b components. Although H3B-8800 modulates WT and mutant spliceosome activity, the preferential killing of spliceosome-mutant cells is due to retention of short, GC-rich introns, which are enriched for genes encoding spliceosome components. These data demonstrate the therapeutic potential of splicing modulation in spliceosome-mutant cancers.
Heterozygous hotspot mutations in the genes encoding the RNA splicing factors splicing factor 3B subunit 1 (SF3B1), U2 small nuclear RNA auxiliary factor 1 (U2AF1), and serine and arginine rich splicing factor 2 (SRSF2) are among the most common mutations in patients with myelodysplastic syndromes (MDS)1,12, chronic myelomonocytic leukemia (CMML)1, and chronic lymphocytic leukemia (CLL)2, and they also occur in a number of solid tumors (including breast6, lung4, uveal melanoma3, and pancreatic carcinomas5). RNA splicing is the process through which introns are removed from precursor mRNAs (pre-mRNAs) to produce mature mRNAs13,14. Introns typically feature a short consensus sequence at the 5′ splice site and three elements, namely the branchpoint sequence (BPS), a polypyrimidine tract (Py-tract), and an AG dinucleotide at the intron–exon junction, at the 3′ splice site. Recent work has demonstrated that mutations in the genes encoding SF3B1, U2AF1, and SRSF2 alter their functions in a manner distinct from that underlying loss of function15–17. Specifically, SF3B1 mutations induce cryptic 3′ splice site selection through reduction of branchpoint fidelity17,18. In contrast, mutations in SRSF2 alter its RNA-binding preferences in a sequence-specific manner, thereby changing the efficiency of exon inclusion15,19. Although the mechanisms through which these effects in RNA splicing promote cancer are still being investigated, several groups have demonstrated that cells with a mutant splicing factor are exquisitely sensitive to genetic or pharmacologic perturbation of splicing7–11. However, the splice modulators used in these preclinical studies were not optimized to induce preferential lethality in spliceosome-mutant cells, are not orally bioavailable, and are not progressing for clinical development11,20.
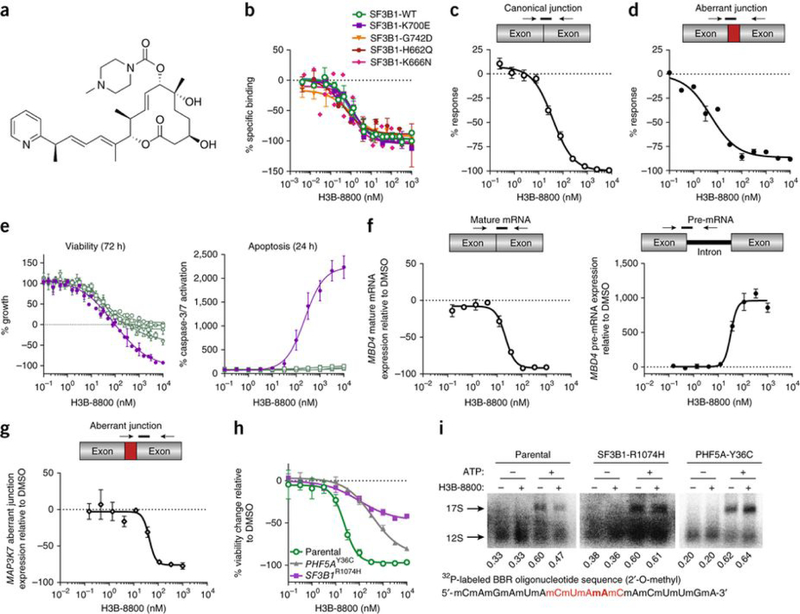
To address these issues, we specifically set out to identify orally bioavailable small molecules with preferential cytotoxicity in spliceosome-mutant cells. Through an iterative medicinal chemistry effort starting from a scaffold based on pladienolide, we identified the small molecule H3B-8800, which potently competed with pladienolide for binding to SF3b complexes containing either WT or mutant SF3B1 (Fig. 1a and Supplementary Fig. 1a). The competitive binding between pladienolide and H3B-8800 was similar for SF3b complexes containing WT SF3B1 and those containing each of eight different cancer-associated SF3B1 mutants (Fig. 1b, Supplementary Fig. 1b, and Supplementary Table 1). In accordance with these binding data and similarly to E7107, a pladienolide analog20, H3B-8800 demonstrated dose-dependent inhibition of both canonical splicing and aberrant splicing induced by mutant SF3B1 in in vitro assays of biochemical splicing (Fig. 1c,d and Supplementary Fig. 1c,d). These results indicate that H3B-8800 potently binds and inhibits splicing catalysis by both WT SF3B1 and its mutant forms in vitro.
Figure 1.
H3B-8800 modulates splicing of WT and mutant SF3B1 spliceosomes in vitro and preferentially kills SF3B1-mutant cells. (a) The chemical structure of H3B-8800. (b) Competitive binding of increasing concentrations of H3B-8800 to SF3b complexes isolated from cells overexpressing WT or mutant SF3B1 in the presence of 1 nM pladienolide B. The y axis represents the percentage change (% response) of specific binding relative to the DMSO control of 0%. Data are represented as mean ± s.d.; n = 3 experimental replicates. (c,d) Quantification of canonical (c) and aberrant (d) junctions using the Ad2-ZDHHC16 pre-mRNA substrate, in which the original Ad2 intron was substituted with intron 9 of ZDHHC16, in an in vitro splicing assay using nuclear extracts from 293FT cells overexpressing SF3B1K700E and treated with increasing concentrations of H3B-8800. The y axis represents the percent response relative to the DMSO control (0%). Data are represented as mean ± s.d.; n = 3 technical replicates. (e) Viability and apoptosis of pancreatic cells bearing WT (Panc10.05, HAPFII, CFPAC1, and Panc04.03 in green) or mutant (Panc05.04 in violet) SF3B1 treated with increasing concentrations of H3B-8800. Data are represented as mean ± s.d.; n = 3 technical replicates. (f,g) Effect of increasing concentrations of H3B-8800 on splicing of MBD4 mature mRNA (open circles) and MBD4 pre-mRNA (black circles) (f) and MAP3K7 aberrant junction expression (open diamonds) (g) in Panc05.04 cells as quantified by TLDA after 6 h of treatment with H3B-8800. Data are represented as mean ± s.e.m.; n = 3 technical replicates. (h) Viability of HCT116 parental cells and clones carrying the pladienolide-resistance mutation SF3B1R1074H or PHF5AY36C. Data are represented as mean± s.d.; n = 3 technical replicates. (i) Binding of U2 snRNA to the canonical branchpoint sequence (2′-O-methyl BBR oligonucleotide in red with the branchpoint adenosine bolded) in nuclear extracts from the same HCT116 cells as in h. Nuclear extracts were incubated in the presence or absence of ATP and H3B-8800; band quantification is shown below each lane. The functional 17S U2 snRNP was formed via ATP-dependent binding of the SF3b complex to the nonfunctional 12S U2 snRNP particle.
We next investigated the cellular activity of H3B-8800 using a panel of SF3B1 WT or mutant pancreatic cancer cell lines. H3B-8800 treatment for 72 h resulted in cellular lethality only in the SF3B1-mutant cell line Panc05.04; this lethality was accompanied by caspase-3/7 cleavage as early as 24 h following incubation (Fig. 1e). In contrast, E7107 did not show clear differential effects in the same cell panel (Supplementary Fig. 1e), suggesting that H3B-8800 has increased selectivity for SF3B1-mutant cells. Next, we tested the ability of H3B-8800 to modulate canonical and aberrant splicing in Panc05.04 (SF3B1-K700E) cells using a TaqMan Low Density Array (TLDA), which is designed to evaluate expression of pre-mRNA and mature mRNA isoforms impacted by SF3B1 modulation as well as aberrant RNA splicing events induced by mutations in SF3B117 (Online Methods). H3B-8800 modulated both canonical and aberrant splicing catalysis as revealed by a dose-dependent decrease in the levels of mature methyl-CpG binding domain 4 (MBD4) mRNA and an accumulation of MBD4 pre-mRNA in addition to decreased SF3B1-mutant-induced aberrant splicing of MAP3K7 (Fig. 1f,g). Similar results were observed for additional mRNAs in the pancreatic cell lines tested as well as for K562 cells expressing SF3B1WT or SF3B1K700E at the SF3B1 endogenous locus (Supplementary Fig. 2). In addition, H3B-8800, but not E7107, demonstrated preferential killing of K562 cells bearing SF3B1K700E as compared to their isogenic SF3B1WT counterparts (Supplementary Fig. 1f).
To verify the selectivity of H3B-8800 for the SF3b complex, we tested the activity of H3B-8800 in HCT116 cells with acquired resistance to pladienolides21 due to mutations in SF3B1 or PHF5A, which encode components of the SF3b complex involved in branchpoint recognition22. SF3B1-R1074H or PHF5A-Y36C mutants conferred resistance to H3B-8800, whereas resistance to bortezomib, a compound whose mechanistic effects are not splicing dependent, was not observed in cells with these mutants (Fig. 1h and Supplementary Fig. 1g). These data indicate the selectivity of H3B-8800 for the SF3b complex and suggest that H3B-8800 may function through interfering with the interaction between the SF3b complex and the branchpoint. To test this hypothesis, we incubated nuclear extract from HCT116 parental cells with H3B-8800, ATP, and a32P-2′-O-methyl oligonucleotide complementary to the U2 small nuclear RNA (snRNA) branchpoint binding region (BBR). H3B-8800 inhibited ATP-dependent 17S U2 small nuclear ribonucleoprotein (snRNP) complex formation (Fig. 1i), providing evidence that H3B-8800 abolishes precatalytic spliceosome assembly, as was previously proposed for E7107 (ref. 23). This inhibition was not observed for nuclear extracts from the two H3B-8800-resistant cell lines (Fig. 1i), suggesting that H3B-8800 modulates splicing through interfering with the interaction between the SF3b complex and the branchpoint region.
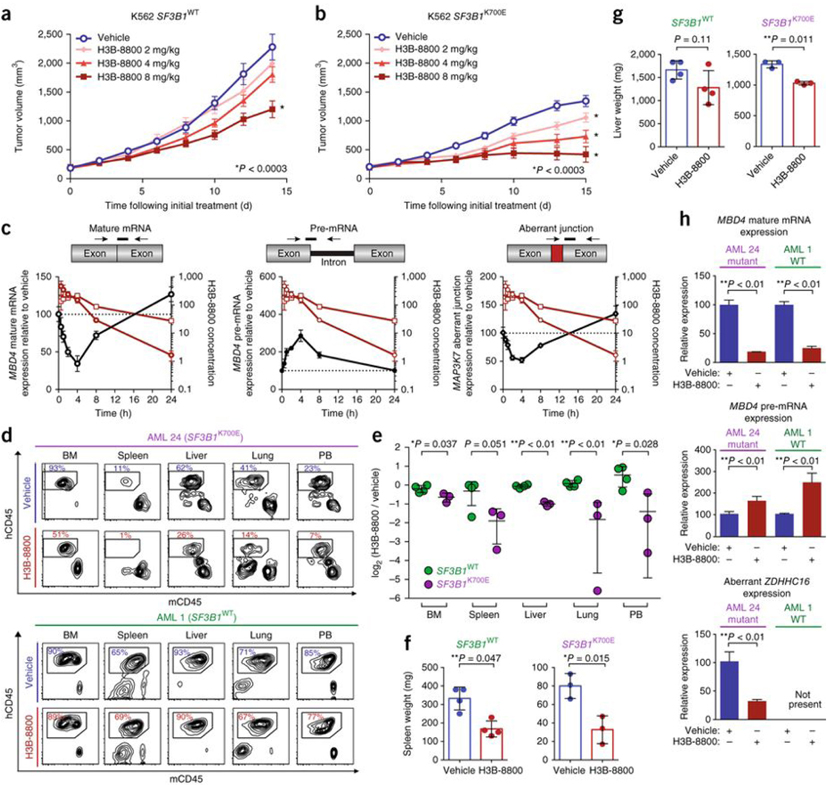
We next tested the antitumor effects of H3B-8800 in vivo in NODSCID interleukin-2 receptor gamma (IL2Rγ)null (NSG) mice bearing xenografts of isogenic K562 cells with SF3B1WT or SF3B1K700E (p.K700E was introduced through genetic modification of the endogenous SF3B1 locus). Oral administration of 2 or 4 mg H3B-8800 per kg body weight (mg/kg) daily slowed the growth of xenografts with SF3B1K700E (P < 0.003, two-way ANOVA followed by Dunnett’s multiple comparison test) but had no effect on SF3B1WT xenografts. Although a dose of 8 mg/kg did slow the growth of SF3B1WT tumors, it completely abrogated growth of SF3B1K700E tumors (P < 0.003; Fig. 2a,b). H3B-8800 similarly demonstrated significant antitumor activity against xenografts of human HNT-34 acute myeloid leukemia (AML) cells bearing the endogenous mutation encoding SF3B1-K700E (Supplementary Fig. 3a). H3B-8800 reached similar concentrations in plasma as well as in tumors of mice harboring SF3B1WT or SF3B1K700E xenografts, and H3B-8800 modulated both canonical and aberrant splicing in a dose-dependent manner (Fig. 2c and Supplementary Fig. 4). Splicing modulation was observed as early as 1 h following treatment, and splicing returned to pretreatment levels within 24 h following treatment, in accordance with the H3B-8800’s pharmacokinetic profile (Fig. 2c).
Figure 2.
H3B-8800 modulates splicing and selectively kills SF3B1-mutant leukemia cells in vivo. (a,b) Mean tumor volume in NSG mice subcutaneously implanted with K562 isogenic cells with (b) or without (a) endogenous mutations in SF3B1. Mice were orally treated with vehicle or H3B-8800 daily at the indicated doses. Mean tumor volumes ± s.e.m. are shown; n = 8 mice per group. (c) Splicing of MBD4 mature mRNA (black open circles), MBD4 pre-mRNA (black circles), and MAP3K7 aberrant junction expression (black open diamonds) in K562 SF3B1K700E tumors after a single oral dose of H3B-8800 (8 mg/kg) as quantified by TLDA. The drug concentration in H3B-8800-treated mice was determined in plasma (red open circles) and tumor (red open squares). Data are represented as mean ± s.e.m.; n = 4 mice per group. The H3B-8800 concentration is shown in ng/ml for plasma and ng/g for tumor. (d) Results from flow cytometry analysis of human versus mouse hematopoietic cells (hCD45 cells versus mCD45 cells) in bone marrow (BM), spleen, liver, lung, and peripheral blood (PB) of PDX-bearing mice with (top) or without (bottom) mutation in SF3B1 following 10 d of vehicle or H3B-8800 treatment (8 mg/kg). Cells were gated on DAPI−mTer119− cells, and the percentages of live Ter119–cells are shown. (e) Quantification of human leukemic cells (hCD45), expressed as log2 of the percentage of hCD45+ cells in HRB-8800-treated mice divided by the percentage of hCD45+ cells in vehicle-treated mice, in each tissue compartment in all PDX models treated with vehicle or H3B-8800. Data are represented as mean ± s.d.; n = 3 mice for SF3B1K700E and 4 for SF3B1WT. (f,g) Spleen (f) and liver (g) weight in PDX mice treated with vehicle or H3B-8800 (8 mg/kg). Data are represented as mean ± s.d.; n = 3 mice for SF3B1K700E and 4 for SF3B1WT. (h) Effects of H3B-8800 (8 mg/kg) on mature mRNA and pre-mRNA MBD4 levels (canonical splicing) and aberrant ZDHHC16 splicing in human cells isolated from PDX mice. The cells were isolated 3 h after the final dose of H3B-8800. Data are represented as mean ± s.d.; n = 3 mice per group. All P values were calculated using a two-tailed Student’s t-test.
H3B-8800 treatment resulted in antileukemic efficacy and splicing modulation in mice bearing AML patient-derived xenografts (PDXs) with a mutation in SF3B1 but had little effect in mice bearing SF3B1WT AML PDXs. In this experiment, NSGS mice (NSG mice with transgenic expression of human granulocyte-macrophage colony-stimulating factor (GM-CSF), IL-3, and SCF) were engrafted with AML cells from a patient with AML harboring SF3B1K700E, NRASA59T, and inv(3)(q21q26.2) or from a patient with AML harboring variations in KRAS, WT1, and MLL1 but lacking any mutations in spliceosome-related genes (Supplementary Table 2). In mice engrafted with the SF3B1K700E PDX, 10 d of H3B-8800 treatment significantly reduced leukemic burden relative to that in vehicle-treated mice, but this treatment had negligible effects in the SF3B1WT PDX recipients (Fig. 2d–g and Supplementary Fig. 5a–c). Analysis of human leukemic cells purified from the mice 3 h after the last dose of H3B-8800 revealed reduced mature mRNA levels and accumulation of pre-mRNA resulting from canonical splicing events in all H3B-8800-treated mice, regardless of SF3B1 genotype. In contrast, reduced levels of aberrant splicing events induced by a mutation in SF3B1 were observed only in the SF3B1-mutant model (Fig. 2h and Supplementary Fig. 5d).
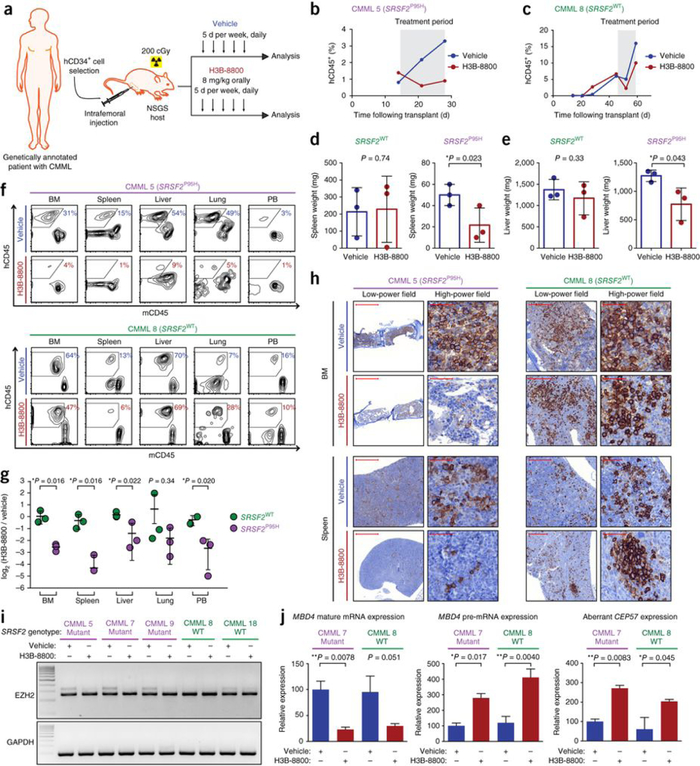
Given the activity and preferential efficacy of H3B-8800 on SF3B1-mutant human AML xenografts in vivo, we next tested H3B-8800 in mouse models with mutated SRSF2. After observing clear anti-tumor activity of H3B-8800 in mice bearing xenografts comprising SRSF2P95H K052 cells (Supplementary Fig. 3b), we generated six PDX models derived from patients with CMML, three of whom harbored SRSF2 mutations (Supplementary Table 2). A PDX model from each patient was generated, and the models were randomized to receive vehicle or H3B-8800 (8 mg/kg orally for 10 d) once human CD45 (hCD45)+ cells in the peripheral blood made up >3% of total mononuclear cells or the concentration of hemoglobin was ≤11 g/dl (Fig. 3a). Of note, leukemic burden was equivalent in mice bearing SRSF2P95H tumors and those with SRSF2WT tumors at the onset of treatment (Supplementary Fig. 6a). H3B-8800 treatment reduced human leukemic burden in peripheral blood and all organs examined in PDX models with SRSF2P95H but not in those with SRSF2WT (Fig. 3b–h and Supplementary Figs. 6b and 7). Moreover, we observed splenomegaly and hepatomegaly due to leukemic cell infiltration, and H3B-8800 treatment for 10 d substantially reduced spleen and liver size only in the SRSF2P95H PDX models (Fig. 3d,e). In accordance with the reduced number of mature leukemic cells observed in H3B-8800-treated PDX models of CMML bearing a mutant spliceosome, but not in those with a WT spliceosome, H3B-8800 treatment reduced the number of immunophenotypically defined leukemia-initiating cells only in spliceosome-mutant PDX models of CMML (Supplementary Fig. 8). Evaluation of RNA splicing in hCD45+ cells 3 h after the last dose of vehicle or H3B-8800 revealed that in vivo H3B-8800 treatment modulated aberrant splicing mediated by mutant SRSF2 (Fig. 3i,j). In addition, H3B-8800 treatment resulted in decreased levels of mature MBD4 mRNA and accumulation of MBD4 pre-mRNA (Fig. 3j) as noted for the cell lines treated with H3B-8800 (Fig. 1f). These data further suggest the efficacy and preferential activity H3B-8800 in treating spliceosome-mutant malignancy in vivo.
Figure 3.
H3B-8800 demonstrates preferential activity on SRSF2-mutant leukemia in PDX mice. (a) Schematic of H3B-8800 testing in CMML PDX mice. Mice were treated orally with 8 mg/kg of H3B-8800 or vehicle for 10 d (in 2 intervals of 5 d each, separated by 2 d). Each sample was engrafted into two NSGS recipient mice, which were then treated with vehicle or H3B-8800 once human CD45+ (hCD45+) cells in peripheral blood were >3% of total peripheral blood mononuclear cells or hemoglobin was ≤11 g/dl. The mice were killed 2 h after the last dose and were analyzed. (b,c) The effect of H3B-8800 on the percentage of peripheral blood hCD45+ cells in spliceosome-mutant (b) and WT (c) PDX models engrafted with an individual spliceosome-mutant or WT tumor (patient numbering is shown in Supplementary Table 2). The treatment period is marked in gray. (d,e) The effect of H3B-8800 on spleen (d) and liver (e) weight in spliceosome-mutant and WT PDX models bearing tumors derived from 3 patients per genotype. Data are represented as mean ± s.d.; n = 3 mice per group. (f) Flow cytometric analysis of hCD45 versus mCD45 cells in bone marrow, spleen, liver, lung, and peripheral blood of vehicle- or H3B-8800-treated mice engrafted with an individual spliceosome-mutant or WT tumor (cells were gated on DAPI−mTer119− cells, and the percentages of live Ter119− cells are shown). (g) Quantification of the proportion of human leukemic cells in H3B-8800-versus vehicle-treated mice (expressed as log2 of the percentage of hCD45+ cells in H3B-8800-treated mice divided by the percentage of hCD45+ cells in vehicle-treated mice) in each tissue compartment in all PDX models. Data are represented as mean ± s.d.; n = 3 mice per group. (h) Immunohistochemical stains for hCD45+ cells in bone marrow or spleen from H3B-8800- versus vehicle-treated mice. Scale bars, 1,000 μm (CMML 5 low-power field), 200 μm (CMML 8 low-power field) and 50 μm (high-power field). (i,j) EZH2 aberrant junction formation as assessed through RT-PCR (i), and mature mRNA and pre-mRNA MBD4 levels as well as levels of CEP57 aberrant junction expression (j) in cDNA from hCD45+ cells isolated 3 h following the last dose of vehicle or H3B-8800. Data are represented as expression relative to GAPDH; mean ± s.d.; n = 3 technical replicates. All P values were calculated using a two-tailed Student’s t test.
To further understand the preferential effects of H3B-8800 on spliceosome-mutant cells, we performed RNA-seq analysis of isogenic SF3B1WT and SF3B1K700E K562 cells treated with DMSO or H3B-8800. We observed enrichment of alternative 3′ splice sites in DMSO-treated K562 SF3B1K700E cells as compared to DMSO-treated SF3B1WT cells (Supplementary Fig. 9a and Supplementary Table 3), consistent with prior reports on the effects of SF3B1 mutations on splicing17,18. At a concentration of 13 nM, the half-maximal inhibitory concentration (IC50) of H3B-8800 in a cell viability assay at 72 h following treatment (Supplementary Fig. 1f), H3B-8800 treatment for 6 h primarily induced exon-skipping and intron-retention events in K562 cells expressing SF3B1-K700E as compared to untreated (DMSO control) cells (Supplementary Fig. 9c and Supplementary Table 3). Genes in which splicing events in these categories occurred (n = 4,353) were more likely to show downregulated expression following H3B-8800 treatment as compared to DMSO treatment (P = 2.1 × 10−26, binomial proportion test; Supplementary Fig. 9d,e and Supplementary Table 4). Expression of genes with intron-retention events that were predicted to induce degradation through the nonsense-mediated decay (NMD) pathway were further downregulated as compared to genes with other types of intron-retention events (P = 6.37 × 10−22, Kruskal–Wallis H test; Supplementary Fig. 9h). Notably, when we compared the effects of treatment with 13 nM H3B-8800 in K562 cells with SF3B1K700E and in those with SF3B1WT, we observed few effects on alternative 3′ splice site usage but a clear increase in exon-skipping and intron-retention events; however, the magnitude of these changes was minimal overall (75th quartile < (2 × log2 (fold change)); Supplementary Fig. 9b and Supplementary Table 4). These results suggest that the splicing changes induced by H3B-8800 treatment are similar in WT and SF3B1K700E K562 cells.
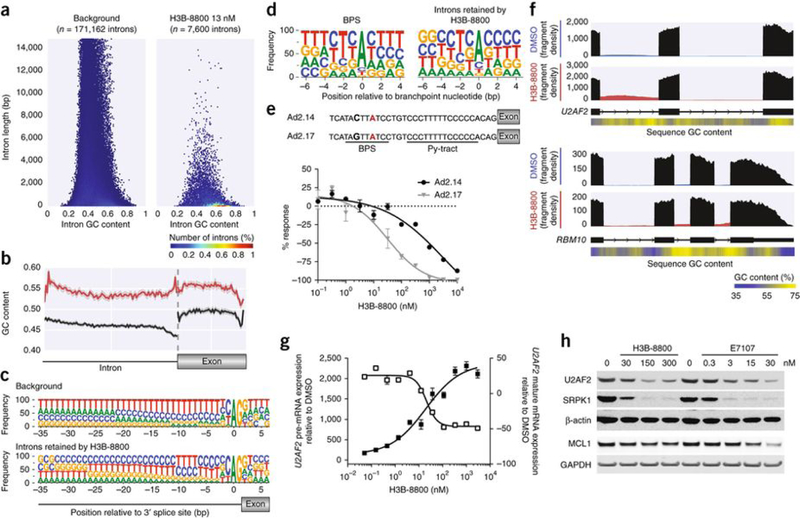
Considering that H3B-8800 has preferential cytotoxic effects on spliceosome-mutant cells as compared to spliceosome-WT cells despite similar overall effects on splicing as observed in the RNA-seq analysis, we next sought to understand the effects of H3B-8800 on splicing in more detail. Characterization of introns retained in K562 SF3B1K700E cells following treatment with H3B-8800 revealed that 27.2% of all significantly retained introns (q < 0.05) were short (≤300 nt) and GC rich (≥60% GC content) relative to 6.1% in all genes with detectable spliced introns (P = 2.2 × 10−16; Fisher’s exact test; Fig. 4a). A similar preferential retention of short, GC-rich introns was also seen following H3B-8800 treatment of NALM-6 SF3B1K700E cells, an effect that was much less evident for E7107 treatment at a similar concentration (Supplementary Fig. 9f). A recent report suggested that short, GC-rich introns represent a specialized class of introns characterized by minimal differences in GC content between the intron and the following exon24. We therefore examined these features in introns retained by H3B-8800 versus all introns from expressed genes. In the results from this analysis, introns retained after H3B-880 treatment were enriched for GC content along their length, and there was an equal enrichment for GC content along the length of the following exon, with local enrichment in GC content at the 5′ and 3′ splice sites (Fig. 4b). The largest divergence in GC content from all genes with detectable spliced introns occurred at the 3′ splice site, where the U2AF2-binding Py-tract is found25. This result implies that the Py-tract of introns retained by H3B-8800 might be predominantly cytosine-rich although it was expected to be thymine-rich; we confirmed this inference via sequence motif analysis (Fig. 4c). In contrast, introns retained by E7107 treatment were less GC-rich than those retained by H3B-8800, and they demonstrated a clear increase in GC content in the following exon (Supplementary Fig. 9g).
Figure 4.
H3B-8800 inhibits splicing of short, GC-rich introns and affects splicing of mRNAs encoding spliceosome components. (a) Hexbin plot showing relative abundance of introns based on length (y axis) and GC content (x axis) of all introns of expressed genes in K562 cells (background, left) versus introns retained in SF3B1K700E cells after H3B-8800 treatment (13 nM, right). The percentage of the total intron count contained within each hex is indicated by its color. (b) GC content within introns retained after H3B-8800 treatment of K562 SF3B1K700E cells (red; n = 5,788 introns) versus a randomly selected subset of introns from background (black; n = 10,000 introns). The GC content within the exons that followed is also shown. The solid line represents the mean GC content, and the shaded area surrounding the solid line represents the 95% confidence interval of that measurement. (c) The 3′ splice site motif of introns retained in K562 SF3B1K700E cells following H3B-8800 treatment as compared to background. (d) Branchpoint sequence motif (extending from 6 nt upstream of the branchpoint nucleotide to 4 nt downstream of the branchpoint) representing all branchpoints sequenced by Mercer et al.26 (left) and only those branchpoints within introns retained in in K562 SF3B1K700E cells following H3B-8800 treatment (right). (e) Top, depiction of two Ad2-derived pre-MRNA substrates: Ad2.14, in which the Py-tract contains five cytosines and five thymines, and Ad2.17, which also contains an alteration in the BPS to reduce binding affinity to U2 snRNP. The branchpoint nucleotide is labelled in red, and the nucleotide that was changed between Ad2.14 and Ad2.17 to alter branchpoint strength is bolded. Bottom, the results from in vitro splicing using these two pre-mRNA substrates. The y axis represents the percent response relative to DMSO control (0%). Data are represented as mean ± s.d.; n = 3 technical replicates. (f) Sashimi plot representing the average read density of three technical replicates in introns 3 and 4 of U2AF2 (top) and introns 20, 21, and 22 of RBM10 (below) of RNA-seq samples from K562 SF3B1K700E cells treated with DMSO or H3B-8800. Exons are shown in black, and introns are shown in blue (DMSO treated) or red (H3B-8800 treated). Relative GC content (derived using a 100-bp sliding window) is depicted in a heat map below the reference sequence. (g) The effect of an increasing concentration of H3B-8800 on the level of U2AF2 pre-mRNA (black squares) and mature mRNA (open squares) in K562 SF3B1K700E cells as quantified by qPCR. Data are represented as mean ± s.e.m.; n = 3 technical replicates. (h) Western blot analysis of U2AF2, SRPK1, and MCL1 protein levels following treatment of K562 SF3B1K700E cells with H3B-8800 or E7107 at the indicated concentrations. β-actin and GAPDH were used as loading controls.
As the 3′ region of the intron also contains the BPS, we next explored whether the BPS in introns retained following H3B-8800 treatment might also be GC rich. Using branchpoint location data from deep sequencing of K562 cells in Mercer et al.26, we discovered that, within introns retained by H3B-8800, the branchpoint nucleotide was less likely to be the preferred adenosine (68.8% versus 78.7%; P = 4.32 × 10−9; Fisher’s exact test; Fig. 4d). This result strongly suggests that the BPS might play a role in determining which introns are more likely to be retained following H3B-8800 treatment. To test this hypothesis, we designed two substrates, Ad2.14 and Ad2.17, based on the Ad2 construct27; both contain a partially cytosine-rich Py-tract conforming to the observed 3′ splice site sequence motif (Fig. 4e). In addition, a base substitution in the BPS of in Ad2.17 was created to reduce binding fidelity to the U2 snRNP. Using these constructs, we observed that H3B-8800 exhibited a greater modulation of splicing for Ad2.17, the substrate with a weakened BPS (Fig. 4e), suggesting that H3B-8800 selectively modulates splicing of introns with a GC-rich 3′ splice site and a weak BPS. In contrast, in the same experiment, E7107 showed a much smaller shift in IC50 values (Supplementary Fig. 10a). Results from similar experiments using two other sets of Ad2 constructs, each with identical pairs of weak and strong BPS sequences but with differing 3′ splice sites, revealed selective modulation by H3B-8800 of the construct with a weakened BPS in sequences with a Py-tract composed completely of cytosine (Supplementary Fig. 10c) or with a partially cytosine-rich Py-tract that was dissimilar to the observed 3′ splice site sequence motif (Supplementary Fig. 10e). In contrast, E7107 inhibited splicing of both sets of constructs equally (Supplementary Fig. 10b,d). Moreover, in constructs in which the BPS was changed to increase fidelity for binding with the U2 snRNP, E7107 showed potent splicing inhibition (IC50, 8.8 nM), whereas H3B-8800 showed no inhibition (Supplementary Fig. 11a,b). Taken together, these results demonstrate that H3B-8800 exhibits sequence-specific properties that modulate splicing that are distinct from those of E7107.
To test whether genes containing short, GC-rich introns retained following H3B-8800 treatment are involved in pathways that might explain the preferential killing of spliceosome-mutant cells exhibited by the drug, we performed pathway enrichment analysis of genes whose splicing was affected by H3B-8800 treatment in the K562 isogenic cell lines. Pathways involved in mRNA splicing were among those significantly enriched for such genes (Supplementary Table 5). Considering the prior data showing that WT spliceosome components are necessary for the survival of spliceosome-mutant cells7–11, we focused on the disruption of mRNA splicing as a possible mechanism underlying the preferential lethality of H3B-8800 against spliceosome-mutant cells. First, we curated a complete list of genes encoding spliceosome components using information from several studies28–30 (Supplementary Table 6 and Online Methods). Short, GC-rich introns retained by H3B-8800 were present in 41 of 404 genes encoding proteins in spliceosome complexes (P = 0.017; binomial proportion test; Supplementary Fig. 11c), including genes encoding RNA binding motif protein 10 (RBM10) and the essential splicing factor U2AF2 (Fig. 4f and Supplementary Fig. 11d,f). H3B-8800 treatment induced intron retention of U2AF2 in a dose-dependent manner (Fig. 4g), indicating that H3B-8800 directly modulated the splicing of genes encoding proteins required for splicing catalysis, and this modulation resulted in a corresponding loss of U2AF2 protein expression in K562 cells (Fig. 4h). Despite the effects of H3B-8800 on U2AF2 splicing and expression, stable reintroduction of full-length U2AF2 cDNA was unable to rescue H3B-8800-induced cell death (Supplementary Fig. 12), likely because H3B-8800 treatment affects expression of many additional splicing factors. For example, H3B-8800 treatment also induced a dose-dependent reduction in protein expression of SRPK1, a kinase involved in splicing factor regulation. Although both H3B-8800 and E7107 treatment reduced U2AF2 and SRPK1 protein expression, only E7107 treatment induced intron retention of MCL1 and reduced MCL1 protein expression (Fig. 4h and Supplementary Fig. 13), further indicating that E7107 has a more pleiotropic effect on splicing modulation than H3B-8800.
The data presented here identify H3B-8800 as a small molecule that selectively modulates splicing to induce preferential lethality in cancer cells bearing a mutant spliceosome. Recent work has shown that short, GC-rich introns are recognized by an intron-defined splicing mechanism that is associated with intron retention observed in cells under normal conditions24. We demonstrate that H3B-8800 preferentially induces retention of these introns, which are characterized by short length, high GC content, and low differentiation in GC content compared with the following exon. Together, these findings suggest that H3B-8800 treatment selectively modulates a pre-existing mechanism for splicing recognition, and this selectivity is not shared with other small-molecule modulators of splicing. The enrichment for intron retention in mRNAs encoding RNA splicing factors provides an explanation for H3B-8800’s preferential killing of spliceosome-mutant tumor cells, which are already deficient in splicing, as compared to spliceosome-WT cells. As the expression of numerous RNA splicing factors was influenced by H3B-8800 treatment, the killing effects of the drug are not attributable to altered expression of any single target.
Despite the fact that the global alterations in splicing conferred by mutant SF3B1 (refs. 17,18), SRSF2 (refs. 15,19), and U2AF1 (ref. 16) are distinct from one another, cells bearing any of the corresponding mutations have been shown to exhibit dependency on the remaining WT spliceosome7–11. It is therefore predicted that expression of any of these cancer-associated RNA splicing factor mutations would confer sensitivity to H3B-8800. The preferential vulnerability of SRSF2- and SF3B1-mutant cells to H3B-8800 results in the differential lethality observed in vitro and in vivo. The selective mechanism of action of H3B-8800 in combination with its unique pharmacological profile among previously described direct modulators of splicing supports clinical testing of H3B-8800 in genetically defined subsets of cancer with RNA splicing factor mutations.
METHODS
Methods, including statements of data availability and any associated accession codes and references, are available in the online version of the paper.
ONLINE METHODS
Cell culture.
The cell lines HCT-116, Panc05.04, Panc10.05, HPAF-II, CFPAC1, and Panc04.03 were obtained from the American Type Culture Collection (ATCC) or from RIKEN BioResource Center and cultured as instructed. The isogenic paired cell lines (K562 and NALM-6 SF3B1K700E and SF3B1WT) were generated at Horizon Discovery and were cultured in the presence of Geneticin (0.7 mg/ml, Life Technologies) for selection. HCT116 clones resistant to H3B-8800 were generated by treating cells with 4 nM of splicing modulator for 2 weeks. Splicing modulator was refreshed every 4 d. When needed, confluent cells were split in a 1:3 ratio and were allowed to recover overnight without splicing modulator treatment after reseeding. At the end of the 2 weeks of compound selection, individual clones were transferred to 12-well plates and further expanded without splicing modulator. Genomic DNA was extracted using the DNeasy Blood & Tissue Kit from Qiagen. The resistance mutations SF3B1R1074H and PHF5AY36C were identified by whole exome sequencing.
Cell viability and apoptosis assays.
Pancreatic cancer cells were seeded at 750 cells per well in a 384 well plate and treated with compound on day 2. K562 isogenic cells (K562-SF3B1K700E and K562-SF3B1K700K) were seeded at 10,000 cells per well in 96-well plates and treated with compound 4 h later. The relative numbers of viable and apoptotic cells were measured via luminescence using CellTiter-Glo or Caspase-Glo 3/7 (Promega) at the indicated time points as instructed. Bortezomib was obtained from Selleckchem.
Expression plasmids.
The plasmids used in the scintillation proximity assay were generated through cloning the cDNA of mxSF3B1 (cloned by ligating the N-terminal half of the product of the mouse Sf3b1 open reading frame (ORF) and the C-terminal half of the product of the Xenopus Sf3b1 ORF, followed by replacement of three amino acid residues in the product of the Xenopus ORF with three human amino acid residues) into the HindIII and KpnI sites of pFlag-CMV2 (Sigma). The mutations corresponding to mxSF3B1-E622D, mxSF3B1-R625L, mxSF3B1-H662Q, mxSF3B1-K666E, mxSF3B1-K666N, mxSF3B1-K666R, mxSF3B1-K700E, and mxSF3B1-G742D were introduced using a site-directed mutagenesis kit (QuikChange II XL, Agilent). Mammalian U2AF2 expression plasmids were generated using Gateway cloning technology (Life Technologies). N-terminal hemagglutinin (HA)-tagged U2AF2 (NP_009210.1) was cloned into the pLenti6.3-V5 Dest vector (Life Technologies), which was used to transfect LentiX293 cells (Promega). Viral supernatants from these cells were then used to infect K562 isogenic cells. These isogenic K562 cells were selected for construct expression using 4 μg/ml blasticidin.
Nuclear extract preparation.
Nuclear extracts were prepared from either HeLaS3 or 293F cells expressing pFLAG-mxSF3B1 plasmids, 293FT cells expressing mxSF3B1, or HCT116 cells. Expression of mxSF3B1 was achieved in 293F cells by transfecting pFLAG-CMV-2-mxSF3B1 plasmids using Lipofectamine 2000 (Life Technologies) and in 293FT cells using the PiggyBac transposon system. Cell pellets were resuspended in hypotonic buffer (10 mM HEPES pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.2 mM phenylmethylsulfonyl fluoride (PMSF), and 0.5 mM dithiothreitol (DTT)). The suspension was brought up to a total of 5 packed cell volumes (PCV). After centrifugation (3,000 r.p.m. for 5 min at 4 °C), the supernatant was discarded, and the cells were brought up to 3 PCV with hypotonic buffer and incubated on ice for 10 min. Cells were lysed using a dounce homogenizer and then centrifuged (4,000 r.p.m. for 15 min at 4 °C). The supernatant was discarded, and the pellet was resuspended with 1/2 packed nuclear volume (PNV) of low-salt buffer (20 mM HEPES pH 7.9, 1.5 mM MgCl2, 20 mM KCl, 0.2 mM EDTA, 25% glycerol, 0.2 mM PMSF, 0.5 mM DTT), followed by 1/2 PNV of high-salt buffer (same as low salt buffer except 1.4M KCl was used). The nuclei were gently mixed by rotating for 30 min before centrifuging (10,000 r.p.m. for 30 min at 4 °C). The supernatant (nuclear extract) was then dialyzed into storage buffer (20 mM HEPES pH 7.9, 100 mM KCl, 0.2 mM EDTA, 20% glycerol, 0.2 mM PMSF, 0.5 mM DTT). Protein concentration was determined using a NanoDrop 8000 UV-Vis spectrophotometer (Thermo Scientific).
In vitro splicing reaction.
The Ad2-derived31 sequence was cloned into the pGEM-3Z vector (Promega) using 5′ EcoRI and 3′ XbaI restriction sites. The Ad2 variants (Ad2.12–Ad2.17; all modifications are described in Fig. 4e and Supplementary Fig. 10) were cloned into the pcDNA3.1(+) vector (Promega) using the same restriction sites. The Ad2-ZDHHC16 hybrid substrate, which contained exons from the Ad2 vector and intron 9 from ZDHHC16, was cloned into the pcDNA3.1(+) vector using the same restriction sites. The plasmids were linearized using XbaI and then were used as DNA templates in the in vitro transcription reactions. The Ftz plasmid that lacked introns (FtzΔi) was obtained from Luo & Reed32 and linearized using EcoRI. All RNAs were in vitro transcribed using the MEGAScript T7 kit and then purified using the MegaClear kit (Invitrogen).
For splicing reactions using pre-mRNAs of Ad2 variants, reactions were prepared using 8 μg of HeLa S3 nuclear extract, 2 ng pre-mRNA, 0.2 ng control, and varying concentrations of H3B-8800, E7107, or DMSO to reach a final reaction volume of 1 μl in this high-throughput assay. After a 15-min preincubation at 30 °C, 1 μl of splicing-activation buffer (0.5 mM ATP, 20 mM creatine phosphate, 1.6 mM MgCl2) was added, and the reactions were incubated for 90 min at 30 °C. The reactions were then quenched with 13 μl DMSO, and 25 nl was used for RT-qPCR.
For splicing reactions using pre-mRNA of the Ad2-ZDHHC16 hybrid, 1 μl reactions were prepared using 10 μg nuclear of extract prepared from 293FT PiggyBac cells, 8 ng pre-mRNA, and varying concentrations of H3B-8800, E7107, or DMSO. After a 15-min preincubation at 30 °C, 2 μl splicing-activation buffer was added, and the reactions were incubated for 90 min at 30 °C. RNA was extracted using a modified protocol from a RNeasy 96 Kit (Qiagen). The splicing reactions were quenched in 350 μl Buffer RLT Plus (Qiagen), and 1.5 volume of ethanol (555 μl) was added. 100 nl of quenched splicing reactions was used for RT-qPCR.
RT-qPCR reactions were prepared using the TaqMan RNA-to-CT 1-step kit (Life Technologies), RNA from splicing reactions, Ad2 canonical junction (forward: 5′-ACTCTCTTCCGCATCGCTGT-3′; reverse: 5′-CCGACGGGTTTCCGATCCAA-3′; probe: 5′-CTGTTGGGCTCGCGGTTG-3′) or Ad2 aberrant junction (probe: 5′-CCAGCTGTTGGGTCTTCGCCC-3′) and Ftz (forward: 5′-TGGCATCAGATTGCAAAGAC-3′; reverse: 5′-ACGCCGGGTGATGTATCTAT-3′; probe: 5′-CGAAACGCACCCGTCAGACG-3′) mRNA primer–probe sets.
Competitive binding assay.
SF3B complexes were immunoprecipitated from nuclear extracts prepared from 293F cells overexpressing Flag-tagged SF3B1. First, batch immobilization of antibody to beads was performed through incubating 80 μg of anti-SF3B1 antibody (MBL International D221–3, Anti-Sap155 monoclonal antibody) and 24 mg anti-mouse PVT scintillation proximity assay (SPA) beads (PerkinElmer) for 30 min. After centrifugation (14,000 r.p.m. for 5 min at 4 °C), the antibody–bead mixture was resuspended in PBS supplemented with PhosSTOP phosphatase inhibitor cocktail (Roche) and cOmplete ULTRA protease inhibitor cocktail (Roche). Nuclear extracts were prepared through diluting 40 mg into a total volume of 16 ml PBS with phosphatase and protease inhibitors, and the mixture was centrifuged (14,000 r.p.m. for 10 min at 4 °C). The supernatant was transferred into a clean tube, and the antibody–bead mixture was added and incubated for 2 h. The beads were collected via centrifuging, washed twice with PBS + 0.1% Triton X-100, and resuspended with 4.8 ml of PBS. 100 μl binding reactions were prepared using slurry and varying concentrations of H3B-8800. After 15 min preincubation at room temperature, 1 nM 3H-probe used in Kotake et al.20 was added. The mixture was incubated at room temperature for 15 min, and luminescence signals were read using a MicroBeta2 Plate Counter (PerkinElmer).
RNA-seq sample preparation.
NALM-6 and K562 isogenic cell lines were treated with H3B-8800 or E7107 at the indicated doses (in Fig. 4) for 6 h. After treatment, the cells were washed with PBS, and RNA was isolated using PureLink (Life Technology) according to the manufacturer’s instructions. cDNA library preparation, sequencing, and raw read filtering methods were described previously17.
U2 snRNP complex assembly assay.
This assay was performed as previously described by Folco23. The BBR 2′-O-methyl oligonucleotide complementary to the branch point binding region (5′-mCmAmGmAmUmAmCmUmAmAmCmAmCmUmUmGmA-3′) and the control oligonucleotide (5′-mAmCmUmGmGmCmAmGmCmAmAmUmGmAmCmUmG-3′) were32P-labeled using γ-ATP (6,000 Ci/mmol, Perkin Elmer) and T4 poly-nucleotide kinase. For all assays, ATP was depleted from nuclear extracts through incubation for 20 min at room temperature. Reaction mixtures (25 μl) containing the depleted nuclear extract (15 μl) were preincubated with or without 1 μM H3B-8800 for 5 min at 30 °C, followed by incubation with 0.5 mM ATP, 3.2 mM MgCl2, and 20 mM creatine phosphate (di-Tris salt) for 5 min at 30 °C. For reactions carried out in the absence of ATP, water was used to supplement the reaction mixture. The radiolabeled BBR or control 2′-O-methyl oligonucleotide (30 ng of each) was added to the reaction mixture last, and the reaction mixture was incubated for 5 min at 30 °C. G50 columns (GE Healthcare) were used to purify the samples before adding 6× DNA loading dye and running 1.2% native low-melting agarose (Invitrogen) gels in Tris/borate/EDTA (TBE) buffer at 70 V for 2 h at room temperature to resolve the 17S and 12S U2 snRNPs. Gels were dried, exposed overnight to a PhosphorImager screen, and imaged.
Real time qPCR and RT-PCR.
RNA was extracted from K562 cells using PureLink (Life Technology) according to the manufacturer’s instructions. cDNA was synthesized using the SuperScript VILO II (Life Technologies) according to the manufacturer’s instructions. cDNA was diluted to 20 ng input for qPCR using the Viia7 system (Life Technologies). For detection of U2AF2 mature mRNA, the following primer–probe set was used: forward: 5′-GAGCACGGTGGACTGATTC-3′; reverse: 5′-GGTGATGTGCTCAAAGCCT-3′; and probe: 5′-CCGCCACGAGAAGAAGAAGAAGGT-3′. For detection of U2AF2 pre-mRNA, the following primer–probe set was used: forward: 5′-TGTTGGTCAGACTGAGGTTG-3′; reverse: 5′-GTGGCACGTCCCAGT ATTTA-3′; and probe: 5′-CCGCCACGAGAAGAAGAAGAAGGT-3′.
RNA was extracted from patient material and PDX models and was used to generate cDNA as previously described11. RT-PCR analyses for expression of EZH2 and GAPDH were also performed using methods and primers as previously described11. For detection of the CEP57 aberrant junction, the following primer–probe set was used: forward: 5′-TGCAGACTGGTCTAGAAACAAA-3′; reverse: 5′-GCTAGGGCTTGTGGACTTT-3′; and probe: 5′-ACTCCGTG TGTTCCCAATGCAAGA-3′. Statistical comparison of the expression of mRNA isoforms as determined through qPCR data was performed using the Student’s t-test.
TaqMan low-density arrays.
K562 isogenic cell lines were seeded at 1 × 105 cells per well in 96-well plates, and Panc05.04 and Panc10.05 cells were seeded at 1.5 × 104 cells per well in a 96-well plate and treated with various concentrations of H3B-8800 for 6 h. RNA (0.5 μg for in vitro studies and 2 μg for in vivo studies) was reverse transcribed using the SuperScript VILO cDNA Synthesis Kit (Thermo Fisher Scientific, CA, USA) following the standard protocol. The resulting cDNA was diluted, and the equivalent of 100 ng was used to prepare a sample specific PCR mix by combining with 50 μl TaqMan Gene Expression Master Mix (Thermo Fisher Scientific, CA, USA) and water up to a final volume of 100 μl. The PCR mix was loaded into the sample fill port of a 24-gene custom developed TaqMan Low-Density Array Card (Thermo Fisher Scientific, CA, USA), which contained two printed technical replicates of each assay per fill port. The TLDA was loaded with eight individual samples and prepared and run on the ViiA7 Real-Time PCR system following the manufacturer’s recommendations (Thermo Fisher Scientific, CA, USA). Threshold PCR cycle (Ct) was determined by the ViiA7 RUO software v 1.2.3 (Thermo Fisher Scientific, CA, USA) using a relative threshold. A Ct greater than or equal to 35 was considered undetectable. Two technical replicates were averaged, and the geometric mean of the expression of five housekeeping genes (B2M, EEF1A1, GAPDH, RPLP0, TUBB) was calculated and used to normalize data as the first step in the comparative ΔΔCt method followed for analysis. Data shown represent three experimental replicates. The list of primers and probes included in the TLDA are reported in Supplementary Table 7.
K562 isogenic lines, HNT-34 and K052 xenograft models efficacy and pharmacokinetic/pharmacodynamic modeling.
1 × 107 K562 isogenic, HNT-34, or K052 cells were subcutaneously implanted into the flank of female NSG or CB17-SCID mice of 6–8 weeks of age. Mice were treated with H3B-8800 (10% ethanol, 5% Tween-80, 85% saline) or vehicle control. For the efficacy studies, the mice were orally dosed daily, and the mice were monitored until they reached either of the following endpoints: (i) excessive tumor volume (≥20 mm in its longest diameter), which was measured three times a week (tumor volume calculated by using the ellipsoid formula: (length × width2) / 2), or (ii) development of any health problem, such as paralysis or excessive body weight loss. The differences in tumor volume during the study period between the vehicle-treated and H3B-8800-treated groups were analyzed by two-way analysis of variance (ANOVA) followed by the Dunnett’s multiple comparison test. For the pharmacokinetic/pharmacodynamic modeling (PK/PD) studies, the mice were treated with one dose, and the tumors were collected at the indicated times (in Figs. 2 and 3) after treatment for further analysis. RNA was isolated using RiboPure RNA purification kit (Ambion) and used for TLDA. All mouse studies were carried out under protocols approved by the Institutional Animal Care and Use Committee at H3 Biomedicine.
Patient samples.
Diagnostic bone marrow or peripheral blood aspirates were obtained from a total of 6 patients with CMML and 2 patients with AML. This includes 5 patients with CMML who were treated at the Memorial Sloan Kettering Cancer Center (MSKCC) and 1 from the Moffitt Cancer Center as well as 2 patients with AML treated at the MSKCC. Patient samples were collected after obtaining written informed consent. The use of human materials was approved by the Institutional Review Board of MSKCC, the Moffitt Cancer Center Scientific Review Committee, and the University of South Florida Institutional Review Board in accordance with the Declaration of Helsinki. Detailed patient characteristics are listed in Supplementary Table 2.
Generation of PDX models.
Generation of CMML PDX models was performed as described in detail by Yoshimi et al.33. Briefly, 6- to 10-week-old NSGS (stock no. 013062) female mice were purchased from the Jackson Laboratory and sublethally irradiated (200 cGy) 3–24 h before transplantation followed by direct intrafemoral injection. CD34+ cells were purified from viably frozen patient bone marrow mononuclear cells (MNCs) through immunomagnetic selection (Miltenyi Biotec). CBC analysis was performed on peripheral blood collected from submandibular bleeding or cardiac puncture post-euthanasia using a Procyte Dx Hematology Analyzer (IDEXX Veterinary Diagnostics). Each of the CMML PDX models used has been previously described33. Genomic analysis of patient bone marrow MNCs and FACS-purified hCD45+ cells from corresponding xenografts was performed using a targeted next-generation sequencing assay as previously described34.
AML PDX models were generated as previously described11, with the exception that NSGS mice were used as recipients (as was done for CMML PDX models described above). All mouse studies were carried out according to the MSKCC Guide for the Care and Use of Laboratory Animals.
Flow cytometric analysis of PDX models.
All flow cytometric antibodies (for both human and mouse studies) were purchased from BD Pharmingen, Biolegend, or eBioscience. Mononuclear cells (MNCs) from patient samples, xenografted tissues, or PB were stained with a combination of antibodies against human CD3 (hCD3, clone: OKT3, purchased from eBioscience), hCD14 (61D3, eBioscience), hCD16 (3G8, Biolegend), hCD19 (HIB19, eBioscience), hCD33 (WM-53, eBioscience), hCD34 (4H11, eBioscience), hCD38 (HB-7, Biolegend), hCD45 (2D1, eBioscience), hCD45RA (HI100, Biolegend), hCD90 (5E10, Biolegend), and hCD123 (7G3, BD Bioscience). Antibodies against mouse CD45.1 (A20, eBioscience) and Ter119 (catalog no. 116223, Biolegend) were used to exclude host-derived cells.
Quantification of RNA-seq data.
Raw sequence reads were aligned to the human reference sequence hg19 by STAR 2.4.2a using two-pass alignment35, and isoform quantification was performed using Kallisto 0.42.4 (ref. 36) against GENCODE annotation v 25 mapped to GRCh37. Estimation of the count of retained introns was performed by defining 6 nt (−3 nt from the splice site and +3 following the splice site) exon–intron boundary regions and counting alignments, which fully overlapped these regions, as previously described17. All raw junction counts, including those from exon–intron boundaries, were pooled over all technical replicates per cohort.
Identification of differential splicing events.
Differential junction usage was assessed using a binomial z test for differences in proportion between treated and untreated sample pools. Proportions were calculated on the basis of all splice junctions (and exon–intron boundaries) which shared a splice site, similar to the percent spliced in (PSI) measurement17. In order to preserve Gaussian assumptions, only those shared splice sites with total junction count (sum raw count over all junctions sharing that splice site) greater than or equal to 10 reads in both treated and untreated cohorts were considered for analysis. z-scores corresponding to a false discovery rate (FDR)-corrected q value of ≤0.05 for treated (‘aberrant’) junctions and ≤0.20 for untreated (‘canonical’) junctions were considered significant.
Two or more splice junctions that share a splice site and have at least one junction that is significantly upregulated in treated and/or mutant samples or at least one junction significantly upregulated in untreated WT samples were considered aberrant or canonical junctions, respectively. Both aberrant and canonical junctions were required to be present in an event for it to be considered differential splicing. In the case of intron-retention events, both exon–intron boundaries (5′ and 3′ splice sites) as well as the junction bridging the two were required to be significant. We report all differential splicing events in Supplementary Table 3.
Gene expression analysis.
Raw gene counts provided by Kallisto v 0.42.4 (ref. 36) were compiled into total gene counts, then analyzed using EdgeR 3.20.2 (ref. 37) to assess the significance of changes between cohorts. Gene changes with a q value < 0.05 were considered significant. Significant gene expression changes are provided in Supplementary Table 4. Prediction of splice variation leading to degradation via the nonsense mediated decay (NMD) pathway was performed as previously described17. For each gene, all associated coding transcripts were assessed to determine whether any annotated exons shared a splice site with a junction found to be significant in treated samples that would modify the protein sequence. If the modified sequence of any transcript would result in a stop codon found >50 nt from the last exon–exon junction—the so-called ‘NMD rule’17 —that transcript is predicted to be targeted by the NMD pathway. Genes for which all transcripts were predicted to be degraded were marked as NMD targeted, and genes for which no transcripts were predicted to be targeted were not. Ambiguous genes, for which some, but not all, transcripts were predicted to be degraded, were ignored. All NMD calls for genes with splicing events are found in Supplementary Table 4.
Analysis of short, GC-rich introns.
A hexbin plot was produced using the matplotlib v 2.0.2 library38. Each hex corresponds to a region of length (y axis) and average overall GC content (x axis) space where introns are found. All colored hexes have at least one intron that matches length and GC content within the bounds defined by the hexagon shape. The color of each hex corresponds to the fraction of the total number of introns represented by introns found in that hex alone.
GC content across an intron and following an exon was assessed by fractionating all intronic and exonic sequences into 100 and 50 bins, respectively, of equal sequence length. The average GC content of each bin was assessed, and then the 95% confidence interval was derived using a permutation test by bootstrapping these averages 5,000 times each. Only introns with sequence length ≥100 nt with at least one known following exon (via RefSeq) of length ≥50 nt were considered. If more than one following exon could be used, one was randomly selected. The resulting figure was drawn using the tsplot function from the seaborn library (https://zenodo.org/record/12710#.WmeMaHlOmUk).
For all sequence analyses, background introns were sampled from a random subset of all genes with at least one intron expressed in the untreated cellular context (n = 6,405 and n = 5,500 for K562 and NALM-6, respectively) that have an expression distribution that is not significantly different from those genes that contained intron retention events in the treated context, measured by Kruskal H test (P = 0.12 and P = 0.55 for K562 and NALM-6, respectively). Genes with matching expression levels were identified using Matching Quantiles Estimation (MQE) in seaborn39. Ideally, background transcripts would also be matched on the basis of transcript turnover rate, however to our knowledge, a comprehensive database in the presented cellular contexts is not currently available.
Pathway analysis.
Overrepresentation analysis of genes containing short (≤300 nt), GC-rich (≥60%) introns retained by H3B-8800 in K562 SF3B1-K700E cells in the most current Reactome40 pathway reference database was accomplished using PANTHER 11.1 (ref. 41). Pathways significant after correction (q < 0.1) using the Benjamini–Hochberg procedure with at least a twofold enrichment over background are reported in Supplementary Table 5. To build a complete database of spliceosome-related genes, we collected 1,512 spliceosome and splicing-related proteins from three sources: (i) 244 spliceosome proteins provided by Hegele et al.29, (ii) 255 spliceosome proteins annotated by Barbosa-Morais et al.30, and (iii) 1,100 proteins from SpliceosomeDB28. Genes were considered splicesome-related if they were found in (i) or (ii). Genes found in (iii) were considered only if they were found in the same protein family as any protein in (i) or (ii) or they were clearly annotated with specific splicing roles. The final table of 404 spliceo-some genes is given in Supplementary Table 6. Enrichment of intron-retention events among 404 spliceosome genes was accomplished by adding these genes as a separate pathway to the existing Reactome database and performing a binomial proportion test followed by Bonferroni correction as previously described41.
Western blotting.
SF3B1-WT or mutant SF3B1-K700E K562 isogenic cell lines were lysed in RIPA buffer (Boston BioProducts) or IP Lysis Buffer (Thermo Fisher Scientific) plus protease-inhibitor cocktail (Mini-complete, EDTA-free, Roche). Forty to fifty micrograms of protein were loaded per well in SDS-PAGE gels (Novex, Life Technologies). Gels were transferred to nitrocellulose membranes using iBlot (Life Technologies). Membranes were blocked in Odyssey Blocking Buffer (LiCor) or 5% milk in Tris-buffered saline and 0.1% Tween20 and were probed overnight at 4 °C with antibodies against the following targets: U2AF2 (1:5,000, Sigma, U4758), HA-Tag (1:1,000, Abcam, 18181), SRPK1 (1:1,000, Bethyl, A302–461A), MCL1 (1:1,000, Cell Signaling Technologies, 5453), β-Actin (1:1,000; Sigma-Aldrich, A5441) or GAPDH (1:5,000, Sigma, G8795 and G9545). After washing, blots were probed at room temperature with secondary antibodies labeled with LI-COR IRDye fluorescent dye or conjugated to HRP. Target proteins were detected using the LI-COR system or the ImageQuant (GE) for enhanced chemiluminescence. Uncropped blots are available in Supplementary Figure 14.
Statistical analysis.
For all results, the relevant statistical tests are provided in the figure legends as well as the total number of samples (n) in each case and control condition. Unless otherwise specified, P < 0.05 was considered significant. Multiple testing correction was performed where applicable using the Benjamini–Hochberg FDR correction, and q < 0.05 was considered significant unless otherwise specified.
Life Sciences Reporting Summary.
Further information on experimental design is available in the Life Sciences Reporting Summary.
Data availability statement.
All RNA sequencing data used in this manuscript are available in GEO under accession number GSE95011.
Supplementary Material
ACKNOWLEDGMENTS
We thank H3 Biomedicine employees for their support in this project. A.Y. was supported by grants from the Aplastic Anemia and Myelodysplastic Syndromes (MDS) International Foundation and the Lauri Strauss Leukemia Foundation. O.A.-W. was supported by grants from the Edward P. Evans Foundation, the Taub Foundation, the Department of Defense Bone Marrow Failure Research Program (BM150092 and W81XWH-12-1-0041), National Institutes of Health National Heart, Lung and Blood Institute (R01 HL128239), the Josie Robertson Investigator Program, an award from the Starr Foundation (I8-A8-075), the Leukemia and Lymphoma Society (2314–17) and the Pershing Square Sohn Cancer Research Alliance.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests: details are available in the online version of the paper.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
References
- 1.Yoshida K et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 478, 64–69 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Wang L et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N. Engl. J. Med 365, 2497–2506 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harbour JW et al. Recurrent mutations at codon 625 of the splicing factor SF3B1 in uveal melanoma. Nat. Genet 45, 133–135 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Imielinski M et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell 150, 1107–1120 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bailey P et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 531, 47–52 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Ciriello G et al. Comprehensive molecular portraits of invasive lobular breast cancer. Cell 163, 506–519 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou Q et al. A chemical genetics approach for the functional assessment of novel cancer genes. Cancer Res. 75, 1949–1958 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Fei DL et al. Wild-Type U2AF1 antagonizes the splicing program characteristic of U2AF1-mutant tumors and is required for cell survival. PLoS Genet 12, e1006384 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Obeng EA et al. Physiologic expression of Sf3b1(K700E) causes impaired erythropoiesis, aberrant splicing, and sensitivity to therapeutic spliceosome modulation. Cancer Cell 30, 404–417 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shirai CL et al. Mutant U2AF1-expressing cells are sensitive to pharmacological modulation of the spliceosome. Nat. Commun 8, 14060 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee SC et al. Modulation of splicing catalysis for therapeutic targeting of leukemia with mutations in genes encoding spliceosomal proteins. Nat. Med 22, 672–678 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Papaemmanuil E et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N. Engl. J. Med 365, 1384–1395 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee Y & Rio DC Mechanisms and regulation of alternative pre-mRNA splicing. Annu. Rev. Biochem 84, 291–323 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nguyen TH et al. CryoEM structures of two spliceosomal complexes: starter and dessert at the spliceosome feast. Curr. Opin. Struct. Biol 36, 48–57 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim E et al. SRSF2 mutations contribute to myelodysplasia by mutant-specific effects on exon recognition. Cancer Cell 27, 617–630 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ilagan JO et al. U2AF1 mutations alter splice site recognition in hematological malignancies. Genome Res 25, 14–26 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Darman RB et al. Cancer-Associated SF3B1 hotspot mutations induce cryptic 3′ splice site selection through use of a different branch point. Cell Rep 13, 1033–1045 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Alsafadi S et al. Cancer-associated SF3B1 mutations affect alternative splicing by promoting alternative branchpoint usage. Nat. Commun 7, 10615 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang J et al. Disease-associated mutation in SRSF2 misregulates splicing by altering RNA-binding affinities. Proc. Natl. Acad. Sci. USA 112, E4726–E4734 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kotake Y et al. Splicing factor SF3b as a target of the antitumor natural product pladienolide. Nat. Chem. Biol 3, 570–575 (2007). [DOI] [PubMed] [Google Scholar]
- 21.Teng T et al. Splicing modulators act at the branch point adenosine binding pocket defined by the PHF5A-SF3b complex. Nat. Commun 8, 15522 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cretu C et al. Molecular Architecture of SF3b and structural consequences of its cancer-related mutations. Mol. Cell 64, 307–319 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Folco EG, Coil KE & Reed R The anti-tumor drug E7107 reveals an essential role for SF3b in remodeling U2 snRNP to expose the branch point-binding region. Genes Dev 25, 440–444 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amit M et al. Differential GC content between exons and introns establishes distinct strategies of splice-site recognition. Cell Rep 1, 543–556 (2012). [DOI] [PubMed] [Google Scholar]
- 25.Jenkins JL, Agrawal AA, Gupta A, Green MR & Kielkopf CL U2AF65 adapts to diverse pre-mRNA splice sites through conformational selection of specific and promiscuous RNA recognition motifs. Nucleic Acids Res 41, 3859–3873 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mercer TR et al. Genome-wide discovery of human splicing branchpoints. Genome Res 25, 290–303 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Corrionero A, Miñana B & Valcárcel J Reduced fidelity of branch point recognition and alternative splicing induced by the anti-tumor drug spliceostatin A. Genes Dev 25, 445–459 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cvitkovic I & Jurica MS Spliceosome database: a tool for tracking components of the spliceosome. Nucleic Acids Res 41, D132–D141 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hegele A et al. Dynamic protein–protein interaction wiring of the human spliceosome. Mol. Cell 45, 567–580 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Barbosa-Morais NL, Carmo-Fonseca M & Aparício S Systematic genome-wide annotation of spliceosomal proteins reveals differential gene family expansion. Genome Res 16, 66–77 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pellizzoni L, Kataoka N, Charroux B & Dreyfuss G A novel function for SMN, the spinal muscular atrophy disease gene product, in pre-mRNA splicing. Cell 95, 615–624 (1998). [DOI] [PubMed] [Google Scholar]
- 32.Luo MJ & Reed R Splicing is required for rapid and efficient mRNA export in metazoans. Proc. Natl. Acad. Sci. USA 96, 14937–14942 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoshimi A et al. Robust patient-derived xenografts of MDS/MPN overlap syndromes capture the unique characteristics of CMML and JMML. Blood 130, 397–407 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng DT et al. Memorial Sloan Kettering-integrated mutation profiling of actionable cancer targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J. Mol. Diagn 17, 251–264 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dobin A et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bray NL, Pimentel H, Melsted P & Pachter L Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol 34, 525–527 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Robinson MD, McCarthy DJ & Smyth GK edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hunter JD Matplotlib: A 2D Graphics Environment. Computing in Science and Engineering 9, 90–95 (2007). [Google Scholar]
- 39.Sgouropoulos N, Yao Q & Yastremiz C Matching a Distribution by Matching Quantiles Estimation. J. Am. Stat. Assoc 110, 742–759 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Croft D et al. The Reactome pathway knowledgebase. Nucleic Acids Res 42, D472–D477 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomas PD et al. PANTHER: a browsable database of gene products organized by biological function, using curated protein family and subfamily classification. Nucleic Acids Res 31, 334–341 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All RNA sequencing data used in this manuscript are available in GEO under accession number GSE95011.