Abstract
We previously established a two-step protocol for differentiation of human pluripotent stem cells (hPSC) into trophoblast, using a StemPro-based minimal media (EMIM) with Bone Morphogenetic Protein-4 (BMP4). However, this protocol was suboptimal, resulting in induction of mixed mesoderm and trophoblast markers. Furthermore, adapting hPSCs to StemPro has proven difficult, and prolonged culture in this media has been shown to promote genomic instability. Therefore, we moved on to use of new media, including E8, and most recently, StemFlex, for rapid adaptation from feeder to non-feeder conditions. In the new protocol, we have incorporated the WNT inhibitor IWP2 into the first step, resulting in uniform differentiation of hPSCs into cytotrophoblast-like cells, without induction of the mesoderm lineage. We also show that, at the end of the second step, there are distinct populations of terminally-differentiated multinucleated hCG-producing syncytiotrophoblast- and HLAG+ extravillous trophoblast-like cells.
Keywords: human pluripotent stem cell, cytotrophoblast, syncytiotrophoblast, extravillous trophoblast, BMP4, IWP2
Introduction
The placenta is an essential transient organ, which supports the growth and development of the fetus in utero. The key cell types in this organ are called trophoblast, whose proper differentiation is essential for normal placental development. In the human placenta, there are three trophoblast cell types. Cytotrophoblast (CTB) are a self-renewing multipotent trophoblast stem cell population. Within floating chorionic villi, the functional units of the human placenta, CTB fuse to form syncytiotrophoblast (STB), cells which are involved in nutrient/gas exchange, and produce the pregnancy hormone human chorionic gonadotropin (hCG). At the interface with the uterus, within anchoring villi, CTB give rise to extravillous trophoblast (EVT), cells which invade the uterine lining and wall and remodel maternal spiral arterioles, establishing the blood supply to the feto-placental unit (Bischof and Finger, 2005). Abnormal trophoblast differentiation is associated with numerous pregnancy complications, including miscarriage, preeclampsia, and fetal growth restriction (Jauniaux et al, 2006; Norwitz ER, 2006; Romero et al, 2011). Study of human trophoblast differentiation has been limited due to the lack of proper model systems. Many studies have used human choriocarcinoma cell lines, which do not represent normal placental cell types (Apps R et al, 2009; Bilban et al, 2010). Mouse models also have limitations due to interspecies differences (Soncin et al, 2018). Primary CTB, isolated directly from placental tissues, have been the gold standard; however, these cells have a limited life span in culture, and, unless isolated from early gestation tissues, can only differentiate into STB (Genbacev et al, 2000; Morrish et al, 1997; Daoud et al, 2005). Most recently, human trophoblast stem cells (hTSC) have been derived from both blastocyst-stage embryos and early placental tissues (Okae et al, 2018) and self-replicating organoids have been established from first trimester CTB (Haider S, 2018); however, due to their derivation from interrupted pregnancies, the disease potential of these cells/organoids is unknown.
In the last two decades, multiple groups, including ours, have used human pluripotent stem cells (hPSCs), both embryonic stem cells (hESCs) and induced pluripotent stem cells (iPSCs), as a model to study trophoblast differentiation (Amita et al, 2013; Erb et al, 2011; Horii et al, 2016; Li et al, 2013; Sudheer et al, 2012; Warmflash et al, 2014; Wu et al, 2008; Xu et al, 2002). We previously published an improved two-step protocol for trophoblast differentiation of these cells (Horii et al, 2016). Unlike other protocols, which aimed to rapidly produce terminally differentiated STB and EVT, our protocol separated trophoblast lineage specification (differentiation into CTB stem cell-like cells) from terminal differentiation (differentiation into STB- and EVT-like cells). However, some disadvantages remained in this protocol. First, the protocol required hPSC to be adapted to StemPro, a step which has proven to take a significant amount of time. Additionally, continuous culture of hPSCs in this media, particularly using enzymatic passage, has been shown to cause genetic instability (Garitaonandia et al, 2015). Another shortcoming of this protocol was that, even though the majority of markers induced following the first step were CTB-associated, some mesoderm-associated markers were also induced, particularly early following addition of BMP4, leaving this model open to criticism from groups that claimed hPSCs are unable to give rise to bona fide trophoblast (Bernardo AS et al, 2011). To limit induction of mesoderm, we had introduced a two-day “rest” in EMIM, a StemPro-based minimal media (Erb et al, 2011; Horii et al, 2016; Li et al, 2013), which led to a decrease in expression of T/Brachyury, a mesoderm marker (Horii et al, 2016). However, recently, two groups have shown that mesoderm induction downstream of BMP4 is WNT-dependent, and can be inhibited using the inhibitor IWP2 (Kurek et al, 2015; Martyn et al, 2018). Here, we have incorporated this inhibitor into an E8-based media and show a modified, improved two-step method for trophoblast differentiation of hPSCs.
Figure 1 shows a schematic of the overall two-step method, with its improvements. Basic protocol 1 explains expansion of hPSCs in feeder-free conditions. Basic protocol 2 describes the details of the first-step, differentiation into CTB-like cells. Basic protocol 3 describes the second-step, differentiation into STB- and EVT-like cells. Briefly, this protocol takes 4 days of BMP4 and IWP2 treatment to obtain over 90% EGFR+ CTB-like cells, yielding up to 1 million cells from one well of a 6-well plate. These cells can then be replated in BMP4 in the presence of feeder-conditioned media, and following an additional 4 to 6 days of culture, will lead to formation of distinct populations of hCG-secreting multinucleated STB-like cells and surface-HLAG+ EVT-like cells. While we have used WA09/H9 hESCs in the experiments shown here, we have successfully applied this protocol to both hESC and iPSC lines. The latter can be established from both normal and diseased tissues, and therefore, offer the advantage of modeling early stages of abnormal human placental development, stages which are normally inaccessible in an ongoing pregnancy.
Figure 1.
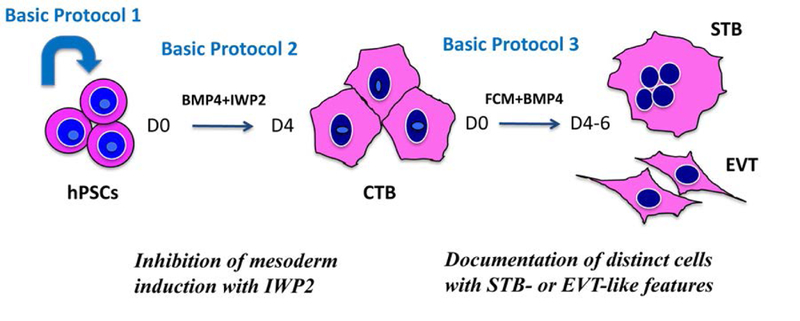
Schematic diagram of two-step trophoblast differentiation protocol using human pluripotent stem cells (hPSCs). Basic protocol 1 describes the expansion of hPSCs to prepare the starting material for differentiation. Basic Protocol 2 focuses on the first step, which is induction of hPSC into cytotrophoblast (CTB) stem cell-like cells. Basic Protocol 3 is focused on the second step, which results in terminal differentiation into distinct populations of multinucleated syncytiotrophoblast (STB)- and invasive extravillous trophoblast (EVT)-like cells.
Basic Protocol 1. Expansion in feeder-free conditions
Introduction
This protocol focuses on the expansion of hPSCs in StemFlex media, before starting trophoblast differentiation.
Material list
hPSC line(s). Note: We have used both hESC (including WA01/H1, WA09/H9) and several of our own iPSC lines (derived from human dermal fibroblast or human umbilical cord mesenchymal stem cells) in this protocol. Prior to use, cells should be adapted to StemFlex media, following protocol provided by ThermoFisher.
0.5 mM EDTA (see Reagents and solutions)
StemFlex (ThermoFisher Cat#A3349401)
Geltrex coated 6-well plate (see Support Protocol 1)
PBS (Corning Cat# 21–040-CV)
Serological pipettes
Conical tubes
Steps and Annotations
-
1)
Aliquot StemFlex media required for the day in room temperature or 37 °C water bath.
-
2)
Place Geltrex-coated plate (either freshly made or stored plate) in the incubator for at least 30 minutes to ensure the plate is warmed up.
-
3)
Take out cryopreserved vial of hPSCs from the liquid nitrogen storage and thaw immediately in the 37 °C water bath.
-
4)
Transfer the cells into 15 ml conical tube with 2–5 ml of pre-warmed StemFlex media and centrifuge 5 minutes at 200 x g, at room temperature.
-
5)
Aspirate the supernatant, and re-suspend with 2 ml of StemFlex. Aspirate the Geltrex from one well, and add 2 ml of cell suspension, making sure to evenly distribute the cells in the well, and place the plate in the 37 °C incubator.
-
6)
Change media the next day, then repeat every other day until cells are sub-confluent.
-
7)
To passage the cells, first prepare the Geltrex-coated plate. Once the Geltrex plate is ready, aspirate the media and wash the cells once with 1ml of PBS. After removing PBS, add 1 ml of EDTA and incubate 3 minutes at 37 °C. Aspirate EDTA and harvest cells with 1–2 ml StemFlex by gently re-suspending cells with 5-ml pipette, then split the cell suspension between the desired number of wells. As an example, 1:9 is often optimal for WA09/H9 hESCs; in this case, use a 15-ml conical tube, add 7–8 ml of StemFlex media to 1–2 ml of harvested cells, for a total of 9 ml cell suspension. Remove the Geltrex from the plate you are passaging into, add 1.5 ml StemFlex media to each well, and, finally, add 1 ml of cell suspension into each well. Other hPSCs, especially iPSC lines, might need some optimization to determine the optimal split ratio.
Note: If the cells dissociate from the plate after the 3 minute-incubation in EDTA, collect them into a 15-ml conical tube and wash the well with 2–5 ml of StemFlex media. Centrifuge the cells for 5 minutes at 200 x g at room temperature to pellet the cells. Then proceed with resuspension in media and splitting as described above.
-
8)
Change the media every other day until the cells are ready for the experiment.
Basic Protocol 2. First-step differentiation (into CTB-like cells)
Introduction
This protocol describes detailed step-wise differentiation of hPSCs into cytotrophoblast (CTB)-like cells. An efficient CTB induction should yield >80% EGFR+ cells (see Figure 2B).
Figure 2.
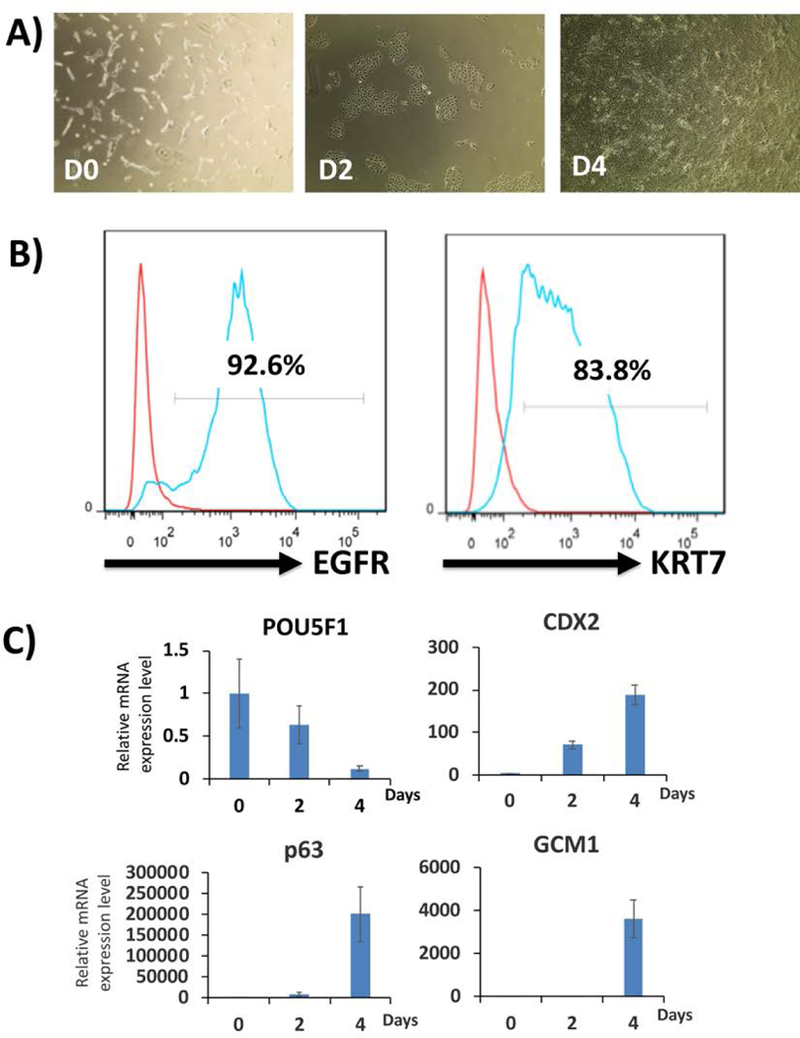
Representative two-step differentiation data from StemFlex-adapted WA09/H9 hESCs.
A) Morphology of the cells during the first step of differentiation (day 0 to day 4). Cells are plated (day −1) in StemFlex media with Rock inhibitor or RevitaCell; this is why they have a spiky morphology on day 0. Once BMP4 and IWP2 are added, differentiation begins uniformly, resulting in flattened epithelial morphology throughout the entire well by day 4.
B) Flow cytometric analysis of cells at the end of the first step of differentiation (day 4) for the CTB marker, EGFR. Over 90% of cells express EGFR compare to the isotype control. These cells are also over 80% positive for pan-trophoblast marker, KRT7.
C) qPCR for lineage-specific markers during the first step of trophoblast differentiation. The pluripotency marker, POU5F1, is decreased, while CTB markers (p63, CDX2 and GCM1) are increased throughout the 4-day period. Data are normalized to 18S and expressed as fold-change above day 0 (undifferentiated state).
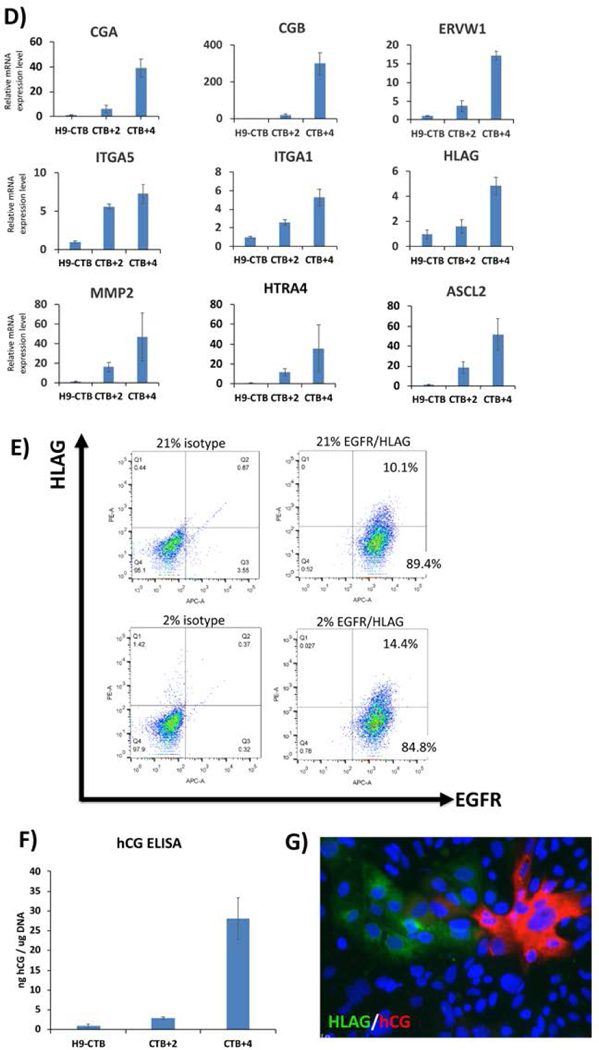
D) qPCR for lineage-specific markers during the second step of trophoblast differentiation. STB markers (CGA, CGB, ERVW1) and EVT markers (ITGA5, ITGA1, HLAG, MMP2, HTRA4, ASCL2) are increased throughout differentiation. Data are normalized to 18S and expressed as fold-change over H9-derived CTB (cells differentiated to day 4 of the first-step differentiation). Time points indicate 2 or 4 days post-replating (CTB+2 and CTB+4, respectively).
E) Flow cytometric analysis of cells following 3 days after second-step of differentiation (CTB+3). Note that while all cells are positive for the CTB marker, EGFR, a portion also co-express the EVT marker, HLAG. Cells differentiated under 2% oxygen showed enhanced HLAG expression compared to those differentiated under 20% oxygen.
F) hCG ELISA confirms differentiation into syncytiotrophoblast (STB)-like cells. Data are normalized to total genomic DNA from the cells in the same well from which supernatant samples were collected.
G) Cells are stained with STB marker hCG and EVT marker HLAG on day 3 following the second-step of differentiation (CTB+3). The lack of overlap between these two markers indicates that STB- and EVT-like cells co-exist in the same culture at the end of the second step.
Material list
hPSCs
0.5 mM EDTA (see Reagents and solutions)
TryPLE (Express ThermoFisher; Cat# 12604021, or Select ThermoFisher cat#12563011; both work for this purpose)
StemFlex (ThermoFisher Cat# A3349401)
Knockout Serum Replacement (ThermoFisher Cat#10828–028)
Differentiation Base Media (see Reagents and Solutions)
First-step Differentiation Media (see Reagents and Solutions)
RevitaCell (100x, ThermoFisher Cat# A2644501) or Y27632 (10mM stock, Selleckchem Cat# S1049, see Reagents and solutions for additional information)
PBS (Corning Cat# 21–040-CV)
Geltrex coated 6 well plate (see Support Protocol 1)
Serological pipettes
Conical tubes
Hemocytometer
Steps and Annotations
-
1)
Culture hPSCs in StemFlex until sub-confluent (~80%). Passage the cells (splitting between 1:6 and 1:9) using 0.5 mM EDTA every 2–3 days to expand the cells (described in detail in Basic Protocol 1, above).
-
2)On day −1, seed cells in Geltrex-coated 6-well plate.
- Pre-warm the Geltrex-coated plate (either freshly made or stored plate) at 37 °C for at least 30 minutes.
- Aspirate the media from sub-confluent well of hPSCs and wash the well with 1 ml PBS.
- Aspirate PBS and add 1 ml TryPLE to dissociate the cells.
- Incubate in 37 °C incubator for 3 minutes.
-
Aspirate TryPLE from the well; add 1 ml of StemFlex into the well using a P1000 pipette and harvest cells, placing in conical tube. Repeat this step with new StemFlex to collect the as many of the cells possible. Re-suspend the cells in a total of 15 ml of StemFlex media and count cells. You should be able to get roughly 2 million cells from one well of a 6-well plate.Note: If the cells dissociate from the plate after the 3 minute-incubation in TryPLE, collect them into a 15 ml conical tube and wash the well with 2–5 ml of StemFlex media. Centrifuge the cells for 5 minutes at 200 x g at room temperature to pellet the cells. Then proceed with resuspension in 15 ml of StemFlex media and count the cells as described above.
-
Remove Geltrex from the well and place 1.5 ml StemFlex media in each well. Seed 60,000 cells per well; add RevitaCell (to final concentration of 1x) or Y27632 (to final concentration of 5 M) and shake the plate in order to have the cells spread and attach evenly on the plate.Note: 60,000 cells per well is optimal for H9 cells, however, seeing density will need to be optimized for each hESC/iPSC line. (i.e. for our iPSCs, we plate 100,000 cells per well). At these seeding densities, cells will grow to roughly 1 million per well by day 4 of differentiation. For passaging and moving forward with the second step of differentiation, you will plate 250,000 cells/one well of a 12-well plate, and 750,000 cells/one well of a 6-well plate. Depending on how many, and what size, wells you need for the second step, you will need to plate a sufficient number of wells for the first step of differentiation.
-
3)
On day 0, remove StemFlex media from the well, and add 1.5 ml of First-step Differentiation Media (see Reagents and Solutions) to each well, and change media every day until day 4. Make Differentiation Media fresh every day by mixing Differentiation Base Media, IWP2, and BMP4. Collect samples on desired days.
-
4)
On day 4, check surface EGFR expression using the flow cytometer. First, remove the media, and wash the cells with 1ml of PBS. Aspirate PBS, and add 1ml of TryPLE express or select in the well. Incubate in 37 °C incubator for 3 minutes. Wash with 1 ml of KSR and 3 ml of PBS (or 25% KSR in PBS), then collect the cells in 15ml conical tube. Centrifuge 5 minutes at 200 x g, at room temperature. (For detailed flow cytometry protocol, see Support Protocol 2.)
-
5)
If flow cytometric analysis shows over 80% EGFR+ cells, proceed to the second-step of differentiation. If the EGFR+ cells compose less than 80% of the cell population, feed the cells with First-step Differentiation Media, let differentiate for another day, and measure the EGFR level again.
Note: For this purpose, be sure to plate an additional well on day −1 in case the first-step requires one more day of differentiation.
-
6)
At the end of this step, successful differentiation should show over 80% surface EGFR+ cells, with the induction of CTB markers (p63, CDX2 and GCM1) and reduction of pluripotency marker (POU5F1). [see Figures 2A–C]. A list of qPCR primers used for evaluation of cells at the end of this step is provided in Table 1.
Table 1.
List of Quantitative Real-Time PCR Primers
| Primer Name | Primer sequence |
|---|---|
| POU5F1 | F 5’- TTG GCT CGA GAA GGA -TGT G
3’ R 5’ – GCA TAG TCG CTG CTT GAT CG −3’ |
| SP5 | F 5’- TCG GAC ATA GGG ACC CAG TT
−3’ R 5’- CTG ACG GTG GGA ACG GTT TA −3’ |
| P63 | F 5’- CTG GAA AAC AAT GCC CAG A
−3’ R 5’- AGA GAG CAT CGA AGG TGG A −3’ |
| CDX2 | F 5’- TTC ACT ACA GTC GCT ACA TCA CC
−3’ R 5’- TTG ATT TTC CTC TCC TTT GCT C −3’ |
| T | F 5’- AGT TGA GGA CAG CAG CTT TTA GTT
−3’ R 5’- GCT GAT TGT CTT CTA CT-3’ |
| GCM1 | F 5’- TTT TTC CAG TCA AAG GGA GAG CA
−3’ R 5’- AGG AAG GTG CTG TGT TCA CTT −3’ |
| CGA | F 5’- CAA CCG CCC TGA ACA CAT CC -
3’ R 5’ – CAG CAA GTG GAC TCT GAG GTG −3’ |
| CGB | F 5’- ACC CTG GCT GTG GAG AAG G
−3’ R 5’- ATG GAC TCG AAG CGC ACA −3’ |
| ERVW1 | F 5’- GCT ACT GTC TGT TGG ACT TAC T
−3’ R 5’- CGG GTG AGT TGG GAG ATT AC −3’ |
| ITGA5 | F 5’- GGC TTC AAC TTA GAC GCG CAG-
3’ R 5’ – TGG CTG GTA TTA GCC TTG GGT −3’ |
| ITGA1 | F 5’- CTG GAC ATA GTC ATA GTG CTG GA -
3’ R 5’ – ACC TGT GTC TGT TTA GGA CCA −3’ |
| HLAG | F 5’- ACT GAG TGG CAA GTC CCT TT -
3’ R 5’ – TGG GGA AGG AAT GCA GTT CAG −3’ |
| MMP2 | F 5’- CCA AGG AGA GCT GCA ACC TG -
3’ R 5’ – TGG GCT TGC GAG GGA AGA ACT −3’ |
| HTRA4 | F 5’- AAA GAA CTG GGG ATG AAG GAT -
3’ R 5’ – TGA CGC CAA TCA CAT CAC CAT −3’ |
| ASCL2 | F 5’- CAC TGC TGG CAA ACG GAG AC -
3’ R 5’ – AAA ACT CCA GAT AGT GGG GGC −3’ |
| L19 | F 5’- AAA ACA AGC GGA TTC TCA TGG A -
3’ R 5’ – TGC GTG CTT CCT TGG TCT TAG −3’ |
Basic Protocol 3. Second-step differentiation (of CTB-like cells into STB- and EVT-like cells)
Introduction
This protocol describes the details of the second-step of trophoblast differentiation. Successful differentiation will result in presence of a mixed population of hCG+ STB- and HLA-G+ EVT-like cells (see Figure 2G)
Material list
hPSC-derived CTB from first-step differentiation
TryPLE (Express ThermoFisher; cat# 12604021, or select ThermoFisher Cat#12563011; both work for this purpose)
Feeder-conditioned media (FCM; see Reagents and Solutions)
Second-step Differentiation Media (see Reagents and Solutions)
Geltrex-coated plate (see Support Protocol 1)
PBS (Corning Cat# 21–040-CV)
Conical tube
Serological pipettes
Steps and Annotations
-
1)
Before you begin, warm up FCM required for passaging either at room temperature or in the 37 °C water bath. Pre-warm the Geltrex-coated plate (either freshly made or stored plate) at 37 °C for at least 30 minutes.
-
2)
Aspirate media from the well containing hPSC-derived CTB, and add 1 ml PBS to wash the cells. Remove the PBS and add 1 ml TryPLE to dissociate the cells. Put the plate in 37 °C incubator for 3 minutes.
-
3)
Without removing TryPLE, add 1 ml of KSR to the well using 5-ml pipette and collect cells into conical tube. Repeat this step with 3 ml PBS to collect as many of the cells as possible.
Note: The addition of KSR helps to form a tight cell pellet. Alternatively, a solution of 25% KSR in PBS may be used.
-
4)
Centrifuge 5 minutes at 200 x g at room temperature. Aspirate the supernatant and resuspend in Second-step Differentiation Media to count the cells.
Note: Usually, we dissociate all 6 wells of one 6-well plate, resulting in ~5–6 million cells, and resuspend with 6 ml of media, which results in sufficiently-high cell density for splitting (see step 5 below). For counting, we take a small aliquot and dilute it further (~1:7).
-
5)
Remove Geltrex from the well and add the appropriate amount of Second-step Differentiation Media (250 l for 12-well plate and 750 l for 6-well plate). Seed 250,000 cells per well for a 12-well plate, and 750,000 cells per well for a 6-well plate; add the appropriate amount of Second-step Differentiation Media for a total volume of 500 l and 1.5 ml per well of a 12-well or 6-well dish, respectively. Agitate the plate in order to have the cells spread and attach evenly on the plate.
-
6)
Change media every day. Collect samples at the desired time points.
-
7)
At the end of this step, cells should show increased STB markers (CGA, CGB, ERVW1) and EVT markers (ITGA5, ITGA1, HLAG, MMP2, HTRA4, ASCL2) by qPCR, compared to the starting material at the end of the first step [Figure 2D]. Flow cytometry should show some EGFR/HLAG double-positive (EVT-like) cells; the proportion of these cells can be highly variable, although, within each hESC/iPSC line, it can be increased 1.5–2 fold by performing the second step under low oxygen (2%) conditions [Figure 2E] (Horii et al. 2016). At the end of the second step, a portion of the cells will also secrete hCG, confirming differentiation into STB-like cells [Figure 2F]. Finally, immunostaining of the cells shows distinct populations of hCG-producing STB-, and HLA-G+ EVT-like cells [Figures 2G]. A list of qPCR primers used for evaluation of lineage-specific markers at the end of this step is provided in Table 1. Flow cytometry protocol is described in Support protocol 2.
Note: For the second-step, we normally use 12-well plates for RNA, DNA, and supernatant collection. A 6-well plate is used for collecting cells to perform flow cytometry or coverslips for immunocytochemistry.
Support Protocols
Support Protocol 1:
Geltrex plate preparation
Material list
Geltrex (Thermo Fisher Cat# A14133–02)
DMEM/F12 (Thermo Fisher Cat# 11330–032)
6-well plate (Corning Cat# 3506)
Serological pipettes
Conical tubes
Parafilm
Steps and Annotations
-
1)
Put 100 l Geltrex aliquot on ice and wait for 30–45 minutes until completely thawed. Prepare 20 ml of ice-cold DMEM/F12 in a 50-ml conical tube, and add 100 l Geltrex to make a 1:200 dilution. Mix gently.
-
2)
Add Geltrex mixture to the plate (2 ml per well for 6-well plate, and 500 l per well for 12-well plate). Incubate the plate at 37 °C for 30 minutes for immediate use. For preparation for later use, incubate for 2 hours in either room temperature or 37 °C incubator. After the incubation, cover with parafilm and store the plate in 4 for up to 4 weeks.
Support Protocol 2:
Flow cytometry on day 4 (end of first-step differentiation) and day+3 (3 days after start of second step differentiation)
Material list
APC conjugated mouse anti-human EGFR Antibody (clone AY13) (BioLegend, Cat#352906)
APC mouse IgG1 isotype (clone MOPC-21) (BioLegend, Cat#400122)
PE conjugated mouse anti-HLA-G (clone MEM-G9) (EXBIO, Cat#1P-292_C100)
PE mouse IgG1 isotype (clone MOPC-21) (BioLegend, Cat#400112)
Flow cytometry washing buffer (see Reagents and Solutions)
Flow cytometry blocking buffer (see Reagents and Solutions)
5 mL polystyrene round bottom test tubes (BD Falcon, Cat#352058)
Steps and Annotations
-
1)
Aspirate supernatant from the tube from Basic Protocol 2 (step 4) for EGFR staining at the end of first-step differentiation, or from Basic Protocol 3 (step 7) for EGFR/HLAG staining at the end of the second-step differentiation. Add 400 l of blocking buffer and break down the cells using a P1000 pipette. Split the cell mixture into 2 test tubes, one for isotype control and one for EGFR or EGFR/HLAG antibody staining.
-
2)
For the day 4 EGFR staining, add 5 l of APC-EGFR or APC-isotype antibody; for the day+3 EGFR/HLAG staining, add 5 l of APC-EGFR or APC-isotype antibody, and 5 g/ml of PE-HLAG or PE-isotype antibody (according to the manufacturer’s protocol) to each tube and incubate for one hour on ice, protected from light.
-
3)
Add 3 ml of washing buffer, and centrifuge 5 minutes at 200 x g, at room temperature. Aspirate the supernatant and repeat this washing step two more times.
-
4)
After the third centrifugation, aspirate the supernatant, and add 250 l of washing buffer. Protect from light. Flow cytometric analysis can be done on BD LSRFortessa, BD FACS Canto II, or an equivalent cytometer.
Reagents and Solutions
bFGF (24 g/mL)
In a sterile environment, first prepare 0.1% (w/v) BSA (Gemini Bio-Products, Cat#700–110) in PBS (Corning Cat# 21–040-CV) solution. To make 24 g/mL bFGF (Biopioneer), combine 2.083 ml of 0.1% (w/v) BSA in PBS solution to 50 g of bFGF. Gently pipette up and down to reconstitute the protein and prepare 100 l aliquots. Store at −20 °C for up to 6 months. Once thawed, keep in 4 °C. Avoid freeze-thaw cycles.
Blocking buffer for flow cytometry
The final composition of the blocking buffer is 1% (w/v) BSA (Cell Signaling Cat#9998S) 10% (v/v) FBS (Omega Scientific cat# FB-02) in PBS. To make 1 ml of blocking buffer, add 800 l of wash buffer, 100 l 10% BSA and 100 l FBS into a single tube. Mix well and keep on ice until use.
BMP4 (10 g/mL)
In a sterile environment, first, prepare 0.1% (w/v) BSA (Gemini Bio-Products Cat#700–110) in PBS (Corning Cat# 21–040-CV) solution. To make a 10 g/mL BMP4 solution, combine 5 ml of 0.1% (w/v) BSA in PBS solution to 50 g of BMP4. Gently pipette up and down to reconstitute the protein. Thoroughly mix and prepare 100 l aliquots. Store at −80 °C for up to a year. Once thawed, keep in 4 °C. Avoid freeze-thaw cycles.
0.5 mM EDTA
In a sterile environment, add 50 l of 0.5M EDTA (ThermoFisher Cat# 15575020) into 50 ml PBS (Corning Cat# 21–040-CV). Filtering is recommended. Store at room temperature.
Differentiation Base Media
1X KnockOut DMEM (ThermoFisher Cat#10829–018)
1X F12 (ThermoFisher Cat#11765–054)
1X ITS (Sigma Cat#I1884–1VL); alternatively, ITS from ThermoFisher (Cat# 41400045) can be used as well.
64 g/ml L- ascorbic acid 2-phosphate (Sigma Cat#A8960–5G)
543 g/ml NaHCO3 7.5% solution (ThermoFisher Cat#25080094)
2% (w/v) BSA (30% solution, Gemini Bio-Products Cat#700–110)
Filtering is recommended for the media preparation.
Store at 4 °C. Use within a week, for one differentiation experiment.
See below for the exact quantities.
| Component | Amount of stock for
final volume of 50 ml |
Stock
concentration |
Final concentration |
|---|---|---|---|
| KO DMEM | 24.5 ml | -- | -- |
| F12 | 24.5 ml | -- | -- |
| ITS | 500 μl | 100X | 1X |
| L ascorbic acid | 96 μl | 33.3 mg/mL | 64 μg/mL |
| NaHCO3 | 362 μl | 75 mg/mL | 543 μg/mL |
| BSA | 3.3 ml | 30% | 2% |
Feeder conditioned media (FCM)
To avoid batch-to-batch variation, which potentially changes the differentiation efficiency in the second-step, it is recommended that large amounts of FCM are prepared and frozen for later use, particularly for comparison of different hESC/iPSC lines. Preparation of FCM takes a total of 7 days. The following procedure will use five T-175 flasks and makes a total of 1750 ml of FCM.
-
1)
Before starting, prepare a sterile bottle that can hold 2L of liquid (in order to be able to hold and mix all the FCM prepared here).
-
2)
In a sterile environment, first, coat each flask with 5 ml of 0.1% gelatin (EMD Millipore, Cat# ES-006-B) and incubate for 30 minutes at 37 °C.
-
3)
Take out 25 million irradiated mouse embryonic fibroblasts/MEFs (5 million cells per flask purchased from ThermoFisher cat# A34180) from the liquid nitrogen storage and thaw immediately in the 37 °C water bath.
-
4)
Transfer the cell mixture to 10 ml of MEF media (see below) in a 15-ml conical tube. Centrifuge the cells for 5 minutes at 200 x g at room temperature.
-
5)
Aspirate the supernatant and resuspend the cells in 25 ml of MEF media.
-
6)
Take out the T-175 flasks from the incubator, aspirate the 0.1% gelatin, and add 25 ml of MEF media. Then, add 5 ml resuspended cells to each flask.
-
7)
Place the cells in the incubator and gently shake the flask to have the cells distribute evenly.
-
8)
The next day (day 0), aspirate the MEF media from the flasks and add 50 ml of WiCell media (see below). After 24 hours (on day 1), collect 50 ml of conditioned media per flask into the sterile 2L bottle. Then add another 50 ml of WiCell media to each MEF flask. Repeat every day for 7 days. Keep the collected media (in the sterile 2L bottle) in the 4 °C.
-
9)
On day 7, mix all the media well in the 2L bottle. Filter the media using a 0.22 μm bottle-top filter (Stericup-GP Sterile Vacuum Filtration System, Millipore Sigma, Cat#SCGPU11RE) and make 40 ml aliquots. Stored at −20 . Once thawed, keep at 4 °C for up to 2 weeks. Avoid freeze-thaw cycles.
First-step Differentiation Media
In a sterile environment, prepare Differentiation Base Media with final concentration of 10 ng/ml BMP4 (1/1000 from the stock solution) and 2 M IWP2 (1/1000 from the stock solution). 1.5ml of media is required for one well of 6-well plate. Prepare fresh. Filtering is not recommended.
| Component | Amount for 1 ml | Stock
concentration |
Final concentration |
|---|---|---|---|
| Differentiation Base Media | 1 ml | -- | -- |
| BMP4 | 1 μl | 10 μg/mL | 10 ng/mL |
| IWP2 | 1 μl | 2 mM | 2 μM |
ITS (Insulin-transferrin-sodium selenite) (100X)
In a sterile environment, add 50 ml of sterile distilled water to the ITS vial (Sigma. Cat#I1884–1VL). Mix well and make 1 ml aliquots. Store in −20 °C for up to one year. Once thawed, store in 4 °C. Avoid freeze-thaw cycles.
IWP2
In a sterile environment, reconstitute 10 mg IWP2 (Selleck Chemicals, Cat# S7085) with 10.72 ml of DMSO to prepare a 2 mM stock solution. Make 100 μl aliquots and store at −80 °C for up to two years. Once thawed, store at 4 °C. Avoid freeze-thaw cycles.
L-Ascorbic acid 2-phosphate
In a sterile environment, aliquot 6 ml of sterile DMEM/F12 media (Thermo Fisher Cat# 11330–032) in a 15 ml conical tube and warm up in 37 °C water bath. Weigh out 200 mg of L-ascorbic acid (Sigma, Cat# A8960–5G) and add into warm DMEM/F12, then vortex to make 33.3 mg/ml L-ascorbic acid. If the powder does not dissolve completely, repeat vortex and continue warming in water bath until fully dissolved. Filter the solution and make 1 ml aliquots. Store at −20 °C. Once thawed, store at 4 °C. Avoid freeze-thaw cycles.
MEF media
DMEM/GlutaMax (ThermoFisher, Cat# 10569010)
10% (v/v) heat-inactivated FBS (Omega Scientific Cat# FB-02)
1X NEAA (ThermoFisher, Cat# 11140–050)
Filtering is recommended for media preparation.
Store at 4 °C and use within 4 weeks.
See below for the exact quantities.
| Component | Amount for 50ml |
Stock
concentration |
Final concentration |
|---|---|---|---|
| DMEM/Glutamax | 44.5 ml | -- | -- |
| FBS | 5 ml | 100% | 10% |
| NEAA | 500 μl | 100X | 1X |
RevitaCell Supplement (ThermoFisher Cat# A2644501)
The 100X solution is ready to use. Prepare 500 μl aliquots and store at −20 °C for up to 1 year. Protect from light.
Second-step Differentiation Media
In a sterile environment, mix FCM and BMP4 to a final concentration of 10 ng/ml (1/1000 from the 10ug/ml stock solution). 1.5ml or 500 l of media is required per well for a 6-well or 12-well plate, respectively. Prepare this media fresh every day. Filtering is not recommended.
| Component | Amount for 1 ml | Stock
concentration |
Final concentration |
|---|---|---|---|
| FCM | 1 ml | -- | -- |
| BMP4 | 1 μl | 10μg/mL | 10ng/mL |
StemFlex media
Thaw frozen 10x supplement (ThermoFisher, Cat# A3349401) at room temperature or 4 °C overnight. Make 5 ml aliquot of this supplement and store at −20 °C for up to 6 months. Protect from light. For making 50ml of StemFlex media, thaw 1 tube of aliquoted supplement at room temperature or 4 °C overnight and mix with 45 ml StemFlex basal media. Filtering is recommended. Protect from light.
Wash buffer for flow cytometry
On a clean work bench, add 500 μl of FBS (Omega Scientific cat# FB-02) to 50 ml PBS (house-made or Corning Cat# 21–040-CV). Mix well and keep on ice until use.
WiCell media
DMEM/F12 (ThermoFisher, Cat# 11330–032)
1X NEAA (ThermoFisher, Cat# 11140–050)
1X GlutaMax (ThermoFisher, Cat# 35050061)
20% Knockout Serum Replacement (ThermoFisher, Cat# 10828–028)
0.1 mM 2-Mercaptoethanol (ThermoFisher, Cat# 21985–023)
Filtering is recommended.
Store at 4 °C and use within 4 weeks.
See below for the exact quantities.
| Component | Amount of stock for
final volume of 50 ml |
Stock
concentration |
Final concentration |
|---|---|---|---|
| DMEM/F12 | 39 ml | -- | -- |
| NEAA | 500 μl | 100X | 1X |
| Glutamax | 500 μl | 100X | 1X |
| Knockout Serum Replacement | 10 ml | 100% | 20% |
| 2-Mercaptoethanol | 91 μl | 55mM | 0.1 mM |
Y27632 (Selleckchem Cat# S1049)
The 10 mM stock solution is already dissolved in DMSO. Prepare 100 μl aliquots and store at −80 °C for up to 2 years. After thawing, store at 4 °C. Avoid freeze thaw cycles.
Commentary
Background information
We had previously established a protocol for stepwise differentiation of hPSCs, first into cytotrophoblast (CTB) stem cell-like cells, and then into terminally differentiated syncytiotrophoblast (STB)- and extravillous trophoblast (EVT)-like cells (Horii et al, 2016). Using that protocol, we showed that hPSC-derived CTB-like cells can recapitulate the behavior of primary CTB, in the setting of both normal and diseased pregnancy (Horii et al, 2016). In particular, we showed the hPSC-derived CTB have a similar response as primary CTB to low oxygen tension, namely, inhibition of STB differentiation, and enhanced EVT differentiation (Horii et al, 2016). In addition, we showed that, similar to CTB isolated from Trisomy 21 placentae, CTB-like cells derived from iPSC with an extra chromosome 21 showed defects in STB differentiation (Horii et al, 2016). This was a significant step toward the development of “disease-in-a-dish” models for studying human pregnancy disorders, such as preeclampsia.
Recently, Sheridan et al. (PNAS, 2019) have used iPSCs derived from placentas from both normal and preeclamptic pregnancies and shown that the preeclamptic iPSCs exhibit a reduced ability to invade when cultured in 20% oxygen, an indication that, there is an underlying defect in trophoblast invasion in preeclamptic placentae (McMaster et al, 2004; Pijnenborg et al, 2011). However, this group uses a different protocol for trophoblast differentiation of hPSCs, namely, a combination of BMP4, TGF inhibitor (A83–01), and FGF receptor inhibitor (PD173074), called “BAP” for short (Amita et al, 2013). It is not clear whether the cells that result from this protocol are distinct groups of STB and EVT (Amita et al, 2013). Furthermore, whether the preeclamptic iPSCs have defects in either trophoblast lineage specification, terminal trophoblast differentiation, or both, remain to be seen. Our two-step protocol offers the advantage of ushering the hPSCs through developmentally-accurate stages of CTB progenitor state to morphologically-distinct subpopulations of STB- or EVT-like cells, both with appropriate secretory function.
Critical Parameters
It is essential to have good starting material. If WA09/H9 ESCs (or other hPSCs) are not growing well, consider thawing a new vial, changing to a new batch of StemFlex, or a new batch of Geltrex. It is also very important to plate the cells very sparsely at the beginning of the first step of differentiation (see Figure 2A). If the hPSC colonies are too big at the start, the middle part of the colony will not differentiate well. This is one reason to use TryPLE, instead of EDTA, to dissociate the cells. In addition, daily cell culture work for differentiation should be done around a similar time in the day, in order to ensure collection of samples and media changes are done every 24 hours.
Troubleshooting
Most of the time, at least 80% of the cells will express EGFR following 4 days of treatment with BMP4 and IWP2. If after 4 days, EGFR expression is between 60–80%, it is best to extend the differentiation by one more day, re-test the EGFR level by flow cytometry, and move to the second step on day 5 if the EGFR level is over 80%. Occasionally, the first step of differentiation has to be extended by two days (a total of 6 days). If this is consistently the case, it is possible that this is the true phenotype of a particular hESC/iPSC line, as we previously noted with the Trisomy 21 hESC/iPSC lines (Horii et al. 2016). However, with a genetically normal hESC/iPSC, it is more likely an issue of either plating too many cells or a specific reagent. If such cells routinely require 6 days to reach >80% EGFR expression, it is best to re-start the differentiation from the beginning, making sure all reagents are prepared fresh and initial plating is sparse enough (see Basic Protocol 2 above and Figure 2A). BMP4 potency is occasionally an issue, and thus consideration should be given to the source of this reagent. Our group has been using BMP4 from R&D or HumanZyme. Roberts et al. (2018) reported various sources of BMP4 used by them and other groups. The efficacy of IWP2 can also be tested by the level of suppression of SP5 (see Anticipated Results below). A detailed troubleshooting guide is provided in Table 2.
Table 2.
Trouble-shooting
| Problem | Possible cause(s) | Solution(s) |
|---|---|---|
| Poor attachment of the hPSCs on Geltrex-coated plate | Matrix issues | Check matrix. Be sure to plate Geltrex in a cold environment (e.g. keep on ice until plating). Gels in 5–10 minutes above 15°C. |
| No Y27632 or RevitaCell | hPSCs attach less when they are single cells. Include Y27632 or RevitaCell when plating. | |
| hPSCs are differentiating | Media issues | Remake media if necessary. Check if the reagents used in the media are expired and used at the correct concentration. |
| Matrix issues | Check matrix. Be sure to plate Geltrex in a cold environment (e.g. keep on ice until plating). Gels in 5–10 minutes above 15°C. Heating the Geltrex could result in premature formation of gel. | |
| Cells lift from plate | Matrix Issues | Check matrix. Be sure to plate Geltrex in a cold environment (e.g. keep on ice until plating). Gels in 5–10 minutes above 15°C. Heating the Geltrex could result in premature formation of gel. |
| Cells do not lift when using EDTA or TryPLE | Too short incubation time | Collect cells as much as possible after 3 minutes, then repeat the dissociation step. |
| Cells lifted too much when using EDTA or TryPLE | Too long incubation time or weak adherence | Do not aspirate the media. Collect the cell mixture with media in a tube. Centrifuge the cells for 5 minutes at 200 x g at room temperature. Aspirate supernatant and resuspend with media. |
| Low EGFR at the first-step differentiation | Overseeding | Re-start the experiment. Seed in a fewer concentration if necessary. |
| Cells are in big clusters | When treating cells with TrypLE, try to break up the cells to single cells. Use a P1000, pipette up and down harshly. | |
| Reagent issues | Check differentiation factors with a positive control to ensure that reagents are working. | |
| High passage number | Thaw earlier passaged hPSCs. | |
| Cells too sparse at the end of the first-step differentiation | Cells are not collected well, or starting material was too small | Add more wells at the beginning of first step of differentiation. |
Anticipated Results
The addition of IWP2 was previously reported to inhibit mesoderm induction downstream of BMP4 (Kurek et al. 2015). In our hands, the addition of IWP2 during the first step of trophoblast differentiation also appears to accelerate/increase efficiency of the CTB induction (Figure 3A). In the presence of IWP2, the edges of the colonies begin to flatten after only one day of differentiation, and by day 4, there is a uniformly-flattened (epithelial) morphology present throughout the culture, a phenotype not consistently achieved by BMP4 alone even after 6 days (Figure 3A). By flow cytometry, cells easily achieve >90% EGFR positivity after 4–6 days (Figure 3B). By qPCR, CTB markers CDX2 and p63 are significantly elevated, peaking by day 4 after beginning of the first step of differentiation; nevertheless, CDX2 (which is also a mesendoderm marker (Mendjan et al, 2014) is significantly suppressed in the presence of IWP2 (Figure 3C). The suppression of the mesoderm lineage, as well as inhibition of endogenous WNT signaling, are confirmed by significantly-decreased expression of T/BRACHYURY and SP5, respectively (Figure 3C).
Figure 3.
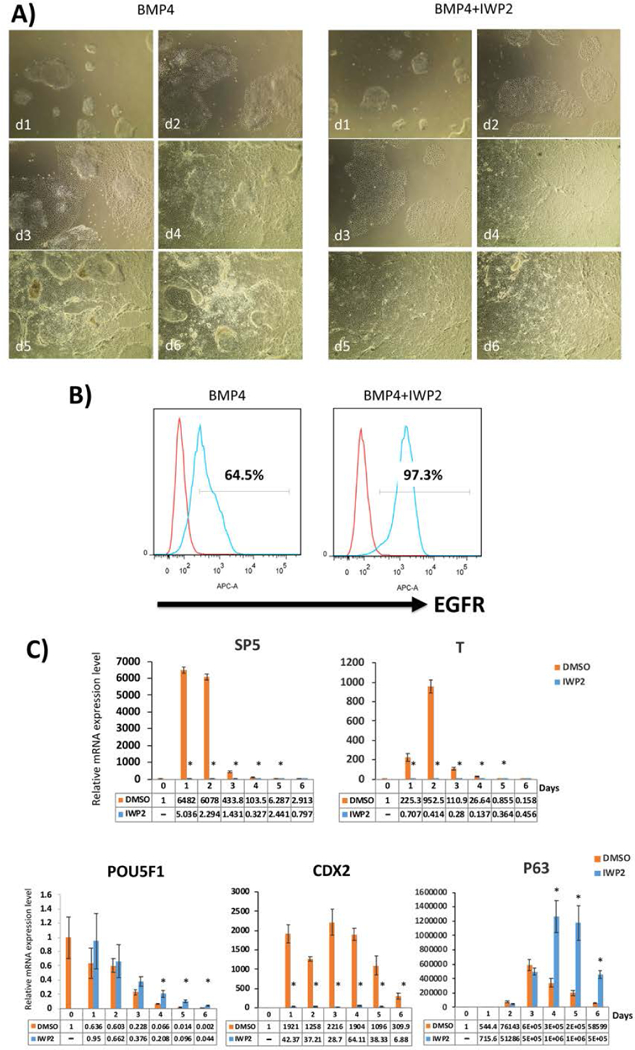
Comparison of trophoblast differentiation using BMP4, with and without IWP2, starting with E8-adapted WA09/H9 hESCs.
A) Morphology of the cells in IWP2 shows more uniform flattening starting from one day post-BMP4 treatment, in comparison to BMP4 alone. This morphological difference was consistent through all 6 days of the first step differentiation.
B) Expression of CTB marker, EGFR, as measured by flow cytometry on day 6 following BMP4 treatment. The addition of IWP2 leads to a higher number of EGFR+ cells.
C) qPCR for lineage-specific marker expression during the first-step differentiation with and without IWP2. Compared to the DMSO vehicle control, marker of WNT signaling, SP5, is suppressed in the presence of IWP2, as expected. T/Brachury, an early marker of mesoderm induction, is also suppressed. At the same time, CTB markers (p63 and CDX2) are significantly increased both with and without IWP2, although the fold change for CDX2 (also a marker of mesoderm) is lower in the presence of IWP2. Pluripotency marker, POU5F1, is appropriately decreased during differentiation. Data are normalized to 18S and expressed as fold-change above day 0 (undifferentiated state). *indicates P<0.05 compared to DMSO control at the same time-point.
As compared to StemPro and EMIM, the StemPro-based minimal media previously used in the first step of our two-step protocol (Horii et al. 2016), passage of hPSC in E8/StemFlex and E8-based minimal media used in the first step do not show a significant difference in induction of trophoblast lineage, either by flow cytometry or qPCR (Figure 4A–C; compare to Figure 2A–C). However, compared to StemPro, we have found that adaptation of hPSCs to E8 and StemFlex is significantly easier, with adaptation to StemFlex taking the least amount of time. In addition, the two-day “rest” in minimal media prior to start of CTB differentiation is not required with E8 or StemFlex. Finally, it is important to note that, in addition to a peak in the CTB progenitor markers CDX2 and p63, the end of the first step of differentiation is characterized by a peak in GCM1 (Figures 2C and 4C), a transcription factor, expressed in both villous and extravillous trophoblast (Wakeland et al. 2017), which is required for initiation of terminal differentiation.
Figure 4.
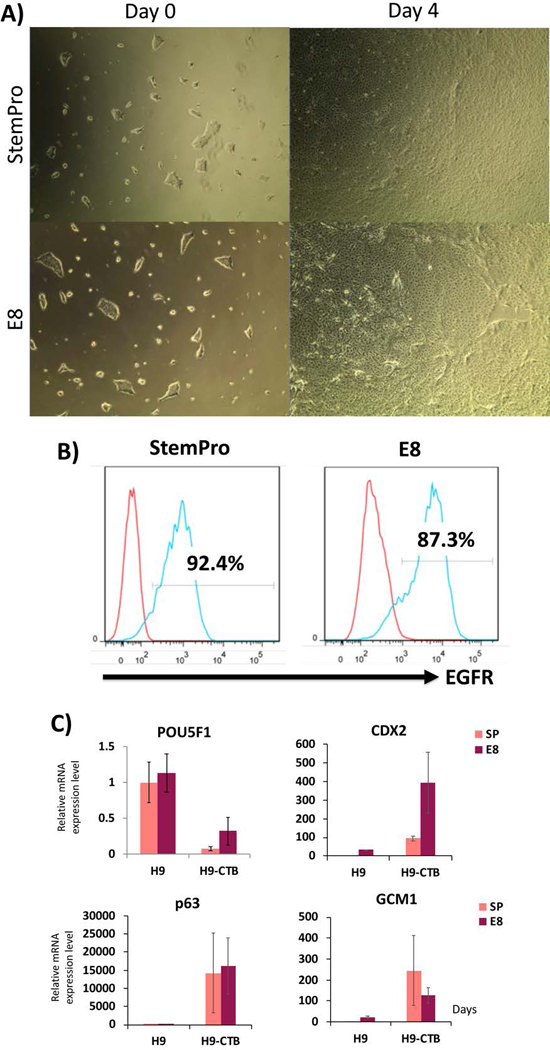
Comparison of CTB differentiation from StemPro- vs. E8-adapted WA09/H9 hESCs, using BMP4+IWP2.
A) First-step differentiation of the StemPro- and E8-adapted H9s do not show any morphological differences.
B) Flow cytometric analysis at the end of the first step (day 4) did not show a difference in EGFR expression.
C) qPCR for lineage-specific markers. The pluripotency marker, POU5F1, is decreased and CTB markers (p63, CDX2 and GCM1) are increased by day 4 following BMP4+IWP2 treatment, with no difference between the StemPro- and E8-adapted cells. Data are normalized to 18S and expressed as fold-change above StemPro-day 0 (undifferentiated state).
The second step of this protocol is done in the presence of feeder-conditioned media (FCM), leading to induction of a mixture of STB- and EVT-like cells, as shown by a combination of flow cytometry, qPCR, immunostaining, and ELISA (Figure 2D–G and Figure 5). qPCR shows induction of STB (CGA, CGB, and ERVW1) and EVT markers (HLAG, ITGA5, ITGA1, MMP2, HTRA4 and ASCL2) (Figures 2D and 5G); note that most of these markers peak after 4 days of differentiation in FCM and BMP4. Curiously, some of these markers are better induced when the differentiation is performed with E8-adapted cells, including STB markers CGB and ERVW1, and the EVT marker ASCL2, while one EVT marker (MMP2) shows the opposite pattern (Figure 5A). Also, compared to StemPro and StemFlex, E8-adapted cells show enhanced STB secretory function, based on hCG ELISA (average of 60 ng hCG/μg DNA in E8, compared to ~30 ng hCG/μg DNA in StemPro and StemFlex-adapted cells; see Figure 5B, and compare also to Figure 2F). We also confirmed here that performing the second step in hypoxia (2% oxygen) increased the proportion of HLA-G+ cells (Figure 2E). Last, but perhaps most importantly, we report here the formation of distinct populations of STB- and EVT-like cells (Figures 2G and 5C), further confirming that this differentiation protocol leads to a developmentally-accurate replication of the process which occurs in the human placenta in vivo. Nevertheless, we submit that the second step of this differentiation protocol requires further refinement. We recently applied the STB and EVT differentiation protocols proposed in Okae et al. (2018), to both human trophoblast stem cell (hTSC) lines (derived from early gestation placental tissues) and our hPSC-derived CTB (cells at the end of the first step of differentiation). However, we found these protocols to be suboptimal to our current second step protocol, using FCM+BMP4, in promoting induction of STB- and EVT-associated markers (data not shown). We therefore conclude that further optimization of these protocols is needed, in order to establish improved conditions for lineage-specific differentiation of hPSC-derived CTB progenitor cells into either STB or EVT.
Figure 5.
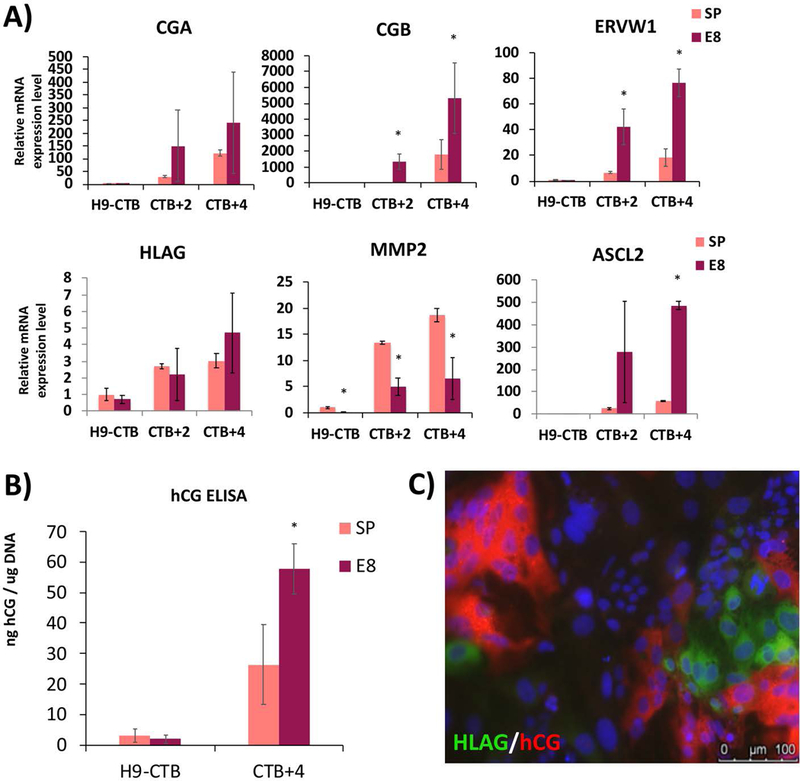
Comparison of second-step differentiation from StemPro- and E8-adapted WA09/H9 hESCs.
A) qPCR for lineage-specific markers. STB markers (CGA, CGB, ERVW1) and EVT markers (HLAG, MMP2, ASCL2) showed increased expression levels by 2 and 4 days post-replating in FCM+BMP4 (CTB+2 and CTB+4, respectively). Data are normalized to 18S and expressed as fold-change above StemPro-adapted H9-CTB (day 4 of the first-step differentiation). *indicates P<0.05 compared to StemPro-adapted cells at the same timepoint.
B) hCG ELISA confirms differentiation into syncytiotrophoblast (STB)-like cells. Data are normalized to total genomic DNA from the cells in the same well from which supernatant samples were collected. *indicates P<0.05 compared to StemPro-adapted cells.
C) Immunostaining using STB marker hCG and EVT marker HLAG on CTB +3 of the second-step differentiation of E8-adapated cells. STB- and EVT-like cells co-exist in the same culture at the end of the second-step differentiation.
Time Considerations
It will require up to 1 week to prepare the cells for the experiment and an additional ~2 weeks to complete the full two-step differentiation and analysis.
Significance Statement.
We previously established a two-step protocol for differentiation of human pluripotent stem cells (hPSCs), first into cytotrophoblast (CTB)-like cells and subsequently into fully differentiated syncytiotrophoblast (STB)- and extravillous trophoblast (EVT)-like cells. However, the first-step differentiation was suboptimal, resulting in 60–80% CTB-like cells, based on surface EGFR expression, and accompanied by simultaneous induction of mesoderm-associated genes. In this article, we present a detailed protocol, where we have incorporated the WNT inhibitor IWP2 into the first step, resulting in consistent and uniform induction of CTB and complete inhibition of mesoderm induction. We also show that following the second step, the resulting culture contains distinct subpopulations of STB- and EVT-like cells.
Acknowledgement
This work was supported by funds from a CIRM Physician Scientist Award (RN3–06396) and NIH/NICHD (R01HD-089537) to M.M.P. M.H. was supported through the California Institute for Regenerative Medicine (CIRM) Research and Training grant (TG2–01154 to the University of California, San Diego) and NIH/NICHD (K99 HD091452). O.T. was supported by the Bridges to Stem Cell Research Internship Program Grant from CIRM (EDUC2–08376). H.Y.C. was supported by the funding from School of Medicine, CHA University, in South Korea.
Footnotes
In this paper, we have shown three important points. First was the development of the two-step method of trophoblast differentiation of hPSCs, first into CTB, then into terminally-differentiated lineages (STB and EVT). Second, we showed that, similar to effect of hypoxia on primary CTB, hypoxia enhances differentiation of hPSC-derived CTB toward the EVT lineage. Finally, we showed that ESC and iPSC line affected by Trisomy 21 demonstrate a defect in STB differentiation, similar to primary CTB isolated from Trisomy 21-affected placentas. These data showed that ESC/iPSC can be used to model trophoblast differentiation under both normal and diseased conditions. This paper formed the basis of the improved protocol, presented here, for trophoblast differentiation of hPSC’s.
Conflict of Interest
The authors have no conflict of interest to report.
Literature Cited
- Amita M, Adachi K, Alexenko AP, Sinha S, Schust DJ, Schulz LC, Roberts RM, Ezashi T. (2013) Complete and unidirectional conversion of human embryonic stem cells to trophoblast by BMP4. Proc Natl Acad Sci U S A. 110(13):E1212–21. 10.1073/pnas.1303094110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apps R, Murphy SP, Fernando R, Gardner L, Ahad T, Moffett A (2009) Human leucocyte antigen (HLA) expression of primary trophoblast cells and placental cell lines, determined using single antigen beads to characterize allotype specificities of anti-HLA antibodies Immunology 127(1): 26–39. doi: 10.1111/j.1365-2567.2008.03019.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardo AS, Faial T, Gardner L, Niakan KK, Ortmann D, Senner CE, Callery EM, Trotter MW, Hemberger M, Smith JC, Bardwell L, Moffett A, Pedersen RA. (2011) BRACHYURY and CDX2 mediate BMP-induced differentiation of human and mouse pluripotent stem cells into embryonic and extraembryonic lineages. Cell Stem Cell. 9(2):144–55. doi: 10.1016/j.stem.2011.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilban M, Tauber S, Haslinger P, Pollheimer J, Saleh L, Pehamberger H, Wagner O, Knöfler M. (2010) Trophoblast invasion: assessment of cellular models using gene expression signatures. Placenta. 31(11):989–96. doi: 10.1016/j.placenta.2010.08.011. [DOI] [PubMed] [Google Scholar]
- Bischof P, Irminger-Finger I. (2005) The human cytotrophoblastic cell, a mononuclear chameleon. Int J Biochem Cell Biol. 37(1):1–16. doi: 10.1089/scd.2010.0281 [DOI] [PubMed] [Google Scholar]
- Daoud G, Amyot M, Rassart E, Masse A, Simoneau L, Lafond J. (2005) ERK1/2 and p38 regulate trophoblasts differentiation in human term placenta. J Physiol 566(Pt 2):409–23 doi: 10.1113/jphysiol.2005.089326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erb TM, Schneider C, Mucko SE, Sanfilippo JS, Lowry NC, Desai MN, Mangoubi RS, Leuba SH, Sammak PJ. (2011) Paracrine and epigenetic control of trophectoderm differentiation from human embryonic stem cells: the role of bone morphogenic protein 4 and histone deacetylases. Stem Cells Dev. 20(9):1601–14. doi: 10.1089/scd.2010.0281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garitaonandia I, Amir H, Boscolo FS, Wambua GK, Schultheisz HL, Sabatini K, Morey R, Waltz S, Wang YC, Tran H, Leonardo TR, Nazor K, Slavin I, Lynch C, Li Y, Coleman R, Gallego Romero I, Altun G, Reynolds D, Dalton S, Parast M, Loring JF, Laurent LC. (2015) Increased risk of genetic and epigenetic instability in human embryonic stem cells associated with specific culture conditions. PLoS One. 10(2):e0118307. doi: 10.1371/journal.pone.0118307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genbacev O, Miller RK. (2000) Post-implantation differentiation and proliferation of cytotrophoblast cells: in vitro models--a review. Placenta 21(Suppl A):S45–S49. doi: 10.1053/plac.1999.0523. [DOI] [PubMed] [Google Scholar]
- Haider S, Meinhardt G, Saleh L, Kunihs V, Gamperl M, Kaindl U, Ellinger A, Burkard TR, Fiala C, Pollheimer J, Mendjan S, Latos PA, Knöfler M (2018) Self-Renewing Trophoblast Organoids Recapitulate the Developmental Program of the Early Human Placenta Stem Cell Reports. 11(2): 537–551. doi: 10.1016/j.stemcr.2018.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horii M, Li Y, Wakeland AK, Pizzo DP, Nelson KK, Sabatini K, Laurent LC, Liu Y, Parast MM. (2016) Human pluripotent stem cells as a model of trophoblast differentiation in both normal development and disease. Proc Natl Acad Sci U S A. 113(27):E3882–91. doi: 10.1073/pnas.1604747113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jauniaux E, Poston L, Burton GJ. (2006) Placental-related diseases of pregnancy: Involvement of oxidative stress and implications in human evolution. Hum Reprod Update. 12(6):747–55, doi: 10.1093/humupd/dml016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurek D, Neagu A, Tastemel M, Tüysüz N, Lehmann J, van de Werken HJ, Philipsen S, van der Linden R, Maas A, van IJcken WF, Drukker M, ten Berge D. (2015). Endogenous WNT signals mediate BMP-induced and spontaneous differentiation of epiblast stem cells and human embryonic stem cells. Stem Cell Reports. 4(1):114–28. doi: 10.1016/j.stemcr.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Moretto-Zita M, Soncin F, Wakeland A, Wolfe L, Leon-Garcia S, Pandian R, Pizzo D, Cui L, Nazor K, Loring JF, Crum CP, Laurent LC, Parast MM. (2013) BMP4-directed trophoblast differentiation of human embryonic stem cells is mediated through a ΔNp63+ cytotrophoblast stem cell state. Development. 140(19):3965–76. doi: 10.1242/dev.092155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martyn I, Kanno TY, Ruzo A, Siggia ED, Brivanlou AH. (2018) Self-organization of a human organizer by combined Wnt and Nodal signalling. Nature. 558(7708):132–135. doi: 10.1038/s41586–018-0150-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMaster MT, Zhou Y, Fisher SJ. (2004) Abnormal placentation and the syndrome of preeclampsia. Semin Nephrol 24(6):540–7. doi:10.1016/j.semnephrol.2004.07.002 [DOI] [PubMed] [Google Scholar]
- Mendjan S, Mascetti VL, Ortmann D, Ortiz M, Karjosukarso DW, Ng Y, Moreau T, Pedersen RA. (2014) NANOG and CDX2 pattern distinct subtypes of human mesoderm during exit from pluripotency. Cell Stem Cell. 15(3):310–325. doi: 10.1016/j.stem.2014.06.006. [DOI] [PubMed] [Google Scholar]
- Morrish DW, Dakour J, Li H, Xiao J, Miller R, Sherburne R, Berdan RC, Guilbert LJ. (1997) In vitro cultured human term cytotrophoblast: a model for normal primary epithelial cells demonstrating a spontaneous differentiation programme that requires EGF for extensive development of syncytium. Placenta. 18(7):577–85. doi.org/10.1016/0143–4004(77)90013–3 [DOI] [PubMed] [Google Scholar]
- Norwitz ER. (2006) Defective implantation and placentation: laying the blueprint for pregnancy complications. Reprod Biomed Online. 13(4):591–599. doi.org/10.1016/S1472–6483(10)60649–9 [DOI] [PubMed] [Google Scholar]
- Okae H, Toh H, Sato T, Hiura H, Takahashi S, Shirane K, Kabayama Y, Suyama M, Sasaki H, Arima T. (2018) Derivation of Human Trophoblast Stem Cells. Cell Stem Cell. 22(1):50–63.e6. doi: 10.1016/j.stem.2017.11.004. [DOI] [PubMed] [Google Scholar]
- Pijnenborg R, Vercruysse L, Hanssens M, Brosens I. (2011) Endovascular trophoblast and preeclampsia: A reassessment. Pregnancy Hypertens 1(1):66–71. doi: 10.1016/j.preghy.2010.10.010. [DOI] [PubMed] [Google Scholar]
- Roberts RM, Ezashi T, Sheridan MA, Yang Y. (2018) Specification of trophoblast from embryonic stem cells exposed to BMP4. Biol Reprod 99(1):212–224. doi: 10.1093/biolre/ioy070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero R, Kusanovic JP, Chaiworapongsa T, Hassan SS (2011) Placental bed disorders in preterm labor, preterm PROM, spontaneous abortion and abruptio placentae Best Pract Res Clin Obstet Gynaecol. 25(3): 313–327. doi: 10.1016/j.bpobgyn.2011.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan MA, Yang Y, Jain A, Lyons AS, Yang P, Brahmasani SR, Dai A, Tian Y, Ellersieck MR, Tuteja G, Schust DJ, Schulz LC, Ezashi T, Roberts RM. (2019) Early onset preeclampsia in a model for human placental trophoblast Proc Natl Acad Sci U S A. 116 (10) 4336–4345. doi: 10.1073/pnas.1816150116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soncin F, Khater M, To C, Pizzo D, Farah O, Wakeland A, Arul Nambi Rajan K, Nelson KK, Chang CW, Moretto-Zita M, Natale DR, Laurent LC, Parast MM. (2018) Comparative analysis of mouse and human placentae across gestation reveals species-specific regulators of placental development. Development. 145(2). doi: 10.1242/dev.156273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudheer S, Bhushan R, Fauler B, Lehrach H, Adjaye J. (2012) FGF Inhibition Directs BMP4-Mediated Differentiation of Human Embryonic Stem Cells to Syncytiotrophoblast. Stem Cells Dev. 21(16):2987–3000. doi: 10.1089/scd.2012.0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakeland AK, Soncin F, Moretto-Zita M, Chang CW, Horii M, Pizzo D, Nelson KK, Laurent LC, Parast MM. (2017) Hypoxia Directs Human Extravillous Trophoblast Differentiation in a Hypoxia-Inducible Factor-Dependent Manner. Am J Pathol. 187(4):767–780. doi: 10.1016/j.ajpath.2016.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warmflash A, Sorre B, Etoc F, Siggia ED, Brivanlou AH. (2014) A method to recapitulate early embryonic spatial patterning in human embryonic stem cells. Nat Methods. 11(8):847–54. doi: 10.1038/nmeth.3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Zhang W, Chen G, Cheng L, Liao J, Jia N, Gao Y, Dai H, Yuan J, Cheng L, Xiao L. (2008) Combinatorial signals of activin/nodal and bone morphogenic protein regulate the early lineage segregation of human embryonic stem cells. J Biol Chem. 283(36):24991–5002 doi: 10.1074/jbc.M803893200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu RH, Chen X, Li DS, Li R, Addicks GC, Glennon C, Zwaka TP, Thomson JA. (2002) BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nat Biotechnol. 20(12):1261–4. doi:10.1038/nbt7 [DOI] [PubMed] [Google Scholar]


